SUMMARY
Multisubunit RNA polymerases (msRNAPs) exhibit high sequence and structural homology, especially within their active sites, which is generally thought to result in msRNAP functional conservation. However, we show that mutations in the trigger loop (TL) in the largest subunit of RNA polymerase I (Pol I) yield phenotypes unexpected from studies of Pol II. For example, a well-characterized gain-of-function mutation in Pol II results in loss-of-function in Pol I [Pol II: rpb1- E1103G; Pol I: rpa190-E1224G]. Studies of chimeric Pol II enzymes hosting Pol I or Pol III TLs suggest that consequences of mutations that alter TL dynamics are dictated by the greater enzymatic context and not solely the TL sequence. Although the rpa190-E1224G mutation diminishes polymerase function, when combined with mutations that perturb Pol I catalysis, it enhances polymerase function, similar to the analogous Pol II mutation. These results suggest that Pol I and Pol II have different rate-limiting steps.
INTRODUCTION
All cells use one or more DNA-dependent RNA polymerases to transcribe their genomes. Although the subunit composition varies between polymerases, obvious sequence and structural homology is preserved in all domains of life (Cramer, 2002). In each enzyme, the catalytic center is formed between the two largest subunits, and these subunits exhibit the highest degree of sequence homology between even highly divergent species.
Prokaryotic cells utilize a single RNA polymerase for synthesis of all RNAs; however, eukaryotes have evolved three specialized nuclear RNA polymerases: RNA polymerase I (Pol I) transcribes the ribosomal DNA, RNA polymerase II (Pol II) transcribes all protein-coding genes and most loci that encode regulatory RNAs, whereas RNA polymerase III (Pol III) primarily synthesizes transfer RNA (tRNA). Of the three nuclear polymerases, Pol II has been studied most extensively, largely due to its diverse portfolio of target genes and its intimate connection to cell differentiation and development. Importantly, the other nuclear polymerases (Pols I and III) account for the vast majority of cellular transcription (Warner, 1999) though they have many fewer transcriptional targets.
Transcription of the rDNA by Pol I accounts for more than 60% of total transcription in growing cells (Warner, 1999). The pre-rRNA is co- and post-transcriptionally processed into mature rRNA species (18S, 5.8S and 25S rRNA in budding yeast) and incorporated into ribosomes. Thus, Pol I transcription is necessarily robust, processive and tightly regulated. Cryo-EM analysis of the Pol I structure and its comparison to Pol II supports high conservation of the active center as predicted from sequence conservation (Cramer et al., 2008; Kuhn et al., 2007). Although ribosome biogenesis and thus Pol I transcription are critical to all cells, little is known about the details of Pol I catalysis.
The catalytic mechanism of transcription is thought to be very similar or identical among msRNAPs stemming from abundant sequence conservation. Structural comparisons between bacterial and archaeal RNA polymerases and Pol II from Saccharomyces cerevisiae (referred to as “yeast” herein) have identified a very high degree of structural homology, especially within the active centers. Coordinated conformational changes in two flexible domains near the active center, the bridge helix and the trigger loop (TL), are proposed to drive each round of nucleotide addition [reviewed in (Brueckner et al., 2009; Kaplan and Kornberg, 2008; Martinez-Rucobo and Cramer, 2013)]. Structural, biochemical and functional studies using both prokaryotic RNA polymerases and eukaryotic Pol II have demonstrated that the involvement of these features in catalysis is conserved across all domains of life (Tan et al., 2008; Vassylyev et al., 2007; Wang et al., 2006).
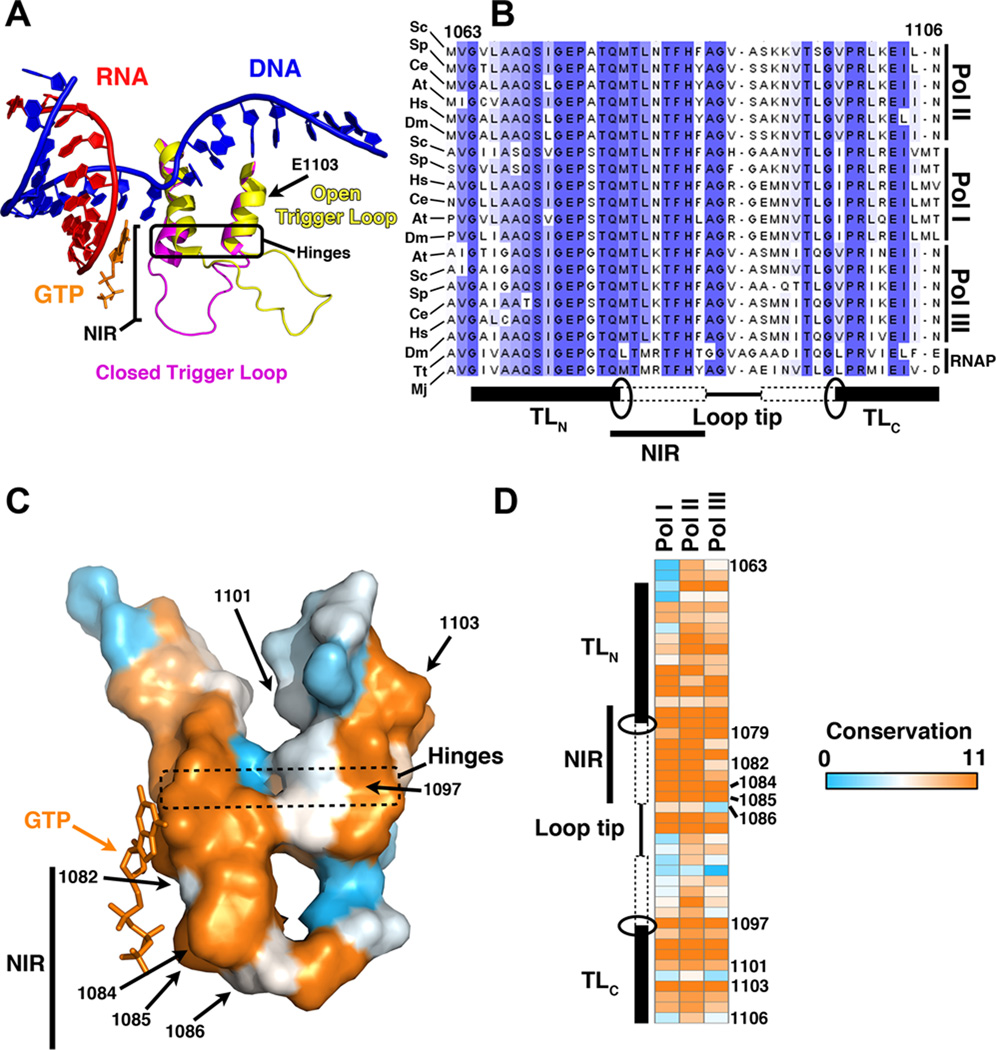
The TL is a small flexible domain in the largest subunit of msRNAPs that plays a critical role in nucleotide addition. The TL has been observed in a number of conformations from unfolded (“open”) to folded (“closed”) that are proposed to promote incorporation of the matched NTP and govern translocation [Figure 1A and (Bar-Nahum et al., 2005; Kaplan, 2010; Kaplan et al., 2008; Larson et al., 2012; Martinez-Rucobo and Cramer, 2013; Yuzenkova et al., 2010)]. Additional studies have shown that alternative intermediate conformations of the TL may contribute to pausing or arrest of elongation complexes (Nayak et al., 2013; Toulokhonov et al., 2007; Zhang et al., 2010a). The dynamic interaction between the TL and other domains within the active center is an area of intense study due to its direct implications for gene expression and regulation thereof [reviewed in (Landick, 2009)]. Although structural and biochemical studies have revealed important roles for the TL in transcription elongation, there is much to learn about the precise mechanism by which this conserved domain of the polymerase functions.
Figure 1. Overview of Pol II trigger loop and conservation among multisubunit RNA polymerases.
A) Schematic of Pol II active site showing TL in open (yellow, PDB 1Y1V (Kettenberger et al., 2004)) or closed conformation (magenta). Closed TL structure is from PDB 2E2H (Wang et al., 2006) and this structure also contained the GTP substrate (orange) shown. NIR = nucleotide interacting region. B) Multiple sequence alignment of RNA polymerase large subunits from three domains of life. Eukaryotic Pol I-III polymerase large subunits are from S. cerevisiae (Sc), Schizosaccharomyces pombe (Sp), Caenorhabditis elegans (Ce), Arabidopsis thaliana (At), Homo sapiens (Hs), and Drosophila melanogaster (Dm). Bacterial polymerase TL is from the β′ subunit of Thermus thermophilus (Tt) and archaeal TL is from the A″ from subunit of Methanocaldococcus jannaschii (Mj). Identical and similar residues are shaded. Alignment generated in MUSCLE (Edgar, 2004) and displayed in Jalview 1.0 (Waterhouse et al., 2009). Schematic below alignment indicates the positions of the N- and C-terminal TL helices (TLN and TLC), ovals represent the positions of the hinges, dashed rectangles indicate the extent of TL helices folding or stabilization upon reaching the closed conformation, and NIR is the TL region directly implicated in substrate interaction. C) Surface representation of isolated TL from closed conformation [PDB 2E2H (Wang et al., 2006)] showing structural arrangement of residues mutated in various constructs and their positions within the folded TL. Conserved residues based on model organism alignment from Figure 1B (eukaryotic subunits only) are color-coded based on conservation as determined by Jalview using a MUSCLE alignment. Most-conserved residues are orange, least-conserved are light blue (scale as in Figure 1D). D) Conservation of TL residues within Pol I, Pol II, Pol III large subunits. Large subunit alignments were generated from eukaryotic Pol large subunit CDD entries (Marchler-Bauer et al., 2013) (cd02584 - Rpb1, cd02735 - Rpa190 and cdk02736 - Rpo31) using MUSCLE and Jalview to generate conservation scores, and scores were plotted as a heat map using GENE-E (http://www.broadinstitute.org/cancer/software/GENE-E/). Most conserved residues are in orange, least conserved are in light blue. Structural figures made using Pymol v. 0.99 (The PyMOL Molecular Graphics System, Schrödinger, LLC).
Point mutations in the TL lead to a wide range of phenotypes including increased or decreased RNA polymerization rates, suppressed or enhanced pausing, enhanced or decreased forward translocation, increased backtracking and altered transcriptional fidelity (Bar-Nahum et al., 2005; Kaplan et al., 2008; Kireeva et al., 2008; Larson et al., 2012; Tan et al., 2008; Yuzenkova et al., 2010; Zhang et al., 2010a). In yeast Pol II, a large collection of mutations in and around the TL have been described (Kaplan et al., 2012; Kireeva et al., 2012). Characterization of the effects of these mutations on Pol II enzymatic properties has enhanced our understanding of Pol II transcription and models for its mechanism.
One of the best-characterized mutations affecting Pol II TL function is rpb1-E1103G. The rpb1-E1103G allele results in a “hyperactive”, or gain-of-function, phenotype (Kaplan et al., 2012; Kaplan et al., 2008; Kireeva et al., 2008; Malagon et al., 2006). The highly conserved E1103 residue is located within base helix C of the TL but distal to the active site and does not make direct contact with substrates (Wang et al., 2006). E1103G mutant polymerases have increased polymerization activity and elevated misincorporation rates. The E1103G mutation promotes active site closure by stabilization of the substrate-interacting “closed” state of the TL (Kireeva et al., 2008). Stabilization of this state is presumed to lead to the observed enhancement of catalysis and misincorporation. The TL must open to release pyrophosphate subsequent to catalysis and it is thought that TL opening may also be required for translocation (Brueckner et al., 2009; Da et al., 2012; Erie and Kennedy, 2009; Larson et al., 2012). Consistent with such a model, single molecule analyses of rpb1-E1103G Pol II show increased rate of nucleotide addition but impaired translocation (Larson et al., 2012). Therefore, E1103 substitution affects multiple steps during transcription, but since catalysis is normally limiting for Pol II, the net effect of E1103G is an increased elongation rate. Examination of the structural consequences of E1103G suggests that it disrupts an important interdomain contact leading to increased flexibility of the TL or destabilization of the open state (Kaplan et al., 2012; Kaplan et al., 2008; Kireeva et al., 2008; Kireeva et al., 2012). Molecular dynamics simulations, using the Thermus thermophilus RNAP structure, support the interpretation that mutation of this conserved acidic residue affects TL mobility (Kireeva et al., 2012). In agreement, substitutions in amino acids nearby the conserved glutamate also result in hyperactive enzymes in bacteria and archaea (Bar-Nahum et al., 2005; Tan et al., 2008; Zhang et al., 2010a).
Based on the TL’s structural and functional conservation, we anticipated functional identity (or at least similarity) between the TLs of yeast Pols I and II. To test this hypothesis we made a series of mutations in the Pol I TL and compared the phenotypes to analogous mutations in RPB1 of Pol II. From these mutations, it was clear that Pol I is less tolerant to mutation of the TL than Pol II. Of the viable mutant strains, we chose to further characterize two alleles analogous to those that resulted in gain-of-function phenotypes for Pol II. To our surprise, both mutations (rpa190-E1224G in base helix C and rpa190-F1205H in the nucleotide interacting region of the TL) reduced the elongation rate of Pol I. Using a set of chimeric alleles of rpb1 (Pol II), fusing TLs of either rpa190 (Pol I) or rpo31 (Pol III) into rpb1, we demonstrate that the divergent functions of the TL residues are not an obligatory feature of the loop sequence, but rather a consequence of the protein context of the RNA polymerase. Furthermore, we show that rpa190-E1224G suppresses phenotypic defects of other rpa190 alleles predicted to alter catalysis, suggesting that impaired transcription elongation by rpa190-E1224G is also context dependent. These data are consistent with a model in which different steps in transcription elongation are rate-limiting for Pol I compared to Pol II. These results demonstrate that assumption of strict conservation of function between msRNAPs can be problematic and that each RNA polymerase system must be carefully scrutinized in its own right.
RESULTS
Pol I is less tolerant to TL mutations than Pol II in vivo
The TL is conserved across all three domains of life (Figure 1B–1D). To characterize the degree to which TL function is conserved between RNA polymerases, we conducted a mutational analysis of the Pol I TL. We constructed eight substitutions in the TL-encoding region of RPA190. These substitutions were located in the substrate proximal, nucleotide interacting region (NIR) as well as in the hinge region of base helix C (TLC) (Figure 1C). Each mutation was analogous to a previously characterized, viable allele of RPB1. Unlike in Pol II, three of these mutations in RPA190 were lethal (superscript indicates position in Rpb1: H1206YH1085Y, N1203SN1082S and G1218DG1097D), as assessed by plasmid shuffle and tetrad analysis (Table 1 and data not shown). Of the five mutations that supported growth, three mutant strains exhibited severe growth defects relative to WT (F1205HF1084H, H1206QH1085Q and L1222SL1101S; Table 1). Both F1207SF1086S and E1224GE1103G exhibited minor growth defects compared to the WT strain under optimal conditions and manifested mild cold-sensitivity. To determine if mutations in the Pol I TL resulted in effects mechanistically similar to those observed for Pol II, we focused subsequent analyses on two mutations that were gain-of-function for Pol II: F1205H and E1224G.
Table 1.
Phenotypic comparison of point mutations in RPA190 and RPB1 TLs
| Pol II | Pol I | ||
|---|---|---|---|
| mutation | Genetic/biochemical classification |
mutation | relative growth rate (% of WT) |
| N1082S | LOF | N1203S | lethal |
| F1084H | GOF | F1205H | 63 ± 7 |
| H1085Y | LOF | H1206Y | lethal |
| H1085Q | LOF | H1206Q | 64 ± 13 |
| F1086S | LOF | F1207S | 100 ± 6; cs |
| G1097D | GOF | G1218D | lethal |
| L1101S | GOF | L1222S | 59 ± 2 |
| E1103G | GOF | E1224G | 89 ± 6; cs |
Table comparing phenotypes of the corresponding TL mutants in Pol I (this study) and Pol II (Kaplan et al., 2012). Mean relative growth rates measured in liquid YEPD media at 30°C for the Pol I mutants are averaged from at least three independent experiments each performed in duplicate, with ± SD value indicated. “cs” stands for cold-sensitivity observed at 23°C and 18°C. GOF and LOF are abbreviation for gain- and loss-of-function phenotypes of Pol II mutants as established in (Kaplan et al., 2012).
F1205H and E1224G mutations in the Pol I TL result in reduced elongation rates
If TL functions in catalysis and translocation by Pol I were similar or identical to that of the Pol II TL, we would predict that mutations in identical residues of the TL would result in similar effects on transcription. To test this hypothesis, we expressed and purified WT, A190-F1205H or A190-E1224G Pol I enzymes from yeast (see Experimental Procedures). Initially we measured enzyme activity using an in vitro multiround transcription assay (Bedwell et al., 2012; Keener et al., 1998). In principle, this assay can detect mutation-dependent effects on a variety of steps in the transcription cycle (initiation, promoter escape or elongation). We observed only minor differences in the activity of the mutant enzymes compared to WT Pol I (Figure S1). Thus, the mutant enzymes were active but not obviously hyperactive relative to WT.
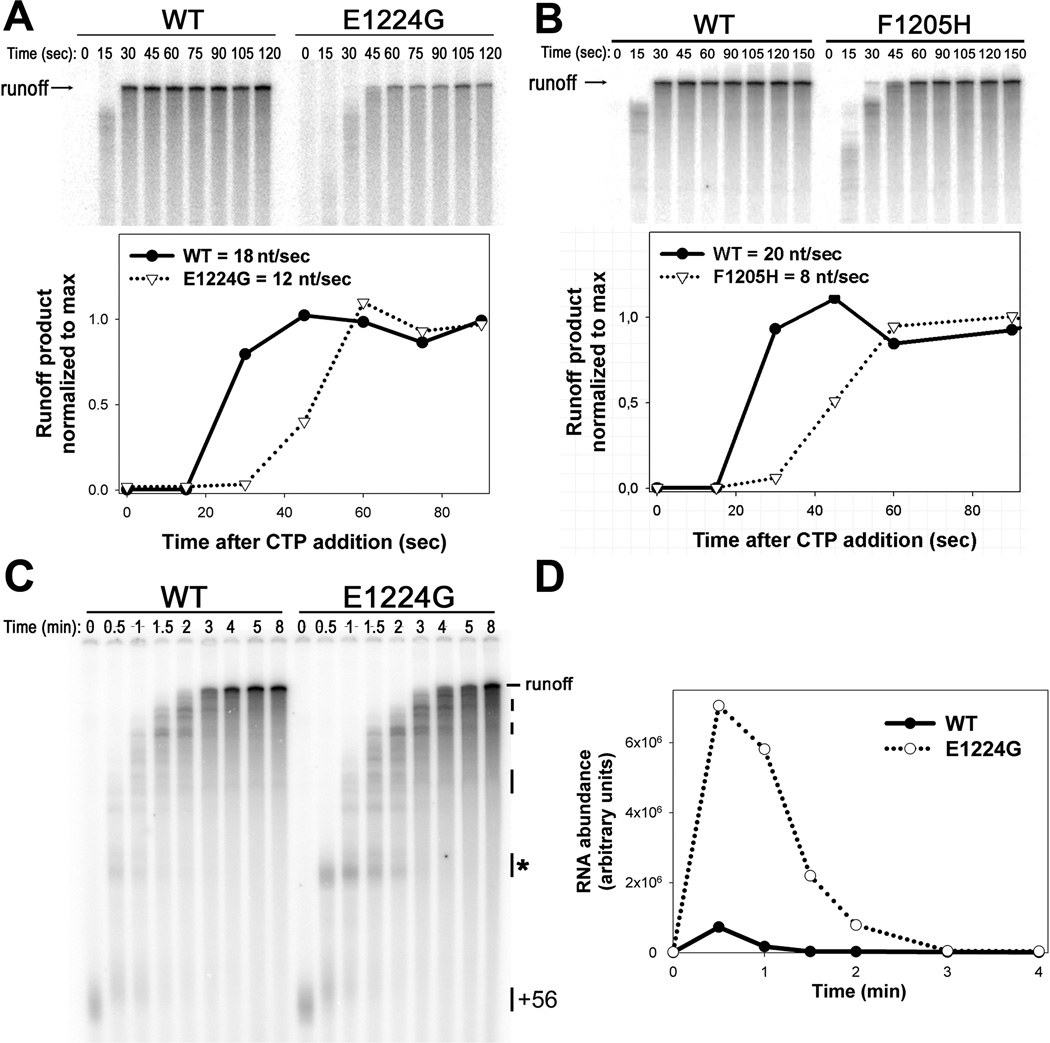
Since the in vitro multiround assay is not sensitive to modest changes in transcription elongation rate, we performed a single-round transcription elongation assay (Schneider, 2012). We determined that at 200 µM NTPs the WT elongation rate was ~18–20 nucleotides per second, whereas the elongation rates of both mutant enzymes were slower (F1205H = ~8 nt/sec and E1224G = ~12 nt/sec; Figure 2A and 2B). Neither mutation in Pol I resulted in hyperactivity, rather both mutant enzymes were impaired for transcription elongation in vitro, contrary to observations for analogous Pol II mutations.
Figure 2. The F1205H and E1224G polymerases have decreased transcription elongation rates in vitro.
A) Transcription elongation assays were performed for WT and E1224G Pol I using 200 µM ATP, UTP, CTP and 20 µM GTP (and 10 µCi α-32P GTP). The 32P-labeled transcripts were separated by gel-electrophoresis and visualized using phosphorimaging. Runoff product accumulation was quantified using ImageQuant software and plotted versus time. The transcription elongation rates for the mutant and WT polymerases were approximated from the lag time required for maximal runoff product accumulation and the length of the runoff product (745 nt). B) Transcription elongation assay for WT and F1205H Pol I enzymes performed as in panel A. C) Transcription elongation assays for WT and E1224G Pol I were performed as for panel A, but with low NTP concentrations (20 µM ATP, UTP, CTP, 2 µM GTP). The positions of the synchronized complexes at +56 (prior to CTP addition), the runoff product and the sequence-specific pauses are indicated. D) Signal intensity for one of the major pause sites (labeled with asterisk, panel C) was quantified using Image Quant software and plotted versus time. The data shown are representative experiments. Each assay was performed at least three independent times. Quantification of pause intensity from C is presented in Figure S2.
A190-E1224G polymerase demonstrates enhanced pausing in vitro
Since the rpb1-E1103G mutation is the best-characterized mutation of the Pol II TL, we extended our comparison, focusing on the analogous mutation in Pol I, rpa190-E1224G. In addition to increased overall elongation rate, the Rpb1-E1103G Pol II mutant enzyme showed decreased pausing in vitro (Kireeva et al., 2008; Malagon et al., 2006). To measure pause-tendency for the WT and mutant Pol I, we used lower NTP concentrations in order to favor pausing. We observed that both WT and mutant Pol I paused at the same sites on the rDNA template. Remarkably, the magnitude and duration of these pauses were significantly increased in the A190-E1224G mutant compared to WT (Figure 2C and 2D and Figure S2). Consistent with assays performed in high NTP concentrations, the net elongation rate of the mutant was also slower than WT when NTP concentration was limiting. Thus, unlike E1103G Pol II, the analogous mutation in Pol I increased pausing rather than decreasing it. The observation of enhanced pausing, though in contrast to Pol II data for the same mutation (Malagon et al., 2006), supports a model that the TL plays a role in pause entry and escape (Nayak et al., 2013; Toulokhonov et al., 2007; Weixlbaumer et al., 2013)
Pol I is not hyperactive in the rpa190-E1224G strain
To test whether our observations in vitro were valid in vivo, we performed a series of analyses. We observed no increase in rRNA synthesis in the E1224G strain relative to WT (Table S1), and we confirmed that any potential overexpression of rRNA was not masked by alteration of the rDNA copy number, decreased transcription initiation by Pol I or exosome-dependent decay of rRNA (Figure S3). Thus, the mutant allele does not result in hyperactive Pol I in vivo.
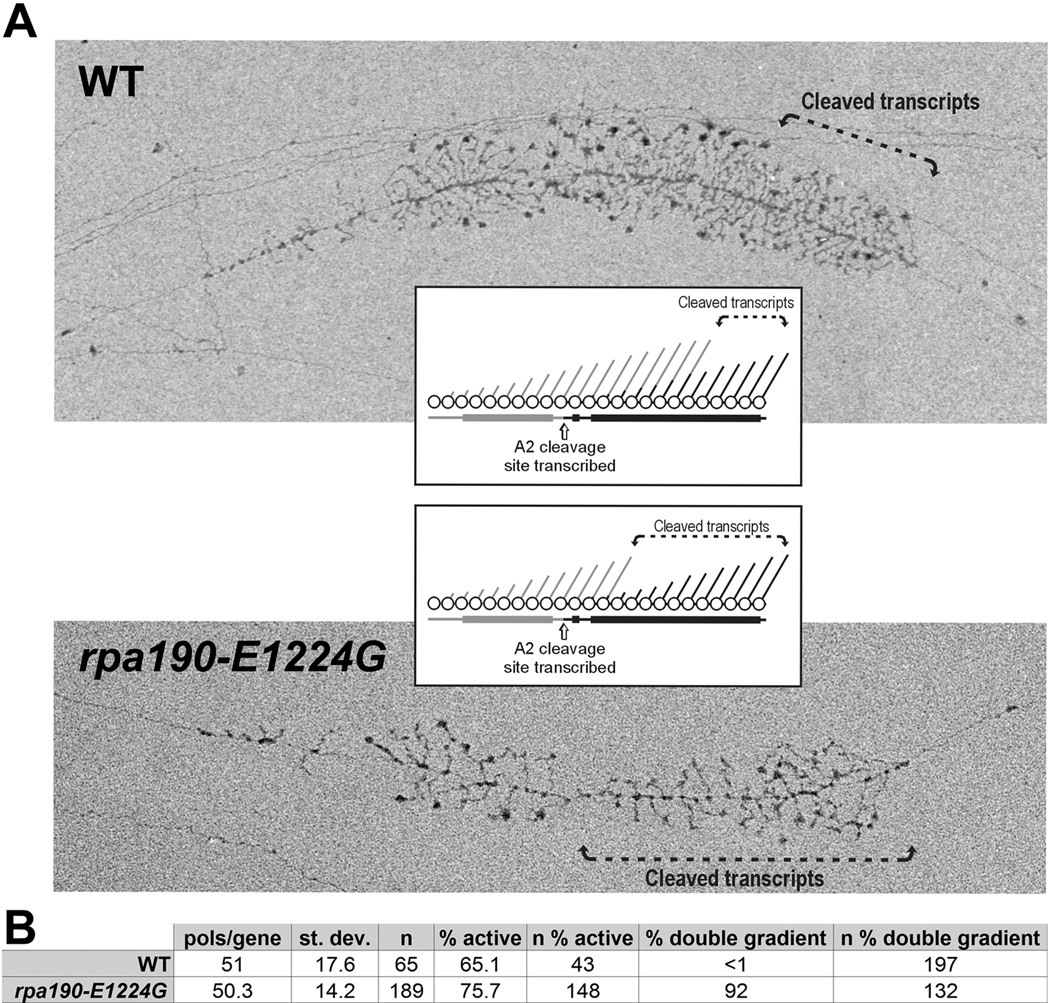
To directly assess the effects of the E1224G mutation on Pol I activity in vivo, we used singlemolecule analysis of transcription by electron microscopy of Miller chromatin spreads. From the Miller spreads, we observed no large defects in polymerase density per gene or any change in the percentage of actively transcribed genes (Figure 3). These data suggest that transcription initiation is not defective for the mutant enzyme. Since this approach results in static images, one cannot directly assess changes in transcription elongation rate; however, analysis of the individual genes and nascent transcripts yielded insights into the enzymes’ elongation properties in vivo. Multiple previous studies using Miller chromatin spreading have identified features of active rRNA genes that are characteristic of moderately impaired elongation rate [e.g. after mycophenolic acid (MPA) treatment or in topoisomerase mutants; (French et al., 2011)]. When compared to rDNA repeats in WT cells, co-transcriptional processing of the rRNA occurs on a greater percentage of nascent rRNA transcripts in the mutant and at a position in which the polymerase is located closer to the 5′-end of the gene, resulting in a distinctive ‘double gradient’ of transcript length. This appearance is interpreted to reflect RNA processing continuing at its normal rate while polymerase elongation rate is reduced. These characteristics were readily observed in the great majority of genes in rpa190-E1224G Miller spreads but were rarely observed in control genes (Figure 3). Thus, rpa190-E1224G has in vivo consequences for Pol I transcription consistent with reduced elongation rate.
Figure 3. EM analysis of Miller chromatin spreads confirms an elongation defect in rpa190-E1224G Pol I.
A) Electron micrographs showing examples of single, active rDNA genes from WT and rpa190-E1224G strains. Gene regions displaying cleaved transcripts are marked on the micrographs with arrowed brackets. The inset schematic illustrates the characteristic cotranscriptional cleavage patterns that correlate with either normal (top) or reduced transcription elongation rate by Pol I. The “double gradient” pattern (bottom schematic) was seen in 92% of 132 rDNA genes in the mutant strain, but in less than 1% of 197 WT genes. B) Multiple rDNA repeats from WT and mutant cells were analyzed and from these counts the average number of polymerases per gene and the percentage of actively transcribed genes were determined. The value “n” reports the number of genes analyzed for each analysis (and within each strain). The standard deviation observed for “pols/gene” is indicated.
Genetic interactions between rpa190-E1224G and Pol I elongation factors
As shown in Table 2, we tested the rpa190-E1224G allele for genetic interactions with mutations in several genes encoding Pol I subunits (rpa49Δ, rpa135-D784G, rpa12Δ), as well as factors involved in Pol I transcription initiation (uaf30Δ, rrn3-S213P), elongation (spt4Δ, spt5(1–797), paf1Δ) and rRNA quality control (trf4Δ, rrp6Δ). We observed synthetic lethality between rpa190-E1224G and rpa12Δ mutations. This result is particularly interesting, since neither rpa190-E1224G nor rpa12Δ single mutations affect the growth rate dramatically under permissive conditions [Table 1; (Nogi et al., 1993)]. The RPA12 gene encodes the A12.2 subunit of Pol I and is involved in polymerase assembly and intrinsic transcript cleavage (Kuhn et al., 2007; (Nogi et al., 1993). This result is analogous to synthetic lethality between rpb1-E1103G and the Pol II cleavage factor deletion dst1Δ (Malagon et al., 2006). Furthermore, we observed that the rpa190-E1224G mutation genetically interacted with three additional alleles: rpa49Δ, rpa135-D784G and paf1Δ. All three of these mutations have been shown to impair Pol I transcription elongation (Kuhn et al., 2007;) Schneider et al., 2007; Zhang et al., 2010b). If rpa190-E1224G were hyperactive, one might expect suppression of these elongation defective alleles based on behavior of rpb1-E1103G (Kaplan et al., 2012). However, we observed synergistic growth defects in the double mutants. These data support a model that E1224G substitution impairs Pol I transcription elongation and sensitizes cells to defects in other factors that promote Pol I elongation.
Table 2.
Genetic interactions between rpa190-E1224G and mutations that influence Pol I transcription suggest a role for E1224 in transcription elongation.
| Function in rRNA synthesis | allele tested | genetic interaction with rpa190-E1224G |
|---|---|---|
| Pol I subunits involved in transcription elongation | rpa12Δ | lethal |
| rpa49Δ | synergistic slow, ts | |
| rpa135-D784G | synergistic slow | |
| Pol I transcription elongation factors | paf1Δ | synergistic slow |
| spt4Δ | additive slow | |
| spt5(1–797) | additive slow | |
| Pol I transcription initiation factors | uaf30Δ | additive slow |
| rrn3-S213P | additive slow | |
| rRNA quality control | trf4Δ | additive slow |
| rrp6Δ | additive slow |
Context-dependent functions of Pol I TL residues revealed by chimeric RPB1-RPA190 alleles
To explain how identical mutations in related polymerases yield divergent outcomes, we can envision several models. These models fall into two general categories. First, subtle substitutions within the TL sequence may alter the responsiveness of the domain to mutations in conserved residues. In this model, the role of Pol II E1103 or analogous residues in the Pol I or Pol III TLs would depend on the TL sequence in which it were placed. Second, amino acids outside of the TL differentially interact with the TL, altering the functional consequence of mutations in conserved residues, such as E1103G.
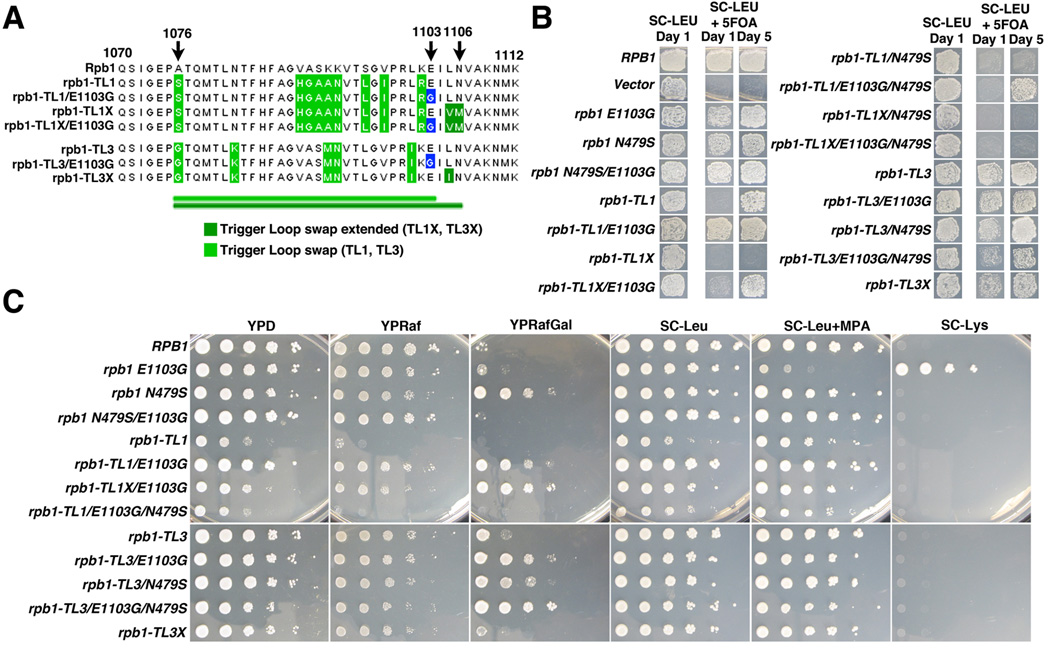
To distinguish between these models we constructed a series of rpb1 chimeric alleles, in which the TL domain from Pol II was substituted with the corresponding sequences from either Pol I or Pol III. We also included E1103G-substituted versions to measure its effect on different TL sequences in the Pol II context (Figure 4A). Expression levels of chimeric Pol II enzymes were measured for additional control (see Methods and Figure S4A–S4D). Almost all of the chimeric rpb1 alleles supported growth, confirming functional conservation of the TL among the three RNA polymerases. The only allele that did not grow was rpb1-TL1X, which carried the largest portion of the Pol I domain inserted into RPB1. Consistent with this observation, a shorter Pol I TL chimera (termed rpb1-TL1) exhibited a severe growth defect relative to the WT RPB1 allele (Figure 4B). The Pol III TL did not dramatically affect growth of the strain (Figure 4A and 4B).
Figure 4. Chimeric alleles of RPB1 support the model that sequences in the Pol I trigger loop impair Pol II function.
A) Summary of chimeric RPB1 alleles used in this study showing amino acid sequence of TL region for each. B) Plasmid shuffle results measuring complementation ability of individual rpb1 alleles. Growth in the presence of 5-FOA indicates proficiency for loss of the RPB1 URA3 plasmid and the resulting growth of chimeric alleles as the sole source of Rpb1. Cells were grown 30°C and images for 1 or 5 days growth are shown. C) 10-fold dilutions of cultured viable strains were plated on indicated growth media. Phenotypes were assessed as follows: growth on YEP with raffinose and galactose as carbon sources (YPRafGal), compared to YEP with raffinose alone (YPRaf), was scored as suppression of galactose toxicity conferred by gal10Δ56. Mycophenolic acid (MPA) sensitivity was scored by comparison of growth in SC-Leu to that on SC-Leu +MPA (20 µg/ml). To assess the Spt phenotype, cells expressing the chimeric alleles of RPB1, all of which also contained the Spt-reporter allele lys2-128δ, were plated on SC-Lys medium. Cells with the Spt- phenotype will grow in the absence of lysine, whereas Spt+ cells (wild type phenotype) will not. Time of growth on each medium were generally based on time required for reasonably sized individual wild type colonies to be observed (for media probing growth defects) or for suppressive phenotypes to be discerned (for media where wild type cells do not grow). See also Table S3 and Figure S4.
Growth defects caused by the rpb1-TL1 allele were partially rescued by E1103G substitution. We additionally observed suppression of rpb1-TL1X lethality by E1103G substitution (Figure 4B). These results suggest that defects in rpb1-TL1 and E1103G counteract each other, similar to native rpb1 gain-of-function (like E1103G) and loss-of-function alleles within the Pol II TL (Kaplan et al., 2012). To demonstrate that the E1103G mutation functions as a gain-of-function allele, irrespective of the TL sequence, it was important to test whether the TL1 chimeras behaved as gain-of-function or loss-of-function mutations in Pol II.
We used several established in vivo assays to determine whether the chimeric constructs induced loss- or gain-of-function phenotypes in Pol II. We found that rpb1-TL1 indeed conferred loss-of-function phenotypes, and these were partially suppressed in rpb1-TL1 E1103G, consistent with suppression of growth defects noted above (Figure 4C). Previously, we have shown that Pol II TL loss-of-function alleles manifest WT phenotypes for Spt-reporter assays (Spt+) and relative resistance to MPA (MPAr), but confer suppression of the gal10Δ56 mutation (e.g., rpb1-N479S in Figure 4C)(Kaplan et al., 2012; Kaplan et al., 2008). Opposite phenotypes (strong Spt−, MPAs but no strong suppression of gal10Δ56 outside of rpb1-G1097D) are common to known Pol II gain-of-function mutants, including the hyperactive rpb1-E1103G strain [Figure 4C and (Kaplan et al., 2012; Kaplan et al., 2008)]. We observed that the rpb1-TL1 chimeric mutant manifested phenotypes similar to known Pol II loss-of function TL alleles (strong suppression of gal10Δ56, Spt+, and MPAr, Figure 4C). Thus, rpb1-TL1 impairs Pol II transcription in vivo; however, the growth defects of rpb1-TL1 were rescued by the E1103G substitution, suggesting that this mutation acts as a gain-of-function in the chimera irrespective of TL sequence.
To further test this conclusion, we examined genetic interactions with a known Pol II loss-of-function allele, rpb1-N479S, to probe rpb1-TL1 and rpb1-TL1/E1103G phenotypes. We observed lethality for the combination of rpb1-N479S with rpb1-TL1. Furthermore, the genetic suppression that E1103G conferred on rpb1-TL1 was lost in combination with rpb1-N479S (Figure 4B and 4C). All of these data are consistent with E1103G acting as a gain-of-function mutation in Pol I/Pol II chimeric alleles, just as it does in a fully Pol II context.
We repeated these assays using constructs that carried Pol III/Pol II chimeric alleles (Figure 4A). We observed little, if any, effect of the Pol III trigger loop on growth phenotype on rich medium, though there is a slight growth defect in our plasmid shuffle assay (Figure 4B). E1103G did not confer strong phenotypes to rpb1-TL3 except increased suppression of gal10Δ56. We note that combination of N479S and E1103G in the rpb1-TL3 context showed a mild enhancement of growth defect and of gal10Δ56 suppression. This result is in contrast to the behavior of E1103G in RPB1 or rpb1-TL1/TL1X backgrounds where they counteract/suppress each other, underscoring the conclusion that the TL makes multiple complex interactions within and outside of the domain. The results gathered from the chimeric rpb1 alleles are summarized in supplemental Table S3. Altogether, these data suggest that the influence of amino acid substitutions on trigger loop function is context dependent and not strictly reliant on the sequence of the TL itself.
One likely site of interaction that could directly influence TL mobility is the bridge helix (BH). The TL is directly adjacent to BH residues and the mobility of both domains has been proposed to be critical for the RNA polymerase nucleotide addition cycle. To test this hypothesis, we constructed chimeric alleles of RPB1 that carried both the Pol I TL and BH. First, we found that Pol II could functionally host Pol I bridge helix constructs (Figure S4E–S4G). Phenotypic analysis suggested that rpb1-BH1 constructs might be loss-of-function for Pol II. However, rather than suppressing growth defects of rpb1-TL1 constructs, rpb1-BH1 domains exacerbated growth defects of rpb1-TL1 constructs, consistent with their single mutant phenotypes, but inconsistent with mutual suppression as might be expected if each Pol I domain increased compatibility for the other within the Pol II context. Thus, the bridge helix does not appear to be the sole mediator of the observed incompatibility of the Pol I TL in Pol II.
Intra-molecular genetic interactions within Pol I TL convert E1224G from a loss-of-function to a gain-of-function mutation
The results above suggested that Pol I TL function is influenced by its enzymatic context and not solely its internal sequence. Furthermore, the data are consistent with the model that a glycine at position 1103/1224 likely has similar effects on TL dynamics regardless of TL sequence. These findings led to the hypothesis that the E1224G mutation in Pol I affects the TL just as the E1103G mutation affects Pol II, but with functionally distinct outcomes. We propose these different outcomes reflect different rate-limiting steps for Pol I and Pol II. It was shown previously that the E1103G mutation improves nucleotide addition by Pol II, but impairs the translocation step in the nucleotide addition cycle (Larson et al., 2012). Since nucleotide addition is apparently rate-limiting for Pol II, rpb1-E1103G results in a gain-offunction phenotype. If the rate-limiting step for Pol I transcription elongation were translocation rather than catalysis, one would expect the rpa190-E1224G mutation to impair transcription elongation. To test this hypothesis, we constructed a series of intra-molecular rpa190 double mutants to assess rpa190-E1224G context-specific behavior in Pol I.
Our analysis is based on the observations that Pol II E1103G substitution shows extensive intra-molecular genetic relationships with substitutions in other Pol II residues, both inside and outside of the TL. These relationships support a general model that increased TL dynamics can compensate for loss-of-function mutations, while exacerbating other hyperactive mutations (Kaplan et al., 2012). We combined several of our originally selected substitutions (Table 1) with E1224G to potentially gain insight into their effects on Pol I transcription. When combined with mutations that are expected to impair catalysis (N1203SN1082S, H1206QH1085Q, F1207SF1086S), the E1224G mutation in Pol I rescued growth defects (Table 3), resulting in a gain-of-function rather than loss-of-function. Also in agreement with Pol II data, combination of the E1224G mutation with another mutation in the hinge (L1222SL1101S) was lethal. These data are consistent with the hypothesis that the primary effects of the rpa190-E1224G mutation on TL motion are similar between Pols I and II; however, the steps in transcription that are rate-limiting may differ between the two enzymes leading to distinct transcriptional outcomes.
Table 3.
Intramolecular genetic interactions with rpa190-E1224G
| rpa190 alleles | % of WT growth rate +/− 1 SD |
observed genetic interactions |
|---|---|---|
| N1203S | inviable | Double mutant is viable, E1224G rescues lethality of N1203S |
| N1203S/E1224G | 64 ± 7 | |
| H1206Q | 64 ± 13 | E1224G rescues growth rate of H1206Q |
| H1206Q/E1224G | 85 ± 3 | |
| F1207S | 100 ± 6 | E1224G does not significantly affect growth of F1207S at 30°C; it rescues cold-sensitivity of F1207S but leads to ts-phenotype of the double mutant (at 37°C) |
| F1207S/E1224G | 88 ± 6 | |
| L1222S | 59 ± 2 | Double mutant lethality |
| L1222S/E1224G | inviable |
DISCUSSION
TL function is not identical between Pol I and Pol II
Most studies of cellular RNA polymerases have focused on Pol II and bacterial RNAP. In this study, we constructed a collection of mutations in the TL region of the largest subunit of Pol I that had been previously characterized in analogous residues in the TL region of Pol II. Using both biochemical and simple phenotypic analyses, we observed substantial differences in the effects of the mutations in Pol I vs. Pol II. To learn more about the root of these observed differences, we focused our analysis on comparisons with the well-characterized, hyperactive rpb1-E1103G mutation in Pol II. In contrast to expectations, the analogous rpa190-E1224G mutation in Pol I led to impaired transcription elongation and loss-of-function.
There is a general assumption that functional data measured in one RNA polymerase system are applicable to most or all other related RNA polymerases. This expectation initially took hold when seminal studies of sequence conservation between divergent RNA polymerases were published (Allison et al., 1985). Based on sequence and subsequent structural data, it has become widely accepted that functional studies of polymerase catalysis can be broadly applied [e.g. (Jennebach et al., 2012; Maoileidigh et al., 2011)]. Our genetic and biochemical data indicate that even minor differences in local protein sequence or structure within RNA polymerase active centers can lead to substantially different functional consequences for otherwise identical perturbations, and we propose that these differences extend to the rate-limiting steps in Pol I and Pol II transcription.
Potential models to explain the different effects of mutations in the trigger loop
Our results are consistent with the model that the TLs in Pol I and Pol II (and Pol III) have dynamics similarly influenced by E1103G and analogous residues, but minor differences in the rates of individual steps in the nucleotide addition cycle by the enzymes render each enzyme differentially sensitive to the mutation. Critically, rpb1-E1103G has been shown to alter both catalysis and translocation, with its net increase in transcription rate coming from effects on catalysis (Larson et al., 2012). Despite an increased transcription rate, E1103G is defective for translocation. This result has been explained by E1103G stabilizing the active site in the closed conformation, as inferred biochemically (Kireeva et al., 2008). The closed conformation should promote catalysis but has also been proposed to inhibit translocation (Feig and Burton, 2010; Larson et al., 2012). According to this interpretation, the E1224G substitution in Pol I would alter Pol I TL dynamics, just as E1103G does in Pol II; but if translocation were more limiting for Pol I than catalysis, the end result would be a slower enzyme. According to this model, any positive effects of rpa190-E1224G on catalysis would be outweighed by negative effects on translocation.
Upon closure of the TL, an almost identical sequence of nucleotide-interacting residues should be positioned for interactions with substrates for Pol I or Pol II. Differences in behavior of identical substitutions between the enzymes thus are quite perplexing. For example, the conserved phenylalanine in the nucleotide-interacting region of the trigger loop (position 1205 in A190; 1084 in Rpb1) would occupy the catalytic center when the trigger loop is closed. We observed opposite effects of mutations at this position in Pol I versus Pol II. It has been shown previously that position 1084 is sensitive to conditional epistasis in Pol II, as double mutant analysis indicated that gain-of-function characteristics of a similar substitution (F1084I) are not maintained in certain double mutant configurations (Kaplan et al., 2012). Residues proximal to F1084 control its gain-of-function or loss-of-function behavior. Consistent with the epistasis studies performed within Pol II, it is likely that minor differences in conformation between the active sites of Pol I and Pol II naturally create distinct enzymatic features that facilitate evolution of specialized metabolic roles.
Unique enzymatic features suit distinct cellular roles
Why would divergent enzymatic features evolve in closely related enzymes? The answer to this question may lie in the unique cellular roles for the RNA polymerases. Unlike most genes transcribed by Pol II, the transcription initiation rate is very high at the rDNA promoter. As a result, the space between transcribing Pol I complexes is small (e.g. Figure 3). This high polymerase density at the rDNA has several important consequences, two of which are highlighted by the mutational analyses performed here.
First, even transient perturbation of transcription elongation could induce potentially catastrophic “traffic jams” on the rDNA. Perhaps Pol I has evolved efficient chemical properties to avoid such catastrophic events. Consistent with this hypothesis, the rpa190-F1205H mutation was dominant (data not shown) and caused a much greater growth defect than the rpa190-E1224G allele (Table 1). We previously characterized a mutation in RPA135 that also reduced transcription elongation rate by impairing nucleotide addition, and like rpa190-F1205H, rpa135(D784G) mutant growth was poor (Schneider, et al., 2007). Almost exactly like the rpa135 mutant, rpa190-F1205H mutants exhibit a series of defects both in transcription and rRNA processing (Figure 2 and data not shown).
Second, Gadal and colleagues proposed that high polymerase density favors Pol I transcription elongation [in essence, trailing Pol I complexes might “push” the neighboring polymerase (Albert et al., 2011)]. Trailing polymerases have been observed to promote elongation by leading polymerases in a number of contexts (Epshtein et al., 2003; Saeki and Svejstrup, 2009). Based on these studies, polymerase clustering would hypothetically suppress defects in Pol I translocation in vivo, explaining the observed in vivo tolerance of E1224G.
Misincorporation by RNA polymerase I
Another well-characterized effect of the rpb1-E1103G mutation involves transcriptional fidelity. The E1103G substitution in Rpb1 increases the rate of incorporation of non-cognate substrates in vivo and in vitro (Kaplan et al., 2008; Kireeva et al., 2008). In vitro, we detected no misincorporation with WT or E1224G mutant Pol I under a variety of experimental conditions. Fidelity mechanisms of RNAPs include substrate selectivity and removal of the misincorporated NTP during proofreading (Sydow and Cramer, 2009). The Cramer lab showed that Pol I has strong intrinsic cleavage activity, unlike Pol II (Kuhn et al., 2007). We suspect that proofreading by Pol I is too efficient to permit detection of misincorporation in vitro. This interpretation is consistent with previous studies using RNA polymerase III, which also possesses intrinsic cleavage capabilities and failed to misincorporate in vitro (Alic et al., 2007).
The A12 subunit is the Pol I homologue of TFIIS (the gene product of DST1). Cotranscriptional cleavage of the nascent transcript during proofreading requires the A12 subunit of Pol I (Kuhn et al., 2007). We observed a synthetic lethal interaction between rpa190-E1224G and rpa12Δ. This interaction is consistent with the observation that rpb1-E1103G is lethal when combined with dst1Δ. TFIIS is known to influence fidelity of transcription by Pol II (Koyama et al., 2007), thus elevated misincorporation by the rpb1-E1103G polymerase might render DST1 essential. A similar model might be possible for Pol I, if the rpa190-E1224G mutation results in increased misincorporation in vivo. However, unlike the E1103G mutation in Pol II, the rpa190-E1224G enzyme is more prone to pausing. If these pauses result in a significant amount of backtracking, A12 would be required for clearance of the resulting arrested complexes, therefore genetically analogous observations (E1103G/E1224G-cleavage factor genetic interactions) could have distinct underlying molecular mechanisms. One or both of these models could explain the lethality observed for the rpa190-E1224G rpa12Δ double mutant.
Growing list of differences between Pol I and Pol II
Despite the high degree of similarity between nuclear RNA polymerases, there is a growing list of functional differences between Pols I and II. Recent evidence has shown that three peripheral subunits of Pol I (A12.2, A34 and A49) are structural equivalents to trans-acting factors for Pol II (TFIIS, TFIIE and TFIIF) (Geiger et al., 2010; Jennebach et al., 2012; Kuhn et al., 2007). Early studies showed that the nuclear polymerases exhibited differential sensitivities to buffer conditions or inhibitors (Roeder and Rutter, 1969). However, most of the highlighted differences between the polymerases were attributed to peripheral subunits or trans-acting factors, whereas the core enzymes were considered to be less diverged (Cramer et al., 2008).
Our study suggests that the list of differences between Pol I and Pol II can now extend into the most conserved domains of the enzymes − the active center. Despite high sequence identity, corresponding mutations in the TL have different consequences for the activity of related enzymes. The data presented here are consistent with the model that different steps in transcription elongation may be rate-limiting for RNA polymerases I and II. It is likely that the functional differences that exist between the nuclear RNA polymerases are critical for their unique, specialized cellular roles. Continued characterization of these differences will lead to a better understanding of eukaryotic RNA metabolism and its regulation.
EXPERIMENTAL PROCEDURES
Yeast strains, plasmids, media and growth conditions
Yeast strains and plasmids used in the study are listed in Supplemental Tables S4–S5. Standard techniques were used for growth and manipulation of yeast (Longtine et al., 1998; Sherman et al., 1986). Media used for analysis of TL swap variants were described previously (Kaplan et al., 2012).
Phenotypic Analysis of the Pol I TL mutants
Viability of the rpa190 TL mutants was assessed by recovery of the segregants in tetrad dissection of the diploid RPA190/rpa190Δ::HIS3Mx6 strain bearing respective rpa190 alleles on a centromeric plasmid. Additionally, survival for all of the rpa190 mutants, except E1224G and F1205H, was confirmed by 5-fluoroorotic acid resistance in the standard “plasmid shuffling” technique (Sikorski and Boeke, 1991). The growth rates of the viable mutants were calculated using the GrowthCurve software (N. Rovinskiy, University of Alabama at Birmingham). Phenotypes were carefully analyzed under various conditions: 30°C, 23°C, 18°C, 37°C. The rpa190-E1224G and rpa190-F1205H mutants were additionally tested for survival on 6-azauracil (250 µg/ml) containing media (SD –Ura). No sensitivity under those conditions was observed unless specified in the text or figures.
Pol I purification
Either RPA190 or rpa190 mutant alleles were expressed from a low copy CEN plasmid (1–2 copies/cell, pRS315 derivative) in a strain carrying a chromosomal deletion of RPA190 and a triple hemaggluttinin (HA)3-hexahistidine (His)6 C-terminal epitope tag on the A135 subunit. Expression level of the mutant alleles and assembly into stable A135/A190 subcomplexes were verified by immunoprecipitation and Western blot (not shown). The cells were grown in 25 L of YEPD until their growth rate started to slow and harvested immediately. The WT or mutant polymerase was purified using a three step purification protocol (Ni-chelate, heparin sepharose and mono-Q) as described in (Schneider, 2012).
In vitro activity assays for Pol I elongation and pausing
The transcription elongation assay was performed as per (Schneider, 2012). Each reaction contained 10 µCi α-32P GTP and unlabeled NTPs at the concentrations indicated in the text or figure legends.
Electron Microscopy
Electron microscopy of Miller chromatin spreads was performed and analyzed as described previously (French et al., 2003).
Phenotypic Analysis of Chimeric RPO21/RPB1 Strains
Sequences encoding variants of the TL domains from RNA Pol I and III and or bridge helixes from Pol I were cloned in place of the analogous region of RPO21/RPB1 (RPO21 is official designation, for simplicity we have chosen to use RPB1 as is common practice) using in vivo gap repair or standard molecular techniques subsequent to PCR amplification. Plasmids were isolated, the sequences of the constructs were verified, and then retransformed into yeast. Plasmid shuffling permitted expression of the chimeric RPB1 alleles in a strain carrying a chromosomal deletion of RPB1. Phenotypic analyses of the resulting strains were performed exactly as described previously (Kaplan et al., 2012). To control for expression of the chimeric Pol II complexes, Rbp1 levels relative to Rpb3 were measured by Western blot (Figure S4A–S4C). Since increases in cellular levels of Rpb1 were detectable after plasmid shuffle for chimera mutants, genetic analyses were performed with overexpression of RPB1 on a 2µ (high copy) plasmid. Overexpression of Rpb1 from 2µ RPB1 was determined to be equal to or greater than all rpb1 chimera mutants (Figure S4B–S4C) while in vivo phenotypes of high copy RPB1 were much weaker or dissimilar to those of rpb1 chimeras, arguing against excess rpb1 generically being responsible for observed chimera phenotypes.
Supplementary Material
Highlights.
-
·
RNA polymerase I poorly tolerates mutations in its trigger loop
-
·
Identical mutations result in divergent phenotypes in RNA polymerases I and II
-
·
Consequences of mutations in polymerase trigger loops depend on enzyme context
-
·
RNA polymerases I and II may have different rate-limiting steps for elongation
ACKNOWLEDGEMENTS
We thank Dr. N.P. Higgins for use of his CHEF electrophoresis system. We thank Woody Robbins and Kim Hardy in the UAB Fermentation Facility for cell preparation for protein purification. We also thank Dr. Charles Turnbough for encouragement and critical discussions during the development of this study. We thank Lindsey Hall (lab of A.L.B.) for excellent technical assistance. This work was supported by the National Institute of General Medical Sciences (NIGMS) of National Institutes of Health (NIH), award numbers R01GM084946 (to D.A.S.), R01GM063952 (to A.L.B.) and R01GM097260 (to C.D.K.). Mechanistic studies on Pol II transcription in the Kaplan lab are also supported by a grant from the Welch Foundation (A-1763). The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Albert B, Léger-Silvestre I, Normand C, Ostermaier MK, Pérez-Fernández J, Panov KI, Zomerdijk JC, Schultz P, Gadal O. RNA polymerase I-specific subunits promote polymerase clustering to enhance the rRNA gene transcription cycle. J Cell Biol. 2011;192:277–293. doi: 10.1083/jcb.201006040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alic N, Ayoub N, Landrieux E, Favry E, Baudouin-Cornu P, Riva M, Carles C. Selectivity and proofreading both contribute significantly to the fidelity of RNA polymerase III transcription. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:10400–10405. doi: 10.1073/pnas.0704116104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allison LA, Moyle M, Shales M, Ingles CJ. Extensive homology among the largest subunits of eukaryotic and prokaryotic RNA polymerases. Cell. 1985;42:599–610. doi: 10.1016/0092-8674(85)90117-5. [DOI] [PubMed] [Google Scholar]
- 4.Bar-Nahum G, Epshtein V, Ruckenstein AE, Rafikov R, Mustaev A, Nudler E. A ratchet mechanism of transcription elongation and its control. Cell. 2005;120:183–193. doi: 10.1016/j.cell.2004.11.045. [DOI] [PubMed] [Google Scholar]
- 5.Bedwell GJ, Appling FD, Anderson SJ, Schneider DA. Efficient transcription by RNA polymerase I using recombinant core factor. Gene. 2012;492:94–99. doi: 10.1016/j.gene.2011.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brueckner F, Ortiz J, Cramer P. A movie of the RNA polymerase nucleotide addition cycle. Curr Opin Struct Biol. 2009;19:294–299. doi: 10.1016/j.sbi.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 7.Cramer P. Multisubunit RNA polymerases. Curr Opin Struct Biol. 2002;12:89–97. doi: 10.1016/s0959-440x(02)00294-4. [DOI] [PubMed] [Google Scholar]
- 8.Cramer P, Armache KJ, Baumli S, Benkert S, Brueckner F, Buchen C, Damsma GE, Dengl S, Geiger SR, Jasiak AJ, et al. Structure of eukaryotic RNA polymerases. Annu Rev Biophys. 2008;37:337–352. doi: 10.1146/annurev.biophys.37.032807.130008. [DOI] [PubMed] [Google Scholar]
- 9.Da LT, Wang D, Huang X. Dynamics of pyrophosphate ion release and its coupled trigger loop motion from closed to open state in RNA polymerase II. J Am Chem Soc. 2012;134:2399–2406. doi: 10.1021/ja210656k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic acids research. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Epshtein V, Toulme F, Rahmouni AR, Borukhov S, Nudler E. Transcription through the roadblocks: the role of RNA polymerase cooperation. The EMBO journal. 2003;22:4719–4727. doi: 10.1093/emboj/cdg452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Erie DA, Kennedy SR. Forks, pincers, and triggers: the tools for nucleotide incorporation and translocation in multi-subunit RNA polymerases. Curr Opin Struct Biol. 2009;19:708–714. doi: 10.1016/j.sbi.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feig M, Burton ZF. RNA polymerase II with open and closed trigger loops: active site dynamics and nucleic acid translocation. Biophysical journal. 2010;99:2577–2586. doi: 10.1016/j.bpj.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.French SL, Osheim YN, Cioci F, Nomura M, Beyer AL. In exponentially growing Saccharomyces cerevisiae cells, rRNA synthesis is determined by the summed RNA polymerase I loading rate rather than by the number of active genes. Mol Cell Biol. 2003;23:1558–1568. doi: 10.1128/MCB.23.5.1558-1568.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.French SL, Sikes ML, Hontz RD, Osheim YN, Lambert TE, ElHage A, Smith MM, Tollervey D, Smith JS, Beyer AL. Distinguishing the roles of Topoisomerases I and II in relief of transcription-induced torsional stress in yeast rRNA genes. Mol Cell Biol. 2011;31:482–494. doi: 10.1128/MCB.00589-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geiger SR, Lorenzen K, Schreieck A, Hanecker P, Kostrewa D, Heck AJ, Cramer P. RNA polymerase I contains a TFIIF-related DNA-binding subcomplex. Molecular cell. 2010;39:583–594. doi: 10.1016/j.molcel.2010.07.028. [DOI] [PubMed] [Google Scholar]
- 17.Jennebach S, Herzog F, Aebersold R, Cramer P. Crosslinking-MS analysis reveals RNA polymerase I domain architecture and basis of rRNA cleavage. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaplan CD. The architecture of RNA polymerase fidelity. BMC Biol. 2010;8:85. doi: 10.1186/1741-7007-8-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaplan CD, Jin H, Zhang IL, Belyanin A. Dissection of Pol II Trigger Loop Function and Pol II Activity-Dependent Control of Start Site Selection In Vivo. Plos Genet. 2012;8 doi: 10.1371/journal.pgen.1002627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaplan CD, Kornberg RD. A bridge to transcription by RNA polymerase. J Biol. 2008;7:39. doi: 10.1186/jbiol99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaplan CD, Larsson KM, Kornberg RD. The RNA polymerase II trigger loop functions in substrate selection and is directly targeted by alpha-amanitin. Molecular cell. 2008;30:547–556. doi: 10.1016/j.molcel.2008.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keener J, Josaitis CA, Dodd JA, Nomura M. Reconstitution of yeast RNA polymerase I transcription in vitro from purified components. TATA-binding protein is not required for basal transcription. J Biol Chem. 1998;273:33795–33802. doi: 10.1074/jbc.273.50.33795. [DOI] [PubMed] [Google Scholar]
- 23.Kettenberger H, Armache KJ, Cramer P. Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol Cell. 2004;16:955–965. doi: 10.1016/j.molcel.2004.11.040. [DOI] [PubMed] [Google Scholar]
- 24.Kireeva ML, Nedialkov YA, Cremona GH, Purtov YA, Lubkowska L, Malagon F, Burton ZF, Strathern JN, Kashlev M. Transient reversal of RNA polymerase II active site closing controls fidelity of transcription elongation. Molecular cell. 2008;30:557–566. doi: 10.1016/j.molcel.2008.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kireeva ML, Opron K, Seibold SA, Domecq C, Cukier RI, Coulombe B, Kashlev M, Burton ZF. Molecular dynamics and mutational analysis of the catalytic and translocation cycle of RNA polymerase. BMC Biophys. 2012;5:11. doi: 10.1186/2046-1682-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koyama H, Ito T, Nakanishi T, Sekimizu K. Stimulation of RNA polymerase II transcript cleavage activity contributes to maintain transcriptional fidelity in yeast. Genes Cells. 2007;12:547–559. doi: 10.1111/j.1365-2443.2007.01072.x. [DOI] [PubMed] [Google Scholar]
- 27.Kuhn CD, Geiger SR, Baumli S, Gartmann M, Gerber J, Jennebach S, Mielke T, Tschochner H, Beckmann R, Cramer P. Functional architecture of RNA polymerase I. Cell. 2007;131:1260–1272. doi: 10.1016/j.cell.2007.10.051. [DOI] [PubMed] [Google Scholar]
- 28.Landick R. Transcriptional pausing without backtracking. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:8797–8798. doi: 10.1073/pnas.0904373106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Larson MH, Zhou J, Kaplan CD, Palangat M, Kornberg RD, Landick R, Block SM. Trigger loop dynamics mediate the balance between the transcriptional fidelity and speed of RNA polymerase II. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:6555–6560. doi: 10.1073/pnas.1200939109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 31.Malagon F, Kireeva ML, Shafer BK, Lubkowska L, Kashlev M, Strathern JN. Mutations in the Saccharomyces cerevisiae RPB1 gene conferring hypersensitivity to 6-azauracil. Genetics. 2006;172:2201–2209. doi: 10.1534/genetics.105.052415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maoileidigh DO, Tadigotla VR, Nudler E, Ruckenstein AE. A unified model of transcription elongation: what have we learned from single-molecule experiments? Biophysical journal. 2011;100:1157–1166. doi: 10.1016/j.bpj.2010.12.3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marchler-Bauer A, Zheng C, Chitsaz F, Derbyshire MK, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Lanczycki CJ, et al. CDD: conserved domains and protein three-dimensional structure. Nucleic acids research. 2013;41:D348–D352. doi: 10.1093/nar/gks1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martinez-Rucobo FW, Cramer P. Structural basis of transcription elongation. Biochim Biophys Acta. 2013;1829:9–19. doi: 10.1016/j.bbagrm.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 35.Nayak D, Voss M, Windgassen T, Mooney RA, Landick R. Cys-pair reporters detect a constrained trigger loop in a paused RNA polymerase. Molecular cell. 2013;50:882–893. doi: 10.1016/j.molcel.2013.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nogi Y, Yano R, Dodd J, Carles C, Nomura M. Gene RRN4 in Saccharomyces cerevisiae encodes the A12.2 subunit of RNA polymerase I and is essential only at high temperatures. Mol Cell Biol. 1993;13:114–122. doi: 10.1128/mcb.13.1.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roeder RG, Rutter WJ. Multiple forms of DNA-dependent RNA polymerase in eukaryotic organisms. Nature. 1969;224:234–237. doi: 10.1038/224234a0. [DOI] [PubMed] [Google Scholar]
- 38.Saeki H, Svejstrup JQ. Stability, flexibility, and dynamic interactions of colliding RNA polymerase II elongation complexes. Molecular cell. 2009;35:191–205. doi: 10.1016/j.molcel.2009.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schneider DA. Quantitative analysis of transcription elongation by RNA polymerase I in vitro. Methods Mol Biol. 2012;809:579–591. doi: 10.1007/978-1-61779-376-9_37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schneider DA, Michel A, Sikes ML, Vu L, Dodd JA, Salgia S, Osheim YN, Beyer AL, Nomura M. Transcription elongation by RNA polymerase I is linked to efficient rRNA processing and ribosome assembly. Molecular cell. 2007;26:217–229. doi: 10.1016/j.molcel.2007.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sherman F, Fink GR, Hicks JB. The Laboratory Course Manual for Methods in Yeast Genetics. NY: 1986. [Google Scholar]
- 42.Sikorski RS, Boeke JD. In vitro mutagenesis and plasmid shuffling: from cloned gene to mutant yeast. Methods Enzymol. 1991;194:302–318. doi: 10.1016/0076-6879(91)94023-6. [DOI] [PubMed] [Google Scholar]
- 43.Sydow JF, Cramer P. RNA polymerase fidelity and transcriptional proofreading. Curr Opin Struct Biol. 2009;19:732–739. doi: 10.1016/j.sbi.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 44.Tan L, Wiesler S, Trzaska D, Carney HC, Weinzierl RO. Bridge helix and trigger loop perturbations generate superactive RNA polymerases. J Biol. 2008;7:40. doi: 10.1186/jbiol98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Toulokhonov I, Zhang J, Palangat M, Landick R. A central role of the RNA polymerase trigger loop in active-site rearrangement during transcriptional pausing. Molecular cell. 2007;27:406–419. doi: 10.1016/j.molcel.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 46.Vassylyev DG, Vassylyeva MN, Zhang J, Palangat M, Artsimovitch I, Landick R. Structural basis for substrate loading in bacterial RNA polymerase. Nature. 2007;448:163–168. doi: 10.1038/nature05931. [DOI] [PubMed] [Google Scholar]
- 47.Wang D, Bushnell DA, Westover KD, Kaplan CD, Kornberg RD. Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis. Cell. 2006;127:941–954. doi: 10.1016/j.cell.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- 49.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weixlbaumer A, Leon K, Landick R, Darst SA. Structural basis of transcriptional pausing in bacteria. Cell. 2013;152:431–441. doi: 10.1016/j.cell.2012.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yuzenkova Y, Bochkareva A, Tadigotla VR, Roghanian M, Zorov S, Severinov K, Zenkin N. Stepwise mechanism for transcription fidelity. BMC Biol. 2010;8 doi: 10.1186/1741-7007-8-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang J, Palangat M, Landick R. Role of the RNA polymerase trigger loop in catalysis and pausing. Nature structural & molecular biology. 2010a;17:99–104. doi: 10.1038/nsmb.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang Y, Smith ADt, Renfrow MB, Schneider DA. The RNA polymeraseassociated factor 1 complex (Paf1C) directly increases the elongation rate of RNA polymerase I and is required for efficient regulation of rRNA synthesis. J Biol Chem. 2010b;285:14152–14159. doi: 10.1074/jbc.M110.115220. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.