Abstract
Kinesin motor proteins comprise an ATPase superfamily that goes hand in hand with microtubules in every eukaryote. The mitotic kinesins, by virtue of their potential therapeutic role in cancerous cells, have been a major focus of research for the past 28 years since the discovery of the canonical Kinesin-1 heavy chain. Perhaps the simplest player in mitotic spindle assembly, Kinesin-5 (also known as Kif11, Eg5, or kinesin spindle protein, KSP) is a plus-end-directed motor localized to interpolar spindle microtubules and to the spindle poles. Comprised of a homotetramer complex, its function primarily is to slide anti-parallel microtubules apart from one another. Based on a multi-faceted analysis of this motor from numerous laboratories over the years, we have learned a great deal about the function of this motor at the atomic level for catalysis and as an integrated element of the cytoskeleton. These data have, in turn, informed the function of motile kinesins on the whole, as well as spearheaded integrative models of the mitotic apparatus in particular and regulation of the microtubule cytoskeleton in general. We review what is known about how this nanomotor works, its place inside the cytoskeleton of cells, and its small-molecule inhibitors that provide a toolbox for understanding motor function and for anticancer treatment in the clinic.
Keywords: kinesin, motor protein, phylogeny, evolution, cytoskeletal motor, catalysis, ATP hydrolysis, mitosis, mitotic spindle, microtubule, mitotic motor protein, targeted inhibitor, Eg5, KSP, Kif11, structural biology, ispinesib, monastrol, allosteric inhibition, L5 loop, review
1. INTRODUCTION
Kinesins are the smallest and most abundant nanomotor in cells and the only canonical motor protein that is ubiquitous in all eukaryotes. Like dynein and myosin, these proteins hydrolyze ATP and convert chemical energy into mechanical energy. This permits transport and movement along the cytoskeletal track. Of the >16 different kinesin isoforms (Wickstead and Gull, 2006), Kinesin-5 (BimC/Eg5/N-2/Kif11) family members were the first identified to be essential in mitosis. Genetic analysis provided the initial insight into the key mitotic role of this motor family. In the search for strains that were defective in cellular division at a restrictive temperature, screens of fungal libraries and fission yeast uncovered BimC (Enos and Morris, 1990) and Cut7 (Hagan and Yanagida, 1990), respectively, in the early 1990s. These mutants had malfunctions in spindle pole body separation and nuclear division and were unable to undergo mitosis. Cloning and sequencing of these mutants (Hagan and Yanagida, 1990; Kashina et al., 1997) revealed that the gene product encoded a 130 kDa protein with high similarity to the conventional kinesin (KHC/Kinesin-1) involved in motility in squid and mammalian brains. Similarly, simultaneous loss of function in Cin8p and Kip1p in S. cerevisiae revealed a redundant function of these Kinesin-5 family members for spindle assembly (Hoyt et al., 1992; Roof et al., 1992).
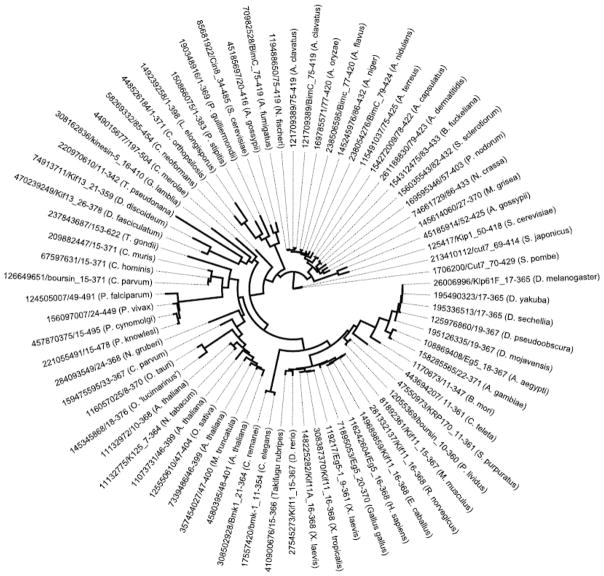
The gene family has expanded from the early days of the molecular and bioinformatics era. Many groups (Hoyt et al., 1992; Sawin et al., 1992; Tihy et al., 1992; Heck et al., 1993; Blangy et al., 1995; Bishop et al., 2005; Bannigan et al., 2007; Chauviere et al., 2008) initially identified orthologs in Xenopus, S. cerevisiae, D. melanogaster, H. sapiens, M. musculus, and C. elegans (Table 1). Currently, there are over 70 different Kinesin-5 proteins identified by sequence homology in 66 eukaryotes (Figure 1). Subsequently classified as the Kinesin-5 family (Lawrence et al., 2004), this group of related kinesins localizes to spindle microtubules and structures present at spindle poles.
Table 1.
Members of the Kinesin-5 family from different eukaryotes and their roles.
| Organism | Protein abbreviation | Subcellular localization | Function | % Identity to human ortholog | NCBI Gene ID |
|---|---|---|---|---|---|
| A. nidulans | BimC | Not determined | Assembly of spindle microtubule array & separation of spindle pole bodies | 54% | 2874347 |
| D. discoideum | Kif13 | Not determined | Not essential for mitosis, but mutation slightly increases rate of spindle elongation | 33% | 8626583 |
| S. pombe | Cut7 | Near spindle pole bodies & spindle MTs | Assembly of mitotic spindle microtubule arrays | 48% | 2542732 |
| S. cerevisiae | Kip1 | Spindle MTs | Redundant with Cin8 | 36% | 852216 |
| S. cerevisiae | Cin8 | Spindle MTs | Separation of spindle pole bodies & assembly of spindle MTs | 41% | 856648 |
| X. laevis | Eg5-1 | Meiotic spindle MTs | Bipolar spindle assembly in vitro at meiotic and mitotic extracts | 89% | 379112 |
| C. elegans | BMK-1 | Spindle MTs | Not essential, but mutation results in reduced fecundity/meiotic defects | 52% | 179948 |
| H. sapiens | HsEg5 | Spindle, poles | Mitotic centrosome separation and bipolar spindle assembly | 100% | 3832 |
| D. melanogaster | Klp61F | Mitotic spindle MTs | Mitotic centrosome separation and bipolar spindle assembly | 60% | 38135 |
| M. musculus | Kif11/Knsl1 | Not determined | Essential in early mouse development | 97% | 16551 |
Figure 1. Phylogenetic relationship between Kinesin-5 proteins.
The phylogenetic analysis is shown as a polar dendrogram with individual sequences labels arranged radially. Seventy-four Kinesin-5 motor domain protein sequences were analyzed by the maximum likelihood, co-estimation method, SATé (Liu et al., 2009). The sequences are labeled with an NCBI GI identifier, protein name (if known), residues corresponding to the motor domain, followed by genus and species. Sequences included were identified from kinesin phylogenies (Wickstead et al., 2010) and by the National Center for Biotechnology Information (NCBI) protein database Reference Sequence (RefSeq). The multiple sequence alignment and maximum likelihood phylogeny were co-calculated by SATé (Liu et al., 2009; Liu et al., 2012). SATé called user defined sequence alignment [MAFFT 6.717; (Katoh et al., 2002)], merger [OPAL 1.0.3; (Wheeler and Kececioglu, 2007)], and phylogeny algorithms [FASTTREE 2.1.4; (Price et al., 2010)]. The decomposition strategy was set to centroid with a maximum subproblem size of 37 sequences. Calculations were allowed to run for a total of 20 iterations without improvement in the maximum likelihood score. Following the final iteration, a final RAxML (Stamatakis, 2006) phylogeny was calculated. Final maximum likelihood score for the phylogeny was −22525.88. Fig Tree v1.3 was used to for visualization.
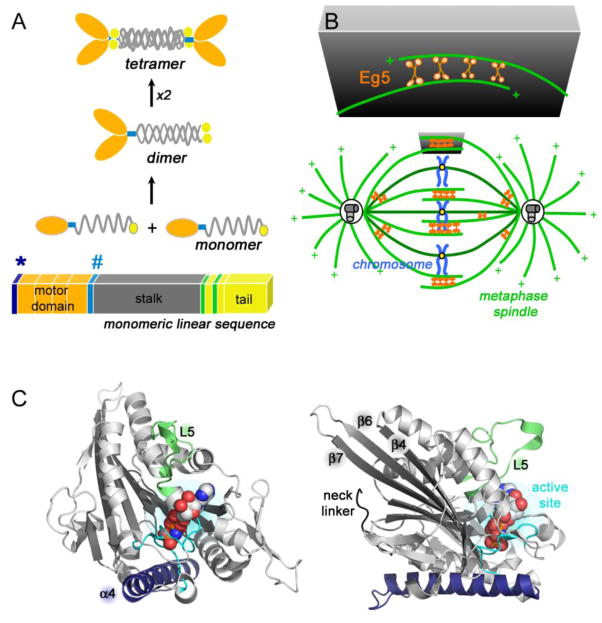
The Kif11 gene product has four domains (Figure 2A), three of which are assigned to roles resulting in differentiation of kinesin function. An N-terminal globular motor domain performs conserved functions of binding microtubules (MTs) and nucleotide hydrolysis. Variations in sequence in the tail, interrupted coiled-coil region, and neck-linker/cover neck (Hesse et al., 2013) are thought to dictate how these motor proteins bind specific cargo, have particular oligomerization states, and control directionality of net motion of the motor domain, respectively. Unlike the canonical dimeric kinesin motors, electron microscopy showed that native Kinesin-5 proteins are bipolar homotetramers (Cole et al., 1994; Blangy et al., 1995), with their motor domains positioned at the ends of the tetramer’s long axis. The arrangement of two sets of antiparallel dimers is hypothesized to result in Kinesin-5 motors crosslinking and sliding antiparallel microtubules (Figure 2B).
Figure 2. Sequence, function, and structure of Kinesin-5 proteins.
(A) Linear sequence organization and general structure of the domains within one kinesin molecule are shown at the bottom of the panel. The asterisk and pound signs highlight the position of the cover neck and the necklinker, respectively. Dimer and tetrameric organization of the Kinesin-5 proteins is also drawn. (B) Cartoon representation of the mitotic spindle and the tetrameric Eg5 molecules cross-bridging spindle microtubules. (C) Two views of the HsEg5 motor domain crystal structure (PDB 3HQD) rotated approximately 120° relative to each other. A non-hydrolyzable AMPPNP molecule bound in the active site. Highlighted are the L5 loop (green), central beta sheet (dark grey), neck linker (black), and the α4 helix (blue). The left and right panels orient the active-site and the microtubule-binding site of HsEg5 to the reader, respectively.
The human Eg5 gene product (HsEg5) is of particular interest amongst the Kinesin-5 proteins, because of its potential as a therapeutic target for cancer treatment. It is sensitive to a battery of small-molecule inhibitors (Hotha et al., 2003; DeBonis et al., 2004; Luo et al., 2007) that allosterically block Eg5 activity (Maliga et al., 2002; DeBonis et al., 2003). The specificity of these synthetic inhibitors is exceptional: although all eukaryotic organisms examined to date contain at least one Kinesin-5 protein, not all Kinesin-5 proteins are sensitive to these compounds (Maliga et al., 2002; DeBonis et al., 2003). Of the inhibitors most frequently employed, monastrol (Mayer et al., 1999) and S-trityl-L-cysteine [STC; (DeBonis et al., 2003)], were uncovered from independent chemical screens, inclusive for mitotic inhibition and exclusive for microtubule interactions. They induced mitotic arrest in human tissue culture cells by inhibiting Eg5-dependent MT motility (Mayer et al., 1999; Kapoor et al., 2000) and resulted in a spectacular reorganization of the mitotic spindle to aberrant monoastral form with no apparent effect on interphase MT arrays (Figure 3). Failure either in chromosome segregation or in the timing of cell division events can result in aneuploidy that is strongly linked with developmental defects and cancer (Kops et al., 2005).
Figure 3. Inhibition of Kinesin-5 by small-molecule inhibitor or knockdown in eukaryotic cells.
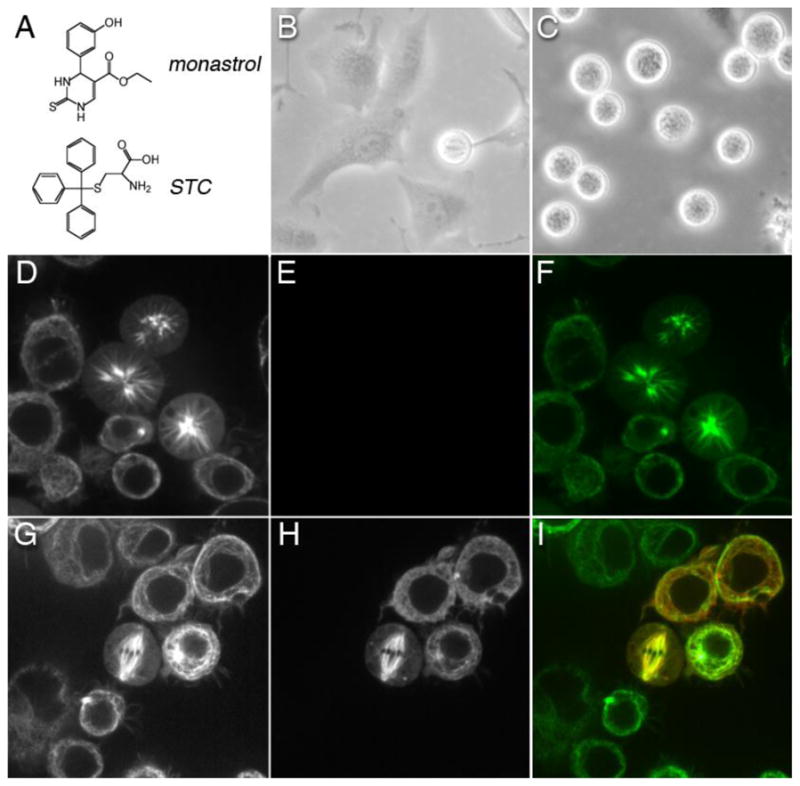
(A) Chemical structure of monastrol and S-trityl-L-cysteine (STC), which are two allosteric inhibitors of human Kinesin-5. After seeding at a density of 1x106 cells and a 24 hr incubation, human HeLa cells were treated with either (B) DMSO or (C) 1 mM STC. Rounded cell shape is diagnostic of live cells in metaphase. From cell counts, 4% of DMSO-treated cells were in metaphase, whereas 43% of STC-treated cells were in metaphase after a 12 hour-long incubation with this drug. Images (B–C) were acquired using a Nikon ELWD 0.3 phase contrast microscope under 10X magnification. Kinesin-5 (Klp61F) dsRNAi knockdown in Drosophila S2 cells expressing tubulin-GFP prevented morphogenesis of bipolar spindles and, instead, exhibited mono-polar arrays. (D) Confocal fluorescence images of living S2 cells expressing tubulin-GFP after dsRNAi knockdown of native Klp61F. The green (GFP) channel of cells displaying aberrant mono-polar mitosis is shown. (E) Red channel of cells in Panel D showing no detectable expression of Klp61F-mKATE chimera. (F) Merge of panels D and E. (G) Tubulin-GFP expression in Klp61F dsRNAi cells transfected with Klp61F-mKATE chimera. Shown is a confocal image of a rescued bipolar spindle in a living transfected cell. (H) Red channel of cells in panel G showing Klp61F-mKATE localization in transfected cells, with untransfected cells nearby. (I) Merge of panels G and H. Images (D–I) were acquired using a Zeiss Axiovert 200 inverted microscope equipped with a Yokogawa spinning disk confocal accessory. 10 X 63x/1.4 oil DIC.
The discovery of these chemical inhibitors of HsEg5 is important on two fronts. First, they can be used as tools to dissect mechanotransduction in this mitotic kinesin and provide answers to still open questions of how catalysis is used and converted into force and motion. Second, numerous small-molecule agents that solely target this human mitotic Kinesin-5 protein with high specificity are leads for anti-cancer therapy; several are in trials as clinical anti-cancer agents [for example, see (Kathman et al., 2007; Carol et al., 2009; Purcell et al., 2010)].
2. CELLULAR FUNCTIONS OF KINESIN-5
Kinesin-5 motors assemble into a bipolar homotetrameric structure that is capable of modulating the dynamics and organization of eukaryotic microtubule arrays (Kashina et al., 1996). Although an essential role for this enzyme in mitosis has been the focus of considerable research effort, recent data also implicate this motor in certain processes within non-dividing cells, such as neurons. Although classical genetic analysis of Kinesin-5 family members has pioneered the investigation of the mitotic role of this motor, recently developed small chemical inhibitors of this enzyme have both enabled finer dissection of this motor’s role in mitosis and opened new avenues of investigation in post-mitotic cells.
Mitotic roles of Kinesin-5
The mitotic function of Kinesin-5 in higher eukaryotes began to be established through groundbreaking analyses including in vitro experiments performed in Xenopus oocyte extracts (Sawin et al., 1992) and in vivo genetic mutational analysis of the Klp61F locus in Drosophila (Heck et al., 1993; Wilson et al., 2004; Garcia et al., 2009). Both approaches yielded the first observations of the now classical Kinesin-5 loss-of-function phenotype including a mono-polar spindle due to failure of centrosome separation, and the resulting radial array of microtubules with chromosomes distributed along the circumference (for example, Figure 3). To date, for all eukaryotes examined, with the exceptions of C. elegans and D. discoideum wherein the motor is required for normal cell division, but not essential, Kinesin-5 activity is essential to assemble the mitotic spindle. A BMK-1 locus of C. elegans Kinesin-5, truncated downstream of the N-terminal motor domain, was found to be dispensible for mitosis, while somehow negatively impacting meiosis to result in reduced fecundity of mutant animals (Bishop et al., 2005). A more complete deletion within the D. discoideum Kinesin-5 gene (kif13) showed a measurable perturbation of spindle assembly, but otherwise did not appear to impair mitosis, or colony growth, in this organism (Tikhonenko et al., 2008).
The discovery that Kinesin-5 motors assemble into bipolar homotetramers (Kashina et al., 1996) gave birth to the idea that these motors support spindle pole separation by sliding antiparallel microtubules, emanating from the two poles, relative to one another (Sawin and Mitchison, 1991b; Sawin et al., 1992; Kashina et al., 1996; Kashina et al., 1997; Sharp et al., 1999; Kapoor and Mitchison, 2001; Kwok et al., 2004; Ferenz et al., 2010). Furthermore, loss of function in Kinesin-5 would therefore lead to unseparated spindle poles or centrosomes, and concomitant collapse of spindle microtubule bundles with chromosomes radially arranged. However, these loss-of-function mutation or inhibitory antibody injection experiments suffer from limitations in the timing and extent of inhibition of motor activity, and thereby are limited in their ability to tease more detailed functional information. Note, however, that a divergent Kinesin-5 in budding yeast, Cin8p, can depolymerize microtubules from their plus ends (Gardner et al., 2008). While such an activity has not yet been described in Kinesin-5 motors of higher eukaryotes, if it exists it would have a significant impact on the development of mitotic spindle assembly models.
Kinesin-5 localization in the mitotic spindle in a variety of systems supports a direct role for this motor in driving the assembly of bipolar spindles (Sawin et al., 1992; Blangy et al., 1995; Sharp et al., 1999). The association of Kinesin-5, in most cases, is regulated by phosphorylation of a C-terminal cassette containing a CDK1 kinase consensus (Sawin and Mitchison, 1995; Sharp et al., 1999), or, in the case of C. elegans, an Aurora B kinase consensus (Bishop et al., 2005). Closer inspection of Kinesin-5 within the spindle finds the motor likely associated preferentially with antiparallel microtubules (Kapitein et al., 2005; Wildenberg et al., 2008). However, the observed affinity for Kinesin-5 with antiparallel microtubules appears to be somewhat at odds with its physical localization within the spindle, wherein its concentration increases towards the poles that are thought to harbor a lower density of antiparallel microtubules.
Furthermore, analysis by speckle microscopy finds Eg5 to dynamically associate with the lattice, with quite short dwell times, and exhibits relatively little motility with respect to the spindle microtubules (Kapoor and Mitchison, 2001; Cheerambathur et al., 2008; Groen et al., 2008; Gable et al., 2012). These observations suggest the possibility that Kinesin-5 may interact with as-of-yet undefined spindle matrix, which forms an independent scaffold that supports the motor while sliding apart microtubules in the spindle (Kapoor and Mitchison, 2001; Tsai et al., 2006; Uteng et al., 2008). In contrast, the unphosphorylated motor does not appear to interact strongly with microtubules in cells and assumes a diffuse localization through the cytoplasm.
Taken together, these data imply a complex regulation of Kinesin-5 activity within sub-domains of the spindle, which currently remains an area of intense speculation. Intriguing hints for a regulatory machinery that may interact with Kinesin-5 and regulate its function is suggested by experiments that isolate Kinesin-5 in complex with a variety of microtubule associated proteins (Gable et al., 2012; Iwakiri et al., 2013; Ma et al., 2011; Uteng et al., 2008; Wang et al., 2010), modified by protein kinases (Garcia et al., 2009; Rapley et al., 2008), or in association with other cell division protein complexes (Tan et al., 2012). The putative functional relationships of these players remains to be determined, although in each case some interplay or synergism with mitotic outcome is apparent.
In addition, the Kinesin-5 motor domain displays two different types of interactions with microtubules. Through a round of its mechanochemical cycle, Kinesin-5 alternately exhibits a high affinity bound state to microtubules as well as a low affinity release state. However, several labs have found that it can also assume an intermediate bound state that is permissive for one-dimensional diffusion along the microtubule lattice and that does not require the motor to consume ATP (Crevel et al., 2004; Kwok et al., 2006; Weinger et al., 2011). It is possible that this unusual state plays a role in the activation of Kinesin-5 activity upon binding across anti-parallel microtubules (Kapitein et al., 2008). How these behaviors contribute to the motor’s function in the mitotic spindle has yet to be elucidated.
A pioneering approach utilizing chemical genetics provided the tools needed to more carefully probe the role of Kinesin-5 in mitosis. Mayer et al. (1999) used a novel chemical genetic approach to discover monastrol, the first of many small chemical inhibitors of human Kinesin-5. Exhibiting exquisite selectivity and high potency, monastrol provides a tool capable of stopping HsEg5 ATPase activity at any point during mitosis. Spurred by the potential of HsEg5 inhibitors as anti-cancer agents, numerous alternative compounds with even higher potency and selectivity have since been developed [reviewed in (El-Nassan, 2013)]. Using these new tools to probe HsEg5 function in mitosis, several investigators found that while HsEg5 function is critical for the separation of centrosomes in mammalian cells during prophase, it appears to be dispensable for the maintenance of the metaphase spindle and subsequent anaphase chromosome movements (Blangy et al., 1995; Kapoor et al., 2000). In contrast, in yeast temperature-sensitive mutants or spindles assembled in Xenopus extracts, inhibition of Kinesin-5 function at metaphase results in spindle collapse (Saunders and Hoyt, 1992; Kapoor et al., 2000). These different outcomes suggest differences between model systems in the degree to which certain functions of Kinesin-5 can be compensated by other machinery. For example, in mammalian cells, Kinesin-12 appears to be critical for maintaining spindle integrity at metaphase in the background of Kinesin-5 inhibition, while this redundancy is lacking in Xenopus spindles assembled in vitro (Tanenbaum et al., 2009; Florian and Mayer, 2011). A recent report finds that mitotic spindle bipolarity and function can be restored in mammalian cells suffering from knockdown of residual Kinesin-5 by RNAi (Raaijmakers et al., 2012). However, in this case the cells had undergone selective pressure in the presence of increasing Kinesin-5 inhibitor prior to the knockdown, thus accumulating unknown mutations permitting their survival. Still, the cells can divide in the near absence of Kinesin-5 activity, perhaps using a mechanism similar to that operating in C. elegans (Bishop et al., 2005).
These experiments led to models of ‘force balance’ within the spindle composed of opposing motor proteins striving to reach a balance while simultaneously coupled to machinery that regulates microtubule assembly and disassembly (Dumont and Mitchison, 2009). The idea that microtubule-sliding mechanisms may be crucial for mitotic spindle assembly was first described by Mclntosh et al. (1969). The anti-parallel microtubule sliding capability of Kinesin-5 rapidly became a cornerstone of refined versions of this model. Kinesin-5 had initially been proposed to promote spindle assembly and stability upon reaching a mechanical force ‘balance point’ against equivalent counteracting minus-end directed microtubule sliding motors, including both Kinesin-14 and dynein complex (Cytrynbaum et al., 2005). This proposed interaction predicts a mechanism for the assembly of a bipolar spindle from the bundling and sliding of antiparallel microtubules, but does not explain overall steady-state spindle length or size. Currently, this model remains controversial, and a convincing link between force-producing motors and spindle size has proven inconsistent across different systems (Miyamoto et al., 2004; Goshima et al., 2005; Mitchison et al., 2005; Saunders et al., 1997). Furthermore, recent advances in imaging technology suggests that much of the data supporting models that incorporate microtubule motors as engines providing opposing mechanical force may be an artifact of the ‘end point’, or snapshot, analysis of fixed cells typically used in these studies. For example, recent live-cell, time-lapse analyses revisiting this question conclude that dynein and Kinesin-5 instead operate independently of one another in spindle morphogenesis and exhibit additive rather than synergistic effects, and specifically rules out a simple mechanical ‘force balance’ explanation (Florian and Mayer, 2012).
While the contribution of a mechanical balance of opposing forces in spindle morphogenesis has become unclear, the coupling of motors to spindle microtubule dynamics has solidified. Originally suggested as a key mechanism for spindle morphogenesis by Inoué and Sato, (1967), the idea that microtubule dynamics and a mechanism that controls the polymerization and depolymerization of microtubules would be crucial for spindle morphogenesis initially took a back seat due to the discovery of microtubule sliding motors. Support for this model came initially from the discovery of microtubule flux towards the poles (Sawin and Mitchison, 1991a). Analogous to tubulin treadmilling, microtubules move towards the spindle poles, where they are depolymerized via their minus ends by depolymerase complexes, and maintain a steady-state length or composition by a matching net polymerization of microtubules at the spindle midzone. Kinesin-5 plays a well-established role in driving microtubule flux towards spindle poles in many systems, flux that is matched by microtubule depolymerizing activity at the spindle poles (Desai et al., 1998; Dhonukshe et al., 2006; Mitchison, 1989; Sawin and Mitchison, 1991a; Mitchison and Salmon, 1992; Zhai et al., 1995; Maddox et al., 2002; Gaetz and Kapoor, 2004; LaFountain et al., 2004; Miyamoto et al., 2004; Ferenz and Wadsworth, 2007; Yang et al., 2008). A functional linkage between Kinesin-5 mediated microtubule flux and spindle pole microtubule depolymerase has been reinforced by the observation that exogenous microtubule depolymerizing agents can compensate for inhibition of Kinesin-5 and restore spindle bipolarity (Kollu et al., 2009; Florian and Mayer, 2011).
Taken altogether, it appears that the function of Kinesin-5 in the mechanism of assembly of the mitotic spindle is likely rather more complicated than currently imagined, and will require a more detailed anatomy of the spindle and additional tools to decipher. Although several spindle assembly models have been developed that attempt to integrate the regulation of microtubule dynamics together with force-producing motors (Dumont and Mitchison, 2009; Goshima and Scholey, 2010; Mogilner and Craig, 2010), lacking are comprehensive anatomy and construction details of mitotic spindles which are crucial to model refinement. However, fine dissection of microtubule dynamics throughout the mitotic spindle is a current focus of spindle research [e.g. (Brugués et al., 2012)], which should help inform such integrative models to help reach a consensus.
Less clear is any functional involvement by Kinesin-5 in post-metaphase aspects of mitosis. Localization data, including time lapse imaging of dynamic Kinesin-5 behavior, clearly show dynamic Kinesin-5 localization in the spindle midzone, and subsequently in the midbody, of dividing insect, and mammalian culture cells (Blangy et al., 1995; Sawin and Mitchison, 1995; Whitehead and Rattner, 1998; Wilson, 1999; Brust-Mascher et al., 2009; Gable et al., 2012). However, correlating these localization data of Kinesin-5 to various central spindle and midbody structures to specific functional deficits has proved elusive. At issue is that mutations in Kinesin-5 in Drosophila or budding yeast (Saunders and Hoyt, 1992; Heck et al., 1993; Wilson et al., 2004; Hildebrandt et al., 2006; Garcia et al., 2009) fail to progress past metaphase to reveal any post-metaphase functionality for this motor. Later studies that reduced Kinesin-5 function by antibody injection into Drosophila embryos found an effect on anaphase-B movements and chromosome motility (Sharp et al., 1999; Sharp et al., 2000; Brust-Mascher et al., 2009) but these data rely on the assumption that low antibody concentrations do not subtly disrupt the mitotic spindles in these studies, thereby altering subsequent anaphase behavior. That subtle spindle defects in this system could alter post-metaphase events are evident in dynein perturbations (Robinson et al., 1999; Sharp et al., 1999; Sharp et al., 2000). Moreover, allosteric inhibition of mammalian Kinesin-5 with drug administration to metaphase cells does not subsequently exhibit aberrant mitotic completion or exit (Blangy et al., 1995; Kapoor et al., 2000). Elucidation of any role for Kinesin-5 in post-metaphase events of mitosis awaits an analysis of post-metaphase loss of function in Kinesin-5, perhaps facilitated by allosteric chemical inhibitors.
Kinesin-5 in post-mitotic cells
Although the function of Kinesin-5 in mitosis has dominated the literature, a small yet growing line of investigation of potential non-mitotic roles for Kinesin-5 has been accumulating. Due, in part, to being masked by its essential role in mitosis in most eukaryotes, potential post-mitotic functions of Kinesin-5 remain difficult to ascertain with classical genetic approaches. The advent of small chemical inhibitors of Kinesin-5 opened this new avenue of investigation by permitting Kinesin-5 inhibition within post-mitotic cells. Mammalian Kinesin-5 exhibits diffuse cytoplasmic localization in non-dividing cells without any clear enrichment on microtubule bundles (Levesque and Compton, 2001; Rapley et al., 2008a). Indeed, upon the discovery of the first small chemical inhibitor of Kinesin-5, monastrol, it was observed that the compound failed to elicit any clear interphase microtubule abnormalities within an immortalized epithelial kidney cell line (BS-C-1), which showed no ill effects of exposure until initiation of mitosis (Mayer et al., 1999). It was argued, based on these observations, that Kinesin-5 played a limited role, if any, in the organization or function of the interphase microtubule cytoskeleton.
However, an earlier study found HsEg5 to be enriched within dividing and, importantly, a variety of post-mitotic developing neurons (Ferhat et al., 1998). Subsequent investigations using the newly developed small molecule inhibitors revealed a variety of developmental effects ranging from neuronal growth cone extension and navigation, to neuronal migration (Haque et al., 2004; Yoon et al., 2005). Bolstered by complementary approaches using RNAi, overexpression of wildtype and mutant Kinesin-5, and optical ablation studies, mounting evidence supports important roles for Kinesin-5 in neurons (Myers and Baas, 2007; Nadar et al., 2008; Falnikar et al., 2011; Nadar et al., 2012). It is currently proposed that Kinesin-5 function is required to modulate the transport of microtubules along axons and into growth cones in collaboration with cytoplasmic dynein. Loss of function in Kinesin-5 promotes abnormal transport of microtubules and their extension into growth cones, thereby inhibiting growth cone movement and navigation.
Taken together, the data offer a cautionary tale concerning the potential use of HsEg5 inhibitors as anticancer agents, warning of potential collateral neuropathies that may result (see Translational Studies below). However, the majority of data linking Kinesin-5 to neuronal function is taken from in vitro experiments. Thus far, peripheral neuropathy is reported in a small fraction of total toxicity analyses of Kinesin-5 inhibitors used in recent trials (Huszar et al., 2009; El-Nassan, 2013) with the dose-limiting toxicity in all the trials to date associated instead with severe neutropenia. As a result, this line of investigation awaits a system to extend the in vitro observed role of Kinesin-5 in neuronal development to a living system so the in vivo role for neuronal Kinesin-5 will be evident.
Regulation of Kinesin-5 function
It is estimated that there are nearly 90,000 experimentally identified posttranslational modifications and 230,000 putative modifications existing on nearly 540,000 proteins (Khoury et al., 2011). Of the 431 identified posttranslational modifications, phosphorylation is most frequently found. Nearly 30% of all proteins in the human proteome are subject to phosphorylation at some point in the cell cycle (Cohen, 2000). Many of these phosphorylation events occur in mitosis, which activate or inactivate proteins temporally and spatially in the cell. Since Kinesin-5 is required for normal mitotic progression, it is no surprise that it requires some form of regulation either by phosphorylation or otherwise to perform its proper tasks.
Regulation via post-translational modifications in the C-terminal tail domain
In the initial characterization of Kinesin-5, a TGXTPXK/RR motif was identified within the evolutionarily conserved BimC box and hypothesized to be a site for phosphorylation by several potential serine/threonine protein kinases (Heck et al., 1993). Further characterization of this conserved motif showed that either T937A and T937E mutations within the motif abolished XlEg5 localization to the mitotic spindle in Xenopus A6 cells, while, in contrast, a T937S mutation preserved this localization, arguing the importance of this threonine to the regulation of this motor (Sawin and Mitchison, 1995). Thr937 was postulated to be a Cdk1 (cyclin-dependent kinase 1) consensus site and its phosphorylation may regulate Kinesin-5 localization to spindle microtubules in a cell cycle-dependent manner. Subsequent work in HeLa cells extended these results to the human enzyme, HsEg5, which was found to be phosphorylated at an equivalent site, Thr926, by Cdk1 (Blangy et al., 1995; Olsen et al., 2010). Phosphorylation by this kinase was determined to be directly responsible for the motor’s association with the spindle apparatus in early prophase. These findings were also later confirmed in several Kinesin-5 family members both in vivo and in vitro with the exceptions of BMK-1 and Cut7 (Giet et al., 1999; Sharp et al., 1999; Cahu et al., 2008; Chee and Haase, 2010; Smith et al., 2011). In the case of Cut7, phosphorylation of this domain is not required for its association with the spindle (Drummond and Hagan, 1998). On the other hand, BMK-1 lacks a BimC box, but nonetheless phosphorylated at analogous C-terminal residues that regulate its localization to the spindle (Bishop et al., 2005).
Kinesin-5 was determined to have additional sites of phosphorylation within its tail domain; however, only one of which has had a function ascribed. Phosphorylation at Ser1033 was identified as necessary for normal mitotic progression (Rapley et al., 2008b). Mutating this residue did not interfere with Kinesin-5 binding to the mitotic spindle but did affect spindle bipolarity. Notably, phosphorylation at Ser1033 occurred on only a small subset (~3%) of the total spindle-associated Kinesin-5. This supports the possibility that there are distinct pools of Kinesin-5 that perform different tasks in cells, and is consistent with prior speculation (Uteng et al., 2008). Finally, proteomics studies identified several phosphosites (Figure 4) beyond residues corresponding to Thr926 and Ser1033 in HsEg5, but no further analysis has been performed to characterize their potential role in functional regulation (Nousiainen et al., 2006; Bodenmiller et al., 2007; Zhai et al., 2008).
Figure 4. Experimentally identified phosphorylation sites on Kinesin-5.
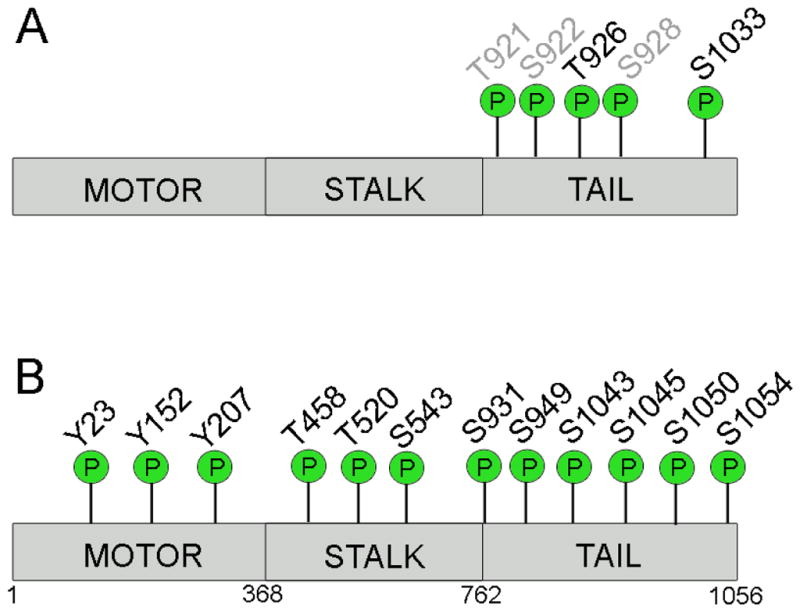
(A) Phosphosites of HsEg5 (black text) and BMK-1 (grey text) that are required to regulate Kinesin-5 function. (B) Additional phosphosites identified in human, Drosophila, and Xenopus in which their role in regulation is undetermined. Cartoon representation of Kinesin-5 linear sequence (grey rectangle) with phosphorylation sites (green spheres) in approximate location with motor, stalk or tail domains.
Regulation via post-translational modifications of the stalk/oligomerization domain
Regulation via the stalk domain was reported in Xenopus studies (Giet et al., 1999; Cahu et al., 2008). In these studies, a non-conserved serine residue within the Kinesin-5 stalk was reported to be the target of Aurora A kinase phosphorylation. In both studies, a regulatory role for this phosphorylation event could not be established. Proteomic analysis in Drosophila (Bodenmiller et al., 2007) confirmed phosphorylation within the stalk at Thr520 (Figure 4), however phosphorylated residues within the stalk were absent in U2OS cells (Rapley et al., 2008). Interestingly, in human cells Thr458 was identified as a phosphosite for two protein kinases with an established cellular role in response to DNA damage (Matsuoka et al., 2007). Taken together, these conflicting results suggest that phosphorylation within the stalk may be cell-type specific or that modification is the result of a response to cell stress.
Regulation via post-translational modifications of the N-terminal motor domain
In an early study of Kinesin-5, it was proposed that post-translational modification of the N-terminal motor domain is also necessary for localization to the mitotic spindle, although no consensus sequence or residue(s) were identified as potential modification sites at the time (Sawin and Mitchison, 1995). Later, three tyrosine residues within the D. melanogaster KLP61F motor domain were determined as being phosphorylated (Figure 4) within close proximity to regions of the protein involved in nucleotide sensing and microtubule interactions, leading the authors to postulate that modification of these residues could alter Kinesin-5 activity (Garcia et al., 2009). Mutation to phenylalanine interfered with protein folding, but no direct effect on Kinesin-5 function was determined. Similarly, in yeast, three residues in close proximity to motor domain regulatory regions were identified as phosphorylated in anaphase, and determined to be required for spindle elongation (Avunie-Masala et al., 2011). It is possible that modification of these or other residues in the motor domain could regulate microtubule dynamics or ATP hydrolysis, just as Wee1 phosphorylation of the Cdk1 ATP-binding domain inhibits its kinase activity (Atherton-Fessler et al., 1993).
Indirect regulation of Kinesin-5
It is likely that mechanisms other than phosphorylation regulate Kinesin-5 function in vivo. For example, one report describes regulation of Kinesin-5 expression via ubiquitination. The E3 ubiquitin ligase, Parkin, indirectly down-regulated Kinesin-5 gene transcription (Liu et al., 2008a). By blocking the binding of c-Jun NH2-terminal kinase to the Kinesin-5 AP1 (activator protein 1) site in its promoter, Parkin could regulate the level of mitotic Kinesin-5 in HEK293 cells (Liu et al., 2008a).
Protein-protein interactions that regulate Kinesin-5
The intricate regulation of Kinesin-5 throughout the cell cycle suggests that it does not act alone but rather in a complex with one or more proteins. As a result, many proteomic screens have been done to try to identify novel protein partners and intricate signaling pathways in which Kinesin-5 is involved (Table 2). Kinesin-5 knock-down by siRNA or chemical inhibition has afforded researchers insights into cellular phenotypes that can occur (Goshima and Vale, 2003), its role in diseases (Formstecher et al., 2006; Liu et al., 2008a; Dharmapuri et al., 2011; Groth-Pedersen et al., 2012; Marra et al., 2013; Martens-de Kemp et al., 2013; Tabernero et al., 2013), and proteins that are either up-regulated or down-regulated as a result (Tsui et al., 2009). While screens that identify protein networks that are affected by Kinesin-5 have been informative in describing potential regulatory networks for this motor, the challenge of elucidating the specific players and biochemical interactions linking these networks remains a goal of future research.
Table 2.
Protein partners.
| Protein interactor | Functional role | Organism | Experimental approach for detection | Reference |
|---|---|---|---|---|
| p150glued | Largest subunit of dynactin, a large microtubule-binding complex involved in ER-to-Golgi transport, spindle formation, and chromosome movement | Yeast Human | 2-hybrid Immunoprecipitaton | (Blangy et al., 1997) |
| Ran-GTP | Essential for translocation of RNA and proteins through nuclear pore complex; involved the control of DNA synthesis and cell cycle progression | Xenopus | Chemical inhibition Microtubule gliding assays | (Wilde et al., 2001; Koffa et al., 2006) |
| Aurora AIR-2 | Protein kinase associated with chromosomes and midbody microtubules and required for polar body extrusion | C. elegans Yeast | RNAi 2-hybrid Coimmunoprecipitation Kinase assays | (Bishop et al., 2005) |
| Traf4 | TNF receptor-associated factor 4 scaffold protein | Yeast Human | 2-hybrid Immunoprecipitaton | (Rozan and El-Deiry, 2006) |
| TPX2 | spindle pole segregation | Xenopus | Protein overexpression | (Eckerdt et al., 2008) |
| proper organization and spindle stability and for targeting Eg5 to spindle microtubules | Pig (LLC-PKl cells) | siRNA Protein overexpression Microtubule gliding assays | (Ma et al., 2010; Ma et al., 2011) | |
| XPF | involved in nucleotide excision repair (NER) and interstrand DNA cross-linking (ICL) | Human | Protein overexpression siRNA | (Tan et al., 2012) |
| NuMA | plays a role in spindle fiber assembly; It remains in the nucleus during interphase and locates to the spindle poles during early mitosis | Xenopus Yeast | RNAi 2-hybrid Protein overexpression Binding assays | (Iwakiri et al., 2013) |
Direct interaction of Kinesin-5 with several proteins involved in mitotic regulation have been identified and confirmed through multiple experimental means. However, consequences on Kinesin-5 function in vivo have not been established. For example, TPX2 has been identified as a Kinesin-5 binding partner. A discrete domain within the C-terminus of TPX2 was identified as the site of its interaction with Kinesin-5 (Eckerdt et al., 2008). This interaction was determined as necessary for spindle pole segregation and spindle stability as well as for targeting Kinesin-5 to spindle microtubules (Eckerdt et al., 2008; Ma et al., 2010; Ma et al., 2011). Kinesin-5 was also shown to directly interact with NuMA (nuclear mitotic apparatus) protein and required for NuMA localization to the mitotic spindle (Iwakiri et al., 2013). However, the authors cautioned that additional studies would need to be performed to elucidate the exact role that this interaction plays in functional regulation of both proteins. In studies to characterize Ran-GTP function, Kinesin-5 was postulated to be a potential protein binding partner (Wilde et al., 2001) and participate in a Ran-dependent complex with HURP and additional proteins (Koffa et al., 2006). However, a direct protein-protein interaction was not determined. In a separate study, Kinesin-5 colocalized with the dynactin subunit p150Glued in vivo and interacted via the Kinesin-5 tail domain in vitro, suggesting that these two proteins may interact in vivo at the conserved Cdk1 site (Blangy et al., 1997). Lastly, in C. elegans, Kinesin-5 was determined to interact directly with Aurora B kinase AIR-2 and it is thought that this interaction mediates localization of Kinesin-5 to the spindle (Bishop et al., 2005).
Proteins that have not previously been shown to participate in events relating to mitosis have also been identified as potential binding partners of Kinesin-5. As such, Traf4 (tumor necrosis factor receptor associated factor 4), a protein involved in the extracellular response to various cell surface stimuli was shown to interact with Kinesin-5 in a yeast two-hybrid screen (Rozan and El-Deiry, 2006). Though Kinesin-5 also coimmunoprecipitated with Flag-tagged Traf4 in H460 cells, evidence showing this interaction is important for the function of either protein has yet to be determined. The nuclear XPF (Xeroderma pigmentosum group F) protein, which plays a role in DNA repair, was also concluded to interact with Kinesin-5 (Tan et al., 2012). In this study, XPF was shown to colocalize with microtubules and Kinesin-5 though a role for regulation in mitosis could not be established. Finally, several additional proteomic screens have afforded knowledge about the potential protein complexes/signaling pathways in which Kinesin-5 may be involved (Paulsen et al., 2009; Maliga et al., 2013), which should aid future studies in determining Kinesin-5’s regulation throughout the cell cycle.
3. MECHANISM OF KINESIN-5
Knowledge of motor mechanism, or mechanotransduction, relies heavily on two types of in vitro investigations. In the first class, chemical-kinetic measurements of molecular motor ensembles in bulk solution largely define our current understanding of the various biochemical processes in kinesin proteins. Such experiments conclude that the catalytic cycle of kinesin includes multiple conformational states coupled to a complex biochemical network. In the second class, single-molecule experiments directly assess physical properties of these molecular machines: micromechanical and optical techniques, which measure piconewton forces generated by motor proteins, provide quantitative information on the dynamic structure of molecular machines at the level of intermolecular interactions. These investigations revealed previously unknown microscopic details, and their results have stimulated discussions of the mechanisms underlying the dynamics of molecular motors.
Described below are the tremendous research efforts that have focused on the mechanism of the human Kinesin-5 motor protein. Studies of a single motor domain of HsEg5, which is approximately 42 kDa in size, comprise the simplest and most amenable biological system to dissect chemistry and structure; as a result, many research labs have contributed to our collective understanding of the biochemical and structural parameters of this ‘business end’ of kinesin proteins. On the other hand, studies of a dimeric HsEg5 and tetrameric kinesin increase in complexity and difficulty, resulting from emergent properties such as processivity of movement, ‘gating’ or synchronization of catalysis and motion between the two/four motor heads, and coordination of antiparallel microtubule gliding.
Catalysis in monomeric HsEg5
Biochemical data discriminate between the Kinesin-5 proteins and other kinesin counterparts. In vitro ATPase activity of human Kinesin-5 is slower than other kinesin motors (Maliga et al., 2002; DeBonis et al., 2003; Cochran et al., 2004). Using chemical-kinetic methods, a series of papers from the Gilbert group (Cochran et al., 2004; Cochran and Gilbert, 2005; Cochran et al., 2006) illuminate key differences displayed by HsEg5 in the coupling of the ATPase cycle to force generation. Like conventional kinesin, binding of monomeric HsEg5 to microtubules results in rapid release of the ADP product. ATP binding forms a MT Eg5* ATP intermediate in at least two kinetic steps, which includes initial substrate collision and a conformational change. ATP hydrolysis proceeds to yield ADP and Pi, with few available details via kinetics; however, real-time measurement of catalysis by vibrational spectroscopy showed that conformational changes must also occur during passage through the transition state (Jun and Kim, 2010). Phosphate release may occur when Eg5 is bound to the microtubule, followed by dissociation from the cytoskeletal track as an Eg5 ADP intermediate, or detachment from the MT may occur as the Eg5 ADP Pi intermediate. Dissociation from the tubulin polymer is relatively slow, whereas reassociation is rapid, as in conventional kinesin.
X-ray crystallography has defined a classic core structure of the kinesin motor domain (Figure 2): a central 8-stranded β-sheet, with 3 helices on each side. Common for all motor proteins, conformational ‘switching,’ or a change in localized secondary structure, is required to convert and amplify small biochemical changes at the nucleotide-binding site into larger movements that manifest in cargo transport. Within the motor domain, kinesins possess three distinct conserved ‘switches’ (Vale, 1996; Vale and Milligan, 2000; Kikkawa et al., 2001; Sablin and Fletterick, 2001; Kull and Endow, 2002): the P loop, switch I, and switch II/helix regions (Figure 2), which undergo changes in the nucleotidease cycle. The P loop is responsible for the binding of the α- and β-phosphate groups of the nucleotide, whereas switch I and switch II loops interact selectively with the γ-phosphate moiety. In addition, a water molecule serves as the nucleophile to attack the terminal phosphate of ATP and must be close to the substrate in the closed active site (Parke et al., 2010). Kinesins, in common with myosins, are thought to undergo an ATP-induced conformational transition in which the switch regions flanking the active site shift between ‘open’ and ‘closed’ states (Kull and Endow, 2002). Indeed, numerous families of nucleotide triphosphatases (NTPases) employ a ‘γ-phosphate’ sensing mechanism to change their molecular conformations between different nucleotide states. The closed state of the active-site switches in NTPases is catalytically competent, whereas the open state is not.
The Protein DataBank has a surprisingly large number of reported HsEg5 crystal structure determinations, compared to other kinesin isoforms. To date, 39 of the 125 kinesin x-ray structures in the PDB are of HsEg5. Only two are not co-crystal structures with bound drug candidates: these are the HsEg5•AMPPNP complex (Parke et al., 2010) and the HsEg5•ADP complex (Turner et al., 2001). We focus this discussion on the 2.2 Å crystal structure of HsEg5•AMPPNP complex (Parke et al., 2010), as it is the catalytic intermediate poised to break ATP down into its products and it is unique amongst all the determinations of kinesin motor domain structure. First, this structure is the first demonstration of a kinesin in which both switch I and switch II regions are fully ordered and in closed conformations around the substrate analog. Second, this is the first kinesin structure to have a nucleophilic water in the active site. Surprisingly, there is not one, but two, water molecules in close association. The apparent lytic water is poised for attack on the γ-phosphate and oriented by a second water molecule (Parke et al., 2010).
This crystallographic ‘snapshot’ of two-waters in the HsEg5•AMPPNP complex was confirmed by a second experimental technique; real-time measurement of catalysis in HsEg5 kinesin by vibrational spectroscopy showed transient formation of a two-water cluster (Jun and Kim, 2010). Moreover, this two-water model in HsEg5 kinesin has been corroborated in another member of the kinesin superfamily (Chang et al., 2013), suggesting that all members of this motor family may share this novel catalytic mechanism for NTP hydrolysis. It is important to note that a two-water catalytic model also has been proposed in a divergent motor protein, myosin (Onishi et al., 2004), and observed experimentally as well in one of its crystal structures (Smith and Rayment, 1996). Together, these data argue that protein-bound water molecules are as essential for biological ATP hydrolysis as amino acids.
Catalysis in dimeric HsEg5
The above chemical-kinetic and structural studies for monomeric HsEg5 delineate the simplest system for understanding the fundamental steps in catalysis and in mechanotransduction. However, the Gilbert group and others also have performed careful and extensive experiments on HsEg5-513, a dimeric form of the motor, with equilibrium and transient kinetic approaches. For this dimeric Kinesin-5, microtubule association, ADP release and ATP binding were found to be fast steps in the catalytic cycle (Krzysiak and Gilbert, 2006). However, product measurement was limited by the rate of ATP hydrolysis. Thus, the rate-limiting step for dimeric Kinesin-5 is ATP hydrolysis, a novel finding for any dimeric kinesin motor measured thus far and clearly different from the canonical Kinesin-1 motor. Each HsEg5 motor head in the dimer contains tightly bound ADP, which is rapidly released upon MT contact. The first ADP from one head is released at >200 s-1, while the second ADP is released more slowly; moreover, the release of the second ADP is dependent upon nucleotide state and is stimulated with ATP bound in the active site. Thus, models of mechanotransduction (discussed in more detail below) support a proposal that ATP-binding precedes force-dependent translocation and HsEg5 stepping is controlled by ATP hydrolysis (Valentine and Gilbert, 2007; Krzysiak et al., 2008). There is no structural data for dimeric HsEg5 to support catalytic understanding at the atomic level; to date, the attainment and resolution of a dimeric kinesin crystal structure has been reported only once in the literature (Kozielski et al., 1997).
Structural transduction of force and work
Biological motors all function by consuming chemical energy (described above), each step of which is coupled to a series of conformational changes. The resultant structural transitions allow sequential interactions with microtubules, which in turn power force generation and movement. Required components for such chemo-mechanical transduction beyond a fuel source (ATP hydrolysis) are a force-generating element and an energy transducer.
As the motor domain houses the site of interaction with the cytoskeletal track, it must possess structural components that serve as a force generator, or the ‘mechano-component’ that produces a power stroke upon energy input. For conventional Kinesin-1 and other homologs, whose motor domain is encoded at the N-terminus of the full-length kinesin protein, this is often cited to be the neck linker (Rice et al., 1999; Schief and Howard, 2001; Kull and Endow, 2002; Mather and Fox, 2006), but recent experiments instead point this role to the kinesin cover neck (Hwang et al., 2008; Khalil et al., 2008). The necklinker is a long, flexible C-terminal extension to the motor domain proper (Figure 2) that allows plus-end processive motion; in contrast, other homologs have an N-terminal extension, termed the neck, that is correlated with minus-end processive motion. The cover neck is the N-terminal portion of the motor domain that has not been resolved in x-ray structures of N-terminal kinesin motor domains.
The HsEg5•ADP structure (Turner et al., 2001) showed, for the first time, an ordered neck linker in a docked state along the motor domain. In comparison, the HsEg5•AMPPNP structure showed an ordered neck linker that was swung away from the motor domain (Parke et al., 2010), supporting the suggestion of a ratchet-like mechanism for translocating the kinesin along the microtubule and their biological activity. Furthermore, high-quality, cryo-EM reports of Drosophila and human Kinesin-5 complexed with microtubules (Bodey et al., 2009; Goulet et al., 2012) supported these above crystallographic studies of motor domains in isolation. The difference in neck linker orientation between the prehydrolysis and product states of HsEg5 was corroborated in the cryo-EM data (Bodey et al., 2009; Goulet et al., 2012).
Two independent groups have provided experimental evidence that suggests the cover neck in Kinesin-5 acts as a force-generating element. Cryoelectron microscopy reconstructions at subnanometer resolution also structurally identified the cover neck in HsEg5 and its parallel docking to the neck linker (Goulet et al., 2012), when HsEg5 bound to microtubules. This complete and consistent structural data for HsEg5 is not available for any other kinesin protein, much less any Kinesin-5 ortholog. These data cumulatively suggest that both the neck linker and cover neck work in tandem as the force generator in Kinesin-5 proteins (Goulet et al., 2012; Hesse et al., 2013), as postulated for Kinesin-1.
Also missing in our understanding are atomic details of the ‘coupling’ component that links the breakdown of ATP to the force generator. In the absence of direct data for most kinesins, the nature of the kinesin transducer is still unresolved. The transducer has been postulated to be an Arg-Glu salt bridge between switch I and II, and its rearrangement is linked to changes at the MT-binding surface (Hirokawa et al., 2009). A second hypothesis is that the α4 relay helix serves as the transducer: the α4 relay helix is proposed to be invariant in its interaction with MTs and is a fixed contact point around which the motor domain flexes in response to the nucleotide bound (Schief and Howard, 2001; Sindelar and Downing, 2010). Comparison of the Eg5•AMPPNP prehydrolysis complex (Parke et al., 2010) with the HsEg5•ADP product complex (Turner et al., 2001) shows that the α4 helix is elongated in the prehydrolysis state and has a characteristic tilt, expected for proper insertion and intercalation between the alpha- and beta-tubulin subunits in the microtubule track. The structure of the kinesin-microtubule interface, as well as the overall conformational shape of the motor domain, is well-matched between the HsEg5-AMPPNP crystal structure and cryo-EM studies (Bodey et al., 2009; Goulet et al., 2012). Although both models correlate chemical state with neck linker positioning (Asenjo et al., 2003), neither provides atomic-level details of how catalytic intermediates are transduced into motion.
A third hypothesis is that the central beta-sheet and the surface L5 loop comprise the transducer in kinesins (Hirose et al., 2006; Kim et al., 2010); homologous motifs in myosins have been accepted to serve as the transducer (Coureux et al., 2004) in this divergent motor protein. Like the models above, beta-sheet torque (Kim et al., 2010) can be correlated with neck linker positioning. Although not discussed in the paper (Parke et al., 2010), the HsEg5•AMPPNP structure (PDB ID 3HQD) showed the L5 loop adopted a ‘closed’ conformation, in comparison to the ‘open’ state in the HsEg5•ADP structure (Turner et al., 2001). These data led to the conclusion that conformational changes in the L5 loop, like the necklinker, are correlated with native catalysis. Both prior and subsequent to these studies, the open and closed conformations of the L5 loop have been deemed important in HsEg5 [e.g., (Cochran and Gilbert, 2005; Rosenfeld et al., 2005; Bodey et al., 2009; Larson et al., 2010; Parke et al., 2010), as the L5 loop is the allosteric drug target site and its closed state cradled inhibitors (Yan et al., 2004). It has even been reported that inhibition of its conformational motion can reverse ATP hydrolysis (Cochran et al., 2005).
However, unlike the interswitch salt-bridge and α4 relay helix transducer models, proposals of how the L5 loop and beta-sheet mediate conversion of catalysis into motion have been put forth. As examples, the L5 loop has been argued to directly control nucleotide binding (Larson et al., 2010; Goulet et al., 2012), an unusual hypothesis for an allosteric site. Beta-sheet torque (Kim et al., 2010) can be correlated with L5 loop conformation and necklinker positioning. In doing so, it serves as a responsive, central spine that leverages and modulates functional outcomes at the adjacent microtubule-binding site. As such, beta-sheet torque can be correlated with drug sensitivity and serve as a structural biomarker for drug efficacy (Kim et al., 2010).
Force measurements: processivity, single-molecule, and optical trap studies
Formulation of models for the cellular role of Kinesin-5 is dependent on understanding the mechanical properties of the motor itself. Consequently, several laboratories have been using a variety of in vitro assays to better understand the motile properties of Kinesin-5 proteins. While Kinesin-5 shares a number of conserved features found in conventional and other kinesins, there are unique characteristics that presumably are suited to its specific cellular role(s).
Kinesins that transport cellular cargo must be able to take multiple steps along a microtubule without dissociating – a property known as processivity. Conventional kinesin-1 takes >100 steps before dissociating (Block et al., 1990; Svoboda and Block, 1994). While this type of behavior would be necessary for a transport motor, Kinesin-5 may not need to be processive to generate forces in the mitotic spindle, as ensemble effects of multiple motors crosslinked in the antiparallel mitotic array may be sufficient. Indeed, while early experiments showed that Eg5 and Klp61F are slow motors that move toward the plus-end of the microtubule (Sawin et al., 1992; Cole et al., 1994; Kashina et al., 1996), solution kinetics examining ATP hydrolysis per microtubule interaction suggested that Eg5 was less processive than kinesin-1 monomers (Crevel et al., 1997). Multiple motor microtubule gliding assays have shown that Kinesin-5 is capable of gliding microtubules in ensemble, albeit at rates ~10-fold slower than conventional kinesin (Table 3). Tetrameric Eg5 was also shown to crosslink microtubules and move toward the plus end at a rate of ~20 nm/sec (Table 4) – with the net effect of sliding anti-parallel microtubules at a rate of ~40 nm/sec (Kapitein et al., 2005). Subsequent work from the same laboratory showed that cross-linking anti-parallel microtubules activates the directional motility of Eg5 (Kapitein et al., 2008). However, each of these experimental setups does not require single Kinesin-5 tetramers (or dimers) to be processive individually, as multiple motors are present in each experimental scenario.
Table 3. In vitro microtubule gliding rates of Kinesin-5 proteins.
Using purified kinesin protein, bulk-measurement rates of microtubule motion were recorded in motility assays. Noted are the oligomeric state of the kinesin protein, buffer, pH, and salt/osmolyte concentration.
| Kinesin-5isoform | Oligomeric State | Buffer (pH) | Salt/Other (Osmolyte) | Velocity (nm/s) | Reference |
|---|---|---|---|---|---|
| X. laevis | native tetramer | 80 mM PIPES (6.8) | - | 12.3 | (Kwok et al., 2004) |
| native tetramer | 80 mM PIPES (6.8) | - | 24.0 | (Kapitein et al., 2005) | |
| native tetramer | 70 mM PIPES (6.8) | 80 mM KCl | 34.4 | (Weinger et al., 2011) | |
| native tetramer | 70 mM PIPES (6.8) | - | 16.8 | (Weinger et al., 2011) | |
| native dimer (1-513) | 70 mM PIPES (6.8) | - | 30.1 | (Weinger et al., 2011) | |
| chimeric dimer (1-373 with KHC 345-426) | 80 mM PIPES (6.8) | - | 61.0 | (Duselder et al., 2012) | |
| chimeric dimer (1-373 with KHC 345-559) | 80 mM PIPES (6.8) | - | 64.0 | (Shastry and Hancock, 2011) | |
| H. sapiens | native dimer (1-513) | 100 mM PIPES (6.8) | - | 26.7 | (Ma et al., 2011) |
| native dimer (1-513) | 80 mM PIPES (6.9) | 150 mM sucrose | 32.9 | (Waitzman et al., 2011) | |
| native dimer (1-513) | 10 mM PIPES (6.9) | - | 47.2 | (Krzysiak and Gilbert, 2006) | |
| native dimer (1-439) | 80 mM PIPES (6.8) | - | 37.0 | (Yajima et al., 2008) | |
| chimeric dimer (1-370)* with KHC 337-432 | 80 mM PIPES (6.9) | 100 mM K-acetate | 104.0 | (Kaseda et al., 2008) | |
| chimeric monomer (1-370)* with KHC 337-432 | 80 mM PIPES (6.9) | 100 mM K-acetate | 92.0 | (Kaseda et al., 2008) | |
| native monomer (1-372) | 80 mM PIPES (6.8) | - | 42.0 | (Yajima et al., 2008) | |
| native monomer (1-367) | 80 mM PIPES (6.9) | 150 mM sucrose | 20.5 | (Waitzman et al., 2011) | |
| D. melanogaster | native tetramer | 100 mM PIPES (6.9) | 50-75 mM KCl | 40.0 | (Cole et al., 1994) |
| native tetramer | 80 mM PIPES (6.8) | - | 32.0 | (van den Wildenberg et al., 2008) | |
| native tetramer | 20 mM Tris (8.0) | 75 mM KCl | 40.0 | (Tao et al., 2006) |
Table 4. Antiparallel microtubule gliding rates of Kinesin-5 proteins.
Using purified kinesin protein, bulk-measurement rates of microtubule motion were recorded in motility assays. Noted are the oligomeric state of the kinesin protein, buffer, pH, and salt/osmolyte concentration.
| Kinesin-5isoform | Oligomeric State | Buffer (pH) | Salt/Other (Osmolyte) | Velocity (nm/s) | Reference |
|---|---|---|---|---|---|
| X. laevis | native tetramer | 80 mM PIPES (6.8) | 80 mM KCl | 28.0 | (Kapitein et al., 2008) |
| native tetramer | 80 mM PIPES (6.8) | - | 40.2 | Kapitein et al., 2006 | |
| native tetramer | 70 mM PIPES (6.8) | 80 mM KCl | 45.6 | (Weinger et al., 2011) | |
| native tetramer | 70 mM PIPES (6.8) | 60 mM KCl | 35.0 | (Kapitein et al., 2008) |
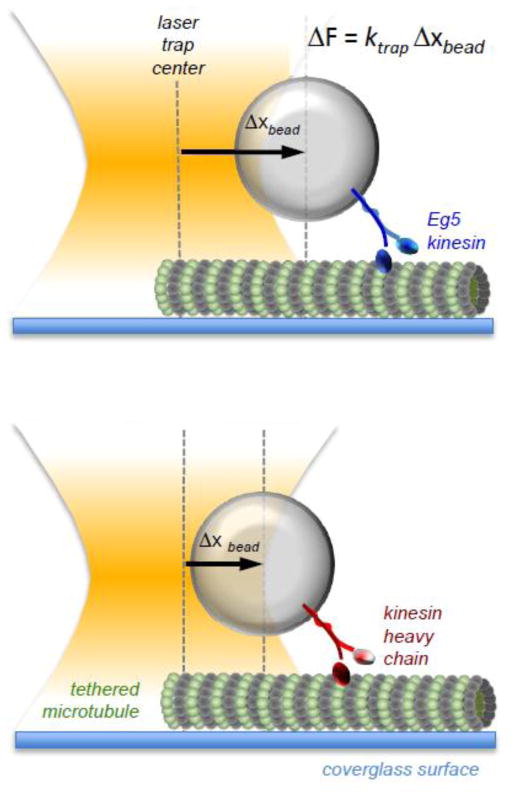
In 2006, two separate groups showed for the first time that Kinesin-5 proteins were indeed processive motors and could take several steps along a microtubule before dissociating (Table 5). Using HsEg5-coated polystyrene beads in an optical trap setup in which microtubules were attached to a functionally modified glass surface to prevent non-specific adherence of the beads (Figure 5), Valentine and colleagues showed that dimeric HsEg5 (Eg5-513) displayed discrete, sequential steps along the microtubule lattice with an average step size of 8.1 nm, taking on average ~8 steps per excursion (Valentine et al., 2006). This setup also allowed for the observation of other unexpected behaviors of HsEg5. First, HsEg5 dimers had a velocity of ~100 nm/sec at saturating ATP levels in the absence of load (2–3 times faster than observed motility in ensemble assays) and a velocity of ~60 nm/sec at saturating ATP levels with an applied constant rearward (opposing) load of 4 pN. Second, while conventional kinesin slows down as it approaches its maximal force-producing level (stall force), HsEg5 dissociates from the microtubule before slowing, with the highest force measured at ~5 pN. Further analysis of their own work led the authors to point out in a later publication that extrapolation of their force-velocity curve to zero velocity for HsEg5 should give a stall force of 9 pN (Valentine et al., 2006), which is significantly higher (Figure 5) than the 6 pN stall force of conventional kinesin heavy chain (Svoboda et al., 1994; Block et al., 2003). Shortly after publication of the optical trap work with HsEg5, Kwok and colleagues provided the first single molecule motility data for fluorescently-tagged Eg5 tetramers moving processively along immobilized microtubules (Kwok et al., 2006). While tetramers had a significantly slower motility rate of 14.2 nm/sec (compared to 100 nm/sec for Eg5 dimers in the optical trap studies described above), the run length was significantly longer (580 nm for the tetramer in single molecule assays compared to 67 nm in the optical trap), yielding an average of >70 steps per run.
Table 5.
Kinesin-5 velocity, measured by single molecule experiments.
| Kinesin-5isoform | Oligomeric State | Buffer (pH) | Salt/Other (Osmolyte) | Velocity (nm/s) | Reference |
|---|---|---|---|---|---|
| Single-molecule measurements (trap) | |||||
| X. laevis | native tetramer | 80 mM PIPES (6.8) | - | 35.0 | (Korneev et al., 2007) |
| X. laevis | chimeric dimer (1-373 with KHC 345-426) | 80 mM PIPES (6.8) | - | 59.0 | (Duselder et al., 2012) |
| H. sapiens | native dimer (1-513) | 80 mM PIPES (6.9) | 200 mM KCl, 50 mM K-acetate | 68.0 | (Valentine and Block, 2009) |
| H. sapiens | native dimer (1-513) | 80 mM PIPES (6.9) | 200 mM KCl, 50 mM K-acetate | 96.0 | (Valentine et al., 2006) |
| Single-molecule measurements (fluorescence) | |||||
| X. laevis | native tetramer | 80 mM PIPES (6.8) | - | 14.6 | (Kwok et al., 2006) |
| X. laevis | native tetramer | 70 mM PIPES (6.8) | - | 8.5 | (Weinger et al., 2011) |
| X. laevis | native tetramer | 70 mM PIPES (6.8) | - | 8.9 | (Kapitein et al., 2008) |
| X. laevis | native tetramer | 70 mM PIPES (6.8) | 20 mM KCl | 10.1 | (Kapitein et al., 2008) |
| X. laevis | native tetramer | 70 mM PIPES (6.8) | 40 mM KCl | 0.0 | (Kapitein et al., 2008) |
| X. laevis | native tetramer | 70 mM PIPES (6.8) | 60 mM KCl | 0.3 | (Kapitein et al., 2008) |
| X. laevis | chimeric dimer (1-373 with KHC 345-426) | 80 mM PIPES (6.8) | - | 112.0 | (Duselder et al., 2012) |
Figure 5. Measuring kinesin stall force with an optical trap.
Using a focused laser line, a sole polystyrene bead can be held in place with high precision. With a single kinesin attached by its tail to the bead, the bead can be placed in close enough proximity to a tethered microtubule that the kinesin can bind to, and begin stepping along, the microtubule. As the kinesin pulls the bead away from the trap center, the tendency of the bead to remain in the center exerts a rearward force on the kinesin molecule, which becomes greater as the kinesin attempts to pull the bead further from the center. Thus, force (F) can be measured as a function of distance (x) if the spring constant of the trap (k) is known. Here, we show Eg5 (Kinesin-5) generating a greater force than kinesin heavy chain (Kinesin-1) as evidenced by its ability to pull the bead further from the trap center (Δx), given the same trap stiffness.
While run length of Kinesin-5 is a useful readout of processivity, analysis of linear excursions in single molecule studies does not necessarily give an accurate picture of processivity, particularly relative to other families of kinesin motors. Building on the finding that Eg5 monomers are capable of carrying out microtubule gliding in ensemble assays (Kaseda et al., 2008), Yajima and colleagues performed a modified form of microtubule gliding by sparsely attaching quantum dots to microtubules and performing three dimensional analysis of the location of each quantum dot, with particular emphasis on the rotational pitch of the microtubule (Yajima et al., 2008). Since conventional kinesin walks along a single protofilament (Ray et al., 1993) it contributes very little rotational torque to a microtubule. Thus, microtubules in a gliding assay exhibit a long-pitch rotation. Conversely, single-headed constructs of conventional kinesin that are non-processive rotate the microtubule with a constant short-pitch (Yajima and Cross, 2005). Analysis of rotational pitch of microtubules with single-headed Eg5 showed a similar short-pitch rotation, indicative of non-processive motility. Dimeric Eg5 showed a significantly longer pitch rotation of microtubules than monomeric Eg5, but still significantly shorter than dimeric conventional kinesin. These results further point to a weakly processive motility for Eg5.
Direct observation of Eg5 tetramers in single molecule assays along immobilized microtubules showed that Eg5 exhibits two distinct types of movement: ATP-dependent directional motility, as well as ATP-independent diffusive movement (Kwok et al., 2006). The bias toward one type of movement or the other can be regulated in motility assays by varying the salt concentration in vitro; low salt buffers show a higher proportion of directed motility events, while increasing ionic strength causes a greater frequency of diffusional events (Kapitein et al., 2005). However, when Eg5 tetramers crosslink two separate microtubules, there is a bias toward directional movement even at high (i.e. physiological) ionic strength. This transition may be mediated by a nonmotor microtubule binding site in the Eg5 tail domain, as deletion of the C-terminal portion of Eg5 decreased the relative sliding of microtubules in antiparallel gliding assays (Weinger et al., 2011). It may well be that this transition from diffusive movement to directional movement contributes to the activity of Kinesin-5 in the mitotic spindle and prevents ATP hydrolysis unless the molecule is attached to two antiparallel microtubules.
More recent studies have attempted to ascertain the cause of the decreased processivity of Eg5 relative to the families of transport motors. Conventional kinesin is believed to be able to carry out hand-over-hand motility as a result of a built-in tension-sensing mechanism coordinated by the neck linker that keeps the two heads out of phase; one stays tightly bound to the microtubule while the other releases from the microtubule and undergoes a diffusive search for its next binding site (Rosenfeld et al., 2003; Hackney, 2005; Yildiz et al., 2008). Perhaps the Kinesin-5 neck linker, which is significantly longer than most other kinesins (Hariharan and Hancock, 2009), is not optimized to communicate this inter-head tension and as a result the two heads are more apt to both enter the weakly bound stage. To test this hypothesis, Shastry and Hancock made sequential deletions of the amino acids in the neck linker of Eg5 from 18 to 14 (the length of processive, conventional kinesin’s neck linker) and tested for changes in velocity and run length (Shastry and Hancock, 2011). While Eg5 with full-length neck linker or 17 residues had minimal processivity, constructs with neck linkers of 15 and 14 amino acids showed increasing run lengths averaging 950 nm and 1770 nm (~120 steps and >200 steps), respectively. Similar results were subsequently shown using a combination of single molecule, microtubule gliding, and optical trap experiments to examine changes in Eg5 neck linker length, with the striking result that while run length changes as the neck linker length changes, velocity and stall force remain relatively constant (Duselder et al., 2012). This data suggests that the increased neck linker length of wild type Eg5 may cause it to be a less processive motor.
Of course, one other obvious regulator of Eg5 motility in the mitotic spindle is the presence of accessory binding proteins that interact with Eg5. One such potential partner is the Ran-regulated mitotic spindle assembly factor TPX2. In vivo, the interaction between Eg5 and TPX2 is required for kinetochore fiber formation and proper location of Eg5. In microtubule gliding assays, TPX2 inhibited Eg5-dependent microtubule gliding in a dose-dependent manner, pointing to a role for TPX2 in regulating Eg5 function (Ma et al., 2011). It is reasonable to believe that other binding partners for Eg5 will be identified, or that post-translational modification of Eg5 may also play a role in its microtubule-dependent activity. Future studies will certainly investigate these and other possibilities.
It is worth noting that a recent study of the major Kinesin-5 isoform from budding yeast, Cin8, showed that it moves toward the minus-end of the microtubule in single molecule studies and at low concentration in microtubule gliding assays, but shows plus-end-directed movement at high concentration in microtubule gliding assays and in anti-parallel microtubule sliding assays (Roostalu et al., 2011). While this type of directional switching has not been reported for other Kinesin-5 proteins, it remains to be seen whether this is an adaptation specific to yeast (that has a closed mitosis and lacks a nuclear-localized dynein for minus-end motility) or is a property of other Kinesin-5 proteins.
4. TRANSLATIONAL STUDIES
Mitosis is a validated point of intervention for cancer therapy and a variety of antimitotic drugs against cytoskeleton proteins are successfully being use in the clinic (Rath and Kozielski, 2012). There is one report that little or no HsEg5 is detectable in normal non-proliferating cells, while expression is prominent in proliferating cells (Hegde et al., 2003). In addition, over-expression of HsEg5 has been noted in a variety of human solid tumors implicating the role of the enzyme in tumorigenesis (Hansen and Justice, 1999; Hegde et al., 2003; Castillo et al., 2007). Therefore, drugs that specifically and solely inhibit Eg5 are potential alternatives to the taxanes and vinca alkaloids that target microtubules and may have fewer clinical side effects. However, development of new antimitotic drugs that are targeted therapies are challenged by a number of scientific unknowns, such as our ignorance of cell stress and death mechanisms upon mitotic arrest of tumor cells (Jackson et al., 2007).
Inhibitors of Kinesin-5
The first inhibitor of Kinesin-5 to be reported was monastrol and it was identified in a cell-based screen for compounds that caused mitotic arrest without direct effects on microtubule dynamics (Mayer et al., 1999). Since its discovery, over 100 different chemical classes of allosteric inhibitors against HsEg5 have been identified in the public scientific literature (and dozens more in the patent literature). These include carbolines, quinazolines, thiazolopyrimidines, thiadiazoles, dihydropyrazoles, isoquinolines, imidazoles, and benzimidazoles. They exhibit 107 differences in potency against HsEg5 [reviewed in (Sarli and Giannis, 2008; Huszar et al., 2009; El-Nassan, 2013)]. More than three have entered into clinical trials (Table 6). The majority of the inhibitors reported are selective for HsEg5 because they bind to an allosteric site, comprised by loop 5 (L5), which is not conserved in sequence or length in other kinesins and even amongst the Kinesin-5 family. While HsEg5 inhibitors are identified at rapid pace for clinical purposes, it is formally possible that more than one inhibitor binding site exists on HsEg5 (Zhang, 2011). Although examination of whether there is a single or multiple, allosteric ‘hot spots’ for these compounds has been neglected by the field at large, several HsEg5 inhibitors have been uncovered that do not bind within the L5 loop. For example, inhibitors that are ATP-competitive (Groen et al., 2008) and MT-competitive (Learman et al., 2009) have been reported. Other papers (Luo et al., 2007; Ulaganathan et al., 2013) have shown that alternate allosteric sites are present within the HsEg5 motor domain, outside the L5 pocket.
Table 6. Clinical results with Kinesin-5 targeted inhibitors in phase I and phase II trials.
Listed is a representative sampling of the clinical data on the outcomes of HsEg5 inhibitors on patients who were not responsive to taxanes. Inhibitors and data are cataloged from greatest to least efficacy in trials.
| Anti-mitotic drug | Tested cancer type | No. patients treated | Clinical results | Significant adverse events | Ref |
|---|---|---|---|---|---|
| SB-743921 | advanced solid tumors or refractory lymphoma | 44 | prolonged stable disease observed in one patient and 14% of patients had stable disease for over 24 weeks; promising efficacy | prolonged neutropenia, hepatic enzyme elevation and hyperbilirubinaemia | (Holen et al., 2011) |
|
| |||||
| MK-0731 | advanced solid tumors | 22 | prolonged stable disease observed in patients with cervical, non-small cell lung, and ovarian cancers | myelosuppression | (Holen et al., 2012) |
|
| |||||
| Ispinesib (SB-715992 | advanced or metastatic breast cancer | 16 | 60% patients had stable disease for at least 42 days and 27% patients lasting for at least 90 days | neutropenia,increased alanine aminotransferase | (Gomez et al., 2012) |
| advanced solid tumors | 30 | 33% of patients had stable disease | neutropenia, nausea, fatigue | (Burris et al., 2011) | |
| pediatric solid tumors | 24 | Prolonged stable disease observed in 15% of patients, but there was substantial patient variation in drug disposition | neutropenia and hyperbilirubinemia | (Souid et al., 2010) | |
| 24 | 33% of patients had stable disease ≥18 weeks | prolonged neutropenia and febrile neutropenia | (Blagden et a l., 2008) | ||
|
| |||||
| AZD4877 | advanced solid tumors | 21 | stable disease in 25% of patients, but little evidence for clinical efficacy | neutropenia and leucopenia | (Esaki et al., 2011) |
| advanced solid tumors | 29 | no report of stable disease | neutropenia | (Infante et al., 2012) | |
|
| |||||
| ARRY-520 | advanced myeloid leukemia | 36 | lack of clinical activity | mucositis, myelosuppression | (Khoury et al., 2012) |
Mechanism of Kinesin-5 inhibition
Monastrol is best characterized out of all the HsEg5 inhibitors. X-ray crystallography was key in identifying the monastrol allosteric binding site in HsEg5. In crystallographic determination of HsEg5:ADP complexed with monastrol, the most significant structural change is the binding of the compounds to an induced-fit site, principally created by the insertion loop (L5) of helix α2 (Turner et al., 2001; Yan et al., 2004). The L5 loop adopts a ‘closed’ conformation and cradled monastrol, in contrast with the ‘open’ conformation observed in the absence of allosteric inhibitor. Additional alterations in HsEg5 were observed in positions of the α2 and α3 helices, as well as a conformational change in switch I from a flexible loop to a short helix. Similar conformational changes were observed in crystal structures of HsEg5 with S-trityl-L-cysteine and other allosteric inhibitors [e.g., (Cox et al., 2005; Fraley et al., 2006; Garbaccio et al., 2006)].
Although there are a few experiments reported on how L5-directed inhibitors alter the kinetics and biochemistry of HsEg5, only the kinetic studies on monastrol were comprehensive; it has been generally presumed by other investigators that other inhibitors that bind to the same site have the same kinetic properties. Steady-state and presteady-state measurements on how monastrol impacts HsEg5 (Cochran et al., 2005) showed that ATPase activity is inhibited through slowed product release and that there is weakened binding and affinity between Eg5 and microtubules. Thus, irregular HsEg5-microtubule interactions alter the ability of the Kinesin-5 protein to generate force and to establish or maintain the bipolar spindle. Aberrations in HsEg5-microtubule interactions in the presence of monastrol were more pronounced in the dimeric form of the kinesin; a significant reduction in microtubule lattice decoration was measured (Krzysiak et al., 2006). Allosteric inhibition of tetrameric Kinesin-5 by monastrol (Kwok et al., 2006) uncovered that Eg5 has an ATP-dependent directional motion, as well as a diffusive component that does not require ATP hydrolysis. Monastrol suppresses the former, but not the later, form of Eg5 motility along microtubules (Kwok et al., 2006).
The role of specific residues in the monastrol-binding pocket (L5, α2, and α3) has been tested, but not yet been systematically explored. In most cases, mutation of specific monastrol-binding pocket residues yielded a negative result: loss of HsEg5 sensitivity to the inhibitor. In contrast, the Drosophila Kinesin-5 protein motor domain (Klp61F) is insensitive to monastrol and STC (Learman et al., 2009), despite containing an L5 loop of equivalent length that has 52% sequence identity to HsEg5. Thus, it has been used as a model system to test for ‘gain-of-allosteric inhibition’ and a chimeric Drosophila Kinesin-5 protein motor domain with a human L5 loop was constructed. The ATPase activity of this chimeric Klp61F kinesin was not inhibited by monastrol, but was inhibited by STC (Liu et al., 2011). This experiment yielded positive proof that the L5 residues in the HsEg5 loop are sufficient to serve as a protein cassette for small-molecule sensitivity, and thus ‘druggability.’ These experiments also suggest that the interactions of this loop with the core motor domain are responsible for the allosteric interruption of ATP hydrolysis.
Successes and lessons of antimitotic strategies
The research and biomedical community have clearly embraced a new era in anti-mitotic therapy with the identification and clinical translation of new targets in mitosis outside of tubulin. Targeted agents against HsEg5 have shown promise in preclinical studies. The effect of HsEg5 inhibitors on proliferating cells in culture is to cause mitotic arrest with a monopolar spindle. The centrosomes do not separate, so the microtubules are organized from a single site in the cells and chromosomes orient in a ring around this site. This phenotype is ubiquitous to all proliferating cells treated with Kinesin-5 inhibitors; there are no reported effects on nonproliferating cells using relevant concentrations of these compounds. Mitotic arrest induced by HsEg5 inhibitors has been shown to result in apoptosis in some tumor cell lines (Liu et al., 2006; Liu et al., 2008b; Orth et al., 2008; Shi et al., 2008).
Ispinesib (SB 715992; Cytokinetics/GSK) inhibited growth in a wide range of human and murine cell lines with IC50 values of 1.2–9.5 nM. Treatment of SKOV3 ovarian tumor cells in vitro with 20 nM ispinesib caused mitotic arrest in cells, which displayed unseparated centrosomes and monopolar mitotic spindles. It also showed favorable clinical activity with a significantly lower toxicity profile in SKOV3 human tumor xenograft models, compared to tubulin targeted drugs (Miller et al., 2005). In addition, it exhibited anti-proliferative activity in tumor cell lines and significant efficacy in several murine tumor models (Johnson et al., 2002).
Ispinesib was the first small molecule inhibitor of HsEg5 that advanced to clinical trials and early clinical trial data highlighted that it was well tolerated at biologically active doses, with no evidence of neurotoxicity, but displaying dose-limiting neutropenia. However, advanced in vivo studies do not match in vitro expectations. Ispinesib (one of the most potent Eg5 inhibitors with a Ki of 1.7 nM) has limited success in phase II clinical trials (Beer et al., 2008; Lee et al., 2008; Tang et al., 2008). Additional HsEg5 inhibitors, SB-743921 from Cytokinetics/GSK (Holen et al., 2011) and MK-0731 from Merck (Cox et al., 2008), are also currently subjects of phase II clinical trials. Other pharmaceutical companies, including Array BioPharma Inc. (ARRAY-520) and Eli Lilly (LY2523355), have also invested candidate HsEg5 inhibitors to treat different forms of cancer. These second-generation HsEg5 inhibitors have had variable success in comparison to ispinesib (Table 6).
In light of this data, three main reasons have been identified for the surprisingly modest impact of mitosis-selective approaches in the clinic [reviewed in (Chan et al., 2012; Mitchison, 2012)]. The first is our limited knowledge of how the mitotic machinery and the apoptotic pathway are linked and how occurrence of mitotic slippage allows arrested cells to adopt different cell fates, such as viable multiploidy. The second is the underestimation of the slowness in doubling time of cells in human tumors, which results in HsEg5 inhibitors only targeting a limited fraction of cells within the tumor population. Third is the half-life, or retention time, of drugs in tumor cells; paxitaxel has a half-life in the range of a week, whereas HsEg5 inhibitors have a median half-life of 12 hours. Finally, identifying patients who might best respond to these agents goes hand-in-hand with the expected development of drug resistance before tumor removal, due to adaptive mutations and drug efflux pumps (Longley and Johnston, 2005). Indeed, L5 loop point mutations conferring resistance to HsEg5 inhibitors have been introduced into cancer cell lines, resulting in their regaining proliferative capability in the presence of drug (Tcherniuk et al., 2010).
Thus, although the HsEg5 drugs thus far were not the desired clinical watershed, these compounds are important in advancing anti-cancer therapy. First and foremost, these small-molecule inhibitors have provided better understanding of the translational barrier in clinical applications, which is key to evolving better treatments. In addition, identification or design of more specific or potent drugs will come, when allosteric communication within the transducer and its effects on Eg5 mechanotransduction are better elucidated. Moreover, examples of approaches to circumvent current clinical complications are to antagonize a cancer-specific target that has oncogenic roles outside mitosis and/or to combine targeted inhibitors to maximize mitotic arrest-cell death and minimize mitotic exit (Chen et al., 2012).
HIGHLIGHTS.
Kinesin-5 is primarily a mitotic microtubule motor that exhibits complex regulation.
Biophysical studies of the motor core alter the paradigm of kinesin catalysis.
Toolbox of small-molecule inhibitors opens basic and clinical research avenues.
Kinesin-5 inhibitors provide translational lessons about targeted patient therapies.
Acknowledgments
We thank members of the Huckaba, Kim, Wojcik, and Worthylake laboratories for their intellectual input and criticism. This work was funded by grants from the National Institutes of Health (R01GM097350 to S.K.; R01GM066328 to E.W.; and P20GM103424 and 5G12RR026260 to T.H.) and the School of Graduate Studies at LSU Health Sciences Center – New Orleans (R.B. and J.R.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Asenjo AB, Krohn N, Sosa H. Configuration of the two kinesin motor domains during ATP hydrolysis. Nat Struct Biol. 2003;10:836–42. doi: 10.1038/nsb984. [DOI] [PubMed] [Google Scholar]
- Atherton-Fessler S, Parker LL, Geahlen RL, Piwnica-Worms H. Mechanisms of p34cdc2 regulation. Mol Cell Biol. 1993;13:1675–85. doi: 10.1128/mcb.13.3.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avunie-Masala R, Movshovich N, Nissenkorn Y, Gerson-Gurwitz A, Fridman V, Koivomagi M, Loog M, Hoyt MA, Zaritsky A, Gheber L. Phospho-regulation of kinesin-5 during anaphase spindle elongation. J Cell Sci. 2011;124:873–8. doi: 10.1242/jcs.077396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannigan A, Scheible WR, Lukowitz W, Fagerstrom C, Wadsworth P, Somerville C, Baskin TI. A conserved role for kinesin-5 in plant mitosis. J Cell Sci. 2007;120:2819–27. doi: 10.1242/jcs.009506. [DOI] [PubMed] [Google Scholar]
- Beer T, Goldman B, Synold T, Ryan C, Vasist L, Van Veldhuizen P, Dakhil S, Lara P, Drelichman A, Hussain M, Crawford E. Southwest Oncology Group phase II study of ispinesib in androgen-independent prostate cancer previously treated with taxanes. Clinical Genitourinary Cancer. 2008;6:103–109. doi: 10.3816/CGC.2008.n.016. [DOI] [PubMed] [Google Scholar]
- Bishop JD, Han Z, Schumacher JM. The Caenorhabditis elegans Aurora B kinase AIR-2 phosphorylates and is required for the localization of a BimC kinesin to meiotic and mitotic spindles. Mol Biol Cell. 2005;16:742–56. doi: 10.1091/mbc.E04-08-0682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagden SP, Molife LR, Seebaran A, Payne M, Reid AH, Protheroe AS, Vasist LS, Williams DD, Bowen C, Kathman SJ, Hodge JP, Dar MM, de Bono JS, Middleton MR. A phase I trial of ispinesib, a kinesin spindle protein inhibitor, with docetaxel in patients with advanced solid tumours. Br J Cancer. 2008;98:894–9. doi: 10.1038/sj.bjc.6604264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blangy A, Arnaud L, Nigg EA. Phosphorylation by p34cdc2 protein kinase regulates binding of the kinesin-related motor HsEg5 to the dynactin subunit p150. J Biol Chem. 1997;272:19418–24. doi: 10.1074/jbc.272.31.19418. [DOI] [PubMed] [Google Scholar]
- Blangy A, Lane HA, d’Herin P, Harper M, Kress M, Nigg EA. Phosphorylation by p34cdc2 regulates spindle association of human Eg5, a kinesin-related motor essential for bipolar spindle formation in vivo. Cell. 1995;83:1159–69. doi: 10.1016/0092-8674(95)90142-6. [DOI] [PubMed] [Google Scholar]
- Block SM, Goldstein LSB, Schnapp BJ. Bead movement by single kinesin molecules studied with optical tweezers. Nature. 1990;348:348–352. doi: 10.1038/348348a0. [DOI] [PubMed] [Google Scholar]
- Block SM, Asbury CL, Shaevitz JW, Lang MJ. Probing the kinesin reaction cycle with a 2D optical force clamp. Proc Natl Acad Sci USA. 2003;100:2531–2536. doi: 10.1073/pnas.0436709100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenmiller B, Mueller LN, Pedrioli PG, Pflieger D, Junger MA, Eng JK, Aebersold R, Tao WA. An integrated chemical, mass spectrometric and computational strategy for (quantitative) phosphoproteomics: application to Drosophila melanogaster Kc167 cells. Mol Biosyst. 2007;3:275–86. doi: 10.1039/b617545g. [DOI] [PubMed] [Google Scholar]
- Bodey AJ, Kikkawa M, Moores CA. 9-Angstrom structure of a microtubule-bound mitotic motor. J Mol Biol. 2009;388:218–24. doi: 10.1016/j.jmb.2009.03.008. [DOI] [PubMed] [Google Scholar]
- Brugués J, Nuzzo V, Mazur E, Needleman DJ. Nucleation and transport organize microtubules in metaphase spindles. Cell. 2012;149:554–564. doi: 10.1016/j.cell.2012.03.027. [DOI] [PubMed] [Google Scholar]
- Brust-Mascher I, Sommi P, Cheerambathur DK, Scholey JM. Kinesin-5-dependent poleward flux and spindle length control in Drosophila embryo mitosis. Molecular Biology of the Cell. 2009;20:1749–1762. doi: 10.1091/mbc.E08-10-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burris HA, 3rd, Jones SF, Williams DD, Kathman SJ, Hodge JP, Pandite L, Ho PT, Boerner SA, Lorusso P. A phase I study of ispinesib, a kinesin spindle protein inhibitor, administered weekly for three consecutive weeks of a 28-day cycle in patients with solid tumors. Invest New Drugs. 2011;29:467–72. doi: 10.1007/s10637-009-9374-x. [DOI] [PubMed] [Google Scholar]
- Cahu J, Olichon A, Hentrich C, Schek H, Drinjakovic J, Zhang C, Doherty-Kirby A, Lajoie G, Surrey T. Phosphorylation by Cdk1 increases the binding of Eg5 to microtubules in vitro and in Xenopus egg extract spindles. PLoS One. 2008;3:e3936. doi: 10.1371/journal.pone.0003936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carol H, Lock R, Houghton PJ, Morton CL, Kolb EA, Gorlick R, Reynolds CP, Maris JM, Keir ST, Billups CA, Smith MA. Initial testing (stage 1) of the kinesin spindle protein inhibitor ispinesib by the pediatric preclinical testing program. Pediatr Blood Cancer. 2009;53:1255–63. doi: 10.1002/pbc.22056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo A, Morse HC, III, Godfrey VL, Naeem R, Justice MJ. Overexpression of Eg5 causes genomic instability and tumor formation in mice. Cancer Res. 2007;67:10138–10147. doi: 10.1158/0008-5472.CAN-07-0326. [DOI] [PubMed] [Google Scholar]
- Chan KS, Koh CG, Li HY. Mitosis-targeted anti-cancer therapies: where they stand. Cell Death Dis. 2012;3:e411. doi: 10.1038/cddis.2012.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Q, Nitta R, Inoue S, Hirokawa N. Structural basis for the ATP-induced isomerization of kinesin. J Mol Biol. 2013;425:1869–80. doi: 10.1016/j.jmb.2013.03.004. [DOI] [PubMed] [Google Scholar]
- Chauviere M, Kress C, Kress M. Disruption of the mitotic kinesin Eg5 gene (Knsl1) results in early embryonic lethality. Biochem Biophys Res Commun. 2008;372:513–9. doi: 10.1016/j.bbrc.2008.04.177. [DOI] [PubMed] [Google Scholar]
- Chee MK, Haase SB. B-cyclin/CDKs regulate mitotic spindle assembly by phosphorylating kinesins-5 in budding yeast. PLoS Genet. 2010;6:e1000935. doi: 10.1371/journal.pgen.1000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheerambathur DK, Brust-Mascher I, Civelekoglu-Scholey G, Scholey JM. Dynamic partitioning of mitotic kinesin-5 cross-linkers between microtubule-bound and freely diffusing states. J Cell Biol. 2008;182:429–436. doi: 10.1083/jcb.200804100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Chow J, Poon R. Inhibition of Eg5 acts synergistically with checkpoint abrogation in promoting mitotic catastrophe. Mol Can Res. 2012;10:626–635. doi: 10.1158/1541-7786.MCR-11-0491. [DOI] [PubMed] [Google Scholar]
- Cochran JC, Gatial JE, 3rd, Kapoor TM, Gilbert SP. Monastrol inhibition of the mitotic kinesin Eg5. J Biol Chem. 2005;280:12658–67. doi: 10.1074/jbc.M413140200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochran JC, Gilbert SP. ATPase mechanism of Eg5 in the absence of microtubules: insight into microtubule activation and allosteric inhibition by monastrol. Biochemistry. 2005;44:16633–48. doi: 10.1021/bi051724w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochran JC, Krzysiak TC, Gilbert SP. Pathway of ATP hydrolysis by monomeric kinesin Eg5. Biochemistry. 2006;45:12334–44. doi: 10.1021/bi0608562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochran JC, Sontag CA, Maliga Z, Kapoor TM, Correia JJ, Gilbert SP. Mechanistic analysis of the mitotic kinesin Eg5. J Biol Chem. 2004;279:38861–70. doi: 10.1074/jbc.M404203200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen P. The regulation of protein function by multisite phosphorylation--a 25 year update. Trends Biochem Sci. 2000;25:596–601. doi: 10.1016/s0968-0004(00)01712-6. [DOI] [PubMed] [Google Scholar]
- Cole DG, Saxton WM, Sheehan KB, Scholey JM. A slow homotetrameric kinesin-related motor protein purified from Drosophila embryos. J Biol Chem. 1994;269:22913–22916. [PMC free article] [PubMed] [Google Scholar]
- Coureux PD, Sweeney HL, Houdusse A. Three myosin V structures delineate essential features of chemo-mechanical transduction. EMBO J. 2004;23:4527–37. doi: 10.1038/sj.emboj.7600458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox CD, Breslin MJ, Mariano BJ, Coleman PJ, Buser CA, Walsh ES, Hamilton K, Huber HE, Kohl NE, Torrent M, Yan Y, Kuo LC, Hartman GD. Kinesin spindle protein (KSP) inhibitors. Part 1: The discovery of 3,5-diaryl-4,5-dihydropyrazoles as potent and selective inhibitors of the mitotic kinesin KSP. Bioorg Med Chem Lett. 2005;15:2041–5. doi: 10.1016/j.bmcl.2005.02.055. [DOI] [PubMed] [Google Scholar]
- Cox CD, Coleman PJ, Breslin MJ, Whitman DB, Garbaccio RM, Fraley ME, Buser CA, Walsh ES, Hamilton K, Schaber MD, Lobell RB, Tao W, Davide JP, Diehl RE, Abrams MT, South VJ, Huber HE, Torrent M, Prueksaritanont T, Li C, Slaughter DE, Mahan E, Fernandez-Metzler C, Yan Y, Kuo LC, Kohl NE, Hartman GD. Kinesin Spindle Protein (KSP) Inhibitors. 9. Discovery of (2S)-4-(2,5-Difluorophenyl)-N-[(3R,4S)-3-fluoro-1-methylpiperidin-4-yl]-2-(hydroxymethyl)-N-methyl-2-phenyl-2,5-dihydro-1H-pyrrole-1-carboxamide (MK-0731) for the Treatment of Taxane-Refractory Cancer. J Med Chem. 2008;51:4239–4252. doi: 10.1021/jm800386y. [DOI] [PubMed] [Google Scholar]
- Crevel IMTC, Alonso MC, Cross RA. Monastrol stabilises an attached low-friction mode of Eg5. Curr Biol. 2004;14:R411–2. doi: 10.1016/j.cub.2004.05.030. [DOI] [PubMed] [Google Scholar]
- Crevel IMTC, Lockhart A, Cross RA. Kinetic evidence for low chemical processivity in ncd and Eg5. J Mol Biol. 1997;273:160–170. doi: 10.1006/jmbi.1997.1319. [DOI] [PubMed] [Google Scholar]
- Cytrynbaum EN, Sommi P, Brust-Mascher I, Scholey JM, Mogilner A. Early spindle assembly in Drosophila embryos: role of a force balance involving cytoskeletal dynamics and nuclear mechanics. Mol Biol Cell. 2005;16:4967–4981. doi: 10.1091/mbc.E05-02-0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBonis S, Simorre JP, Crevel I, Lebeau L, Skoufias DA, Blangy A, Ebel C, Gans P, Cross R, Hackney DD, Wade RH, Kozielski F. Interaction of the mitotic inhibitor monastrol with human kinesin Eg5. Biochemistry. 2003;42:338–49. doi: 10.1021/bi026716j. [DOI] [PubMed] [Google Scholar]
- DeBonis S, Skoufias DA, Lebeau L, Lopez R, Robin G, Margolis RL, Wade RH, Kozielski F. In vitro screening for inhibitors of the human mitotic kinesin Eg5 with antimitotic and antitumor activities. Mol Cancer Ther. 2004;3:1079–90. [PubMed] [Google Scholar]
- Desai A, Maddox PS, Mitchison TJ, Salmon ED. Anaphase A chromosome movement and poleward spindle microtubule flux occur at similar rates in Xenopus extract spindles. J Cell Biol. 1998;141:703–713. doi: 10.1083/jcb.141.3.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmapuri S, Peruzzi D, Marra E, Palombo F, Bett AJ, Bartz SR, Yong M, Ciliberto G, La Monica N, Buser CA, Toniatti C, Aurisicchio L. Intratumor RNA interference of cell cycle genes slows down tumor progression. Gene Ther. 2011;18:727–33. doi: 10.1038/gt.2011.27. [DOI] [PubMed] [Google Scholar]
- Dhonukshe P, Vischer N, Gadella TWJ. Contribution of microtubule growth polarity and flux to spindle assembly and functioning in plant cells. J Cell Sci. 2006;119:3193–3205. doi: 10.1242/jcs.03048. [DOI] [PubMed] [Google Scholar]
- Drummond DR, Hagan IM. Mutations in the bimC box of Cut7 indicate divergence of regulation within the bimC family of kinesin related proteins. J Cell Sci. 1998;111 ( Pt 7):853–65. doi: 10.1242/jcs.111.7.853. [DOI] [PubMed] [Google Scholar]
- Dumont S, Mitchison TJ. Force and length in the mitotic spindle. Curr Biol. 2009;19:749–761. doi: 10.1016/j.cub.2009.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duselder A, Thiede C, Schmidt CF, Lakamper S. Neck-linker length dependence of processive Kinesin-5 motility. J Mol Biol. 2012;423:159–68. doi: 10.1016/j.jmb.2012.06.043. [DOI] [PubMed] [Google Scholar]
- Eckerdt F, Eyers PA, Lewellyn AL, Prigent C, Maller JL. Spindle pole regulation by a discrete Eg5-interacting domain in TPX2. Curr Biol. 2008;18:519–25. doi: 10.1016/j.cub.2008.02.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Nassan HB. Advances in the discovery of kinesin spindle protein (Eg5) inhibitors as antitumor agents. Eur J Med Chem. 2013;62:614–631. doi: 10.1016/j.ejmech.2013.01.031. [DOI] [PubMed] [Google Scholar]
- Enos A, Morris N. Mutation of a gene that encodes a kinesin-like protein blocks nuclear division in A. nidulans. Cell. 1990;60:1019–1027. doi: 10.1016/0092-8674(90)90350-n. [DOI] [PubMed] [Google Scholar]
- Esaki T, Seto T, Ariyama H, Arita S, Fujimoto C, Tsukasa K, Kometani T, Nosaki K, Hirai F, Yagawa K. Phase I study to assess the safety, tolerability and pharmacokinetics of AZD4877 in Japanese patients with solid tumors. Arch Drug Inf. 2011;4:23–31. doi: 10.1111/j.1753-5174.2011.00034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falnikar A, Tole S, Baas PW. Kinesin-5, a mitotic microtubule-associated motor protein, modulates neuronal migration. Mol Biol Cell. 2011;22:1561–1574. doi: 10.1091/mbc.E10-11-0905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferenz NP, Gable AD, Wadsworth P. Mitotic functions of kinesin-5. Semin Cell Dev Biol. 2010;21:255–259. doi: 10.1016/j.semcdb.2010.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferenz NP, Wadsworth P. Prophase microtubule arrays undergo flux-like behavior in mammalian cells. Mol Biol Cell. 2007;18:3993–4002. doi: 10.1091/mbc.E07-05-0420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferhat L, Cook C, Chauviere M, Harper M, Kress M, Lyons GE, Baas PW. Expression of the mitotic motor protein Eg5 in postmitotic neurons: implications for neuronal development. J Neurosci. 1998;18:7822–7835. doi: 10.1523/JNEUROSCI.18-19-07822.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florian S, Mayer TU. Modulated microtubule dynamics enable Hklp2/Kif15 to assemble bipolar spindles. Cell Cycle. 2011;10:3533–3544. doi: 10.4161/cc.10.20.17817. [DOI] [PubMed] [Google Scholar]
- Florian S, Mayer TU. The functional antagonism between Eg5 and dynein in spindle bipolarization is not compatible with a simple push-pull model. Cell Reports. 2012;1:408–416. doi: 10.1016/j.celrep.2012.03.006. [DOI] [PubMed] [Google Scholar]
- Formstecher E, Reverdy C, Cholay M, Planquette C, Trouplin V, Lehrmann H, Aresta S, Calabrese A, Arar K, Daviet L, Colland F. Combination of active and inactive siRNA targeting the mitotic kinesin Eg5 impairs silencing efficiency in several cancer cell lines. Oligonucleotides. 2006;16:387–94. doi: 10.1089/oli.2006.16.387. [DOI] [PubMed] [Google Scholar]
- Fraley ME, Garbaccio RM, Arrington KL, Hoffman WF, Tasber ES, Coleman PJ, Buser CA, Walsh ES, Hamilton K, Fernandes C, Schaber MD, Lobell RB, Tao W, South VJ, Yan Y, Kuo LC, Prueksaritanont T, Shu C, Torrent M, Heimbrook DC, Kohl NE, Huber HE, Hartman GD. Kinesin spindle protein (KSP) inhibitors. Part 2: The design, synthesis, and characterization of 2,4-diaryl-2,5-dihydropyrrole inhibitors of the mitotic kinesin KSP. Bioorg Med Chem Lett. 2006;16:1775–1779. doi: 10.1016/j.bmcl.2006.01.030. [DOI] [PubMed] [Google Scholar]
- Gable AD, Qiu M, Titus J, Balchand S, Ferenz NP, Ma N, Collins ES, Fagerstrom C, Ross JL, Yang G, Wadsworth P. Dynamic reorganization of Eg5 in the mammalian spindle throughout mitosis requires dynein and TPX2. Mol Biol Cell. 2012;23:1254–1266. doi: 10.1091/mbc.E11-09-0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaetz J, Kapoor TM. Dynein/dynactin regulate metaphase spindle length by targeting depolymerizing activities to spindle poles. J Cell Biol. 2004;166:465–471. doi: 10.1083/jcb.200404015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbaccio RM, Fraley ME, Tasber ES, Olson CM, Hoffman WF, Arrington KL, Torrent M, Buser CA, Walsh ES, Hamilton K, Schaber MD, Fernandes C, Lobell RB, Tao W, South VJ, Yan Y, Kuo LC, Prueksaritanont T, Slaughter DE, Shu C, Heimbrook DC, Kohl NE, Huber HE, Hartman GD. Kinesin spindle protein (KSP) inhibitors. Part 3: Synthesis and evaluation of phenolic 2,4-diaryl-2,5-dihydropyrroles with reduced hERG binding and employment of a phosphate prodrug strategy for aqueous solubility. Bioorg Med Chem Lett. 2006;16:1780–1783. doi: 10.1016/j.bmcl.2005.12.094. [DOI] [PubMed] [Google Scholar]
- Garcia K, Stumpff J, Duncan T, Su TT. Tyrosines in the kinesin-5 head domain are necessary for phosphorylation by Wee1 and for mitotic spindle integrity. Curr Biol. 2009;19:1670–6. doi: 10.1016/j.cub.2009.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner MK, Bouck DC, Paliulis LV, Meehl JB, O’Toole ET, Haase J, Soubry A, Joglekar AP, Winey M, Salmon ED, Bloom K, Odde DJ. Chromosome congression by Kinesin-5 motor-mediated disassembly of longer kinetochore microtubules. Cell. 2008;135:894–906. doi: 10.1016/j.cell.2008.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giet R, Uzbekov R, Cubizolles F, Le Guellec K, Prigent C. The Xenopus laevis aurora-related protein kinase pEg2 associates with and phosphorylates the kinesin-related protein XlEg5. J Biol Chem. 1999;274:15005–13. doi: 10.1074/jbc.274.21.15005. [DOI] [PubMed] [Google Scholar]
- Gomez HL, Philco M, Pimentel P, Kiyan M, Monsalvo ML, Conlan MG, Saikali KG, Chen MM, Seroogy JJ, Wolff AA, Escandon RD. Phase I dose-escalation and pharmacokinetic study of ispinesib, a kinesin spindle protein inhibitor, administered on days 1 and 15 of a 28-day schedule in patients with no prior treatment for advanced breast cancer. Anticancer Drugs. 2012;23:335–41. doi: 10.1097/CAD.0b013e32834e74d6. [DOI] [PubMed] [Google Scholar]
- Goshima G, Scholey JM. Control of mitotic spindle length. Ann Rev Cell Dev Biol. 2010;26:21–57. doi: 10.1146/annurev-cellbio-100109-104006. [DOI] [PubMed] [Google Scholar]
- Goshima G, Vale RD. The roles of microtubule-based motor proteins in mitosis: comprehensive RNAi analysis in the Drosophila S2 cell line. J Cell Biol. 2003;162:1003–16. doi: 10.1083/jcb.200303022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Wollman R, Stuurman N, Scholey JM, Vale RD. Length control of the metaphase spindle. Curr Biol. 2005;15:1979–1988. doi: 10.1016/j.cub.2005.09.054. [DOI] [PubMed] [Google Scholar]
- Goulet A, Behnke-Parks WM, Sindelar CV, Major J, Rosenfeld SS, Moores CA. The structural basis of force generation by the mitotic motor kinesin-5. J Biol Chem. 2012;287:44654–66. doi: 10.1074/jbc.M112.404228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groen AC, Needleman DJ, Brangwynne C, Gradinaru C, Fowler B, Mazitschek R, Mitchison TJ. A novel small-molecule inhibitor reveals a possible role of kinesin-5 in anastral spindle-pole assembly. J Cell Sci. 2008;121:2293–2300. doi: 10.1242/jcs.024018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth-Pedersen L, Aits S, Corcelle-Termeau E, Petersen NH, Nylandsted J, Jaattela M. Identification of cytoskeleton-associated proteins essential for lysosomal stability and survival of human cancer cells. PLoS One. 2012;7:e45381. doi: 10.1371/journal.pone.0045381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackney DD. The tethered motor domain of a kinesin-microtubule complex catalyzes reversible synthesis of bound ATP. Proc Natl Acad Sci U S A. 2005;102:18338–43. doi: 10.1073/pnas.0505288102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan I, Yanagida M. Novel potential mitotic motor protein encoded by the fission yeast cut7+ gene. Nature. 1990;347:563–6. doi: 10.1038/347563a0. [DOI] [PubMed] [Google Scholar]
- Hansen GM, Justice MJ. Activation of Hex and mEg5 by retroviral insertion may contribute to mouse B-cell leukemia. Oncogene. 1999;18:6531–9. doi: 10.1038/sj.onc.1203023. [DOI] [PubMed] [Google Scholar]
- Haque S, Hasaka T, Brooks A, Lobanov P, Baas P. Monastrol, a prototype anti-cancer drug that inhibits a mitotic kinesin, induces rapid bursts of axonal outgrowth from cultured postmitotic neurons. Cell Mot Cyto. 2004;58:10–16. doi: 10.1002/cm.10176. [DOI] [PubMed] [Google Scholar]
- Hariharan V, Hancock WO. Insights into the mechanical properties of the kinesin neck linker domain from sequence analysis and molecular dynamics simulations. Cell Mol Bioeng. 2009;2:177–189. doi: 10.1007/s12195-009-0059-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck MM, Pereira A, Pesavento P, Yannoni Y, Spradling AC, Goldstein LS. The kinesin-like protein KLP61F is essential for mitosis in Drosophila. J Cell Biol. 1993;123:665–79. doi: 10.1083/jcb.123.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde PS, Cogswell J, Carrick K, Jackson J, Wood KW, Eng WK, Brawner M, Huang PS, Bergsma D. Differential gene expression analysis of kinesin spindle protein in human solid tumors. Proc Am Soc Clin Oncol. 2003;22:abstr 535. [Google Scholar]
- Hesse WR, Steiner M, Wohlever ML, Kamm RD, Hwang W, Lang MJ. Modular aspects of kinesin force generation machinery. Biophys J. 2013;104:1969–1978. doi: 10.1016/j.bpj.2013.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt ER, Gheber L, Kingsbury T, Hoyt MA. Homotetrameric form of Cin8p, a Saccharomyces cerevisiae kinesin-5 motor, is essential for its in vivo function. J Biol Chem. 2006;281:26004–26013. doi: 10.1074/jbc.M604817200. [DOI] [PubMed] [Google Scholar]
- Hirokawa N, Nitta R, Okada Y. The mechanisms of kinesin motor motility: lessons from the monomeric motor KIF1A. Nat Rev Mol Cell Biol. 2009;10:877–84. doi: 10.1038/nrm2807. [DOI] [PubMed] [Google Scholar]
- Hirose K, Akimaru E, Akiba T, Endow SA, Amos LA. Large conformational changes in a kinesin motor catalyzed by interaction with microtubules. Mol Cell. 2006;23:913–23. doi: 10.1016/j.molcel.2006.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holen K, DiPaola R, Liu G, Tan AR, Wilding G, Hsu K, Agrawal N, Chen C, Xue L, Rosenberg E, Stein M. A phase I trial of MK-0731, a kinesin spindle protein (KSP) inhibitor, in patients with solid tumors. Invest New Drugs. 2012;30:1088–95. doi: 10.1007/s10637-011-9653-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holen KD, Belani CP, Wilding G, Ramalingam S, Volkman JL, Ramanathan RK, Vasist LS, Bowen CJ, Hodge JP, Dar MM, Ho PT. A first in human study of SB-743921, a kinesin spindle protein inhibitor, to determine pharmacokinetics, biologic effects and establish a recommended phase II dose. Cancer Chemother Pharmacol. 2011;67:447–54. doi: 10.1007/s00280-010-1346-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotha S, Yarrow JC, Yang JG, Garrett S, Renduchintala KV, Mayer TU, Kapoor TM. HR22C16: a potent small-molecule probe for the dynamics of cell division. Angew Chem Int Ed Engl. 2003;42:2379–82. doi: 10.1002/anie.200351173. [DOI] [PubMed] [Google Scholar]
- Hoyt MA, He L, Loo KK, Saunders WS. Two Saccharomyces cerevisiae kinesin-related gene products required for mitotic spindle assembly. J Cell Biol. 1992;118:109–20. doi: 10.1083/jcb.118.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huszar D, Theoclitou ME, Skolnik J, Herbst R. Kinesin motor proteins as targets for cancer therapy. Cancer Metastasis Rev. 2009;28:197–208. doi: 10.1007/s10555-009-9185-8. [DOI] [PubMed] [Google Scholar]
- Hwang W, Lang MJ, Karplus M. Force generation in kinesin hinges on cover-neck bundle formation. Structure. 2008;16:62–71. doi: 10.1016/j.str.2007.11.008. [DOI] [PubMed] [Google Scholar]
- Infante JR, Kurzrock R, Spratlin J, Burris HA, Eckhardt SG, Li J, Wu K, Skolnik JM, Hylander-Gans L, Osmukhina A, Huszar D, Herbst RS. A Phase I study to assess the safety, tolerability, and pharmacokinetics of AZD4877, an intravenous Eg5 inhibitor in patients with advanced solid tumors. Cancer Chemother Pharmacol. 2012;69:165–72. doi: 10.1007/s00280-011-1667-z. [DOI] [PubMed] [Google Scholar]
- Inoué S, Sato H. Cell motility by labile association of molecules. The nature of mitotic spindle fibers and their role in chromosome movement. J General Phys. 1967;50(Suppl):259–92. [PMC free article] [PubMed] [Google Scholar]
- Iwakiri Y, Kamakura S, Hayase J, Sumimoto H. Interaction of NuMA protein with the kinesin Eg5: its possible role in bipolar spindle assembly and chromosome alignment. Biochem J. 2013;451:195–204. doi: 10.1042/BJ20121447. [DOI] [PubMed] [Google Scholar]
- Jackson JR, Patrick DR, Dar MM, Huang PS. Targeted anti-mitotic therapies: can we improve on tubulin agents? Nat Rev Cancer. 2007;7:107–17. doi: 10.1038/nrc2049. [DOI] [PubMed] [Google Scholar]
- Johnson RK, McCabe FL, Cauder E, Inlow-Porter L, Whitacre M, Winkler JD, Bergnes G, Feng B, Morgans D, Wood KW, Jackson JR. SB-715992, a potent and selective inhibitor of KSP mitotic kinesin, demonstrates broad-spectrum activity in advanced murine tumors and human tumor xenografts. Proc Annu Meet Am Assoc Cancer Res. 2002;43:abst 269. [Google Scholar]
- Jun B, Kim S. Real-time structural transitions are coupled to chemical steps in ATP hydrolysis by Eg5 kinesin. J Biol Chem. 2010;285:11073–7. doi: 10.1074/jbc.C110.103762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitein LC, Kwok BH, Weinger JS, Schmidt CF, Kapoor TM, Peterman EJ. Microtubule cross-linking triggers the directional motility of kinesin-5. J Cell Biol. 2008;182:421–8. doi: 10.1083/jcb.200801145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitein LC, Peterman EJG, Kwok BH, Kim JH, Kapoor TM, Schmidt CF. The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature. 2005;435:114–118. doi: 10.1038/nature03503. [DOI] [PubMed] [Google Scholar]
- Kapoor TM, Mayer TU, Coughlin ML, Mitchison TJ. Probing spindle assembly mechanisms with monastrol, a small molecule inhibitor of the mitotic kinesin, Eg5. J Cell Biol. 2000;150:975–988. doi: 10.1083/jcb.150.5.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor TM, Mitchison TJ. Eg5 is static in bipolar spindles relative to tubulin: evidence for a static spindle matrix. J Cell Biol. 2001;154:1125–1133. doi: 10.1083/jcb.200106011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaseda K, Crevel I, Hirose K, Cross RA. Single-headed mode of kinesin-5. EMBO Rep. 2008;9:761–5. doi: 10.1038/embor.2008.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashina AS, Baskin RJ, Cole DG, Wedaman KP, Saxton WM, Scholey JM. A bipolar kinesin. Nature. 1996;379:270–272. doi: 10.1038/379270a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashina AS, Rogers GC, Scholey JM. The bimC family of kinesins: essential bipolar mitotic motors driving centrosome separation. Biochim Biophys Acta. 1997;1357:257–71. doi: 10.1016/s0167-4889(97)00037-2. [DOI] [PubMed] [Google Scholar]
- Kathman SJ, Williams DH, Hodge JP, Dar M. A Bayesian population PK-PD model of ispinesib-induced myelosuppression. Clin Pharmacol Ther. 2007;81:88–94. doi: 10.1038/sj.clpt.6100021. [DOI] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–66. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AS, Appleyard DC, Labno AK, Georges A, Karplus M, Belcher AM, Hwang W, Lang MJ. Kinesin’s cover-neck bundle folds forward to generate force. Proc Natl Acad Sci U S A. 2008;105:19247–52. doi: 10.1073/pnas.0805147105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury GA, Baliban RC, Floudas CA. Proteome-wide post-translational modification statistics: frequency analysis and curation of the swiss-prot database. Sci Rep. 2011;1 doi: 10.1038/srep00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury HJ, Garcia-Manero G, Borthakur G, Kadia T, Foudray MC, Arellano M, Langston A, Bethelmie-Bryan B, Rush S, Litwiler K, Karan S, Simmons H, Marcus AI, Ptaszynski M, Kantarjian H. A phase 1 dose-escalation study of ARRY-520, a kinesin spindle protein inhibitor, in patients with advanced myeloid leukemias. Cancer. 2012;118:3556–64. doi: 10.1002/cncr.26664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikkawa M, Sablin EP, Okada Y, Yajima H, Fletterick RJ, Hirokawa N. Switch-based mechanism of kinesin motors. Nature. 2001;411:439–45. doi: 10.1038/35078000. [DOI] [PubMed] [Google Scholar]
- Kim ED, Buckley R, Learman S, Richard J, Parke C, Worthylake DK, Wojcik EJ, Walker RA, Kim S. Allosteric drug discrimination is coupled to mechanochemical changes in the kinesin-5 motor core. J Biol Chem. 2010;285:18650–61. doi: 10.1074/jbc.M109.092072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koffa MD, Casanova CM, Santarella R, Kocher T, Wilm M, Mattaj IW. HURP is part of a Ran-dependent complex involved in spindle formation. Curr Biol. 2006;16:743–54. doi: 10.1016/j.cub.2006.03.056. [DOI] [PubMed] [Google Scholar]
- Kollu S, Bakhoum SF, Compton DA. Interplay of microtubule dynamics and sliding during bipolar spindle formation in mammalian cells. Current Biol. 2009;19:2108–2113. doi: 10.1016/j.cub.2009.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kops GJ, Weaver BA, Cleveland DW. On the road to cancer: aneuploidy and the mitotic checkpoint. Nat Rev Cancer. 2005;5:773–85. doi: 10.1038/nrc1714. [DOI] [PubMed] [Google Scholar]
- Korneev MJ, Lakamper S, Schmidt CF. Load-dependent release limits the processive stepping of the tetrameric Eg5 motor. Eur Biophys J. 2007;36:675–81. doi: 10.1007/s00249-007-0134-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozielski F, Sack S, Marx A, Thormahlen M, Schonbrunn E, Biou V, Thompson A, Mandelkow EM, Mandelkow E. The crystal structure of dimeric kinesin and implications for microtubule-dependent motility. Cell. 1997;91:985–94. doi: 10.1016/s0092-8674(00)80489-4. [DOI] [PubMed] [Google Scholar]
- Krzysiak TC, Gilbert SP. Dimeric Eg5 maintains processivity through alternating-site catalysis with rate-limiting ATP hydrolysis. J Biol Chem. 2006;281:39444–54. doi: 10.1074/jbc.M608056200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzysiak TC, Grabe M, Gilbert SP. Getting in sync with dimeric Eg5. Initiation and regulation of the processive run. J Biol Chem. 2008;283:2078–87. doi: 10.1074/jbc.M708354200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzysiak TC, Wendt T, Sproul LR, Tittmann P, Gross H, Gilbert SP, Hoenger A. A structural model for monastrol inhibition of dimeric kinesin Eg5. EMBO J. 2006;25:2263–73. doi: 10.1038/sj.emboj.7601108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kull FJ, Endow SA. Kinesin: switch I & II and the motor mechanism. J Cell Sci. 2002;115:15–23. doi: 10.1242/jcs.115.1.15. [DOI] [PubMed] [Google Scholar]
- Kwok BH, Kapitein LC, Kim JH, Peterman EJ, Schmidt CF, Kapoor TM. Allosteric inhibition of kinesin-5 modulates its processive directional motility. Nat Chem Biol. 2006;2:480–5. doi: 10.1038/nchembio812. [DOI] [PubMed] [Google Scholar]
- Kwok BH, Yang JG, Kapoor TM. The rate of bipolar spindle assembly depends on the microtubule-gliding velocity of the mitotic kinesin Eg5. Curr Biol. 2004;14:1783–1788. doi: 10.1016/j.cub.2004.09.052. [DOI] [PubMed] [Google Scholar]
- LaFountain JR, Cohan CS, Siegel AJ, LaFountain DJ. Direct visualization of microtubule flux during metaphase and anaphase in crane-fly spermatocytes. Mol Biol Cell. 2004;15:5724–5732. doi: 10.1091/mbc.E04-08-0750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson AG, Naber N, Cooke R, Pate E, Rice SE. The conserved L5 loop establishes the pre-powerstroke conformation of the Kinesin-5 motor, Eg5. Biophys J. 2010;98:2619–27. doi: 10.1016/j.bpj.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence CJ, Dawe RK, Christie KR, Cleveland DW, Dawson SC, Endow SA, Goldstein LS, Goodson HV, Hirokawa N, Howard J, Malmberg RL, McIntosh JR, Miki H, Mitchison TJ, Okada Y, Reddy AS, Saxton WM, Schliwa M, Scholey JM, Vale RD, Walczak CE, Wordeman L. A standardized kinesin nomenclature. J Cell Biol. 2004;167:19–22. doi: 10.1083/jcb.200408113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Learman SS, Kim CD, Stevens NS, Kim S, Wojcik EJ, Walker RA. NSC 622124 inhibits human Eg5 and other kinesins via interaction with the conserved microtubule-binding site. Biochemistry. 2009;48:1754–1762. doi: 10.1021/bi801291q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Bélanger K, Rao S, Petrella T, Tozer R, Wood L, Savage K, Eisenhauer E, Synold T, Wainman N, Seymour L. A phase II study of ispinesib (SB-715992) in patients with metastatic or recurrent malignant melanoma: a National Cancer Institute of Canada Clinical Trials Group trial. Investigational New Drugs. 2008;26:249–255. doi: 10.1007/s10637-007-9097-9. [DOI] [PubMed] [Google Scholar]
- Levesque AA, Compton DA. The chromokinesin Kid is necessary for chromosome arm orientation and oscillation, but not congression, on mitotic spindles. J Cell Biol. 2001;154:1135–1146. doi: 10.1083/jcb.200106093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Raghavan S, Nelesen S, Linder CR, Warnow T. Rapid and accurate large-scale coestimation of sequence alignments and phylogenetic trees. Science. 2009;324:1561–4. doi: 10.1126/science.1171243. [DOI] [PubMed] [Google Scholar]
- Liu K, Warnow TJ, Holder MT, Nelesen SM, Yu J, Stamatakis AP, Linder CR. SATe-II: very fast and accurate simultaneous estimation of multiple sequence alignments and phylogenetic trees. Syst Biol. 2012;61:90–106. doi: 10.1093/sysbio/syr095. [DOI] [PubMed] [Google Scholar]
- Liu L, Parameswaran S, Liu J, Kim S, Wojcik EJ. Loop 5-directed compounds inhibit chimeric kinesin-5 motors: implications for conserved allosteric mechanisms. J Biol Chem. 2011;286:6201–10. doi: 10.1074/jbc.M110.154989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Aneja R, Liu C, Sun L, Gao J, Wang H, Dong JT, Sarli V, Giannis A, Joshi HC, Zhou J. Inhibition of the mitotic kinesin Eg5 up-regulates Hsp70 through the phosphatidylinositol 3-kinase/Akt pathway in multiple myeloma cells. J Biol Chem. 2006;281:18090–7. doi: 10.1074/jbc.M601324200. [DOI] [PubMed] [Google Scholar]
- Liu M, Aneja R, Sun X, Xie S, Wang H, Wu X, Dong JT, Li M, Joshi HC, Zhou J. Parkin regulates Eg5 expression by Hsp70 ubiquitination-dependent inactivation of c-Jun NH2-terminal kinase. J Biol Chem. 2008a;283:35783–8. doi: 10.1074/jbc.M806860200. [DOI] [PubMed] [Google Scholar]
- Liu M, Yu H, Huo L, Liu J, Li M, Zhou J. Validating the mitotic kinesin Eg5 as a therapeutic target in pancreatic cancer cells and tumor xenografts using a specific inhibitor. Biochem Pharmacol. 2008b;76:169–78. doi: 10.1016/j.bcp.2008.04.018. [DOI] [PubMed] [Google Scholar]
- Longley DB, Johnston PG. Molecular mechanisms of drug resistance. J Pathol. 2005;205:275–92. doi: 10.1002/path.1706. [DOI] [PubMed] [Google Scholar]
- Luo L, Parrish CA, Nevins N, McNulty DE, Chaudhari AM, Carson JD, Sudakin V, Shaw AN, Lehr R, Zhao H, Sweitzer S, Lad L, Wood KW, Sakowicz R, Annan RS, Huang PS, Jackson JR, Dhanak D, Copeland RA, Auger KR. ATP-competitive inhibitors of the mitotic kinesin KSP that function via an allosteric mechanism. Nature Chem Biol. 2007;3:722–726. doi: 10.1038/nchembio.2007.34. [DOI] [PubMed] [Google Scholar]
- Ma N, Titus J, Gable A, Ross JL, Wadsworth P. TPX2 regulates the localization and activity of Eg5 in the mammalian mitotic spindle. J Cell Biol. 2011;195:87–98. doi: 10.1083/jcb.201106149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma N, Tulu US, Ferenz NP, Fagerstrom C, Wilde A, Wadsworth P. Poleward transport of TPX2 in the mammalian mitotic spindle requires dynein, Eg5, and microtubule flux. Mol Biol Cell. 2010;21:979–88. doi: 10.1091/mbc.E09-07-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox P, Desai A, Oegema K, Mitchison TJ, Salmon ED. Poleward microtubule flux is a major component of spindle dynamics and anaphase a in mitotic Drosophila embryos. Curr Biol. 2002;12:1670–1674. doi: 10.1016/s0960-9822(02)01183-1. [DOI] [PubMed] [Google Scholar]
- Maliga Z, Junqueira M, Toyoda Y, Ettinger A, Mora-Bermudez F, Klemm RW, Vasilj A, Guhr E, Ibarlucea-Benitez I, Poser I, Bonifacio E, Huttner WB, Shevchenko A, Hyman AA. A genomic toolkit to investigate kinesin and myosin motor function in cells. Nat Cell Biol. 2013;15:325–34. doi: 10.1038/ncb2689. [DOI] [PubMed] [Google Scholar]
- Maliga Z, Kapoor TM, Mitchison TJ. Evidence that monastrol is an allosteric inhibitor of the mitotic kinesin Eg5. Chem Biol. 2002;9:989–996. doi: 10.1016/s1074-5521(02)00212-0. [DOI] [PubMed] [Google Scholar]
- Marra E, Palombo F, Ciliberto G, Aurisicchio L. Kinesin spindle protein SiRNA slows tumor progression. J Cell Physiol. 2013;228:58–64. doi: 10.1002/jcp.24103. [DOI] [PubMed] [Google Scholar]
- Martens-de Kemp SR, Nagel R, Stigter-van Walsum M, van der Meulen IH, van Beusechem VW, Braakhuis BJ, Brakenhoff RH. Functional genetic screens identify genes essential for tumor cell survival in head and neck and lung cancer. Clinical Canc Res. 2013;19:1994–2003. doi: 10.1158/1078-0432.CCR-12-2539. [DOI] [PubMed] [Google Scholar]
- Mather WH, Fox RF. Kinesin’s biased stepping mechanism: amplification of neck linker zippering. Biophys J. 2006;91:2416–26. doi: 10.1529/biophysj.106.087049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, Shiloh Y, Gygi SP, Elledge SJ. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–6. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- Mayer TU, Kapoor TM, Haggarty SJ, King RW, Schreiber SL, Mitchison TJ. Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen. Science. 1999;286:971–4. doi: 10.1126/science.286.5441.971. [DOI] [PubMed] [Google Scholar]
- Mclntosh JR, Hepler PK, Van Wie DG. Model for mitosis. Nature. 1969;224:659–663. [Google Scholar]
- Miller K, Ng C, Ang P, Brufsky AM, Lee SC, Dees EC, Piccart M, Verrill M, Wardley A, Loftiss J, Bal J, Yeoh S, Hodge J, Williams D, Dar M, Kathman S, Ho PC. Phase II, open label study of SB-715992 (ispinesib) in subjects with advanced or metastatic breast cancer [abstract 1089]; San Antonio (TX). 28th Annual San Antonio Breast Cancer Symposium.2005. [Google Scholar]
- Mitchison TJ. Polewards microtubule flux in the mitotic spindle: evidence from photoactivation of fluorescence. J Cell Biol. 1989;109:637–652. doi: 10.1083/jcb.109.2.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison TJ. The proliferation rate paradox in antimitotic chemotherapy. Mol Biol Cell. 2012;23:1–6. doi: 10.1091/mbc.E10-04-0335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison TJ, Maddox PS, Gaetz J, Groen AC, Shirasu M, Desai A, Salmon ED, Kapoor TM. Roles of polymerization dynamics, opposed motors, and a tensile element in governing the length of Xenopus extract meiotic spindles. Mol Biol Cell. 2005;16:3064–3076. doi: 10.1091/mbc.E05-02-0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison TJ, Salmon ED. Poleward kinetochore fiber movement occurs during both metaphase and anaphase-A in newt lung cell mitosis. J Cell Biol. 1992;119:569–582. doi: 10.1083/jcb.119.3.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto DT, Perlman ZE, Burbank KS, Groen AC, Mitchison TJ. The kinesin Eg5 drives poleward microtubule flux in Xenopus laevis egg extract spindles. J Cell Biol. 2004;167:813–818. doi: 10.1083/jcb.200407126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogilner A, Craig E. Towards a quantitative understanding of mitotic spindle assembly and mechanics. J Cell Sci. 2010;123:3435–3445. doi: 10.1242/jcs.062208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers KA, Baas PW. Kinesin-5 regulates the growth of the axon by acting as a brake on its microtubule array. J Cell Biol. 2007;178:1081–1091. doi: 10.1083/jcb.200702074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadar VC, Ketschek A, Myers KA, Gallo G, Baas PW. Kinesin-5 is essential for growth-cone turning. Curr Biol. 2008;18:1972–1977. doi: 10.1016/j.cub.2008.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadar VC, Lin S, Baas PW. Microtubule redistribution in growth cones elicited by focal inactivation of kinesin-5. J Neurosci. 2012;32:5783–5794. doi: 10.1523/JNEUROSCI.0144-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nousiainen M, Sillje HH, Sauer G, Nigg EA, Korner R. Phosphoproteome analysis of the human mitotic spindle. Proc Natl Acad Sci U S A. 2006;103:5391–6. doi: 10.1073/pnas.0507066103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen JV, Vermeulen M, Santamaria A, Kumar C, Miller ML, Jensen LJ, Gnad F, Cox J, Jensen TS, Nigg EA, Brunak S, Mann M. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Science Signaling. 2010;3 doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- Onishi H, Mochizuki N, Morales MF. On the myosin catalysis of ATP hydrolysis. Biochemistry. 2004;43:3757–63. doi: 10.1021/bi040002m. [DOI] [PubMed] [Google Scholar]
- Orth JD, Tang Y, Shi J, Loy CT, Amendt C, Wilm C, Zenke FT, Mitchison TJ. Quantitative live imaging of cancer and normal cells treated with Kinesin-5 inhibitors indicates significant differences in phenotypic responses and cell fate. Mol Cancer Ther. 2008;7:3480–9. doi: 10.1158/1535-7163.MCT-08-0684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parke CL, Wojcik EJ, Kim S, Worthylake DK. ATP hydrolysis in Eg5 kinesin involves a catalytic two-water mechanism. J Biol Chem. 2010;285:5859–5867. doi: 10.1074/jbc.M109.071233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen RD, Soni DV, Wollman R, Hahn AT, Yee MC, Guan A, Hesley JA, Miller SC, Cromwell EF, Solow-Cordero DE, Meyer T, Cimprich KA. A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol Cell. 2009;35:228–39. doi: 10.1016/j.molcel.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell JW, Davis J, Reddy M, Martin S, Samayoa K, Vo H, Thomsen K, Bean P, Kuo WL, Ziyad S, Billig J, Feiler HS, Gray JW, Wood KW, Cases S. Activity of the kinesin spindle protein inhibitor Ispinesib (SB-715992) in models of breast cancer. Clinical Canc Res. 2010;16:566–576. doi: 10.1158/1078-0432.CCR-09-1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaijmakers JA, van Heesbeen RGHP, Meaders JL, Geers EF, Fernandez-Garcia B, Medema RH, Tanenbaum ME. Nuclear envelope-associated dynein drives prophase centrosome separation and enables Eg5-independent bipolar spindle formation. EMBO J. 2012;31:4179–4190. doi: 10.1038/emboj.2012.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapley J, Nicolas M, Groen A, Regue L, Bertran MT, Caelles C, Avruch J, Roig J. The NIMA-family kinase Nek6 phosphorylates the kinesin Eg5 at a novel site necessary for mitotic spindle formation. J Cell Sci. 2008;121:3912–3921. doi: 10.1242/jcs.035360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath O, Kozielski F. Kinesins and cancer. Nat Rev Cancer. 2012;12:527–39. doi: 10.1038/nrc3310. [DOI] [PubMed] [Google Scholar]
- Ray S, Meyhofer E, Milligan RA, Howard J. Kinesin follows the microtubule’s protofilament axis. J Cell Biol. 1993;121:1083–93. doi: 10.1083/jcb.121.5.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice S, Lin AW, Safer D, Hart CL, Naber N, Carragher BO, Cain SM, Pechatnikova E, Wilson-Kubalek EM, Whittaker M, Pate E, Cooke R, Taylor EW, Milligan RA, Vale RD. A structural change in the kinesin motor protein that drives motility. Nature. 1999;402:778–84. doi: 10.1038/45483. [DOI] [PubMed] [Google Scholar]
- Robinson JT, Wojcik E, Sanders MA, McGrail M, Hays TS. Cytoplasmic dynein is required for the nuclear attachment and migration of centrosomes during mitosis in Drosophila. J Cell Biol. 1999;146:597–608. doi: 10.1083/jcb.146.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roof DM, Meluh PB, Rose MD. Kinesin-related proteins required for assembly of the mitotic spindle. J Cell Biol. 1992;118:95–108. doi: 10.1083/jcb.118.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roostalu J, Hentrich C, Bieling P, Telley I, Schiebel A, Surrey T. Directional switching of the kinesin Cin8 through motor coupling. Science. 2011;332:94–99. doi: 10.1126/science.1199945. [DOI] [PubMed] [Google Scholar]
- Rosenfeld SS, Fordyce PM, Jefferson GM, King PH, Block SM. Stepping and stretching. How kinesin uses internal strain to walk processively. J Biol Chem. 2003;278:18550–6. doi: 10.1074/jbc.M300849200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld SS, Xing J, Jefferson GM, King PH. Docking and rolling, a model of how the mitotic motor Eg5 works. J Biol Chem. 2005;280:35684–95. doi: 10.1074/jbc.M506561200. [DOI] [PubMed] [Google Scholar]
- Rozan LM, El-Deiry WS. Identification and characterization of proteins interacting with Traf4, an enigmatic p53 target. Cancer Biol Ther. 2006;5:1228–35. doi: 10.4161/cbt.5.9.3295. [DOI] [PubMed] [Google Scholar]
- Sablin EP, Fletterick RJ. Nucleotide switches in molecular motors: structural analysis of kinesins and myosins. Current Opinion in Structural Biology. 2001;11:716–724. doi: 10.1016/s0959-440x(01)00265-2. [DOI] [PubMed] [Google Scholar]
- Sarli V, Giannis A. Targeting the kinesin spindle protein: basic principles and clinical implications. Clin Cancer Res. 2008;14:7583–7. doi: 10.1158/1078-0432.CCR-08-0120. [DOI] [PubMed] [Google Scholar]
- Saunders WS, Hornack D, Lengyel V, Deng C. The Saccharomyces cerevisiae kinesin-related motor Kar3p acts at preanaphase spindle poles to limit the number and length of cytoplasmic microtubules. J Cell Biol. 1997;137:417–431. doi: 10.1083/jcb.137.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders WS, Hoyt MA. Kinesin-related proteins required for structural integrity of the mitotic spindle. Cell. 1992;70:451–458. doi: 10.1016/0092-8674(92)90169-d. [DOI] [PubMed] [Google Scholar]
- Sawin KE, Leguellec K, Philippe M, Mitchison TJ. Mitotic spindle organization by a plus-end-directed microtubule motor. Nature. 1992;359:540–543. doi: 10.1038/359540a0. [DOI] [PubMed] [Google Scholar]
- Sawin KE, Mitchison TJ. Mitotic spindle assembly by two different pathways in vitro. TJ Cell Biol. 1991a;112:925–940. doi: 10.1083/jcb.112.5.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin KE, Mitchison TJ. Poleward microtubule flux mitotic spindles assembled in vitro. J Cell Biol. 1991b;112:941–954. doi: 10.1083/jcb.112.5.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin KE, Mitchison TJ. Mutations in the kinesin-like protein Eg5 disrupting localization to the mitotic spindle. Proc Natl Acad Sci U S A. 1995;92:4289–4293. doi: 10.1073/pnas.92.10.4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schief WR, Howard J. Conformational changes during kinesin motility. Curr Opin Cell Biol. 2001;13:19–28. doi: 10.1016/s0955-0674(00)00169-1. [DOI] [PubMed] [Google Scholar]
- Sharp DJ, Brown HM, Kwon M, Rogers GC, Holland G, Scholey JM. Functional coordination of three mitotic motors in Drosophila embryos. Mol Biol Cell. 2000;11:241–253. doi: 10.1091/mbc.11.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp DJ, McDonald KL, Brown HM, Matthies HJ, Walczak C, Vale RD, Mitchison TJ, Scholey JM. The bipolar kinesin, KLP61F, cross-links microtubules within interpolar microtubule bundles of Drosophila embryonic mitotic spindles. J Cell Biol. 1999;144:125–138. doi: 10.1083/jcb.144.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shastry S, Hancock WO. Interhead tension determines processivity across diverse N-terminal kinesins. Proc Natl Acad Sci U S A. 2011;108:16253–8. doi: 10.1073/pnas.1102628108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Orth JD, Mitchison T. Cell type variation in responses to antimitotic drugs that target microtubules and kinesin-5. Cancer Res. 2008;68:3269–76. doi: 10.1158/0008-5472.CAN-07-6699. [DOI] [PubMed] [Google Scholar]
- Sindelar CV, Downing KH. An atomic-level mechanism for activation of the kinesin molecular motors. Proc Natl Acad Sci U S A. 2010;107:4111–6. doi: 10.1073/pnas.0911208107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CA, Rayment I. X-ray structure of the magnesium(II).ADP.vanadate complex of the Dictyostelium discoideum myosin motor domain to 1.9 A resolution. Biochemistry. 1996;35:5404–17. doi: 10.1021/bi952633+. [DOI] [PubMed] [Google Scholar]
- Smith E, Hegarat N, Vesely C, Roseboom I, Larch C, Streicher H, Straatman K, Flynn H, Skehel M, Hirota T, Kuriyama R, Hochegger H. Differential control of Eg5-dependent centrosome separation by Plk1 and Cdk1. EMBO J. 2011;30:2233–45. doi: 10.1038/emboj.2011.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souid AK, Dubowy RL, Ingle AM, Conlan MG, Sun J, Blaney SM, Adamson PC. A pediatric phase I trial and pharmacokinetic study of ispinesib: a Children’s Oncology Group phase I consortium study. Pediatr Blood Cancer. 2010;55:1323–8. doi: 10.1002/pbc.22609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–90. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Svoboda K, Block SM. Force and velocity measured for single kinesin Molecules. Cell. 1994;77:773–784. doi: 10.1016/0092-8674(94)90060-4. [DOI] [PubMed] [Google Scholar]
- Tabernero J, Shapiro GI, LoRusso PM, Cervantes A, Schwartz GK, Weiss GJ, Paz-Ares L, Cho DC, Infante JR, Alsina M, Gounder MM, Falzone R, Harrop J, White AC, Toudjarska I, Bumcrot D, Meyers RE, Hinkle G, Svrzikapa N, Hutabarat RM, Clausen VA, Cehelsky J, Nochur SV, Gamba-Vitalo C, Vaishnaw AK, Sah DW, Gollob JA, Burris HA., 3rd First-in-humans trial of an RNA interference therapeutic targeting VEGF and KSP in cancer patients with liver involvement. Cancer Discov. 2013;3:406–17. doi: 10.1158/2159-8290.CD-12-0429. [DOI] [PubMed] [Google Scholar]
- Tan LJ, Saijo M, Kuraoka I, Narita T, Takahata C, Iwai S, Tanaka K. Xeroderma pigmentosum group F protein binds to Eg5 and is required for proper mitosis: implications for XP-F and XFE. Genes Cells. 2012;17:173–85. doi: 10.1111/j.1365-2443.2012.01582.x. [DOI] [PubMed] [Google Scholar]
- Tanenbaum ME, Macůrek L, Janssen A, Geers EF, Alvarez-Fernández M, Medema RH. Kif15 cooperates with eg5 to promote bipolar spindle assembly. Curr Biol. 2009;19:1703–1711. doi: 10.1016/j.cub.2009.08.027. [DOI] [PubMed] [Google Scholar]
- Tang P, Siu L, Chen E, Hotte S, Chia S, Schwarz J, Pond G, Johnson C, Colevas A, Synold T, Vasist L, Winquist E. Phase II study of ispinesib in recurrent or metastatic squamous cell carcinoma of the head and neck. Invest New Drugs. 2008;26:257–264. doi: 10.1007/s10637-007-9098-8. [DOI] [PubMed] [Google Scholar]
- Tao L, Mogilner A, Civelekoglu-Scholey G, Wollman R, Evans J, Stahlberg H, Scholey JM. A homotetrameric kinesin-5, KLP61F, bundles microtubules and antagonizes Ncd in motility assays. Curr Biol. 2006;16:2293–302. doi: 10.1016/j.cub.2006.09.064. [DOI] [PubMed] [Google Scholar]
- Tcherniuk S, van Lis R, Kozielski F, Skoufias DA. Mutations in the human kinesin Eg5 that confer resistance to monastrol and S-trityl-L-cysteine in tumor derived cell lines. Biochem Pharmacol. 2010;79:864–72. doi: 10.1016/j.bcp.2009.11.001. [DOI] [PubMed] [Google Scholar]
- Tihy F, Kress M, Harper M, Dutrillaux B, Lemieux N. Localization of the human kinesin-related gene to band 10q24 by fluorescence in situ hybridization. Genomics. 1992;13:1371–2. doi: 10.1016/0888-7543(92)90075-4. [DOI] [PubMed] [Google Scholar]
- Tikhonenko I, Nag DK, Martin N, Koonce MP. Kinesin-5 is not essential for mitotic spindle elongation in Dictyostelium. Cell Motil Cytoskeleton. 2008;65:853–862. doi: 10.1002/cm.20307. [DOI] [PubMed] [Google Scholar]
- Tsai MY, Wang S, Heidinger JM, Shumaker DK, Adam SA, Goldman RD, Zheng Y. A mitotic lamin B matrix induced by RanGTP required for spindle assembly. Science. 2006;311:1887–1893. doi: 10.1126/science.1122771. [DOI] [PubMed] [Google Scholar]
- Tsui M, Xie T, Orth JD, Carpenter AE, Rudnicki S, Kim S, Shamu CE, Mitchison TJ. An intermittent live cell imaging screen for siRNA enhancers and suppressors of a kinesin-5 inhibitor. PLoS One. 2009;4:e7339. doi: 10.1371/journal.pone.0007339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner J, Anderson R, Guo J, Beraud C, Fletterick R, Sakowicz R. Crystal structure of the mitotic spindle kinesin Eg5 reveals a novel conformation of the neck-linker. J Biol Chem. 2001;276:25496–502. doi: 10.1074/jbc.M100395200. [DOI] [PubMed] [Google Scholar]
- Ulaganathan V, Talapatra SK, Rath O, Pannifer A, Hackney DD, Kozielski F. Structural insights into a unique inhibitor binding pocket in kinesin spindle protein. J Am Chem Soc. 2013;135:2263–72. doi: 10.1021/ja310377d. [DOI] [PubMed] [Google Scholar]
- Uteng M, Hentrich C, Miura K, Bieling P, Surrey T. Poleward transport of Eg5 by dynein-dynactin in Xenopus laevis egg extract spindles. J Cell Biol. 2008;182:715–726. doi: 10.1083/jcb.200801125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale RD. Switches, latches, and amplifiers: Common themes of G proteins and molecular motors. J Cell Biol. 1996;135:291–302. doi: 10.1083/jcb.135.2.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale RD, Milligan RA. The way things move: Looking under the hood of molecular motor proteins. Science. 2000;288:88–95. doi: 10.1126/science.288.5463.88. [DOI] [PubMed] [Google Scholar]
- Valentine MT, Block SM. Force and Premature Binding of ADP Can Regulate the Processivity of Individual Eg5 Dimers. Biophys J. 2009;97:1671–1677. doi: 10.1016/j.bpj.2009.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine MT, Fordyce PM, Krzysiak TC, Gilbert SP, Block SM. Individual dimers of the mitotic kinesin motor Eg5 step processively and support substantial loads in vitro. Nat Cell Biol. 2006;8:470–U89. doi: 10.1038/ncb1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine MT, Gilbert SP. To step or not to step? How biochemistry and mechanics influence processivity in Kinesin and Eg5. Curr Opin Cell Biol. 2007;19:75–81. doi: 10.1016/j.ceb.2006.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Wildenberg SM, Tao L, Kapitein LC, Schmidt CF, Scholey JM, Peterman EJ. The homotetrameric kinesin-5 KLP61F preferentially crosslinks microtubules into antiparallel orientations. Curr Biol. 2008;18:1860–4. doi: 10.1016/j.cub.2008.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waitzman JS, Larson AG, Cochran JC, Naber N, Cooke R, Jon Kull F, Pate E, Rice SE. The loop 5 element structurally and kinetically coordinates dimers of the human kinesin-5, Eg5. Biophys J. 2011;101:2760–9. doi: 10.1016/j.bpj.2011.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Gao X, Huang Y, Yao Z, Shi Q, Wu M. Nucleophosmin/B23 inhibits Eg5-mediated microtubule depolymerization by inactivating its ATPase activity. J Biol Chem. 2010;285:19060–19067. doi: 10.1074/jbc.M110.100396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinger JS, Qiu MH, Yang G, Kapoor TM. A nonmotor microtubule binding site in Kinesin-5 Is required for filament crosslinking and sliding. Current Biology. 2011;21:154–160. doi: 10.1016/j.cub.2010.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler TJ, Kececioglu JD. Multiple alignment by aligning alignments. Bioinformatics. 2007;23:i559–68. doi: 10.1093/bioinformatics/btm226. [DOI] [PubMed] [Google Scholar]
- Whitehead CM, Rattner JB. Expanding the role of HsEg5 within the mitotic and post-mitotic phases of the cell cycle. J Cell Sci. 1998;111:2551–2561. doi: 10.1242/jcs.111.17.2551. [DOI] [PubMed] [Google Scholar]
- Wickstead B, Gull K. A “holistic” kinesin phylogeny reveals new kinesin families and predicts protein functions. Mol Biol Cell. 2006;17:1734–1743. doi: 10.1091/mbc.E05-11-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickstead B, Gull K, Richards TA. Patterns of kinesin evolution reveal a complex ancestral eukaryote with a multifunctional cytoskeleton. BMC Evol Biol. 2010;10:110. doi: 10.1186/1471-2148-10-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilde A, Lizarraga SB, Zhang L, Wiese C, Gliksman NR, Walczak CE, Zheng Y. Ran stimulates spindle assembly by altering microtubule dynamics and the balance of motor activities. Nat Cell Biol. 2001;3:221–7. doi: 10.1038/35060000. [DOI] [PubMed] [Google Scholar]
- Wildenberg SMJLvd, Tao L, Kapitein LC, Schmidt CF, Scholey JM, Peterman EJG. The homotetrameric kinesin-5 KLP61F preferentially crosslinks microtubules into antiparallel orientations. Curr Biol. 2008;18:1860–1864. doi: 10.1016/j.cub.2008.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson PG. BimC motor protein KLP61F cycles between mitotic spindles and fusomes in Drosophila germ cells. Curr Biol. 1999;9:923–926. doi: 10.1016/s0960-9822(99)80400-x. [DOI] [PubMed] [Google Scholar]
- Wilson PG, Simmons R, Saighal S, Shigali S. Novel nuclear defects in KLP61F-deficient mutants in Drosophila are partially suppressed by loss of Ncd function. J Cell Sci. 2004;117:4921–4933. doi: 10.1242/jcs.01334. [DOI] [PubMed] [Google Scholar]
- Yajima J, Cross RA. A torque component in the kinesin-1 power stroke. Nat Chem Biol. 2005;1:338–41. doi: 10.1038/nchembio740. [DOI] [PubMed] [Google Scholar]
- Yajima J, Mizutani K, Nishizaka T. A torque component present in mitotic kinesin Eg5 revealed by three-dimensional tracking. Nat Struct Mol Biol. 2008;15:1119–21. doi: 10.1038/nsmb.1491. [DOI] [PubMed] [Google Scholar]
- Yan Y, Sardana V, Xu B, Homnick C, Halczenko W, Buser CA, Schaber M, Hartman GD, Huber HE, Kuo LC. Inhibition of a mitotic motor protein: where, how, and conformational consequences. J Mol Biol. 2004;335:547–54. doi: 10.1016/j.jmb.2003.10.074. [DOI] [PubMed] [Google Scholar]
- Yang G, Cameron LA, Maddox PS, Salmon ED, Danuser G. Regional variation of microtubule flux reveals microtubule organization in the metaphase meiotic spindle. J Cell Biol. 2008;182:631–639. doi: 10.1083/jcb.200801105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz A, Tomishige M, Gennerich A, Vale RD. Intramolecular strain coordinates kinesin stepping behavior along microtubules. Cell. 2008;134:1030–41. doi: 10.1016/j.cell.2008.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon SY, Choi JE, Huh JW, Hwang O, Lee HS, Hong HN, Kim D. Monastrol, a selective inhibitor of the mitotic kinesin Eg5, induces a distinctive growth profile of dendrites and axons in primary cortical neuron cultures. Cell Motility Cytoskel. 2005;60:181–190. doi: 10.1002/cm.20057. [DOI] [PubMed] [Google Scholar]
- Zhai B, Villen J, Beausoleil SA, Mintseris J, Gygi SP. Phosphoproteome analysis of Drosophila metanogaster embryos. J Proteome Research. 2008;7:1675–1682. doi: 10.1021/pr700696a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Y, Kronebusch PJ, Borisy GG. Kinetochore microtubule dynamics and the metaphase-anaphase transition. J Cell Biol. 1995;131:721–734. doi: 10.1083/jcb.131.3.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W. Exploring the intermediate states of ADP-ATP exchange: a simulation study on Eg5. J Phys Chem B. 2011;115:784–95. doi: 10.1021/jp107255t. [DOI] [PubMed] [Google Scholar]