Abstract
Rotavirus B (RVB) in the genus Rotavirus of the family Reoviridae is known to be a cause of acute gastroenteritis among children and adults in parts of Asia including China, India, Bangladesh and Myanmar. In a 15-month surveillance programme between March 2007 and May 2008, 3,080 stool specimens were collected from children and adults with acute gastroenteritis in an infectious disease hospital in Kathmandu, Nepal. In 25 (0.8%) specimens RVB was detected, for the first time in Nepal, by the use of polyacrylamide gel electrophoresis followed by confirmation with reverse-transcription PCR and sequencing. The strains detected in this study had very similar electropherotypes, and their VP7 sequences were almost identical and phylogenetically belonged to the Indo-Bangladeshi lineage which was distinct from the Chinese lineage. Thus, this study showed the circulation of RVB strains belonging to the Indo-Bangladeshi lineage in a broader region than previously documented, suggesting that this phylogenetic divide corresponded to the geographic divide created by the Himalayan Mountains. Further studies may be warranted to identify and characterize the RVB strains in northern Vietnam which is adjacent to southern China with a long and less mountainous border.
Keywords: Rotavirus B, Acute diarrhoea, Nepal
Introduction
Rotavirus is classified in the genus Rotavirus of the family Reoviridae, and comprises icosahedral, non-enveloped viruses with 11 segments of double-stranded RNA as the genome [1]. Genus Rotavirus is further classified into species, and species Rotavirus A (RVA), Rotavirus B (RVB) and Rotavirus C (RVC) have been detected in humans [1]. RVB is also detected in various animals including lambs, pigs, cattle, goats and rats [2–7]. However, no interspecies transmission of RVB between humans and animals is evident, with the exception of an earlier report providing serological evidence that human infections with rat RVB were prevalent in the Baltimore area, USA [8].
RVB was initially established as a human enteric pathogen in large epidemics of acute diarrhoea primarily, but not exclusively, among individuals older than 15 years of age in China during the 1980s [9, 10]. With an interval of 16 years since the first detection in China, five adult cases of severe gastroenteritis caused by RVB were reported in Kolkata, India [11], and the circulation of RVB was retrospectively identified in western India as early as 1993 [12]. Since 2000, RVB was also identified among children and adults with severe diarrhoea in Bangladesh [13–15], and more recently in 2008 a single case of RVB was reported from Myanmar [16].
Previously in Nepal, a country neighbouring China, Bangladesh, India and Myanmar where RVB was shown to be endemic, Pandey et al. [17] carried out a short-term surveillance programme to determine the diversity of aetiological agents causing diarrhoea in adult patients in an infectious and tropical disease hospital in Kathmandu. They identified bacterial pathogens such as diarrhoeagenic Escherichia coli and Shigella species in one third of the cases. Additionally, in this surveillance programme, common enteric helminths and protozoa were also examined, but no viral pathogens were sought for. We therefore set up a 15-month surveillance programme in which stool specimens from patients in all age groups attending the same hospital for the treatment of diarrhoea were examined for the presence of the RVB genome by polyacrylamide gel electrophoresis to further define the geographic spread of RVB and to determine the aetiological role of RVB in this population.
Materials and Methods
Patient enrolment and specimen collection
Patients with acute gastroenteritis who either attended the oral rehydration unit (referred to as outpatients) or were admitted to the wards (referred to as inpatients) of Sukraraj Tropical and Infectious Disease Hospital, Kathmandu, Nepal were eligible for inclusion in this study. Acute gastroenteritis was defined as three or more watery or looser-than-usual stool passages in a preceding 24-hour period with onset within the previous 2 weeks. Enrolment was carried out from 15 March, 2007 to 31 May, 2008, a period just 2 weeks short of 15 months in duration. One stool specimen was obtained for the study from each patient when informed consent was obtained.
Identification of RVB by electropherotyping
The presence of rotavirus in stool specimens from patients with acute gastroenteritis was determined by the detection of the RNA migration pattern of the 11 segments of the rotavirus genome (electropherotypes) upon 10% polyacrylamide gel electrophoresis (PAGE) followed by straining with silver nitrate as described previously [18]. The presence of a 4-2-1-1-1-1-1 pattern was regarded as indicating the presence of RVB, whereas a 4-2-3-2 or 4-3-2-2 pattern was regarded as indicating the presence of RVA or RVC respectively. The genomic RNA of rotavirus was prepared by extracting from 300 µl of 10% stool suspension with sodium dodecyl sulfate (SDS) and phenol, and precipitating with ethanol as described previously [19].
Confirmation of RVB by reverse-transcription (RT)-PCR and nucleotide sequencing
Viral RNAs were extracted from the specimens positive for RVB by electropherotyping using the QIAamp Viral RNA Mini Kit (QIAGEN Sciences, Germantown, MD, USA) and were reverse transcribed and amplified with a pair of the VP7 consensus primers, B5-2 (5'-GGCAATAAAATGGCTTCATTGC-3') and B5-3 (5'-GGGTTTTTACAGCTTCGGCT-3') and with another pair of the NSP2 consensus primers, GB8-1 (5'-GGTAGAAATTAATCTATTCAGTG-3') and GB8-3 (5'-GGGTTTTAAAAATAGCCGTTAAC-3') as described previously [20] using the AcessQuick RT-PCR system (Promega Corporation, Madison, WI, USA). The amplified products were then purified using a QIAquick PCR purification kit (QIAGEN) according to the manufacturer’s instructions. Nucleotide sequencing reactions were performed using the BigDye Terminator Cycle Sequencing Ready Reaction Kit, version 3.1 (Applied Biosystems, Foster City, CA, USA), and the nucleotide sequence was determined with the ABI Prism 3730 genetic analyzer (Applied Biosystems). The nucleotide sequences thus obtained were aligned with the Megalign programme in the Lasergene 8 software package (DNAstar, Inc. Madison, WI, USA).
Phylogenetic analysis of the VP7 genes
Phylogenetic analysis was performed using the MEGA5 software package [21]. The nucleotide sequences of the VP7 gene of Nepalese RVB strains and reference sequences obtained from the DNA databases were aligned using the CLUSTALW programme, and the genetic distances between sequences were calculated by the Kimura two-parameter method. A phylogenetic tree was then constructed using the neighbour-joining method. The statistical significance at the branching point was calculated with 1,000 pseudo-replicate data sets.
Nucleotide sequence accession numbers
The sequences (n = 3) of the VP7 genes of Nepalese RVB were submitted to the DDJB/GenBank/EMBL database and were assigned accession numbers AB821399 to AB821401.
Results
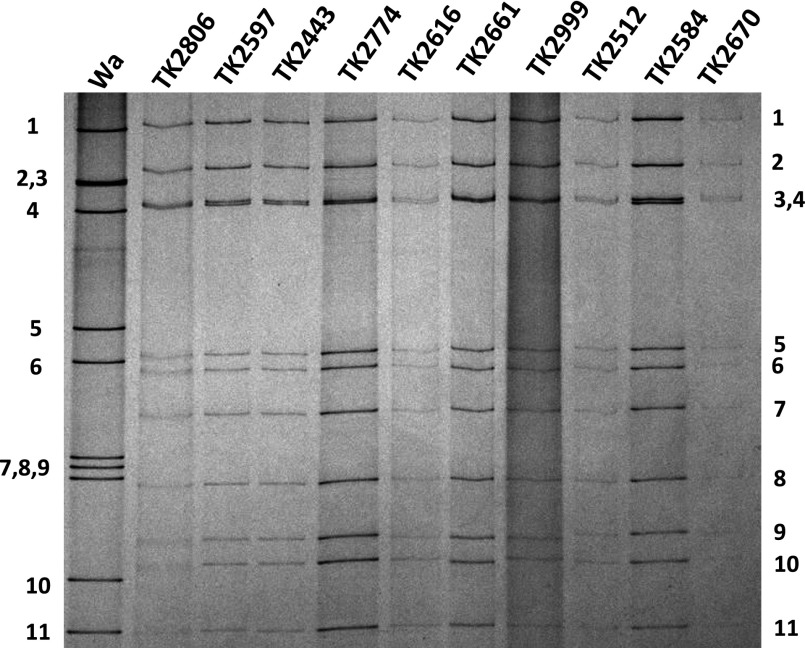
When a total of 3,080 diarrhoeic stool samples were examined by PAGE for the presence of a 4-2-1-1-1-1-1 genomic RNA migration pattern typical of RVB, 25 specimens contain electropherotypes indicating the presence of RVB. The electropherotypes of these RVB specimens were very similar to each other (Fig. 1). There were 57 specimens that contained RVA and one specimen that contained an electropherotype suggestive of RVC. Since one specimen contained both RVB and RVA electropherotypes, the overall detection rate of rotaviruses was 2.7% (82/3,080) of which 1.9% (57) was attributed to RVA and 0.8% (25) to RVB. RVB was detected in 19 of 2,642 (0.68%) specimens collected from hospitalized patients, whereas it was detected in 6 of 438 (1.4%) patients treated in the oral rehydration unit (outpatients). However, the difference in the RVB detection rates between the two groups was not statistically significant.
Fig. 1.
The electropherotypes of the representative RVB strains detected from the faeces of patients with acute diarrhoea in Nepal. The names of strains are indicated on the top of the lane. The approximate position of each of the 11 genome segments for the prototype RVA human strain Wa is indicated to the left of the panel, and that for RVBs is indicated to the right.
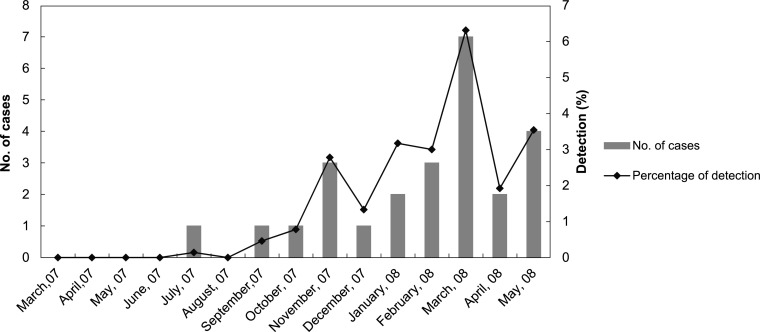
With regard to the age of patients, RVB was detected from patients as young as 2 years of age and as old as 60 years of age, but the majority (20/25) of RVB cases were found in adult patients aged between 15 and 45 years, with no statistical difference between males and females. When the occurrence of the RVB strains was examined along the timeline of the 15 months of surveillance, it was noted that the distribution was skewed toward the latter half of the surveillance period (Fig. 2).
Fig. 2.
Monthly distribution of RVB infections in patients with acute gastroenteritis in Nepal from June 2007 to May 2008.
When the VP7 based RT-PCR was applied to the 25 RVB specimens, there was a band of an expected size (814 bp) in 20 specimens, confirming that these specimens contained RVB. In the remaining 5 specimens in which there was no band suggestive of the presence of RVB, however, another NSP2-based RT-PCR produced a band with an expected size (1006 bp) in every one of the 5 specimens.
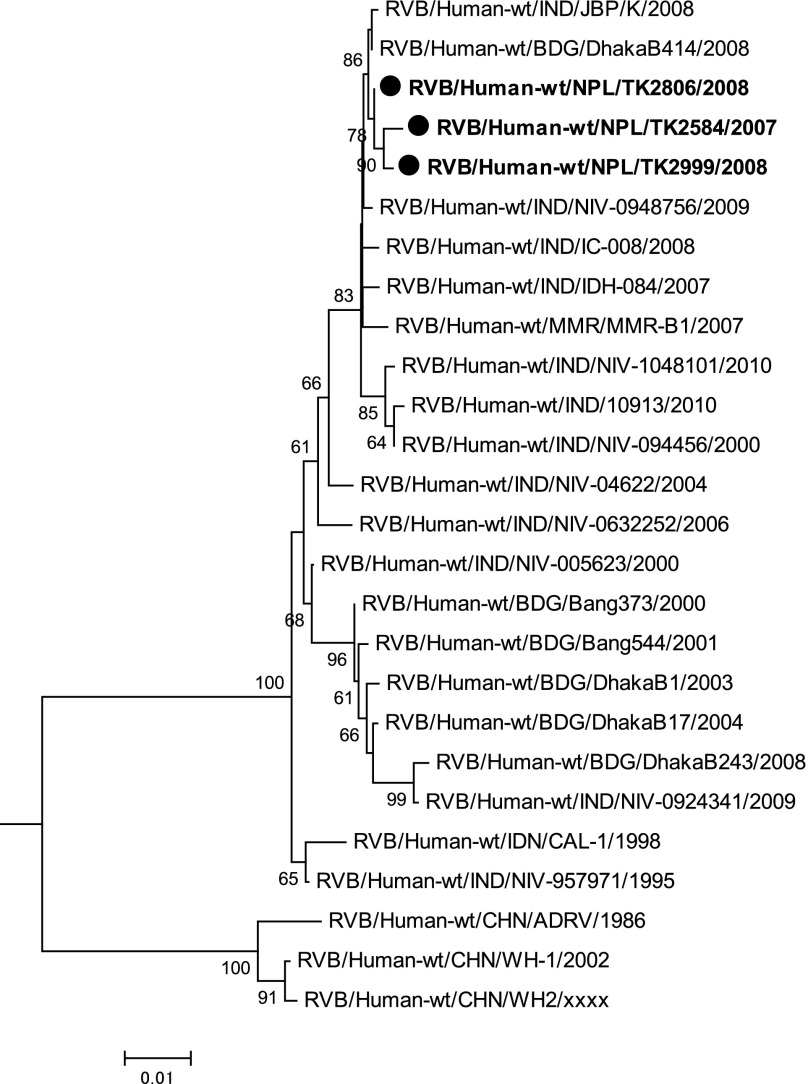
We then determined the nucleotide sequence of the partial VP7 genes derived from TK2584, TK2806, and TK2999, which were detected from a 19-year-old patient in December 2007, a 15-year-old patient in March 2008, and an 18-year-old patient in May 2008, respectively. The VP7 genes of these three RVB strains had a virtually an identical sequence (99.6–99.9%) with 1-3 nucleotide substitutions among the three sequences of which only one was non-synonymous. In addition, they showed high nucleotide sequence identities with Indian, Bangladeshi and Myanmarese human RVB strains (96.7–100%). On the other hand, the sequences of these three Nepalese strains showed fewer nucleotide identities with Chinese human RVB strains (90.5–91.6%). Indeed, the phylogenetic tree shown in Figure 3 indicates that the VP7 sequences of human RVB strains were divided into two lineages, namely the Indo-Bangladeshi and Chinese lineages, and that the Nepalese RVB strains belonged to the Indo-Bangladeshi lineage.
Fig. 3.
A phylogenetic tree for VP7 genes of Nepalese human RVB strains (indicated in boldface with closed circular dots) along with other human RVB strains obtained from the DNA database. The tree was constructed using the neighbour-joining method included in the MEGA 5 software package with bootstrap probabilities after 1,000 replicate trials. The genetic distance is indicated at the bottom. Percent bootstrap support is indicated by the value at each node when the value was 60% or larger. Two porcine RVB strains (accession numbers AB490441, JQ043795) were used as the outgroup but are not shown in the tree. Only the node where the root is located is shown.
Discussion
This study documented for the first time the multiple detection and characterization of RVB in Kathmandu, Nepal. RVB was regarded as an emerging pathogen for humans but its detection was limited in China for many years [9, 10]. Since 1998, however, RVB has been detected sporadically in various locations in India, Bangladesh, and more recently in Myanmar [16]. This expansion of the number of countries where RVB was detected is considered to result from increased awareness and surveillance efforts rather than a true increase in the incidence of acute diarrhoea caused by RVB. While this paper is the first to report the presence of RVB in Nepal, the detection of RVB was not unexpected in view of the geographic location of the country: Nepal being sandwiched between China and India and is located upstream from Bangladesh.
With regard to the detection rate of RVB among patients with sporadic acute diarrhoea, the detection rate of 0.8% (25/3,080) obtained in this study was lower than that reported in other studies. In one study carried out in Bangladesh, RVB was detected by PAGE in 5.5% (12/220) of adult patients and in 3% (6/67) of children less than 2 years of age [13], and in another study also carried out in Bangladesh, RVB was detected by a more sensitive RT-PCR assay in 2.4% (12/220) of both adult patients and children [14]. In a more systematic survey in Bangladesh in which PAGE detected both RVB and RVA in every one in 100 patients admitted to the hospital with acute diarrhoea, RVB and RVA were detected in 2.4% (26/1106) and 23.4% (259/1106) of patients, respectively [15]. In Pune, Alappuzha, and Belgaum, India during the 2004–2010 period, RVB was detected using RT-PCR in 4.1% (36/870), 7.3% (8/110) and 4.1% (8/197), respectively, of patients with sporadic acute gastroenteritis [22]. In Myanmar, RVB was detected using PAGE in one patient (0.5%) among 195 patients with acute diarrhoea comprising 90 children and 105 adults in which RVA was detected in 16 (18%) and 3 (2.9%) patients, respectively [16]. Although a simple direct comparison of these detection rates of RVB in different settings may be tenuous, the average detection rate obtained after weighting with the inverse of the variance of the detection rate in each study was 2.3% (95% confidence interval [CI]: 1.67–3.00%). Thus, the overall detection rate of 0.8% (95% CI: 0.53–1.2%) obtained in this study appears to be significantly lower than that in other countries where RVB was reported as an aetiological agent of sporadic acute gastroenteritis. The detection rate in Myanmar (0.5%) was lower as a point estimate, but, since the upper 95% confidence limit was 2.9% and the lower 0.01%, the difference is not statistically significant when the weighted average detection rate described above is taken into account.
RVB infections were detected in both genders and in all age groups, but it was noted that 75% (20/25) of RVB were detected in patients aged between 15 and 45 years (adolescents and adults). These observations were similar to those reported from India [22], and the cause was attributed to as a low exposure of children to RVB or a lower shedding of RVB in faecal specimens [22]. The seasonality of RVB infections in the endemic countries is not clear, but outbreaks were occasionally noted. In western India, for example, an outbreak of RVB infections affecting >200 patients occurred in the summer months (April-May) in 2009 [23]. The skewed monthly distribution of RVB detection observed in the present study (Fig. 2) suggests the possibility of a prolonged outbreak in the latter part of the surveillance period (September 2007 to May 2008) or a substantial variation in the circulation of RVB from year to year in Nepal. In this regard, it is noteworthy that the electropherotypes of the RVB strains detected in the present study were very similar to each other and that at least the partial VP7 sequences of three RVB strains detected at 2–3 month intervals were almost identical, suggesting that most of these infections were caused by only a few strains, or perhaps a single strain. However, caution needs to be exercised. Saiada et al. [15] also reported that an almost identical electropherotype was observed in 6 out of 8 RVB strains in their study as well as nearly identical VP4 sequences for the strains detected in hospitalized patients in Bangladesh during a one- year period. Thus, it may be possible that the genetic diversity among locally-circulating RVB strains is very small.
Phylogentic analysis confirmed the similarity of the VP7 sequence of the three Nepalese strains with that of the Indo-Bangladeshi strains, in that they clustered together with a 100% bootstrap probability. The distinctness of the VP7 sequence of Chinese strains is unlikely to be due to the fact that ADRV was isolated much earlier than the Indo-Bangladeshi strains because even a more recent isolate, WH-1, detected in Wuhan, China in 2002 also clustered together with ADRV detected in 1986. This phylogenetic divide appears to correspond to the geographic divide created by the Himalayan Mountains and impeding direct human communications. Among the countries that share a national border with China, Vietnam may be noteworthy in that the northern region is adjacent to China and shares a long and less mountainous border. It will be worthwhile, therefore, to explore the circulation of the RVB strains in northern Vietnam, and, if they exist, to identify and characterize them in order to determine to which lineage the strains belong.
In conclusion, this study described for the first time, the detection of RVB in 0.8% of patients with acute gastroenteritis in Nepal, revealing the circulation of RVB in a broader geographic region than previously documented. The strains detected in this study were very similar to each other in their electropherotypes, and their VP7 sequences were almost identical and phylogenetically belonged to the Indo-Bangladeshi lineage which was distinct from the Chinese lineage. This study indicates the need for further studies to explore the boundary of the two distinct lineages to which the human RVB strains belong.
Acknowledgements
S. B. Pun was the recipient of a post-doctoral fellowship under the Global Centre of Excellence Programme on Integrated Global Control Strategy for Tropical and Emerging Infectious Diseases from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
This study was supported in part by grants in-aid for scientific research from the Ministry of Health, Labour, and Welfare, Japan.
Conflicts of interest
There is no conflict of interest for any author to declare regarding this study.
References
- 1.Cunliffe NA, Nakagomi O. Reoviruses. In: Greenwood D, Slack RCB, Barer MR, Irving WB, eds. Medical Microbiology. 18th ed. London: Churchill Livingstone; 2012. pp. 559–56522160312
- 2.Chasey D, Banks J. The commonest rotaviruses from neonatal lamb diarrhoea in England and Wales have atypical electropherotypes. Vet Rec 1984; 115(13): 326–327 [DOI] [PubMed] [Google Scholar]
- 3.Eiden J, Vonderfecht S, Theil K, Torres-Medina A, Yolken RH. Genetic and antigenic relatedness of human and animal strains of antigenically distinct rotaviruses. J Infect Dis 1986; 154(6): 972–982 [DOI] [PubMed] [Google Scholar]
- 4.Kuga K, Miyazaki A, Suzuki T, Takagi M, Hattori N, Katsuda K, Mase M, Sugiyama M, Tsunemitsu H. Genetic diversity and classification of the outer capsid glycoprotein VP7 of porcine group B rotaviruses. Arch Virol 2009; 154(11): 1785–1795 [DOI] [PubMed] [Google Scholar]
- 5.Marthaler D, Rossow K, Gramer M, Collins J, Goyal S, Tsunemitsu H, Kuga K, Suzuki T, Ciarlet M, Matthijnssens J. Detection of substantial porcine group B rotavirus genetic diversity in the United States, resulting in a modified classification proposal for G genotypes. Virology 2012; 433(1): 85–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Munoz M, Alvarez M, Lanza I, Carmenes P. An outbreak of diarrhoea associated with atypical rotaviruses in goat kids. Res Vet Sci 1995; 59(2): 180–182 [DOI] [PubMed] [Google Scholar]
- 7.Parwani AV, Lucchelli A, Saif LJ. Identification of group B rotaviruses with short genome electropherotypes from adult cows with diarrhea. J Clin Microbiol 1996; 34(5): 1303–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eiden J, Vonderfecht S, Yolken RH. Evidence that a novel rotavirus-like agent of rats can cause gastroenteritis in man. Lancet 1985; 2(8445): 8–11 [DOI] [PubMed] [Google Scholar]
- 9.Hung T, Chen GM, Wang CG, Yao HL, Fang ZY, Chao TX, Chou ZY, Ye W, Chang XJ, Den SS, et al. Waterborne outbreak of rotavirus diarrhoea in adults in China caused by a novel rotavirus. Lancet 1984; 1(8387): 1139–1142 [PubMed] [Google Scholar]
- 10.Fang ZY, Ye Q, Ho MS, Dong H, Qing S, Penaranda ME, Hung T, Wen L, Glass RI. Investigation of an outbreak of adult diarrhea rotavirus in China. J Infect Dis 1989; 160(6): 948–953 [DOI] [PubMed] [Google Scholar]
- 11.Krishnan T, Sen A, Choudhury JS, Das S, Naik TN, Bhattacharya SK. Emergence of adult diarrhoea rotavirus in Calcutta, India. Lancet 1999; 353(9150): 380–381 [DOI] [PubMed] [Google Scholar]
- 12.Kelkar SD, Zade JK. Group B rotaviruses similar to strain CAL-1, have been circulating in Western India since 1993. Epidemiol Infect 2004; 132(4): 745–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanekata T, Ahmed MU, Kader A, Taniguchi K, Kobayashi N. Human group B rotavirus infections cause severe diarrhea in children and adults in Bangladesh. J Clin Microbiol 2003; 41(5): 2187–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rahman M, Hassan ZM, Zafrul H, Saiada F, Banik S, Faruque AS, Delbeke T, Matthijnssens J, Van Ranst M, Azim T. Sequence analysis and evolution of group B rotaviruses. Virus Res 2007; 125(2): 219–225 [DOI] [PubMed] [Google Scholar]
- 15.Saiada F, Rahman HN, Moni S, Karim MM, Pourkarim MR, Azim T, Rahman M. Clinical presentation and molecular characterization of group B rotaviruses in diarrhoea patients in Bangladesh. J Med Microbiol 2011; 60(Pt 4): 529–536 [DOI] [PubMed] [Google Scholar]
- 16.Aung TS, Kobayashi N, Nagashima S, Ghosh S, Aung MS, Oo KY, Win N. Detection of group B rotavirus in an adult with acute gastroenteritis in Yangon, Myanmar. J Med Virol 2009; 81(11): 1968–1974 [DOI] [PubMed] [Google Scholar]
- 17.Pandey BD, Thapa LB, Sherchand JB, Rimal N, Bhattara A, Morita K. Etiology of diarrhoea among adult patients during the early monsoon period in Kathmandu, Nepal. Jpn J Trop Med Hyg 2002; 30(2): 133–137 [Google Scholar]
- 18.Herring AJ, Inglis NF, Ojeh CK, Snodgrass DR, Menzies JD. Rapid diagnosis of rotavirus infection by direct detection of viral nucleic acid in silver-stained polyacrylamide gels. J Clin Microbiol 1982; 16(3): 473–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kobayashi N, Lintag IC, Urasawa T, Taniguchi K, Saniel MC, Urasawa S. Unusual human rotavirus strains having subgroup I specificity and “long” RNA electropherotype. Arch Virol 1989; 109(1-2): 11–23 [DOI] [PubMed] [Google Scholar]
- 20.Ahmed MU, Kobayashi N, Wakuda M, Sanekata T, Taniguchi K, Kader A, Naik TN, Ishino M, Alam MM, Kojima K, Mise K, Sumi A. Genetic analysis of group B human rotaviruses detected in Bangladesh in 2000 and 2001. J Med Virol 2004; 72(1): 149–155 [DOI] [PubMed] [Google Scholar]
- 21.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 2011; 28(10): 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lahon A, Maniya NH, Tambe GU, Chinchole PR, Purwar S, Jacob G, Chitambar SD. Group B rotavirus infection in patients with acute gastroenteritis from India: 1994–1995 and 2004–2010. Epidemiol Infect 2013; 141(5): 969–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chitambar SD, Lahon A, Tatte VS, Maniya NH, Tambe GU, Khatri KI, Desai HS, Ugare MR, Kulkarni SV, Waghmare AP. Occurrence of group B rotavirus infections in the outbreaks of acute gastroenteritis from western India. Indian J Med Res 2011; 134: 399–400 [PMC free article] [PubMed] [Google Scholar]