Abstract
The JHK virus (JHKV) was previously described as a type C retrovirus that has some distinctive ultrastructural features and replicates constitutively in a human B-lymphoblastoid cell line, JHK-3. In order to facilitate the cloning of sequences from JHKV, a series of partially degenerate consensus retroviral PCR primers were created by a data-driven design approach based on an alignment of 14 diverse gammaretroviral genomes. These primers were used in the PCR amplification of purified JHK virion cDNA, and ana lysis of the resulting amplified sequence indicates that the JHKV is in the murine leukemia virus (MLV) family. The JHK sequence is nearly identical to the corresponding region of the Bxv-1 endogenous mouse retrovirus (GenBank accession AC115959) and distinct from XMRV. JHKV gag-specific amplification was demonstrated with nucleic acids from uncultivated, frozen, peripheral blood mononuclear cells (PBMCs) of the index patient, but not in PBMCs from nine healthy blood donors. Unlike earlier reports, in which MLV-like sequences were identified in human source material, which may have been due to murine contamination, budding retrovirions were demonstrated repeatedly by electron microscopy in uncultivated lymphocytes of the index patient that were morphologically identical in their development to the virions in the JHK-3 cells, and immunological evidence was obtained that the index patient produced IgG antibodies that bound to the budding viral particles in patient PBMCs and in the JHK-3 cells. These data indicate that the patient had been infected by JHKV, lending significance to the demonstration of JHKV amplicons in nucleic acids of the patient’s PBMCs. In future studies, the PCR primer sets described herein may expand the detection of an amplifiable subset of viruses related to MLV.
Keywords: B-lymphoblastoid cells, electron microscopy, Epstein–Barr virus, gammaretrovirus, murine leukemia virus
Retroviruses uniquely require integration into host cell genomic DNA during replication through reverse transcription of their RNA. They can cause a variety of diseases in humans and animals; the search and possible roles for novel retroviruses in human diseases were reviewed in depth by Voisset et al., who listed the four confirmed human viral pathogens, HTLV-1 and -2 and HIV-1 and -2, as well as 25 diseases putatively associated with retroviruses [1]. In 2006, a retrovirus (designated XMRV) related to the murine leukemia virus (MLV) group, was described and reported to be linked to prostate carcinoma [2], and in 2009, to chronic fatigue syndrome (CFS) [3]. The MLVs comprise a group of mouse genome-integrated endogenous proviruses derived from exogenous retrovirus infections earlier in the history of the species; the MLVs are classified by sequence and host-range receptor-binding affinity as, for example, xenotropic (indicating an inability to bind to cells of most inbred and some wild mice) or polytropic (able to bind to multiple species’ receptors) [4]. XMRV-related studies generated considerable literature, including initially confirmatory and later negative reports of human infection, leading to considerable controversy and manuscript retraction. Subsequent reports confirmed that the sources of XMRV infection of samples from patients could be traced to contamination from mouse tissue [5,6], murine nucleic acids [7], or from different cell lines chronically infected with murine retroviruses, such as the XMRV-producer 22Rv1 prostate carcinoma [8], and other prostate LAPC4 and VCaP cell lines [9]. The origin of XMRV was shown to be the recombination of two provirus genomes, PreXMRV-1 and PreXMRV-2 in prostate cancer xenografts passaged in nude mice [4,6]. An MLV-like sequence has also been detected in an Epstein–Barr virus (EBV)-positive human B-cell line, JY [10]. In 2010, Lo et al. reported the detection of polytropic MLV-related gene sequences in the blood of patients with CFS and healthy blood donors (a report later withdrawn) [11,12]; and more recently Lee et al. described very sensitive PCR assays using various primers to detect MLV-like gag sequences and mouse contaminants in human blood samples, some from CFS patients [13]. Most recently, Lee et al. definitively excluded XMRV as etiologic in prostatic cancer in archival and newly collected samples [14], and Alter et al. excluded XMRV and polytropic MLV as etiologic in CFS in a large, controlled, clinical study using specific primers [15]. In these various studies, the detection of almost all such sequences by PCR has depended on the availability of specific MLV-related primers.
In 1997 our laboratory described a human B-lymphoblastoid cell line, JHK-3, that constitutively produces both EBV and a relatively fragile, enveloped RNA virus, containing manganese-dependent reverse transcriptase (RT) activity and resembling C-type retrovirus particles with somewhat distinctive ultrastructural features [16]. The JHK-3 cell line had been established in 1989 by cocultivating the peripheral blood mononuclear cells (PBMCs) from a healthy subject with the PBMCs from a patient with a 3-year history of a viral-like, ill-defined, subacute illness. After 2 months’ incubation of this culture, the cell-free supernatant medium was added to fresh, phytohemagglutinin-treated PBMCs that had been taken from the same healthy donor; the outgrowth of these cells was designated as the JHK-3 line. Many previous attempts to obtain molecular clones of the JHK virus (JHKV) using a variety of published retroviral PCR primers and established protocols were not successful in obtaining bona fide retroviral sequences. Other approaches, including collaborative efforts by others using Virochip DNA microarray techniques, also did not identify retroviral sequence. We then undertook to design retrovirus-specific, consensus PCR primers, using a sequence homology-driven approach described herein. To obtain purified JHK viral RNA from the JHK-3 cells for PCR, we employed a urea-nuclease procedure [17] to eliminate any extra-virion, PCR-amplifiable, cellular nucleic acids and the large amount of microvesicles that lymphoblastoid cells produce from virion preparations. These primers permitted the detection of a partial nucleotide sequence of the 5′ (gag) region of the JHKV that has near identity (99%) to Bxv-1-related MLV-like gag sequences (GenBank accession AC115959) and distinct from XMRV. We showed by high-resolution transmission electron microscopy (EM) that freshly obtained, uncultivated PBMCs from the patient contained developing retrovirions ultrastructurally indistinguishable from those in the JHK-3 cells that constitutively produce virions with the Bxv-1-like sequence. We further showed by quantitative immunogold EM techniques that IgG antibodies in the serum of the index patient (designated as IP) bound to budding viral particles in the patient’s uncultivated PBMCs and in the JHK-3 cell cultures, whereas IgG in serum from healthy subjects did not bind significantly to JHK virions, but IgG from the patient did bind.
Materials & methods
Cells
The B-lymphoblastoid suspension cell cultures, including JHK-3, K-3II (developed from a normal, healthy donor) [16] and the DG-75 cell sublines (UW [which produces DG-75 retrovirus constitutively] and HAD [which is virus-free]) [18] were propagated in RPMI-1640 medium containing 10–20% fetal bovine serum and ciprofloxacin. The JHK-3 cell line was deposited with the American Type Culture Collection (ATCC; MD, USA) as CRL10991. Anchorage-dependent human A549 bronchioloalveolar carcinoma cells were grown in minimal essential medium with 10% calf serum. PBMCs from unidentified healthy blood donors were obtained from the Blood Center of Wisconsin (WI, USA). Ficoll-Hypaque gradient-purified PBMCs from heparinized blood samples of the IP were obtained at various times beginning in January 1989, and either fixed in glutaraldehyde for EM or stored frozen at −80°C or in liquid N2. For virus isolation, the patient’s September 1989 PBMCs were cocultivated with healthy donor lymphocytes in medium supplemented with IL-2 (a technique used to isolate HIV, HTLV and HHV6) [19]. After 2 months’ cultivation the cells began to degenerate, and cell-free supernatant medium from this culture was added to phytohemagglutinin-stimulated PBMCs from the same healthy donor, whose lymphocytes did not show viral particles. The cell outgrowth of this culture was designated the JHK-3 cell line. Repeated attempts to establish a cell line from PBMCs of the IP were unsuccessful. We derived from the healthy donor a B-lymphoblastoid cell line K-3II that produced no detectable virions of either JHKV or EBV [16]. This study (#21-91) was approved by the Medical College of Wisconsin’s Human Research Review Committee.
Viruses
For JHKV, supernatant fluid collected from JHK-3 cell cultures 5 days after medium change was clarified by centrifugation at 1000 g, passed through 0.45- and 0.22-μm filters, and ultra-centrifuged at 100,000 g through a 20% sucrose cushion. The viral pellet was resuspended in DEPC-treated water, and processed through the urea-nuclease procedure described by Sun et al. such that viral RNA was preserved and cellular PCR-amplifiable RNA and DNA were eliminated [17]; in brief, this consists of suspending the viral pellet in water, adding urea (to 1.5–2.0 M), micro-coccal nuclease and ribonuclease A, followed by RNA extraction and repeated deoxyribonuclease I treatments. DG-75 virus was harvested from the B-lymphoblastoid DG-75 (strain UW) cell subline, and the MLV LP-BM5 virus (GenBank accession D14687) from SC-1 cells [16].
PCR amplification
PCR amplification reactions were carried out as follows: 4 mM Tris HCl (pH 8.6), 60 mM KCl, 2 mM MgSO4, 1.5 mM MgCl2, 10 mM (NH4)2SO4, 0.1% Triton X-100, 0.1 mg/ml nuclease-free bovine serum albumin (BSA), 2 mM dNTP, 20 pMol forward primer, 20 pMol reverse primer, 0.5 U Taq DNA polymerase (Invitrogen®; Life Technologies, NY, USA) and 0.5 U cloned Pfu DNA polymerase (Stratagene®; Agilent Technologies, CA, USA). Reactions also contained 500 ng of template DNA, or an equal volume of water as a control. Reactions were run with five cycles of 2 min at 94°C, 30 s at 52°C and 1 min at 72°C, followed by 30 cycles of 30 s at 94°C, 30 s at 55°C and 1 min at 72°C. A final extension reaction of 10 min at 72°C was carried out, and the sample held at 4°C.
Electron microscopy
For high-resolution transmission EM, fresh, uncultivated PBMCs from IP and cultured JHK-3 cells were prefixed with glutaraldehyde in a cacodylate buffer, fixed in 2% OsO4, embedded in Epon, sectioned and processed for viewing as previously described [16,20]. For negative staining, an ultracentrifuged (100,000 g for 60 min) viral suspension of partially purified viral pellets from JHK-3 cell supernatant fluid was placed on carbon-coated, Formvar holey grids prepared as described [21] and stained with 2% sodium phosphotungstate. For antibody studies, the direct and indirect immunogold standard techniques of Nermut et al. were employed [22]. In the direct method, the IP’s serum IgG was purified, labeled with 10-nm colloidal gold particles by E-Y Laboratories (CA, USA), and diluted 1:50 in saline containing 0.1% BSA and 0.05% Tween-20. The ultrathin sections of Lowicryl-K4M-embedded JHK-3 cells or patient’s lymphocytes were pretreated with 0.1% BSA to diminish nonspecific binding, incubated for 60 min with the gold-labeled IgG, washed thoroughly with 0.1% BSA in saline and stained with 2% uranyl acetate. The sections were observed in a Phillips EM-300 electron microscope at direct magnifications from 17,000× to 60,000× and the labeling density as number of 10-nm gold particles/μm2 determined as described [23].
For the indirect immunogold method, Formvar-covered grids loaded with suspensions of saline-washed, sucrose-gradient-purified JHKV were incubated for 90 min at 20°C with the serum samples diluted 1:10 (also at 1:30 and 1:100), then exposed to goat antihuman IgG antibody conjugated with 10-nm colloidal gold particles (E-Y Laboratories), extensively washed, fixed and negatively stained with potassium phosphotung-state; 30–50 virions were counted in each of four to five randomly selected grid openings of each of two grids per serum sample at a magnification of 28,000×, the number of gold-labeled and non-labeled virions were recorded and the percentage of labeled virions were calculated. The double-blind protocol used to compare the JHKV binding of IgG in sera from different donors was as follows: technician one coded the serum sample tubes and distributed the diluted serum samples into them; after the samples were processed by the EM facility (technician two), the EM operator recorded the measurements for each sample and controls, and the results were delivered to technician three who assembled the coded results.
Clinical history
The IP was a 39-year-old woman with a 3-year history of an ill-defined, viral-like complex of symptoms. An initial, mild febrile illness was followed by a relatively acute onset of marked easy fatigability, and then by severe, delayed post exertional malaise and fatigue, somnolence requiring prolonged bed rest, impaired cognition and verbal expression, forgetfulness, lower extremity muscle weakness, myalgias and paresthesias, occasional arthralgias and drenching night sweats, lymphadenopathy, and immunologic anergy. She slowly regained full activity and function.
Results
Construction of consensus primers
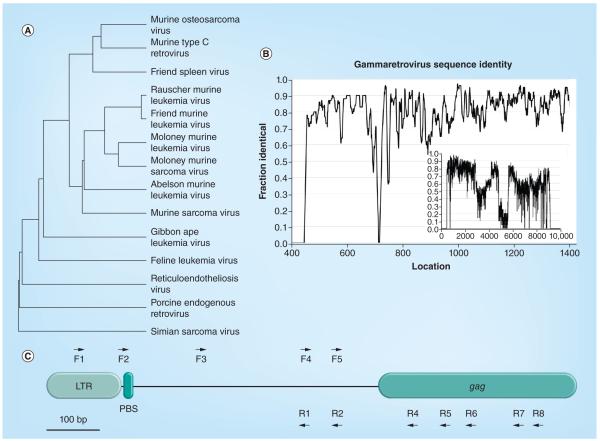
In order to PCR-amplify the JHKV, a multivirus homology approach was devised to design primers for the amplification of unsequenced retroviruses. This method uses conserved regions or ‘islands’ of sequence identity conserved between a wide range of gammaretroviruses to create PCR primer sets with limited degeneracy that can be used to amplify sequences from most gammaretroviruses, including the JHKV. A diverse selection of the available gammaretroviral genomes was first aligned (Figure 1A). To search for potential primer locations, we measured the sequence conservation along the length of the genomes by calculating the ‘fraction identical’, which is defined as the fraction of of the aligned sequences that matched to the most common residue at each location. To smooth the distribution, a running average of the conservation across a 10-bp window was calculated (Figure 1B). The inset graph in Figure 1B shows the distribution of conservation across the entire genome, indicating that the 5′ end showed the greatest number of conserved islands from which to choose potential primers.
Figure 1. Selection of primers.
(A) Dendrogram of retroviral sequences used to construct PCR primer sequences. Sequences were aligned with the ClustalW program. (B) Graph of sequence identity across aligned retroviral sequences. A moving average (10-bp window) of the fraction of sequences with the most abundant residue at each location was calculated across the retroviral genome. The inset graph shows the entire genome and the main graph shows 1400 residues of the 5′ region. (C) Diagram of the 5′ end retroviral primers. The locations of the 5′ end F and R primers are shown with respect to each other and to biological features (LTR, PBS and the gag gene). Distances are to scale.
F: Forward; LTR: Long terminal repeat; PBS: Primer binding site; R: Reverse.
To increase the chance of successful amplification, one criterion for choosing primer sets required the amplified products to be <1 kB in length, since amplifying a limited region relaxes the requirements not only for the processivity of first-strand cDNA and PCR synthesis reactions, but also for the quality of input virus RNA. A second primer design criterion required that the primers had compatible Tm values, allowing forward and reverse primers from different sets to be used interchangeably. Target locations for the degenerate primers were chosen by generating a consensus sequence from the alignment of the sequences of the collection of gammaretroviruses known to infect a diverse group of species. From this alignment shown in Figure 1B, several islands of conserved sequence were selected to create degenerate primer sequences that would match the greatest number of virus sequences with the least amount of degeneracy. Islands of sequence conservation as potential primer sites were identified to be: within the long terminal repeat (LTR) region of the viruses; between the LTR and gag protein coding region; and within the 5′ end of the gag coding region. With these sequence islands, the primers shown in Figure 1C were determined, using a mixture of automated primer design software and hand selection (Table 1).
Table 1.
Primer sequences.
| Primer | Sequence | Start | End |
|---|---|---|---|
| F1 | CCTCTTGCTGTTGCATCCG | 54 | 73 |
| F2 | TTTCATTTGGGGGCTCGTC | 139 | 158 |
| F3 | ATGCGYCTGNDTCKGTACTAGTT | 284 | 307 |
| F4 | AYGTGGTTCTGGTAGGAGACG | 480 | 501 |
| F5 | TTTGCTTTCGGTTTTTCGC | 539 | 558 |
| R1 | TCCTACCAGAACCACATATCC | 476 | 497 |
| R2 | TCGGARCCAAACCGAAAGC | 538 | 557 |
| R4 | GCTCGACATCTTTCCAGTG | 680 | 699 |
| R5 | TGCAGAGCAGAAGGTRACC | 742 | 761 |
| R6 | GGTTAAARGTRCCRTCTCG | 791 | 810 |
| R7 | YTCCCAGGTSACSATGTAK | 880 | 899 |
| R8 | AAAGGGTTTGACCCAGGGAG | 918 | 938 |
| Feature | |||
| 5′ long terminal repeat | 1 | 73 | |
| Unique 5′ sequence | 74 | 143 | |
| Primer binding site | 146 | 163 | |
F: Forward; R: Reverse.
PCR amplification of JHKV RNA with consensus primers
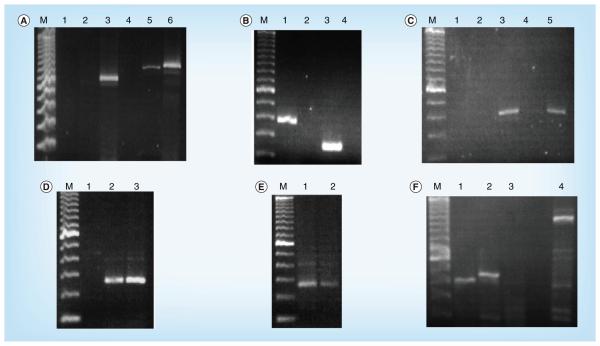
To test the primers, cDNA produced from JHK viral RNA purified by the urea-nuclease procedure was used as a template in an amplification reaction with the initial sets of primers in combination. Figure 2A shows the successful application of JHK viral cDNA with the F1 forward primer and the R5, R7 and R8 reverse primers (lanes 3, 5 and 6). The sizes of the observed amplified products were consistent with the predicted sizes of the amplified products (729, 848 and 888 bp, respectively). Cloning and sequencing of the fragments showed that they shared significant sequence identity with the gammaretrovirus sequences. To extend this finding, additional primer pairs were used in PCR reactions with the JHK viral cDNA as a template. Figure 2B shows the successful amplification with the F5 and F6 forward primers and the R9 reverse primer (lanes 1 and 3). Again, the predicted fragment sizes (365 and 236, respectively) are consistent with the amplified products, and sequences of the cloned fragments show a high degree of conservation with gammaretroviruses.
Figure 2. PCR amplification of JHK viral sequences in virions and cells.
(A) PCR amplification of JHK cDNA. PCR reactions were carried out using JHK cDNA as a template. The F1 forward primer was used in all reactions. Reverse PCR primers used were: lane 1, R1; lane 2, R4; lane 3, R5; lane 4, R6; lane 5, R7; and lane 6, R8. Lane M contains a 100-bp marker DNA sample. (B) PCR amplification of JHK cDNA. PCR primers used were: lane 1, F5 and R9; lane 2, F5 and R10; lane 3, F6 and R9; and lane 4, F6 and R10. Lane M contains a 100-bp marker DNA sample. (C) PCR amplification of cDNA from JHK virions and JHK-3 cell total RNA. PCR reactions were carried out using either cDNA prepared from JHK virions (lanes 1–3) or JHK-3 cell total RNA (lanes 4 and 5) as a template. PCR primers used were: lane 1, F5 alone; lane 2, R8 alone; and lanes 3–5, F5 and R8. Lane 4 did not contain Taq polymerase. Lane M contains a 100-bp marker DNA sample. (D) PCR amplification of JHK total RNA with and without reverse transcriptase. PCR reactions were carried out using either JHK total RNA (lanes 2 and 3) as a template or no template (lane 1). The F5 and R8 PCR primers were used in all reactions. Lane 2 contained reverse transcriptase and lane 3 did not. Lane M contains a 100-bp marker DNA sample. (E) PCR amplification of index patient total peripheral blood mononuclear cell-extracted RNA with and without reverse transcriptase. PCR reactions were carried out using total RNA derived from peripheral blood lymphocytes isolated from the index patient (lanes 1 and 2) as a template. The F5 and R8 PCR primers were used in all reactions. Lane 1 contained reverse transcriptase and lane 2 did not; the band in lane 2 is most probably due to residual DNA contaminating cellular RNA extracts requiring DNase treatment [17]. Lane M contains a 100-bp marker DNA sample. (F) PCR amplification of total RNA from cell lines. PCR reactions were carried out using total RNA derived from JHK cells (lanes 1 and 4), DG-75 (strain UW) containing DG-75 murine leukemia virus-like virus cells (lane 2) or A549 cells (lane 3) as a template. The F5 and R8 PCR primers were used in lanes 1–3, and the F1 and R8 PCR primers were used in lane 4. Lane M contains a 100-bp marker DNA sample.
Figure 2C demonstrates that the amplification of the JHKV cDNA was dependent on the presence of both the forward and reverse primers as well as Taq polymerase. Individual forward or reverse primers (lanes 1 and 2), as well as reactions that lacked Taq polymerase (lane 4), did not produce amplified product. Amplified product of the correct size was observed with both primers and Taq polymerase, using either cDNA prepared from purified JHKV or total RNA prepared from JHK-3 cells. The amplification of total JHK-3 RNA indicates the potential presence of integrated DNA copies of the sequence. To examine that possibility more directly, JHK-3 total cellular RNA was used as template with the F5 forward and R8 reverse primers. No amplification was detected in the absence of template (lane 1), but amplification of a DNA fragment was observed with JHK total cellular RNA that was either treated (lane 3) or not treated (lane 2) with RT. A reproducible increase in product was detected with the treatment of the total cellular RNA with RT, indicating that both RNA and DNA copies of the sequence are present in the total cellular RNA preparation.
JHKV amplicon in nucleic acid of PBMCs of the IP
Since the JHK-3 cells were originally developed by coculturing peripheral lymphocytes isolated from the IP with normal human PBMCs, we wished to determine if the sequence could be amplified directly from total cellular RNA extracted from PBMCs of the IP that were stored frozen at −80°C without having been opened since collection in 1992. Figure 2E shows the amplification of the patient’s RNA with the F5 forward and R8 reverse primers. Amplified product is present in lanes 1 and 2 both with and without incubation of the crude RNA fraction with RT. Figure 2E demonstrates the same amplified fragments and sequences observed with this template, reducing the possibility of contamination with murine sequences. We also screened total cellular RNA from the PBMCs of nine normal lymphocyte blood donors and observed no amplification (data not shown).
Testing of consensus primers in other human cell lines
To determine if other human cell lines carried the same sequence, amplifications (Figure 2F) were carried out with the F5 forward and R8 reverse primers (lanes 1–3) using total cellular RNA from JHK-3 cells (lanes 1 and 4), human B-lymphoblastoid cells DG-75 (strain UW), a subline chronically infected with the DG-75 gammaretrovirus (lane 2) [18,24]; or virus-free human bronchioloalveolar A549 cells. Amplified products were observed with the total cellular RNA from JHK-3 and DG-75 cells, but not from A549 cells. The amplified product from the retrovirus-infected DG-75 cell line was slightly larger than the JHK-3 product. Cloning and sequencing of amplified products produced inserts with sequences that matched the putative JHK sequence obtained with JHK cellular RNA template (lane 1), and the DG-75 viral sequence with the DG-75 cellular RNA template (in lane 2). In lane 3, although there was not a significant band, some clones were obtained that corresponded to human genomic sequences with A549 cellular RNA template. In addition, F1 forward and R8 reverse primers were used with the JHK template (lane 4) to obtain a DNA fragment with a size corresponding to the predicted 888 bp, indicating that the sequence extending from the putative LTR to the gag coding region was present as a single piece.
Relationship of JHKV to other retroviral genomes
The sequences of the amplified JHK products were assembled to give several-fold redundant sequence coverage in both directions in order to construct a consensus sequence. A search using the basic local alignment search tool of the NCBI nr nucleotide database with the consensus sequence revealed that the most similar sequences included gammaretroviruses and integrated murine proviruses.
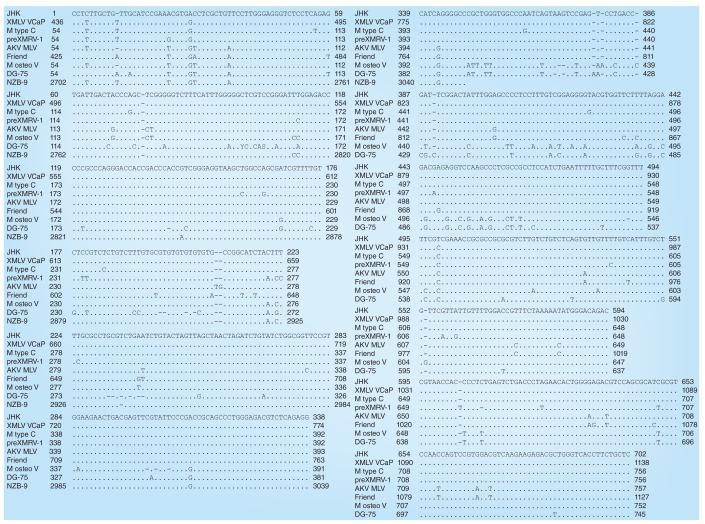
The most similar sequences to JHKV with >99% identity were those of the xenotropic MLV isolate VCaP (GenBank accession JF908815.1), MLV N417 (GenBank accession HQ246218.1), xenotropic MLV isolate LAPC4 (GenBank accession JF908816.1) and the murine retrovirus C genome (GenBank accession X94150). The DG-75 virus sequence (GenBank accession AF221065.1) [24] is 94% identical to JHKV. The highest region identity between JHKV and xenotropic MuLV-related virus VP62 (GenBank accession EF185282.1) is 84%, and JHKV does not have the characteristic 24-bp deletion found in XMRV [2]. The alignment of the JHKV sequence with the genomic sequences of the murine type C retrovirus, DG-75 MLV, and xenotropic MLV isolate VCaP are shown in Figure 3.
Figure 3. Alignment of the JHK sequence with other gammaretrovirus-like sequences (cont.).
Alignment of the JHK virus sequence (GenBank accession number HM119591) with other retroviral sequences (M type C retrovirus [GenBank accession number X94150.1], DG-75 MLV [GenBank accession number AF221065.1] and XMLV isolate VCaP [GenBank accession number JF908815.1]) using the CLUSTAL 2.1 algorithm provided on the European Bioinformatics Institute website [101].
M: Murine; MLV: Murine leukemia virus; XMLV: Xenotropic murine leukemia virus.
The isolation of a new viral sequence by a PCR approach raises the possibility of potential artifacts due to contamination with extraneous viral sequences. Since our laboratory had previously isolated and sequenced the entire DG-75 retrovirus genome [24], it was possible that the amplified product was DG-75 virus, but the results show that this was not the case. First, as seen in Figure 2F, the size of the amplified products was observed to be slightly different from those observed with the JHK and DG-75 cellular RNA templates. Second, the JHKV and DG-75 sequences are approximately only 94% identical, a difference unlikely to be due to sequencing errors since there are a number of other retroviruses and proviral inserts that have higher levels of identity to the JHK sequence, in particular Bxc-1.
A second potential source of contamination would be mouse sequences, although since the sequence was also amplified from material directly isolated from the IP, this would appear to be unlikely. To directly examine the possibility that the JHK cells were contaminated with mouse cells, PCR primers specific for mouse and human repetitive DNA sequences were used in the method described by Pelz et al. [25]. Only human-specific sequences were amplified from human A549 and JHK-3 cells and only mouse-specific sequences were amplified from murine L929 cells, indicating that any mouse cell contamination of the JHK-3 cell line was below the limit of detection for the assay (0.01% cell contamination; data not shown).
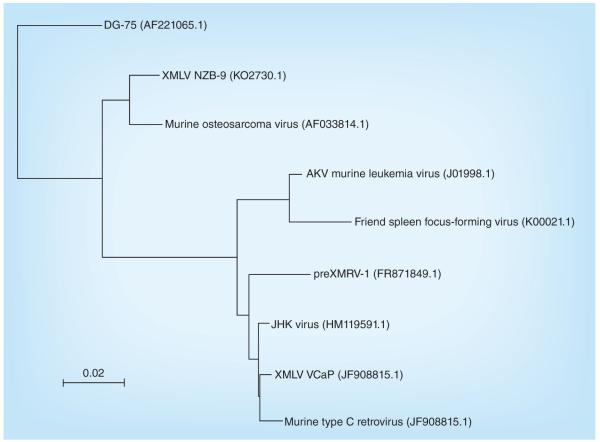
Phylogenetic sequence relationships to JHKV are shown in Figure 4.
Figure 4. Phylogenetic sequence tree diagram.
Created with NCBI Blast Tree View. GenBank accession numbers are shown in parentheses.
XMLV: Xenotropic murine leukemia virus.
Ultrastructural similarity of budding virions in uncultivated PBMCs of the IP & in JHK-3 cell cultures
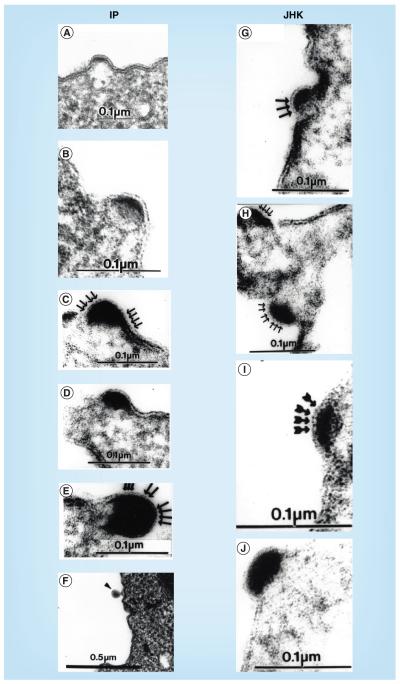
Figure 5 shows budding virion structures observed by transmission EM of ultrathin sections of Epon-embedded, uncultivated PBMCs taken from the blood of the IP in side-by-side comparison with the developing JHK virion forms previously described in the JHK-3 cell cultures [16]. The IP’s mononuclear cells, many with distorted nuclei, showed 75–85-nm enveloped viral particles budding from the plasma membranes of B lymphocytes (see ‘Results’ section) in the four samples taken January, April and September of 1989 and February 1990, but not subsequently. Free mature virions were not observed in PBMC preparations, probably attributable to the operations involved in separation of mononuclear cells from other blood components, followed by washing prior to suspension in glutaraldehyde fixative. These data indicate, but do not prove, that the IP had been infected by JHKV or a very similar virus.
Figure 5. Budding retrovirion formation in peripheral blood mononuclear cells from the index patient and in JHK-3 cell cultures.
Transmission electron microscopy of Epon-embedded ultrathin sections of (A–F) freshly obtained, uncultivated, Ficoll-Hypaque-separated peripheral blood mononuclear cells from the IP, and (G–J) JHK-3 cultivated cells. Note the similarity of the retrovirus-like budding in the IP’s peripheral blood mononuclear cells and the budding virions in JHK-3 cells [16], with the progressive accumulation in the forming core of highly electron-dense material, which is sometimes crescentic, but more usually biconvex (lenticular) or semilunar, under the plasma membrane with apparent scalloping (arrows), in comparison with the adjacent, less osmiophilic cytoplasm.
IP: Index patient.
Binding of antibodies to JHK virions by immunogold labeling EM
Due to the lack of adequate amounts of purified JHKV for reliably detecting JHKV-specific antigen by common immunologic tests, direct visualization by EM of binding of antibodies to JHK virions was considered to be most specific.
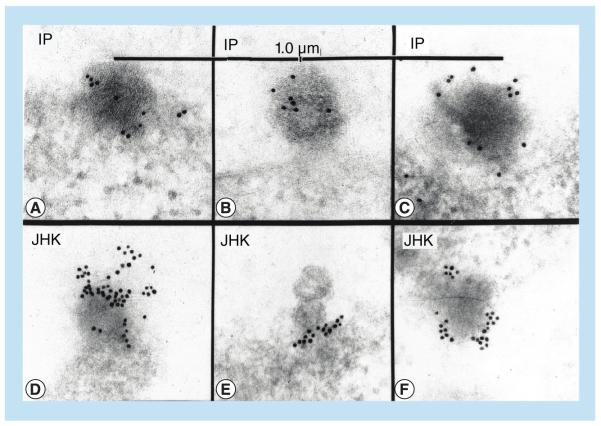
Figure 6 demonstrates by direct immunogold EM on Lowicryl-K4M-embedded ultrathin sections that the patient’s IgG conjugated with 10-nm colloidal gold bound to budding virions in the patient’s own B lymphocytes as well as to the budding virions in the JHK-3 cells. Lowicryl-K4M was used because it preserves antigens better than the Epon procedure, while providing the necessary resolution. Table 2 shows the specificity and statistical significance of the patient’s IgG binding to budding virions compared with adjacent plasma membrane. As further tests of specificity, controls were employed with directly gold-labeled patient IgG to show that binding was specific to JHKV and not to MLV LP-BM5 retrovirus, and pretreatment of JHK virions with unlabeled IP’s IgG prevented subsequent binding of the patient’s gold-labeled IgG (data not shown).
Figure 6. Binding of colloidal gold-labeled IgG from the index patient to budding virions in peripheral blood mononuclear cells of the index patient and in JHK-3 cells by the direct electron microscopic immunogold method.
The IP’s IgG, conjugated with 10-nm colloidal gold, was used to label budding viral particles in Lowicryl-K4M-embedded ultrathin sections of JHK virions in the JHK-3 cells (D–F), and in the IP’s uncultivated lymphocytes (A–C), as described in the ‘Materials & methods’ section. The difference between labeling was determined stereologically [23], as the labeling density in terms of number of 10-nm gold particles per μm2 of the plasma membrane plus the adjacent cytoplasm versus budding structures, and evaluated by the nonparametric Mann–Whitney two-tailed test.
IP: Index patient.
Table 2.
Specificity of labeling of budding virus particles in thin sections of JHK-3 cells and the index patient’s lymphocytes with index patient IgG conjugated with colloidal gold.
| Budding structures (± SEM) |
Plasma membrane (± SEM) |
Significance† | |
|---|---|---|---|
| JHK-3 cells | 1.52 ± 0.19 (n = 31) |
0.72 ± 0.08 (n = 184) |
p < 0.001 |
| Index patient’s lymphocytes |
5.00 ± 0.70 (n = 12) |
1.50 ± 0.18 (n = 107) |
p < 0.02 |
Differences between labeling densities were determined stereologically as the number of 10-nm gold particles per μm2 [23] of the plasma membrane plus the adjacent cytoplasm versus budding structures (nonparametric Mann–Whitney two-tailed p-value).
SEM: Standard error of the mean.
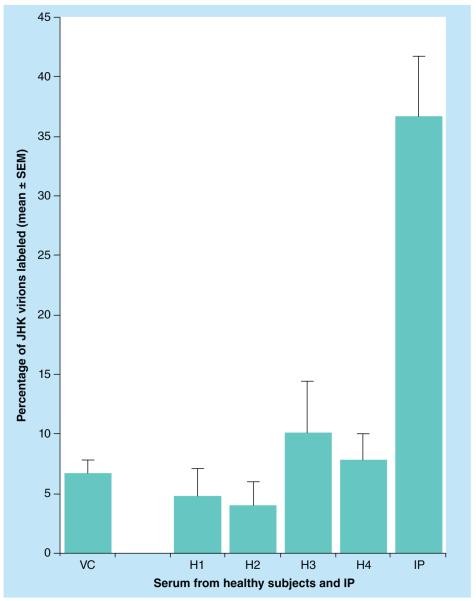
Figure 7 shows the results of a double-blinded quantitation by the indirect immunogold EM method. There was significant binding of anti-JHKV IgG in the patient’s serum to JHK virions as compared with IgG in serum samples from control healthy subjects. JHKV virions were bound to EM grids, treated with serum samples, then exposed to goat antihuman IgG conjugated with 10-nm colloidal gold, and the percentages of gold-labeled virions were determined as described in the ‘Materials & methods’ section. These data obtained by the two immunogold EM methods suggest that the IP had been infected by JHKV or an antigenically very similar virus.
Figure 7. Measurement of binding to negatively stained JHK virions of IgG in the sera from healthy control subjects and the index patient by the quantitative indirect electron microscopic immunogold method.
The method of procedure and double-blinded protocol are as described in the ‘Materials & methods’ section. H1 is the normal donor whose peripheral blood mononuclear cells were cocultivated in the original mixed lymphocyte culture with peripheral blood mononuclear cells of the IP to produce the JHK-3 cell line and from whom the K-3II cell line was derived [16]. The p-values determined by the nonparametric Mann–Whitney two-tailed test compared each healthy person’s serum result against measurements in the IP’s serum.
p-value range: <0.002 to <0.0001.
H: Healthy subject; IP: Index patient; SEM: Standard error of the mean;
VC: JHK virion control without serum.
Discussion
Identification of the JHKV
The previously described JHKV [16] is now identified as a MLV based on its gag sequence homology most similar to Bxv-1, and quite distinct from XMRV. The identification of the JHKV was made possible by the use of the consensus PCR primers created by the approach described herein, following many years of failure using a variety of molecular methods to clone different portions of the JHK retroviral genome. The following findings lend some significance to the demonstration of the JHKV MLV-like amplicon in nucleic acids of PBMCs of the IP: the apparent ultrastructural identity of budding retrovirus particles in ultrathin sections of the JHK-3 cell cultures and of uncultivated PBMCs from the blood of IP revealed by transmission EM; the binding of IP’s gold-conjugated IgG to budding virions in both the ultrathin sections of cultivated JHK-3 B-lymphoblastoid cells and of uncultivated PBMCs from IP; and the significant binding of IgG from the IP to partially purified virions from JHK-3 cells compared with the IgG from healthy subjects, also demonstrated by immunogold EM techniques.
The matter of MLV-related sequence contamination
Possible sources of contamination
The most likely source of retroviral contamination in our laboratory would be from the two human B-lymphoblastoid cell strains, UW or KAR, chronically infected with DG-75 virus [18], a xenotropic MLV-like virus, the entire genome of which we had previously sequenced [24]. Another possible contaminant could be the MLV-like LP-BM5 virus produced in SC-1 cells propagated in our laboratory [16]. The sequences of each of these viruses are distinct from that of the JHKV (see ‘Results’ section), both with 94% identity. One formal possibility is that the JHKV MLV-like gag sequence is a mouse contaminant. Although it is known, or has been suspected, that other cell lines of different cellular origins have been propagated in the peritoneum of mice, either to help establish the cell line or to rid cells of mycoplasma (at one time not uncommon practices), the JHK-3 cells were never propagated in mice. Our PCR assays with primers directed to repetitive DNA sequences from mouse and human did not detect mouse cell contamination of the JHK-3 cells (see ‘Results’ section).
Approaches to showing sequence is not a contaminant
The use of our PCR primers demonstrated bona fide JHKV gag sequence amplicons in the IP’s PBMCs that had been stored at −80°C in ampoules never opened from the time that the IP’s blood was immediately processed after collection in 1992 from the patient until opened for nucleic acid extraction for PCR. Furthermore, JHKV-like budding virions were demonstrated in the patient’s PBMCs freshly obtained on four occasions over a 13-month period (Figure 5). Finally, IgG antibodies in the patient’s serum were shown by direct immunogold EM to bind to JHK virions in the JHK-3 cells as well as to the budding virions in the patient’s PBMCs. These findings suggest that the patient had been infected with JHKV or a similar virus. From this we conclude that the JHKV sequence was most likely amplified from a virus either infecting or integrated into the JHK-3 cells and not from contamination with other viruses, cells or sequences. Although it may be possible to use some of these nearly identical sequences to design primers so as to amplify additional genome sequence, this was not consonant with the goal of using primers that are potentially capable of amplifying a class of sequences rather than a specific one. Sequencing the entire genome, especially the env region, and analysis of viral integration sites should further resolve an additional formal possibility that the viral sequence we have cloned is not from the virus in the patient’s lymphocytes.
Very recently, Das Gupta et al. listed the multiple possible sources of mouse virus or nucleic acid contamination; these include mouse DNA in trace amounts in some commercial Taq polymerases (especially so-called hot-start preparations containing mouse antibody), PCR master mix preparations or DNA extraction kits in PCR assays, in addition to retrovirus-contaminated cell lines [8]. Such laboratory contamination led to the retraction of the original report of association of XMRV in CFS patients [26]. It has been pointed out that some of the positive findings that involved non-PCR-based methods, such as serology, immunohistochemistry and fluorescence in situ hybridization (methods that apparently detected XMRV or similar viruses in human samples), do not have an adequate explanation. It was further indicated that such evidence leaves open the possibility that either mouse DNA or an XMRV- or other MLV-like virus may be present in at least some human beings. In contrast to most studies detecting MLV-like sequences by PCR, we attempted to support molecular cloning of the JHKV by evidence obtained by EM and serology with the patient’s IgG.
Reported association of MLV-like sequences in human patients
Other investigators had previously identified MLV-like gag sequences following PCR of nucleic acid samples from whole blood, PBMCs or plasma from human subjects [11–13]. Of particular interest are the reports by Lo et al. of genetically diverse groups of MLV-related sequences with a closer relationship to those of polytropic mouse endogenous retroviruses than to either XMRVs or ecotropic MLVs [11,12]. More recently, Lee et al. have identified MLV-like gag sequences using very sensitive PCR assays with 15 sets of primers that can amplify conserved regions of murine endogenous and exogenous retrovirus sequences from blood and PBMCs of CFS/ myalgic encephalopathy (ME) patients and healthy controls [13]. They, and others, have pointed out, however, that most of the primer sets that had been used to study CFS/ME would not allow detection of all groups of MLVs or even other members of the gammaretrovirus family [27].
The JHKV cannot yet be claimed to be linked to the IP’s illness as etiological cause or consequence, although the amplification by PCR from the PBMCs of the IP, the budding virions in the patient’s lymphocytes, and the demonstration by EM of the patient’s IgG antibodies binding to JHK virions provide evidence for infection by the JHK virus. Establishing a possible etiologic relationship would require a rather large clinical study of patients and controls. Although the history of the patient’s ill-defined, viral-like illness, from which the patient recovered completely some years later, bears a resemblance to CFS/ME (among other disease complexes), no definite diagnostic marker yet exists for CFS/ME. Negative serologic studies with purified XMRV antigen, either by western blot [28] or chemiluminescent assay [29], have shown that XMRV has not caused infection in CFS/ME or healthy subjects, most recently substantiated by Lee et al. [14] and Alter et al. [15]. As previously noted, a number of relevant PCR studies have made clear the possible association of mouse virus contamination either as MLV-like viruses or nucleic acids in materials from patients and healthy donors; and Lee et al. could not conclude that the sequences they detected by PCR originated in the blood samples tested [13]. No study to our knowledge has shown a direct relationship to viral particles infecting the patient, such as we have shown by the ultrastructural similarity of the budding viral forms in the uncultivated PBMCs of IP to those in the JHK-3 cells.
The very recent study by Alter et al. of a large, well-controlled clinical study of CFS/ME patients and healthy subjects clearly failed to show, with the use of their PCR primers, any connection to XMRV or polytropic MLV-like viruses [15]. A sequence homology search comparing the PCR primers their study utilized with the primers described herein showed that their sequences were not a good match with ours. In our estimation, the primers used in those studies would not have been able to detect MLV-related JHKV-like strains, if present. The PCR primers we describe may expand the possibly detectable subsets of viruses related to MLV and may be helpful for use in future studies. As noted by Weiss, the absence of evidence is not evidence of absence [30].
Future perspective
JHKV has somewhat unusual morphologic and morphogenetic features [16]. In ultrathin sections of JHK-3 cells JHK virions had been measured to be 85.3 nm (± 2.8 standard error of the mean [SEM]), compared with ultrathin sections of HTLV-1 that measured 112 nm (± 2.2 SEM), and of MLV LP-BM5 119 nm (± 2.1 SEM) (p < 0.0001). Negatively stained mature JHK virions had an overall diameter of 72 nm (0.95 SEM), further indicating that they are smaller than those of at least some classical retroviruses. Although it is generally considered that retroviruses do not have icosahedral shape, Markham rotational image enhancement [31] had demonstrated five- and sixfold axes of symmetry of JHK virions in both ultrathin sections or negatively stained preparations, indicating an icosahedral shape of the virion and nucleocapsid. During viral budding, the forming core was more usually lenticular, sometimes crescentic or semilunar under the plasma membrane, with apparent indentations or scalloping over the budding virus membrane [16], very similar to the budding forms (Figure 5) observed in uncultivated PBMC samples taken repeatedly from the patient over a 13-month period.
Based on the evidence presented, we do not conclude that the JHK virus is a ‘human retrovirus’. Whether the JHK virions observed, the amplicons sequenced or possibly JHKV-related gammaretroviruses may be involved in identifiable human infection awaits further study, for which our PCR primers may enable detection.
Executive summary.
▪The JHK virus (JHKV) was previously described as a retrovirus constitutively produced in a B-lymphoblastoid human cell line JHK-3 established by cocultivation of blood mononuclear cells from a patient and a healthy subject.
▪Consensus retroviral primers were designed by a data-driven approach for use in PCR with purified JHK virion RNA to identify JHKV as murine leukemia virus (MLV)-like, with a gag sequence most similar to MLV-Bxv-1, but dissimilar to XMRV and other MLV-like variants.
▪With these primers, JHKV gag-specific amplification was demonstrated in frozen, freshly obtained, peripheral blood mononuclear cells (PBMCs) of the patient, but not in nine healthy blood donors.
▪Electron microscopy demonstrated budding virions in uncultivated PBMCs of the index patient that appeared essentially identical to those in JHK-3 cell cultures.
▪Quantitative direct immunogold electron microscopy showed the binding of the patient’s labeled IgG to JHK virions in the JHK-3 cells as well as to the budding virions in the patient’s lymphocytes.
▪Quantitative indirect immunogold electron microscopy showed significant binding of JHK virions to the patient’s IgG antibodies, but not the IgG of healthy subjects.
▪The data indicate that the patient had been infected by the MLV-like JHKV, lending significance to the demonstration of the JHKV amplicon in PBMCs of the patient.
Acknowledgements
We thank M Casey for excellent technical assistance. We gratefully acknowledge insightful advice from D Miller and RH Silverman, and we thank LW Cashdollar for stimulating discussions.
The work was supported by grants from the NIH RO1-AI32710, the Centers for Disease Control and Prevention, the American Cancer Society, the CFIDS Association of America, and the Council for Tobacco Research USA, as well as an American Cancer Society Scholar in Cancer Research award to SE Grossberg.
Ethical conduct of research The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
Footnotes
For reprint orders, please contact: reprints@futuremedicine.com
Financial & competing interests disclosure The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
- 1.Voisset C, Weiss RA, Griffiths DJ. Human RNA “rumor” viruses: the search for novel human retroviruses in chronic disease. Microbiol. Mol. Biol. Rev. 2008;72(1):157–196. doi: 10.1128/MMBR.00033-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Urisman A, Molinaro RJ, Fischer N, et al. Identification of a novel gammaretrovirus in prostate tumors of patients homozygous for R462Q RNASEL variant. PLoS Pathog. 2006;2(3):e25. doi: 10.1371/journal.ppat.0020025. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 3.Lombardi VC, Ruscetti FW, Das Gupta J, et al. Detection of an infectious retrovirus, XMRV, in blood cells of patients with chronic fatigue syndrome. Science. 2009;326(5952):585–589. doi: 10.1126/science.1179052. [DOI] [PubMed] [Google Scholar]
- 4.Cingoz O, Coffin JM. Endogenous murine leukemia viruses: relationship to XMRV and related sequences detected in human DNA samples. Adv. Virol. 2011;2011:940210. doi: 10.1155/2011/940210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Knox K, Carrigan D, Simmons G, et al. No evidence of murine-like gammaretroviruses in CFS patients previously identified as XMRV-infected. Science. 2011;333(6038):94–97. doi: 10.1126/science.1204963. [DOI] [PubMed] [Google Scholar]
- 6.Paprotka T, Delviks-Frankenberry KA, Cingoz O, et al. Recombinant origin of the retrovirus XMRV. Science. 2011;333(6038):97–101. doi: 10.1126/science.1205292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kearney MF, Spindler J, Wiegand A, et al. Multiple sources of contamination in samples from patients reported to have XMRV infection. PLoS ONE. 2012;7(2):e30889. doi: 10.1371/journal.pone.0030889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Das Gupta J, Luk KC, Tang N, et al. Absence of XMRV and closely related viruses in primary prostate cancer tissues used to derive the XMRV-infected cell line 22Rv1. PLoS ONE. 2012;7(5):e36072. doi: 10.1371/journal.pone.0036072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sfanos KS, Aloia AL, Hicks JL, et al. Identification of replication competent murine gammaretroviruses in commonly used prostate cancer cell lines. PLoS ONE. 2011;6(6):e20874. doi: 10.1371/journal.pone.0020874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin Z, Puetter A, Coco J, et al. Detection of murine leukemia virus in the Epstein–Barr virus-positive human B-cell line JY, using a computational RNA-Seq-based exogenous agent detection pipeline, PARSES. J. Virol. 2012;86(6):2970–2977. doi: 10.1128/JVI.06717-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lo SC, Pripuzova N, Li B, et al. Detection of MLV-related virus gene sequences in blood of patients with chronic fatigue syndrome and healthy blood donors. Proc. Natl Acad. Sci. USA. 2010;107(36):15874–15879. doi: 10.1073/pnas.1006901107. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Lo SC, Pripuzova N, Li B, et al. Retraction for Lo et al., Detection of MLV-related virus gene sequences in blood of patients with chronic fatigue syndrome and healthy blood donors. Proc. Natl Acad. Sci. USA. 2012;109(1):346. doi: 10.1073/pnas.1119641109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee LL, Lin L, Bell DS, Levine S, Hanson MR. Sensitivity of PCR assays for murine gammaretroviruses and mouse contamination in human blood samples. PLoS ONE. 2012;7(5):e37482. doi: 10.1371/journal.pone.0037482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee D, Das Gupta J, Gaughan C, et al. In-depth investigation of archival and prospectively collected samples reveals no evidence for XMRV infection in prostate cancer. PLoS ONE. 2012;7(9):e44954. doi: 10.1371/journal.pone.0044954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alter HJ, Mikovits JA, Switzer WM, et al. A multicenter blinded analysis indicates no association between chronic fatigue syndrome/myalgic encephalomyelitis and either xenotropic murine leukemia virus-related virus or polytropic murine leukemia virus. MBio. 2012;3(5):e00266–12. doi: 10.1128/mBio.00266-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grossberg SE, Kushnaryov VM, Cashdollar LW, Raisch KP, Miller G, Sun HY. A human B-lymphoblastoid cell line constitutively producing Epstein–Barr herpesvirus and JHK retrovirus. Res. Virol. 1997;148(3):191–206. doi: 10.1016/s0923-2516(97)83989-1. [DOI] [PubMed] [Google Scholar]
- 17.Sun HY, McNally MT, Jackson VE, Grossberg SE. Urea-nuclease treatment of concentrated retrovirions preserves viral RNA and removes polymerase chain reaction-amplifiable cellular RNA and DNA. J. Virol. Methods. 2006;137(2):304–308. doi: 10.1016/j.jviromet.2006.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raisch KP, Kushnaryov VM, Grossberg SE, Cashdollar LW. Constitutive production of a murine retrovirus in the human B-lymphoblastoid cell line, DG-75. Virology. 1998;250(1):135–139. doi: 10.1006/viro.1998.9363. [DOI] [PubMed] [Google Scholar]
- 19.Carrigan DR, Knox KK, Tapper MA. Suppression of human immunodeficiency virus type 1 replication by human herpesvirus-6. J. Infect. Dis. 1990;162(4):844–851. doi: 10.1093/infdis/162.4.844. [DOI] [PubMed] [Google Scholar]
- 20.Hayat MA, Miller SE. Negative Staining. McGraw-Hill; NY, USA: 1990. pp. 188–211. [Google Scholar]
- 21.Fassel TA, Raisch KP, Chetty N, Grossberg SE, Kushnaryov VM. Ruthenium red preserves glycoprotein peplomers of C-type retroviruses for transmission electron microscopy. Biotech. Histochem. 1998;73(4):222–227. doi: 10.3109/10520299809141113. [DOI] [PubMed] [Google Scholar]
- 22.Nermut MV, Green NM, Eason P, Yamada SS, Yamada KM. Electron microscopy and structural model of human fibronectin receptor. EMBO. 1988;7(13):4093–4099. doi: 10.1002/j.1460-2075.1988.tb03303.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Russ JC. Practical Stereology. Plenum Press; NY, USA: 1986. [Google Scholar]
- 24.Raisch KP, Pizzato M, Sun HY, Takeuchi Y, Cashdollar LW, Grossberg SE. Molecular cloning, complete sequence, and biological characterization of a xenotropic murine leukemia virus constitutively released from the human B-lymphoblastoid cell line DG-75. Virology. 2003;308(1):83–91. doi: 10.1016/s0042-6822(02)00074-0. [DOI] [PubMed] [Google Scholar]
- 25.Pelz O, Wu M, Nikolova T, et al. Duplex polymerase chain reaction quantification of human cells in a murine background. Stem Cells. 2005;23(6):828–833. doi: 10.1634/stemcells.2004-0206. [DOI] [PubMed] [Google Scholar]
- 26.Alberts B. Retraction. Science. 2011;334(6063):1636. doi: 10.1126/science.334.6063.1636-a. [DOI] [PubMed] [Google Scholar]
- 27.Elfaitouri A, Shao X, Mattsson Ulfstedt J, et al. Murine gammaretrovirus group G3 was not found in Swedish patients with myalgic encephalomyelitis/chronic fatigue syndrome and fibromyalgia. PLoS ONE. 2011;6(10):e24602. doi: 10.1371/journal.pone.0024602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Satterfield BC, Garcia RA, Jia H, Tang S, Zheng H, Switzer WM. Serologic and PCR testing of persons with chronic fatigue syndrome in the United States shows no association with xenotropic or polytropic murine leukemia virus-related viruses. Retrovirology. 2011;8:12. doi: 10.1186/1742-4690-8-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oakes B, Qiu X, Levine S, Hackett J, Jr, Huber BT. Failure to detect XMRV-specific antibodies in the plasma of CFS patients using highly sensitive chemiluminescence immunoassays. Adv. Virol. 2011;2011:854540. doi: 10.1155/2011/854540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weiss RA. A cautionary tale of virus and disease. BMC Biol. 2010;8:124. doi: 10.1186/1741-7007-8-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nermut MV, Grief C, Hashmi S, Hockley DJ. Further evidence of icosahedral symmetry in human and simian immunodeficiency virus. AIDS Res. Hum. Retroviruses. 1993;9(10):929–938. doi: 10.1089/aid.1993.9.929. [DOI] [PubMed] [Google Scholar]
Website
- 101.The European Bioinformatics Institute Multiple Sequence Alignment. www.ebi.ac.uk/Tools/msa/clustalw2.