Abstract
Long non-coding RNAs (lncRNAs) are a novel class of regulatory genes that play critical roles in various processes ranging from normal development to human diseases such as cancer progression. Recent studies have shown that lncRNAs regulate the gene expression by chromatin remodelling, transcription, splicing and RNA decay control, enhancer function, and epigenetic regulation. However, little is known about translation regulation by lncRNAs. We identified a translational regulatory lncRNA (treRNA) through genome-wide computational analysis. We found that treRNA is upregulated in paired clinical breast cancer primary and lymph-node metastasis samples, and that its expression stimulates tumour invasion in vitro and metastasis in vivo. Interestingly, we found that treRNA downregulates the expression of the epithelial marker E-cadherin by suppressing the translation of its mRNA. We identified a novel ribonucleoprotein (RNP) complex, consisting of RNA-binding proteins (hnRNP K, FXR1, and FXR2), PUF60 and SF3B3, that is required for this treRNA functions. Translational suppression by treRNA is dependent on the 3′UTR of the E-cadherin mRNA. Taken together, our study indicates a novel mechanism of gene regulation by lncRNAs in cancer progression.
Keywords: non-coding RNA, translation, metastasis
Introduction
RNAs have long been considered as an intermediate between DNA sequences and proteins that execute cellular functions. However, protein-coding genes represent only 2% of the human genome. Recent genome-wide analyses suggest that there are at least thousands of non-coding RNAs (ncRNAs) transcribed from mammalian genomes (Orom et al, 2010; Cabili et al, 2011). There are two major classes of ncRNAs, the small ncRNA, such as microRNAs (miRNAs) and long ncRNAs (lncRNAs), which consists of ncRNAs >200 nucleotides. Although ribosome profiling showed that lncRNAs are bound by ribosomes, the large majority of lncRNAs are not translated into peptides or proteins (Banfei et al, 2012; Guttman et al, 2013). Extensive studies have shown that small ncRNAs have diverse biological functions in development, physiological, and pathological processes, and are critical regulators in human diseases such as cancer progression (Calin and Croce, 2006; He et al, 2007; Bartel, 2009; Ebert and Sharp, 2010). Unlike the small ncRNAs, very little is known about the expression and function of lncRNAs.
lncRNAs have been shown to be involved in cellular processes such as apoptosis and cell cycle (Khalil et al, 2009; Mattick, 2009; Gupta et al, 2010; Huarte et al, 2010; Kino et al, 2010; Poliseno et al, 2010; Cesana et al, 2011; Guttman et al, 2011) and development including differentiation, X chromosome inactivation, and genomic imprinting (Mancini-Dinardo et al, 2006; Tian et al, 2010; Guttman et al, 2011). They have also been implicated in human diseases such as coronary artery disease, amyotrophic lateral sclerosis, and Alzheimer’s disease (Faghihi et al, 2008; Wang et al, 2008; Harismendy et al, 2011). In addition, lncRNAs have been shown to have oncogenic and tumour suppression functions in cancer development (Petrovics et al, 2004; Calin et al, 2007; Beltran et al, 2008; Perez et al, 2008; Li et al, 2009; Mourtada-Maarabouni et al, 2009; Benetatos et al, 2010; Gupta et al, 2010; Huarte et al, 2010). LncRNAs exert their regulatory functions through a variety of mechanisms, including chromosome remodelling, splicing, and mRNA stability (Azzalin et al, 2007; Martianov et al, 2007; Rinn et al, 2007; Faghihi et al, 2008; Nagano et al, 2008; Wang et al, 2008; Mercer et al, 2009; Ponting et al, 2009; Bernard et al, 2010; Kim et al, 2010; Liu et al, 2010; Orom et al, 2010; Schmitz et al, 2010; Tian et al, 2010; Tripathi et al, 2010; Tsai et al, 2010; Yap et al, 2010; Flynn et al, 2011; Gong and Maquat, 2011; Heo and Sung, 2011; Khalil and Rinn, 2011; Kotake et al, 2011; Maison et al, 2011; Wang and Chang, 2011; Wapinski and Chang, 2011). Although most of the steps in gene expression can be regulated by lncRNAs, little is known about translational regulation by lncRNAs.
Previously, we identified human translational regulatory lncRNA (treRNA) (originally named as ncRNA-a7) through genome-wide computational analysis, and showed that treRNA can function as an enhancer and regulate Snail transcription in cis (Orom et al, 2010). TreRNA consists of two exons and its transcript is spliced and polyadenylated. TreRNA contains three putative open reading frames (ORFs); however, ribosome profiling experiment failed to detect the treRNA association with ribosomes and the putative peptides encoded by treRNA were not detected by the ENCODE proteomic study (Guo et al, 2010; Banfei, et al, 2012; Guttman et al, 2013). In addition, in vitro translation of treRNA did not generate any detectable peptide (Orom et al, 2010). Thus, treRNA is considered as an lncRNA. Here, we report the identification of a novel RNA–protein complex that mediates the metastasis-promoting function of treRNA by translational regulation.
Results
treRNA is overexpressed in lymph-node metastatic human breast cancer samples
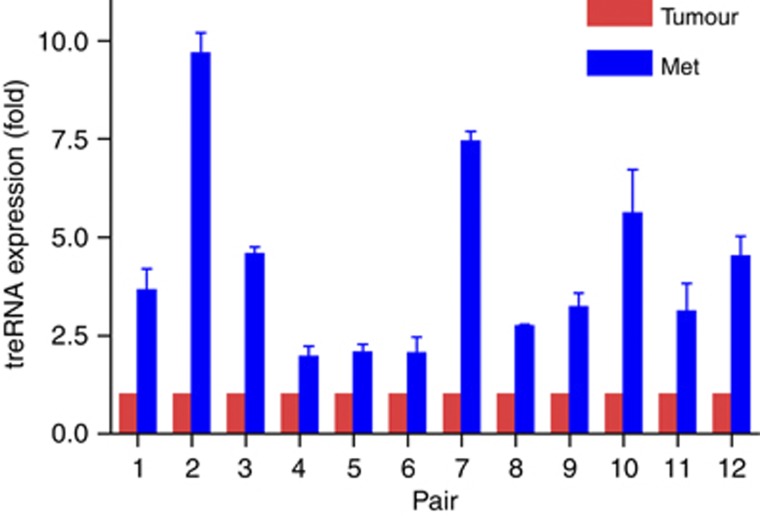
To explore the relevance of treRNA in cancer, we examined the treRNA expression in the matched primary and lymph-node metastatic tumour specimen from the same patient by quantitative RT–PCR (qRT–PCR). Expression analysis of treRNA in 12 primary breast-cancer carcinoma and the corresponding lymph-node metastases pairs, each from the same patient, showed markedly higher treRNA levels in the metastases of all 12 pairs (Figure 1). The average levels of log-transformed gene expression between lymph-node metastasis and primary tumour were significantly different (P<0.0001). The geometric mean of the ratio of gene expression levels in metastasis versus that in primary tumour is 3.72 (95% confidence interval of 2.79–4.96), indicating a significant association between the expression level of treRNA and metastatic progression. We also determined that the expression of treRNA expression was significantly higher in colon cancer specimens than in healthy controls (Supplementary Figure S1). This suggested that increased expression of treRNA may be a common feature involved in tumour progression and metastasis.
Figure 1.
treRNA expression in clinical breast cancer specimens. Each bar represents the percentage of GAPDH-normalized treRNA expression in 12 pairs of primary breast cancer and lymph-node metastasis samples (each pair came from the same patient). The expression of treRNA in the primary breast cancer sample in each pair was scaled to 100% to allow comparison. treRNA expression is significantly higher in the metastasis samples compared with the primary samples (P<0.0001). The geometric mean of the ratio of gene expression levels in the metastatic versus the primary tumour is 3.72 (95% confidence interval is 2.79–4.96).
treRNA promotes tumour invasion in vitro and metastasis in vivo
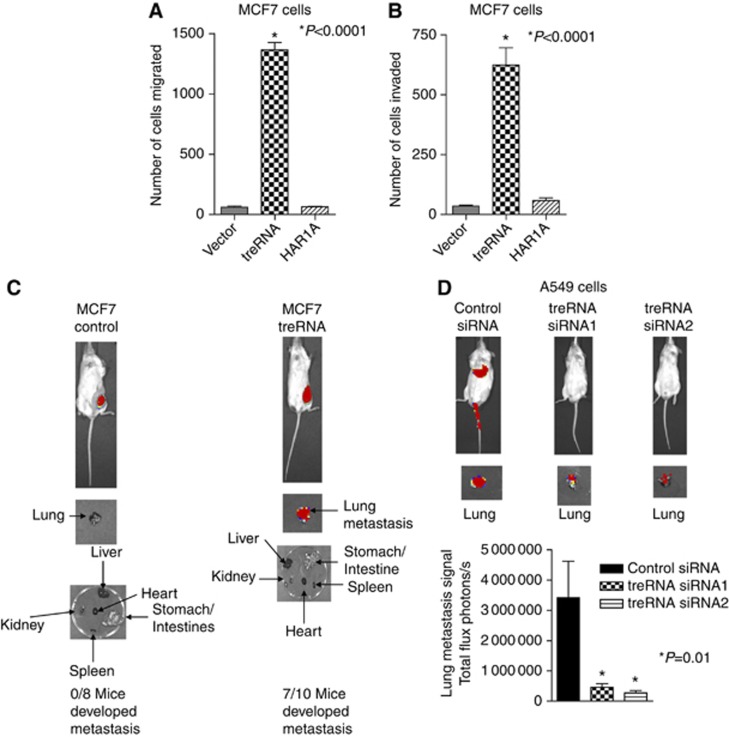
Because treRNA is upregulated in metastatic clinical samples, we examined the functional role of treRNA in tumour invasion and metastasis. Enforced expression of treRNA in non-invasive and non-metastatic human breast cancer MCF7 cells significantly increased cell migration and cell invasion through Matrigel, whereas control lncRNA HAR1A did not (Figure 2A and B). The enforced expression level of treRNA in MCF7 cells was within the range of treRNA expression in clinical samples (Supplementary Figure S2). The expression of treRNA or HAR1A in MCF7 cells did not affect cell proliferation in vitro (Supplementary Figure S3). To determine its functions in vivo, luciferase-tagged MCF7 cells (MCF7-luc) expressing treRNA or a vector control were transplanted into mouse mammary fat pads. The growth of primary tumours in MCF7-luc cells expressing treRNA is slightly higher than MCF7-luc cells expressing a vector control (Supplementary Figure S4), indicating that treRNA may affect primary tumour growth. Lung metastasis developed in mice following the transplantation of MCF-luc cells expressing treRNA (Figure 2C), whereas no lung metastasis developed following the transplantation of MCF7-luc cells expressing a vector control (Figure 2C). To determine the functions of endogenous treRNA in migration, invasion, and metastasis, we used two different small-interfering RNAs (siRNAs) to knockdown the expression of treRNA in A549 cells (Supplementary Figure S5). Knockdown of treRNA did not affect cell proliferation in vitro (Supplementary Figure S6). Knockdown of treRNA in A549 cells dramatically suppressed cell migration and invasion (Supplementary Figure S7). To determine its function in vivo, luciferase-tagged A549 cells (A549-luc) expressing treRNA siRNAs or a control siRNA were transplanted into mice via tail vein injection. Although there was no clear luciferase signals in the animals transplanted with A549-luc cells expressing treRNA siRNAs when the entire animals were imaged, imaging of the lungs dissected from the animals showed that metastasis was developed in these animals (Figure 2D). However, the luciferase signals of lung metastasis in A549-luc cells expressing treRNA siRNAs were significantly lower than cells expressing a control siRNA (Figure 2D), demonstrating that knockdown of endogenous treRNA suppressed metastasis in vivo. Taken together, these results indicated that treRNA was capable of promoting tumour invasion in vitro and metastasis in vivo.
Figure 2.
treRNA promotes migration and invasion in vitro and metastasis in vivo. (A, B) Quantitative analysis of MCF7 cells expressing a vector control, treRNA, or control long non-coding RNA HAR1A in migration (A) and invasion (B) assays. Data represent mean and s.d. (triplicates). (C) Transplantation of MCF7 cells expressing luciferase and treRNA in mouse mammary fat pads led to lung metastasis. Transplantation of MCF7 cells expressing a vector control does not lead to metastasis. (D) Knockdown of endogenous treRNA suppressed lung metastasis in vivo. A549 cells expressing luciferase and two different treRNA siRNAs or a control siRNA were transplanted into mice via tail vein injection. Lung metastasis was significantly reduced in A549 cells with treRNA knockdown. The data were collected from three independent experiments and presented as mean±s.d.
treRNA suppresses epithelial markers
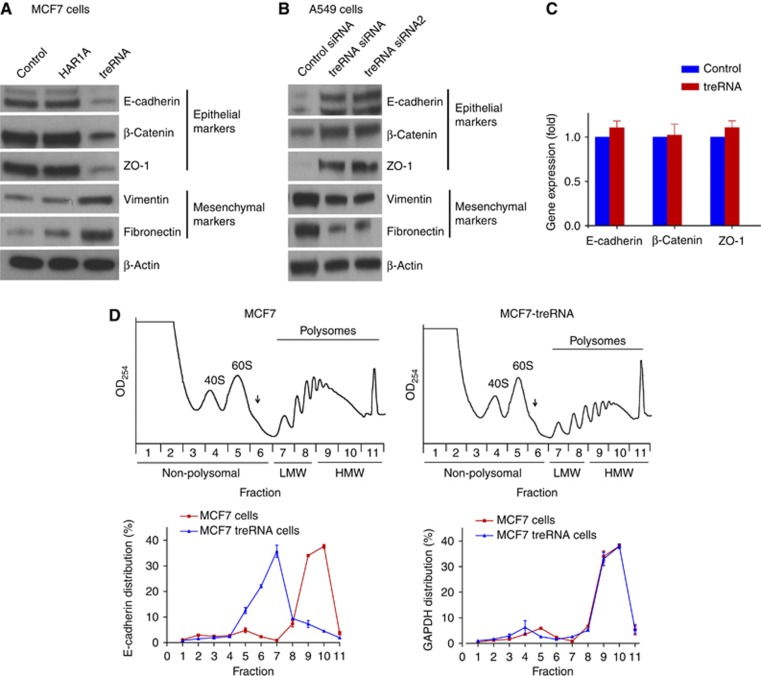
Because we have shown that treRNA downregulation decreases the expression of metastasis-promoting gene Snail (Orom et al, 2010), it is possible that the effect of enforced treRNA expression on invasion and metastasis is modulated by Snail. To address this issue, we evaluated the impact of enforced treRNA expression on Snail expression. To our surprise, treRNA upregulation had no effect on Snail expression (Supplementary Figure S8), which indicated that treRNA is required but not sufficient for Snail expression. Thus, we concluded that the increase in invasion and metastasis caused by enforced treRNA expression was not modulated by changes in Snail expression. Because the loss of epithelial markers is frequently associated with metastasis (Steeg, 2006; Chang et al, 2008; Cook et al, 2011; Valastyan and Weinberg, 2011), we assessed the effects of treRNA expression on epithelial markers. Enforced expression of treRNA in MCF7 cells significantly suppressed E-cadherin (Figure 3A), and to a lesser extent zonula occludens-1 (ZO-1) and β-catenin (Figure 3A). It also increased the expression of fibronectin and to a lesser extent vimentin (Figure 3A). Thus, treRNA expression in MCF7 cells induced a partial epithelial–mesenchymal transition phenotype without affecting Snail expression. Conversely, knockdown of treRNA in A549 cells increased the expression of E-cadherin, ZO-1, and β-catenin and decreased the expression of vimentin and fibronectin (Figure 3B). qRT–PCR was performed to determine whether the suppression of epithelial markers by treRNA was a result of reduced mRNA expression, reflecting an effect on transcription and/or stability of the mRNA. qRT–PCR showed that there was no change in the mRNA levels of E-cadherin, ZO-1, and β-catenin, indicating that treRNA regulates the expression of epithelial markers independent of changes in mRNA levels (Figure 3C; Supplementary Figure S9). In addition, the lack of effect on mRNA levels was consistent with a Snail-independent mechanism. We then determined the cellular location of spliced treRNA. Nuclear and cytoplasm fractionation was isolated from A549 cells, and the expression of endogenous treRNA was quantified by qRT–PCR. Approximately 75% of spliced treRNA was located in the cytoplasm (Supplementary Figure S10).
Figure 3.
treRNA suppresses the expression of epithelial markers at the translation step. (A) Immunoblots of epithelial and mesenchymal markers in MCF7 cells expressing treRNA or a vector control or a control long non-coding RNA HAR1A. MCF7 cells expressing treRNA showed the inhibition of the expression of epithelial markers E-cadherin, ZO-1, and β-catenin; and the increase in the expression of mesenchymal markers fibronectin and vimentin. (B) Immunoblots of epithelial and mesenchymal markers in A549 cells that express treRNA endogenously. A549 cells expressing two different treRNA siRNAs showed the increase in the expression of epithelial markers E-cadherin, ZO-1, and β-catenin. (C) Quantitative RT–PCR in MCF7 cells expressing treRNA and MCF7 cells expressing a vector control showed no difference in the RNA expression of epithelial markers E-cadherin, ZO-1, and β-catenin. Experiment was carried out in triplicate. Data were presented as mean±s.d. (D) Polysome profiles of MCF7 cells expressing treRNA or a vector control. The polysomal distribution of the E-cadherin and GAPDH mRNAs was determined by isolating the RNA from each fraction collected from a 10–50% sucrose gradient. The arrow indicates the location of the 80S ribosome. The amount of the E-cadherin and GAPDH mRNAs was determined by qRT–PCR. The percentage of the mRNA level in each fraction from MCF7 cells expressing treRNA or a vector control is shown. Experiment was carried out in triplicate. Data were represented as mean±s.d.
Source data for this figure is available on the online supplementary information page.
TreRNA suppresses the translation of the E-Cadherin mRNA
To determine whether treRNA suppressed the translation of the E-cadherin mRNA, we used polysome analysis to evaluate the translational status of E-cadherin mRNA in MCF7 cells expressing treRNA or a vector control. Basically, polysome analysis involves separating mRNA/protein complexes through a 10–50% sucrose gradient, isolating RNA from the fractions, and determining the distribution of E-cadherin mRNA within the gradient. The distribution of an mRNA within the gradient reflects its translational efficiency with efficiently translated RNAs associated with high-molecular weight (HMW, fractions 9–11) polysomes (heavy polysomes) and with inefficiently translated mRNAs associated with low-molecular weight (LMW, fractions 7–8) polysomes (light polysomes) or in the non-polysomal portion of the gradient (fractions 1–6). Polysome analysis showed that in MCF7 cells expressing a vector control, over 80% of the E-cadherin mRNA was associated with HMW polysomes (fractions 9–11), indicating efficient translation (Figure 3D). In contrast, enforced expression of treRNA resulted in the redistribution of over 80% of the E-cadherin mRNA to the LMW and non-polysomal portion of the gradient (Figure 3D). This effect on E-cadherin did not appear to be a general phenomenon as the distribution of the GAPDH mRNA was unaffected with over 80% of this mRNA associated with HMW polysomes in both cases (Figure 3D). Together, these data indicated that treRNA altered the polysomal distribution of the E-cadherin mRNA without affecting global translation.
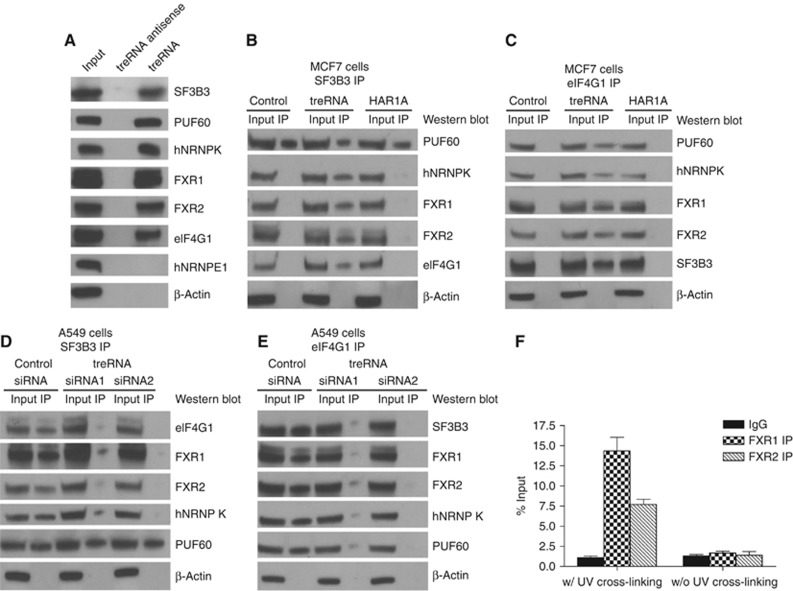
Identification of treRNA ribonucleoprotein complex(s) that mediates treRNA function
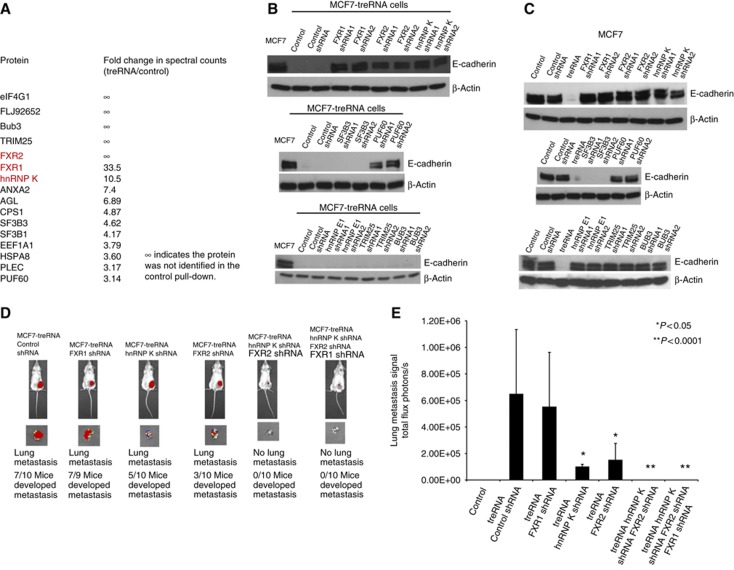
To elucidate the molecular mechanisms of translation suppression by treRNA, we utilized an affinity purification method by tagging treRNA with an MS2 RNA at its 3′ end to isolate protein complex(es) that bind to treRNA and mediate its functions (Supplementary Figure S11). This tagged treRNA had the same translational suppression function as an untagged treRNA (Supplementary Figure S12). We generated an affinity matrix by expressing a fusion protein of bacteriophage MS2 binding protein and maltose binding protein (MBP) that was bound to amylose beads. Cytoplasmic extracts were isolated from the MCF7 cells expressing MS2-tagged treRNA or MS2 tag control and applied to the affinity matrix. Proteins binding to the affinity matrix were then isolated and subjected to mass spectrometry analysis. We selected proteins that have had a higher number of spectral counts in the mass spectrometry analysis and ranked them based on the fold increase in spectral counts of tagged treRNA versus MS2 tag alone (Figure 4A). There were three RNA-binding proteins among the top hits from the analysis, hnRNP K, FXR1, and FXR2. hnRNP K shares similar structure with other poly(c)-binding proteins (PCBPs) such as hnRNP E1 and hnRNP E2. It is expressed in both the cytoplasm and the nucleus, and is involved in multiple steps of gene expression, such as chromatin remodelling, transcription, splicing, mRNA stability, and translation (Tomonaga and Levens, 1995; Ostareck et al, 1997; Ostareck-Lederer et al, 1998; Kim et al, 2000; Shnyreva et al, 2000; Evdokimova et al, 2001; Ostareck-Lederer et al, 2002; Ostrowski et al, 2003; Bomsztyk et al, 2004; Eiring et al, 2010; Hussey et al, 2011). FXR1 and FXR2 belong to a family of RNA-binding proteins, which also includes FMR1, the absence of which is the major cause of the Fragile X syndrome (Jin and Warren, 2000; Hoogeveen et al, 2002). These proteins play critical roles in development (Hoogeveen et al, 2002). FXR1, FXR2, and FMR1 can interact with each other and with themselves (Ceman et al, 1999). Although they can shuttle between cytoplasm and nucleus, their main localization in cells is the cytoplasm where they associate with ribosomes and form ribonucleoprotein (RNP) complexes (Hoogeveen et al, 2002). hnRNP K, FXR1, and FXR2 all contain RNA-binding KH domains, which are important for their functions (Valverde et al, 2008) and all have been shown to play a critical role in post-transcriptional gene regulation (Vasudevan and Steitz, 2007; Glisovic et al, 2008; van Kouwenhove et al, 2011). We also selected two other proteins for validation, RNP-binding protein PUF60 and SF3B3, which is a subunit of splicing factor 3b protein complex that has been shown to interact with PUF60 (Kielkopf et al, 2004; Corsini et al, 2009; Prigge et al, 2009). PUF homologue has also been shown to attenuate translation in C. elegans (Friend et al, 2012).
Figure 4.
Identification of proteins associated with treRNA. (A) Top protein candidates from mass spectrometry analysis are listed. The fold ratios of spectral counts of protein candidates in cells expressing MS2-tagged treRNA or MS2 tag control vector were calculated. ∞ indicates that these proteins were detected only in cells expressing treRNA, but not in cells expressing a vector control. (B–E) The components of treRNA RNP complex are required for treRNA functions. (B) The effects on E-cadherin expression in MCF7 cells expressing treRNA by the knockdown of the protein candidates from mass spectrometry analysis. Knockdown of hnRNP K, FXR1, FXR2, and PUF60 expression by two different short-hairpin RNAs (shRNAs) in MCF7-treRNA cells rescued the suppression of E-cadherin by treRNA. (C) The effects on E-cadherin expression in MCF7 cells by the knockdown of the protein candidates. Knockdown of hnRNP K, FXR1, FXR 2, BUB3, TRIM25 and hnRNP E1 expression individually in MCF7 cells does not affect E-cadherin expression. Knockdown of PUF60 expression partially affects E-cadherin expression. Knockdown of SF3B3 expression significantly decreases E-cadherin expression, suggesting that SF3B3 is required for E-cadherin expression in MCF7 cells. (D) MCF7-luc-treRNA cells expressing shRNAs targeting hnRNP K, FXR1, and FXR2 individually or in combination were transplanted into mouse mammary fat pads. The number of mice that developed lung metastasis was reduced when hnRNP K or FXR2 expression was knocked down. No lung metastasis was developed when both hnRNP K and FXR2 expressions were knocked down, or hnRNP K, FXR1, and FXR2 triple expression knockdown. (E) Lung metastatic tumour signals were measured using a luminescence Xenogen imaging system. In the mice that developed lung metastases, tumour signals in the lung were significantly reduced when hnRNP K or FXR2 expression was knocked down when compared with that of cells expressing a control shRNA. There is no lung metastasis signal when both hnRNP K and FXR2 expressions were knocked down, or in the hnRNP K, FXR1 and FXR2 triple expression knockdown. The graph showed the mean photon flux ±s.d. of three independent experiments.
Source data for this figure is available on the online supplementary information page.
We next examined the effect of these proteins on E-cadherin expression. The expression of treRNA does not alter the expression of hnRNP K, FXR1, FXR2, PUF60, and SF3B3 in MCF7 cells (Supplementary Figure S13). We used two different short-hairpin RNAs (shRNAs) to knockdown the expression of each of the five genes we selected (Supplementary Figure S14). Knockdown of hnRNP K, FXR1, and FXR2 expression in MCF7 cells expressing treRNA rescued the suppressed E-cadherin phenotype (Figure 4B). Knockdown of PUF60 expression partially rescued the E-cadherin suppression, whereas SF3B3 expression knockdown did not rescue E-cadherin suppression (Figure 4B). We also tested two other genes that rank high on the list, BUB3 and TRIM25, along with hnRNP E1, an RNA-binding protein that shares similar structure with hnRNP K. Knockdown of BUB33, TRIM25, or hnRNP E1 did not rescue the E-cadherin suppression by treRNA (Figure 4B). To further determine their effects on E-cadherin expression, we knocked down the expression of these genes in MCF7 cells expressing a vector control. Knockdown of hnRNP K or FXR1 or FXR2 or BUB3 or TRIM25 or hnRNP E1 expression did not affect E-cadherin expression, whereas PUF60 is partially required and SF3B3 is required for E-cadherin expression (Figure 4C). Although SF3B3 was not proven to mediate the suppression by treRNA because it was required for E-cadherin expression, it was important for treRNA function because it interacted with PUF60 and treRNA complex. These results suggested that the hnRNP K, FXR1, FXR2 and PUF60 mediated treRNA effect on E-cadherin suppression, whereas SF3B3 appeared to be required for E-cadherin expression.
We then determined whether hnRNP K, FXR1, and FXR2 were required for metastasis-promoting functions of treRNA in vivo. We knocked down the expression of these three genes individually or in combination in luciferase-tagged MCF7 cells expressing treRNA and transplanted these cells into mouse mammary fat pads. Knockdown of FXR1 or FXR2 expression did not significantly affect cell proliferation in vitro (Supplementary Figure S15). Knockdown of FXR2 in MCF7 cells expressing treRNA did not affect primary tumour growth in vivo (Supplementary Figure S16). Knockdown of hnRNP K or FXR1 increased primary tumour growth, whereas double knockdown of hnRNP K and FXR2 or triple knockdown of hnRNP K, FXR1, and FXR2 significantly decreased primary tumour growth (Supplementary Figure S16), indicating that the combination of hnRNP K and FXR2 expression was required for primary tumour growth in vivo. Both metastasis growth in lungs and the number of mice that developed metastasis nodules were similar in mice transplanted with MCF7-treRNA cells expressing FXR1 shRNA or a control shRNA (Figure 4D and E). In contrast, both metastasis growth in lungs and the number of mice that developed lung metastasis were significantly reduced in mice injected with MCF7-treRNA cells expressing hnRNP K shRNA or FXR2 shRNA when compared with cells expressing a control shRNA (Figure 4D and E). Double knockdown of hnRNP K and FXR2 or triple knockdown of hnRNP K, FXR2, and FXR1 in MCF7-treRNA cells completely abolished the metastasis-promoting function of treRNA in vivo (Figure 4D and E). These results demonstrated that hnRNP K and FXR2 were required for the treRNA function on metastasis in vivo.
TreRNA protein complex suppresses translation by binding translation initiation factor EIF4G1
We performed RNA pull-down to determine whether treRNA and hnRNP K, FXR1, FXR2, PUF60, and SF3B3 form a ribonucleoprotein complex. Cytoplasmic MCF7 cell extracts were incubated with in vitro-transcribed biotinylated transcripts of treRNA or antisense treRNA (negative control), and the presence of these proteins was detected by western blotting. Pull-down with treRNA, but not with an antisense treRNA, was able to detect all five proteins but not control RNA-binding protein hnRNP E1 (Figure 5A), demonstrating that all five proteins associate with treRNA. Because treRNA suppressed translation and translation initiation factor eIF4G1 was present in mass spectrometry analysis of treRNA affinity column pull-down (Figure 4A), we hypothesized that eIF4G was associated with the treRNA–protein complex. RNA IP indicated that treRNA was associated with eIF4G1 in the complex (Figure 5A). To investigate whether treRNA expression promoted RNP complex formation and the binding of eIF4G1 to the complex, we immunoprecipitated SF3B3 or eIF4G1 in MCF7 cells expressing treRNA or a vector control or a control lncRNA HAR1A. Co-immunoprecipitation experiments showed that SF3B3 was associated with PUF60 in MCF7 cells expressing treRNA or a vector control or HAR1A (Figure 5B), confirming the results from previous publications that splicing factor 3B protein complex binds to PUF60 (Corsini et al, 2009; Prigge et al, 2009). Interestingly, SF3B3 interacted with hnRNP K, FXR1, FXR2, and eIF4G1 in MCF7 cells expressing treRNA, but not in MCF7 cells expressing a vector control or HAR1A, demonstrating that treRNA was required for the formation of treRNA RNP complex and its binding to eIF4G1 (Figure 5B). Immunoprecipitation (IP) of eIF4G1 further confirmed the observation that treRNA promoted treRNA–protein complex formation and its binding to eIF4G1 (Figure 5C). To further validate these results, we performed the converse experiments with treRNA siRNAs to knockdown treRNA expression in A549 cells that express endogenous treRNA. Co-immunoprecipitation of SF3B3 or eIF4G1 in A549 cells transfected with treRNA siRNAs or a control siRNA confirmed that SF3B3 associated with PUF60 regardless of treRNA expression (Figure 5D). Knockdown of endogenous treRNA expression abolished the association of SF3B3 with the treRNA RNP complex, hnRNP K, FXR1, FXR2, and its binding to eIF4G1 (Figure 5D and E). In order to further validate that endogenous treRNA interacted with treRNA RNP complex, we performed UV cross-linking IP. A549 cells were UV cross-linked and immunoprecipitated using an IgG control or antibodies of FXR1 and FXR2, members of the treRNA RNP complex. Quantitative real-time PCR analysis indicated that treRNA was associated with FXR1 and FXR2 endogenously (Figure 5F). In addition, when biotinylated treRNA was incubated with MCF7 cytosolic cell extracts, the E-cadherin mRNA was present in the pull-down (Supplementary Figure S17), indicating their association. Together, these results suggested that treRNA expression promoted the binding of PUF60–SF3B3 complex to hnRNP K, FXR1, and FXR2 to form a treRNA RNP complex and its’ binding to eIF4G1 affected translation.
Figure 5.
TreRNA RNP complex binds to the translation initiation factor eIF4G1. (A) RNA pull-down was performed by incubating cytoplasmic extracts of MCF7 cells with biotinylated treRNA or antisense treRNA (negative control) followed by detection of the presence of the treRNA RNP complex by western blotting. All the treRNA RNP complex components, hnRNP K, FXR1, FXR2, PUF60, and SF3B3 were detected in treRNA pull-down, but not in treRNA antisense pull-down. Translation initiation factor eIF4G1 was also detected in the treRNA pull-down, demonstrating that it binds to treRNA RNP complex. RNA-binding protein hnRNP E1 that shares similar structure with hnRNP K was not detected in treRNA pull-down. (B, C) SF3B3 or eIF4G1 was immunoprecipitated in MCF7 cells expressing treRNA or a vector control or control long non-coding RNA HAR1A, and the proteins in the treRNA RNP complex were detected by western blotting. SF3B3 binds to PUF60 in the absence or presence of treRNA. Upon treRNA expression, SF3B3 and PUF60 bind to hnRNP K, FXR1, FXR2, and eIF4G1. eIF4G1 immunoprecipitation further confirmed the interaction of these proteins in the presence of treRNA. (D, E) SF3B3 or eIF4G1 was immunoprecipitated in A549 cells that endogenously express treRNA and was transfected with treRNA siRNAs. Knockdown of treRNA disrupts the binding of SF3B3 to hnRNP K, FXR1, FXR2, and eIF4G1, but not binding to PUF60. Immunoprecipitation of eIF4G1 further confirmed that the interaction of eIF4G1 with hnRNP K, FXR1, FXR2, PUF60, and SF3B3 requires treRNA expression. (F) Endogenous treRNA was associated with FXR1 and FXR2. A549 cells were UV cross-linked or without UV cross-linking, and immunoprecipitated using an IgG control or antibodies against FXR1 and FXR2. RNAs were extracted from the immunoprecipitation and quantitative RT–PCR indicated that treRNA was associated with FXR1 and FXR2 endogenously. The data were collected from three independent experiments and presented as mean±s.d.
Source data for this figure is available on the online supplementary information page.
Translation suppression by treRNA RNP complex is dependent on the 3′UTR of its target genes
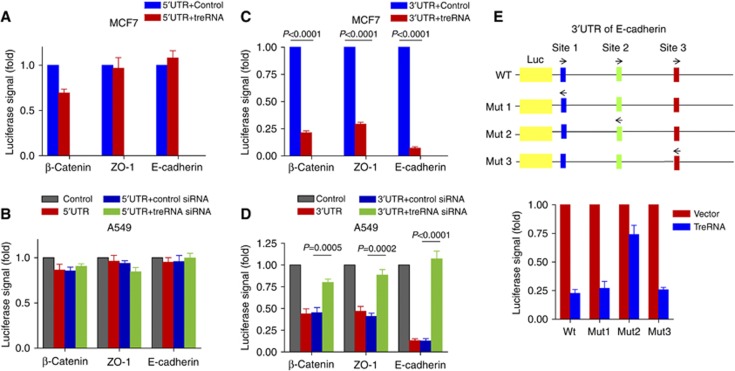
It has been shown that RNA-binding proteins associate with UTR regions of genes (Chaudhury et al, 2010; Kedde et al, 2010; Hussey et al, 2011). To identify the regions of the E-cadherin mRNA that were required for the treRNA-mediated translational suppression, we constructed reporter plasmids by cloning the 3′UTR of E-cadherin downstream of a luciferase gene and its 5′UTR upstream of a luciferase gene. As ZO-1 and β-catenin expression levels were suppressed by treRNA, we also analysed the UTRs of these mRNAs. Enforced expression of treRNA in MCF7 cells or knockdown of endogenous treRNA in A549 cells did not affect luciferase activity when 5′UTR reporter plasmids were transfected into these cells. This indicated that the translation suppression by treRNA was not dependent on the 5′UTRs of target genes (Figure 6A and B). Interestingly, treRNA expression in MCF7 cells significantly decreased the luciferase signal of the 3′UTR reporter plasmid, and conversely knockdown of treRNA in A549 cells considerably increased the luciferase activity when 3′UTR reporter plasmids were transfected (Figure 6C and D). Together, these data indicated that elements in the 3′UTR of E-cadherin were required for the translational suppression mediated by treRNA. In order to delineate the sequences in the E-cadherin 3′UTR that mediate this effect, we used catRAPID computational program (Bellucci et al, 2011) to identify potential binding sites of hnRNPK, FXR1, and FXR2 and found three regions (site 1: 12–80, site 2: 946–1019; site 3: 1325–1396) each of which is predicted to contain potential binding sites for all three proteins. We constructed mutant reporter plasmids with the replacement of site 1, site 2, or site 3 with their respective reverse sequences (Figure 6E). Site 2 mutant partially rescued the suppression of luciferase activity by treRNA (Figure 6E), whereas site 1 or 3 mutants had no effect (Figure 6E), indicating that site 2 of the E-cadherin 3′UTR was critical for treRNA complex functions. These results suggested that translation suppression by treRNA RNP complex was dependent on the 3′UTRs of its target genes.
Figure 6.
Translational suppression by treRNA RNP complex is dependent on the 3′UTRs of its target genes, but not on the 5′UTRs. Reporter plasmids were constructed by cloning the 3′UTRs or 5′UTRs of E-cadherin, ZO-1, β-catenin downstream or upstream of a luciferase gene. These reporter plasmids were co-transfected with treRNA or a vector control, and Renilla luciferase (pRL-TK) (normalizing control) into MCF7 cells. The reporter plasmids were also co-transfected with treRNA siRNAs or a control siRNA, and Renilla luciferase (pRL-TK) (normalizing control) into A549 cells. Firefly luciferase activity was normalized against the Renilla luciferase activity. Experiment was carried out in triplicate. Data were presented as the mean±s.d. (A) There is no significant change in luciferase signal of 5′UTR reporter plasmids when treRNA expression is upregulated in MCF7 cells or (B) knocked down in A549 cells, indicating that 5′UTR is not required for treRNA functions. (C) The luciferase activity of the 3′UTR reporter plasmids was significantly decreased in MCF7 expressing treRNA. (D) Conversely, the luciferase activity of the 3′UTR reporter plasmids increased when treRNA is knocked down in A549 cells. (E) The 3′UTR of E-cadherin (wt) was cloned downstream of a luciferase gene to generate a reporter construct. The mutant reporter plasmids were generated by replacing the respective binding sites with their respective reverse sequences in the 3′UTR. MCF7 cells were transfected with the reporter constructs, a Renilla luciferase construct (normalizing control), and treRNA (or a vector control). Luciferase activity was then determined. The site 2 mutant in the 3′UTR of E-cadherin partially rescued the suppression of luciferase activity by treRNA.
Discussion
Our previous study demonstrated that treRNA controls gene expression by enhancing the expression of neighbouring genes in the nucleus (Orom et al, 2010). The present study indicated that treRNA reduces the level of E-cadherin protein without affecting the level of its mRNA. In addition, we showed that the expression of treRNA results in the redistribution of the E-cadherin mRNA from HMW polysomes to LMW polysomes. Together, these data strongly indicate that treRNA suppresses the translation of the E-cadherin mRNA. Thus, treRNA employs different molecular mechanisms of gene regulation in the nucleus and the cytoplasm. It has been suggested that lncRNAs use various molecular machineries to regulate gene expression (Azzalin et al, 2007; Martianov et al, 2007; Rinn et al, 2007; Faghihi et al, 2008; Nagano et al, 2008; Wang et al, 2008; Bernard et al, 2010; Kim et al, 2010; Liu et al, 2010; Orom et al, 2010; Schmitz et al, 2010; Tian et al, 2010; Tripathi et al, 2010; Tsai et al, 2010; Yap et al, 2010; Flynn et al, 2011; Gong and Maquat, 2011; Heo and Sung, 2011; Kotake et al, 2011; Maison et al, 2011; Wang and Chang, 2011; Yoon et al, 2012). Our results indicated that even one lncRNA, treRNA, can use different mechanisms at multiple steps during the transcription and translation cycles.
Our results showed that treRNA RNP complex mediates the translational suppression of E-cadherin by treRNA. The exact mechanism by which treRNA affects E-cadherin translation is unknown, but it is important to note that it is likely not due to direct hybridization of the target mRNA. Although we found that the 3′UTR of E-cadherin is required for the translational suppression by treRNA, there is no sequence homology between the sequences of treRNA and the 3′UTR of E-cadherin. Thus, unlike LincRNA-p21, which suppresses translation through direct binding to its target mRNA (Yoon et al, 2012), it is unlikely that direct hybridization between treRNA and E-cadherin is responsible for this regulation. However, we found potential binding sites in the 3′UTR of the E-cadherin mRNA for hnRNP K, FXR1, and FXR2. These proteins are part of the treRNA protein complex and it may be through interactions with these binding proteins that treRNA elicits its effects. As our knockdown data indicate that hnRNP K, FXR1, and FXR2 do not directly affect E-cadherin expression (Figure 4C), it is unlikely that treRNA regulates the E-cadherin expression by competing for these RNA-binding proteins. Further studies are required to confirm a direct model where treRNA protein complex acts directly on E-cadherin mRNA. E-cadherin expression is regulated by multiple cellular pathways. Its regulation by treRNA is one of the mechanisms that control its expression. As the treRNA RNP complex components are expressed in most cell types, it is possible that the mechanism we have identified is shared by other lncRNAs to regulate translation. Such a translational regulation by lncRNAs provides another layer of complexity in gene regulation beyond that described for miRNA-mediated silencing of gene expression (Sonenberg and Dever, 2003; Gebauer and Hentze, 2004; Filipowicz et al, 2008; Sonenberg and Hinnebusch, 2009; Jackson et al, 2010; Silvera et al, 2010; Fukaya and Tomari, 2012; Meijer et al, 2013).
On the basis of the data from our study, we propose the following model for the molecular mechanism of translation suppression by treRNA. TreRNA promotes the formation of a treRNA-associated protein (treRNP) complex consisting of hnRNP K, FXR1, FXR2, PUF60, and SF3B3. This treRNP complex interacts, directly or indirectly, with the 3′UTR of E-cadherin mRNA, and reduces the translation efficiency. Further studies are required to evaluate this model and to elucidate the precise mechanism in which the treRNA regulates the translation of E-cadherin mRNA.
In addition to the epithelial markers regulated by treRNA, hnRNP K, FXR1, and FXR2 bind other mRNAs which are potential targets of treRNA regulation. Whether other genes regulated by treRNA RNP complex play a role in tumour invasion and metastasis remains to be investigated. Identification of these genes regulated by treRNA will reveal additional treRNA functions and the cellular processes that are regulated by treRNA.
TreRNA is upregulated in clinical metastatic breast cancer samples and primary colon cancer samples, suggesting that treRNA is involved in cancer development including metastasis. It is possible that cellular context is also important for gene regulation by treRNA and other lncRNAs. Identification of lncRNAs involved in cancer progression will further improve our understanding of aberrant gene regulations operative in cancer. It will also be interesting to investigate whether lncRNAs and RNA-binding proteins can serve as potential diagnostic and prognostic markers as well as therapeutic targets in cancer. Elucidation of the functions of lncRNAs and RNA-binding proteins with which they associate will further improve our understanding of their roles in biological processes and human diseases.
Materials and methods
Lentivirus transfection and transduction
To generate MCF7 cells stably overexpressing treRNA, human treRNA was amplified by PCR using cDNA from A549 cells as a template and cloned into lentiviral vector pLu. Lentivirus was produced by co-transfecting subconfluent human embryonic kidney (HEK) 293T cells with treRNA expression plasmid and packaging plasmids pMDLg/pRRE and RSV-Rev) using Lipofectamine 2000. Infectious lentiviruses were collected 48 h after transfection, centrifuged to remove cell debris, and filtered through 0.45 μm filters (Millipore). MCF7 cells were transduced with the treRNA lentivirus. Efficiency of overexpression was determined by real-time PCR.
MCF7 or MCF-treRNA cells stably expressing FXR1 shRNA, FXR2 shRNA, hnRNP K shRNA, SF3B3 shRNA, PUF60 shRNA, TRIM25 shRNA, BUB3 shRNA, hNRNPE1 shRNA, or control non-target shRNA were established using a vector-based shRNA technique. All shRNAs were cloned into pLKO lentiviral vector and purchased from Sigma. The lentiviruses were processed as described above and transduced into respective cells. The knockdown efficiency was determined by western blotting.
treRNA knockdown
To knockdown treRNA expression, siRNA oligos (siRNA1: sense 5′-GAAGGGAACCAGUGCUAAAUU-3′, antisense 5′-UUUAGCACUGGUUCCCUUCUU-3′; siRNA2: sense 5′-CCGAUUUGAGAGAGUGAGAUU-3′, antisense 5′-CUCACUCUCUCAAAUCGGUU-3′) were used. A control siRNA (sense: 5′-GCUUCCUGCUACACAGAAUUU-3′, antisense 5′-UUCUGUGUAGCAGGAAGCUU) was used as a negative control. siRNAs were purchased from Dharmacon. A549 cells were transfected with 100 nM of siRNAs using Lipofectamine RNAiMAX reagent (Invitrogen) following the manufacturer’s instructions. Cells were collected after 48 h of transfection for western blot analysis and RNA isolation. Knockdown was confirmed by qRT–PCR.
Tumour transplantation in mice
The MCF7 human breast cancer cell line stably expressing Firefly Luciferase gene with treRNA or respective shRNAs were routinely maintained at 37 °C in a humidified atmosphere of 5% CO2 and 95% air in DMEM supplemented with 10% fetal bovine serum (FBS). For orthotopic injections, 7 × 106 cells/mouse were transplanted into the mammary fat pad of the female NOD/SCID mice (6–8 weeks old). A slow-release pellet of 17β-estradiol (1.7 mg, 90-day release; Innovative Research of America, Sarasota, FL) was implanted subcutaneously in the dorsal interscapular region before the transplantation of MCF7 cells. A549 cells stably expressing Firefly Luciferase gene were transfected with treRNA siRNAs or a control siRNA. In all, 0.5 × 106 cells/mouse were injected into the lateral tail veins of 6- to 8-week-old NOD/SCID mice. Mice bearing luciferase-positive tumours were imaged by an IVIS 200 Imaging system (Xenogen Corporation, Hopkinton, MA, USA). Bioluminescent flux (photons/s/sr/cm2) was determined for the primary tumours or lung metastasis. Animal experiment protocols were approved by the Institutional Animal Care and Use Committee (IACUC) of the Wistar Institute. Animal procedures were conducted in compliance with the IACUC.
Transwell migration and invasion assay
In vitro cell migration assays were performed as described previously (Gumireddy et al, 2009) using Trans-well chambers (8 μM pore size; Costar). Cells were allowed to grow to subconfluency (∼75–80%) and were serum starved for 24 h. After detachment with trypsin, cells were washed with PBS, resuspended in serum-free medium, and 250 μl of cell suspensions (2 × 105 cells ml−1) was added to the upper chamber. Complete medium was added to the bottom wells of the chambers. The cells that had not migrated were removed from the upper face of the filters using cotton swabs, and the cells that had migrated to the lower face of the filters were fixed with 5% glutaraldehyde solution and stained with 0.5% solution of Toluidine Blue in 2% sodium carbonate. Images of three random × 10 fields were captured from each membrane and the number of migratory cells was counted. The mean of triplicate assays for each experimental condition was used. Similar inserts coated with Matrigel were used to determine the invasive potential in the invasion assay.
Cell proliferation assay
To measure the proliferative activity of cells, each group of cells was seeded in 96-well plates at a density of 4 × 103 cells per well in triplicate. Cell proliferation was quantified at day 0, day 1, day 2, and day 3 with a CellTiter 96 AQeous Non-radioactive Cell Proliferation Assay system (Promega, Madison, WI, USA) according to the manufacturer’s protocol. Briefly, cells were incubated with 20 μl of MTS/PMS solution for 4 h at 37 °C in a humidified 5% CO2 atmosphere. Cell proliferation was then measured at 490 nm using a multi-well plate reader (Wallac Victor, PerkinElmer). Cell growth curves were plotted with OD value versus time. Three independent experiments were performed.
RNA isolation, reverse transcription, and real-time PCR analysis
Total RNA was extracted from cell lines, frozen primary and metastasis tissues using Trizol total RNA isolation reagent (Invitrogen), according to the manufacturer’s specifications and treated with Turbo DNase (Ambion). cDNA was synthesized from total RNA using random hexamers with a TaqMan cDNA Reverse Transcription Kit (Applied Biosystems). Gene primers were designed using a Primer Express v3.0 Software and real-time PCR was performed using the SYBR Green Jumpstart Taq ReadyMix (Sigma). Levels of E-cadherin (Assay ID: Hs01023894) and GAPDH (Assay ID: Hs99999905) mRNAs were amplified using TaqMan Gene Expression Assays (Applied Biosystems). All reactions were carried out on the 7500 Fast Real Time PCR system (Applied Biosystems). The average of three independent analyses for each gene and sample was calculated using the ΔΔ threshold cycle (Ct) method and was normalized to the endogenous reference control gene GAPDH.
Polysome analysis
Polysomes were isolated as described (Johannes and Sarnow, 1998). Briefly, 20 million MCF7-vector control MCF7-treRNAs were incubated with 100 μg/ml cycloheximide for 3 min at 37 °C before harvesting and washed with ice-cold PBS containing 100 μg/ml cycloheximide, lysed in 500 μl of polysome lysis buffer (15 mM Tris–HCl pH 7.4, 15 mM MgCl2, 0.3 M NaCl, 1 mg/ml heparin, 0.1 mg/ml cycloheximide, and 1% Triton X-100). Extracts were incubated on ice for 10 min and centrifuged at 13 000 r.p.m. for 10 min at 4 °C to remove nuclei and debris. The supernatants were layered on 10–50% continuous, linear sucrose gradients prepared with lysis buffer without Triton X-100. The gradients were centrifuged in an SW41 Beckman rotor at 35 000 r.p.m. for 190 min at 4 °C. Gradients were collected into eleven 1 ml fractions by monitoring RNA absorbance at 254 nm using an ISCO fractionator (Brandel, Inc.). RNA was extracted by precipitation with guanidinium-HCL and ethanol followed by LiCl precipitation and further purified by RNeasy column (Qiagen) according to the manufacturer’s instructions. RNA was treated with Turbo DNase (Ambion). RNA isolated from the non-polysomal (fractions 1–6), LMW (fractions 7–8) polysomal, and HMW polysomal (fractions 9–11) fractions was pooled. In all, 0.5 μg of pooled RNA was reverse transcribed using the Taqman reverse transcription kit (Applied Biosystems). Resulting cDNA was subjected to qRT–PCR. The mRNA and treRNA distribution across the polysome fractions was graphically presented as the percentage. All polysomal analysis was done a minimum of three times.
MS2 pull-down assay
MCF7 cells were infected with lentivirus expressing 3′tagged treRNA-MS2 or MS2 tag alone, and cells were harvested at 72 h post infection. Cytoplasmic proteins were extracted using a NE-PER Cytoplasmic and Nuclear Protein Extraction Kit (Thermo Scientific) as per the manufacturer’s instructions and then incubated with fusion MS2 binding protein-MBP bound amylose magnetic beads for 3 h at 4 °C. The beads were subsequently washed thrice with wash buffer 1 (20 mM HEPES pH 7.9, 200 mM KCl, and 1 mM EDTA); twice with wash buffer 2 (20 mM HEPES pH 7.9, 20 mM KCl, and 1 mM EDTA) and finally once with ice-cold PBS. Bound proteins were dissociated by boiling with 1 × SDS–PAGE sample buffer and resolved on 15% SDS–PAGE. Samples were run 2 cm on a 1D SDS mini-gel, followed by staining with Colloidal Blue. The 2-cm lanes were sliced into 10 uniform fractions, gel slices were destained overnight, dried in a Speedvac, reduced using 100 μl 20 mM Tris(2-carboxyethyl)phosphine hydrochloride (TCEP) in 25 mM ammonium bicarbonate, pH 8.0, and alkylated using 100 μl 40 mM iodoacetamide in 25 mM ammonium bicarbonate, pH 8.0. After thorough washing, gel pieces were dried, rehydrated with 20 μl of 0.02 μg/μl modified Trypsin (Promega) in 40 mM ammonium bicarbonate, and incubated overnight with shaking at 37°C. The tryptic digest was removed, gel slices were extracted with 20 μl of 40 mM ammonium bicarbonate, supernatants were combined, and digestion was stopped using 4 μl concentrated acetic acid. Tryptic digests were analysed by injecting 5 μl onto a 15-cm nanocapillary reverse-phase column (New Objective PicoFrit 75 μm column terminating in a nanospray 15 μm tip self-packed with Microm Magic C18 AQ 200A, 5 μm resin), which was directly coupled to a ThermoElectron Orbitrap XL mass spectrometer. Peptides were eluted at a flow rate of 200 nl/min using 0.1% formic acid in MilliQ water as solvent A and 0.1% formic acid in acetonitrile as solvent B and a gradient consisting of: 3–28% B over 40 min, 28–50% over 25.5 min, 50–80% B over 5 min, and a 4.5-min hold at 80% B before returning to initial conditions of 3% B. MS and MS/MS data were acquired in data-dependent mode with full MS scans from m/z=350–2000 at 60 000 resolution using a top six method with a minimum MS signal threshold of 1000.
RNA pull-down assay
To prepare a plasmid construct for the use as a template for RNA synthesis, treRNA was amplified by PCR and cloned into a pGEM-T Easy (Promega) cloning vector. The constructs were linearized and transcribed in vitro with biotin RNA labelling mix (Roche) and T7 RNA polymerase, treated with RNase-free DNase I (Roche), and purified with the RNeasy Mini Kit (Qiagen). Biotinylated RNAs of sense or antisense treRNA incubated with cytosolic extracts for 2 h at 4 °C. Proteins bound were recovered by further incubating with streptavidin magnetic beads for 1 h at 4 °C. The beads were washed with binding and washing buffer (10 mM Tris–HCl, 1 mM EDTA, and 2 m NaCl), boiled in SDS–PAGE sample buffer. Samples were separated by SDS–PAGE and the retrieved proteins were detected by the standard western blotting technique. To determine the amount of E-cadherin, biotinylated treRNA or a vector control was incubated with cytosolic cell extracts and pull-downs were performed as described above. Total RNA was extracted from the beads. One microgram of total RNA was transcribed and real-time PCR was performed with gene-specific primers. Relative E-cadherin mRNA level was normalized to GAPDH.
UV cross-linking IP assay
A549 cells (1 × 107) were cultured in 150 mm plates, washed with PBS and UV cross-linked at 400 mJ/cm2 in Stratalinker as described (Ule et al, 2005). UV cross-linked or non-cross-linked cells were lysed in RIPA buffer containing 1X protease inhibitor cocktail and RNase inhibitor and incubated on ice for 10 min. Protein G agarose beads were washed with 1X IP buffer (50 mM Tris, pH7.6, 100 mM NaCl, 0.5 mM DTT, 2 mM MgCl2, and 1X protease inhibitor cocktail) and were coated with FXR1 antibody or FXR2 antibody or control IgG. An aliquot (10%) of lysate was kept aside to use as an input and the rest of the cell lysates was incubated with antibody or IgG-coated beads at 4 °C overnight. The protein–RNA complexes on the beads after IP were washed two times with 1X IP buffer and twice with IP buffer containing 1 M urea (Luo and Reed, 2003). RNA was isolated from the beads and cDNA was synthesized with 0.5 μg of RNA as described above and used for real-time PCR analyses with treRNA primer. The percent input was calculated using a Relative Standard Curve Method.
IPs and immunoblotting
Cell pellets were lysed with lysis buffer (50 mM Tris pH 7.4, 150 mM NaCl, 0.5% Triton X-100, 0.25% sodium deoxycholate, 1 mM EGTA pH 8.0, 50 mM sodium fluoride, 1 mM sodium vanadate, and 10 mM sodium pyrophosphate with complete protease inhibitor; Roche). In all, 200 μg of total cell lysate was incubated with primary antibody and protein G or A sepharose beads (GE Healthcare) overnight at 4 °C. The beads with bound proteins were pelleted and washed six times with lysis buffer and once with ice-cold PBS. The beads were boiled with 1X SDS–PAGE sample buffer. Five percent input and fifty percent of immunoprecipitated proteins were separated using 4–12% SDS–PAGE gels and transferred onto PVDF membrane (Millipore, Bedford, MA, USA). Membranes were probed with specific primary antibodies. Blots were washed and probed with respective secondary peroxidase-conjugated antibodies, and the bands were visualized by chemoluminescence (GE Healthcare). The following antibodies were used: rabbit polyclonal E-cadherin, BUB3, hNRNP E1 (Cell Signaling Technology), mouse monoclonal β catenin (BD Biosciences), rabbit polyclonal ZO-1 (Zymed Laboratories), mouse monoclonal eIF4G (Abcam), mouse monoclonal Fibronectin and β-actin (Sigma-Aldrich), mouse monoclonal Vimentin (Calbiochem), rabbit polyclonal SF3B3, PUF60, hnRNP K, and TRIM25 (Bethyl Laboratories), mouse monoclonal FXR1, FXR2 (Millipore), and secondary rabbit and mouse peroxidase conjugated (GE Healthcare).
Luciferase reporter assay
The DNA fragments of 5′UTR regions of human E-cadherin, β-catenin and ZO1 were PCR amplified and cloned upstream of the firefly luciferase gene in the pGL3 vector (Promega). 3′UTR regions of human E-cadherin, β-catenin, and ZO1 were PCR amplified and cloned downstream of a firefly luciferase gene in pGL4 vector. Cells were co-transfected with 100 ng of treRNA, 0.5 μg of control or reporter DNA, and 0.2 ng of Renilla luciferase (pRL-TK) as a normalizing control. Luciferase activity was determined using a Dual-Luciferase Reporter Assay (Promega) according to the manufacturer’s instructions 48 h after transfections. Firefly luciferase was normalized against Renilla luciferase. Transfections were performed in triplicates and repeated three times. Mutations in wild-type human E-cadherin 3UTR were generated by replacing the three potential binding sites with their respective reverse sequences individually (site 1 from 12 to 80 bp; site 2 from 946 to 1019, bp; site 3 from 1325 to 1396, bp downstream of stop codon), and performed the luciferase assay as described above.
Isolation of nuclear and cytoplasmic RNAs
Nuclear and cytoplasmic RNAs were isolated from A549 cell line, with a Norgen Biotek Corp Cytoplasmic and Nuclear RNA Purification Kit (Cat # 2100) according to the manufacturer’s instructions. To remove genomic DNA contamination, on-column DNase digestion reaction was performed. First-strand cDNA was synthesized from 2.0 μg of cytoplasmic and nuclear RNAs using a Superscript First-Strand Synthesis System kit (Invitrogen). PCRs were performed with the following primer combinations: treRNA (F-5′-AATGAAAACACCAGGCCGGG-3′ and R-5′-TCTCACTCTCTCAA ATCGGC-3′) Nuclear-specific gene U2 snRNA (F-5′-CATCGCTTCTCGGCCTTTTG-3′ and R-5′-TGGAGGTACTGCAATACCAGG-3′) and cytoplasm-specific gene S14 (F-5′-GGCAGACCGAGATGAATCCTC-3′ and R-5′-CAGGTCCAGGGGTCTTGGTCC-3′). The experiment was repeated three times and each sample measured as duplicates.
Statistical analysis
For treRNA expression in breast cancer, two samples were collected from each patient. One sample was from the primary tumour, and the other was from metastasis. The experiments were performed twice for each sample. To assess the reproducibility of quantitative measurements of the studied gene expression, intraclass correlation coefficients (ICCs) were calculated and results show a high degree of consistency between two experiments for both tumour samples (ICC=99% with standard error, s.e.=0.005) and metastasis samples (ICC=96% with s.e.=0.021). To examine whether the level of gene expression in metastasis is different from that in primary tumours, the average of gene expression from the replicates of each sample was calculated and then log-transformed for statistical comparison using a paired t-test. The difference in the log-transformed gene expression levels between paired metastasis and primary tumour is equivalent to the log-transformed ratio of gene expression levels in metastasis versus that in primary tumour. For the treRNA expression in colon cancer samples (n=56) and normal colon tissues (n=19), log-transformed treRNA data were used for two-group t-test. Stata 12.0 (StataCorp LP, College Station, TX, USA) was used for data analysis.
GenBank accession number:
The GenBank accession numbers are FXR1: NM_005087, FXR2: NM_004860, hnRNP K: NM_002140, PUF60: NM_078480, SF3B3: NM_012426, BUB3: NM_004725, andTRIM25: NM_005082.
ShRNA sequences
The shRNA sequences are FXR1shRNA1 CCGGGCTAAAGTTCGGATGATGAAACTCGAGTTTCATCATCCGAACTTTAGCTTTTTTG; FXR1 shRNA2: CCGGGCTAGAGGTTTCTTGGAATTTCTCGAGAAATTCCAAGAAACCTCTAGCTTTTTTG; FXR2 shRNA1: CCGGGCGGATGATGAAGGGAGATTTCTCGAGAAATCTCCCTTCATCATCCGCTTTTT; FXR2 shRNA2: CCGGCGAGATACAATTCTTCATCTACTCGAGTAGATGAAGAATTGTATCTCGTTTTT; SF3B3 shRNA1: CCGGAGGAGGGTGACTGGATAATTACTCGAGTAATTATCCAGTCACCCTCCTTTTTT; SF3B3 shRNA2: CCGGAGACAGATGAAGATATGGTTACTCGAGTAACCATATCTTCATCTGTCTTTTTT; PUF60shRNA1: CCGGCGATGACATCAAGAGCGTGTTCTCGAGAACACGCTCTTGATGTCATCGTTTTT PUF60shRNA2: CCGGAGACTCCATCAAGATGGAGAACTCGAGTTCTCCATCTTGATGGAGTCTTTTTT; hnRNP KshRNA1: CCGGGCCAGTGTTTCAGTCCCAGACCTCGAGGTCTGGGACTGAAACACTGGCTTTTTG; hnRNP KshRNA2: CCGGTGATGTTTGATGACCGTCGCGCTCGAGCGCGACGGTCATCAAACATCATTTTTG; hnRNP E1shRNA1: CCGGCCCATGATCCAACTGTGTAATCTCGAGATTACACAGTTGGATCATGGGTTTTTG; hnRNP E1shRNA2: CCGGCGGGTGTAAGATCAAAGAGATCTCGAGATCTCTTTGATCTTACACCCGTTTTTG; BUB3shRNA1: CCGGCCACCTAATCATCCTGTGAAACTCGAGTTTCACAGGATGATTAGGTGGTTTTTG; BUB3shRNA2: CCGGCCTTCAAATGTCACAGACTAACTCGAGTTAGTCTGTGACATTTGAAGGTTTTTG; TRIM25shRNA1: CCGGCCAGCTCACATCCGAACTCAACTCGAGTTGAGTTCGGATGTGAGCTGGTTTTT; TRIM25shRNA2: CCGGGTGCCCGATTCCTCTTAGAGACTCGAGTCTCTAAGAGGAATCGGGCACTTTTT; and control shRNA: CCGGCAACAAGATGAAGAGCACCAACTCGAGTTGGTGCTCTTCATCTTGTTGTTTTT.
Primer sequences for the construction of 3′UTR and 5′UTR reporter plasmids
The primer sequences are E-cadherin 3′UTR Forward (5′-3′) GGGACTCGAGAGAGGCGGGC; E-cadherin 3′UTR Reverse (5′-3′) ATGGTTTAACAAAATTGTTTAATAAAATTTATAAAAAT; ZO1 3′UTR Forward (5′-3′) CTCTTGAA ATATAGGAAC TTAAATAATGTGAA; ZO1 3′UTR Reverse (5′-3′) TTTATTTTAAAAAGTTTTATTTTGGAGATTTAGAA; Bcatenin 3′UTR Forward (5′-3′) ATCATC CTTTAGGTAA GAAGTTTTAA AAAGC; Bcatenin 3′UTR Reverse (5′-3′) CAATCGAATGAATTAAAAGTTTAATTCTGAAC; E-cadherin 5′UTR Forward (5′-3′) AGCCAATCAGCGGTACGGGGG; E-cadherin 5′UTR Reverse (5′-3′) GGCTGGCCGGGGACGCCG; Bcatenin 5′UTR Forward (5′-3′) AGGATACAGC GGCTTCTGCGCG; Bcatenin 5′UTR Reverse (5′-3′) TGTCCACGCTGGATTTTCAAAACAG; ZO1 5′UTR Forward (5′-3′) CGGGCATGCT CAGTGGGC; and ZO1 5′UTR Reverse (5′-3′) CTTGTCTCTCTCCAGCGCCG.
Supplementary Material
Acknowledgments
We would like to thank Drs Melissa J Moore and Ngan Vo for generously providing the plasmids; Drs Gain Tartaglia and Federico Agostini for the catRAPID analysis. This work was supported by NCIR01CA148759-01; Breast Cancer Alliance; WW Smith Foundation; Edward Mallinckrodt Jr Foundation; National Natural Science Foundation of China (81372328).
Author contributions: KG and QH designed the study. KG, AL, TS, GJJ, UAØ, and DWS performed the experiments. JY, JT, LZ, and GAC provided materials. QL performed the statistical analysis. KG, GJJ, DWS, GAC, and QH analysed the data. KG, GJJ, and QH wrote the manuscript with input from all authors.
Footnotes
The authors declare that they have no conflict of interest.
References
- Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J (2007) Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 318: 798–801 [DOI] [PubMed] [Google Scholar]
- Banfei B, Jia H, Khatun J, Wood E, Risk B, Gundling WE Jr, Kundaje A, Gunawardena HP, Yu Y, Xie L, Krajewski K, Strahl BD, Che X, Bickle P, Giddings MC, Brown JB, Lipovich L (2012) Long noncoding RNAs are rarely translated in two human cell lines. Genome Res 22: 1646–1657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellucci M, Agostini F, Masin M, Tartaglia GG (2011) Predicting protein associations with long noncoding RNAs. Nat Methods 8: 444–445 [DOI] [PubMed] [Google Scholar]
- Beltran M, Puig I, Pena C, Garcia JM, Alvarez AB, Pena R, Bonilla F, de Herreros AG (2008) A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Dev 22: 756–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benetatos L, Hatzimichael E, Dasoula A, Dranitsaris G, Tsiara S, Syrrou M, Georgiou I, Bourantas KL (2010) CpG methylation analysis of the MEG3 and SNRPN imprinted genes in acute myeloid leukemia and myelodysplastic syndromes. Leuk Res 34: 148–153 [DOI] [PubMed] [Google Scholar]
- Bernard D, Prasanth KV, Tripathi V, Colasse S, Nakamura T, Xuan Z, Zhang MQ, Sedel F, Jourdren L, Coulpier F, Triller A, Spector DL, Bessis A (2010) A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J 29: 3082–3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomsztyk K, Denisenko O, Ostrowski J (2004) hnRNP K: one protein multiple processes. Bioessays 26: 629–638 [DOI] [PubMed] [Google Scholar]
- Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL (2011) Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 25: 1915–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Croce CM (2006) MicroRNA signatures in human cancers. Nat Rev Cancer 6: 857–866 [DOI] [PubMed] [Google Scholar]
- Calin GA, Liu CG, Ferracin M, Hyslop T, Spizzo R, Sevignani C, Fabbri M, Cimmino A, Lee EJ, Wojcik SE, Shimizu M, Tili E, Rossi S, Taccioli C, Pichiorri F, Liu X, Zupo S, Herlea V, Gramantieri L, Lanza G et al. (2007) Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell 12: 215–229 [DOI] [PubMed] [Google Scholar]
- Ceman S, Brown V, Warren ST (1999) Isolation of an FMRP-associated messenger ribonucleoprotein particle and identification of nucleolin and the fragile X-related proteins as components of the complex. Mol Cell Biol 19: 7925–7932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I (2011) A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 147: 358–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang XB, Mengos A, Hou YX, Cui L, Jensen TJ, Aleksandrov A, Riordan JR, Gentzsch M (2008) Role of N-linked oligosaccharides in the biosynthetic processing of the cystic fibrosis membrane conductance regulator. J Cell Sci 121: 2814–2823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury A, Hussey GS, Ray PS, Jin G, Fox PL, Howe PH (2010) TGF-beta-mediated phosphorylation of hnRNP E1 induces EMT via transcript-selective translational induction of Dab2 and ILEI. Nat Cell Biol 12: 286–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook LM, Hurst DR, Welch DR (2011) Metastasis suppressors and the tumor microenvironment. Semin Cancer Biol 21: 113–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsini L, Hothorn M, Stier G, Rybin V, Scheffzek K, Gibson TJ, Sattler M (2009) Dimerization and protein binding specificity of the U2AF homology motif of the splicing factor Puf60. J Biol Chem 284: 630–639 [DOI] [PubMed] [Google Scholar]
- Ebert MS, Sharp PA (2010) Emerging roles for natural microRNA sponges. Curr Biol 20: R858–R861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiring AM, Harb JG, Neviani P, Garton C, Oaks JJ, Spizzo R, Liu S, Schwind S, Santhanam R, Hickey CJ, Becker H, Chandler JC, Andino R, Cortes J, Hokland P, Huettner CS, Bhatia R, Roy DC, Liebhaber SA, Caligiuri MA et al. (2010) miR-328 functions as an RNA decoy to modulate hnRNP E2 regulation of mRNA translation in leukemic blasts. Cell 140: 652–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evdokimova V, Ruzanov P, Imataka H, Raught B, Svitkin Y, Ovchinnikov LP, Sonenberg N (2001) The major mRNA-associated protein YB-1 is a potent 5′ cap-dependent mRNA stabilizer. EMBO J 20: 5491–5502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, Finch CE St, Laurent G 3rd, Kenny PJ, Wahlestedt C (2008) Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nat Med 14: 723–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N (2008) Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 9: 102–114 [DOI] [PubMed] [Google Scholar]
- Flynn RL, Centore RC, O’Sullivan RJ, Rai R, Tse A, Songyang Z, Chang S, Karlseder J, Zou L (2011) TERRA and hnRNPA1 orchestrate an RPA-to-POT1 switch on telomeric single-stranded DNA. Nature 471: 532–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friend K, Campbell ZT, Cooke A, Kroll-Conner P, Wickens MP, Kimble J (2012) A conserved PUF-Ago-eEF1A complex attenuates translation elongation. Nat Struct Mol Biol 19: 176–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaya T, Tomari Y (2012) MicroRNAs mediate gene silencing via multiple different pathways in drosophila. Mol Cell 48: 825–836 [DOI] [PubMed] [Google Scholar]
- Gebauer F, Hentze MW (2004) Molecular mechanisms of translational control. Nat Rev Mol Cell Biol 5: 827–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glisovic T, Bachorik JL, Yong J, Dreyfuss G (2008) RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett 582: 1977–1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong C, Maquat LE (2011) lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature 470: 284–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumireddy K, Li A, Gimotty PA, Klein-Szanto AJ, Showe LC, Katsaros D, Coukos G, Zhang L, Huang Q (2009) KLF17 is a negative regulator of epithelial-mesenchymal transition and metastasis in breast cancer. Nat Cell Biol 11: 1297–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP (2010) Mammalian microRNAs predominantly act to decrease target mRNA level. Nature 466: 835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY (2010) Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464: 1071–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, Yang X, Amit I, Meissner A, Regev A, Rinn JL, Root DE, Lander ES (2011) lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477: 295–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Russell P, Ingolia NT, Weissman JS, Lander ES (2013) Ribosome profiling provides evidence that large noncoding RNAs do not code proteins. Cell 154: 240–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harismendy O, Notani D, Song X, Rahim NG, Tanasa B, Heintzman N, Ren B, Fu XD, Topol EJ, Rosenfeld MG, Frazer KA (2011) 9p21 DNA variants associated with coronary artery disease impair interferon-gamma signalling response. Nature 470: 264–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, He X, Lowe SW, Hannon GJ (2007) microRNAs join the p53 network--another piece in the tumour-suppression puzzle. Nat Rev Cancer 7: 819–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo JB, Sung S (2011) Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 331: 76–79 [DOI] [PubMed] [Google Scholar]
- Hoogeveen AT, Willemsen R, Oostra BA (2002) Fragile X syndrome, the Fragile X related proteins, and animal models. Microsc Res Tech 57: 148–155 [DOI] [PubMed] [Google Scholar]
- Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, Attardi LD, Regev A, Lander ES, Jacks T, Rinn JL (2010) A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 142: 409–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussey GS, Chaudhury A, Dawson AE, Lindner DJ, Knudsen CR, Wilce MC, Merrick WC, Howe PH (2011) Identification of an mRNP complex regulating tumorigenesis at the translational elongation step. Mol Cell 41: 419–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, Pestova TV (2010) The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 11: 113–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin P, Warren ST (2000) Understanding the molecular basis of fragile X syndrome. Hum Mol Genet 9: 901–908 [DOI] [PubMed] [Google Scholar]
- Johannes G, Sarnow P (1998) Cap-independent polysomal association of natural mRNAs encoding c-myc, BiP, and eIF4G conferred by internal ribosome entry sites. RNA 4: 1500–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedde M, van Kouwenhove M, Zwart W, Oude Vrielink JA, Elkon R, Agami R (2010) A Pumilio-induced RNA structure switch in p27-3′ UTR controls miR-221 and miR-222 accessibility. Nat Cell Biol 12: 1014–1020 [DOI] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, Regev A, Lander ES, Rinn JL (2009) Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA 106: 11667–11672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AM, Rinn JL (2011) RNA-protein interactions in human health and disease. Semin Cell Dev Biol 22: 359–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kielkopf CL, Lucke S, Green MR (2004) U2AF homology motifs: protein ecognition in the RRM world. Genes Dev 18: 1513–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Hahm B, Kim YK, Choi M, Jang SK (2000) Protein-protein interaction among hnRNPs shuttling between nucleus and cytoplasm. J Mol Biol 298: 395–405 [DOI] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, Markenscoff-Papadimitriou E, Kuhl D, Bito H, Worley PF, Kreiman G, Greenberg ME (2010) Widespread transcription at neuronal activity-regulated enhancers. Nature 465: 182–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP (2010) Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal 3: ra8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M, Xiong Y (2011) Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene 30: 1956–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Feng T, Lian Y, Zhang G, Garen A, Song X (2009) Role of human noncoding RNAs in the control of tumorigenesis. Proc Natl Acad Sci USA 106: 12956–12961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Marquardt S, Lister C, Swiezewski S, Dean C (2010) Targeted 3′ processing of antisense transcripts triggers Arabidopsis FLC chromatin silencing. Science 327: 94–97 [DOI] [PubMed] [Google Scholar]
- Luo MJ, Reed R (2003) Identification of RNA binding proteins by UV cross-linking. Curr Protoc Mol Biol Chapter 27:Unit 27, 22. [DOI] [PubMed] [Google Scholar]
- Maison C, Bailly D, Roche D, Montes de Oca R, Probst AV, Vassias I, Dingli F, Lombard B, Loew D, Quivy JP, Almouzni G (2011) SUMOylation promotes de novo targeting of HP1alpha to pericentric heterochromatin. Nat Genet 43: 220–227 [DOI] [PubMed] [Google Scholar]
- Mancini-Dinardo D, Steele SJ, Levorse JM, Ingram RS, Tilghman SM (2006) Elongation of the Kcnq1ot1 transcript is required for genomic imprinting of neighboring genes. Genes Dev 20: 1268–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martianov I, Ramadass A, Serra Barros A, Chow N, Akoulitchev A (2007) Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature 445: 666–670 [DOI] [PubMed] [Google Scholar]
- Mattick JS (2009) The genetic signatures of noncoding RNAs. PLoS Genet 5: e1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer HA, Kong YW, Lu WT, Wilczynska A, Spriggs RV, Robinson SW, Godfrey JD, Willis AE, Bushell M (2013) Translational repression and eIF4A2 activity are critical for microRNA-mediated gene regulation. Science 340: 82–85 [DOI] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Mattick JS (2009) Long non-coding RNAs: insights into functions. Nat Rev Genet 10: 155–159 [DOI] [PubMed] [Google Scholar]
- Mourtada-Maarabouni M, Pickard MR, Hedge VL, Farzaneh F, Williams GT (2009) GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene 28: 195–208 [DOI] [PubMed] [Google Scholar]
- Nagano T, Mitchell JA, Sanz LA, Pauler FM, Ferguson-Smith AC, Feil R, Fraser P (2008) The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science 322: 1717–1720 [DOI] [PubMed] [Google Scholar]
- Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, Guigo R, Shiekhattar R (2010) Long noncoding RNAs with enhancer-like function in human cells. Cell 143: 46–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostareck DH, Ostareck-Lederer A, Wilm M, Thiele BJ, Mann M, Hentze MW (1997) mRNA silencing in erythroid differentiation: hnRNP K and hnRNP E1 regulate 15-lipoxygenase translation from the 3′ end. Cell 89: 597–606 [DOI] [PubMed] [Google Scholar]
- Ostareck-Lederer A, Ostareck DH, Cans C, Neubauer G, Bomsztyk K, Superti-Furga G, Hentze MW (2002) c-Src-mediated phosphorylation of hnRNP K drives translational activation of specifically silenced mRNAs. Mol Cell Biol 22: 4535–4543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostareck-Lederer A, Ostareck DH, Hentze MW (1998) Cytoplasmic regulatory functions of the KH-domain proteins hnRNPs K and E1/E2. Trends Biochem Sci 23: 409–411 [DOI] [PubMed] [Google Scholar]
- Ostrowski J, Kawata Y, Schullery DS, Denisenko ON, Bomsztyk K (2003) Transient recruitment of the hnRNP K protein to inducibly transcribed gene loci. Nucleic Acids Res 31: 3954–3962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez DS, Hoage TR, Pritchett JR, Ducharme-Smith AL, Halling ML, Ganapathiraju SC, Streng PS, Smith DI (2008) Long, abundantly expressed non-coding transcripts are altered in cancer. Hum Mol Genet 17: 642–655 [DOI] [PubMed] [Google Scholar]
- Petrovics G, Zhang W, Makarem M, Street JP, Connelly R, Sun L, Sesterhenn IA, Srikantan V, Moul JW, Srivastava S (2004) Elevated expression of PCGEM1, a prostate-specific gene with cell growth-promoting function, is associated with high-risk prostate cancer patients. Oncogene 23: 605–611 [DOI] [PubMed] [Google Scholar]
- Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP (2010) A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 465: 1033–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP, Oliver PL, Reik W (2009) Evolution and functions of long noncoding RNAs. Cell 136: 629–641 [DOI] [PubMed] [Google Scholar]
- Prigge JR, Iverson SV, Siders AM, Schmidt EE (2009) Interactome for auxiliary splicing factor U2AF(65) suggests diverse roles. Biochim Biophys Acta 1789: 487–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY (2007) Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129: 1311–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz KM, Mayer C, Postepska A, Grummt I (2010) Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev 24: 2264–2269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shnyreva M, Schullery DS, Suzuki H, Higaki Y, Bomsztyk K (2000) Interaction of two multifunctional proteins. Heterogeneous nuclear ribonucleoprotein K and Y-box-binding protein. J Biol Chem 275: 15498–15503 [DOI] [PubMed] [Google Scholar]
- Silvera D, Formenti SC, Schneider RJ (2010) Translational control in cancer. Nat Rev Cancer 10: 254–266 [DOI] [PubMed] [Google Scholar]
- Sonenberg N, Dever TE (2003) Eukaryotic translation initiation factors and regulators. Curr Opin Struct Biol 13: 56–63 [DOI] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG (2009) Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136: 731–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steeg PS (2006) Tumor metastasis: mechanistic insights and clinical challenges. Nat Med 12: 895–904 [DOI] [PubMed] [Google Scholar]
- Tian D, Sun S, Lee JT (2010) The long noncoding RNA, Jpx, is a molecular switch for X chromosome inactivation. Cell 143: 390–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomonaga T, Levens D (1995) Heterogeneous nuclear ribonucleoprotein K is a DNA-binding transactivator. J Biol Chem 270: 4875–4881 [DOI] [PubMed] [Google Scholar]
- Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, Blencowe BJ, Prasanth SG, Prasanth KV (2010) The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell 39: 925–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY (2010) Long noncoding RNA as modular scaffold of histone modification complexes. Science 329: 689–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J, Jensen K, Mele A, Darnell RB (2005) CLIP: a method for identifying protein-RNA interaction sites in living cells. Methods 37: 376–386 [DOI] [PubMed] [Google Scholar]
- Valastyan S, Weinberg RA (2011) Tumor metastasis: molecular insights and evolving paradigms. Cell 147: 275–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde R, Edwards L, Regan L (2008) Structure and function of KH domains. FEBS J 275: 2712–2726 [DOI] [PubMed] [Google Scholar]
- van Kouwenhove M, Kedde M, Agami R (2011) MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nat Rev Cancer 11: 644–656 [DOI] [PubMed] [Google Scholar]
- Vasudevan S, Steitz JA (2007) AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell 128: 1105–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Chang HY (2011) Molecular mechanisms of long noncoding RNAs. Mol Cell 43: 904–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Arai S, Song X, Reichart D, Du K, Pascual G, Tempst P, Rosenfeld MG, Glass CK, Kurokawa R (2008) Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature 454: 126–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wapinski O, Chang HY (2011) Long noncoding RNAs and human disease. Trends Cell Biol 21: 354–361 [DOI] [PubMed] [Google Scholar]
- Yap KL, Li S, Munoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, Gil J, Walsh MJ, Zhou MM (2010) Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell 38: 662–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JH, Abdelmohsen K, Srikantan S, Yang X, Martindale JL, De S, Huarte M, Zhan M, Becker KG, Gorospe M (2012) LincRNA-p21 suppresses target mRNA translation. Mol Cell 47: 648–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.