Abstract
Filopodia explore the environment, sensing soluble and mechanical cues during directional motility and tissue morphogenesis. How filopodia are initiated and spatially restricted to specific sites on the plasma membrane is still unclear. Here, we show that the membrane deforming and curvature sensing IRSp53 (Insulin Receptor Substrate of 53 kDa) protein slows down actin filament barbed end growth. This inhibition is relieved by CDC42 and counteracted by VASP, which also binds to IRSp53. The VASP:IRSp53 interaction is regulated by activated CDC42 and promotes high-density clustering of VASP, which is required for processive actin filament elongation. The interaction also mediates VASP recruitment to liposomes. In cells, IRSp53 and VASP accumulate at discrete foci at the leading edge, where filopodia are initiated. Genetic removal of IRSp53 impairs the formation of VASP foci, filopodia and chemotactic motility, while IRSp53 null mice display defective wound healing. Thus, IRSp53 dampens barbed end growth. CDC42 activation inhibits this activity and promotes IRSp53-dependent recruitment and clustering of VASP to drive actin assembly. These events result in spatial restriction of VASP filament elongation for initiation of filopodia during cell migration, invasion, and tissue repair.
Keywords: actin dynamics, cell migration, CDC42, filopodia, IRSp53
Introduction
Cells move and interact with the environment by forming migratory structures composed of self-organized polymers of actin. These protrusions can be flat-sheet lamellipodia, or elongated, finger-like filopodia. Cells use lamellipodia as expanding membrane sheets driving cell locomotion (Wu et al, 2012), while filopodia act as compasses navigating cell migration (Faix et al, 2009; Yang and Svitkina, 2011a), as well as mechanical sensors (Chan and Odde, 2008).
In lamellipodia, actin is organized in networks of branched filaments (Svitkina et al, 2003; Yang and Svitkina, 2011b), generated by the concerted action of the Arp2/3 complex, capping proteins, and filament elongation factors (Svitkina and Borisy, 1999; Wiesner et al, 2003; Akin and Mullins, 2008). Conversely, filopodia are composed of parallel actin bundles that emanate from the cell periphery (Adams, 2004). However, in several cell types, filopodia and lamellipodia rapidly interchange during protrusion. Furthermore, filopodia are frequently embedded into, or arise from, pre-existing lamellipodia (Rottner et al, 1999; Svitkina et al, 2003). This finding suggests that filopodia form through a reorganization of the underlying actin network by convergent elongation of pre-existing filaments, or by de novo actin nucleation and elongation (Faix et al, 2009; Yang and Svitkina, 2011a). Whatever the case, the occurrence of lamellipodia-to-filopodia transition suggests commonalities between the two structures. For example, while it appears that capping proteins, including CP and EPS8, determine which actin-based protrusive organelle dominates at the cell periphery (Mejillano et al, 2004; Akin and Mullins, 2008; Vaggi et al, 2011), they do not function in isolation. The ENA/VASP family of proteins (Gertler et al, 1995; Reinhard et al, 1995; Gertler et al, 1996), which in mammals includes MENA, VASP, and EVL, also influence the underlying actin architecture of migratory protrusions (Bear et al, 2002). Notably, by cellular localization alone, ENA/VASP proteins appear to be vital to this lamellipodial–filopodial transition since they localize to active sites of actin assembly, such as the tips of protruding lamellipodia and filopodia (Lanier et al, 1999; Rottner et al, 1999). At the leading edge, the intensity of GFP-VASP has been shown to increase locally in puncta that subsequently give rise to filopodia, suggesting that higher order clustering of this protein may be critical for generating linear filaments to support filopodia protrusions (Lanier et al, 1999; Rottner et al, 1999; Svitkina et al, 2003; Applewhite et al, 2007). Biochemical studies have recently shown that soluble VASP displays weak, processive polymerase activity (Hansen and Mullins, 2010). High-density VASP clustering, however, is required to enhance processive filament elongation, even in the presence of high concentrations of capping proteins (Breitsprecher et al, 2008, 2011). Not surprisingly, genetic evidence in various organisms showed that ENA/VASP proteins are essential players in filopodia formation (Schirenbeck et al, 2006; Gates et al, 2007; Kwiatkowski et al, 2007). Whether and how the activity and higher order clustering of ENA/VASP family proteins is differentially tuned to control lamellipodia-to-filopodia transition remains, however, poorly defined.
Molecules sitting at the actin:membrane interface are predicted to be important in this process, albeit their nature and mechanisms of action remain unclear. One candidate is IRSp53 (Insulin Receptor Substrate of 53 kDa) (also called BAIAP2, brain angiogenic inhibitor interacting protein 2) (Abbott et al, 1999; Oda et al, 1999; Okamura-Oho et al, 1999). IRSp53 possesses an inverted Bin-Amphiphysin-Rvs167 (I-BAR) domain that binds to PI(4,5)P2-rich lipid inducing negative curvatures, such as the ones required for filopodia protrusions (Zhao et al, 2011). Consistently, IRSp53 expression is sufficient to induce filopodia-like structures (Bockmann et al, 2002; Yamagishi et al, 2004; Disanza et al, 2006). Furthermore, IRSp53 binds activated CDC42 and, through its SH3 domain, a number of actin regulatory proteins that are involved in filopodia protrusions (Ahmed et al, 2010). Among them, IRSp53 was reported to bind to MENA in vitro (Krugmann et al, 2001) and to VASP in FRET-based and co-immunoprecipitation assays (Lim et al, 2008; Vaggi et al, 2011). Interestingly, the interaction between VASP and IRSp53 enhances the bundling activity of the former (Lim et al, 2008; Vaggi et al, 2011). However, it is unknown whether IRSp53 affects other key biochemical activities of MENA and VASP, and what the functional consequences of these interactions are.
Here, we show that IRSp53 alone slows down barbed end growth. Binding to CDC42 relieves this inhibition and promotes IRSp53-dependent recruitment and clustering of VASP to the plasma membrane to initiate processive F-actin elongation. These events result in spatial restriction of VASP activity to initiate filopodia, drive cell migration, and promote tissue repair.
Results
Characterization of the IRSp53–VASP interaction
We initially characterized the interaction between IRSp53 and VASP using purified proteins and various binding assays. IRSp53 associates with VASP, as well as EVL, through its SH3 domain (Supplementary Figure S1a–f) that contacts VASP on proline-rich sites partially overlapping, but distinct from the Profilin binding sites (Supplementary Figure S1c and g). Next, since VASP has recently been shown to acquire processive elongation activity upon clustering and IRSp53 can form dimer through its I-BAR domain (Millard et al, 2005), we tested whether in solution the two proteins can form high order oligomer by dynamic light scattering. Purified IRSp53 produced a single major species with a hydrodynamic radius of about 7.2 nm (Figure 1A) consistent with its dimeric structure (similar results were obtained by hydrodynamic measurements; Supplementary Figure S2a), while the VASP tetramer gave rise to a single species with a radius of about 14.3 nm (Figure 1A). Notably, the mixture of both proteins led to the formation of large clusters with an average diameter of about 200 nm (Figure 2A, left). Importantly, the VASP-ΔPRD mutant, which is unable to bind to IRSp53 (Supplementary Figure S1e), and the IRSp53 W413G mutant, which lacks VASP binding ability (Supplementary Figure S1c), did not form heterocomplexes (Figure 1A, right, and data not shown). In contrast, a VASP mutant lacking the central three GP5 motifs (VASP-ΔGP5), which retains IRSp53 binding ability, albeit with reduced affinity (Supplementary Figure S1d and f), was still able to hetero-oligomerize upon addition of IRSp53 (Figure 1A, middle). Thus, IRSp53 promotes clustering of VASP in solution, and might therefore be a decisive factor in the regulation of VASP function in vitro and in vivo.
Figure 1.
IRSp53 slows down barbed end growth: an effect relieved by CDC42. (A) IRSp53 and VASP form large clusters in solution. Intensity weighted diameter distributions for IRSp53 and VASP showed single species for either IRSp53 (red lines) or VASP WT and mutant proteins alone at concentrations of up to 30 μM (green lines). Aggregates formed in mixtures of 10 μM IRSp53 and 10 μM VASP WT or VASPΔ(GP5)3, whereas no clusters were observed with VASP-ΔPRD (blue lines). (B) VASP increases the association rate constant of profilin-actin to barbed ends. Barbed end growth was measured in bulk pyrenyl-actin polymerization assays using 1.25 μM MgATP-G-actin (5% pyrenyl-labelled), 5 μM profilin, and VASP (V) at 0 (blue lines), 64 nM (red lines), and 169 nM (green lines), in the absence (dotted lines) or in the presence (continuous lines) of 0.16 nM spectrin-actin seeds. Inset shows cumulated data from additional assays performed with different VASP concentrations. (C) IRSp53 slows down actin polymerization by weakly capping barbed ends. Kinetics of actin polymerization induced by spectrin-actin seeds, measured as described in (A), in the presence of the indicated concentrations of IRSp53. (D) IRSp53 and VASP inhibit barbed end depolymerization. Depolymerization of actin filaments (2.5 μM F-actin, 50% pyrenyl-labelled) was induced by 50-fold dilution into polymerization buffer in the presence of 0.29 μM IRSp53 (red curve) or 0.34 μM VASP (green curve). (E) Summary of barbed end growth inhibition (Cap) by the indicated IRSp53 constructs and mutants (see also Supplementary Figure S2). (F) Active CDC42 relieves IRSp53 capping activity. Kinetics of actin polymerization induced by spectrin-actin seeds was measured as described in (A) in the presence of 0.58 μM IRSp53, alone or with 10 μM CDC42-GTP or CDC42-GDP. Control (CTR) sample contained no IRSp53. (G) Kinetics of actin polymerization induced by spectrin-actin seeds was measured as described in (A) in the presence of the indicated concentrations of IRSp53 or its truncated mutant 1–375. Right graph represents a magnification of the initial phase of elongation by seeds (inset in the left graph).
Figure 2.
IRSp53 recruits and clusters Ena/VASP proteins to drive processive actin filament elongation in the presence of capping protein. (A) IRSp53 inhibits barbed end elongation. (Left) Polymerization of 1.0 μM G-actin (20% Atto-488 labelled) in the absence or presence of 1 μM IRSp53 in 1 × TIRF buffer monitored by TIRF microscopy. Time is indicated in seconds in the top right corner of each frame. Scale bar, 10 μm. (Right) Quantification of actin filament elongation rates in the absence or presence of IRSp53 as monitored by TIRF microscopy. Elongation rates are presented as mean±s.e.m. from three independent experiments. At least 24 filaments per condition/experiment were measured. *, P<0.05 and **, P<0.01 (T-test, compared to 1 μM actin). (B) TIRF micrographs of the assembly of 1.0 μM G-actin (20% Atto-488 labelled) on uncoated (red arrowhead) or His-IRSp53-saturated Ni-NTA beads in the presence of 50 nM capping protein and 200 nM VASP, EVL, or hVASP DdGAB. Blue arrowheads indicate buckling filaments (also see Supplementary Movie 1). Only filaments that were attached with their barbed ends to the beads were observed, whereas capping protein largely abolished filament growth in solution. Time is indicated in seconds in the upper right corner of each frame. Scale bar, 10 μm. (C) Quantification of actin filament elongation rates from experiment shown in (B). Data are presented as mean±s.d. of three independent experiments, in which at least 16 filaments/experiment were measured. ***, P<0.005 (T-test, compared to His-VASP control).
IRSp53-mediated inhibition of barbed end growth is relieved by CDC42
Given the established role of VASP as an actin elongation factor with a relatively weak filament nucleation activity (Huttelmaier et al, 1999; Samarin et al, 2003; Barzik et al, 2005; Breitsprecher et al, 2008; Pasic et al, 2008), we next assessed whether IRSp53 regulates VASP filament elongation in bulk pyrenyl-actin polymerization assays. In this assay, VASP increased the rate (up to 2.5-fold) of spectrin-actin seeded barbed end growth from both ATP-G-actin (data not shown) and profilin-ATP-G-actin in an identical and dose-dependent fashion, under conditions in which its nucleating effect was negligible (Figure 1B). In contrast, IRSp53 slowed down barbed end growth by ∼10-fold, in a substoichiometric concentration range with respect to G-actin, indicating that the interaction of IRSp53 with barbed ends, rather than with G-actin, mediates this inhibitory effect (Figure 1C). The interaction of VASP and IRSp53 with barbed ends also inhibited dilution-induced depolymerization of filaments (Figure 1D).
A structure/function analysis of IRSp53 domains mediating barbed end growth inhibition revealed that the SH3 domain binding activity is dispensable. Indeed, an IRSp53-W413G mutant inhibited barbed end growth to the same extent as wild-type (WT) IRSp53, but bound to barbed ends instantaneously with a Kd of 0.29 μM (Figure 1E, scheme; Supplementary Figure S2b and c). In contrast, the isolated I-BAR domain of IRSp53 had no effect on barbed end growth (Figure 1E-scheme; Supplementary Figure S2b). We determined that the minimal region that mediates barbed end growth inhibition includes the I-BAR domain and a stretch of 30 amino acids just before the SH3 domain (Figure 1E, scheme; Supplementary Figure S2d). These results suggest that IRSp53 uses multiple interaction surfaces for inhibiting barbed end growth (e.g., the I-BAR domain to bind to filaments and additional surfaces to dock onto the barbed end protomers) or that it adopts an appropriate three-dimensional conformation centred on the dimeric I-BAR domain. Indeed, IRSp53 has recently been shown by the Dominguez group (in a work submitted elsewhere) to fold into a closed conformation held together by an intramolecular interaction between the SH3 domain and a proline-rich region that is part of an atypical CDC42 binding interface. Mutation of these prolines (P278DA, P281D) leads to an ‘open’ IRSp53 conformation, which was capable of inhibiting barbed end growth, albeit slightly less efficiently than WT IRSp53 (Supplementary Figure S2d). CDC42-GTPγS abolished the slowing down of barbed end growth by IRSp53-WT and the proline mutant, but not by a CDC42 binding-deficient mutant (IRSp53-I267A-S268A; Figure 1E; Supplementary Figure S2e). Importantly, only active GTP-loaded CDC42, but not the GDP-bound form, relieved IRSp53-mediated inhibition of barbed end elongation (Figure 1F). Thus, IRSp53 can inhibit barbed end growth in both the ‘closed’ and ‘open’ conformation, and CDC42 relieves this inhibition presumably by sterically hindering the IRSp53:barbed end interaction.
Notably, inhibition of barbed end elongation by IRSp53 developed slowly during filament growth (Figure 1C). This finding is further supported by data showing that steady-state amounts of assembled actin were unaffected by the presence of IRSp53 and/or VASP at the concentrations tested (Supplementary Figure S2f). Furthermore, an IRSp53 mutant retaining the minimal surfaces for barbed end inhibition, but lacking the entire SH3 domain (and thus presumably adopting an open conformation) instantaneously inhibited barbed end growth (IRSp53-1–374; Figure 1G), similarly to IRSp53-W413G (Supplementary Figure S2b).
Finally, we monitored filament elongation in real time by in vitro TIRF microscopy in the presence of IRSp53. Similarly to our bulk spectrin-actin seed assays, addition of increasing concentrations of IRSp53 significantly slowed down barbed growth by ∼30% (Figure 2A). This effect was reverted by the addition of activated CDC42 (Supplementary Figure S2g). It must be noted, however, that TIRF data were collected in the early phase of filament elongation, between 0 and 400 s. In this time frame, we also detected partial inhibition of growth in bulk polymerization spectrin-actin seed assays. These data provide further evidence for the slow kinetics of IRSp53 association to barbed ends. To detect more extensive barbed end growth inhibition by TIRF, we would need to use concentrations of IRSp53 much higher than those we can achieve given the relatively low Kd (∼0.3 μM) of IRSp53 for barbed ends (Supplementary Figure S2c). The low affinity of IRSp53 for barbed ends could also result in short t1/2 of the IRSp53:barbed end complex, which may allow growth of barbed ends that are only transiently occupied by IRSp53. Alternatively, IRSp53 may bind slowly to barbed ends because it needs to undergo a slow conformational change for efficient binding, or a slow conformational change might follow IRSp53 binding to the sides of filaments, close to the barbed ends, resulting in a reduction in the rate of filament growth.
Although VASP and IRSp53 bind to each other, they have independent and opposing effects on barbed end growth. The addition of VASP to IRSp53-blocked barbed ends resulted in restoration of fast filament growth to rates obtained with VASP alone (Supplementary Figure S3a). Hence, in this assay, the possible association of VASP and IRSp53 to barbed ends does not change the kinetic behaviour of VASP-bound barbed ends. Thus, the formation of a VASP::IRSp53 complex has negligible effects on the filament elongation rate of VASP in solution.
IRSp53 promotes VASP recruitment and clustering on supported surfaces to drive processive filament elongation
High-density clustering of VASP onto functionalized beads allows processive, WH2 domain-mediated actin filament elongation, even in the presence of high concentrations of capping protein (Breitsprecher et al, 2008, 2011). Thus, we assessed whether beads saturated with IRSp53 could recruit and cluster VASP to drive processive actin assembly in the presence of capping protein using TIRF microscopy. To facilitate the visualization of actin filament growth, we initially used a chimaeric VASP (VASP-DdGAB), bearing the GAB motif of the Dictyostelium discoideum VASP homologue. This DdGAB motif, due to its high affinity for G-actin, markedly enhances filament elongation compared to WT mammalian VASP at the low G-actin concentration (1 μM) used in the TIRF assay (Figure 2B and C; Supplementary Figure S3d; Breitsprecher et al, 2011). We observed that high-density crowding or clustering of VASP-DdGAB on IRSp53-coated beads relieved inhibition of actin growth by capping protein and promoted marked processive filament elongation (Figure 2B and C; Supplementary Figure S3d). Buckling actin filaments grew away from the bead surfaces with elongation rates ranging from 37.1±7 to 45.4±5.5 actin subunits/s, which is comparable to the rate obtained with VASP-DdGAB-coated control beads (Figure 2B and C; Supplementary Figure S3d; Supplementary Movie 1). This elongation rate is about four times faster than the elongation rate (∼10 actin subunits/s) of spontaneously growing actin control filaments (Breitsprecher et al, 2008, 2011). The recruitment of VASP-DdGAB to the beads was specifically mediated by IRSp53, since no filament growth was observed when uncoated beads were incubated with soluble VASP (Figure 2B and C). Likewise, no filament growth was observed when beads were coated with IRSp53-W413G, which possesses a defective SH3 domain (Supplementary Figure S3d). We extended these observations to human VASP (hVASP) and EVL (hEVL). In both cases, the recruitment of the proteins to IRSp53-coated beads in the presence of capping proteins promoted processive filament elongation in the presence of capping proteins (Figure 2B and C). As expected, the rate of filament elongation in the presence of hVASP or hEVL was 3- to 4-fold slower compared to that of hVASP-DdGAB, due to the lower affinities of their GAB domains for G-actin (Breitsprecher et al, 2011). Collectively, these results demonstrate that bead-immobilized IRSp53 is necessary and sufficient to recruit and cluster VASP from solution, which in turn drives processive actin filament assembly in the presence of CP.
CDC42 favours the formation of an IRSp53–VASP complex in vivo
IRSp53 can bind both activated RAC (Miki et al, 2000) and CDC42 (Govind et al, 2001; Krugmann et al, 2001). MENA, another member of the ENA/VASP family protein, has been shown to bind to IRSp53 in a CDC42-dependent manner in vitro (Krugmann et al, 2001). Using recombinant purified proteins, we first examined whether IRSp53 serves as a link between VASP and active CDC42. Binding of IRSp53 to GTP-CDC42 was readily detected, while VASP interacted with GTP-CDC42 in an IRSp53-dependent manner, indicating the existence of a CDC42–IRSp53–VASP complex (Figure 3A). Furthermore, the addition of saturating amounts of GTP-CDC42 increased the ability of IRSp53 to interact with VASP in vitro (Figure 3B). Similar results were obtained in co-immunoprecipitation experiments. The ectopic expression of active CDC42, but not of a dominant-negative mutant, significantly enhanced the amount of endogeneous IRSp53 co-immunoprecipitating with VASP (Figure 3C). The CDC42-dependent increase in the VASP:IRSp53 association was accompanied by a diminution of the EPS8:IRSp53 interaction (Figure 3C). Notably, VASP and EPS8 compete for binding to the SH3 domain of IRSp53, and in unstimulated conditions EPS8, which contrary to VASP acts as a capper, binds more efficiently than VASP to IRSp53 (Menna et al, 2009; Vaggi et al, 2011). Thus, CDC42 binding promotes a switch in IRSp53-based molecular complexes that favours IRSp53 association with VASP, while reducing the interaction with EPS8, leading to the formation of an actin assembly promoting CDC42:IRSp53:VASP complex.
Figure 3.
CDC42 modulates the interaction between IRSp53 and VASP. (A) IRSp53 connects VASP with CDC42. Equimolar concentrations (200 nM) of IRSp53and/or VASP were incubated with 1 μM GDP- or GTPγS-loaded GST-CDC42 immobilized on beads, as indicated. Inputs (10% of total) and bound material were analysed by immunoblotting to visualize VASP or by Ponceau staining to detect IRSp53 and CDC42. (B) Equal amounts of VASP were incubated with Ni-NTA agarose beads coating with increasing concentrations of recombinant purified His-IRSp53 (0, 100, 250, and 500 nM), in the absence or presence of saturating concentrations of GTP-loaded GST-CDC42. Inputs (10% of total) and bound material were immunoblotted with the indicated antibodies (Ponceau staining visualizes IRSp53). Right panel: Quantification of the fraction of VASP bound to His-IRSp53-Ni-NTA beads was determined by analysing blots with the ImageJ software. The fraction (bound/total) of VASP was plotted against the concentrations of IRSp53 (data are the mean±s.d. of three independent experiments). (C) Lysates (2 mg) of HeLa cells, transfected with empty vector as a control (EV) or with either CDC42 dominant-negative (N17) or constitutively active (L61) mutants, were immunoprecipitated with an anti-VASP (VASP) or control antibody (CTR). Input lysates (40 μg) and immunoprecipitates (IPs) were immunoblotted with the indicated antibodies. Right panel: The amount of IRSp53 bound to VASP was determined using the ImageJ software. Relative binding between IRSp53 and VASP (data are the mean±s.d. of five independent experiments).
Source data for this figure is available on the online supplementary information page.
IRSp53-mediated association of VASP to PIP2-enriched membrane is required for proper clustering of VASP at the PM and filopodia formation
Filopodia initiation is preceded by localized accumulation of VASP into discrete foci along the PM (Rottner et al, 1999; Svitkina et al, 2003; Yang et al, 2007). IRSp53 also binds and localizes at the PM, accumulating at PIP2-rich sites (Zhao et al, 2011). Thus, IRSp53 may control and enhance VASP activity by recruiting and spatially restricting its localization on PIP2-rich, curved PM. We first tested this hypothesis using FACS-based in vitro assays that monitor the ability of fluorescently labelled (Alexa-488) proteins to bind PIP2-rich liposomes in solution (Temmerman and Nickel, 2009). Fluorescently labelled IRSp53 and IRSp53-W413G bound efficiently to liposomes only when PIP2 was included in the lipid mixture (Figure 4A; Supplementary Figure S4a). Alexa-488-VASP instead did not associate with liposomes. However, when PIP2 liposomes were incubated first with unlabelled IRSp53, Alexa-488-VASP readily bound to PIP2 lipids (Figure 4A). This interaction was abolished when unlabelled IRSp53-W413G was used instead of WT IRSp53 (Figure 4A), or by the addition of an excess of unlabelled EPS8 that competes with VASP for binding to IRSp53 (Disanza et al, 2006) (Figure 4A).
Figure 4.
IRSp53 is required for VASP recruitment and clustering at the PM. (A) VASP is recruited to PIP2-containing liposomes through IRSp53. Unlabelled or labelled (*Alexa-488) IRSp53-WT, IRSp53-W413G, EPS8, and VASP, at the indicated concentrations and combinations, were added to 1 mM of 10% PIP2 containing liposomes. Interactions were analysed by FACS and data were processed with the CellQuest Pro software (Becton Dickinson) and expressed as mean±s.e.m. of at least three independent experiments performed in duplicate. **Indicates P<0.01 (t-test, comparing each condition to control, buffer). (B) IRSp53 removal diminishes VASP clustering at the PM. Still images from differential interference contrast (DIC) and fluorescence time-lapse analysis of migrating cells. MEFs transfected with empty vector (KO-pBABE), KO-pBABE-IRSp53 WT, and KO-pBABE-IRSp53 W413G were electroporated with GFP-VASP and plated on laminin (40 μg/ml). Twenty-four hours after electroporation, cells were analysed by time-lapse imaging (5 min, time interval 5 s). Bar is 5 μm (2 μm for the insets). Arrows indicate VASP puncta (see also Supplementary Movies 2, 3, 4, 5). (C) Quantification of the number (N) of clusters (mean±s.e.m.) formed by GFP-VASP was obtained by analysing 20 cells for each condition. (D) The expression levels of EGFP-VASP, IRSp53, and vinculin were analysed by immunoblotting in KO-pBABE, KO-pBABE-IRSp53 WT, and KO-pBABE-IRSp53 W413G transfected MEFs.
We next explored whether IRSp53 is required for the formation of VASP foci at the leading edge. We took advantage of MEF cells derived from IRSp53 null mice (Sawallisch et al, 2009; Weiss et al, 2009) stably infected with mock empty vector, or vectors expressing IRSp53-WT or -W413G to levels similar to the endogeneous protein (Figures 4D and 5C). We analysed by time-lapse imaging the cellular dynamics of GFP-VASP in MEFs plated on laminin (Figure 4B–D; Supplementary Movies 2, 3, 4, 5). In reconstituted WT IRSp53 MEFs, VASP was distributed to focal adhesions and prominently enriched along lamellipodia (Figure 4B). At this latter location, bright foci of GFP-VASP frequently moved laterally, merged, and fused. These foci invariably preceded the extension of filopodia that were also positive for fascin (Figure 4B; Supplementary Figure S4b and c; Supplementary Movies 7 and 8). Conversely, in MEFs devoid of IRSp53 or reconstituted with IRSp53-W413G mutant, the formation of leading edge-localized GFP-VASP foci was significantly reduced, while VASP localization and dynamics in lamellipodia and focal adhesion remained unaltered (Figure 4B; Supplementary Movies 2, 3, 4, 5). We also tested whether IRSp53 accumulates along the leading edge in discrete foci together with VASP by dual-colour time-lapse microscopy using GFP-VASP and mCherry-IRSp53. Notably, both proteins accumulated into bright, dynamic puncta at sites of filopodia initiation (Supplementary Figure S4b; Supplementary Movie 7). Time-resolved analysis of the accumulation of cherry-IRSp53 and GFP-VASP into foci revealed that IRSp53 preceded the recruitment of VASP by 1.4 s on average. (Supplementary Figure S4c; Supplementary Movie 8). Notably, mDia1, another putative interactor of IRSp53 with filament elongation activity that is implicated in filopodia (Goh et al, 2012), as well as N-WASP and WAVE (data not shown), did not display any accumulation into foci at the leading edge (Supplementary Figure S4d). This finding suggests that these proteins are not required in the initial phase of filopodia formation. Thus, IRSp53 is specifically implicated in the recruitment of VASP to PIP2-rich membrane and in its apparent clustering along PM sites where filopodia are initiated, implying a requirement of the IRSp53:VASP complex in the formation of these protrusions.
Figure 5.
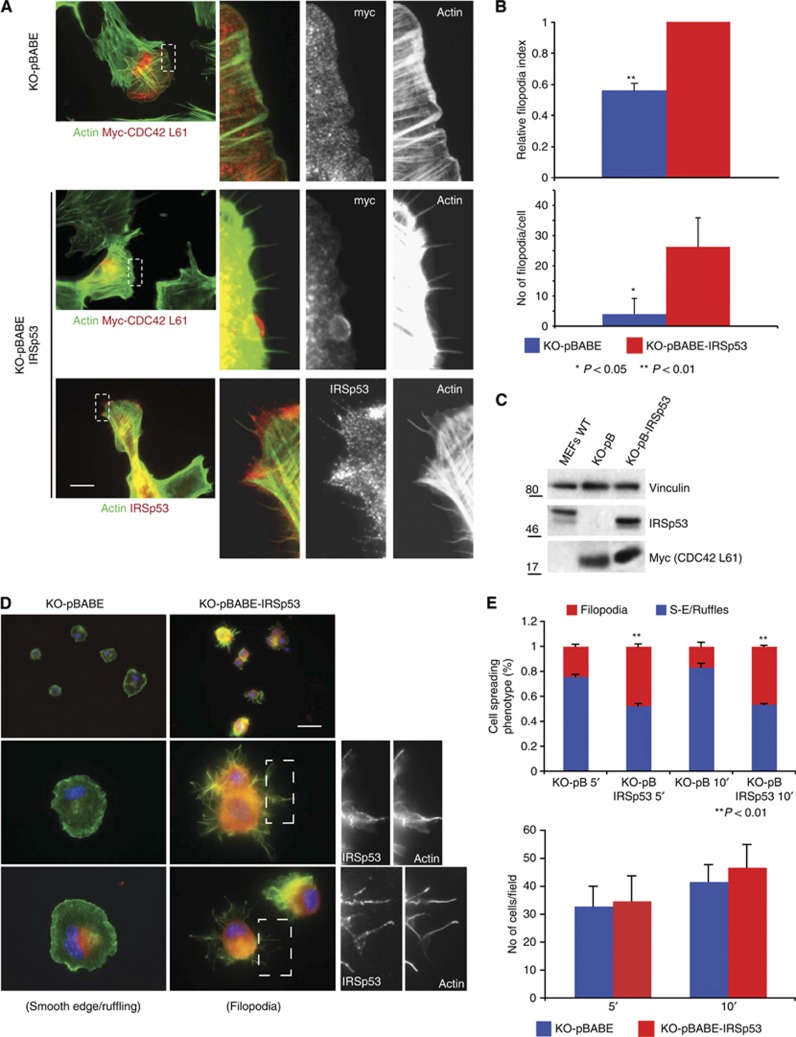
IRSp53 is required for CDC42-induced filopodia formation. (A) MEF KO-pB and KO-pB-IRSp53 cells, transfected with a constitutively active Myc-tagged CDC42 (Myc-CDC42 L61), were fixed and stained with anti-Myc or anti-IRSp53 antibodies and FITC-phalloidin to detect myc-CDC42 L61, IRSp53 (KO-pB-IRSp53), or F-actin, respectively. The right panels represent magnifications of the indicated areas of each corresponding image. Scale bar is 10 μm (2.5 μm for the magnifications). (B) Quantification of filopodia induced by CDC42 L61 in cells shown in (A). Upper panel. The number of cells expressing CDC42 L61 and presenting filopodia was counted. Data are expressed as the fraction of CDC42 L61-expressing cells presenting filopodia with respect to CDC42 L61-expressing KO-pB-IRSp53 cells (relative filopodia index). CDC42 L61 induced filopodia in ∼80% of transfected KO-pB-IRSp53 cells. Lower panel: The number of filopodia/cell was counted. At least 100 cells were analysed in each experiment and three independent experiments were performed. Data are the mean±s.e.m. (C) The expression levels of IRSp53, Myc-CDC42-L61, and vinculin were analysed by immunoblotting. There are four distinct splicing isoforms of IRSp53 (Scita et al, 2008), which differ only in the very C-terminal region, displaying either a WH2 domain or a PDZ binding motif. The isoform we introduced into IRSp53 KO MEFs was BAIAP2-S carrying a PDZ motif at its C-terminus. (D) IRSp53 controls the mode of cell spreading and increases the number of filopodia forming cells. MEFs KO-pB and KO-pB-IRSp53 were seeded on fibronectin-coated coverslips and fixed at 5 or 10 min after seeding. Cells were stained with FITC-phalloidin (green), anti-IRSp53 antibody (red) and Dapi (blue) to detect actin filaments, IRSp53 (pB-KO-IRSp53) and nuclei, respectively. The right panels represent magnifications of the indicated areas. Scale bar is 20 μm for the upper panels, 10 μm for the lower panels, and 5 μm for magnifications. (E) Upper panel: Quantification of spreading phenotypes. Normalized data±s.e.m. of the number of cells displaying smooth-edged/ruffle (S-E/Ruffles) and filopodia spreading phenotypes in KO-pBABE and KO-pBABE-IRSp53 MEF cells, 5 and 10 min after seeding. At least 50 cells/experiment were analysed in three independent experiments. Lower panel: Quantification of adhering cells. The number of adhering cells/field±s.e.m. in KO-pBABE and KO-pBABE-IRSp53 MEF cells, 5 and 10 min after seeding. At least 10 fields in each experiment were counted.
To test the latter hypothesis, we monitored filopodia induced by the expression of active CDC42 or following the initial phases of cell spreading on extracellular matrix (ECM) substrates (Kozma et al, 1995; Applewhite et al, 2007). A >40% reduction in CDC42-induced filopodia was observed in MEFs devoid of IRSp53 compared with cells reconstituted with IRSp53-WT (Figure 5A–C). Of note, when IRSp53 is expressed, it displays a diffuse punctate distribution throughout the cytoplasm, but also decorates the shafts of protruding filopodia (Figure 5A–C). Similarly, in IRSp53-WT-reconstituted MEFs, VASP, as expected, was confined to filopodia tips. Conversely, removal of IRSp53 or re-expression of IRSp53-W413G in IRSp53 null MEFs prevented the proper localization of VASP (Supplementary Figure S5a–d). VASP localized at the tips of filopodia in ∼80% of IRSp53-WT reconstituted cells, but only in 20 and 30% of empty vector and IRSp53-W314G reconstituted cells, respectively.
Filopodia protrusions are also typically formed during cell spreading onto ECM. Three different spreading modes have been described for mouse fibroblasts: smooth-edged, filopodial or ruffling (Applewhite et al, 2007). ENA/VASP proteins are known to increase the number of cells that form filopodia (Applewhite et al, 2007). We performed the same analysis in IRSp53 null MEFs infected with empty vector or IRSp53-WT shortly after plating on fibronectin (Figure 5D). We then scored cells into one of the phenotypic spreading modes (Figure 5E). Removal of IRSp53 does not affect cell adhesion (Figure 5E), but favours a smooth-edged/ruffling mode of spreading and significantly reduces the number of cells forming filopodia, mimicking VASP loss-of-function (Applewhite et al, 2007). Finally, functional interference of CDC42 by expression of a dominant-negative mutant significantly reduced VASP clustering at filopodia initiation sites and its subsequent localization at filopodia tips during cell spreading (Supplementary Figure S5e). Thus, IRSp53 is required for efficient filopodia formation during CDC42-dependent cell spreading, strengthening the notion that IRSp53 and VASP act in the same pathway.
IRSp53 drives cell directional migration and invasion, and is required for efficient epidermal wound healing in vivo
Filopodia drive directional migration and invasion into ECM (Vignjevic and Montagnac, 2008). Thus, we tested the impact of IRSp53 on these processes using our genetically modified MEFs in a battery of migration and invasion assays (Figure 6). In wound healing assays, IRSp53 null MEFs were defective in closing the wound when compared with IRSp53-reconstituted cells. Defective wound closure was due to reduced speed and impaired directional migration (Figure 6A; Supplementary Movie 9). Importantly, IRSp53 null MEFs were impaired in extending polarized protrusions in the wound space and moved randomly rather than directionally (Supplementary Movie 9). Similar results were obtained in trans-well migration and invasion assays. IRSp53 removal also impaired chemotactic migration towards serum or PDGF (Figure 6B) and directed cell invasion in Matrigel-coated transwells (Figure 6C) as well as into native collagen type I chamber (Sabeh et al, 2009) (Supplementary Figure S6b; Supplementary Movie 10).
Figure 6.
IRSp53 removal affects directional cell migration and invasion. (A) Wound healing assay. KO-pBABE and KO-pBABE-IRSp53 MEFs, seeded and grown to confluence, were scratched with a tip and analysed by DIC time-lapse imaging (20 h, 5 min time interval; see Supplementary Movie 9). Left panels: Still images taken at the indicated time points. Bar is 50 μm. White-dashed lines mark the front of migrating cells. Right panels: Single-cell migration was analysed using the ImageJ software. The directional migration index (upper panel) and the average speed (central panel) of cells were calculated. The directional migration index represents the ratio between the total distance covered by cells and the distance covered longitudinally towards the centre of the wound. At least 20 cells were analysed for each condition. Lower panel: Total wound area was measured at the indicated time points using the ImageJ software and relative wound closure was determined as described in Materials and methods. Data are the mean±s.e.m. from n=5 fields/experiment. Data are from three independent experiments. (B) Transwell assay. KO-pBABE and KO-pBABE-IRSp53 MEFs were seeded on fibronectin-coated chambers. Twenty percent serum (left panel) or 10 ng/ml PDGF (right panel) was added in the lower chamber to generate a chemo-attractive gradient. Cells were fixed after 6 h and stained with crystal violet. Bar is 50 μm. The number of cells passing the semi-permeable membrane was counted. At least 20 different fields were analysed for each condition. Data are the mean±s.e.m. from three independent experiments. (C) Invasion assay. KO-pBABE and KO-pBABE-IRSp53 MEFs were seeded on Matrigel-coated chambers. PDGF was added in the lower chamber to generate a chemo-attractive gradient. Cells were fixed after 16 h and stained with Dapi to detect nuclei. Left panel: Quantification of the number of cells passing the semi-permeable membrane. At least 10 fields were analysed for each condition. Data are the mean±s.e.m. from three independent experiments. Right panel: Representative images of invading cells. Bar is 10 μm.
To assess whether IRSp53 removal also resulted in an organismal migratory phenotype, we subjected 10-week-old IRSp53 KO and WT littermate mice to the dermal punch wound model of wound healing. During tissue repair keratinocytes, endothelial cells, macrophages, and mesenchymal cells, such as fibroblasts or myofibroblasts, migrate into the wound area (Gabbiani, 2003; Stappenbeck and Miyoshi, 2009). To assess epidermal wound healing, we punched four wounds on the back skin of 10-week-old IRSP53 KO and WT mice and monitored wound closure over a 7-day period. Microphotographs, morphometric quantification, and H&E analysis of wounds revealed that IRSp53 KO animals displayed a marked delay in wound closure relative to control animals (Figure 7A and B). Indeed, a thick clot was still present at 7 days after wounding and the wound limits were larger, while the re-epithelialization process was incomplete (Figure 7B). Notably, we detected no differences in the cellular composition and overall morphology between IRSP53 KO and WT mice, indicating that IRSp53 is dispensable for skin development, but required for wound re-epithelialization (Figure 7C). Importantly, IRSp53 is expressed in epidermal keratinocytes, macrophages, isolated dermal fibroblasts, and endothelial cells (Figure 7D and E). Removal of IRSp53 delayed wound closure by keratinocytes in vitro (data not shown). Furthermore, quantitative analysis of cells at wound sites revealed that while endothelial cells were recruited in similar numbers and generated blood vessels of similar size in IRSp53 KO and WT mice, macrophages were instead significantly reduced in IRSp53 KO mice (Supplementary Figure S7). This result is consistent with previous findings showing that IRSp53 removal impairs macrophage chemotactic cell motility (Abou-Kheir et al, 2008). Collectively, these data suggest that IRSp53 removal impairs the re-population of wound sites by epithelial keratinocytes, macrophages, and dermal fibroblasts, which are the major cells involved in wound healing in vivo.
Figure 7.
Genetic removal of IRSp53 in mice impairs wound repair. (A) Time course of wound closure. (Left panels) Microphotographs of wounds were captured on days 0, 1, 2, 4, 5, 6, and 7 after wounding to determine the degree of wound closure in IRSp53 KO and control WT mice. Representative images from days 0, 2, and 5 are shown. Right panel. Total wound area on days 0, 2, and 5 after wounding was measured using the ImageJ software and relative wound closure was quantified as described in Materials and methods. Data are the mean±s.e.m. from n=6 animals/group. Scale bar is 2 mm. (B) H&E analysis of back skin from IRSp53 KO and WT mice at day 7 after wounding. Black arrows denote the wound limits. White arrowheads denote the re-epithelialization regions. Lower panels represent × 5 magnifications of the insets (dashed red lines) in upper panels. C, clot; E, epithelium. Scale bar is 200 μm (higher magnifications, 40 μm). (C) H&E analysis of back skin from IRSp53 KO (left) and WT (right) mice. Lower panels represent × 5 magnifications of the insets (dashed red lines) in upper panels. Scale bar is 200 μm (lower panels, 40 μm). (D) IHC analysis of IRSp53 in back skin from IRSp53 KO (left) and WT (right) mice. Lower panels represent × 5 magnifications of the insets (dashed red lines) in upper panels. Scale bar is 200 μm (lower panels, 40 μm). (E) IRSp53 is expressed in mouse keratinocytes, dermal fibroblasts, macrophages, and endothelial cells. The expression levels of IRSp53 and vinculin were analysed by immunoblotting in MEFs and mouse keratinocyte (C50), dermal fibroblast (M Dunni, M-D) cell lines. Bone marrow macrophages (Mϕ) and lung endothelial cells (EC) derived from IRSp53 KO or WT mice were also analysed.
Source data for this figure is available on the online supplementary information page.
Discussion
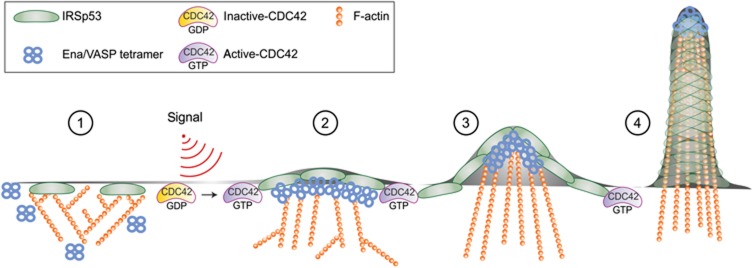
Our study identifies a signalling and molecular mechanism that controls, in a spatially confined manner, VASP actin regulatory activity during the initial phases of filopodia formation from lamellipodia sheets, in response to CDC42 activation. We show that the membrane deforming I-BAR-domain containing protein IRSp53 plays a critical role in this process and characterize the physiological relevance of IRSp53 regulation of filopodia in tissue repair and wound healing in mice. While CDC42 and VASP are established critical factors in filopodia formation, the signalling mechanisms regulating VASP activity and, in particular, its clustering-dependent processive elongation function, have remained elusive. Here, we show that the CDC42-IRSp53 axis is critical in mediating a switch from inhibition of actin barbed end growth (by IRSp53) to processive filament elongation following IRSp53-mediated clustering of VASP. Moreover, this signalling axis spatially restricts VASP activity along the leading edge of migrating cells, resulting in filopodia formation (Figure 8).
Figure 8.
Model depicting how CDC42 switches IRSp53 from inhibition of actin assembly to filament elongation by clustering VASP during filopodia formation. A series of distinct signalling and biochemically defined steps characterize the initial phases of filopodia formation, mediated by the membrane deforming protein IRSp53 and its interactor VASP. Step 1: in the absence of stimuli, IRSp53 binds to the PM and slows down barbed end elongation. The constitutive association of IRSp53 with the capping protein EPS8 likely reinforces EPS8 capping activity (not shown) (Disanza et al, 2006; Vaggi et al, 2011). Step 2: following filopodia-inducing stimuli, activated CDC42 binds to IRSp53. This interaction relieves IRSp53-mediated inhibition of filament growth and promotes the formation of a CDC42:IRSp53:VASP complex, and simultaneously reduces the formation of the IRSP53:EPS8 complex. Step 3: binding to CDC42 further facilitates the formation of IRSp53 and VASP foci at the leading edge of the PM, in which VASP clustering allows processive actin filament elongation in the presence of capping protein. Step 4: membrane deforming activity of IRSp53 and processive filament elongation by VASP work in concert to extend actin filaments beyond the x, y plane of the membrane. During this process, the filaments become cross-linked by fascin (not shown), to resist counter membrane tension and extracellular compression forces.
At the molecular level, IRSp53 exerts this function through a number of essential, biochemical, and signalling activities. First, via the I-BAR domain, IRSP53 can bind, sense, and promote PM deformation (Zhao et al, 2011). This property is entirely independent from forces generated by actin polymerization or protein:protein interactions (Zhao et al, 2011), suggesting that an initial step in the assembly of the machinery for filopodia formation is localized membrane evagination (Figure 8, step 1). These initial protrusions are subsequently, or concomitantly, stabilized and extended by actin filaments that elongate in a linear fashion (Figure 8, step 2). Importantly, in the absence of stimuli, IRSp53 slows down barbed end growth, thus safeguarding against unwanted growth of actin filaments that would occupy the free space of outwardly curved PM, leading to unrestrained, isotropic filopodia extensions. In this respect, it is well established that the expression of the isolated IRSp53 I-BAR domain, which is devoid of barbed end activity, elicits the formation of aberrant, isometrically distributed filopodia-like structures that are nearly devoid of F-actin (Yang et al, 2009). Intriguingly, the weak capping activity of IRSp53 may be reinforced by its association with EPS8 (Disanza et al, 2006), which caps barbed ends with nanomolar affinity (Disanza et al, 2004), suggesting that an EPS8:IRSp53 complex may be critical for proper control of filament growth at the leading edge. The inhibition of barbed end growth by IRSp53 is relieved by activated CDC42, which appears to compete with EPS8 for binding to IRSp53, as evidenced in co-immunoprecipitation experiments. This latter finding is remarkable from different points of view. At the biochemical level, it suggests that IRSp53 adopts different conformations controlled by CDC42 binding. In the absence of active CDC42, IRSp53 folds into a ‘closed state’ that negatively regulates actin polymerization. Binding of CDC42 induces a conformational switch that not only impedes barbed end association, but also promotes IRSp53 binding with some of its downstream targets (see below). Structural/biochemical evidence in this direction are provided by recent findings by Roberto Dominguez and colleagues (manuscript submitted elsewhere) that reveal how an SH3-CRIB-based intramolecular interaction is critical in mediating the switch from a ‘closed’ to an ‘open/active’ state of IRSp53. One additional important feature of IRSp53 is that the conformational switch appears to be dependent on consecutive association with EPS8 and CDC42 that eventually assemble into an open heterohexamer. Under these conditions, the barbed capping activity of IRSp53 is prevented by CDC42, while SH3 interactors, such as VASP, may bind to IRSp53, compete out EPS8, and eventually switch the activity of the complex towards filament elongation. Notably, we have previously shown that IRSp53 may contribute to filopodia elongation by promoting the bundling activity of a CDC42–IRSp53–VASP complex through oligomerization (Vaggi et al, 2011). Thus, the formation of this complex may not only be required to promote processive filament elongation, an activity that is essential to initiate the growth of actin filaments and of filopodia, but also the bundling of filaments to sustain filopodia extension. This latter activity is thought to be primarily mediated by fascin (Vignjevic et al, 2006). Nevertheless, bundling particularly close to the barbed ends, where the IRSp53::VASP complex is restricted, may subsequently favour tighter cross-linking by fascin. This cross-linking may confer sufficient stiffness to filopodia that then allows them to resist counter PM tension and extracellular forces.
The regulation of IRSp53 barbed end capping activity by CDC42 is also important in the context of RHO family GTPase signalling, and more generally, in the regulation of filament barbed end growth since little is known about the mechanisms and signalling pathways that target this critical activity. Indeed, blockade of barbed end growth is viewed as a ‘default process’ that must immediately take place to avoid uncontrolled filament elongation that would otherwise occur on free barbed ends, given the high concentration of G-actin (or G-actin::profilin complex) in cells (Koestler et al, 2009). During filopodia initiation, instead, CDC42 actively and locally inhibits IRSp53 capping, further promoting a switch towards the formation of an active IRSP53::VASP complex driving filopodia formation.
Our data also shed light on how VASP activity is regulated in cells. First, we provide evidence that VASP, via IRSp53, is directly linked to CDC42, thus defining a CDC42–IRSp53–VASP signalling relay axis for actin assembly underlying filopodia formation. Second, we show that IRSp53 drives VASP clustering, which is essential for promoting sustained processive actin filament elongation of VASP at the PM in the presence of capping proteins (Breitsprecher et al, 2008, 2011). It has recently been shown, using single molecule TIRF analysis, that isolated VASP is also capable of promoting processive elongation in the presence of profilin (Hansen and Mullins, 2010). However, in solution, individual VASP tetramers are only weakly processive (Hansen and Mullins, 2010). Clustering of VASP into dense foci appears, therefore, to be necessary to enhance this activity for the extension of spatially restricted and timely coordinated filopodia. Consistently, we show that VASP is recruited to foci at the leading edge primarily via specific IRSp53-mediated protein:protein interactions. We cannot exclude that barbed end binding by VASP may also contribute to localization at the PM. However, VASP association to free barbed ends does not explain signalling-dependent VASP clustering as satisfactorily as IRSp53-mediated recruitment of VASP. In this latter respect, it is worth emphasizing that the binding of VASP to membrane-associated proteins, such as Mig-10/RIAM/Lamellipodin or CXCR2, has previously been reported (Krause et al, 2004; Neel et al, 2009). However, none of these proteins localized dynamically into discrete foci at filopodia initiation sites. Rather, they were distributed along the entire leading edge of lamellipodia, suggesting that they more likely contribute to the regulation of VASP activity in these migratory protrusions. Thus, it appears likely that distinct VASP-based protein complexes participate in the control of its multiple activities in different migratory structures, ultimately regulating the transition between lamellipodia-to-filopodia. It must be also pointed out that for this transition to be complete other activities, such as nucleation by formin family proteins (Peng et al, 2003; Pellegrin and Mellor, 2005; Schirenbeck et al, 2005) and bundling by fascin (Faix et al, 2009) are likely to be required to cooperate with the IRSp53::VASP filopodia initiation complex. Reconstituting the interplay between these factors will lead to a better understanding of how cells dynamically regulate the architecture of functional actin networks and their biological consequences.
Materials and methods
All animal experiments were conducted in accordance with national guidelines and were approved by the ethics committee of the Animal Welfare Office of the Italian Work Ministry and conformed to the legal mandates and Italian guidelines for the care and maintenance of laboratory animals.
Expression vectors, antibodies, reagents, and cells
Cytomegalovirus (CMV)-promoter-based and elongation factor-1 (EF1)-promoter-based eukaryotic expression vectors and GST bacterial expression vectors were generated by recombinant PCR. pEGFP–IRSp53 was from H Nakagawa (Austrian Academy of Sciences, Vienna, Austria), pmCherry-Fascin was from K Rottner (Institute of Genetics, University of Bonn, Germany) and JV Small (Institute of Molecular Biotechnology, Vienna, Austria), pmCherry-Eps8 (Addgene, One Kendall Sq, Cambridge, MA). All constructs were verified by sequencing.
The antibodies used were monoclonal anti-Eps8 (Transduction Laboratories, Lexington, KY); rabbit polyclonal anti-GST (Santa Cruz Biotechnology, Santa Cruz, CA); anti-Myc 9E10 (Babco, Berkeley, CA); anti-tubulin and rabbit polyclonal anti-IRSp53 (Sigma-Aldrich, St Louis, MO); rat monoclonal anti-F4/80 (AbD Serotec, Oxford, UK); rabbit polyclonal anti-CD31 (Abcam, Cambridge, UK). The monoclonal anti-IRSp53 was generated as described (Disanza et al, 2006). Immortalized fibroblasts from IRSp53+/+ and IRSp53–/– embryos were as described (Weiss et al, 2009). pBABE control, pBABE IRSp53 WT, and pBABE IRSp53 W413G re-expressing cells were obtained by infection of immortalized fibroblasts from IRSp53–/– embryos.
For additional detailed information on Materials and methods, see Supplementary data.
Actin polymerization assays
Actin polymerization was monitored by the increase in fluorescence of 10% pyrenyl-labelled actin. Seeded polymerization was induced by addition of 0.05 M KCl, 1 mM MgCl2 and 0.2 mM EGTA to a solution of Ca-ATP–G-actin (1.25 μM) containing spectrin-actin seeds, 5 μM profilin, and various concentrations of purified proteins. Fluorescence measurements were performed at 20°C in a Safas Sfx or a Spex Fluorolog 2 spectrofluorimeter. Samples from seeded-polymerization assay were pelleted for 30 min at 400 000 g in a Beckman TL-100 tabletop ultracentrifuge. Equal amounts of starting materials, supernatants, and pellets were solubilized in loading buffer, boiled, and resolved on an SDS-PAGE gel. Each single assay was repeated at least five times with similar results. Representative experiments are shown.
Dilution-induced depolymerization of pyrenyl F-actin was monitored in a Spex Fluorolog 2 instrument. A 50% labelled F-actin solution (2.5 μM F-actin) was diluted 50-fold into polymerization buffer (5 mM Tris–HCl pH 7.8, 0.1 mM CaCl2, 0.2 mM ATP, 1 mM DTT, 0.01% NaN3, 50 mM KCl, 1 mM MgCl2, and 0.5 mM EGTA) containing the desired amount of IRSp53 or VASP.
TIRF microscopy for monitoring of actin polymerization and elongation was performed with flow cells, essentially as described in Breitsprecher et al (2011) and Jegou et al (2011) (see Supplementary Experimental Procedures for details).
Statistical analysis
All data are presented as the mean±standard errors of mean (s.e.m.), except when specified otherwise. A Student’s t-test was used to calculate the P-values.
Supplementary Material
Acknowledgments
We are indebted to Rosalind Gunby for critically editing the manuscript, to Frank B Gertler and Dorothy Schafer for providing VASP reagents, peptide arrays and critically reading the manuscript, and to Vic Small and Klemens Rottner for providing pmCherry-Fascin. Work in the authors’ laboratory is supported by grants from the Associazione Italiana per la Ricerca sul Cancro (GS and AD), the European Research Council (Advanced-ERC-268836 to GS), the Italian Ministry of Education-University-Research (MIUR) (GS), the Association for International Cancer Research (GS), and the CARIPLO Foundation (GS and AD). MFC acknowledges grants from the ERC (Advanced-ERC-249982), from the EU (FP7 program MitoSys-# 241548, and from the Ligue Nationale contre le Cancer (équipe labellisée). CA is supported by fund G.0441.10N from the FWO-Vlaanderen. JF was supported by grants from the Deutsche Forschungsgemeinschaft (330/9-1 and 330/5-1). RD was supported by the National Institute of Health grant R01 MH087950. DK was supported by training grants T32 AR053461 from National Institute of Health and PF-13-033-01-DMC from the American Cancer Society.
Author contributions: AD, SB and M-FC wrote part of the manuscript, conceived and performed the experiments, and analysed the data. MW, FM, DSU, PM, H-MM, JL, DW, SC, and AP performed experiments and analysed the data. DJK and GR-L conceived and performed the experiments, and analysed the data. WN and CA conceived experiments and analysed the data. RD conceived and designed the experiments, and analysed the data. JF and GS wrote the manuscript, conceived and designed the experiments, and analysed the data.
Footnotes
The authors declare that they have no conflict of interest.
References
- Abbott MA, Wells DG, Fallon JR (1999) The insulin receptor tyrosine kinase substrate p58/53 and the insulin receptor are components of CNS synapses. J Neurosci 19: 7300–7308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abou-Kheir W, Isaac B, Yamaguchi H, Cox D (2008) Membrane targeting of WAVE2 is not sufficient for WAVE2-dependent actin polymerization: a role for IRSp53 in mediating the interaction between Rac and WAVE2. J Cell Sci 121: 379–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams JC (2004) Roles of fascin in cell adhesion and motility. Curr Opin Cell Biol 16: 590–596 [DOI] [PubMed] [Google Scholar]
- Ahmed S, Goh WI, Bu W (2010) I-BAR domains, IRSp53 and filopodium formation. Semin Cell Dev Biol 21: 350–356 [DOI] [PubMed] [Google Scholar]
- Akin O, Mullins RD (2008) Capping protein increases the rate of actin-based motility by promoting filament nucleation by the Arp2/3 complex. Cell 133: 841–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Applewhite DA, Barzik M, Kojima SI, Svitkina TM, Gertler FB, Borisy GG (2007) Ena/VASP proteins have an anti-capping independent function in filopodia formation. Mol Biol Cell 18: 2579–2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann C, Fischer L, Walter U, Reinhard M (1999) The EVH2 domain of the vasodilator-stimulated phosphoprotein mediates tetramerization, F-actin binding, and actin bundle formation. J Biol Chem 274: 23549–23557 [DOI] [PubMed] [Google Scholar]
- Barzik M, Kotova TI, Higgs HN, Hazelwood L, Hanein D, Gertler FB, Schafer DA (2005) Ena/VASP proteins enhance actin polymerization in the presence of barbed end capping proteins. J Biol Chem 280: 28653–28662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear JE, Svitkina TM, Krause M, Schafer DA, Loureiro JJ, Strasser GA, Maly IV, Chaga OY, Cooper JA, Borisy GG, Gertler FB (2002) Antagonism between Ena/VASP proteins and actin filament capping regulates fibroblast motility. Cell 109: 509–521 [DOI] [PubMed] [Google Scholar]
- Bockmann J, Kreutz MR, Gundelfinger ED, Bockers TM (2002) ProSAP/Shank postsynaptic density proteins interact with insulin receptor tyrosine kinase substrate IRSp53. J Neurochem 83: 1013–1017 [DOI] [PubMed] [Google Scholar]
- Breitsprecher D, Kiesewetter AK, Linkner J, Urbanke C, Resch GP, Small JV, Faix J (2008) Clustering of VASP actively drives processive, WH2 domain-mediated actin filament elongation. EMBO J 27: 2943–2954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitsprecher D, Kiesewetter AK, Linkner J, Vinzenz M, Stradal TE, Small JV, Curth U, Dickinson RB, Faix J (2011) Molecular mechanism of Ena/VASP-mediated actin-filament elongation. EMBO J 30: 456–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CE, Odde DJ (2008) Traction dynamics of filopodia on compliant substrates. Science 322: 1687–1691 [DOI] [PubMed] [Google Scholar]
- Disanza A, Carlier MF, Stradal TE, Didry D, Frittoli E, Confalonieri S, Croce A, Wehland J, Di Fiore PP, Scita G (2004) Eps8 controls actin-based motility by capping the barbed ends of actin filaments. Nat Cell Biol 6: 1180–1188 [DOI] [PubMed] [Google Scholar]
- Disanza A, Mantoani S, Hertzog M, Gerboth S, Frittoli E, Steffen A, Berhoerster K, Kreienkamp HJ, Milanesi F, Di Fiore PP, Ciliberto A, Stradal TE, Scita G (2006) Regulation of cell shape by Cdc42 is mediated by the synergic actin-bundling activity of the Eps8-IRSp53 complex. Nat Cell Biol 8: 1337–1347 [DOI] [PubMed] [Google Scholar]
- Faix J, Breitsprecher D, Stradal TE, Rottner K (2009) Filopodia: Complex models for simple rods. Int J Biochem Cell Biol 41: 1656–1664 [DOI] [PubMed] [Google Scholar]
- Ferron F, Rebowski G, Lee SH, Dominguez R (2007) Structural basis for the recruitment of profilin-actin complexes during filament elongation by Ena/VASP. EMBO J 26: 4597–4606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabbiani G (2003) The myofibroblast in wound healing and fibrocontractive diseases. J Pathol 200: 500–503 [DOI] [PubMed] [Google Scholar]
- Gates J, Mahaffey JP, Rogers SL, Emerson M, Rogers EM, Sottile SL, Van Vactor D, Gertler FB, Peifer M (2007) Enabled plays key roles in embryonic epithelial morphogenesis in Drosophila. Development 134: 2027–2039 [DOI] [PubMed] [Google Scholar]
- Gertler FB, Comer AR, Juang JL, Ahern SM, Clark MJ, Liebl EC, Hoffmann FM (1995) Enabled, a dosage-sensitive suppressor of mutations in the Drosophila Abl tyrosine kinase, encodes an Abl substrate with SH3 domain-binding properties. Genes Dev 9: 521–533 [DOI] [PubMed] [Google Scholar]
- Gertler FB, Niebuhr K, Reinhard M, Wehland J, Soriano P (1996) Mena, a relative of VASP and Drosophila Enabled, is implicated in the control of microfilament dynamics. Cell 87: 227–239 [DOI] [PubMed] [Google Scholar]
- Goh WI, Lim KB, Sudhaharan T, Sem KP, Bu W, Chou AM, Ahmed S (2012) mDia1 and WAVE2 proteins interact directly with IRSp53 in filopodia and are involved in filopodium formation. J Biol Chem 287: 4702–4714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govind S, Kozma R, Monfries C, Lim L, Ahmed S (2001) Cdc42Hs facilitates cytoskeletal reorganization and neurite outgrowth by localizing the 58-kD insulin receptor substrate to filamentous actin. J Cell Biol 152: 579–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen SD, Mullins RD (2010) VASP is a processive actin polymerase that requires monomeric actin for barbed end association. J Cell Biol 191: 571–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttelmaier S, Harbeck B, Steffens O, Messerschmidt T, Illenberger S, Jockusch BM (1999) Characterization of the actin binding properties of the vasodilator-stimulated phosphoprotein VASP. FEBS Lett 451: 68–74 [DOI] [PubMed] [Google Scholar]
- Jegou A, Niedermayer T, Orban J, Didry D, Lipowsky R, Carlier MF, Romet-Lemonne G (2011) Individual actin filaments in a microfluidic flow reveal the mechanism of ATP hydrolysis and give insight into the properties of profilin. PLoS Biol 9: e1001161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koestler SA, Rottner K, Lai F, Block J, Vinzenz M, Small JV (2009) F- and G-actin concentrations in lamellipodia of moving cells. PLoS One 4: e4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozma R, Ahmed S, Best A, Lim L (1995) The Ras-related protein Cdc42Hs and bradykinin promote formation of peripheral actin microspikes and filopodia in Swiss 3T3 fibroblasts. Mol Cell Biol 15: 1942–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause M, Leslie JD, Stewart M, Lafuente EM, Valderrama F, Jagannathan R, Strasser GA, Rubinson DA, Liu H, Way M, Yaffe MB, Boussiotis VA, Gertler FB (2004) Lamellipodin, an Ena/VASP ligand, is implicated in the regulation of lamellipodial dynamics. Dev Cell 7: 571–583 [DOI] [PubMed] [Google Scholar]
- Krugmann S, Jordens I, Gevaert K, Driessens M, Vandekerckhove J, Hall A (2001) Cdc42 induces filopodia by promoting the formation of an IRSp53:Mena complex. Curr Biol 11: 1645–1655 [DOI] [PubMed] [Google Scholar]
- Kwiatkowski AV, Rubinson DA, Dent EW, Edward van Veen J, Leslie JD, Zhang J, Mebane LM, Philippar U, Pinheiro EM, Burds AA, Bronson RT, Mori S, Fassler R, Gertler FB (2007) Ena/VASP is required for neuritogenesis in the developing cortex. Neuron 56: 441–455 [DOI] [PubMed] [Google Scholar]
- Lanier LM, Gates MA, Witke W, Menzies AS, Wehman AM, Macklis JD, Kwiatkowski D, Soriano P, Gertler FB (1999) Mena is required for neurulation and commissure formation. Neuron 22: 313–325 [DOI] [PubMed] [Google Scholar]
- Lim KB, Bu W, Goh WI, Koh E, Ong SH, Pawson T, Sudhaharan T, Ahmed S (2008) The Cdc42 effector IRSp53 generates filopodia by coupling membrane protrusion with actin dynamics. J Biol Chem 283: 20454–20472 [DOI] [PubMed] [Google Scholar]
- Mejillano MR, Kojima S, Applewhite DA, Gertler FB, Svitkina TM, Borisy GG (2004) Lamellipodial versus filopodial mode of the actin nanomachinery: pivotal role of the filament barbed end. Cell 118: 363–373 [DOI] [PubMed] [Google Scholar]
- Menna E, Disanza A, Cagnoli C, Schenk U, Gelsomino G, Frittoli E, Hertzog M, Offenhauser N, Sawallisch C, Kreienkamp HJ, Gertler FB, Di Fiore PP, Scita G, Matteoli M (2009) Eps8 regulates axonal filopodia in hippocampal neurons in response to brain-derived neurotrophic factor (BDNF). PLoS Biol 7: e1000138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki H, Yamaguchi H, Suetsugu S, Takenawa T (2000) IRSp53 is an essential intermediate between Rac and WAVE in the regulation of membrane ruffling. Nature 408: 732–735 [DOI] [PubMed] [Google Scholar]
- Millard TH, Bompard G, Heung MY, Dafforn TR, Scott DJ, Machesky LM, Futterer K (2005) Structural basis of filopodia formation induced by the IRSp53/MIM homology domain of human IRSp53. EMBO J 24: 240–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neel NF, Barzik M, Raman D, Sobolik-Delmaire T, Sai J, Ham AJ, Mernaugh RL, Gertler FB, Richmond A (2009) VASP is a CXCR2-interacting protein that regulates CXCR2-mediated polarization and chemotaxis. J Cell Sci 122: 1882–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda K, Shiratsuchi T, Nishimori H, Inazawa J, Yoshikawa H, Taketani Y, Nakamura Y, Tokino T (1999) Identification of BAIAP2 (BAI-associated protein 2), a novel human homologue of hamster IRSp53, whose SH3 domain interacts with the cytoplasmic domain of BAI1. Cytogenet Cell Genet 84: 75–82 [DOI] [PubMed] [Google Scholar]
- Okamura-Oho Y, Miyashita T, Ohmi K, Yamada M (1999) Dentatorubral-pallidoluysian atrophy protein interacts through a proline-rich region near polyglutamine with the SH3 domain of an insulin receptor tyrosine kinase substrate. Hum Mol Genet 8: 947–957 [DOI] [PubMed] [Google Scholar]
- Pasic L, Kotova T, Schafer DA (2008) Ena/VASP proteins capture actin filament barbed ends. J Biol Chem 283: 9814–9819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrin S, Mellor H (2005) The Rho family GTPase Rif induces filopodia through mDia2. Curr Biol 15: 129–133 [DOI] [PubMed] [Google Scholar]
- Peng J, Wallar BJ, Flanders A, Swiatek PJ, Alberts AS (2003) Disruption of the Diaphanous-related formin Drf1 gene encoding mDia1 reveals a role for Drf3 as an effector for Cdc42. Curr Biol 13: 534–545 [DOI] [PubMed] [Google Scholar]
- Reinhard M, Jouvenal K, Tripier D, Walter U (1995) Identification, purification, and characterization of a zyxin-related protein that binds the focal adhesion and microfilament protein VASP (vasodilator-stimulated phosphoprotein). Proc Natl Acad Sci USA 92: 7956–7960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottner K, Behrendt B, Small JV, Wehland J (1999) VASP dynamics during lamellipodia protrusion. Nat Cell Biol 1: 321–322 [DOI] [PubMed] [Google Scholar]
- Sabeh F, Shimizu-Hirota R, Weiss SJ (2009) Protease-dependent versus -independent cancer cell invasion programs: three-dimensional amoeboid movement revisited. J Cell Biol 185: 11–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samarin S, Romero S, Kocks C, Didry D, Pantaloni D, Carlier MF (2003) How VASP enhances actin-based motility. J Cell Biol 163: 131–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawallisch C, Berhorster K, Disanza A, Mantoani S, Kintscher M, Stoenica L, Dityatev A, Sieber S, Kindler S, Morellini F, Schweizer M, Boeckers TM, Korte M, Scita G, Kreienkamp HJ (2009) The insulin receptor substrate of 53 kDa (IRSp53) limits hippocampal synaptic plasticity. J Biol Chem 284: 9225–9236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirenbeck A, Arasada R, Bretschneider T, Schleicher M, Faix J (2005) Formins and VASPs may co-operate in the formation of filopodia. Biochem Soc Trans 33: 1256–1259 [DOI] [PubMed] [Google Scholar]
- Schirenbeck A, Arasada R, Bretschneider T, Stradal TE, Schleicher M, Faix J (2006) The bundling activity of vasodilator-stimulated phosphoprotein is required for filopodium formation. Proc Natl Acad Sci USA 103: 7694–7699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scita G, Confalonieri S, Lappalainen P, Suetsugu S (2008) IRSp53: crossing the road of membrane and actin dynamics in the formation of membrane protrusions. Trends Cell Biol 18: 52–60 [DOI] [PubMed] [Google Scholar]
- Stappenbeck TS, Miyoshi H (2009) The role of stromal stem cells in tissue regeneration and wound repair. Science 324: 1666–1669 [DOI] [PubMed] [Google Scholar]
- Svitkina TM, Borisy GG (1999) Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J Cell Biol 145: 1009–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkina TM, Bulanova EA, Chaga OY, Vignjevic DM, Kojima S, Vasiliev JM, Borisy GG (2003) Mechanism of filopodia initiation by reorganization of a dendritic network. J Cell Biol 160: 409–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temmerman K, Nickel W (2009) A novel flow cytometric assay to quantify interactions between proteins and membrane lipids. J Lipid Res 50: 1245–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaggi F, Disanza A, Milanesi F, Di Fiore PP, Menna E, Matteoli M, Gov NS, Scita G, Ciliberto A (2011) The Eps8/IRSp53/VASP network differentially controls actin capping and bundling in filopodia formation. PLoS Comput Biol 7: e1002088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignjevic D, Kojima S, Aratyn Y, Danciu O, Svitkina T, Borisy GG (2006) Role of fascin in filopodial protrusion. J Cell Biol 174: 863–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignjevic D, Montagnac G (2008) Reorganisation of the dendritic actin network during cancer cell migration and invasion. Semin Cancer Biol 18: 12–22 [DOI] [PubMed] [Google Scholar]
- Weiss SM, Ladwein M, Schmidt D, Ehinger J, Lommel S, Stading K, Beutling U, Disanza A, Frank R, Jansch L, Scita G, Gunzer F, Rottner K, Stradal TE (2009) IRSp53 links the enterohemorrhagic E. coli effectors Tir and EspFU for actin pedestal formation. Cell Host Microbe 5: 244–258 [DOI] [PubMed] [Google Scholar]
- Wiesner S, Helfer E, Didry D, Ducouret G, Lafuma F, Carlier MF, Pantaloni D (2003) A biomimetic motility assay provides insight into the mechanism of actin-based motility. J Cell Biol 160: 387–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Asokan SB, Berginski ME, Haynes EM, Sharpless NE, Griffith JD, Gomez SM, Bear JE (2012) Arp2/3 is critical for lamellipodia and response to extracellular matrix cues but is dispensable for chemotaxis. Cell 148: 973–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi A, Masuda M, Ohki T, Onishi H, Mochizuki N (2004) A novel actin bundling/filopodium-forming domain conserved in insulin receptor tyrosine kinase substrate p53 and missing in metastasis protein. J Biol Chem 279: 14929–14936 [DOI] [PubMed] [Google Scholar]
- Yang C, Czech L, Gerboth S, Kojima S, Scita G, Svitkina T (2007) Novel roles of formin mDia2 in lamellipodia and filopodia formation in motile cells. PLoS Biol 5: e317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Hoelzle M, Disanza A, Scita G, Svitkina T (2009) Coordination of membrane and actin cytoskeleton dynamics during filopodia protrusion. PLoS ONE 4: e5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Svitkina T (2011a) Filopodia initiation: focus on the Arp2/3 complex and formins. Cell Adh Migr 5: 402–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Svitkina T (2011b) Visualizing branched actin filaments in lamellipodia by electron tomography. Nat Cell Biol 13: 1012–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Pykalainen A, Lappalainen P (2011) I-BAR domain proteins: linking actin and plasma membrane dynamics. Curr Opin Cell Biol 23: 14–21 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.