Abstract
In the last few years, genome-wide association studies (GWASs) have identified hundreds of predisposition loci for several types of human cancers. Recent progress has been made in determining the underlying mechanisms through which different single-nucleotide polymorphisms (SNPs) affect predisposition to cancer. Although there has been much debate about the clinical utility of GWASs, less attention has been paid to how GWASs and post-GWASs functional analysis have contributed to understanding the aetiology of cancer. Most common variants associated with cancer risk are localized in non-protein-coding regions highlighting transcriptional regulation as a common theme in the mechanism of cancer predisposition. Here, we outline strategies to functionally dissect predisposition loci and discuss their limitations as well as challenges for future studies.
Keywords: cancer predisposition, eQTL, GWAS, SNPs, transcription
INTRODUCTION
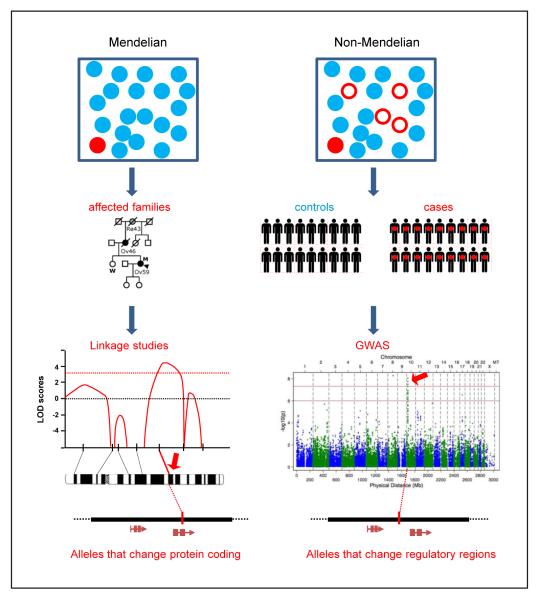
One of the central goals of human genetics is to understand the genes and pathways underlying traits. Gene mapping of disorders with a Mendelian pattern of inheritance using linkage analysis has been highly successful. Genetic variants underlying these single-gene Mendelian disorders tend to be highly penetrant – i.e. a high percentage of carriers of the genotype manifest the phenotype – and rare in the population (Fig. 1). Mapping of non-Mendelian (or complex) traits, in which variants in multiple genes contribute to the trait, had to await the sequencing of the human genome and cataloguing of human genetic variants [1–3]. In contrast to Mendelian disorders, inherited variants underlying complex diseases have modest penetrance but higher frequency in the population (Fig. 1). Over the past several years, genome-wide association studies (GWASs) of complex traits, including cancer, have successfully identified thousands of chromosomal loci associated with hundreds of traits (National Human Genome Research Institute GWASs catalogue) (Table 1) [4]. Typically, in GWASs, thousands to millions of individual single-nucleotide polymorphisms (SNPs) are genotyped in a large number of individuals with and without the trait [4]. Thus, we now have the ability to understand the underlying genetic causes of common diseases.
Fig. 1.
Different strategies to identify predisposition loci. Overall strategies to identify loci in Mendelian and in complex (non-Mendelian) diseases. In Mendelian diseases, individuals carrying high penetrance variants that confer risk (red circles) tend to express the disease phenotype (red filled circles) but are rare in the population. In complex diseases, individuals carrying common risk variants (red circles) tend to have low penetrance and many carriers will not manifest the disease phenotype (open red circles). Mendelian diseases can be investigated with family-based linkage studies and risk loci (red arrow) are positionally identified [typically a logarithm of odds (LOD) score >3; dashed red line]. Complex diseases can be investigated with (genome-wide) association studies and risk loci (red arrow) are positionally identified (typically P ≤ 5 × 10−8).
Table 1.
Web resources and databases used in post-GWAS analysis
| Web resources | Descriptiona | URL |
|---|---|---|
| National Human Genome Research Institute (NHGRI) GWAS catalogue |
Includes studies assaying ≥100,000 SNPs in the initial stage (excludes candidate gene studies). SNP–trait associations are limited to those with P- values <1.0 × 10−5 |
http://www.genome.gov/gwastudies/ |
| Genetic Associations and Mechanisms in Oncology (GAME- ON) consortium |
The overall goal of GAME-ON is to foster an interdisciplinary and collaborative approach to the translation of promising research leads deriving from the initial wave of cancer GWAS. It is limited to breast, ovarian, prostate, lung and colorectal cancers |
http://epi.grants.cancer.gov/gameon/ |
| Collaborative Oncological Gene- environment Study (COGS) |
The central focus of the project is to define individual risk of breast, ovarian and prostate cancer; i.e. to identify individuals at an increased risk of these three cancers. It also aims to identify genetic and lifestyle factors that are associated with certain tumour subtypes and affect clinical outcome |
http://www.cogseu.org/ |
| 1000 Genomes | The goal of the 1000 Genomes project is to find most genetic variants that have frequencies of at least 1% in the populations studied |
http://www.1000genomes.org/ |
| The Cancer Genome Atlas (TCGA) |
The goal is to identify the changes in the genome in each cancer and to understanding how such changes interact to drive the disease |
http://cancergenome.nih.gov/ |
| Encyclopedia of DNA Elements (ENCODE) |
The overall aim is to identify all functional elements in the human genome sequence |
http://www.genome.gov/10005107
http://genome.ucsc.edu/ENCODE/ http://www.nature.com/encode/#/threads |
| Catalogue of Somatic Mutations in Cancer (COSMIC) |
COSMIC is designed to store and display somatic mutation information and related details, and contains information relating to human cancers |
http://cancer.sanger.ac.uk/cancergenome/projects/cosmic/ |
| Missense Prediction Tool Catalogue – National Genetics Reference Laboratory (NGRL) |
The aim is to review and catalogue available tools for the evaluation and classification of missense variants |
http://www.ngrl.org.uk/Manchester/page/missense-prediction-tool-catalogue |
| University of California Santa Cruz (UCSC) Human Genome Browser |
Contains the reference sequence and working draft assemblies for a large collection of genomes. It also provides portals to the ENCODE project |
http://genome.ucsc.edu/ |
| Genotype-Tissue Expression (GTex) |
The aim is to study human gene expression and regulation in multiple tissues, providing insights into the mechanisms of gene regulation |
http://commonfund.nih.gov/GTEx/ |
| Gene Expression Omnibus (GEO) |
GEO is a public functional genomics data repository supporting MIAME (Minimum Information About a Microarray Experiment)-compliant data submissions |
http://www.ncbi.nlm.nih.gov/geo/ |
Adapted from the original descriptions provided on the websites.
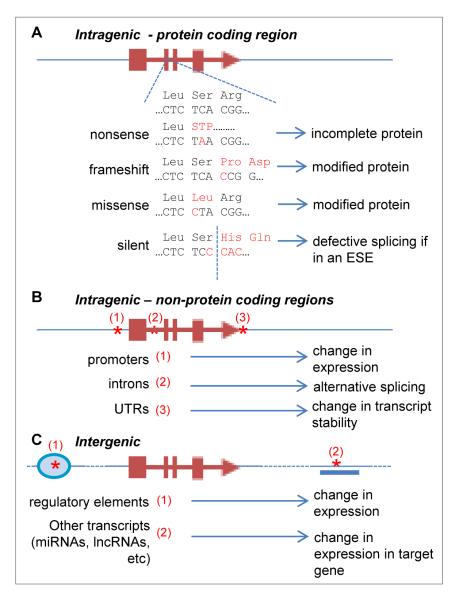
Another important distinction between Mendelian disorders and complex traits is the location of the variant in the human genome; most Mendelian variants are located in protein-coding regions whereas most variants underlying complex traits discovered to date are located outside these regions [5] (Fig. 1). This observation presents a key challenge to understanding how the variant influences the trait (Fig. 2). When a variant is located in a protein-coding sequence, its effect on the protein can, in many cases, be readily inferred from the genetic code. This is the case for frameshift and nonsense mutations. In other cases, such as missense or splice site changes, robust prediction tools have been developed [6–10]. However, when a variant is in a non-protein-coding region, there is a less developed framework to decipher whether these changes have functional impact. The lack of a defined genetic code for the non-protein-coding region of the genome makes causal allele and gene identification difficult. Despite several technical and conceptual advances in the last few years, determination of the molecular mechanisms behind the detected associations continues to be a significant challenge [11]. The aim of this review is to outline strategies to address the challenges emerging when exploring functional mechanisms linked to trait-associated SNPs.
Fig. 2.
Potential mechanisms through which SNP variation can influence a target gene. Examples of how single-nucleotide variation can influence target genes. A. In protein-coding regions, SNPs can lead to changes in protein sequence leading to truncated or modified proteins with defective function or stability. Silent variation (i.e. nucleotide variation codes for the same amino acid residue) may disrupt exonic splicing enhancers (ESEs) leading to illegitimate or inefficient splicing. B. SNPs in intragenic (but non-protein-coding) regions can modify the activity of promoters, disrupt or create splicing acceptor and donor sites, and modify transcript instability in untranslated regions (UTRs), for example by influencing polyadenylation. C. Most GWAS hits are found in intergenic regions. SNPs in these regions may affect target genes by modifying regulatory sequences such as enhancers or insulator elements. SNPs may also modify other transcripts such as microRNAs (miRNAs) or long non-coding RNAs (lncRNAs) that may regulate other target genes.
The Genetic Associations and Mechanisms in Oncology (GAME-ON) consortium (Table 1) has examined GWASs data for prostate, ovarian, breast, lung and colorectal cancers to develop systematic strategies to determine the functional contribution of SNPs and their target genes [11–20]. The main objective was to apply these strategies to data generated by genetic association studies such as the recently completed Collaborative Oncological Gene-environment Study (COGS) (Table 1) which genotyped an unprecedented number of cancer cases and controls (~200,000) for over 200,000 SNPs, chosen because they represented the top-ranked SNPs associated with cancer in prior GWASs. Here we describe our findings from analyses of cancer predisposition loci.
During the past three years the main challenges to revealing the mechanistic basis of risk became clear. Most of the SNPs implicated in cancer predisposition have so far been shown to have modest effects (as reflected by small odds ratio values) [21]. While this finding was consistent with the common disease/common variant model in which common SNPs are expected to have mild to moderate effects [22], it highlighted the limited sensitivity of molecular biology methods to investigate the effects of moderate changes in gene expression or activity in vivo. A further complication is that these effects might be spread over the lifetime of an organism or may only manifest during specific developmental stages. Moreover, some of the expected effects may be restricted to specific cell types. The latter issue is very significant for some cancers, such as ovarian carcinoma, in which the originating cell and tissue type is an area of intense investigation [23].
There are rapid advances in the field as large consortia such as the 1000 Genomes project (1KG) [24], The Cancer Genome Atlas (TCGA) [25–27], the Encyclopedia of DNA Elements (ENCODE) [28–32] and the Catalogue of Somatic Mutations (COSMIC) [33] (Table 1) have generated much data, thus periodic examination of these databases is essential. In summary, during the past 3 years, the GAME-ON consortium has made much progress in developing comprehensive strategies to analyse results from GWASs to illuminate the mechanism(s) behind the associations. Although in some cases the analytical approaches and tools might be specific to the particular cancer under study, we believe that these strategies can be utilized for different cancers (or indeed for different diseases). Below, we describe the general outline of our systematic approach and highlight information gained from as well as its current limitations and future challenges.
OUTLINE OF THE OVERALL APPROACH
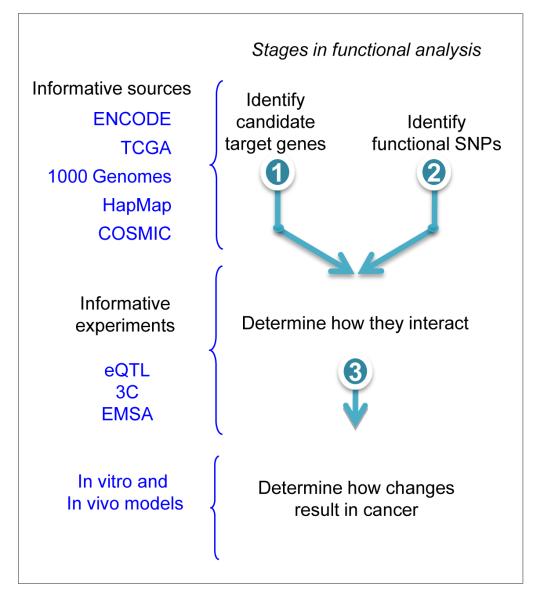
The overall approach can be divided in three stages (Fig. 3). In the first stage, the most significant SNP found in analyses or meta-analyses of GWAS is used as a starting point to examine other candidate functional SNPs in the region, and potential target genes in the region are explored in the second stage. Ideally, the first two stages should be conducted in parallel. The aim of the final stage is to uncover direct evidence for participation of the SNP/target gene pairs in the mechanism of oncogenesis in the context of an organism.
Fig. 3.
Outline of strategies for functional dissection of a predisposition locus. Stages 1 and 2 are normally conducted in parallel.
Stage 1: searching for the causal SNP(s)
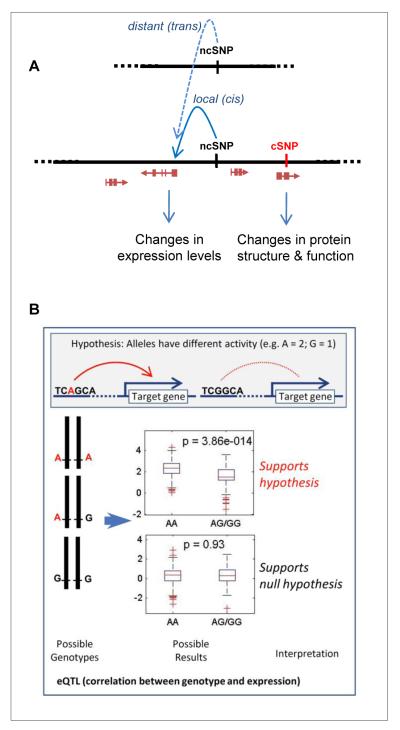
The design of most genotyping chips for GWAS is based on the principle of linkage disequilibrium (LD) in the human genome. LD is essentially the degree of correlation between a set of variants. For example, if many SNPs are in LD, then only a subset of ‘tags’ needs to be genotyped. These tags represent specific proxy markers for other correlated SNPs in a defined chromosomal region [34]. Although the use of tagging SNPs results in reproducible and robust signals and minimizes the number required for testing, it is expected that these SNPs may not necessarily constitute the causal variant, defined as the nucleotide change that results in relevant biological activity responsible for cancer predisposition. As ‘causal’ is a term that, in addition to a number of conceptual issues, requires considerable evidence [35], here we use the term ‘functional’ as the aim at this stage is to record the sources of evidence for or against the SNP having any functional impact on the activity of the locus. For example, coding SNPs might have a functional impact on the activity of the protein product, or alternatively a non-coding SNP might influence the activity of a regulatory region that controls or modulates expression of a target gene located nearby (cis) or at a distance (trans) (Fig. 4).
Fig. 4.
Coding and non-coding SNPs. A. Single-nucleotide polymorphisms (SNPs) may have different actions. Coding SNPs (cSNPs) which are located in the protein-coding region of a gene (genes are indicated by a red arrow and boxes representing different exons) can change the sequence of a protein and therefore its structure and function. Non-coding SNPs (ncSNPs) may be located in a transcriptional regulatory region and act on a target gene that is local (cis) or distant (trans). They may result in changes in protein expression levels. The distant (trans) interaction is shown simply by a single dashed line, to represent an indirect mode of interaction for which the molecular details are largely unknown. B. Expression of quantitative trait locus (eQTL) analysis is a test for association between genotype and transcript abundance. Nucleotide changes corresponding to the different SNP alleles can modify the transcriptional activity of a regulatory region (top panel). If this hypothesis is correct, one should find an association between the genotype and changes in expression levels. The alternative hypothesis, i.e. that the SNP is in LD with the true causal SNP lying in a coding region, can be tested by careful SNP annotation and detailed fine mapping of the region.
In cases in which the SNP is in a coding region, first the potential impact of the variant is assessed using robust prediction algorithms based on multiple sequence alignments [36] (for review and comparisons of prediction algorithms see [37, 38]). The National Genetics Reference Laboratory in Manchester provides a comprehensive missense prediction tool catalogue (Table 1). Of note, it seems that the performance of these tools is dependent not only on the algorithms chosen but also on sequence alignments [39] and therefore the depth of alignments to be used (e.g. human to frog, or human to yeast) should be selected with care; in some cases manual editing of the sequence alignments may be necessary. We have consistently used Polyphen 2 which combines a user-friendly interface to conduct batch queries for our analyses [40]. Structural information is also important to assess the functional impact of variants [8, 41]. Thus, if X-ray diffraction or solution (NMR) structural models are available for the protein or domain in which the variant is located, we recommend incorporating that information. If a functional change for a common risk variant is identified in cancers, and more specifically in the cancer type of interest, the SNP is than considered a good candidate for the causal SNP in the locus.
Next, whether the gene in which the variant was found has been described as a somatic change in cancer tissues can be evaluated. A search in the COSMIC database should reveal somatic mutations that might provide information, i.e. whether they may contribute to cancer [15]. We would consider the evidence to be strengthened if the mutation in the gene targets a similar functional domain as the SNP (e.g. variants in the kinase domain or in the ATP binding pocket). However, it is important to note that COSMIC records variants found in somatic tissues but in many experiments the sequence of matched normal tissue from the same individual is not determined. Thus, the presence of a variant in somatic cancer tissues might be the result of incomplete filtering of germline variation with no predicted functional impact. In addition, a validated somatic change may not contribute (‘driver’ mutation) but rather may be irrelevant (‘passenger’ mutation) to cancer development. Somatic catalogues such as COSMIC are becoming increasingly comprehensive and frequency (or recurrence in multiple tumour types) of a certain variant will also constitute important information to evaluate a candidate variant.
However, when a comprehensive analysis of GWAS hits was conducted it was shown that there is enrichment for trait-associated SNPs in non-coding regions [5]. More specifically, these SNPs tend to be found at DNase I hypersensitive sites, which indicates open chromatin and is usually correlated with regulatory regions [5]. Although such an analysis was conducted with SNPs for all traits, our findings suggest that it is also valid for cancer-related GWAS hits [11]. In fact, analysis of 1KG data showed that the upper boundary for coding variants in GWAS signals is 1/3 indicating that the majority of GWAS hits will involve non-coding variants [24, 42].
Initially, a list of all SNPs in the region with an r2 value (a measure of LD) >0.2 is generated. However, this threshold is arbitrary. We developed an automated bioinformatics tool (unpublished data) that annotates SNPs according to their location in relation to genes or previously annotated regions in the locus and their possible functional impact. Next, we determine whether these SNPs are predicted to be located in coding regions or in candidate DNA regulatory regions according to a number of DNA sequence elements, chromatin marks or other protein-binding sites (called ‘biofeatures’). It is important to note that while some regulatory regions are found in many cell and tissue types, many are cell-type specific [30]. Thus, when the SNP is located in a regulatory region found in a cell line relevant for the cancer under study we consider strong evidence. This is done by visually inspecting information generated by the ENCODE project [28–32] and currently available through its portal at the University of California, Santa Cruz Human Genome Browser (Table 1). Of importance, several tools have been developed to automate this process and integrate SNP annotation with additional data [43–45]. Because annotation databases are being constantly updated, it is advisable to schedule periodic searches during the project. At the end of this stage, a list of candidate SNPs should be available, ranked by the degree of evidence to support a functional impact.
Stage 2: indentifying candidate target genes
In parallel to the SNP annotation process, we use an arbitrary-sized window (typically 1– 2 megabases) centered on the most significant SNP, and determine the number of all transcripts (both protein coding and non-coding) within this interval. Because interactions between SNPs in regulatory regions and target genes can occur across long distances, this region does not depend on LD structure.
Analysis of expression quantitative trait loci (eQTLs) has emerged as an important method to determine the priority of candidate target gene(s) for a given non-protein-coding risk locus (Fig. 4). eQTLs are polymorphisms that are associated with transcript levels. In other words, eQTLs are variants within regulatory elements that control the abundance of transcript(s). The initial work in this field was performed in the HapMap lymphoblastoid cell lines as this provided a source of both genotypes and transcript levels [46–49]. These studies unambiguously demonstrated that RNA levels are under genetic control. Soon after these studies were reported, other research groups began applying this method to link specific risk-associated loci with transcript levels. The hypothesis is that a risk-associated SNP is a regulatory element that is correlated with mRNA levels. Transcripts that are associated with genotypic status become strong candidates for further functional testing.
eQTL analyses are typically stratified by whether the transcripts being tested are near the risk SNP (local) or at a large distance from the risk SNP (distant) (Fig. 4); analyses for local transcripts are often referred to as cis-eQTLs and for distant transcripts are referred to as trans-eQTLs. However, cis and trans have mechanistic connotations, so we prefer to use the terms local and distant, respectively. As stated above, for local eQTL analyses a window size of 1–2 megabases is usually selected and is centered on the SNP under investigation. All of the transcripts in this interval are tested for correlation with the genotypic status of the risk allele. For distant eQTL studies, all transcripts outside the window defined in the local analysis are tested. When performing distant (essentially genome-wide) studies, the statistics must be appropriately adjusted to reflect the much larger testing burden.
Due to power considerations, most of the cancer risk eQTL studies performed to date have been local. Our group has primarily evaluated prostate cancer risk loci in both normal and prostate cancer tissues. To date, we have tested 12 prostate cancer risk loci and have identified four SNPs that are strongly associated with candidate genes [50].
Recently, we made use of the multilayer data in the TCGA and ENCODE databases to perform genome-wide eQTL analyses. The breast cancer dataset was selected because it contained the greatest number of samples. Because tumours somatically acquire alterations, such as copy number and methylation changes, that are known to affect RNA expression, we first developed a method to adjust for these factors. Gene expression was modelled as having inputs from germline variants, somatic copy number changes, and promoter methylation. Next we tested 15 previously described risk variants that were strongly associated with breast cancer risk. Three risk loci and candidate target genes were implicated in a local-based analysis. A novel distant (trans) analysis was then performed, which revealed an additional three candidate genes [51].
It is important to consider the possible meanings of a negative result. A negative result may be a true negative; that is, the transcript under consideration is not influenced by the polymorphism. For most expression experiments, RNA levels are measured at one point; however, expression varies across space and time. Indeed, a key issue is which tissue to use for eQTL analyses. Although most investigators use the target tissue to perform eQTL studies, it is entirely possible that the eQTL–target gene association acts in a non-cell autonomous manner. For example, inflammatory cells, stroma and/or the microenvironment may act upon the target tissue. In addition, in cancer, tumour tissue and normal tissue can be studied. Information gained from databases with both genotype and expression data, such as TCGA and Genotype-Tissue Expression (Table 1), will contribute to understanding these issues. In addition, an eQTL–target gene relationship may only be operative during a particular developmental time point. If this is the case, animal models will be required. Lastly, eQTLs may regulate other types of transcripts, such as non-protein-coding transcripts and/or be involved in splicing.
Alternatively, a negative result may be falsely negative whereby the transcript is associated with genotypic status, but the assay results are negative. Reasons for false-negative results include power and assay sensitivity. As transcript levels are quantitative traits, platform precision and accuracy are crucial. Furthermore, similar to disease GWASs, more associations are being discovered as sample sizes are increasing.
Additional information
Positive eQTL results are considered strong evidence for further analysis because the logical link between the risk locus and the candidate gene is maintained. However, a null result does not exclude a possible association, due to the reasons discussed above. Additional information can also support the analysis and help determine the priority when two or more candidate genes are implicated by eQTL analysis. For example, using TCGA data we can extract expression information for tumours and control normal tissues and determine the genes in which expression levels are significantly changed when these two tissue types are compared. Another approach that involves more ‘hands-on’ processing is to extract similar data (tumour versus normal comparisons of expression levels) from the Gene Expression Omnibus (GEO) data repository (Table 1) [52], which provides independent datasets. Of note, GEO profiles might provide several probe sets for each gene which may not be consistent. These results should be interpreted with caution because the number of control samples available for analysis is generally very small. This analysis may identify genes that display significant changes in expression in tumour tissues. However, these results should also be interpreted carefully because the observed changes may not necessarily be linked to the risk locus and may instead be due to other confounding somatic changes occurring during the development of the tumour.
A list of genes in the locus should be available at the end of this stage, ranked by the strength of evidence obtained from different data sources. This information is then integrated with information derived from stage 1 to generate testable hypotheses for stage 3 (Fig. 3). For example, it can now be directly tested whether SNPs that have been found to coincide with chromatin contexts (suggesting the presence of a regulatory region), and are associated by eQTL analysis with a target gene that is a target of mutation in somatic tissues, indeed regulate the target gene in human cells. It is not unusual to find evidence for multiple genes in a locus being implicated in cancer. In this case, all candidates should be considered for further analyses.
Annotation-poor cancer loci
It is possible that information may not be available for the tissue relevant to the cancer in question about biofeatures that can be used to identify regulatory regions. In this case, data need to be generated to annotate the region and identify potential regulatory sites. The formaldehyde-assisted isolation of regulatory sequences (FAIRE-Seq) [53] or DNAse I hypersensitive site sequencing, DNase-Seq [54] techniques, which identify open chromatin regions, can be performed. Chromatin immunoprecipitation using several post-translational histone modification markers (e.g. histone acetylation or mono-, di-, and trimethylation) is also informative, but more costly.
A parallel functional analysis can also help in the identification of cell type-specific regulatory regions. We have developed a method to generate tiling clones (spanning 2kb per clone), by polymerase chain reaction using a bacterial artificial chromosome (BAC) containing the locus, which then undergo recombinational cloning in a vector containing a luciferase gene driven by a minimal promoter (Pharaoh et al. 2013; Nat Genet in press). These clones are then transfected into the cells of interest, and regions that show enhancer activity for the luciferase gene can be studied further. In particular, if an associated SNP is located in the tiling clone that shows activity, site-directed mutagenesis can be performed to introduce the minor allele of the SNP in question (usually the BAC contains the major allele).
Stage 3: linking functional SNP to candidate target gene
Once we have a series of candidate SNPs that are hypothesized to work by modifying the activity of a regulatory region that acts on certain genes, we can attempt to demonstrate that the region and the target gene are in physical association, which indicates the formation of an active enhancer complex. Chromosome conformation capture (3C) is used to demonstrate this link [55, 56]. Whether two linearly distant regions are in physical proximity in the nucleus can be specifically determined using the 3C technique. Ideally, the two regions being tested should be farther than 10 kb apart. At short distances, it is difficult to distinguish a true interaction from the background noise. Again, for all of these studies, choice of cell type is an important consideration.
Once the physical proximity has been determined, the next step is to identify the transcription factor that is predicted to bind to the enhancer region. This can be done by the use of algorithms that predict the binding of transcription factors given the underlying DNA sequence. However, the sequence may belong to a binding site to an unidentified transcription factor. In this case, an electrophoretic mobility assay (EMSA) may be conducted, in which the DNA sequence is used as a probe and is incubated with a nuclear extract of the cell line in question. If the cell expresses the transcription factor that binds the DNA sequence present in the extract, it is expected that migration of the the probe will be retarded. The shifted (retarded) band indicates the presence of the factor but gives no indication of its identity. If the use of a prediction algorithm generates a hypothesis of which factor is binding, the antibody that recognizes the hypothesized factor can be added to the EMSA. Finally, if the factor corresponds to the hypothesis, then the shifted band is expected to be further retarded in its migration (supershift).
LIMITATIONS
Although the strategies presented here have been successful in providing information on several predisposition loci [13–15], they have several limitations that need to be addressed in order to examine efficiently all predisposition loci identified so far.
Cell autonomous mechanisms
Most of the simplistic experimental models used to validate a target gene are based on detecting exclusively cell autonomous activities. Non-cell autonomous activities are not evaluated in most model systems used. For example if changes in the expression of a gene induce the stroma to be more permissive to the growth of a tumour from a pre-cancerous lesion, the effect of modulation by the gene in the cell of origin of the cancer will not be seen. It is important to develop and refine 3D co-culture models [57], but also to start exploring in vivo models that can highlight non-cell autonomous processes. These approaches are relatively expensive and labour intensive, therefore the ability to perform systematic analyses of a large number of candidate genes or SNPs in the near future is unlikely. However, once candidate genes have been prioritized it might be important to take the observations to an animal model.
Indirect mechanisms
An extension of the non-cell autonomous model is one in which an SNP may modify behaviour. It is conceivable that a certain SNP might regulate a gene that does not have a direct effect on cancer predisposition as a regulator of apoptosis or an oncogene. Rather this gene might influence behaviour, for example predispose to addiction or to inhale more deeply when smoking in the case of lung cancer. SNPs that contribute to different behavioural traits may increase or decrease the exposure to environmental risk factors.
Single target
The recent studies using ENCODE have revealed among other things that enhancers may regulate the expression of more than one target gene [30, 31]. The mechanism of association might be related to changes in expression in a cassette of genes. Conversely, a target gene might be influenced by more than one enhancer. This is supported by the high degree of connectivity in networks generated by co-expression [58]. Thus, models should be developed to test hypotheses in which the contribution to a certain association might be multifactorial (for example, a change in an SNP might change the activity or expression in more than one gene in the locus, or in some cases of other genes outside the locus).
CONCLUSION
A change in approach for the analysis of common variants linked to cancer predisposition was required, from family-based linkage studies to population-based association studies. A concerted effort of collaborative consortia, such as GAME-ON, has accelerated the discovery of loci implicated in cancer. Due to its smaller effects (as judged by smaller OR values), the analysis of these variants also required a change in the analytical framework used to characterize these regions [11]. Here, we presented strategies for the systematic analysis of these loci and discussed their limitations. These analyses have already revealed a common mechanistic theme; many of these variants associated with disease act through modification of transcriptional regulation. This highlights the importance of analysis of GWASs in understanding the biology of cancer. Future work is needed to concentrate on critically evaluating the strategies and data sources presented here and on refining in vitro and in vivo models to avoid the experimental problems discussed. The large number of already identified loci will be the source of investigations for many years.
ACKNOWLEDGEMENTS
The GAME-ON consortium is funded by the National Institutes of Health (NIH) post-GWASs initiative. Work in the authors’ laboratories is funded by NIH awards (1U19CA148112, 1U19CA148537 and 1U19CA148065). We thank all investigators in the GAME-ON consortium for helpful discussions. We also sincerely thank all individuals who have contributed to cancer GWASs by providing DNA, tissues and clinical information.
Footnotes
Conflict of interest statement No conflict of interest was declared.
References
- 1.Venter JC, Adams MD, Myers EW, et al. The sequence of the human genome. Science. 2001;291:1304–51. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 2.Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 3.Altshuler D, Brooks LD, Chakravarti A, Collins FS, Daly MJ, Donnelly P. A haplotype map of the human genome. Nature. 2005;437:1299–320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manolio TA. Genomewide association studies and assessment of the risk of disease. N Engl J Med. 2010;363:166–76. doi: 10.1056/NEJMra0905980. [DOI] [PubMed] [Google Scholar]
- 5.Maurano MT, Humbert R, Rynes E, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–5. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sunyaev S, Ramensky V, Koch I, Lathe W, III, Kondrashov AS, Bork P. Prediction of deleterious human alleles. Human Molecular Genetics. 2001;10:591–7. doi: 10.1093/hmg/10.6.591. [DOI] [PubMed] [Google Scholar]
- 7.Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11:863–74. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karchin R, Diekhans M, Kelly L, et al. LS-SNP: large-scale annotation of coding non-synonymous SNPs based on multiple information sources. Bioinformatics. 2005;21:2814–20. doi: 10.1093/bioinformatics/bti442. [DOI] [PubMed] [Google Scholar]
- 9.Reese MG, Eeckman FH, Kulp D, Haussler D. Improved splice site detection in Genie. J Comput Biol. 1997;4:311–23. doi: 10.1089/cmb.1997.4.311. [DOI] [PubMed] [Google Scholar]
- 10.Yeo G, Burge CB. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol. 2004;11:377–94. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- 11.Freedman ML, Monteiro AN, Gayther SA, et al. Principles for the post-GWAS functional characterization of cancer risk loci. Nat Genet. 2011;43:513–8. doi: 10.1038/ng.840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pooley KA, et al. A genome-wide association scan (GWAS) for mean telomere length within the COGS project. Nat Genetics. 2013 doi: 10.1093/hmg/ddt355. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bojesen SE, et al. Multiple independent variants at the TERT locus are associated with telomere length and risks of breast and ovarian cancer. Nat Genet. 2013 doi: 10.1038/ng.2566. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eeles R, et al. Identification of 23 new prostate cancer susceptibility loci using the iCOGS custom genotyping array. Nat Genet. 2013 doi: 10.1038/ng.2560. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pharoah P, et al. GWAS meta-analysis and replication identifies three new susceptibility loci for ovarian cancer. Nat Genet. 2013 doi: 10.1038/ng.2564. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Michailidou K, et al. Large-scale genotyping identifies more than 40 novel breast cancer susceptibility loci. Nat Genet. 2013 doi: 10.1038/ng.2563. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia-Closas M, et al. Genome-wide association studies identify four ER-negative specific breast cancer risk loci. Nat Genet. 2013 doi: 10.1038/ng.2561. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.French JD, et al. Functional Variants at the 11q13 Breast Cancer Risk Loci Regulate Cyclin D1 Expression through Long-Range Enhancers. Am J Hum Genet. 2013 doi: 10.1016/j.ajhg.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaudet MM, et al. Identification of a BRCA2-specific Modifier Locus at 6p24 Related to Breast Cancer Risk. PLoS Genet. 2013 doi: 10.1371/journal.pgen.1003173. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Couch FJ, et al. Genome-Wide Association Study in BRCA1 Mutation Carriers Identifies Novel Loci Associated with Breast and Ovarian Cancer Risk. PLoS Genet. 2013 doi: 10.1371/journal.pgen.1003212. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Varghese JS, Easton DF. Genome-wide association studies in common cancers--what have we learnt? Curr Opin Genet Dev. 2010;20:201–9. doi: 10.1016/j.gde.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 22.Reich DE, Lander ES. On the allelic spectrum of human disease. Trends in Genetics. 2001;17:502–10. doi: 10.1016/s0168-9525(01)02410-6. [DOI] [PubMed] [Google Scholar]
- 23.Berns EM, Bowtell DD. The changing view of high-grade serous ovarian cancer. Cancer Res. 2012;72:2701–4. doi: 10.1158/0008-5472.CAN-11-3911. [DOI] [PubMed] [Google Scholar]
- 24.A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–15. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hammerman PS, Hayes DN, Wilkerson MD, et al. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–25. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neph S, Vierstra J, Stergachis AB, et al. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature. 2012;489:83–90. doi: 10.1038/nature11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thurman RE, Rynes E, Humbert R, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dunham I, Kundaje A, Aldred SF, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–13. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Djebali S, Davis CA, Merkel A, et al. Landscape of transcription in human cells. Nature. 2012;489:101–8. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Forbes SA, Bindal N, Bamford S, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945–50. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gabriel SB, Schaffner SF, Nguyen H, et al. The Structure of Haplotype Blocks in the Human Genome. Science. 2002;296:2225–9. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 35.Shipley B. Cause and correlation in biology : a user’s guide to path analysis, structural equations, and causal inference. Cambridge University Press; Cambridge, UK ; New York, NY. USA: 2000. [Google Scholar]
- 36.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–81. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 37.Karchin R. Next generation tools for the annotation of human SNPs. Brief Bioinform. 2009;10:35–52. doi: 10.1093/bib/bbn047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thusberg J, Olatubosun A, Vihinen M. Performance of mutation pathogenicity prediction methods on missense variants. Hum Mutat. 2011;32:358–68. doi: 10.1002/humu.21445. [DOI] [PubMed] [Google Scholar]
- 39.Hicks S, Wheeler DA, Plon SE, Kimmel M. Prediction of missense mutation functionality depends on both the algorithm and sequence alignment employed. Hum Mutat. 2011;32:661–8. doi: 10.1002/humu.21490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karchin R, Monteiro AN, Tavtigian SV, Carvalho MA, Sali A. Functional Impact of Missense Variants in BRCA1 Predicted by Supervised Learning. PLoS ComputBiol. 2007;3:e26. doi: 10.1371/journal.pcbi.0030026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abecasis GR, Auton A, Brooks LD, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Coetzee SG, Rhie SK, Berman BP, Coetzee GA, Noushmehr H. FunciSNP: an R/bioconductor tool integrating functional non-coding data sets with genetic association studies to identify candidate regulatory SNPs. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boyle AP, Hong EL, Hariharan M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–7. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–4. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stranger BE, Forrest MS, Clark AG, et al. Genome-wide associations of gene expression variation in humans. PLoSGenet. 2005;1:e78. doi: 10.1371/journal.pgen.0010078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cheung VG, Spielman RS. The genetics of variation in gene expression. Nat Genet. 2002;32(Suppl):522–5. doi: 10.1038/ng1036. [DOI] [PubMed] [Google Scholar]
- 48.Cheung VG, Conlin LK, Weber TM, Arcaro M, Jen KY, Morley M, Spielman RS. Natural variation in human gene expression assessed in lymphoblastoid cells. Nat Genet. 2003;33:422–5. doi: 10.1038/ng1094. [DOI] [PubMed] [Google Scholar]
- 49.Schadt EE, Monks SA, Drake TA, et al. Genetics of gene expression surveyed in maize, mouse and man. Nature. 2003;422:297–302. doi: 10.1038/nature01434. [DOI] [PubMed] [Google Scholar]
- 50.Grisanzio C, Werner L, Takeda D, et al. Genetic and functional analyses implicate the NUDT11, HNF1B, and SLC22A3 genes in prostate cancer pathogenesis. Proc Natl Acad Sci U S A. 2012;109:11252–7. doi: 10.1073/pnas.1200853109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Freedman M. Cell. 2013 Jan 31; [Google Scholar]
- 52.Barrett T, Wilhite SE, Ledoux P, et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Giresi PG, Kim J, McDaniell RM, Iyer VR, Lieb JD. FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res. 2007;17:877–85. doi: 10.1101/gr.5533506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Crawford GE, Holt IE, Whittle J, et al. Genome-wide mapping of DNase hypersensitive sites using massively parallel signature sequencing (MPSS) Genome Res. 2006;16:123–31. doi: 10.1101/gr.4074106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–11. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 56.Dekker J. The three ‘C’ s of chromosome conformation capture: controls, controls, controls. Nat Methods. 2006;3:17–21. doi: 10.1038/nmeth823. [DOI] [PubMed] [Google Scholar]
- 57.Lawrenson K, Benjamin E, Turmaine M, Jacobs I, Gayther S, Dafou D. In vitro three-dimensional modelling of human ovarian surface epithelial cells. Cell Proliferation. 2009 doi: 10.1111/j.1365-2184.2009.00604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gerstein MB, Kundaje A, Hariharan M, et al. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]