Abstract
The RNA chaperone Hfq is a key regulator of the function of small RNAs (sRNAs). Hfq has been shown to facilitate sRNAs binding to target mRNAs and to directly regulate translation through the action of sRNAs. Here, we present evidence that Hfq acts as the repressor of cirA mRNA translation in the absence of sRNA. Hfq binding to cirA prevents translation initiation, which correlates with cirA mRNA instability. In contrast, RyhB pairing to cirA mRNA promotes changes in RNA structure that displace Hfq, thereby allowing efficient translation as well as mRNA stabilization. Because CirA is a receptor for the antibiotic colicin Ia, in addition to acting as an Fur (Ferric Uptake Regulator)-regulated siderophore transporter, translational activation of cirA mRNA by RyhB promotes colicin sensitivity under conditions of iron starvation. Altogether, these results indicate that Fur and RyhB modulate an unexpected feed-forward loop mechanism related to iron physiology and colicin sensitivity.
Keywords: cirA , colicin, Hfq, sRNA, translation activation
Introduction
To successfully occupy their specific niche, bacteria have developed strategies to prevent growth of competitors. One of these strategies is the production of bacteriocins that are protein toxins produced by most bacteria to compete with similar or closely related strains (Riley and Wertz, 2002). Colicins are E. coli-specific bacteriocins released in the environment to kill other E. coli strains. They have been shown to play key roles in promoting diversity and coexistence of bacterial populations in the mammalian colon (Kerr et al, 2002; Kirkup and Riley, 2004). Colicins are divided into two subgroups according to the energy-transducing system they use to invade targeted cells. Whereas group A colicins use the Tol system, group B colicins use the Ton system (Lazdunski et al, 1998; Braun et al, 2002). Colicins exert their activity through a variety of mechanisms ranging from pore formation in the inner membrane to nuclease activity against DNA or RNA (Kleanthous, 2010).
Colicin gene clusters are generally carried on a plasmid and typically encode an immunity protein, which is expressed constitutively to protect the cell from its own colicin attack. The cluster also encodes a lysis protein that allows the release of the colicin in the environment through cell lysis (Riley, 1993a, 1993b). Once released, colicins bind with high affinity to outer membrane receptors of the target cells. Proteins recognized by colicins range from vitamin transporter to siderophore receptors (e.g., CirA, FepA, and FhuA).
One of the most extensively studied pore-forming colicin is colicin Ia, which targets the outer membrane protein (OMP) CirA (Buchanan et al, 2007; Jakes and Finkelstein, 2010). Colicin Ia is a 69-kDa protein that inserts into the inner membrane of the target cell to form a channel responsible for cell death (Wiener et al, 1997). CirA is a TonB-dependent transporter involved in the uptake of ferric iron (Fe3+) complexed with catechol siderophores such as dihydroxybenzoate (DHB) and dihydroxybenzoyl serine (DHBS), which are respectively precursor and breakdown product of the siderophore enterobactin (Hantke, 1990). As for most genes involved in iron uptake, cirA transcription is repressed by the Fur (Ferric Uptake Regulator) protein bound to ferrous iron (Fe2+) (Griggs et al, 1987). Moreover, two redundant small RNAs (sRNAs), OmrA and OmrB, have also been shown to repress cirA translation under conditions of high osmolarity (Guillier and Gottesman, 2006, 2008).
Bacterial sRNAs are key regulators of cellular functions by modulating gene expression in response to various environmental cues (Gottesman and Storz, 2010). These sRNAs regulate target mRNAs by direct base pairing to positively or negatively affect their translation and stability (Storz et al, 2011). In most cases, sRNAs require the RNA chaperone Hfq for optimal regulation by promoting sRNA-mRNA pairing and by stabilizing some sRNAs in vivo (Vogel and Luisi, 2011; Andrade et al, 2012). Hfq can also act as a translational repressor by competing directly with initiating 30S ribosomal subunit for accessibility to the ribosome binding site (RBS) (Vytvytska et al, 2000; Desnoyers and Massé, 2012).
The sRNA RyhB, which regulates iron homeostasis, is one of the most studied Hfq-dependent sRNAs. Under iron-rich conditions, Fe2+-Fur represses ryhB transcription. In contrast, during iron starvation conditions, Fur becomes inactive and relieves repression of ryhB (Massé and Gottesman, 2002; Salvail and Massé, 2011). Under these conditions, RyhB directly regulates ∼20 different mRNAs encoding iron-using proteins. By binding to those mRNAs, RyhB shuts down translation and stimulates their rapid degradation through the action of RNase E (Massé et al, 2003; Massé et al, 2005). RyhB also promotes siderophore production through repression of cysE mRNA that encodes a serine acetyltransferase, which results in an increased flux of serine into enterobactin production (Salvail et al, 2010).
In addition, RyhB can act as a gene activator. Indeed, RyhB activates translation of the Fur-independent shiA mRNA that encodes a transporter of shikimate, an intermediate in the synthesis of enterobactin (Prévost et al, 2007). Hfq binds the 5′-untranslated region (UTR) of shiA and potentially promotes the formation of an inhibitory structure that sequesters the translation initiation region (TIR) and limits translation. However, under conditions of iron starvation, RyhB base pairs with shiA mRNA to disrupt the inhibitory structure, thereby favouring translation and transcript stabilization. This mechanism is reminiscent of similar cases of translational activation by sRNAs such as activation of rpoS by DsrA (Sledjeski et al, 1996; McCullen et al, 2010) and activation of glmS mRNA by GlmZ sRNA (Urban and Vogel, 2008).
In this work, we present evidence that RyhB is a novel regulator of cirA expression. We showed that RyhB expression was essential for CirA synthesis during iron starvation. Our data further suggested that in the absence of RyhB, Hfq repressed cirA mRNA translation, thereby causing rapid transcript turnover through the action of RNase E and low accumulation of CirA. RyhB pairing to cirA mRNA activated its translation and prevented its destabilization by RNase E. The resulting increased levels of CirA transporter made the cells sensitive to the bactericidal action of colicin Ia. This new RyhB-mediated regulation was unexpected considering that cirA is, to our knowledge, the first Fur-regulated gene to require post-transcriptional activation by RyhB to be expressed during iron starvation. The Fur-RyhB-cirA regulatory circuit forms a coherent feed-forward loop (Fur represses cirA and ryhB while RyhB activates cirA), predicted to confer altered regulatory dynamics in comparison to direct regulation by Fur (Mangan and Alon, 2003; Beisel and Storz, 2010). In addition to its role as a gene silencer, our results confirmed the role of RyhB in gene activation.
Results
RyhB expression is essential for CirA production
Previous microarray results have shown that RyhB expression resulted in a 3.3-fold increase in cirA transcript levels in a Δfur ΔryhB background (Massé et al, 2005). To extend these results, we used quantitative real-time PCR (qRT–PCR) to monitor cirA mRNA levels after a 20-min expression of RyhB in ΔryhB and Δfur ΔryhB backgrounds, as in the case of the microarray study. Results showed that cirA transcript levels increased 4.7-fold upon RyhB expression in the Δfur ΔryhB background (Figure 1A). This increase was higher than the previously characterized RyhB-positive target shiA (2.3-fold increase) (Prévost et al, 2007). Intriguingly, RyhB expression in a ΔryhB background resulted in a 3.3-fold decrease in cirA transcript levels. We have previously reported that RyhB expression under these conditions promoted an increase in free intracellular iron concentration (Jacques et al, 2006). Thus, the decrease in cirA mRNA levels observed here may result from Fe2+-Fur repression, as observed in the case of Fur-regulated genes (Massé et al, 2005). In contrast, RyhB still upregulated the expression of the Fur-independent shiA gene in the ΔryhB background (Figure 1A).
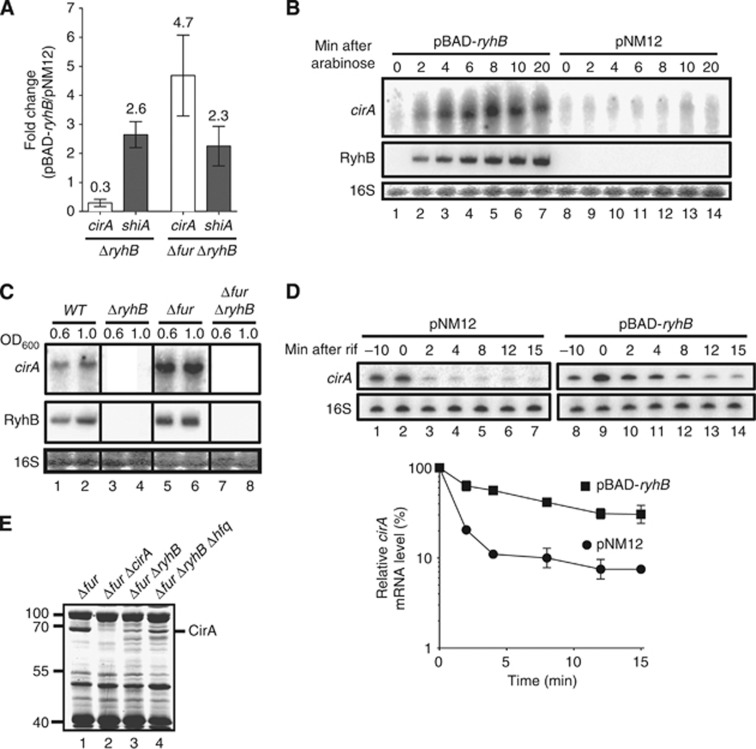
Figure 1.
RyhB expression is essential for CirA production. (A) Quantitative RT-PCR (qRT-PCR) analysis of RyhB effect on cirA and shiA mRNA levels. Strains EM1455 (ΔryhB) and EM1493 (Δfur ΔryhB), each carrying either pBAD-ryhB or control plasmid pNM12, were grown in LB medium. Arabinose was added (0.1%, total concentration) to an OD600 of 0.5, followed by total RNA extracted after 20 min. Primers complementary to cirA and shiA open reading frames (ORFs) were used for qRT–PCR assays. Mean and standard deviation (s.d.) values of triplicate samples are shown. (B) Northern blot analysis of RyhB effect on cirA mRNA levels. Strain EM1493 (Δfur ΔryhB) carrying pBAD-ryhB or control plasmid pNM12 was grown in LB medium. Arabinose was added (0.1%, total concentration) to an OD600 of 0.5 and total RNA was extracted at the indicated time points. A probe complementary to cirA ORF was used. (C) Northern blot analysis of RyhB and Fur effects on cirA mRNA levels in cells grown under iron-poor conditions. Strains EM1055 (WT), EM1238 (ΔryhB), KP392 (Δfur) and KP393 (Δfur ΔryhB) were grown in M63 iron-free glucose minimal medium and total RNA was extracted at the indicated values of OD600. A probe complementary to cirA ORF was used. (D) Primer extension analysis of RyhB effect on cirA mRNA stability. Strain EM1493 (Δfur ΔryhB) carrying pBAD-ryhB or control plasmid pNM12 was grown in LB medium and arabinose was added (0.1%, total concentration) to an OD600 of 0.5 (−10 time point). Rifampicin (250 μg/ml, final concentration) was added 10 min after the addition of arabinose (0 time point) and total RNA was extracted at the indicated time points. Primers complementary to cirA (EM1408) and 16S rRNA (EM345) were used to perform primer extension reactions on total RNA samples. Primer extension signals of two independent experiments were quantified by densitometry and normalized to time zero (0) for both pNM12 and pBAD-ryhB. Mean and s.d. values of duplicate experiments are shown. (E) Coomassie-stained SDS gel of outer membrane proteins (OMPs) extracts from KP392 (Δfur), HS437 (Δfur ΔcirA), KP393 (Δfur ΔryhB) and MPC253 (Δfur ΔryhB Δhfq) cells grown in LB medium at an OD600 of 1.0. Molecular sizes of co-migrating proteins (L) are indicated at the left in kDa.
Source data for this figure is available on the online supplementary information page.
Full-length cirA mRNA expression was monitored by northern blot following pulse expression of RyhB in the Δfur ΔryhB background. This showed that cirA transcript accumulation following RyhB expression was swift with maximal transcript accumulation following 8 min of sRNA induction (Figure 1B, lane 5). However, despite high cirA promoter activity in the Δfur ΔryhB background (Supplementary Figure S1A, see pNM12), cirA transcript poorly accumulated in the absence of RyhB (Figure 1B, lanes 8–14). Moreover, RyhB had no significant effect on cirA promoter (Supplementary Figure S1A, pNM12 and pBAD-ryhB), suggesting that RyhB regulated cirA post-transcriptionally.
Since RyhB is naturally expressed during iron starvation, we next studied the endogeneous effect of RyhB on cirA expression in iron-free minimal medium. Results showed that cirA mRNA levels in WT cells correlated with RyhB levels (Figure 1C, lanes 1 and 2). In contrast, ΔryhB cells did not accumulate cirA transcripts at any time (Figure 1C, lanes 3 and 4), despite promoter activity similar to WT cells (Supplementary Figure S1B, WT and ΔryhB). Inactivation of fur resulted in increased levels of cirA mRNA as compared to WT cells (Figure 1C, lanes 1 and 2, and lanes 5 and 6). This behaviour probably resulted from increased cirA promoter activity (Supplementary Figure S1B, WT and Δfur) due to the absence of Fe2+-Fur repression. The absence of Fur repression also resulted in higher levels of RyhB in Δfur cells as compared to WT cells (Figure 1C, lanes 1 and 2, and lanes 5 and 6) which may also contribute to increased levels of cirA mRNA. Strikingly, inactivation of ryhB in a Δfur background resulted in a severe decrease in cirA transcript levels (Figure 1C, compare lanes 5 and 6 with lanes 7 and 8), despite an increase in cirA promoter activity (Supplementary Figure S1B, Δfur and Δfur ΔryhB, OD600 of 0.6). Taken together, these results suggested that RyhB expression was essential to promote cirA transcript accumulation upon iron starvation.
Because our data indicated that RyhB was essential for cirA expression, we hypothesized that RyhB could stabilize cirA mRNA. To address this, we determined the half-life of cirA mRNA under conditions where RyhB was expressed (pBAD-ryhB) or not (pNM12). Because full-length cirA accumulated poorly in the Δfur ΔryhB background in LB medium (Figure 1B), we performed a primer extension using a reverse primer complementary to the beginning of cirA coding sequence. This approach was found to be sensitive enough to observe cirA mRNA expression in the absence of RyhB (Figure 1D, lane 2). While significant amounts of cirA transcripts were detected after 8 min in the presence of RyhB (Figure 1D, lane 12), most of the cirA mRNA disappeared after 2 min in the absence of RyhB (Figure 1D, lane 3). Densitometry analysis revealed that cirA mRNA had a half-life of 5 min when RyhB was expressed (pBAD-ryhB) but of less than 2 min in the absence of RyhB (Figure 1D).
Since RyhB promoted cirA mRNA accumulation, we next sought to determine the effect on CirA protein expression. Because CirA is an OMP, we compared the OMP profiles of Δfur, Δfur ΔcirA, Δfur ΔryhB, and Δfur ΔryhB Δhfq cells grown in LB media. Comparison of the profiles between Δfur and Δfur ΔryhB cells revealed the presence of a protein of ∼65 kDa for which the levels of expression were very low in Δfur ΔryhB cells as compared to Δfur cells (Figure 1E, lanes 1 and 3). Intriguingly, the absence of Hfq in Δfur ΔryhB Δhfq cells appears to improve the expression of the 65-kDa protein as compared to Δfur ΔryhB cells (Figure 1E, compare lanes 3 and 4). Because this protein was absent in Δfur ΔcirA cells (Figure 1E, lane 2) and since the estimated molecular weight of CirA is between 54 and 74 kDa (Buchanan et al, 2007), we assumed this protein to be CirA. The bulk of these results suggested that RyhB was essential for CirA production.
RyhB directly activates cirA in vivo
Data reported above (Figure 1) suggested that RyhB activated cirA through direct pairing with its mRNA. Bioinformatic analysis using the TargetRNA software (Tjaden et al, 2006) suggested potential pairing of RyhB with cirA 5′-UTR in the region −58 to −41 relative to the AUG start codon (Figures 2 and 3A). We then proceeded to investigate this potential RyhB pairing site by chemical in vitro analysis. A 5′-end-labelled cirA transcript containing the whole 5′-UTR and 79 additional nucleotides (nt) from the coding region was incubated in the presence or absence of a 20-fold molar excess of unlabelled RyhB, followed by a partial digestion by lead acetate (PbAc, cleaves unpaired residues). Results showed that the addition of RyhB (R) protected from cleavage (Figure 3B, lanes 6 and 7) in the region −42 to −51 that corresponded to the predicted RyhB pairing core (Figures 2 and 3A).
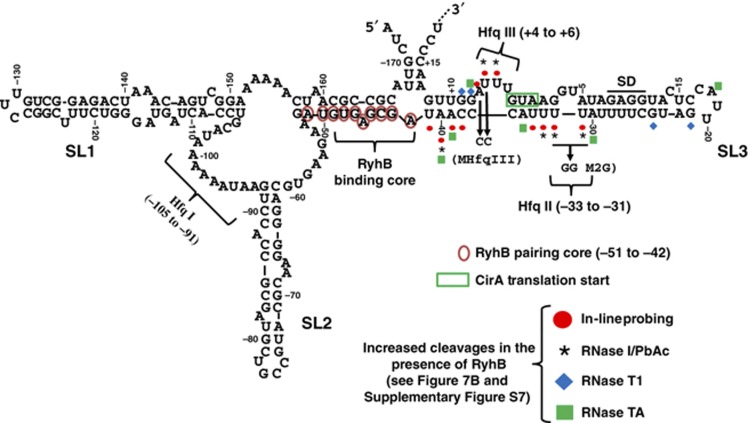
Figure 2.
The 5′-UTR of cirA mRNA potentially forms an inhibitory structure preventing translation initiation. RNA-fold prediction of cirA 5′-UTR secondary structure (nucleotides −173 to +18 relative to the AUG start codon). The RyhB pairing core, the Hfq binding sites, the M2G mutation of Hfq binding site II and the MHfqIII mutation of Hfq binding site III are indicated. Probing results presented in Figure 7B and Supplementary Figure S7 are summarized here. Stem-loop structures are numbered (SL1, SL2, and SL3).
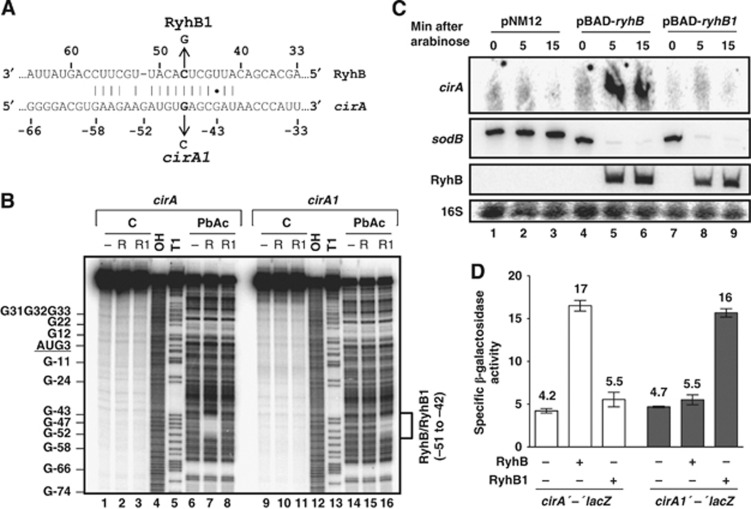
Figure 3.
RyhB base pairs with cirA mRNA in vivo. (A) TargetRNA prediction of pairing between RyhB and cirA mRNA. The cirA1 and RyhB1 mutations used in (B–D) are indicated. (B) Lead acetate (PbAc) probing of RyhB pairing with cirA mRNA. 5′-end-labelled cirA or cirA1 mRNAs (nucleotides −173 to +78 relative to the AUG start codon) were incubated in the absence or in the presence of RyhB (R) or RyhB1 (R1) before addition of PbAc. C, non-reacted controls; OH, alkaline ladder; T1, RNase T1 ladder (guanine residues). (C) Northern blot analysis of arabinose-induced RyhB1 effect on cirA mRNA levels. Strain EM1493 (Δfur ΔryhB) carrying pBAD-ryhB, pBAD-ryhB1 or control plasmid pNM12 was grown in LB medium. Arabinose (0.1%, total concentration) was added to an OD600 of 0.5 and total RNA was extracted at the indicated time points. Probes complementary to cirA and sodB open reading frames (ORFs) were used. (D) Effect of arabinose-induced RyhB and RyhB1 on cirA′-′lacZ and cirA1′-′lacZ translational fusions in a Δfur ΔryhB background. Cells were grown in LB medium and arabinose (0.1%, total concentration) was added to an OD600 of 0.1. Specific β-galactosidase activity from three independent cultures was then measured 3 h later. Mean and standard deviation (s.d.) values are shown. Empty vector pNM12 was used as a control.
Source data for this figure is available on the online supplementary information page.
To further confirm that RyhB paired directly with cirA mRNA in vivo, we constructed the RyhB1 allele in which nucleotide C47 was exchanged for G47 to decrease the strength of pairing with cirA (see details in Figure 3A). The cleavage assay showed that the addition of RyhB1 (R1) did not protect bases that were determined to be part of the RyhB pairing core (Figure 3B, lanes 6 and 8). These data strongly suggest that RyhB1 was not able to base pair with cirA mRNA in vitro. RyhB1 was then expressed using an arabinose-inducible vector and its effect on cirA mRNA levels was monitored by northern blot analysis. Results showed that in spite of similar levels of accumulation compared to RyhB (Figure 3C, lanes 5 and 6, and lanes 8 and 9), RyhB1 failed to promote cirA expression within 15 min as opposed to RyhB expression that resulted in rapid accumulation of cirA transcripts (Figure 3C, lanes 4–6, and lanes 7–9). However, RyhB1 was still able to repress sodB, an RyhB-repressed gene (Figure 3C, lanes 7–9). This observation suggested that RyhB1 remained functional and that the loss of cirA regulation probably reflected a disruption of target-specific base pairing.
To further characterize RyhB-cirA interaction, we constructed the cirA1 allele in which nucleotide G47 was changed to C47 to restore base pairing with RyhB1 (see Figure 3A for details). Results indicated that RyhB addition did not promote cleavage protection of cirA1 (Figure 3B, lanes 14 and 15), indicating that RyhB was not able to base pair with cirA1 mRNA. As predicted, the compensatory mutation introduced in RyhB1 was able to restore base pairing with cirA1 since the nucleotides of cirA1 that are part of the RyhB pairing core were protected from cleavage (Figure 3B, lane 14 and lane 16). We next performed in vivo covariation mutagenesis using cirA′-′lacZ and cirA1′-′lacZ translational fusions, harbouring region −280 to +33 with respect to the initiation codon. Results indicated that RyhB expression resulted in a four-fold activation of the cirA′-′lacZ translational construct but failed to activate cirA1′-′lacZ fusion while RyhB1 only activated the complementary cirA1′-′lacZ construct without affecting the cirA′-′lacZ construct (Figure 3D). Taken together, these data confirmed that RyhB promoted CirA synthesis through direct pairing with cirA mRNA.
RyhB protects cirA mRNA from RNase E degradation
Since RNase E is known to be the main endonuclease responsible for RNA turnover in E. coli (Belasco, 2010), we explored the possibility that RyhB protected cirA mRNA from RNase E degradation. Two different mutants of RNase E were used and their effect on cirA transcript levels in the presence or absence of RyhB expression was investigated. One mutant (rne131) lacked residues 586–1061 that constitute the C-terminal domain involved in interaction with other components of the RNA degradosome and with Hfq (Kido et al, 1996; Vanzo et al, 1998). Results showed that cirA mRNA levels were low in ΔryhB as in rne131 ΔryhB cells (Supplementary Figure S2A, lanes 2 and 4). These data led us to conclude that the degradation of cirA mRNA in the absence of RyhB did not require interaction of RNase E with the RNA degradosome components or with Hfq. The other mutant (rne-3071) was a thermosensitive allele where RNase E becomes inactive at non-permissive temperature (44°C for 15 min) (McDowall et al, 1993). In these experiments, cirA transcript accumulated at levels similar to WT in the absence of RyhB expression when RNase E was inactivated following a 15-min exposure of the rne-3071 ΔryhB strain at 44°C (Supplementary Figure S2B, lanes 6 and 8). These data indicate that RNase E was responsible for the turnover of cirA mRNA in the absence of RyhB.
RyhB stabilizes cirA mRNA through translational activation
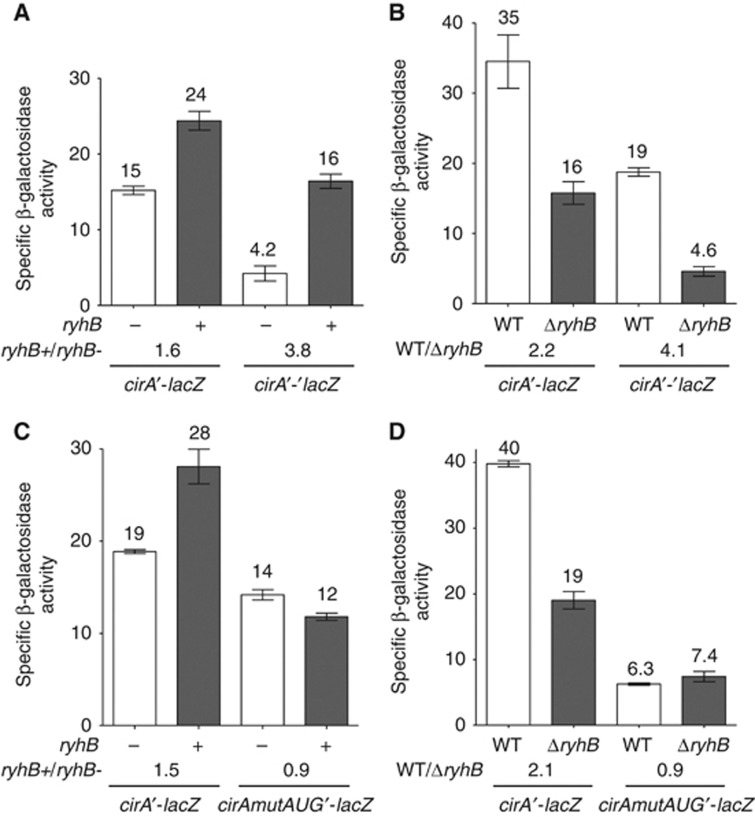
RNase E is considered to be responsible for the rapid decay of translationally impaired mRNAs (Arnold et al, 1998; Deana and Belasco, 2005). We thus tested the possibility that RyhB protected cirA mRNA from RNase E degradation by activating its translation, as reported in the case of shiA mRNA (Prévost et al, 2007). This possibility was investigated by first comparing the effect of RyhB on the activity of cirA′-lacZ transcriptional fusion and cirA′-′lacZ translational fusion. Results demonstrated that RyhB expression resulted in a 1.6-fold activation of transcriptional cirA′-lacZ fusion and a 3.8-fold activation of translational cirA′-′lacZ fusion (Figure 4A). Moreover, we compared the activities of both fusions in WT and ΔryhB cells grown in an iron-free minimal medium. The absence of ryhB resulted in a 2.2-fold decrease in activity for transcriptional cirA′-lacZ fusion and a 4.1-fold decrease in activity for translational cirA′-′lacZ fusion (Figure 4B). The differential effect of RyhB expression on translational fusion as compared to transcriptional fusion suggested that RyhB activated cirA translation.
Figure 4.
RyhB protects cirA mRNA from degradation by activating its translation. (A) Effect of arabinose-induced RyhB on cirA′-lacZ transcriptional fusion and on cirA′-′lacZ translational fusion in a Δfur ΔryhB background. Cells were grown in LB medium and 0.1% arabinose was added at an OD600 of 0.1. Specific β-galactosidase activity from three independent cultures was then measured 3 h later. Mean and standard deviation (s.d.) values are shown. Empty vector pNM12 was used as a control. (B) β-Galactosidase assay of cirA′-lacZ transcriptional fusion and cirA′-′lacZ translational fusion in WT and ΔryhB cells grown in M63 iron-free glucose minimal medium at an OD600 of 0.6. Specific β-galactosidase activity from three independent cultures was measured. Mean and s.d. values are shown. (C) Effect of arabinose-induced RyhB on cirAmutAUG′-lacZ transcriptional fusion in a Δfur ΔryhB background. Cells were grown in LB medium and 0.1% arabinose was added at an OD600 of 0.1. Specific β-galactosidase activity from three independent cultures was then measured 3 h later. Mean and s.d. values are shown. Empty vector pNM12 was used as a control. (D) β-galactosidase assay of cirAmutAUG′-lacZ transcriptional fusion in WT and ΔryhB cells grown in M63 iron-free glucose minimal medium at an OD600 of 0.6. Specific β-galactosidase activity from three independent cultures was measured. Mean and s.d. values are shown.
We next constructed the cirAmutAUG′-lacZ transcriptional fusion in which the cirA initiation codon AUG was changed to CUG to prevent translation initiation. As expected, β-galactosidase activity was not detected in cirAmutAUG′-′lacZ translational fusion (data not shown). RyhB expression did not activate cirAmutAUG′-lacZ transcriptional fusion as opposed to a 1.5-fold activation observed in the case of cirA′-lacZ fusion (Figure 4C). Furthermore, when cells were grown in minimal iron-free medium, there was no effect on cirAmutAUG′-lacZ transcriptional fusion whether RyhB was present or not (WT/ΔryhB=0.9) as opposed to a 2.1-fold increase in cirA′-lacZ activity observed in WT cells as compared to ΔryhB cells (Figure 4D). cirAmutAUG′-lacZ transcriptional fusion activity was six-fold lower than cirA′-lacZ fusion. We attributed the decrease in β-galactosidase activity to the instability of untranslated cirAmutAUG mRNA. Our results suggested that RyhB pairing with cirA mRNA increased translation, protected the ORF from RNase E degradation and therefore resulted in accumulation of full-length cirA mRNA.
Hfq represses cirA translation
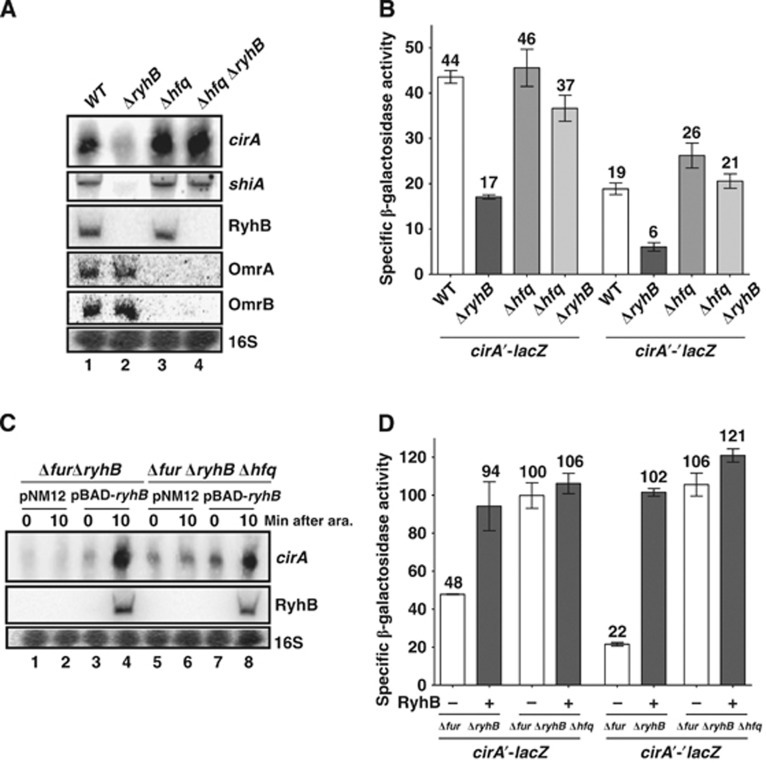
Previously, we have reported that Hfq repressed shiA mRNA translation when RyhB was not expressed (Prévost et al, 2007). We thus investigated whether Hfq played a similar role in cirA activation by RyhB. Northern blots were used to monitor cirA transcript levels in Δhfq and Δhfq ΔryhB cells. Results revealed that hfq inactivation in a ΔryhB background fully restored cirA expression (Figure 5A, lanes 2 and 4), as observed in the case of shiA mRNA. In agreement with previous reports (Wassarman et al, 2001; Holmqvist et al, 2010), we observed that Hfq had an effect on OmrA/B expression as revealed by the fact that both sRNAs were barely detectable in Δhfq and Δhfq ΔryhB cells in comparison with WT and ΔryhB cells (Figure 5A, lanes 1 and 2, and lanes 3 and 4). With respect to this observation, the WT levels of cirA mRNA in Δhfq ΔryhB cells could be attributed to decreased OmrA/B repression. However, this is unlikely because omrAB inactivation in a ΔryhB background failed to increase levels of cirA mRNA (Supplementary Figure S3, lanes 2 and 4).
Figure 5.
Hfq represses cirA mRNA translation. (A) Northern blot analysis of Hfq effect on cirA mRNA levels. Strains EM1055 (WT), EM1238 (ΔryhB), EM1265 (Δhfq) and KP111 (Δhfq ΔryhB) were grown in M63 iron-free glucose minimal medium at an OD600 of 0.6 and total RNA was extracted. Probes complementary to cirA and shiA open reading frames (ORFs) were used. (B) β-Galactosidase assay of cirA′-lacZ transcriptional fusion and cirA′-′lacZ translational fusion in Δhfq and Δhfq ΔryhB cells grown in M63 iron-free glucose minimal medium at an OD600 of 0.6. Specific β-galactosidase activity from three independent cultures was measured. Mean and standard deviation (s.d.) values are shown. (C) Northern blot analysis of arabinose-induced RyhB effect on cirA mRNA levels in Δfur ΔryhB Δhfq cells. Strains HS506 (Δfur ΔryhB) and HS518 (Δfur ΔryhB Δhfq), each carrying pBAD-ryhB or control plasmid pNM12, were grown in M63 iron-free glycerol minimal medium. Arabinose (0.1%) was added at an OD600 of 0.3 and total RNA was extracted 10 min later. A probe complementary to cirA ORF was used. (D) Effect of arabinose-induced RyhB on cirA′-lacZ transcriptional fusion and cirA′-′lacZ translational fusion in a Δfur ΔryhB Δhfq background. Cells were grown in M63 iron-free glycerol minimal medium and 0.1% arabinose was added at an OD600 of 0.1. Specific β-galactosidase activity from three independent cultures was then measured 3 h later. Mean and s.d. values are shown. Empty vector pNM12 was used as a control.
Source data for this figure is available on the online supplementary information page.
We next monitored the activity of transcriptional cirA′-lacZ and translational cirA′-′lacZ fusions in Δhfq ΔryhB cells in comparison to ΔryhB cells. Results showed that inactivation of hfq in ΔryhB cells restored WT levels of activity in both cases, suggesting that RyhB activated cirA mRNA translation by counteracting Hfq repression (Figure 5B). The greater effect of hfq inactivation on cirA′-′lacZ translational fusion (3.5-fold increase) as opposed to cirA′-lacZ transcriptional fusion (2.2-fold increase) suggested that cirA repression by Hfq occurred at the translational level.
The possibility that RyhB activated cirA solely by antagonizing Hfq repression would be incompatible with its capacity to further activate cirA in the absence of Hfq. However, pulse expression of RyhB in the Δfur ΔryhB Δhfq background resulted in a 1.8-fold increase in cirA mRNA levels within 10 min, indicating that RyhB was able to activate cirA in the absence of Hfq (Figure 5C, lanes 5 and 6, and lanes 7 and 8). To determine whether this increase in cirA mRNA resulted from translational activation, we monitored RyhB effect on cirA′-lacZ transcriptional and cirA′-′lacZ translational fusions. Results demonstrated that inactivation of hfq in the Δfur ΔryhB background resulted in a 2.1-fold increase in cirA′-lacZ transcriptional activity and in a 4.8-fold increase in cirA′-′lacZ translational activity (Figure 5D). These results were similar to the effect of RyhB on both fusions in the Δfur ΔryhB background (2-fold activation for cirA′-lacZ transcriptional fusion and 4.6-fold activation for cirA′-′lacZ translational fusion), in agreement with previous results (Figure 5B). These data confirmed that RyhB expression counteracted the repressive effect of Hfq on cirA mRNA. However, RyhB expression in Δfur ΔryhB Δhfq cells did not result in a significant increase in cirA′-lacZ transcriptional activity but a small but significant 1.1-fold increase in cirA′-′lacZ translational activity (Figure 5D). These results contrasted with those of northern blot experiments (Figure 5C) where a 1.8-fold increase in cirA transcript levels was observed following RyhB expression. Together, these results suggested that Hfq destabilized cirA mRNA through translational repression and that RyhB expression abolished this repression.
Hfq inhibits 30S ribosomal subunit binding to cirA mRNA upon translation initiation
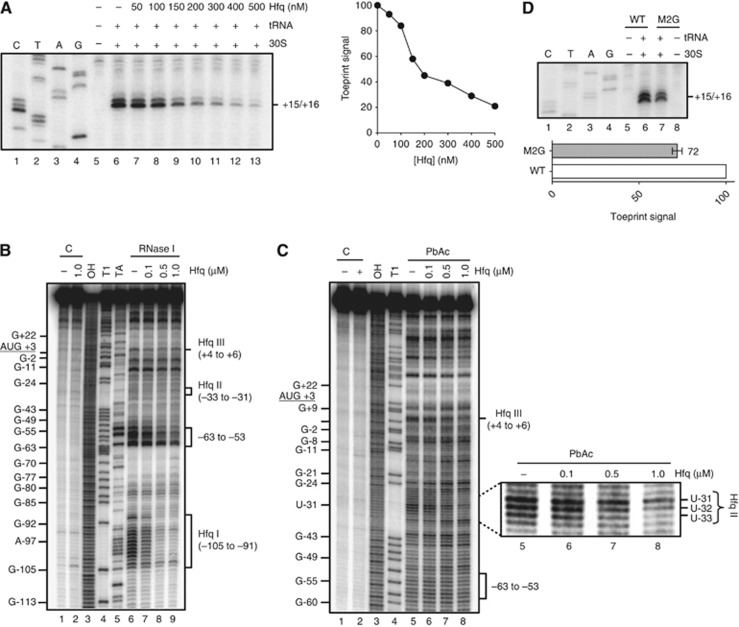
We next sought to decipher the mechanism by which Hfq repressed cirA mRNA translation. Previous studies had unveiled the capacity of Hfq to inhibit 30S translation initiation complex formation on some mRNAs through binding to AU-rich single-stranded regions located in the vicinity of the TIR and acting as translational enhancers (Vytvytska et al, 2000; Desnoyers and Massé, 2012). To determine whether Hfq could repress cirA translation through this mechanism, we first performed toeprinting experiments to study the effect of Hfq on 30S binding to cirA mRNA. We used the Hfq-independent lpp transcript (Vytvytska et al, 1998, 2000) as a control (Supplementary Figure S4A and B). Results indicated that addition of purified 30S ribosomal subunits and tRNAfmet to cirA mRNA (0.2 μM) resulted in a strong block of reverse transcription at positions +15/+16 downstream of the AUG start codon (Figure 6A). These observations were consistent with the formation of a translation initiation complex on cirA mRNA. Addition of increasing amounts of purified Hfq (200 nM) resulted in a 2.2-fold inhibition of initiation complex formation (Hfq:cirA molar ratio, 1:1) and in near complete inhibition at high concentration (500 nM) of Hfq (Hfq:cirA molar ratio, 2.5:1). These results suggested that Hfq binding to cirA mRNA may be sufficient to prevent translation initiation in vivo. As expected, higher concentrations of Hfq were required to prevent ribosome binding to lpp mRNA (Supplementary Figure S4A).
Figure 6.
Hfq represses 30S ribosomal subunit binding to cirA mRNA upon translation initiation. (A) Toeprint analysis of Hfq effect on 30S ribosomal subunit binding to cirA mRNA (200 nM) upon translation initiation. Unlabelled cirA mRNA was incubated in the presence of the indicated amounts of purified Hfq protein. Positions +15/+16 relative to cirA AUG start codon are indicated. CTAG refers to sequencing ladders. Toeprint signals were quantified by densitometry and data are reported in arbitrary units. (B) RNase I probing of Hfq binding sites on cirA mRNA. 5′-end-labelled cirA mRNA (nucleotides −173 to +78 relative to the AUG start codon) was incubated in the presence of the indicated amounts of purified Hfq protein before addition of RNase I. Hfq I, Hfq II, and Hfq III refer to Hfq binding sites I, II, and III (see Figure 2). C, non-reacted controls; OH, alkaline ladder; T1, RNase T1 ladder (guanine residues); TA, RNase TA ladder (adenine residues). (C) PbAc probing of Hfq binding sites on cirA mRNA. 5′-end-labelled cirA mRNA (nucleotides −173 to +78 relative to the AUG start codon) was incubated in the presence of the indicated amounts of purified Hfq protein before addition of PbAc (5 mM). Hfq II and Hfq III refer to Hfq binding sites II and III (see Figure 2). Some guanine and adenine positions are given for clarity. C, non-reacted controls; OH, alkaline ladder; T1, RNase T1 ladder (guanine residues). The region corresponding to Hfq binding site II (Hfq II) is enlarged to illustrate protection from PbAc cleavage of nucleotides −33 to −31 following addition of Hfq. (D) Toeprint analysis of 30S ribosomal subunit binding to cirAM2G mRNA. Unlabelled cirA and cirAM2G mRNAs (0.2 μM final) were incubated in the presence of 30S ribosomal unit and tRNAfmet. Positions +15/+16 relative to cirA and cirAM2G AUG start codons are indicated. CTAG refers to sequencing ladders. Toeprint signals of two independent experiments were quantified by densitometry and normalized to WT toeprint signal (lane 6). Data are reported in arbitrary units. Mean and standard deviation (s.d.) values of duplicate experiments are shown.
Source data for this figure is available on the online supplementary information page.
We next performed enzymatic and chemical probing using RNase I (cleaves unpaired nucleotides) and PbAc to identify potential Hfq binding sites in the 5′-UTR of cirA mRNA. Addition of increasing amounts of Hfq to 5′-end-labelled cirA mRNA resulted in protection of nucleotides −105 to −91 from RNase I cleavage (Figure 6B, Hfq I site, lane 6 and lanes 7–9). This region is AU-rich and single-stranded, according to its susceptibility to RNase I cleavage (Figure 6B, lane 6, Hfq I site). These properties were consistent with previous descriptions of Hfq binding sites (Lorenz et al, 2010; Balbontín et al, 2010). Protection of nucleotides −63 to −53 from RNase I cleavage was also observed at high concentrations of Hfq (Figure 6B, lanes 6 and lanes 7–9) that may result from Hfq binding to site I.
Interestingly, incubation of cirA with Hfq resulted in a weak but noticeable protection of nucleotides +4 to +6 from RNase I cleavage (Figure 6B, Hfq III site, lanes 6–9). These nucleotides are also protected from PbAc cleavage in the presence of Hfq (Figure 6C, Hfq III site, lanes 5–8). This region is U-rich and single-stranded according to secondary structure prediction (Figure 2) and to the susceptibility of nucleotides +4 to +6 to RNase I (Figure 6B, Hfq III site, lane 6) and PbAc (Figure 6C, Hfq III site, lane 5) cleavages in the absence of Hfq. Binding of this U stretch by Hfq could potentially hamper 30S binding to the AUG start codon of cirA mRNA upon translation initiation (Figure 6A).
Nucleotides −33 to −31, which are part of an AU-rich region, were also protected from PbAc cleavage upon addition of Hfq (Figure 6C, Hfq II site, lanes 5–8). These nucleotides were predicted to be part of a stem-loop structure (Figure 2, SL3, nucleotides A-40 to G+12), consistent with their protection from RNase I cleavage (Figure 6B, Hfq II site, lane 6). These nucleotides (−33 to −31) are also part of an AU-rich region (nucleotides A-35 to U-26) located a few nucleotides upstream of the Shine-Dalgarno sequence, suggesting that this region acts as a translational enhancer. To investigate this, we constructed a cirAM2G mRNA in which nucleotides U-32 and U-31 were replaced by two guanine residues (Figure 2) and 30S binding was monitored by toeprinting. Results showed that there was ∼30% less binding of 30S to cirAM2G mRNA than to cirA mRNA (Figure 6D, lanes 6 and 7), suggesting an important role of region A-35 to U-26 in translation initiation.
Despite the fact that Hfq I site was highly AU-rich (Figure 2), we did not expect this region to act as a translational enhancer. The Hfq I site is too far upstream of the Shine-Dalgarno sequence in comparison with translational enhancer elements previously described (Hook-Barnard et al, 2007; Desnoyers and Massé, 2012). Indeed, replacement of nucleotides A-100 and A-98 for cytosine residues (cirAML, Supplementary Figure S5A) did not result in reduced formation of 30S translation initiation complex (Supplementary Figure S5B, lanes 7 and 8). Taken together, our results suggested that Hfq may repress cirA mRNA translation by preventing 30S initiation complex formation through binding in the vicinity of the AUG start codon (Hfq III site) and/or to a translational enhancer element (Hfq II site) located a few nucleotides upstream of the Shine-Dalgarno sequence.
RyhB promotes 30S ribosomal subunit binding to cirA mRNA in the presence of Hfq
Data presented above (Figure 4) strongly suggested that RyhB stabilized cirA mRNA through translational activation. We hypothesized that RyhB pairing with cirA 5′-UTR would result in increased 30S binding to the AUG start codon upon translation initiation. Unexpectedly, addition of increasing amounts of RyhB to cirA mRNA (0.2 μM) resulted in a 12% inhibition of initiation complex formation in the presence of RyhB (0.4 μM, RyhB:cirA molar ratio, 2:1) and a 30% inhibition with a higher concentration (0.8 μM) of RyhB (RyhB:cirA molar ratio, 4:1) (Supplementary Figure S6, lane 6 and lanes 8–10). This negative effect of RyhB on 30S binding to cirA mRNA appeared to be pairing specific since addition of RyhB1 did not repress initiation complex formation (Supplementary Figure S6, lane 6 and lanes 12 and 14).
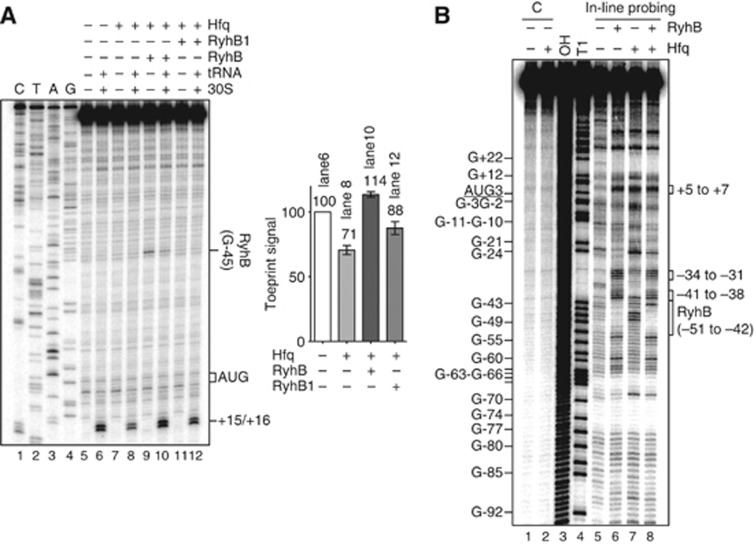
As a follow-up to results presented above (Figure 5B) that suggested that RyhB activated cirA by counteracting Hfq translational repression, we tested the effect of RyhB on 30S binding to cirA mRNA in the presence of Hfq. Addition of Hfq (150 nM, Hfq:cirA molar ratio, 0.75:1) resulted in 30% inhibition of 30S binding to cirA mRNA (Figure 7A, lanes 6 and 8). Consistent with the in vivo data, addition of RyhB (150 nM) in the presence of Hfq (Hfq:RyhB:cirA molar ratio, 0.75:0.75:1) resulted in a 60% increase in the toeprint signal (Figure 7A, lanes 8 and 10), whereas addition of the same amount of RyhB1 only results in a 25% increase in 30S binding (Figure 7A, lanes 8 and 12). The weak RyhB1-dependent activation may be attributable to Hfq displacement from cirA mRNA via RyhB1 poly(U) tail, which has previously been shown to bind Hfq with high affinity (Otaka et al, 2011). We could not exclude the possibility that this titration effect may contribute to RyhB-positive effect on 30S binding to cirA mRNA (Figure 7A, lanes 8 and 10). However, the fact that RyhB induced stronger toeprint signals than those observed upon addition of RyhB1 (Figure 7A, lanes 10 and 12), strongly suggested that the RyhB-dependent activation was base pairing specific. Moreover, RyhB did not affect ribosome binding to lpp mRNA (negative control) in the presence of Hfq (Supplementary Figure S4B, compare lanes 8 and 10). Taken together, toeprinting data supported the interpretation that RyhB directly promoted translational initiation complex formation on cirA mRNA in the presence of Hfq.
Figure 7.
RyhB promotes 30S ribosomal subunit binding to cirA mRNA in the presence of Hfq. (A) Toeprint analysis of RyhB and RyhB1 effects on 30S ribosomal subunit binding to cirA mRNA in the presence of Hfq upon translation initiation. Unlabelled cirA mRNA (0.2 μM final) was incubated in the presence of Hfq (0.15 μM) and RyhB (0.15 μM) or RyhB1 (0.15 μM). Positions +15/+16 relative to cirA AUG start codon are indicated. CTAG refers to sequencing ladders. Toeprint signals of two independent experiments were quantified by densitometry and normalized to the toeprint signal in lane 6. Data are reported in arbitrary units. Mean and standard deviation (s.d.) values of duplicate experiments are shown. (B) In-line probing analysis of Hfq and RyhB effects on cirA mRNA. 5′-end-labelled cirA mRNA (nucleotides −173 to +78 relative to the AUG start codon) was incubated for 40 h in the presence of Hfq (0.3 μM) and RyhB (1 μM). Some guanine positions are illustrated for clarity. C, non-reacted controls; OH, alkaline ladder; T1, RNase T1 ladder (guanine residues).
Source data for this figure is available on the online supplementary information page.
We next investigated the mechanism by which RyhB stimulated ribosome binding to cirA mRNA in the presence of Hfq upon translation initiation. Previous studies of sRNA-mediated translational activation (e.g., RyhB-shiA, DsrA-rpoS, and GlmZ-glmS) have shown that sRNA pairing with its target mRNA usually results in the disruption of an inhibitory structure that sequesters the Shine-Dalgarno sequence (Fröhlich and Vogel, 2009). We performed in-line probing to determine whether cirA regulation by RyhB and Hfq occurred through a similar mechanism. This technique exploits the natural susceptibility of unpaired and unstructured nucleotides of RNA to spontaneous cleavage in solution (Wakeman and Winkler, 2009). Results showed that RyhB pairing with 5′-end-labelled cirA mRNA in the presence of Hfq (confirmed by the cleavage protection of nucleotides −51 to −42) (Figure 7B, lanes 7 and 8) resulted in increased cleavage of nucleotides −41 to −38 and −34 to −31 (Figure 7B, lanes 7 and 8). Notably, these secondary structure changes are also observed when RyhB is added in the absence of Hfq (compare lanes 5 and 6), suggesting that they do not require Hfq to occur. These RyhB effects on cirA mRNA secondary structure were further confirmed by enzymatic and chemical probing (Supplementary Figure S7). Altogether, these structural changes showed that RyhB pairing with cirA mRNA in the presence of Hfq promoted the partial unfolding of SL3 and that resulted in increased accessibility of region −35 to −26, predicted to be bound by Hfq (Figure 2, Hfq II; Supplementary Figure 6B, C) and to act as a potential translational enhancer (Figure 6D). Based on the bulk of these results, it was predicted that this region was more accessible to 30S subunit upon translation initiation.
The addition of RyhB in the absence of Hfq resulted in cleavage induction of nucleotides +5 to +7 (Figure 7B, lanes 5 and 6), which was previously characterized as a Hfq binding site (Figure 2, Hfq III). Unexpectedly, cleavage of this region was observed when cirA mRNA was incubated with Hfq alone (Figure 7B, lanes 5 and 7). This induced cleavage may result from residual RNase activity in the Hfq preparation (Supplementary Figure S8, lanes 1 and 2). However, it is more probable that the high susceptibility of region +5 to +7 to cleavage upon incubation with RyhB and Hfq (Figure 7B, lane 8) may be in large part attributable to structural changes specific to RyhB pairing, as RyhB alone promoted the same cleavage intensity in this region (Figure 7B, lanes 6 and 8). These data then suggested that RyhB pairing with cirA mRNA in the presence of Hfq resulted in the unfolding of Hfq III region (nucleotides +4 to +6), as it was shown for Hfq II. Since this region is located immediately downstream of the AUG start codon, increased accessibility of this region upon RyhB pairing may stimulate initiation complex formation. Unexpectedly, the structure of the Shine-Dalgarno region (nucleotides −11 to −7) remained unchanged upon RyhB pairing with cirA mRNA in the absence or in the presence of Hfq (Figure 7B, lane 5 and lane 6, lane 7 and lane 8), thereby discarding the possibility that RyhB may activate cirA mRNA translation by making the Shine-Dalgarno more accessible to the ribosome.
Taken together, results described above strongly suggested that RyhB pairing with cirA mRNA resulted in increased accessibility of regions predicted to be targeted by Hfq to repress translation when RyhB was not expressed (Hfq II and Hfq III sites). Indeed, when we mutated the Hfq II site (mutant M2G), we observed a significant decrease in expression of cirA′-′lacZ translational fusion (Supplementary Figure S9). Overall, the data provided a plausible mechanism underlying the increased ribosome binding to cirA mRNA upon RyhB pairing in the presence of Hfq.
RyhB promotes colicin Ia sensitivity through cirA activation
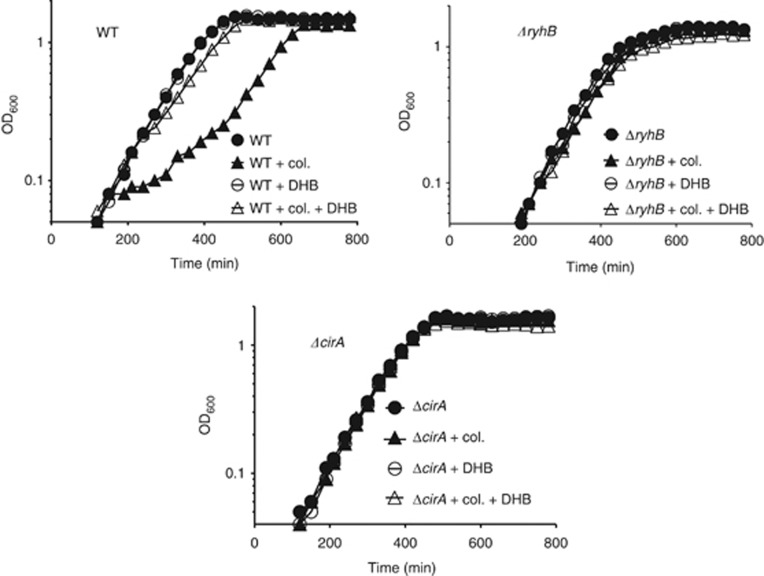
RyhB was expected to modulate colicin sensitivity upon iron starvation by activating cirA translation because colicin Ia is one of the main ligands bound by CirA. WT cells that accumulate high levels of cirA mRNA under conditions of iron starvation should be sensitive to colicin treatment as opposed to ΔryhB cells that are expressing low levels of cirA mRNA. We monitored the growth of WT, ΔryhB and ΔcirA cells in an iron-free minimal medium in the presence or absence of colicin Ia (added at early exponential phase). As expected, WT cells growth was impaired when colicin Ia was added to the medium (Figure 8). In contrast, the growth of ΔryhB and ΔcirA cells was not significantly affected. We explain the observed recovery phenotype of WT+colicin by the active division of a small cell population that was not exposed to colicins at the beginning of the treatment. No suppressor mutations conferring colicin Ia resistance were observed during the course of the experiment (Supplementary Figure S10).
Figure 8.
RyhB expression promotes colicin Ia sensitivity under conditions of iron starvation. Strains EM1055 (WT), EM1238 (ΔryhB), and HS221 (ΔcirA) were grown in M63 iron-free glucose minimal medium and cleared colicin Ia lysate was added at an OD600 of 0.1. When needed, 2,3-dihydroxybenzoic acid (DHB, 33 μM) was added 30 min before addition of colicin Ia. For a determination of the levels of RyhB and cirA RNAs performed under the same growth conditions, please refer to Figure 1C.
Because the siderophore DHB is an additional substrate of CirA, we set up a series of experiments to determine whether prebinding of DHB to CirA could prevent the binding of colicin Ia and protect WT cells from the bactericidal effect of colicin Ia. Results demonstrated that the addition of DHB 30 min before colicin Ia treatment of WT cells conferred nearly complete resistance to colicin Ia (Figure 8). Taken together, these data confirmed that RyhB promoted colicin Ia sensitivity under conditions of iron starvation by activating cirA translation.
Discussion
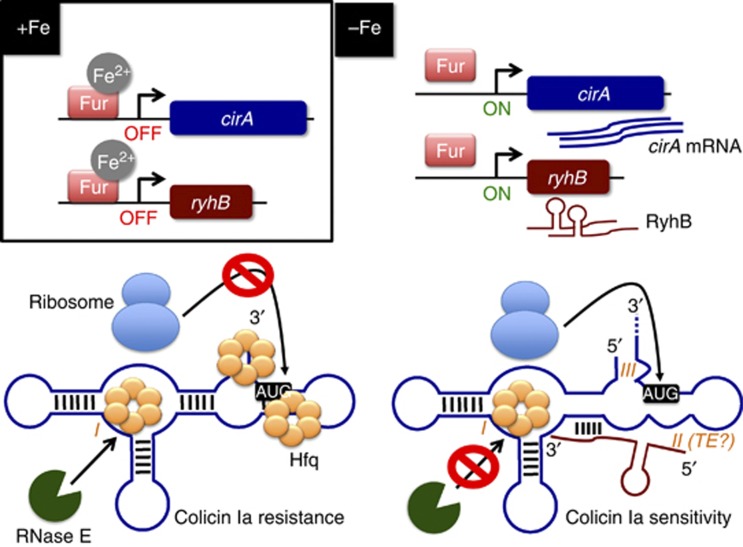
Results reported here unveiled key elements of a complex regulatory network controlling the cellular sensitivity to the antibiotic colicin Ia in an iron-dependent manner (Figure 9). Our data support the interpretation that, under conditions of iron sufficiency, Fe2+-Fur (active Fur) repressed cirA and ryhB transcription, which resulted in low levels of CirA. However, under conditions of iron starvation, Fur became inactive and cirA and ryhB were actively transcribed. Strikingly, although cirA mRNA was expressed, Hfq binding to sites II and III prevented formation of the translation initiation complex, leading to rapid cirA destabilization by RNase E. Hfq-induced translational repression was relieved only when sRNA RyhB base paired with cirA mRNA. RyhB allowed translation initiation of cirA mRNA by promoting structural changes that increased the accessibility of ribosomal subunit 30S to sites II (translation enhancer) and III (RBS). Translational activation of cirA mRNA resulted in increased accumulation of CirA protein in the outer membrane, thereby making cells sensitive to the antibiotic colicin Ia.
Figure 9.
Working model for RyhB-mediated translational activation of cirA mRNA (see the text for details). Fe2+, ferrous iron; TE, translational enhancer. I, II, and III refer to Hfq binding sites I, II, and III characterized in the study, respectively.
Comparison with other positive sRNA-dependent regulations
One may assume that the regulation of cirA mRNA by RyhB and Hfq is highly similar to other cases of sRNA-mediated translational activation previously characterized (e.g., RyhB-shiA; DsrA-rpoS; ans GlmZ-glmS). In such instances, sRNA pairing with the target mRNA typically results in the disruption of a secondary structure that is detrimental to translation initiation (Fröhlich and Vogel, 2009). However, some key differences highlight each mechanism from the others. First, while Hfq represses cirA and shiA translation when RyhB is not expressed, it does not seem to be the case for rpoS and glmS. Indeed, the hfq mutant strain does not exhibit increased levels of rpoS and glmS translation under conditions of low DsrA and GlmZ expression, respectively (Sledjeski et al, 2001; Urban and Vogel, 2008). Second, RyhB and Hfq are not required for the translation of cirA and shiA. In other words, the secondary structure of both mRNAs is not predicted to prevent translation initiation, according to in vitro and in vivo data. Similar levels of 30S binding to cirA mRNA were observed whether RyhB and Hfq were present or not (Figure 7A). Moreover, cirA and shiA transcripts accumulated to WT levels in Δhfq ΔryhB cells (Figure 5A) and the levels of cirA and shiA translation were identical to WT cells in a Δhfq ΔryhB background (Figure 5B; Prévost et al, 2007). These results were in marked contrast with those of DsrA-rpoS and GlmZ-glmS regulations in which case sRNA and Hfq have been shown to be essential for translation of the target mRNA (Sledjeski et al, 2001; Urban and Vogel, 2008; McCullen et al, 2010). Third, whereas RyhB-shiA, DsrA-rpoS and GlmZ-glmS translational activations occurred through increased accessibility of the Shine-Dalgarno sequence following the sRNA pairing (Prévost et al, 2007), it did not appear to be the case for RyhB-cirA regulation.
Indeed, no structural changes in the Shine-Dalgarno region were observed upon RyhB pairing with cirA mRNA in the presence or in the absence of Hfq (Figure 7B; Supplementary Figure S7). Considering that cirA mRNA can be translated to WT levels in the absence of RyhB and Hfq (Figure 5B), we may assume RBS to be available for ribosome binding upon translation initiation, which is supported by probing data (Supplementary Figure S7B, lane 5; note the high susceptibility of nucleotides G-11, G-10, and G-8 to RNase T1 cleavage) and toeprinting experiments (Figure 7A; Supplementary Figure S6). Remarkably, most structural changes observed upon RyhB pairing to cirA mRNA occurred in regions targeted by Hfq for translational repression (sites II and III, see Figure 2). This result suggested that RyhB action may aim to dislodge Hfq from these sites (discussed below) instead of increasing ribosome access to the Shine-Dalgarno sequence. The incapacity of RyhB to increase 30S binding to cirA mRNA in the absence of Hfq further supported this hypothesis (Supplementary Figure S6).
Antagonistic regulation of cirA translation by Hfq and RyhB
Our work demonstrated a situation in which RyhB activated cirA translation by preventing Hfq binding to a potential translational enhancer element (site II, see Figure 2). To our knowledge, a similar mechanism of sRNA-mediated gene activation has not yet been characterized. The presence of AU-rich sequences located several nucleotides upstream of the Shine-Dalgarno sequence has been reported to enhance translation and to stabilize transcripts (Zhang and Deutscher, 1992; Hook-Barnard et al, 2007). These translational enhancer elements have been shown to increase interaction with ribosomal protein S1 (Boni et al, 1991; Komarova et al, 2002) that typically recognizes AU-rich single-stranded sequences (Ringquist et al, 1995; Hajnsdorf and Boni, 2012). We predict the AU-rich region −35 to −26 of cirA mRNA that we identified as a Hfq binding site (Hfq site II) to act as a translational enhancer, given its close proximity to the Shine-Dalgarno sequence (Figure 2) and the high accessibility of nucleotides −33 to −31, according to PbAc probing (Figure 6C, lane 5). Moreover, mutation of nucleotides U-31 and U-32 to guanines reduced 30S binding by 30% (Figure 6D). This decrease corresponded to the level of repression observed upon incubation of cirA mRNA with Hfq (Figure 7A), suggesting that most of the translational repression mediated by Hfq could occur through binding to these nucleotides. Our findings are in agreement with these observations and suggest a potential role of Hfq II site as a translational enhancer.
Although the 30% repression of 30S binding observed upon incubation of cirA mRNA with Hfq is moderate, we suggest that it is sufficient to repress translation and to promote cirA transcript destabilization. The 60% activation by RyhB observed in toeprint experiments (Figure 7A) would then allow translation to resume and cirA mRNA to accumulate. Further repression of ribosome binding observed with higher concentrations of Hfq (Figure 6A) may not occur in vivo due to limiting concentrations of Hfq in the cell (Moon and Gottesman, 2011; Hussein and Lim, 2011). Moreover, the Hfq binding site III (Figure 2, region +4 to +9) of cirA mRNA may be critical for translational regulation. Given that any sRNA pairing immediately upstream of the Shine-Dalgarno sequence or within the first five codons of the open reading frame was able to compete directly with initiating ribosomes (Bouvier et al, 2008), we would predict Hfq binding to site III to interfere with translation initiation complex formation. Our probing data suggested that Hfq may be dislodged from sites II and III following RyhB pairing (Figure 7B; Supplementary Figure S7). Considering this possibility, we hypothesized that structural changes induced by RyhB upon base pairing with cirA mRNA destabilized Hfq interactions with sites II and III, thereby resulting in translational activation.
It is tempting to suggest that structural changes occurring on cirA mRNA following RyhB pairing not to have major effects on translation in the absence of Hfq, as suggested by toeprint experiments (Supplementary Figure S6). However, RyhB expression in Δhfq cells resulted in a significant increase in cirA mRNA levels (Figure 5C). This Hfq-independent effect of RyhB on cirA expression remains elusive. One may assume this activation to result from the subtle 15% increase in cirA translation observed upon RyhB expression in the absence of Hfq (Figure 5D) that could be sufficient to stabilize cirA mRNA. On the other hand, the possibility of an RyhB-mediated stabilization of cirA transcript independent of ribosome protection cannot be excluded.
Introduction of mutations into the Hfq binding site I did not reduce 30S binding to cirA mRNA (Supplementary Figure S5), suggesting that this region may not regulate translation initiation. Based on the observation that Hfq greatly stimulated RyhB pairing to cirA mRNA in vitro (Supplementary Figure S11), we predict Hfq to bind Hfq I site to promote RyhB-cirA duplex formation in vivo. Upon pairing with cirA mRNA, the RyhB poly(U) tail, which was reported to bind Hfq with high affinity (Otaka et al, 2011), is oriented towards Hfq site I. Binding of Hfq to both site I and RyhB 3′-end under these conditions could then promote stabilization of the RyhB-cirA hybrid, thus allowing sustained translational activation during iron starvation.
A feed-forward loop motif controls colicin sensitivity and iron uptake
The Fur-RyhB-cirA regulatory circuit forms a coherent type 2 feed-forward loop in which Fe2+-Fur represses cirA and ryhB while RyhB activates cirA (Supplementary Figure S12). Such a network motif is predicted to delay the induction of the target gene, according to computational modelling on transcription networks (Mangan and Alon, 2003). Application of the model to cirA regulation when cells experience iron starvation predicts cirA expression to be delayed in comparison to direct regulation by Fe2+-Fur. This would be logical, considering that once Fur becomes inactive and cirA promoter is fully expressed, RyhB synthesis must reach a certain threshold for cirA mRNA to be translated and stabilized. Conversely, no lag of repression is expected for transcriptional networks with the same configuration as the Fur-RyhB-cirA loop. This prediction may not apply to cirA repression, considering the fact that feed-forward loops integrating sRNAs often display altered regulatory dynamics when compared to transcriptional feed-forward loops (Shimoni et al, 2007; Beisel and Storz, 2010, 2011). One could imagine that under conditions when cells switch from iron-poor to iron-rich conditions and when Fur becomes active, despite the fact that Fe2+-Fur represses cirA and ryhB transcription, there are still cirA transcripts activated by existing RyhB molecules that were transcribed before Fur repression. Consequently, CirA proteins will be synthesized until the existing pool of cirA transcripts becomes exhausted. We would then expect cirA repression to be delayed in this context as compared to direct repression by Fe2+-Fur.
Physiological relevance of colicin sensitivity upon iron starvation
Colicins are produced following DNA damages as a result of the SOS response as well as various cellular stresses such as thymine starvation, high temperatures or anaerobiosis (Cascales et al, 2007). Under these conditions, colicin-producing strains lyse and released colicins kill surrounding colicin-sensitive cells. Despite the fact that the role of colicins in survival to iron starvation has not yet been addressed, we can speculate colicin sensitivity to confer a selective advantage for the bacterial community when iron is scarce. Cells experiencing iron deficiency and, thereby expressing CirA, would become sensitive to the killing action of colicin Ia and lyse. Then, the released iron could be imported from the environment by the remaining cell population for survival. The DHB molecules expelled in the medium upon cell death could in turn protect the remaining cells from colicin action through competitive binding to CirA, as observed here (Figure 8 in the case of WT cells), thus preventing elimination of the whole bacterial population.
Our work further supports the active role of RyhB in gene activation in addition to its well-known function as a gene silencer. The feed-forward loop by the Fur-RyhB-cirA regulatory circuit also unveils a novel type of iron-dependent regulation. To our knowledge, cirA is the first Fur-regulated gene to require further post-transcriptional activation by RyhB upon iron starvation that has been characterized so far. Further investigation is warranted to uncover similar type of regulation in the future.
Materials and methods
Strains and plasmids
Derivatives of E. coli MG1655 were used in all experiments (Supplementary Table S1). DH5α strain was used for routine cloning procedures. EM1055 (wild type, MG1655 derivative), EM1451 (Δara714 leu+), EM1455 (Δara714 leu+ ΔryhB::cat), EM1238 (ΔryhB::cat), EM1256 (Δfur::kan), EM1237 (DY330 [W3110 ΔlacU169 gal490 λcI857 Δ(cro-bioA)]), EM1265 [hfq-1::Ω(kan;Bcl1)], EM1277 (rne-3071 zce-726_Tn10), EM1280 (rne-3071 zce-726_Tn10 ΔryhB::cat), EM1377 (rne-131 zce-726_Tn10), KP392 (Δfur::kan), KP393 (Δfur::kan ΔryhB::cat), and JW4130-1 (rrnB3 ΔlacZ4787 hsdR514 Δ(araBAD)567 Δ(rhaBAD)568 rph-1 Δhfq-722::kan) have been described earlier (Yu et al, 2000; Massé and Gottesman, 2002; Massé et al, 2003; Baba et al, 2006; Desnoyers et al, 2009; Salvail et al, 2010). Strains constructed by P1 transduction were selected for the appropriate antibiotic-resistant marker. Except as otherwise indicated, for cells carrying plasmid pNM12, pBAD-ryhB, pBAD-ryhB1 and pCP20 (see Supplementary Table S2 for a list of plasmids), ampicillin was used at a final concentration of 50 μg/ml. Deletion/insertion mutations in cirA (ΔcirA::tet), omrAB (ΔomrAB::tet), araB (ΔaraB::kan) and fur (Δfur::tet) were constructed by the method described by Yu et al. Tetracycline resistance cassette was first amplified by PCR from strain EM1053 (oligos EM215-EM216) (see Supplementary Table S3 for a list of oligonucleotides). The PCR fragment obtained was then amplified with oligos containing sequences that are homologous to the 5′ and 3′ ends of the cassette and sequences homologous to cirA (oligos EM1044–EM1045), omrAB (oligos EM1599–EM1600), and fur (oligos EM1989–EM1990). Flippase recognition target (FRT)-flanked kanamycin resistance cassette was generated by PCR from a DH5α strain harbouring pKD4 plasmid with oligos containing sequences that are homologous to the 5′ and 3′ ends of the cassette and sequences homologous to araB (oligos EM1388–EM1389). The resulting PCR products were transformed into EM1237 after induction of λred, according to Yu et al, selecting for tetracycline resistance for cirA and omrAB mutations and for kanamycin resistance for araB mutation. Recombinant products were verified by sequencing. Following P1 transduction of araB::kan, kanamycin resistance cassette was removed using flippase (Flp) encoding helper plasmid pCP20 as described (Datsenko and Wanner, 2000).
RNA extraction and northern blot analysis
Northern blots were performed as described previously (Desnoyers and Massé, 2012) with some modifications. Total RNA was extracted from cells grown at 37°C or 30°C, for experiments with thermosensitive strains (EM1277 and EM1280), in LB or M63 minimal medium containing glucose or glycerol (0.2%) using the hot phenol procedure (Aiba et al, 1981). Arabinose (0.1%), 2,2′-dipyridyl (250 μM) or FeSO4 (5 μM) was added when indicated. Following total RNA extraction, 5–10 μg of total RNA was loaded on polyacrylamide gel (4–6% acrylamide 29:1, 8 M urea) and 15 μg was loaded on agarose gel (1%, 1X MOPS).
Primer extension
Reverse transcription was performed as previously described (Prévost et al, 2007).
OMPs analysis
Cells were grown in 50 ml of LB medium to an OD600 of 1.0 and 10 ml of culture was harvested and OMPs were extracted as previously described (Morona and Reeves, 1982; Guillier and Gottesman, 2006).
β-Galactosidase assays
Kinetic assays for β-galactosidase activity were performed as described (Prévost et al, 2007), using a SpectraMax 250 microtitre plate reader (Molecular Devices).
qRT–PCR analysis
qRT–PCR was performed according to a previous report (Salvail et al, 2010).
RNA secondary structure probing
Secondary structure probing was performed as described earlier (Desnoyers et al, 2009). In-line probing experiments were performed as previously described (Regulski and Breaker, 2008; Wakeman and Winkler, 2009).
Toeprinting assays
Toeprinting assays were carried out according to a previous report (Salvail et al, 2010).
Supplementary Material
Acknowledgments
We would like to thank Charles M Dozois (INRS-Institut Armand-Frappier) for the colicin-producing strains and Eric Cascales (Laboratoire d’Ingénierie des Systèmes Macromoléculaires, CNRS UMR7255), Hanah Margalit (The Hebrew University of Jerusalem), Uri Alon and Avi Mayo (Weizmann Institute of Science) for fruitful discussions. We also thank Fabien Darfeuille (Université Bordeaux Segalen), Karine Prévost and Guillaume Tremblay for excellent technical assistance. This work was funded by an operating grant MOP69005 to EM from the Canadian Institutes for Health Research (CIHR). HS holds an Alexander-Graham-Bell PhD studentship from the Natural Sciences and Engineering Research Council of Canada (NSERC). EM is a Fonds de la Recherche en Santé du Québec (FRSQ) Senior scholar.
Author contributions: HS, M-PC, and EM designed research; HS, M-PC, and JB performed research; HS, M-PC, JB, and EM analysed data; HS and EM wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Aiba H, Adhya S, de Crombrugghe B (1981) Evidence for two functional gal promoters in intact Escherichia coli cells. J Biol Chem 256: 11905–11910 [PubMed] [Google Scholar]
- Andrade JM, Pobre V, Matos AM, Arraiano CM (2012) The crucial role of PNPase in the degradation of small RNAs that are not associated with Hfq. RNA 18: 844–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold TE, Yu J, Belasco JG (1998) mRNA stabilization by the ompA 5′ untranslated region: two protective elements hinder distinct pathways for mRNA degradation. RNA 4: 319–330 [PMC free article] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H (2006) Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2: 2006.0008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balbontín R, Fiorini F, Figueroa-Bossi N, Casadesús J, Bossi L (2010) Recognition of heptameric seed sequence underlies multi-target regulation by RybB small RNA in Salmonella enterica. Mol Microbiol 78: 380–394 [DOI] [PubMed] [Google Scholar]
- Beisel CL, Storz G (2010) Base pairing small RNAs and their roles in global regulatory networks. FEMS Microbiol Rev 34: 866–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisel CL, Storz G (2011) The base-pairing RNA spot 42 participates in a multioutput feedforward loop to help enact catabolite repression in Escherichia coli. Mol Cell 41: 286–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belasco JG (2010) All things must pass: contrasts and commonalities in eukaryotic and bacterial mRNA decay. Nat Rev Mol Cell Biol 11: 467–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boni IV, Isaeva DM, Musychenko ML, Tzareva NV (1991) Ribosome-messenger recognition: mRNA target sites for ribosomal protein S1. Nucleic Acids Res 19: 155–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier M, Sharma CM, Mika F, Nierhaus KH, Vogel J (2008) Small RNA binding to 5′ mRNA coding region inhibits translational initiation. Mol Cell 32: 827–837 [DOI] [PubMed] [Google Scholar]
- Braun M, Killmann H, Maier E, Benz R, Braun V (2002) Diffusion through channel derivatives of the Escherichia coli FhuA transport protein. Eur J Biochem 269: 4948–4959 [DOI] [PubMed] [Google Scholar]
- Buchanan SK, Lukacik P, Grizot S, Ghirlando R, Ali MM, Barnard TJ, Jakes KS, Kienker PK, Esser L (2007) Structure of colicin I receptor bound to the R-domain of colicin Ia: implications for protein import. EMBO J 26: 2594–2604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascales E, Buchanan SK, Duché D, Kleanthous C, Lloubès R, Postle K, Riley M, Slatin S, Cavard D (2007) Colicin biology. Microbiol Mol Biol Rev 71: 158–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA 97: 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deana A, Belasco JG (2005) Lost in translation: the influence of ribosomes on bacterial mRNA decay. Genes Dev 19: 2526–2533 [DOI] [PubMed] [Google Scholar]
- Desnoyers G, Massé E (2012) Noncanonical repression of translation initiation through small RNA recruitment of the RNA chaperone Hfq. Genes Dev 26: 726–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desnoyers G, Morissette A, Prévost K, Massé E (2009) Small RNA-induced differential degradation of the polycistronic mRNA iscRSUA. EMBO J 28: 1551–1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fröhlich KS, Vogel J (2009) Activation of gene expression by small RNA. Curr Opin Microbiol 12: 674–682 [DOI] [PubMed] [Google Scholar]
- Gottesman S, Storz G (2010) Bacterial small RNA regulators: versatile roles and rapidly evolving variations. Cold Spring Harb Perspect Biol 3: pii.a003798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griggs DW, Tharp BB, Konisky J (1987) Cloning and promoter identification of the iron-regulated cir gene of Escherichia coli. J Bacteriol 12: 5343–5352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillier M, Gottesman S (2006) Remodelling of the Escherichia coli outer membrane by two small regulatory RNAs. Mol Microbiol 59: 231–247 [DOI] [PubMed] [Google Scholar]
- Guillier M, Gottesman S (2008) The 5′ end of two redundant sRNAs is involved in the regulation of multiple targets, including their own regulator. Nucleic Acids Res 36: 6781–6794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajnsdorf E, Boni IV (2012) Multiple activities of RNA-binding proteins S1 and Hfq. Biochimie 94: 1544–1553 [DOI] [PubMed] [Google Scholar]
- Hantke K (1990) Dihydroxybenzoylserine--a siderophore for E. coli. FEMS Microbiol Lett 55: 5–8 [DOI] [PubMed] [Google Scholar]
- Holmqvist E, Reimegård J, Sterk M, Grantcharova N, Römling U, Wagner EG (2010) Two antisense RNAs target the transcriptional regulator CsgD to inhibit curli synthesis. EMBO J 29: 1840–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hook-Barnard IG, Brickman TJ, McIntosh MA (2007) Identification of an AU-rich translational enhancer within the Escherichia coli fepB leader RNA. J Bacteriol 189: 4028–4037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussein R, Lim HN (2011) Disruption of small RNA signaling caused by competition for Hfq. Proc Natl Acad Sci USA 108: 1110–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques JF, Jang S, Prévost K, Desnoyers G, Desmarais M, Imlay J, Massé E (2006) RyhB small RNA modulates the free intracellular iron pool and is essential for normal growth during iron limitation in Escherichia coli. Mol Microbiol 62: 1181–1190 [DOI] [PubMed] [Google Scholar]
- Jakes KS, Finkelstein A (2010) The colicin Ia receptor, Cir, is also the translocator for colicin Ia. Mol Microbiol 75: 567–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr B, Riley MA, Feldman MW, Bohannan BJ (2002) Local dispersal promotes biodiversity in a real-life game of rock-paper-scissors. Nature 418: 171–174 [DOI] [PubMed] [Google Scholar]
- Kido M, Yamanaka K, Mitani T, Niki H, Ogura T, Hiraga S (1996) RNase E polypeptides lacking a carboxyl-terminal half suppress a mukB mutation in Escherichia coli. J Bacteriol 178: 3917–3925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkup BC, Riley MA (2004) Antibiotic-mediated antagonism leads to a bacterial game of rock-paper-scissors in vivo. Nature 428: 412–414 [DOI] [PubMed] [Google Scholar]
- Kleanthous C (2010) Swimming against the tide: progress and challenges in our understanding of colicin translocation. Nat Rev Microbiol 8: 843–848 [DOI] [PubMed] [Google Scholar]
- Komarova AV, Tchufistova LS, Supina EV, Boni IV (2002) Protein S1 counteracts the inhibitory effect of the extended Shine-Dalgarno sequence on translation. RNA 8: 1137–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazdunski CJ, Bouveret E, Rigal A, Journet L, Lloubès R, Bénédetti H (1998) Colicin import into Escherichia coli cells. J Bacteriol 180: 4993–5002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz C, Gesell T, Zimmermann B, Schoeberl U, Bilusic I, Rajkowitsch L, Waldsich C, von Haeseler A, Schroeder R (2010) Genomic SELEX for Hfq-binding RNAs identifies genomic aptamers predominantly in antisense transcripts. Nucleic Acids Res 38: 3794–3808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangan S, Alon U (2003) Structure and function of the feed-forward loop network motif. Proc Natl Acad Sci USA 100: 11980–11985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massé E, Gottesman S (2002) A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc Natl Acad Sci USA 99: 4620–4625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massé E, Escorcia FE, Gottesman S (2003) Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev 17: 2374–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massé E, Vanderpool CK, Gottesman S (2005) Effect of RyhB small RNA on global iron use in Escherichia coli. J Bacteriol 187: 6962–6971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullen CA, Benhammou JN, Majdalani N, Gottesman S (2010) Mechanism of positive regulation by DsrA and RprA small noncoding RNAs: pairing increases translation and protects rpoS mRNA from degradation. J Bacteriol 192: 5559–5571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowall KJ, Hernandez RG, Lin-Chao S, Cohen SN (1993) The ams-1 and rne-3071 temperature-sensitive mutations in the ams gene are in close proximity to each other and cause substitutions within a domain that resembles a product of the Escherichia coli mre locus. J Bacteriol 175: 4245–4249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morona R, Reeves P (1982) The tolC locus of Escherichia coli affects the expression of three major outer membrane proteins. J Bacteriol 150: 1016–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon K, Gottesman S (2011) Competition among Hfq-binding small RNAs in Escherichia coli. Mol Microbiol 82: 1545–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otaka H, Ishikawa H, Morita T, Aiba H (2011) PolyU tail of rho-independent terminator of bacterial small RNAs is essential for Hfq action. Proc Natl Acad Sci USA 108: 13059–13064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell BS, Rivas MP, Court DL, Nakamura Y, Turnbough CL Jr (1994) Rapid confirmation of single copy lambda prophage integration by PCR. Nucleic Acids Res 22: 5765–5766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prévost K, Salvail H, Desnoyers G, Jacques JF, Phaneuf E, Massé E (2007) The small RNA RyhB activates the translation of shiA mRNA encoding a permease of shikimate, a compound involved in siderophore synthesis. Mol Microbiol 64: 1260–1273 [DOI] [PubMed] [Google Scholar]
- Regulski EE, Breaker RR (2008) In-line probing analysis of riboswitches. Methods Mol Biol 419: 53–67 [DOI] [PubMed] [Google Scholar]
- Repoila F, Gottesman S (2001) Signal transduction cascade for regulation of RpoS: temperature regulation of DsrA. J Bacteriol 183: 4012–4023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley MA (1993a) Molecular mechanisms of colicin evolution. Mol Biol Evol 10: 1380–1395 [DOI] [PubMed] [Google Scholar]
- Riley MA (1993b) Positive selection for colicin diversity in bacteria. Mol Biol Evol 10: 1048–1059 [DOI] [PubMed] [Google Scholar]
- Riley MA, Wertz JE (2002) Bacteriocins: evolution, ecology, and application. Annu Rev Microbiol 56: 117–137 [DOI] [PubMed] [Google Scholar]
- Ringquist S, Jones T, Snyder EE, Gibson T, Boni I, Gold L (1995) High-affinity RNA ligands to Escherichia coli ribosomes and ribosomal protein S1: comparison of natural and unnatural binding sites. Biochemistry 34: 3640–3648 [DOI] [PubMed] [Google Scholar]
- Salvail H, Lanthier-Bourbonnais P, Sobota JM, Caza M, Benjamin JAM, Mendieta ME, Lépine F, Dozois CM, Imlay J, Massé E (2010) A small RNA promotes siderophore production through transcriptional and metabolic remodeling. Proc Natl Acad Sci USA 107: 15223–15228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvail H, Massé E (2011) Regulating iron storage and metabolism with RNA: an overview of posttranscriptional controls of intracellular iron homeostasis. Wiley Interdiscip Rev RNA 3: 26–36 [DOI] [PubMed] [Google Scholar]
- Shimoni Y, Friedlander G, Hetzroni G, Niv G, Altuvia S, Biham O, Margalit H (2007) Regulation of gene expression by small non-coding RNAs: a quantitative view. Mol Syst Biol 3: 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sledjeski DD, Gupta A, Gottesman S (1996) The small RNA, DsrA, is essential for the low temperature expression of RpoS during exponential growth in Escherichia coli. EMBO J 15: 3993–4000 [PMC free article] [PubMed] [Google Scholar]
- Sledjeski DD, Whitman C, Zhang A (2001) Hfq is necessary for regulation by the untranslated RNA DsrA. J Bacteriol 183: 1997–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz G, Vogel J, Wassarman KM (2011) Regulation by small RNAs in bacteria: expanding frontiers. Mol Cell 43: 880–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjaden B, Goodwin SS, Opdyke JA, Guillier M, Fu DX, Gottesman S, Storz G (2006) Target prediction for small, noncoding RNAs in bacteria. Nucleic Acids Res 34: 2791–2802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban JH, Vogel J (2008) Two seemingly homologous noncoding RNAs act hierarchically to activate glmS mRNA translation. PLoS Biol 6: e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanzo NF, Li YS, Py B, Blum E, Higgins CF, Raynal LC, Krisch HM, Carpousis AJ (1998) Ribonuclease E organizes the protein interactions in the Escherichia coli RNA degradosome. Genes Dev 12: 2770–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J, Luisi BF (2011) Hfq and its constellation of RNA. Nat Rev Microbiol 9: 578–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vytvytska O, Jakobsen JS, Balcunaite G, Andersen JS, Baccarini M, von Gabain A (1998) Host factor I, Hfq, binds to Escherichia coli ompA mRNA in a growth rate-dependent fashion and regulates its stability. Proc Natl Acad Sci USA 95: 14118–14123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vytvytska O, Moll I, Kaberdin VR, von Gabain A, Bläsi U (2000) Hfq (HF1) stimulates ompA mRNA decay by interfering with ribosome binding. Genes Dev 14: 1109–1118 [PMC free article] [PubMed] [Google Scholar]
- Wakeman CA, Winkler WC (2009) Analysis of the RNA backbone: structural analysis of riboswitches by in-line probing and selective 2′-hydroxyl acylation and primer extension. Methods Mol Biol 540: 173–191 [DOI] [PubMed] [Google Scholar]
- Wassarman KM, Repoila F, Rosenow C, Storz G, Gottesman S (2001) Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev 15: 1637–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiener M, Freymann D, Ghosh P, Stroud RM (1997) Crystal structure of colicin Ia. Nature 385: 461–464 [DOI] [PubMed] [Google Scholar]
- Yu DG, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL (2000) An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci USA 97: 5978–5983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Deutscher MP (1992) A uridine-rich sequence required for translation of prokaryotic mRNA. Proc Natl Acad Sci USA 89: 2605–2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.