Abstract
Thrombin stimulates the expression of multiple genes in endothelial cells (ECs), but the trans-acting factors responsible for this induction remain undefined. We have previously described a thrombin-inducible nuclear factor (TINF), which binds to an element in the PDGF B promoter and is responsible for the thrombin inducibility of this gene. Inactive cytoplasmic TINF is rapidly activated and translocated to nuclei of ECs upon stimulation with thrombin. We have now purified TINF from thrombin-treated ECs. Amino acid sequencing revealed it to be a member of the Y-box protein family, and the sole Y-box protein–encoding cDNA we detected in human or bovine ECs corresponded to DNA-binding protein B (dbpB). DbpB translocated to the nucleus after thrombin stimulation of ECs as shown by FACS analysis of nuclei from ECs expressing GFP-dbpB fusion proteins. During thrombin activation, dbpB was found to be cleaved, yielding a 30-kDa NH2-terminal fragment that recognized the thrombin-response element sequence, but not the Y-box consensus sequence. Preincubation of ECs with protein tyrosine phosphatase inhibitors completely blocked dbpB activation by thrombin and blocked induction of endogenous PDGF B–chain mRNA and promoter activation by thrombin. Y-box proteins are known to act constitutively to regulate the expression of several genes. Activation of this class of transcription factors in response to thrombin or any other agonist represents a novel signaling pathway.
Introduction
Thrombin is a coagulation system protease and platelet aggregating substance that would be expected to be present at sites of vascular injury. Thrombin is also a potent endothelial cell (EC) agonist, and its activation of early and late events in these cells has been under investigation for many years (1–7). Thrombin causes the induction of multiple genes in ECs and other cells; however, the molecular mechanism underlying thrombin-induced gene expression in ECs, or in any other cell type, remains poorly understood. Others had earlier demonstrated the induction of the PDGF B–chain gene by thrombin (8–11), and we identified a 9-bp element in the PDGF B–chain promoter that was responsible for thrombin-induced transcription of this gene (12). This element was conserved in the PDGF B–chain promoters of multiple species and was also found in many other thrombin-inducible genes, such as tissue factor and the PDGF A chain. We also identified a thrombin-dependent nuclear protein, thrombin-inducible nuclear factor (TINF), which specifically bound an oligonucleotide corresponding to the sequence of the thrombin-response element (ThRE). TINF was found to be constitutively present in the EC cytoplasm in a non–DNA-binding form, but it became activated (bound an oligonucleotide corresponding to the ThRE) and translocated to the nucleus in response to thrombin (12).
Here we report that TINF belongs to the Y-box protein family. Y-box proteins, which are highly conserved from Escherichia coli through humans, are DNA-binding proteins that participate in both the regulation of transcription and of translation. As regulators of transcription they act as both suppressors and enhancers (13). Several genes have Y-box sequences in their promoter regions and are thought to be regulated by Y-box proteins (MHC class II, MDR1, Rous sarcoma virus [RSV] genes, to name a few) (14–28). The regulation of MHC class II genes by a Y-box protein has been defined using mutational analysis (29–32). To date, Y-box proteins have been shown to be constitutively present in cells and to play a role in the expression of constitutive and induced genes without changes in their nuclear level. No previous reports have described the necessity for activation of a Y-box–binding protein in response to a cellular agonist. Furthermore, Y-box–binding proteins have not been previously implicated in the regulation of transcription by thrombin.
Methods
Materials.
Bovine α-thrombin was purchased from USB Corp. (Cleveland, Ohio, USA). Endotoxin was removed from this thrombin using Acticlean ETOX prepacked columns from Sterogene (Arcadia, California, USA). DMEM/Ham’s F-12 medium (DMEM/F-12) was from Irvine Scientific (Santa Ana, California, USA); FBS was obtained from BioWhittaker Inc. (Walkersville, Maryland, USA). Tissue culture plastic was from Costar (Corning Inc., Corning, New York, USA) and Becton Dickinson and Co. (Franklin Lakes, New Jersey, USA). NH2-D-Phe-Pro-Arg-chloromethylketone (PPACK) was from Bachem (Bubendorf, Switzerland); T4 kinase for end-labeling of oligonucleotides from Boehringer Mannheim (Indianapolis, Indiana, USA); [γ-32P]dATP, [14C]adenine, and [γ-32P]dCTP were from NEN Life Science Products (Boston, Massachusetts, USA); Q-Sepharose was from Amersham Pharmacia Biotech (Piscataway, New Jersey, USA); Spectrozyme TH (H-D-hexahydrotyrosyl-L-alanyl-L-arginine-p-nitroanilide-diacetate salt) was from American Diagnostica (Greenwich, Connecticut, USA); mRNA isolation kit from Boehringer Mannheim; NP-40 from Pierce Chemical Co. (Rockford, Illinois, USA); and Trizol reagent, Lipofectin, and oligo(dT)12–18 were from Life Technologies Inc. (Grand Island, New York, USA). Sodium orthovanadate was purchased from Fisher Scientific (Fair Lawn, New Jersey, USA), phenyl arsine oxide (PAO) and cathepsin G from Calbiochem-Novabiochem Corp. (San Diego, California, USA), trypsin from Worthington (Freehold, New Jersey,USA), factor Xa and plasmin from Enzyme Research Laboratories (South Bend, Indiana, USA), Nytran blotting membrane from Micron Separation (Westboro, Massachusetts, USA), the pSV-β-galactosidase control vector and the luciferase and β-galactosidase assay kits from Promega Corp. (Madison, Wisconsin, USA), paraformaldehyde from Electron Microscopy Sciences (Fort Washington, Pennsylvania, USA), and Vectashield mounting medium with propidium iodide from Vector Laboratories (Burlingame, California, USA). All other chemicals, unless otherwise noted, were purchased from Sigma Chemical Co. (St. Louis, Missouri, USA).
Stimulation of ECs with thrombin and preparation of extracts for electrophoretic mobility shift assays.
Human umbilical vein ECs and bovine aortic ECs were isolated and grown as described previously (12). Confluent cells were washed twice with media without FBS in the case of bovine EC or with media containing 1% FBS in the case of human ECs. ECs were treated with thrombin (3–10 U/mL) for 2 hours at 37°C. At the end of the incubation and before cell lysis, a 100× molar excess of the thrombin inhibitor PPACK was added for 10 minutes at 37°C to inactivate thrombin bound to the cell surface. Cells were washed twice with ice-cold PBS, and then cytosolic and nuclear extracts were prepared as described by Dignam et al. (33).
Electrophoretic mobility shift assay (EMSA) with 8% acrylamide gels was performed as we have described previously (12). Probes represented wild-type or mutated ThREs of the PDGF B–chain promoter (12). The ThRE sequence ctCCACCCACC was changed as follows: CTGATTGGCCAA (Y-box consensus); ctAAATGCACC (4-base mutant); ctAAGTTTGAAG (9-base mutant).
Purification of TINF (DNA binding protein B).
Cytosolic extracts of thrombin-treated bovine aortic ECs (∼108 cells) were prepared as already described here. The extract was heat treated (60°C for 2 hours), centrifuged at 21,000 g in a Micromax centrifuge (International Equipment Co., Needham Heights, Massachusetts, USA) for 15 minutes at room temperature, and applied to Q-Sepharose (2 mL) in 0.1 M salt. The gel was gently mixed with 1 mL extract and 1 mL glycerol for 1 hour at 4°C. Fractions were eluted in a stepwise manner with 4 mL of 0.2, 0.3, 0.4, and 0.5 M KCl and 10 mM Tris (pH 7.5) with 25% glycerol by mixing at 4°C for 30 minutes. TINF eluted at 0.2–0.3 M KCl. Each salt fraction was concentrated 400-fold and desalted by Centriprep and Microcon ultrafiltration (Amicon, Beverly, Massachusetts, USA) with a 10,000 molecular weight cutoff. Fractions were assayed for TINF activity by EMSA. The anion exchange chromatography step resulted in a 15-fold purification and an approximately 40% recovery. The active fraction was concentrated and subjected to SDS-PAGE (15%). To identify the location of TINF, 2 mm (width) × 1 mm (height) gel slices were cut from the edge of the lane. TINF was eluted from a fraction of each gel slice by mixing for 1 hour at room temperature in 25 μL gel elution buffer (1% Triton X-100, 20 mM HEPES [pH 7.6], 1 mM EDTA, 100 mM NaCl, 2 mM DTT, 0.1 mM PMSF, and 1 mg/mL aprotinin) and subjected to EMSA analysis to identify the slice containing TINF. The remainder of the gel slice containing TINF was refractionated on SDS-PAGE, and the band with TINF activity was subjected to tryptic digestion and amino acid sequencing. Near homogeneous TINF with a molecular size of approximately 30 kDa was obtained with greater than 75% recovery. Sequence analysis was performed at the Harvard Microchemistry Facility by tandem mass spectrometry (MS/MS) on a Finnigan LCQ Quadrupole Ion Trap Mass Spectrometer (ThermoQuest, San Jose, California, USA).
mRNA isolation and 5′/3′- rapid amplification of complementary ends.
mRNA was isolated using oligo(dT) affinity purification (mRNA isolation kit from Boehringer Mannheim). 5′/3′-rapid amplification of complementary ends (5′/3′-RACE) with mRNA from human and bovine ECs was performed using a kit from Boehringer Mannheim. Three specific primers were designed for 5′-RACE, all of them corresponding to the conserved region of the known Y-box proteins (no. 1, used for the RT reaction, 5′-GGTAGTTCTGCTGGTAATTGCG-3′; nos. 2 and 3 used for PCR amplification of RT reaction products, 5′-GCGACGTGGATAGCGTCTGTAATGGT-3′ and 5′-GATATCGGTCTGC-TGCGTATTTACTGC-3′). For 3′ RACE, two specific primers were used: no. 4, 5′-CCTAAACCACAAGATGGCAAAGAGAGAC-3′ and no. 5, 5′-GGCTTACCATCTCACCATCATCATCCGGT-3′.
Northern blotting.
Human ECs were grown to confluence, washed twice with medium containing 1% FBS, and incubated with the indicated reagents for 6 hours at 37°C. Medium was then aspirated, the cells were washed with PBS, and total RNA was extracted with Trizol reagent. RNA was separated by electrophoresis on a formaldehyde denaturing gel, transferred to Nytran membrane by capillary transfer, and hybridized with [32P]dCTP-labeled cDNA probes for human PDGF B–chain (2.9 kb) cDNA or GAPDH cDNA. Autoradiograms were quantitated by computerized densitometry.
Analysis of dbpB.
A nearly full-length human dbpB cDNA was obtained by RT-PCR using the published sequence and mRNA isolated from human ECs as the template. A 120-bp oligonucleotide corresponding to the 5′ terminus was synthesized (Midland Certified Reagent Co., Midland, Texas, USA) and ligated to the PCR product using the Bgl1 site of dbpB. The full-length dbpB cDNA was ligated into the pcDNA3 vector. Truncated dbpB was produced using PCR by introduction of a stop codon after 207 amino acids. The pcDNA3 vector without insert was used as a control. In vitro transcription and translation of dbpB were performed using the TNT T7 Quick Coupled Transcription/Translation System from Promega Corp. according to the manufacturer’s instructions. Antibody against a COOH-terminal peptide of dbpB was prepared by Biosynthesis Inc. (Lewisville, Texas, USA). The antigen was identical to the peptide used by Shen et al. (34) to produce a dbpB-specific antibody. For ultraviolet cross-linking of dbpB, 32P-labeled oligonucleotide probe was added to a lysate or in vitro translation mixture, and the sample was irradiated at 254 nm for 45 minutes. After irradiation, the sample was subjected to SDS-PAGE (8–12% gels).
Immunoprecipitation and Western blotting of dbpB.
The dbpB was immunoprecipitated from extracts (100 μL), prepared as already described here, by incubating with constant mixing at 4°C for 2 hours with either antiserum or preimmune serum (10 μL) and buffer (37 μL of 50 mM HEPES [pH 7.4], 150 mM NaCl, 1 mM EDTA, 2.5 mM MgCl2, and 1% NP-40). Immobilized Protein A/G (50 μL; Pierce Chemical Co.) was added to the mixture and incubated for 2 hours at 4°C. After centrifugation for 5 minutes at 12,000 g, the supernatant was discarded, and the gel was washed three times with buffer and once with PBS. The dbpB was eluted by heat treatment at 60°C for 30 minutes in 50 μL PBS. The eluate was subjected to SDS-PAGE. In the case of Western blotting, EC extracts (75 μL) or in vitro translation mixture were subjected to 10% SDS-PAGE and then transferred to a PVDF membrane (Immobilon-P; Millipore Corp., Bedford, Massachusetts, USA). The membrane was treated with anti-dbpB COOH-terminus antiserum followed by a chemiluminescence detection method using an Immune-Star kit (Bio-Rad Laboratories Inc., Hercules, California, USA).
Transfection of bovine aortic ECs and luciferase activity assays.
Bovine ECs were grown to 95% confluence in six-well plates, washed with Optimem medium (Life Technologies Inc.) and transfected using Lipofectin (Life Technologies Inc.) with DNA (1 μg). Luciferase was under the control of a 400-bp promoter fragment of the PDGF B–chain gene in pGL3-Basic Vector (Promega Corp.). The pSV-β-galactosidase control vector (Promega Corp.) was cotransfected with the luciferase construct to correct for transfection efficiency and the specificity of various inhibitors. After a 6-hour transfection, ECs were washed with Optimem medium and incubated with appropriate reagents. Lysates for luciferase and β-galactosidase assays were prepared after 15 hours. β-Galactosidase levels were found to be constant and not affected by inhibitors. The luciferase assay was performed according to the kit’s instructions. All treatment groups were in triplicate, and all experiments were repeated at least three times.
FACS analysis of nuclei from EC expressing green fluorescent protein-dbpB fusion protein.
Full-length dbpB and truncated dbpB cDNA were cloned into the EGFP-C3 vector from CLONTECH Laboratories Inc. (Palo Alto, California, USA), and bovine ECs were transiently transfected with the constructs as already described here. The cells were stimulated for 2 hours with 10 U/mL thrombin 36 hours after transfection, washed with PBS, and lysed by incubation in hypotonic buffer containing protease inhibitors followed by trituration using a no. 26 needle. Nuclei were pelleted at 1,000 g, washed three times with PBS, and analyzed by FACS (Becton Dickinson and Co.).
Detection of GFP-dbpB fusion protein localization by fluorescent microscopy.
Bovine ECs were grown on fibronectin-coated glass slides, transfected with GFP-dbpB constructs as already described here, fixed in 4% paraformaldehyde 36 hours after transfection, permeabilized with 0.3% Triton-X, treated with RNase A, washed, and mounted in mounting medium. Nuclei were stained with propidium iodide.
Results
Identification of TINF as dbpB.
TINF was purified to near homogeneity as just described here. An attempt at NH2-terminal sequencing yielded the finding of a blocked terminus. TINF was then subjected to tryptic digestion, and several HPLC-purified peaks were sequenced by tandem mass spectrometry. Four peptide sequences were obtained (no. 1: NDTKEDVFVHQTAIK; no. 2: NGYGFINR; no. 3: GETVEFDVVEGEK; and no. 4: EDVFVHQTAIK), revealing that this region of TINF had complete homology to multiple members of the Y-box–binding protein family. These proteins are highly conserved from E. coli to human, with individual members of the family differing only in short NH2-terminal and COOH-terminal regions, which have been implicated in DNA and RNA sequence recognition.
To determine whether multiple Y-box–binding proteins were expressed in cultured ECs, we isolated mRNA from human umbilical vein and bovine aortic ECs and performed 5′- and 3′-RACE analysis. The 3′-RACE analysis of human EC mRNA, using specific primers corresponding to sequences within the highly conserved domain (essentially identical in all Y-box–binding proteins), yielded as a sole product the complete 3′-terminal sequence of dbpB. The 5′-RACE resulted in the partial 5′-terminal sequence of the same mRNA, namely, dbpB. Multiple 5′- and 3′-RACE analyses with cloning and sequencing of many cDNAs yielded the finding of no additional Y-box–binding protein cDNAs expressed by either human or bovine ECs. In bovine ECs, a homologue of human dbpB distinct from the only reported bovine Y-box protein EF1A was detected.
Translocation of dbpB to the EC nucleus after thrombin stimulation.
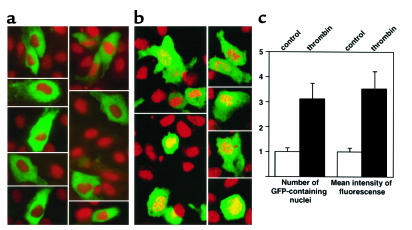
The fusion protein GFP-dbpB transiently expressed in bovine ECs was retained in the cytoplasm (Figure 1a). After stimulation with thrombin, a portion of the fusion protein in the ECs was translocated to the nuclei (Figure 1b). To quantify this observation, we performed FACS analysis of nuclei isolated from nonstimulated and thrombin-stimulated ECs. Both the number of GFP-containing nuclei and the mean intensity of fluorescence per nucleus increased more than threefold in response to thrombin stimulation (Figure 1c).
Figure 1.
Localization of full-length dbpB in endothelial cells with and without thrombin treatment. Bovine ECs were transiently transfected with a cDNA encoding a fusion protein of full-length dbpB and green fluorescent protein, as described in Methods. (a) Without thrombin stimulation. (b) After 2-hour stimulation with thrombin (10 U/mL). Nuclei were stained with propidium iodide. (c) FACS quantitation of this experiment. Nuclei were isolated from nonstimulated and thrombin-stimulated ECs (10 U/mL for 2 hours) and analyzed by FACS. Arbitrary units are used for both the number of GFP-containing nuclei and mean intensity of fluorescence of the nuclei (n = 10; P < 0.01).
Proteolytic cleavage of dbpB during the activation.
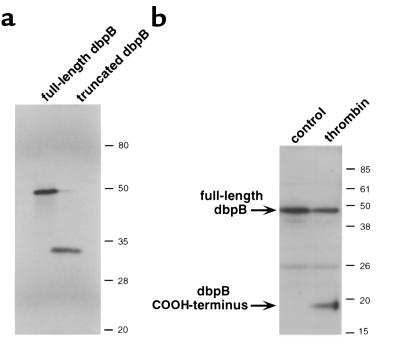
It has been reported that the apparent size of dbpB and other Y-box proteins in SDS-PAGE is 46–56 kDa (34–40). The predicted size from the cDNA sequence is 35 kDa with the difference in sizes explained by possible posttranslational modification of dbpB or an unusual charge distribution in the protein (34). We transcribed and translated dbpB in vitro, and the 35S-labeled dbpB that was produced exhibited an apparent molecular size of approximately 48–49 kDa in SDS-PAGE (Figure 2a). The active DNA-binding form of dbpB that we purified from thrombin-treated ECs had a molecular size of approximately 30 kDa by SDS-PAGE. Antibody made against the 15 COOH-terminal amino acids of dbpB recognized approximately 48- to 49-kDa dbpB in cell lysates (Figure 2b) as well as in vitro–translated dbpB (data not shown); however, it did not recognize the 30-kDa active (DNA-binding) form of dbpB. These results suggested that the active form was shorter than non–DNA-binding dbpB, possibly as the result of proteolytic cleavage of the COOH-terminus of dbpB during the intracellular activation process. This possibility was supported by the fact that some extracts from thrombin-treated ECs, but never control extracts, contained an approximately 18- to 19-kDa peptide that was recognized by the dbpB antibody (Figure 2b). The sum of the sizes of this peptide and activated dbpB (30 kDa) approximated the size of full-length dbpB (48–49 kDa). The 18-kDa fragment generated during the activation of dbpB is apparently unstable in the cell or in the process of extract preparation because it is not always detected in spite of a “cocktail” of protease inhibitors.
Figure 2.
Antibody directed to the COOH-terminus of dbpB recognized a 49-kDa protein in EC lysates as well as in vitro–translated dbpB. (a) In vitro–translated, full-length dbpB labeled with [35S]methionine exhibited an apparent size of 49 kDa in SDS-PAGE (8% gel). The truncated dbpB had an apparent size of 30 kDa (the same size as active dbpB found in extracts from thrombin-stimulated ECs). (b) Western blot of cytosolic extracts of human ECs using an antibody directed to the COOH-terminus of dbpB. In the absence of stimulation, a 49-kDa protein was detected. Stimulation of ECs with thrombin (10 U/mL for 2 hours) resulted in the appearance of a new band of approximately 19 kDa, presumably the COOH-terminus of dbpB cleaved during the process of activation.
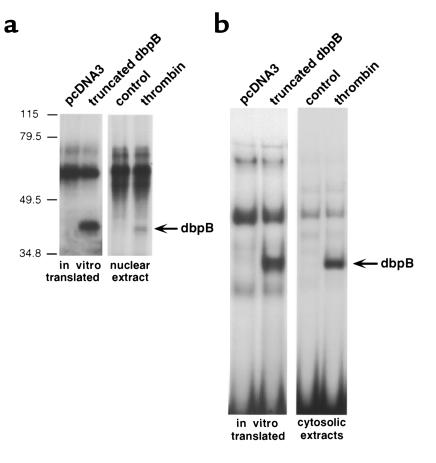
We used in vitro transcription/translation to prepare a truncated dbpB of approximately 30 kDa in SDS-PAGE (Figure 2a). The truncation was made by introducing a stop codon after 207 amino acids. Others had previously shown that the corresponding truncation in the YB-1 protein resulted in its localization to the nucleus versus the full-length protein that remained principally in the cytoplasm (41). Similarly, when we expressed in bovine ECs a fusion protein of GFP and truncated dbpB, it localized to the nucleus unlike full-length dbpB (Figure 1a, and data not shown). We examined the DNA-binding properties of truncated dbpB translated in vitro and compared them to activated dbpB in extracts from thrombin-stimulated ECs. The truncated dbpB bound to an oligonucleotide corresponding to the ThRE of the PDGF B–chain promoter, and it exhibited an electrophoretic mobility very similar to thrombin-activated dbpB purified from ECs both in SDS-PAGE after ultraviolet cross-linking (Figure 3a) and in EMSA (Figure 3b).
Figure 3.
Comparison of truncated dbpB to TINF. A truncated dbpB cDNA was prepared by introducing a stop codon following position 621 and subcloning into the pcDNA3 vector. The truncated dbpB cDNA was transcribed and translated in vitro and subjected to SDS-PAGE (8% gel) following cross-linking to the DNA probe (a) and EMSA (b). (The ThRE oligonucleotide was used as a probe in both assays). pcDNA3, empty pcDNA3 vector used as a control for the in vitro transcription and translation reaction; truncated dbpB, truncated dbpB cDNA in pcDNA3 vector used for an in vitro translation reaction; contro, extract from nonstimulated ECs; thrombin, extract from thrombin-stimulated ECs (10 U/mL for 2 hours).
The appearance of the short dbpB form in the lysates from thrombin-stimulated ECs was not due to thrombin cleavage of this protein during lysate preparation. In all experiments, a large excess of thrombin inhibitor was added to the cells before lysis, and “zero time” controls yielded no active dbpB. Second, we did not detect short dbpB after incubation of cytosolic lysates from control ECs with nonlysed ECs that had been stimulated with thrombin.
To find other EC agonists capable of activating dbpB, we tested a series of agents known to activate some of the same intracellular signaling pathways as thrombin. Stimulation of ECs with a stable analog of thromboxane A2 U46,601 (10 μM and 100 μM), platelet-activating factor (100 nM and 10 μM), histamine (10 μM and 100 μM), TNF-α (10 ng/mL), IL-1 (20 ng/mL), TGF-β (25 ng/mL), bFGF (10 ng/mL), EGF (20 ng/mL), or IFN-γ (100 ng/mL) did not activate dbpB in ECs (data not shown). Other serine proteases were also tested for their ability to activate dbpB in ECs. Cells were incubated with trypsin (0.05–5 U/mL), cathepsin G (0.01 U/mL), factor Xa (0.2–11 U/mL), and plasmin (0.1 U/mL), followed by cytosolic extract preparation and EMSA with the ThRE oligonucleotide. None of these proteases activated dbpB in ECs (data not shown).
DNA-binding specificity of truncated dbpB.
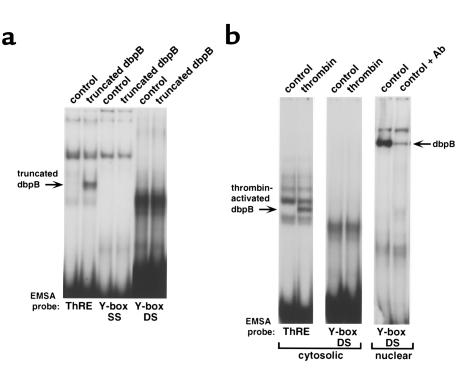
Both in vitro–translated truncated dbpB and thrombin-activated dbpB in cytosolic and nuclear extracts of ECs failed to form a complex with a Y-box consensus sequence oligonucleotide (inverted CCAT box) in EMSA (Figure 4, a and b). The same Y-box consensus probe was recognized by the full-length dbpB present constitutively in nuclear extracts, and this complex was supershifted by anti-dbpB antiserum (Figure 4b).
Figure 4.
DNA-binding properties of thrombin-activated dbpB. A truncated dbpB cDNA corresponding in size to the thrombin-activated dbpB from EC extracts was transcribed and translated in vitro, and the reaction mixture was used for EMSA with the ThRE, a Y-box single-stranded, or a Y-box double-stranded consensus oligonucleotide as the probe (a). As a control, the empty pcDNA3 vector was used in a similar reaction. Cytosolic extracts were prepared from untreated and thrombin-stimulated ECs as described. EMSA was performed using the ThRE or consensus Y-box sequence oligonucleotide as a probe (b). The same Y-box consensus probe was used to detect full-length dbpB in nuclear extracts of ECs, and the complex was supershifted with anti-dbpB antibody. Control, in vitro transcription and translation reaction performed with an empty pcDNA3 vector (a) or cytosolic and nuclear extracts from untreated ECs (b); truncated dbpB, truncated (207–amino acid) dbpB transcribed and translated in vitro; thrombin, extract from EC stimulated with thrombin (10 U/m for 2 hours); control + Ab, nuclear extract from untreated ECs preincubated with antiserum against the COOH-terminus of full-length dbpB before the binding reaction with the radiolabeled Y-box consensus probe; ThRE, ThRE probe used in EMSA; Y-box SS, sense single-stranded Y-box consensus probe used in EMSA; Y-box DS, double-stranded Y-box consensus probe used; cytosolic, cytosolic extract; nuclear, nuclear extract.
Protein tyrosine dephosphorylation in the activation of dbpB.
Alkaline phosphatase nonspecifically dephosphorylates proteins, cleaving phosphates from both tyrosine residues and serine/threonine residues. When cytosolic extracts prepared from nonstimulated human or bovine ECs were incubated with alkaline phosphatase (20 U/mL) for 30 minutes at 37°C, a new DNA-binding activity was detected by EMSA. The protein that was revealed by incubation with alkaline phosphatase exhibited the same electrophoretic mobility and DNA-binding properties as activated dbpB (data not shown).
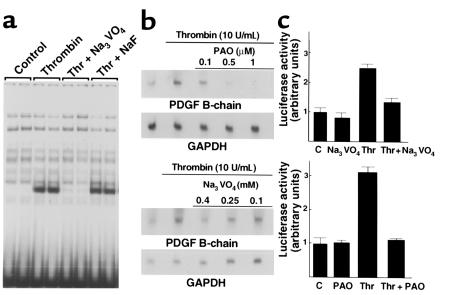
To test further whether phosphatase activity was involved in activation of dbpB by thrombin, we used inhibitors of alkaline phosphatase, protein serine/threonine phosphatase, and protein tyrosine phosphatase (PTP) activities. Pretreatment of human or bovine ECs with two inhibitors of alkaline phosphatase, levamizole and tetramisole, did not prevent activation of dbpB by thrombin (data not shown). The serine/threonine phosphatase inhibitors okadaic acid (10 μM for 2 hours) (data not shown) and sodium fluoride (500 μM for 1 hour) (Figure 5a) also failed to prevent the activation of dbpB. Two well-documented, chemically distinct inhibitors of PTP activity were used to pretreat ECs before stimulation with thrombin: sodium vanadate (50–500 μM for 30 minutes) and phenylarsine oxide (PAO; 1–50 μM for 45 minutes). Neither agent was cytotoxic nor decreased thrombin activity significantly. Both inhibitors completely prevented the activation of dbpB as shown by EMSA (Figure 5a), suggesting the participation of a PTP in dbpB activation. Pretreatment of human ECs with sodium vanadate or with PAO also prevented the induction of PDGF B–chain mRNA level after thrombin treatment (Figure 5b). In complementary experiments, bovine ECs were transiently transfected with a plasmid containing the luciferase reporter gene under the control of a 400-bp PDGF B–chain promoter fragment. Pretreatment with either PAO or sodium vanadate prevented the thrombin-induced transcription of the reporter gene (Figure 5c).
Figure 5.
Effect of tyrosine phosphatase inhibitors on activation of dbpB and PDGF B–chain transcription. (a) Effect of sodium vanadate (Na3VO4) and sodium fluoride (NaF) on dbpB activation by thrombin. ECs were pretreated with Na3VO4 or NaF (400 μM for 30 minutes) and then stimulated with thrombin (10 U/mL for 2 hours). EMSA was performed as described in Methods. Control, extract from untreated EC; Thr, extract from EC stimulated with thrombin. (b) Effect of sodium vanadate (Na3VO4) and phenyl arsine oxide (PAO) on thrombin-induced PDGF B–chain mRNA in ECs. ECs were pretreated with Na3VO4 or PAO and then stimulated with thrombin (10 U/mL for 6 hours). Total RNA was extracted using Trizol reagent and Northern hybridization was performed using either the PDGF B–chain or GAPDH cDNAs as probe. (c) Effect of PTP inhibitors on thrombin-induced transcription driven by the PDGF B–chain promoter. Bovine ECs were transiently transfected (as described in Methods) with a luciferase reporter gene under the control of a 400-bp PDGF B–chain promoter fragment in pGL3-Basic vector (Promega Corp.). Pretreatment (30 minutes) with PTP inhibitors (400 μM Na3VO4, or 200 nM PAO) was initiated after a 6-hour transfection incubation. Lysates were prepared after a 15-hour incubation with thrombin (10 U/mL). Luciferase activity was normalized to β-galactosidase activity derived from cotransfected pSV β-galactosidase cDNA.
Discussion
The role of thrombin as an inducer of gene expression in ECs and other cell types has been well documented by multiple investigators during the past decade. Though much has been learned during this time about the cell-surface receptors and the early signal transduction mediators responsible for thrombin’s action on cells, thrombin-induced nuclear events remain poorly defined. We have previously extended the observation of others that the PDGF B–chain gene is induced by thrombin (8–11) by identifying both a 9-bp ThRE in the PDGF B–chain promoter, as well as a TINF that binds this element (12). Mutations in as few as four bases in this region led to the loss of both the DNA binding of TINF and thrombin-induced transcription driven by the PDGF B promoter. Purification and amino acid sequencing now show that this thrombin-induced protein belongs to the Y-box protein family, which is the most evolutionary conserved family of DNA binding proteins known. Y-box proteins have a highly conserved nucleic acid binding domain (almost identical to the cold shock domain in bacterial Y-box–binding proteins) and short variable NH2-terminal and COOH-terminal domains, which are thought to participate in DNA sequence recognition. The 5′/3′-RACE revealed TINF in human ECs to be dbpB and, in bovine ECs, to be a bovine analog of dbpB, distinct from EF1A, a bovine Y-box protein reported previously (42). DbpB has been shown to play a role in the expression of the major histocompatibility I-A β gene (20, 43), as well as GM-CSF (21).
The predicted size based on the DNA sequence for dbpB is about 35 kDa. However, several groups of investigators have observed that this protein, as well as other Y-box proteins, migrate in SDS-PAGE with an apparent size of 46–50 kDa (34–40). The difference in the predicted and apparent sizes has been postulated to be the result of posttranslational modification or an unusual charge distribution in Y-box proteins (34). We have transcribed and translated dbpB in vitro, and the apparent size of the protein in SDS-PAGE is 48–49 kDa. Given that proteins are not posttranslationally modified in the in vitro transcription-translation system, the difference in the predicted and apparent sizes must be attributed to the structure of dbpB.
Active dbpB in extracts from thrombin-treated ECs has a size of 30 kDa by SDS-PAGE. We believe that the difference in size between inactive and thrombin-activated dbpB is the result of a proteolytic cleavage that occurs during activation. Our results with an antibody directed against the COOH-terminus of dbpB supports this model; the antibody recognizes only the inactive 48- to 49-kDa form of the protein and fails to recognize the active short form, suggesting that the COOH-terminal portion of dbpB is cleaved during activation. Furthermore, an 18- to 19-kDa unstable peptide recognized by the antibody against the COOH terminus of dbpB can often be detected in extracts from thrombin-stimulated ECs. The COOH-terminal portion of another Y-box family member, YB-1, has been shown to contain a cytoplasmic-retention domain (41). When a full-length YB-1 cDNA was transfected into cells, its product was found principally in the cytoplasm; whereas, when a truncated form of YB-1 was expressed in the same cells, the product was rapidly translocated to the nucleus. We observed the same cellular distribution for dbpB and its truncated form.
Truncated dbpB coexpressed in bovine ECs with a PDGF B–chain promoter-luciferase construct did not significantly increase transcriptional activity, either alone or in the presence of a low concentration of thrombin (data not shown). There are at least two explanations for this negative result. The first is that activated dbpB binds DNA but requires an additional thrombin-induced nuclear factor to induce transcription. There are many such examples in the literature in which two, or even three, transcription factors must be expressed in combination in order to stimulate transcriptional activity. Thus, activated dbpB may be necessary, but not sufficient, for thrombin induction of the PDGF B–chain transcription. A second possible explanation is that the short form of dbpB that we have used in our studies is not the true length of in vivo activated dbpB. The exact cleavage site is unknown because purified, activated dbpB is NH2-terminally blocked. The short dbpB that we originally used was chosen because it was approximately the correct size in SDS-PAGE and it bound the thrombin-response oligonucleotide; however, the region around this putative cleavage site is rich in charged and aromatic amino acids, the presence or absence of which may cause dramatic changes in truncated dbpB properties (e.g., translocation to the nucleus, DNA-binding activity, electrophoretic mobility). We are currently testing other expression vectors containing truncated forms of dbpB, but none to date has induced PDGF B–chain promoter activity.
Y-box proteins have been reported to function as repressors of gene expression (20, 21) and as positive regulators of constitutive transcription (22, 30). However, to our knowledge, these proteins have not been previously shown to be activated by extracellular stimuli to promote the induced expression of genes. The Y-box consists of an inverted CCAAT box sequence that was first identified as a regulatory sequence in the promoters of MHC class II genes. The consensus Y box element was defined by mutational analysis (29–32). Although the conserved cold shock domain was thought to provide the binding to DNA, more recent studies have shown that not all members of the Y-box protein family bind this Y-box consensus sequence. A Y-box–binding protein NF-GMb is a GM-CSF promoter repressor that binds to two repeats of 5′-CCTG-3′ (21). Another Y-box binding protein binds an IFN-response element (44). The Y-box protein RSV-EF-1 does not bind the consensus Y-box–binding sequence, but rather recognizes several distinct sequences in the RSV promoter with highest affinity for the sequence AAGGTGG (45). RSV-EF-1 is therefore similar to activated dbpB, which does not recognize the consensus Y-box sequence, but binds to the ThRE in the PDGF B–chain promoter. The DNA-binding specificity of dbpB suggests that expression of other genes that do not have a classical Y-box sequence may also be regulated by this Y-box protein.
Cytosolic thrombin-activated dbpB binds single-stranded DNA, which is consistent with the action of other Y-box proteins (21, 22, 38). Many promoters have single-stranded DNA regions that are thought to be important in the regulation of gene expression (for review, see ref. 46). A single-stranded region has been well-characterized in the PDGF A–chain promoter (47, 48).
In our attempt to find intracellular pathways activated by thrombin that lead to the activation of dbpB, we discovered that PTP activity appears to be required. Thrombin is known to activate phosphatases in platelets, and this activity is implicated in platelet aggregation (49–51). Activation of PTPs by thrombin is implicated in microvesicle formation by platelets (52) and in thrombin-induced fibroblast proliferation. The PTP SHP2 is positively linked to one of the protease-activated receptors, PAR-2 (53), and has a positive regulatory role in the IL-2 signaling cascade (54). PTP activity has been implicated in transcriptional regulation, including participation in the regulation of NF-κB activation in ECs (55). In preliminary experiments, we were unable to detect tyrosine phosphorylation of full-length dbpB, suggesting that a signaling mediator in the pathway to dbpB activation is the substrate of PTP.
We have demonstrated the thrombin-specific activation of a Y-box protein dbpB, which recognizes a nucleotide sequence corresponding to the ThRE in the PDGF B–chain promoter. This activation appears to be mediated via a pathway involving a PTP and an intracellular protease and does not require de novo protein synthesis. This pathway would allow a rapid cellular response to thrombin by activating a constitutive protein dbpB present in the cytoplasm of ECs, causing it to enter the nucleus and modulate target gene expression.
Acknowledgments
We thank A. Bunting and P. Hoang for cell culture assistance, L. Gudipathy for performing the RACE assays on bovine cells, J. Drazba for assistance with fluorescent microscopy, and A. Raber for performing the FACS analysis. This work was supported by NIH grant HL-29582 to P.E. DiCorleto and by a fellowship award to O.I. Stenina from the American Heart Association, Northeast Ohio Affiliate. Human umbilical vein ECs were harvested from cords collected through the Birthing Services Department at the Cleveland Clinic Foundation and the Perinatal Clinical Research Center (NIH General Clinic Research Center award RR-00080) at the Cleveland Metrohealth Hospital.
Footnotes
Olga I. Stenina and Earl J. Poptic contributed equally to this work.
References
- 1.Coughlin SR. How the protease thrombin talks to cells. Proc Natl Acad Sci USA. 1999;96:11023–11027. doi: 10.1073/pnas.96.20.11023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grand RJA, Turnell AS, Grabham PW. Cellular consequences of thrombin-receptor activation. Biochem J. 1996;313:353–368. doi: 10.1042/bj3130353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carveth HJ, et al. Regulation of platelet-activating factor (PAF) synthesis and PAF-mediated neutrophil adhesion to endothelial cells activated by thrombin. Semin Thromb Hemost. 1992;18:126–134. doi: 10.1055/s-2007-1002417. [DOI] [PubMed] [Google Scholar]
- 4.Rabiet MJ, Plantier JL, Dejana E. Thrombin-induced endothelial cell dysfunction. Br Med Bull. 1994;50:936–945. doi: 10.1093/oxfordjournals.bmb.a072935. [DOI] [PubMed] [Google Scholar]
- 5.Molino M, et al. Endothelial cell thrombin receptors and PAR-2. Two protease-activated receptors located in a single cellular environment. J Biol Chem. 1997;272:11133–11141. doi: 10.1074/jbc.272.17.11133. [DOI] [PubMed] [Google Scholar]
- 6.Garcia JG, Verin AD, Schaphorst KL. Regulation of thrombin-mediated endothelial cell contraction and permeability. Semin Thromb Hemost. 1996;22:309–315. doi: 10.1055/s-2007-999025. [DOI] [PubMed] [Google Scholar]
- 7.Brass LF, Molino M. Protease-activated G protein–coupled receptors on human platelets and endothelial cells. Thromb Haemost. 1997;78:234–241. [PubMed] [Google Scholar]
- 8.Daniel TO, Gibbs VC, Milfay DF, Garovoy MR, Williams LT. Thrombin stimulates c-sis gene expression in microvascular endothelial cells. J Biol Chem. 1986;261:9579–9582. [PubMed] [Google Scholar]
- 9.Daniel TO, Gibbs VC, Milfay DF, Williams LT. Agents that increase cAMP accumulation block endothelial c-sis induction by thrombin and transforming growth factor-beta. J Biol Chem. 1987;262:11893–11896. [PubMed] [Google Scholar]
- 10.Kaetzel DM, Coyne DW, Fenstermaker RA. Transcriptional control of the platelet-derived growth factor subunit genes. Biofactors. 1993;4:71–81. [PubMed] [Google Scholar]
- 11.Grandaliano G, et al. Thrombin regulates PDGF expression in bovine glomerular endothelial cells. J Am Soc Nephrol. 1998;9:583–589. doi: 10.1681/ASN.V94583. [DOI] [PubMed] [Google Scholar]
- 12.Scarpati E, DiCorleto P. Identification of a thrombin response element in the human platelet-derived growth factor B-chain (c-sis) promoter. J Biol Chem. 1996;271:3025–3032. doi: 10.1074/jbc.271.6.3025. [DOI] [PubMed] [Google Scholar]
- 13.Matsumoto K, Wolffe AP. Gene regulation by Y-box proteins: coupling control of transcription and translation. Trends Cell Biol. 1998;8:318–323. doi: 10.1016/s0962-8924(98)01300-2. [DOI] [PubMed] [Google Scholar]
- 14.Ladomery M, Sommerville J. A role for Y-box proteins in cell proliferation. Bioessays. 1995;17:9–11. doi: 10.1002/bies.950170104. [DOI] [PubMed] [Google Scholar]
- 15.Sakura H, Maekawa T, Imamoto F, Yasuda K, Ishii S. Two human genes isolated by a novel method encode DNA-binding proteins containing a common region of homology. Gene. 1988;73:499–507. doi: 10.1016/0378-1119(88)90514-8. [DOI] [PubMed] [Google Scholar]
- 16.Lipson K, Chen S, Koniecki J, Ku D-H, Baserga R. Transcriptional activity of the human thymidine kinase gene determined by a method using the polymerase chain reaction and an intron-specific probe. Proc Natl Acad Sci USA. 1989;86:6848–6852. doi: 10.1073/pnas.86.24.9774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Travali S, et al. Structure of the human gene for the proliferating cell nuclear antigen. J Biol Chem. 1989;264:7466–7472. [PubMed] [Google Scholar]
- 18.Pearson B, Nasheuer H-P, Wang T. Human DNA polymerase alpha gene: sequences controlling expression in cycling and serum-stimulated cells. Mol Cell Biol. 1991;11:2081–2095. doi: 10.1128/mcb.11.4.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gai X, Lipson K, Prystowsky M. Unusual DNA binding characteristics of an in vitro translation product of the CCAAT binding protein mYB-1. Nucleic Acid Res. 1992;20:601–606. doi: 10.1093/nar/20.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lloberas J, Maki R, Celada A. Repression of major histocompatibility complex I-A beta gene expression by dbpA and dbpB (mYB-1) proteins. Mol Cell Biol. 1995;15:5092–5099. doi: 10.1128/mcb.15.9.5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coles LS, Diamond P, Occhiodoro F, Vadas MA, Shannon MF. Cold shock domain proteins repress transcription from the GM-CSF promoter. Nucleic Acid Res. 1996;24:2311–2317. doi: 10.1093/nar/24.12.2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mertens PR, Harendza S, Pollock AS, Lovett DH. Glomerular mesangial cell-specific transactivation of matrix metalloproteinase 2 transcription is mediated by YB-1. J Biol Chem. 1997;272:22905–22912. doi: 10.1074/jbc.272.36.22905. [DOI] [PubMed] [Google Scholar]
- 23.Li WW, et al. Suppression of grp78 core promoter element-mediated stress induction by the dbpA and dbpB (YB-1) cold shock domain proteins. Mol Cell Biol. 1997;17:61–68. doi: 10.1128/mcb.17.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pratt SL, Horseman ND. Identification of two Y-box binding proteins that interact with the promoters of columbid annexin I genes. Gene. 1998;214:147–156. doi: 10.1016/s0378-1119(98)00211-x. [DOI] [PubMed] [Google Scholar]
- 25.Ohga T, et al. Direct involvement of the Y-box binding protein YB-1 in genotoxic stress-induced activation of the human multidrug resistance 1 gene. J Biol Chem. 1998;273:5997–6000. doi: 10.1074/jbc.273.11.5997. [DOI] [PubMed] [Google Scholar]
- 26.Mertens PR, Alfonso-Jaume MA, Steinmann K, Lovett DH. A synergistic interaction of transcription factors AP2 and YB-1 regulates gelatinase A enhancer-dependent transcription. J Biol Chem. 1998;273:32957–32965. doi: 10.1074/jbc.273.49.32957. [DOI] [PubMed] [Google Scholar]
- 27.Dhalla AK, Ririe SS, Swamynathan SK, Weber KT, Guntaka RV. chk-YB-1b, a Y-box binding protein activates transcription from rat alpha(I) procollagen gene promoter. Biochem J. 1998;336:373–379. doi: 10.1042/bj3360373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sawaya BE, Khalili K, Amini S. Transcription of the human immunodeficiency virus type 1 (HIV-1) promoter in central nervous system cells: effect of YB-1 on expression of the HIV-1 long terminal repeat. J Gen Virol. 1998;79:239–246. doi: 10.1099/0022-1317-79-2-239. [DOI] [PubMed] [Google Scholar]
- 29.Boss JM, Strominger JL. Regulation of a transfected human class II major histocompatibility complex gene in human fibroblasts. Proc Natl Acad Sci USA. 1986;83:9139–9143. doi: 10.1073/pnas.83.23.9139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dorn A, et al. Conserved major histocompatibility complex class II boxes—X and Y—are transcriptional control elements and specifically bind nuclear proteins. Proc Natl Acad Sci USA. 1987;84:6249–6253. doi: 10.1073/pnas.84.17.6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dorn A, Bollekens J, Staub A, Benoist C, Mathis D. A multiplicity of CCAAT box-binding proteins. Cell. 1987;50:863–872. doi: 10.1016/0092-8674(87)90513-7. [DOI] [PubMed] [Google Scholar]
- 32.Miwa K, Doyle C, Strominger JL. Sequence-specific interactions of nuclear factors with conserved sequences of human class II major histocompatibility complex genes. Proc Natl Acad Sci USA. 1987;84:4939–4943. doi: 10.1073/pnas.84.14.4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dignam J, Lebovitz R, Roeder R. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acid Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shen Q, Wu R, Leonard JL, Newburger PE. Identification and molecular cloning of a human selenocysteine insertion sequence-binding protein. A bifunctional role for DNA-binding protein B. J Biol Chem. 1998;273:5443–5446. doi: 10.1074/jbc.273.10.5443. [DOI] [PubMed] [Google Scholar]
- 35.Ozer J, Faber M, Chalkley R, Sealy L. Isolation and characterization of a cDNA clone for the CCAAT transcription factor EFIA reveals a novel structural motif. J Biol Chem. 1990;265:22143–22152. [PubMed] [Google Scholar]
- 36.Tafuri SR, Familari M, Wolffe AP. A mouse Y box protein, MSY1, is associated with paternal mRNA in spermatocytes. J Biol Chem. 1993;268:12213–12220. [PubMed] [Google Scholar]
- 37.Tafuri SR, Wolffe AP. Selective recruitment of masked maternal mRNA from messenger ribonucleoprotein particles containing FRGY2 (mRNP4) J Biol Chem. 1993;268:24255–24261. [PubMed] [Google Scholar]
- 38.Horwitz EM, Maloney KA, Ley TJ. A human protein containing a “cold shock” domain binds specifically to H-DNA upstream from the human gamma-globin genes. J Biol Chem. 1994;269:14130–14139. [PubMed] [Google Scholar]
- 39.Bouvet P, Matsumoto K, Wolffe AP. Sequence-specific RNA recognition by the Xenopus Y-box proteins. An essential role for the cold shock domain. J Biol Chem. 1995;270:28297–28303. doi: 10.1074/jbc.270.47.28297. [DOI] [PubMed] [Google Scholar]
- 40.Matsumoto K, Meric F, Wolffe AP. Translational repression dependent on the interaction of the Xenopus Y-box protein FRGY2 with mRNA. Role of the cold shock domain, tail domain, and selective RNA sequence recognition. J Biol Chem. 1996;271:22706–22712. doi: 10.1074/jbc.271.37.22706. [DOI] [PubMed] [Google Scholar]
- 41.Koike K, et al. Nuclear translocation of the Y-box binding protein by ultraviolet irradiation. FEBS Lett. 1997;417:390–394. doi: 10.1016/s0014-5793(97)01296-9. [DOI] [PubMed] [Google Scholar]
- 42.Ozer J, Chalkey R, Sealy L. Isolation of the CCAAT transcription factor subunit EFIA cDNA and a potentially functional EFIA processed pseudogene from Bos taurus: insights into the evolution of the EFIA/dbpB/YB-1 gene family. Gene. 1993;124:223–230. doi: 10.1016/0378-1119(93)90397-l. [DOI] [PubMed] [Google Scholar]
- 43.Lloberas J, Soler C, Celada A. Repression mechanisms of the I-A beta gene of the major histocompatibility complex. Immunobiology. 1997;198:249–263. doi: 10.1016/s0171-2985(97)80045-9. [DOI] [PubMed] [Google Scholar]
- 44.Yan C, Tamm I. Molecular cloning and characterization of interferon alpha/beta response element binding factors of the murine (2′-5′) oligoadenylate synthetase ME-12 gene. Proc Natl Acad Sci USA. 1991;88:144–148. doi: 10.1073/pnas.88.1.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kandala JC, Guntaka RV. Cloning of Rous sarcoma virus enhancer factor genes. I. Evidence that RSV-EF-I is related to Y-box (inverted CCAAT) binding proteins and binds to multiple motifs in the RSV enhancer. Virology. 1994;198:514–523. doi: 10.1006/viro.1994.1062. [DOI] [PubMed] [Google Scholar]
- 46.Swamynathan S, Nambiar A, Guntaka R. Role of single-stranded DNA regions and Y-box proteins in transcriptional regulation of viral and cellular genes. FASEB J. 1998;12:515–522. doi: 10.1096/fasebj.12.7.515. [DOI] [PubMed] [Google Scholar]
- 47.Wang Z, Lin X-H, Qiu Q-Q, Deuel TF. Modulation of transcription of the platelet-derived growth factor A–chain gene by a promoter region sensitive to S1 nuclease. J Biol Chem. 1992;267:17022–17031. [PubMed] [Google Scholar]
- 48.Khachigian LM, Williams AJ, Collins T. Interplay of Sp1 and Egr-1 in the proximal platelet-derived growth factor A–chain promoter in cultured vascular endothelial cells. J Biol Chem. 1995;270:27679–27686. doi: 10.1074/jbc.270.46.27679. [DOI] [PubMed] [Google Scholar]
- 49.Luber K, Siess W. Integrin-dependent protein dephosphorylation on tyrosine induced by activation of the thrombin receptor in human platelets. Cell Signal. 1994;6:279–284. doi: 10.1016/0898-6568(94)90032-9. [DOI] [PubMed] [Google Scholar]
- 50.Li RY, Gaits F, Ragab-Thomas JM, Chap H. Tyrosine phosphorylation of an SH2-containing protein tyrosine phosphatase is coupled to platelet thrombin receptor via a pertussis toxin-sensitive heterotrimeric G-protein. EMBO J. 1995;14:2519–2526. doi: 10.1002/j.1460-2075.1995.tb07249.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li RY, et al. Protein tyrosine phosphatase SHP-1 fails to associate with cytoskeleton but is normally phosphorylated upon thrombin stimulation of thrombasthenic platelets. Thromb Haemost. 1997;77:150–154. [PubMed] [Google Scholar]
- 52.Pasquet JM, Dachary-Prigent J, Nurden AT. Microvesicle release is associated with extensive protein tyrosine dephosphorylation in platelets stimulated by A23187 or a mixture of thrombin and collagen. Biochem J. 1998;333:591–599. doi: 10.1042/bj3330591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu Z, Ahmad S, Schwartz JL, Banville D, Shen SH. Protein-tyrosine phosphatase SHP2 is positively linked to proteinase-activated receptor 2–mediated mitogenic pathway. J Biol Chem. 1997;272:7519–7524. doi: 10.1074/jbc.272.11.7519. [DOI] [PubMed] [Google Scholar]
- 54.Gadina M, Stancato LM, Bacon CM, Larner AC, O’Shea JJ. Involvement of SHP-2 in multiple aspects of IL-2 signaling: evidence for a positive regulatory role. J Immunol. 1998;160:4657–4661. [PubMed] [Google Scholar]
- 55.Dhawan S, Singh S, Aggarwal BB. Induction of endothelial cell surface adhesion molecules by tumor necrosis factor is blocked by protein tyrosine phosphatase inhibitors: role of the nuclear transcription factor NF-kappa B. Eur J Immunol. 1997;27:2172–2179. doi: 10.1002/eji.1830270909. [DOI] [PubMed] [Google Scholar]