Abstract
Imaging of nanomaterials in biological tissues provides vital information for the development of nanotherapeutics and diagnostics. Multiplexed imaging of different nanoparticles (NPs) greatly reduces costs, the need to use multiple animals, and increases the biodistribution information that can enhance diagnostic applications and accelerate the screening of potential therapeutics. Various approaches have been developed for imaging NPs; however, the readout of existing imaging techniques relies on specific properties of the core material or surface ligands, and these techniques are limited because of the relatively small number of NPs that can be simultaneously measured in a single experiment. Here, we demonstrate the use of laser desorption/ionization mass spectrometry (LDI-MS) in an imaging format to investigate surface chemistry dictated intra-organ distribution of NPs. This new LDI-MS imaging method enables multiplexed imaging of NPs with potentially unlimited readouts and without additional labeling of the NPs. It provides the capability to detect and image attomole levels of NPs with almost no interferences from biomolecules. Using this new imaging approach we find that the intra-organ distributions of same-sized NPs are directly linked to their surface chemistry.
Various nanoparticles (NPs) with tailorable surface chemistry are widely employed in biomedical applications such as drug delivery,1 sensing,2 and therapeutics.3 For these applications NPs are functionalized with different core materials and hydrophilic monolayers to provide biocompatibility,4 biomolecular targeting5 and recognition.6 The surface chemistry of NPs plays an important role in the absorption, distribution, metabolism, excretion, and toxicity of NPs.7 For example, Arvizo et al. showed that the biodistributions and pharmacokinetics of NPs are dictated by the charged character of the monolayers on the NPs.8
The effective use of NPs requires imaging of NPs in tissues, especially in biomedical applications.9 Various approaches (such as optical, electrical, radioactive, or magnetic measurements) have been developed for imaging NPs;10 however, multiplexed applications of these strategies are limited because it is challenging to engineer the large variety of labels necessary for multiplexed imaging on NPs. Quantum dots (QDs) have been used for multiplexed optical imaging;11 however, this approach has limitations due to QD toxicity.12 Additionally, the required variation of core size to tune emission maxima affects the biological behavior3d of the QDs, a complicating factor in assessing their interactions in vivo.7b Surface-enhanced Raman scattering (SERS) from NPs is another imaging modality that has been used to achieve multiplexed imaging.13 However, for SERS it is challenging to decrease the size of the NPs (typically around 25-100 nm) enough to minimize reticuloendothelial uptake.14 Therefore, more broadly applicable techniques are needed to provide simultaneous site-specific information about multiple same-sized and differently functionalized NPs in biological systems.
Laser desorption/ionization mass spectrometry (LDI-MS) provides an advantageous means for multiplexed detection of NPs in complex media, e.g., it has been employed to quickly screen multiple NPs in cells.15 Specifically, it tracks NPs by measuring their surface ligands, which typically control their chemistry in vivo. The NP cores absorb the laser energy, which selectively ionizes attached ligands and provides highly sensitive detection of NPs. In this method, the ligands that comprise the surface monolayer act as “mass barcodes”, enabling simultaneous detection of multiple NPs with various core sizes and materials.15, 16 The cellular uptake of multiple NPs and NP stability in cells can both be quantified using this technique.15, 16b
Here, we demonstrate LDI-MS in an imaging format and use this powerful new approach to study how surface chemistry dictates NP biodistributions in vivo. Using LDI-MS in this format provides a highly sensitive and selective approach for the simultaneous imaging of different NPs varying only in surface ligand structure in tissues (Scheme 1). This “mass barcode” method minimizes the interference signals from biomolecules in tissues by selectively ionizing the ligands on NPs, and images NPs at attomole level. The value of this approach is illustrated by multiplexed imaging of gold NPs (AuNPs) in mice to allow direct observation of the different intra-organ NP biodistributions dictated by NP surface functionality.
Scheme 1.
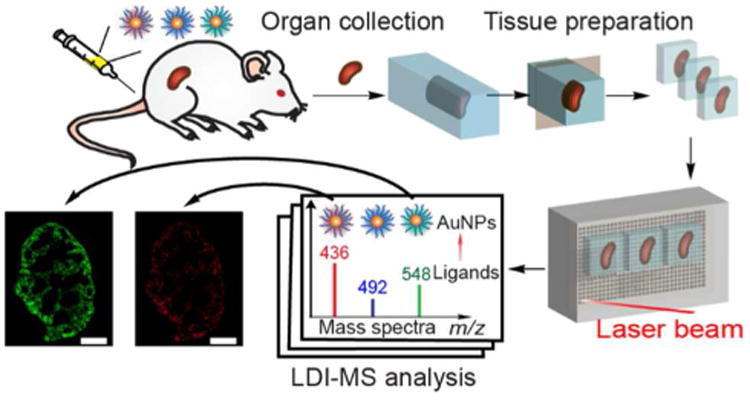
Workflow for the LDI-MS imaging strategy to obtain the biodistributions of multiple AuNPs in mouse tissues.
We chose three AuNPs for this study, focusing on gold cores due to their promise in therapeutic/imaging applications (Figure 1).1a The specific NPs chosen feature similar core diameters ((2.0 ± 0.2) nm, see Supporting Information Figure S1). The NPs were functionalized with different ligands to both alter their surface properties7b and to give the AuNPs unique mass barcodes for detection by LDI-MS16a (Figure S2). Such similar-sized NPs would be difficult or impossible to distinguish in tissues by using other methods described above.11, 13
Figure 1.
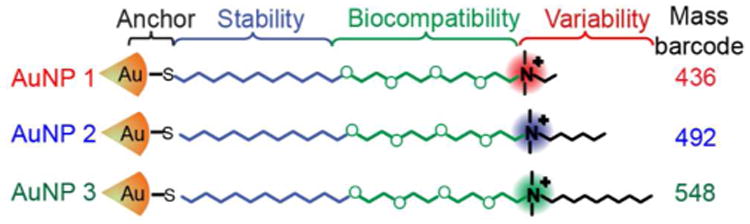
Structures of the surface monolayers on the AuNPs used in this study: the “mass barcode” is the m/z of the AuNP surface ligand.
For the imaging studies, AuNP solutions were intravenously injected into mice that were then sacrificed 24 h after NP administration. The amount of gold in major organs was quantified by using inductively coupled plasma (ICP)-MS (Figure S3), and these measurements showed that the AuNPs accumulated to the greatest degree in the liver and spleen, as expected.8 For our study, the splenic tissues were chosen because they have distinct histological regions (i.e., red pulp and white pulp)17 that allow us to compare these regions in optical images and LDI-MS images side-by-side.
Splenic tissue samples harvested from the mice were embedded, sliced, and placed on an indium tin oxide (ITO) coated glass slide for LDI-MS detection. The “mass barcodes” of injected AuNPs and ions from the gold core (m/z 197) were detected by LDI-MS (Figure 2a, b, and c), indicating the existence of the AuNPs in the tissues 24 h after intravenous administration (see Figure S4 for the detailed identification of ions). Very few ions from the multitude of biomolecules found in tissue are observed in the mass spectra due to the absence of an applied matrix, a selectivity a typical for matrix assisted laser desorption/ionization (MALDI) MS imaging.18 Because only the NP cores absorb the specific wavelength of laser light used (i.e., 355 nm), selective ionization of the ligands occurs. The selective detection of only ligands bound to the AuNPs and not free ligands or other isobaric ions was confirmed by a series of control experiments (see Figure S5 for the LDI-MS analysis of a post-hoc mixture of free ligand and mouse tissues).
Figure 2.
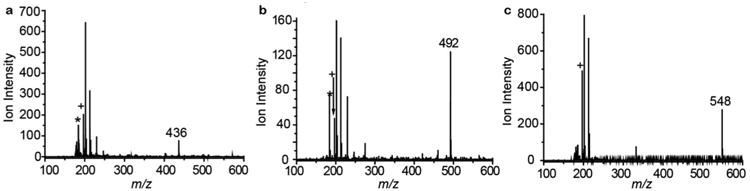
Representative LDI mass spectra of AuNPs in mouse spleens. a, AuNP 1 (molecular ion m/z 436). b, AuNP 2 (molecular ion m/z 492). c, AuNP 3 (molecular ion m/z 548). Other ions in the spectra correspond to common fragments of the phosphatidylcholine headgroup (e.g. m/z 184), one of which is indicated with an *, and Au+ (m/z 197), indicated by a +.
We next explored the imaging capability of LDI-MS. In this process, the laser beam was rastered across tissue sections, and the ion intensities of selected m/z were used to generate an ion intensity map that is indicative of AuNP distribution within the tissue. (Scheme 1 describes the process; see also Figure S6 for the LDI-MS image of AuNP 2 in the liver.) Figure 3 a, b, and c are LDI-MS images of AuNP 1, 2, and 3, respectively. These images clearly show that the functionalized AuNPs accumulate in the red pulp region of the splenic tissues, confirming that the vascular red pulp areas eliminate these AuNPs from circulation in the mouse.19 Importantly, we find attomole levels of AuNPs can be routinely detected (see Figure S7 for details), allowing use of a significantly lower dose of NPs in the in vivo experiments than is required by other imaging methods.
Figure 3.
LDI-MS images of AuNPs in mouse spleens. The biodistributions of AuNPs are shown in panel a (AuNP 1), b (AuNP 2), and c (AuNP 3). Ions selected for image generation: AuNP 1 m/z 436, AuNP 2 m/z 492, and AuNP 3 m/z 548. Scale bar represents 500 μm.
To demonstrate multiplexed imaging, three different AuNPs were concurrently injected into a tumor-bearing mouse. In this experiment, 24 h after the injection, the organs were collected, sliced, and imaged. LDI-MS images of selected tissues from a mouse injected with AuNPs 1, 2, and 3 are shown in Figure 4. The optical image illustrates the regions of the spleen (Figure 4a; see Supporting Information Figure S8 for hematoxylin and eosin staining image). Gold and surface ligand ions from each AuNP were readily detected (Figure 4b); this confirms the multiplexed imaging capability of our approach. (See Figure S9 for list of detected ions.)
Figure 4.
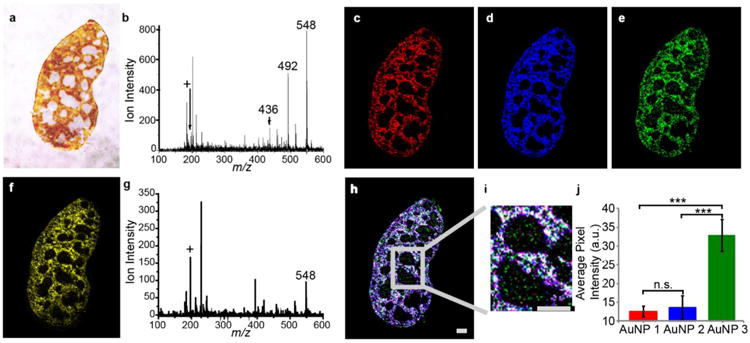
LDI-MS images of spleens from a tumor-bearing mouse injected with a mixture of AuNP 1, 2, and 3. a, Optical image of the mouse splenic tissue. b, Representative mass spectrum of the red pulp region of the spleen, indicating the existence of AuNP 1, 2, and 3. c, Representative mass spectrum of the white pulp region of the spleen that shows the detection of AuNP 3. Ion intensity images of d, gold ion (m/z 197); e, AuNP 1 (m/z 436); f, AuNP 2 (m/z 492); and g, AuNP 3 (m/z 548) are illustrated. h, The overlapped image of AuNP 1, 2, and 3, indicating the co-localization of the three AuNPs. i, Expanded image of the distribution of AuNPs in the white pulp region (scale bar represents 500 μm). j, Statistical evaluation of the surface functionality dictated bio-distribution of AuNPs in the white pulp areas (one-way ANOVA was performed, n =6, and all error bars represent standard deviation). ***, P < 0.001; n.s., P > 0.05.
Given the role of spleen in the immune system, the fact that it is enlarged when tumors are present, and its response to the surface chemistry of AuNPs,20 we closely examined the LDI-MS images of this organ. These images indicate that gold and surface ligand ions were detected mostly in the red pulp region of the spleen (Figure 4c, d, e, and f). The predominance of the AuNPs in the red pulp is due to the role of the spleen in blood filtration and clearance. Significantly, intact AuNPs can also be detected in the white pulp region of the spleen (Figure 4g and h) that contains lymphocytes that are involved in immune responses. An expanded image of one region of the white pulp shows that AuNP 3, with the most hydrophobic monolayer, accumulates to a significantly greater extent in this region than the other two AuNPs (Figure 4i). Statistical evaluation of the average mass spectrometric intensity per pixel shows that AuNP 3 accumulates to a greater extent than the other two AuNPs in the white pulp (Figure 4j; see Figure S10 for detailed statistical evaluation). When we further consider that the relative ionization efficiency of AuNP 3 is very similar to AuNP 1 and lower than AuNP 2 (Figure S11), then it becomes even clearer that the AuNP 3 is accumulated more significantly in the white pulp region. These data indicate that the intra-organ distribution of the NPs is influenced by the surface chemistry of the specific NP. This information is difficult to show by traditional techniques, highlighting the advantage of multiplexed LDI-MS imaging.
In summary, the surface chemistry dependent intra-organ distributions of NPs have been investigated by a new MS-based imaging technique. The LDI-MS imaging method enables the detection of multiple NPs at attomole levels in tissues. Because our approach uses NP surface-ligand mass as the readout, a very large number of NPs can potentially be simultaneously imaged, resulting in a significant multiplexed advantage. This robust multiplexed imaging approach introduces a new way to detect and locate engineered NPs. We have previously shown that LDI-MS can be used to analyze many different types of NPs, e.g., AuNPs, quantum dots, and magnetic NPs15, 16, so imaging of these NPs should also be possible. We can also detect a wide range of surface functionalities with LDI-MS,16a which should make this imaging technique appropriate for a broad scope of nanomaterials. One improvement that needs to be made in the method is the ability to analyze NPs having ligands with higher molecular weights (> ∼ 1000 Da). Using this new “mass barcode“ approach, we find that the intra-organ distribution of same-sized NPs is directly linked to their surface functionality, which is difficult to obtain using any other techniques. We envision that this described method will facilitate our understanding of the role of NP surface chemistry in nanomaterial-biological interactions by simultaneously comparing different NPs in a single model animal. This will minimize individual variances and thus provide insight into NP engineering, therapeutics, and diagnostics. Finally, the high sensitivity of this LDI-MS technique allows minimization of the tissue amount required for the detection, making this method a potential tool for clinical analyses of biopsies and microbiopsies.
Supplementary Material
Acknowledgments
This work was supported by NSF (CMMI-1025020) and the NIH (EB014277 and ES017871). The authors thank Prof. Jerry for assistance with the animal study and valuable discussions.
Footnotes
Author Contributions: B.Y., V.M.R., and R.W.V. conceived and designed the experiments. B.Y., S.T.K., C.S.K., K.S., D.F.M., Y.X., Y.J., A.L.R., and F.S.A. performed the experiments. B.Y. developed the imaging technique, S.T.K., A.L.R., C.S.K., and K.S. did the animal experiments, D.F.M. synthesized the NPs, B.Y., Y.X., Y.J., and F.S.A. quantified the gold amounts in tissues. All authors analyzed and discussed the data. The manuscript was prepared by B.Y., R.W.V., V.M.R., C.S.K, S.T.K., and K.S. All authors have given approval to the final version of the manuscript.
Additional experimental details and results. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.(a) Rana S, Bajaj A, Mout R, Rotello VM. Adv Drug Deliver Rev. 2012;64:200. doi: 10.1016/j.addr.2011.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Zhang J, Yuan ZF, Wang Y, Chen WH, Luo GF, Cheng SX, Zhuo RX, Zhang XZ. J Am Chem Soc. 2013;135:5068. doi: 10.1021/ja312004m. [DOI] [PubMed] [Google Scholar]
- 2.Bajaj A, Miranda OR, Phillips R, Kim IB, Jerry DJ, Bunz UHF, Rotello VM. J Am Chem Soc. 2009;132:1018. doi: 10.1021/ja9061272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.(a) De M, Ghosh PS, Rotello VM. Adv Mater. 2008;20:4225. [Google Scholar]; (b) Petros RA, DeSimone JM. Nat Rev Drug Discov. 2010;9:615. doi: 10.1038/nrd2591. [DOI] [PubMed] [Google Scholar]; (c) Liu J, Yu M, Zhou C, Yang S, Ning X, Zheng J. J Am Chem Soc. 2013;135:4978. doi: 10.1021/ja401612x. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Barreto JA, O'Malley W, Kubeil M, Graham B, Stephan H, Spiccia L. Adv Mater. 2011;23:H18. doi: 10.1002/adma.201100140. [DOI] [PubMed] [Google Scholar]
- 4.Naahidi S, Jafari M, Edalat F, Raymond K, Khademhosseini A, Chen P. J Control Release. 2013;166:182. doi: 10.1016/j.jconrel.2012.12.013. [DOI] [PubMed] [Google Scholar]
- 5.Gao JH, Gu HW, Xu B. Acc Chem Res. 2009;42:1097. doi: 10.1021/ar9000026. [DOI] [PubMed] [Google Scholar]
- 6.Duncan B, Elci SG, Rotello VM. Nano Today. 2012;7:228. doi: 10.1016/j.nantod.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.(a) Zolnik BS, Sadrieh N. Adv Drug Deliver Rev. 2009;61:422. doi: 10.1016/j.addr.2009.03.006. [DOI] [PubMed] [Google Scholar]; (b) Kim ST, Saha K, Kim C, Rotello VM. Acc Chem Res. 2013;46:681. doi: 10.1021/ar3000647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arvizo RR, Miranda OR, Moyano DF, Walden CA, Giri K, Bhattacharya R, Robertson JD, Rotello VM, Reid JM, Mukherjee P. Plos One. 2011;6:e24374. doi: 10.1371/journal.pone.0024374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.(a) He X, Ma Y, Li M, Zhang P, Li Y, Zhang Z. Small. 2013;9:1482. doi: 10.1002/smll.201201502. [DOI] [PubMed] [Google Scholar]; (b) Lee DE, Koo H, Sun IC, Ryu JH, Kim K, Kwon IC. Chem Soc Rev. 2012;41:2656. doi: 10.1039/c2cs15261d. [DOI] [PubMed] [Google Scholar]
- 10.(a) Rojas S, Gispert JD, Martin R, Abad S, Menchon C, Pareto D, Victor VM, Alvaro M, Garcia H, Herance JR. ACS Nano. 2011;5:5552. doi: 10.1021/nn200986z. [DOI] [PubMed] [Google Scholar]; (b) Carregal-Romero S, Caballero-Díaz E, Beqa L, Abdelmonem AM, Ochs M, Hühn D, Suau BS, Valcarcel M, Parak WJ. Annu Rev Anal Chem. 2013;6:53. doi: 10.1146/annurev-anchem-062012-092621. [DOI] [PubMed] [Google Scholar]; (c) Liu Q, Sun Y, Yang T, Feng W, Li C, Li F. J Am Chem Soc. 133:17122. doi: 10.1021/ja207078s. [DOI] [PubMed] [Google Scholar]; (d) Tian Q, Hu J, Zhu Y, Zou R, Chen Z, Yang S, Li R, Su Q, Han Y, Liu X. J Am Chem Soc. 2013;135:8571. doi: 10.1021/ja4013497. [DOI] [PubMed] [Google Scholar]
- 11.Gao XH, Cui YY, Levenson RM, Chung LWK, Nie SM. Nat Biotechnol. 2004;22:969. doi: 10.1038/nbt994. [DOI] [PubMed] [Google Scholar]
- 12.Winnik FM, Maysinger D. Acc Chem Res. 2012;46:672. doi: 10.1021/ar3000585. [DOI] [PubMed] [Google Scholar]
- 13.Wang Y, Seebald JL, Szeto DP, Irudayaraj J. Acs Nano. 2010;4:4039. doi: 10.1021/nn100351h. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y, Hong H, Myklejord DV, Cai WB. Small. 2011;7:3261. doi: 10.1002/smll.201100597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu ZJ, Ghosh PS, Miranda OR, Vachet RW, Rotello VM. J Am Chem Soc. 2008;130:14139. doi: 10.1021/ja805392f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.(a) Yan B, Zhu ZJ, Miranda OR, Chompoosor A, Rotello VM, Vachet RW. Anal Bioanal Chem. 2010;396:1025. doi: 10.1007/s00216-009-3250-6. [DOI] [PubMed] [Google Scholar]; (b) Zhu ZJ, Yeh YC, Tang R, Yan B, Tamayo J, Vachet RW, Rotello VM. Nat Chem. 2011;3:963. doi: 10.1038/nchem.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Yan B, Jeong Y, Mercante LA, Tonga GY, Kim C, Zhu ZJ, Vachet RW, Rotello VM. Nanoscale. 2013;5:5063. doi: 10.1039/c3nr01384g. [DOI] [PubMed] [Google Scholar]; (d) Creran B, Yan B, Moyano DF, Gilbert MM, Vachet RW, Rotello VM. Chem Commun. 2012;48:4543. doi: 10.1039/c2cc30499f. [DOI] [PubMed] [Google Scholar]
- 17.Steiniger B. eLS. John Wiley & Sons, Ltd; Chichester: 2001. [Google Scholar]
- 18.Cornett DS, Reyzer ML, Chaurand P, Caprioli RM. Nat Methods. 2007;4:828. doi: 10.1038/nmeth1094. [DOI] [PubMed] [Google Scholar]
- 19.Khlebtsov N, Dykman L. Chem Soc Rev. 2011;40:1647. doi: 10.1039/c0cs00018c. [DOI] [PubMed] [Google Scholar]
- 20.Moyano DF, Goldsmith M, Solfiell DJ, Landesman-Milo D, Miranda OR, Peer D, Rotello VM. J Am Chem Soc. 2012;134:3965. doi: 10.1021/ja2108905. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



