Abstract
Cholesterol is a critical component of cellular membranes, regulating assembly and function of membrane-based protein/lipid complexes. Many RNA viruses, including enteroviruses, remodel host membranes to generate organelles with unique lipid blueprints on which they assemble replication complexes and synthesize viral RNA. Here we find that clathrin-mediated endocytosis (CME) is harnessed by enteroviruses to traffic cholesterol from the plasma membrane (PM) and extracellular medium to replication organelles where cholesterol then regulates viral polyprotein processing and facilitates genome synthesis. When CME is disrupted, cellular cholesterol pools are instead stored in lipid droplets; cholesterol cannot be trafficked to replication organelles; and replication is inhibited. In contrast, replication is stimulated in cholesterol-elevated cells like those lacking caveolins or those from Niemann-Pick disease patients. Our findings indicate cholesterol as a critical determinant for enteroviral replication and outline roles for the endocytic machinery in both the enteroviral lifecycle and host cell cholesterol homeostasis.
Introduction
Membranes often serve as platforms on which viral replication machinery is assembled and genomes are replicated. Membranes can potentially facilitate replication by limiting diffusion, providing proper orientation of replication machinery and allowing greater sensitivity to changes in substrate/enzyme concentrations (McCloskey and Poo 1986; den Boon and Ahlquist 2010). These membranes utilized for replication, so called “replication organelles”, can originate from the endoplasmic reticulum (ER), the Golgi apparatus, the Trans-Golgi network (TGN), endosomes and even mitochondria (Miller and Krijnse-Locker, 2008).
Enteroviruses are a family of non-enveloped (+) strand RNA viruses that include many important human pathogens such as poliovirus (PV), Coxsackievirus, human rhinovirus (HRV), enterovirus and echovirus. Upon infection, their (+) strand RNA genome is translated into structural proteins and replication machinery. The latter assembles on the cytosolic leaflet of host membranes to synthesize RNA which is then either packaged into virions or used as a template for further translation into structural and replication proteins (Paul et al., 1987). PV, Coxsackievirus B3 (CVB3) and Enterovirus 71 (EV71) all assemble their replication complexes on phosphatidylinositol 4-phosphate (PI4P) lipid enriched replication organelles by selectively recruiting host Type IIIβ phosphatidylinositol 4-kinases (PI4KIIIβ) to membranes derived from ER exit sites (Hsu et al., 2010; Sasaki et al., 2011; Greninger et al., 2011). Inhibiting PI4P production blocks their replication, thus highlighting the critical role of lipids in the enteroviral lifecycle.
Discovery of PI4P lipids prompted us to seek additional lipid signatures of replication organelles. Here we show that multiple different enteroviruses exploit CME pathways and the associated Rab11 recycling endocytic compartment to traffic cholesterol from the PM and extracellular medium to replication organelle membranes. We demonstrate that cholesterol facilitates viral RNA synthesis and regulates the proteolysis of specific viral polyproteins required for initiating viral RNA synthesis and packaging viral RNA. Finally we reveal a broader role for CME machinery in shaping the cholesterol landscape of mammalian cells where disruption of CME triggers storage of PM cholesterol pools within lipid droplets. Although endocytic machinery has been identified in previous host factor screens, these studies have largely focused on endocytic roles in viral attachment, entry and export (Hsu and Spindler, 2012; Mercer et al., 2010; Rowe et al., 2008). Our studies reveal a role for endocytic machinery in both the viral lifecycle and in the maintenance of host cell cholesterol homeostasis; and suggest new panviral therapeutic strategies focused on blocking cholesterol trafficking to replication organelle membranes.
RESULTS
Endocytic machinery regulates enteroviral replication downstream of viral entry
We first screened a subset of human genes with siRNA, targeting those genes with established roles in CME including clathrin, AP2, dynamin 2, Rab5, Rab11, Huntington interacting protein 1 (HIP1), Disabled 2 (DAB2), and Epsin15L for impact on enteroviral replication. To separate impact on replication from viral entry, disassembly or export, siRNA treated HeLa cells were transfected with viral RNA replicons where capsid-encoding sequences had been replaced by Renilla luciferase, allowing us to quantify viral RNA translation and synthesis by monitoring bioluminescence.
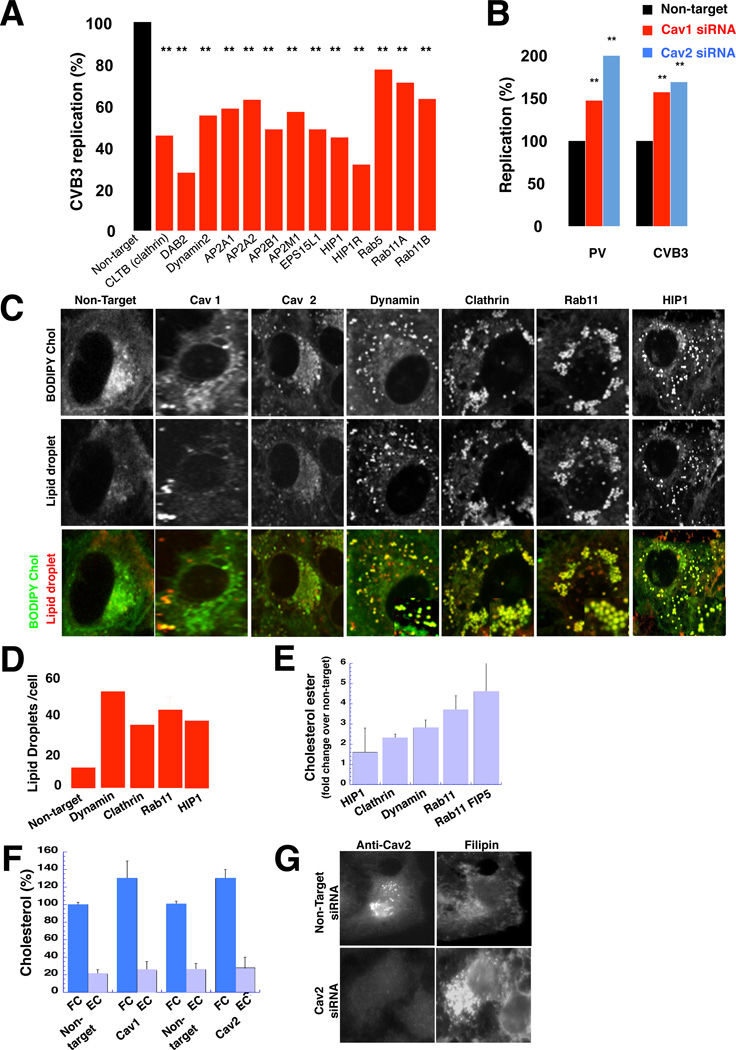
We found that >75% depletion of CME components (Figure S1A), all resulted in significant inhibition of both CVB3 and PV replication (Figure 1A; Figure S1B; Table 1) and replication organelle biogenesis was disrupted (Figures S1C, S1D and S1E). The replication measurements were normalized for cell viability which was largely unaffected by the siRNA treatments (Table 1). In contrast, when we depleted caveolin-1 (Cav1) or caveolin-2 (Cav2) proteins, which participate in non-clathrin-mediated trafficking pathways, viral replication was stimulated by up to 3-fold over non-target siRNA treated cells (Figure 1B; Figure S1F; Table 1). Note that since Cav2 depletion did not affect Cav1 levels, while Cav1 depletion decreased Cav2 levels, this suggested that it is Cav2 that mediates the stimulation of replication.
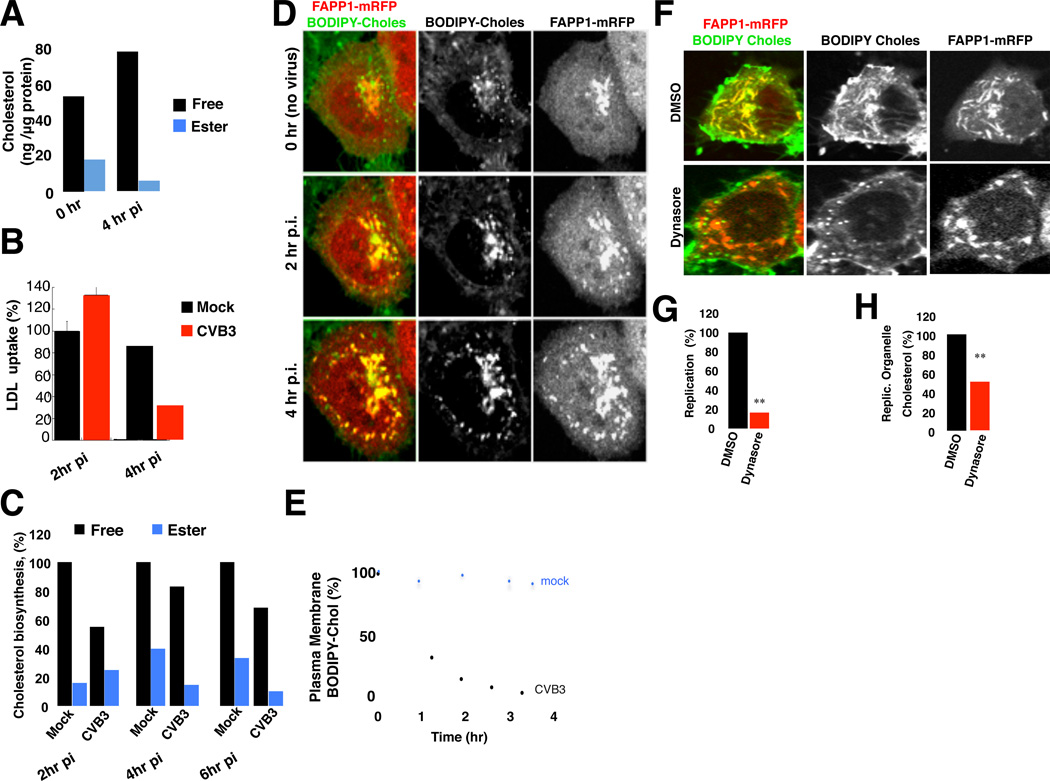
Figure 1. Disrupting host cell endocytic machinery impacts both enteroviral replication and cellular cholesterol landscape.
(A) CVB3 replication when CME components are depleted. Mean peak replication data +/−SEM, from 2 independent experiments with 6 replicates each was normalized with respect to cell viability and plotted as % of non-target siRNA treated cells. **: p<0.01
(B) PV and CVB3 replication when caveolins are depleted. Mean peak replication data +/−SEM from 2 independent experiments with 6 replicates each was normalized with respect to cell viability and plotted as % of non-target siRNA treated cells. **: p<0.01
(C) Plasma membrane cholesterol is stored in lipid droplets when CME components are depleted. Scale bar 5µm.
(D) Quantification of lipid droplets immunolabeled with anti-ADRP. Mean data +/−SEM from n=50 cells for each siRNA treatment is plotted.
(E) Steady state esterified cholesterol levels when CME components are depleted. Mean data +/− SEM of 3 independent experiments for each siRNA.
(F) Steady state free and esterified cholesterol levels when caveolins are depleted. Mean data +/− SEM of 3 independent experiments for each siRNA.
(G) Free cholesterol distribution when Cav 2 is depleted. Free cholesterol is labeled with filipin. Scale bar 5µm.
Figure 1 related to Figure S1 and Table S1.
Notably there was no correlative impact of any of the endocytic perturbations on the transfection/translation efficiency of a reporter mRNA (Table 1), PKR antiviral response or cellular PI4P levels which could account for the effects on replication (Figures S1G and S1H). Furthermore acute treatment of cells with chlorpromazine, an inhibitor of CME, significantly blocked viral replication (Figure S1I). These findings suggested roles for endocytic proteins in the viral lifecycle.
Enhanced esterification and storage of plasma membrane cholesterol pools when CME is disrupted
Cellular cholesterol homeostasis is established by vesicular and non-vesicular cholesterol uptake and distribution; biosynthesis and esterification of free cholesterol (i.e. membrane-bound) at the ER; storage of free and esterified cholesterol within lipid droplets; and cholesterol efflux (Ikonen, 2008). CME traffics subcellular cholesterol pools, the LDL receptor, which binds LDL-cholesterol, and the NPC1L1 receptor, which binds cholesterol micelles (Brown and Goldstein 1986; Chang and Chang 2008). Caveolins also have important roles in regulating the cholesterol landscape of cells: helping organize PM cholesterol domains; regulating cholesterol traffic to and from the PM; facilitating cholesterol efflux; and modulating cholesterol storage in lipid droplets (Parton and Del Pozo 2013; Cohen et al., 2004; Fu et al., 2004).
We first investigated whether there was any common impact on host cell cholesterol homeostasis when any of the CME components were disrupted. In pulse chase experiments with BODIPY-cholesterol, a fluorescent live-cell mimic of free cholesterol, which partitions into the PM bilayer when added exogenously to the cells (Holtta-Vuori et al., 2008), we tracked the dynamics of the PM free cholesterol pool, the largest reservoir for this lipid in mammalian cells (Warnock et al., 1993). In non-target, Cav 1 or Cav 2 siRNA treated cells, within 30 minutes of pulsing (for < 3 min), BODIPY-cholesterol had reached a steady-state distribution among the PM, endocytic and Golgi/TGN compartments (Figure 1C), similar to untagged-free cholesterol (Jansen et al., 2011).
In contrast, when cells had been depleted of CME components, BODIPY-cholesterol was rapidly trafficked from the PM to numerous, large spherical cytoplasmic puncta. These puncta were determined to be lipid droplets, by Nile Red stain (Figure 1C) and antibodies against ADRP (Figure S1J). Lipid droplets, widely believed to originate from ER membranes, are storage organelles for neutral lipids including all esterified as well as some free cholesterol pools (Hsieh et al., 2012; Prinz 2013). At steady state these cells had up to 6-fold more lipid droplets per cell than control cells (Figure 1D; Figure S1J); and bulk measurements revealed a ∼3-fold increase in esterified cholesterol pools (Figure 1E) while free cholesterol pools were minimally decreased (<20%) (Figure S1K). Consistent with activation of cholesterol storage pathways, we also measured a significant increase in cholesterol esterification and storage activities at the ER (Figure S1L).
The ER increases its cholesterol esterification and storage activities when its free cholesterol levels rise, for example as a result of cholesterol being trafficked to it (Brown and Goldstein 1997; 1998). When we depleted clathrin from cells in LDL-cholesterol deficient media (i.e., media containing lipoprotein deficient serum), cholesterol storage was still enhanced and replication inhibited (Figures S1M and S1N). This indicated that when CME was disrupted, the increased storage of cholesterol was not a consequence of enhanced uptake and transfer of LDL derived cholesterol to the ER, rather, given the BODIPY-cholesterol dynamics (Figure 1C) it suggested that intracellular free cholesterol pools, primarily from the PM, were being trafficked to the ER for storage in lipid droplets.
Increased free cholesterol pools in Cav 1 and Cav 2 depleted cells
On the other hand in Cav1 or Cav2 depleted cells esterified cholesterol pool abundances were similar to control cells, but there was a ∼ 40% increase in free cholesterol pools (Figure 1F) which by filipin labeling, a fluorescent reporter that selectively binds and sequesters free cholesterol within membranes, was found to localize to the PM, the Golgi/TGN and recycling endosomes (Figure 1G; Figures S1O and S1P). This increase in free cholesterol pools was not due to increased cholesterol biosynthesis as determined by direct measurements and nuclear SREBP2 levels (Figures S1L and S1Q). However when cells were depleted of caveolins in LDL-cholesterol deficient media, neither free cholesterol levels nor replication was increased (relative to control) (Figures S1R and S1S). This indicated that the free cholesterol pools were being derived from the LDL-cholesterol uptake pathway. Whether clathrin-mediated LDL uptake is stimulated in the absence of caveolins remains to be determined, but given that the ER in caveolin depleted cells does not sense an increase in cholesterol levels- since cholesterol biosynthesis and esterification are unaffected- this indicates that caveolins are modulating LDL-cholesterol transfer to the ER, and/or the latter’s cholesterol storage activities.
Enteroviral replication organelle membranes are enriched in free cholesterol
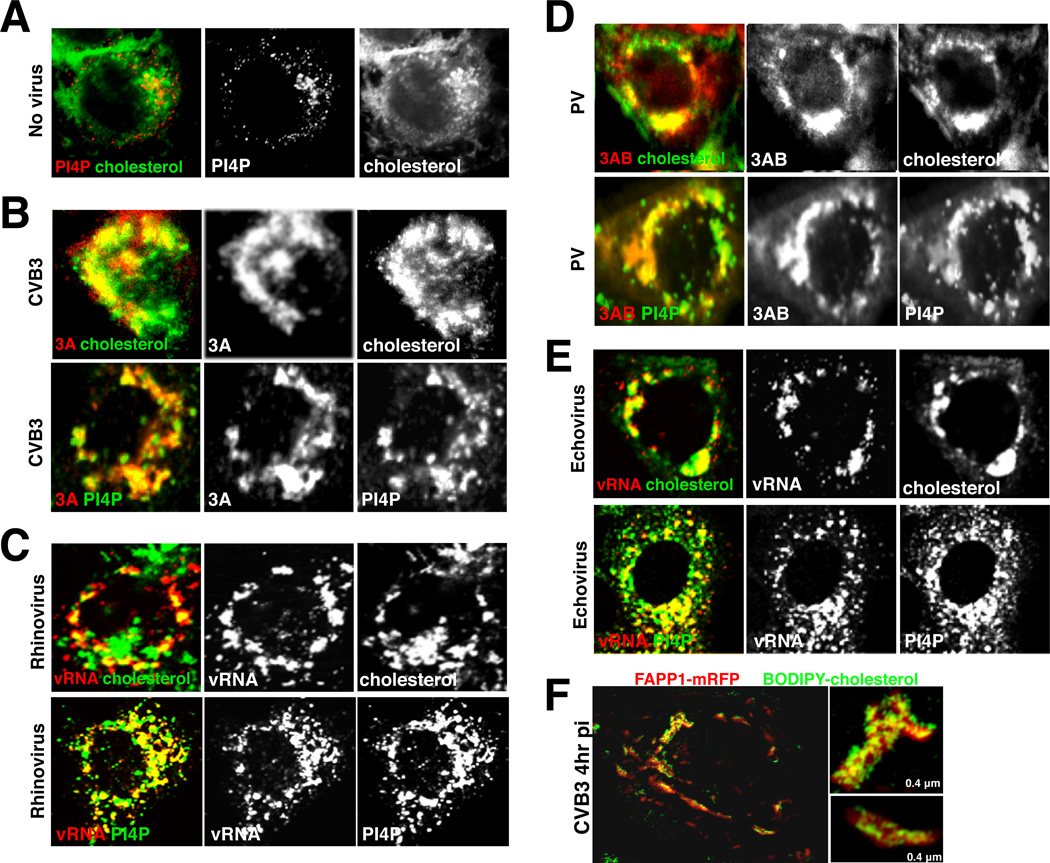
To determine whether the impact on viral replication of CME and caveolin disruption could be through their modulation of the host cholesterol landscape, we first investigated whether replication organelle membranes themselves contained cholesterol. Using filipin, in conjunction with antibodies against enteroviral replication proteins (3A, 3AB) and dsRNA, we obtained confocal images of free cholesterol distribution within uninfected (Figure 2A) and CVB3, PV (Type 1 Mahoney), rhinovirus (type 2) or echovirus (11 strain Gregory) infected cells at peak replication times (Figures 2B–2E). All replication organelles were enriched in free cholesterol (Pearson coefficient of colocalization with replication proteins: CVB3 0.70 +/− 0.03 in n=10 cells; PV 0.79 +/− 0.04 in n=10 cells), as well as PI4P lipids, the latter synthesized by PI4KIIIβ, whose inhibition blocked replication (Figure S2A and S2B; Hsu et al., 2010).
Figure 2. Enteroviral replication organelles are enriched in cholesterol and PI4P lipids.
(A) Free cholesterol and PI4P distribution in mock infected HeLa cell. Scale bar 5µm.
(B)-(E) Free cholesterol and PI4P distribution in CVB3, HRV2, PV and Echovirus infected cell replication organelles. Scale bar 5µm.
(F) SIM imaging of free cholesterol and PI4P distribution at CVB3 replication organelles.
Figure 2 related to Figure S2.
Replication organelles are at the limit of resolution with conventional diffraction limited microscopy (Belov et al., 2012) so to obtain spatial information on cholesterol and PI4P organization within replication organelles we utilized structured illumination microscopy (SIM), which provides (x, y)-axes resolution of ∼ 140nm and z-axis resolution of ∼250nm (Gustafsson et al., 2008). Live cells expressing FAPP1-mRFP to detect PI4P lipids (Hsu et al., 2010) were pulsed with BODIPY-cholesterol and allowed to reach steady-state distribution. Cells were then infected with CVB3 for 4 hrs. SIM imaging demonstrated that replication organelles have a non-uniform distribution of cholesterol, with segregated cholesterol domains of 100–400 nm in size (Figure 2F).
Cholesterol facilitates enteroviral RNA synthesis at replication organelles
We next investigated whether free cholesterol within replication organelle membranes was required for replication. Previous studies have focused on cholesterols’ role in regulating viral attachment, viral entry or mediating signaling for anti-viral immune responses (Howes et al., 2010; Mercer et al., 2010; Mackenzie et al., 2007). We first determined that chronically lowering intracellular free cholesterol pools (>50%) by growing cells in serum-free media, LDL-deficient media, or LDL-deficient media supplemented with Lovastatin, a blocker of cholesterol biosynthesis (Krukemyer and Talbert, 1987) for up to 72 hrs, all inhibited PV, CVB3 and rhinovirus replication (Figures S3A, S3B and S3C).
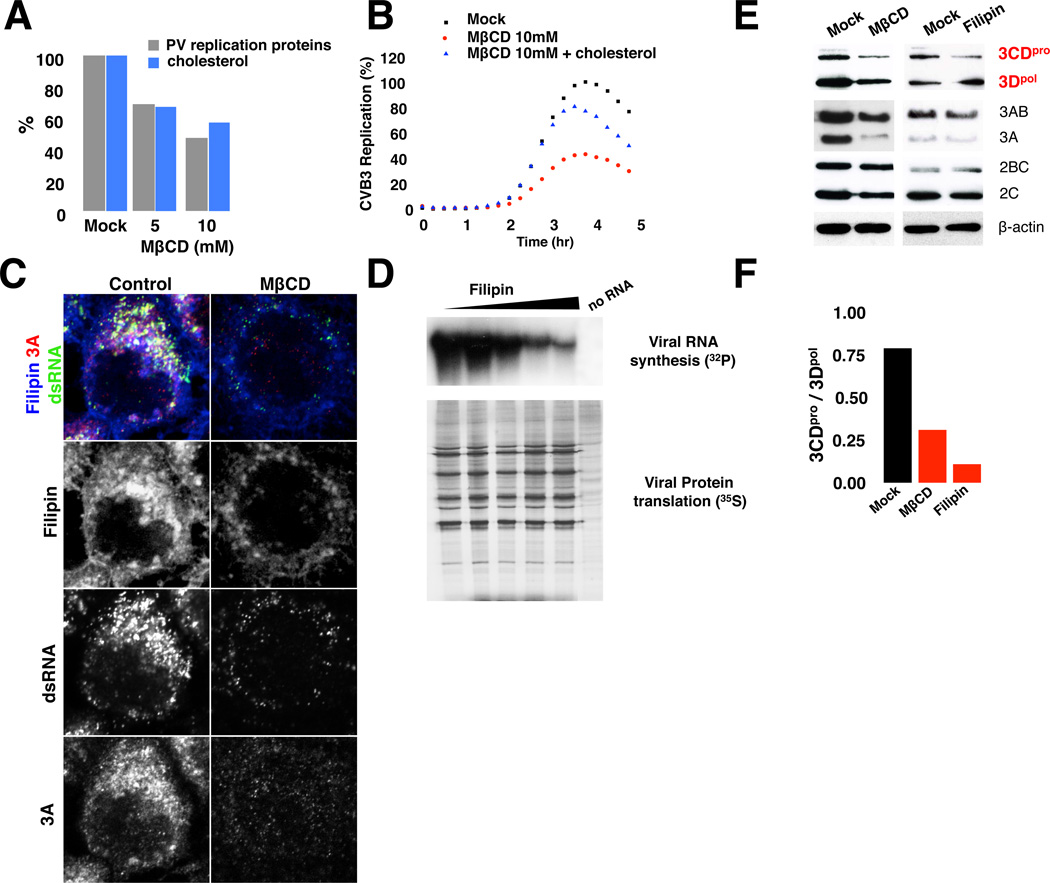
We next acutely depleted PM free cholesterol pools by treating cells with methyl-β-cyclodextrin (MβCD) for 1 hr. Subsequently cells were transfected with CVB3 or PV replicons and replication data normalized by the transfection/translation efficiency of a reporter RNA under similar conditions. Note also that MβCD treatment had no significant effect on PKR response (Figure S3D). After acutely lowering PM free cholesterol levels with MβCD, we found a dose-dependent inhibition of replication and replication organelle biogenesis (Figures 3A–3C). Significantly, replication could be rescued by adding back cholesterol (Figures 3B and Figure S3E), indicating that it had a facilitative role for enteroviral replication.
Figure 3. Cholesterol facilitates viral RNA synthesis and attenuates viral 3CDpro processing.
(A) Acute free cholesterol extraction from PM inhibits PV replication. Mean data +/− SEM of PV replication and cholesterol levels is plotted (n=50 cells for each condition).
(B) Cholesterol rescues CVB3 replicon replication. Mean data +/− SEM, from 6 replicates for each condition is plotted.
(C) Inhibition of replication organelle biogenesis after acute MβCD treatment. Antibodies against dsRNA and 3A proteins detect replication organelles. Scale bar 5um.
(D) Cell-free PV RNA translation and synthesis assay. Viral RNA synthesis assayed by 32P-CTP (top) and translation assayed by 35S-methionine labeling (bottom) on filipin treated membranes.
(E) Immunoblot analysis of 3CDpro, 2BC and 3AB processing after mock; MβCD (1 hr pre-treatment); or filipin treatment (at 3 hr pi). Viral proteins were harvested at 5 hr pi.
(F) 3CDpro/3Dpol protein ratio after cholesterol perturbation. Mean data +/− SEM from 3 independent experiments is plotted.
Figure 3 related to Figure S3.
To then determine the impact of cholesterol on viral RNA translation and/or RNA synthesis, two processes which feedback on each other, we utilized a cell-free assay with isolated membranes where translation and RNA synthesis could be uncoupled (Barton and Flanegan 1993). We disrupted the organization of free cholesterol and its abundance within membranes by either filipin (Nichols et al., 2001) (Figure 3D) or MβCD treatment respectively (Figure S3F). Both methods of cholesterol perturbation inhibited viral RNA synthesis but had no effect on translation (Figure 3D). These findings suggest that free cholesterol organization and abundance within replication organelle membranes are critical for viral RNA synthesis.
Cholesterol attenuates enteroviral 3CDpro polyprotein processing
Enteroviral genome is translated into a 247kD polyprotein, which is proteolytically processed in sequential steps into a repertoire of structural and replication proteins (Paul et al., 1987). Given the small size of the viral genome, each proteolytic intermediate plays a distinct and important role in replication. In particular 3CDpro processing is critical for replication. 3CDpro is required for formation of the replication complex, priming viral RNA synthesis and processing capsids (Andino et al., 1990; Cornell and Semler 2002). 3CDpro is also cleaved autocatalytically in cis to produce 3Cpro protease and 3Dpol proteins, where 3Cpro facilitates processing of other viral proteins and 3Dpol is the RNA-dependent RNA polymerase that replicates the genome.
We found that disrupting cholesterol organization or abundance within membranes with filipin or MβCD respectively stimulated the proteolytic processing of viral 3CDpro by almost 8-fold over mock treated cells (Figures 3E and 3F). This effect was specific for 3CDpro since neither 2BC (Figure S3G) nor 3AB protein processing (not shown) was stimulated. This steep decrease in 3CDpro levels, relative to other replication proteins, would be prohibitive to both the initiation of viral RNA synthesis as well as viral encapsidation. Thus our results suggest that cholesterol is critical for regulating 3CDpro processing kinetics and thereby the levels of 3CDpro, 3Cpro and 3Dpol proteins during infection.
Replication is stimulated in free cholesterol rich cells of Niemann-Pick Type C disease
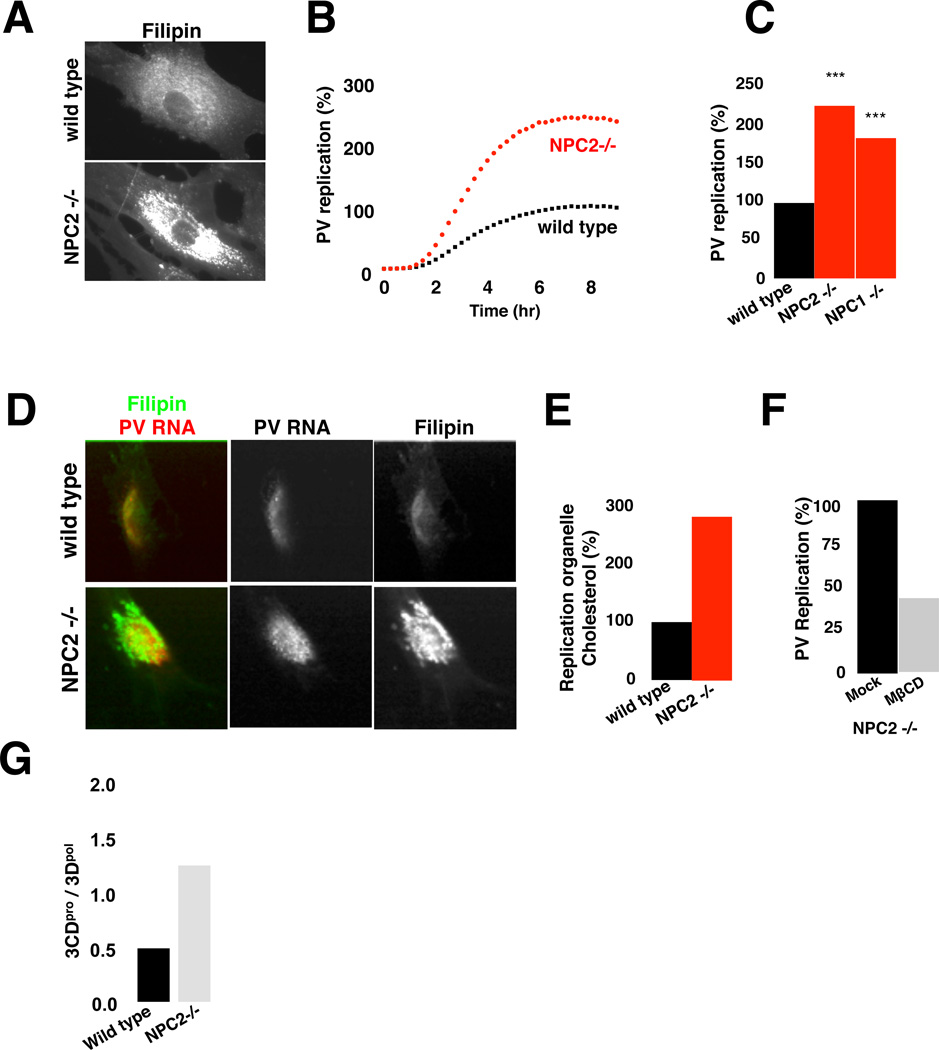
Consistent with the facilitative role for cholesterol in replication, caveolin- depleted cells had ∼40% more cholesterol in their replication organelles than control cells (Figure S4A and S4B). We next investigated the fate of enteroviral replication in primary cells from individuals with Niemann-Pick Type-C (NPC) disease where intracellular free cholesterol levels are very high (Figure 4A) (Rosenbaum and Maxfield, 2011). This disease has been assigned to the loss-of-function mutations in NPC1 and/or NPC2 endosomal cholesterol transporter proteins, which result in disruption of cholesterol export out of endosomes while ER cholesterol biosynthesis is continued.
Figure 4. High free cholesterol pools in Niemann-Pick Type C disease fibroblasts stimulate replication.
(A) Free cholesterol distribution in wild type and NPC2 (−/−) fibroblasts.
(B) PV replicon replication in wild type and NPC2 (−/−) fibroblasts. Mean data +/−SEM with 6 replicates each is plotted.
(C) Peak PV replication levels for wild type, NPC1 (−/−) and NPC2 (−/−) fibroblasts. Mean data +/− SEM from 5 independent experiments, normalized by the expression of reporter luciferase RNA, is plotted. ***: p<0.001
(D) Cholesterol and viral dsRNA distribution in wild type and NPC2 (−/−) fibroblasts.
(E) Free cholesterol levels within wt and NPC2 (−/−) fibroblast replication organelles. Mean data +/−SEM, from wild type (n=30) and NPC2 (−/−) (n=30) fibroblasts plotted.
(F) Acute cholesterol extraction inhibits PV replicon replication in NPC2−/− fibroblasts. Mean data +/− SEM from 2 independent experiments is plotted.
(G) 3CDpro / 3Dpol ratio in wild type and NPC2 (−/−) fibroblasts. Mean data +/−SEM from 3 independent experiments are plotted.
Scale bars 10µm.
Figure 4 related to Figure S4.
Primary wild type (NPC1wt, NPC2wt) and mutant (NPC1−/−, NPC2−/−) fibroblasts were transfected with PV replicons and replication was measured. We found a ∼ 3-fold increase in replication in NPC1−/− and NPC2−/− fibroblasts relative to wild type along with a ∼3-fold increase in replication organelle cholesterol (Figures 4B–4E). Note that replication data were normalized by the transfection/translation efficiency of a reporter RNA; and wild type and mutant cell PKR responses were similar (Figure S4C). Furthermore lowering NPC cells’ cholesterol pools with MβCD, inhibited replication by ∼60% relative to mock-treated cells (Figure 4F), indicating that cholesterol was responsible for stimulating replication in NPC cells. Finally, 3CDpro processing was significantly attenuated in NPC cells (Figure 4G) as well as in caveolin-depleted cells relative to each respective control cell (Figure S4D). This attenuation in 3CDpro processing may account for the enhanced replication in NPC cells or caveolin-depleted cells by promoting initiation of replication complexes and viral RNA encapsidation (Franco et al., 2005; Molla et al., 1994).
Enteroviruses stimulate the net uptake of cholesterol from the PM and extracellular medium by modulating CME
Given our findings, we conjectured that enteroviruses remodel their hosts’ cholesterol landscape in order to enrich for free cholesterol pools during infection. Supporting this conjecture, our bulk cholesterol measurements revealed a net increase of 40% in free cholesterol and a net decrease of 50% in esterified cholesterol pools within PV and CVB3 infected cells at peak replication times relative to mock infected cells (Figure 5A, 4hr pi).
Figure 5. Enteroviruses elevate intracellular free cholesterol pools.
(A) Quantification of free and esterified cholesterol pools at 0 hr and 4 hr post CVB3 infection. Mean data +/−SEM from 3 independent experiments plotted.
(B) LDL-uptake during CVB3 infection. Mean BODIPY-LDL uptake data from infected cells at 2hr (n=30) and 4hr pi (n=30); and for mock-infected cells at 4hr pi (n=30) was plotted as % of uptake of mock-infected cells at 2 hr pi (n=30) +/− SEM.
(C) Free and esterified cholesterol biosynthesis at 0, 2 and 4 hr post CVB3 infection. Mean data +/−SEM from one experiment with 3 replicates are plotted.
(D) Free cholesterol redistributes from PM to replication organelles during CVB3 infection. Cells expressing FAPP1-mRFP and co-labeled with BODIPY-cholesterol were infected and imaged by time-lapse confocal microscopy. See also Movies S1.1–S1.4. Scale bar 5µm.
(E) Quantification of plasma membrane BODIPY-cholesterol levels during CVB3 infection. Mean data +/−SEM from mock (n=10) and CVB3 (n=10) infected cells is plotted.
(F) Dynasore blocks BODIPY-cholesterol trafficking from plasma membrane to replication organelles. HeLa cells expressing FAPP1-mRFP were infected with CVB3 for 3 hrs, pulsed with BODIPY-cholesterol and subsequently chased with either DMSO or Dynasore (80µM) for 1 hr prior to confocal imaging. Scale bar 5µm.
(G) Dynasore blocks CVB3 replication. DMSO or Dynasore (80µM) treated cells were transfected with CVB3 replicons. Mean data +/−SEM of peak replication levels in 3 independent experiments for each condition is plotted.
(H) Dynasore blocks endogenous free cholesterol pools from trafficking to replication organelles. Experimental design similar to (F) but cells were fixed and labeled with filipin and anti- 3A antibodies. Mean data +/−SEM from n=30 cells for each condition is plotted.
Figure 5 related to Figure S5 and Movie 1.
To determine the mechanisms underlying these changes we first measured the clathrin-mediated LDL-cholesterol uptake during infection. We found it to be stimulated ∼30% within the first 2hrs of infection relative to control cells (Figure 5B, 2hrpi). But by 4hrs post infection, LDL-cholesterol uptake was inhibited by > 50% (Figure 5B, 4hr pi). Notably when cells were transferred into LDL-deficient media at 1hr post replicon transfection, replication was significantly inhibited (Figure S3B) highlighting the importance of LDL-cholesterol uptake early in replication. While cholesterol biosynthesis was uninterrupted during infection, the rate of synthesis paralleled the changes in LDL-uptake: decreasing by ∼30% within 2hrs post infection (Figure 5C, 2hr pi) but increasing back to uninfected cell rates by peak replication and later (Figure 5C, 4 hr and 6hr pi). This increase likely reflects the impact of replication organelles emerging from the ER, since they would be predicted to remove cholesterol. In contrast to biosynthesis, cholesterol esterification was inhibited throughout infection (Figure 5C, esterified) and lipid droplets were depleted (Figure S5A). Note that host transcription and translation are largely shut down by enteroviruses, thus these changes in cholesterol homeostasis suggest posttranslational viral modulation of host proteins regulating LDL-uptake, esterification and biosynthesis. Indeed HMG CoA reductase cholesterol biosynthesis enzyme levels and distribution were unchanged during infection (Figures S5B and S5C).
We next investigated the dynamics of the PM free cholesterol pools during infection. Cells expressing FAPP1-mRFP were pulsed with BODIPY-cholesterol and incubated for 1hr in order for the label to reach steady state distribution (Figure 5D, 0hr). Subsequently cells were CVB3 or mock infected and confocal time-lapse movies were taken for the duration of infection. In contrast to mock (Figure 5D, 0hr pi; Movie S1.1), following CVB3 infection, there was rapid vesicular internalization of PM cholesterol pools (Movies S1.2, S1.3). Within 2hrs post-infection >90% of the label had been depleted from the PM (Figure 5D, 2hr pi; Figure 5E). Following internalization many of these vesicles were observed fusing with and transferring their BODIPY- cholesterol label to replication organelles emerging from ER exit sites (Figure 5D, 4hr pi; Movie S1.4).
BODIPY-cholesterol was frequently observed to colocalize with clathrin labeled vesicular structures within the first 2hrs post-infection (Figures S5D and S5E). Acute treatment with dynasore, a non-competitive dynamin GTPase inhibitor (Macia et al., 2006) which disrupts CME (Dutta and Donaldson 2012), blocked both BODIPY-cholesterol as well as native cholesterol trafficking to replication organelles (Figure 5F) and significantly inhibited both replication (Figure 5G) and replication organelle cholesterol content (Figures 5F and 5H). These data, together with stimulation of LDL-cholesterol uptake (Figure 5B) indicate that CME is virally modulated during infection to enrich intracellular free cholesterol pools and redistribute them to replication organelles. One candidate enteroviral protein to increase LDL and PM cholesterol uptake is 2BC. As previously reported (Cornell et al., 2006), when ectopically expressed, 2BC increased the endocytic uptake of AM4–65 lipid tracer from the PM by ∼ 4-fold relative to mock and significantly this uptake was sensitive to dynasore treatment (Figure S5F and S5G).
Enteroviral 3A proteins recruit Rab11 recycling endosomes to target cholesterol to replication organelles and prevent it from recycling back to the PM
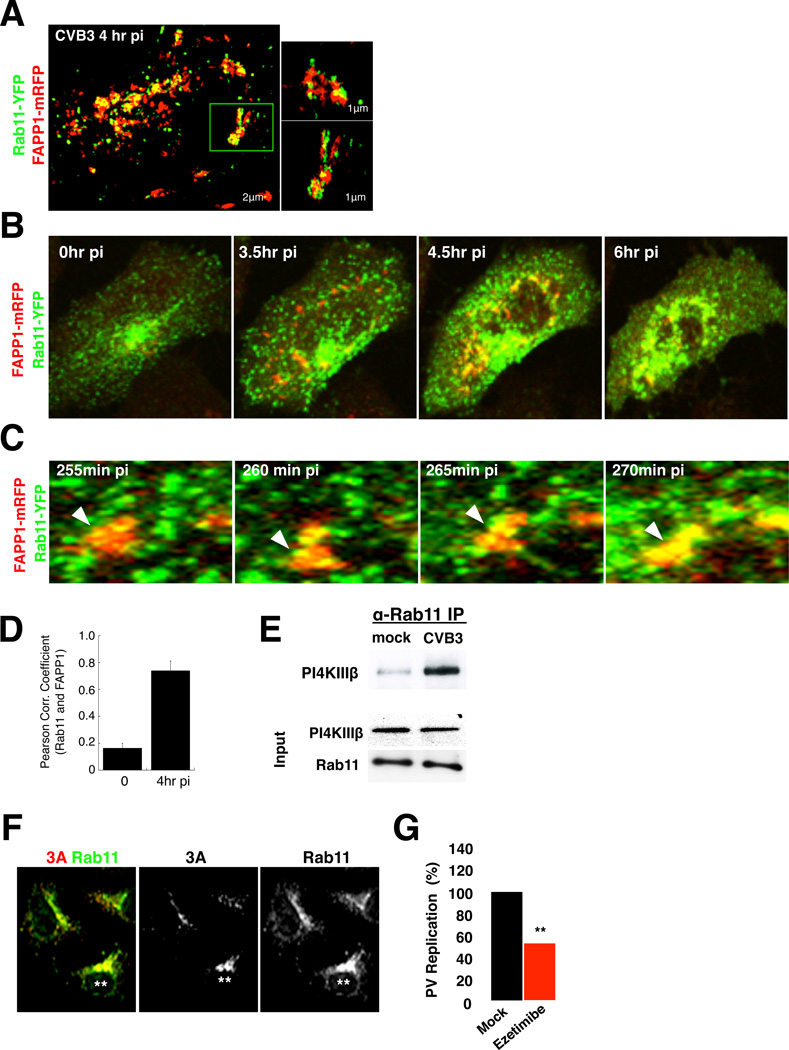
We next investigated how internalized endosomal cholesterol could be targeted to replication organelles given that the latter emerge from ER exit sites (Hsu et al., 2010). The temporal correlation between the resumption of cholesterol biosynthesis at the time of replication organelle emergence (Figure 5C) suggests that some cholesterol is likely transferred from the ER to replication organelles. In addition, within 2hrs of infection, we found by confocal imaging and SIM that PM cholesterol pools also redistributed to recycling endosomes containing Rab11 (Figures S6A and S6B).
By co-expressing Rab11-YFP and FAPP1mRFP, we investigated the fate of recycling endosomes relative to replication organelles during CVB3 infection. Time-lapse microscopy revealed numerous Rab11-YFP recycling endosomes trafficking to and merging with FAPP1-mRFP labeled replication organelles (Figures 6B–6D; Movies S2.1 and S2.2). Recruitment was recycling endosome specific since neither early nor late endosomal markers localized to replication organelles (Figure S6C). Furthermore as assessed by co-immunoprecipitation the physical interaction between Rab11 and PI4KIIIβ was significantly increased by peak replication times even though respective protein abundances were unchanged (Figure 6E).
Figure 6. Enteroviral 3A proteins recruit Rab11 recycling endosomes and target free cholesterol to replication organelles.
(A) Live cell SIM imaging of Rab11-YFP and FAPP1-mRFP distribution in CVB3 infected cells at peak replication. Insets highlight single replication organelles.
(B) Confocal time-lapse images of Rab11-YFP and FAPP1-mRFP dynamics in CVB3 infected cells. See also Movie S2.1. Scale bar 5µm.
(C) Fusion of Rab11-YFP recycling endosomes with replication organelles in boxed region in (B). See also Movie S2.2. Scale bar 1µm.
(D) Quantification of Rab11 colocalization with FAPP1 labeled replication organelles. Mean Pearson correlation coefficients +/−SEM are plotted (n=5 cells for each time point)
(E) Enhanced co-immunoprecipitation of PI4KIIIβ with Rab11 in CVB3 infected cells at peak replication.
(F) Ectopic CVB3 3A expression recruits Rab11 to 3A containing membranes. Scale bar 5µm.
(G) Ezetimibe, inhibits PV replicon replication. Mean peak replication data +/− SEM of replicon transfected cells from 3 independent experiments with 6 replicates each is plotted. ***: p<0.001
Figure 6 related to Figure S6 and Movie 2.
We found that ectopic expression of enteroviral 3A proteins alone, which selectively enhance PI4KIIIβ recruitment to membranes (Greninger et al., 2012; Hsu et al., 2010), were also sufficient to enhance Rab11 recruitment to the same membranes (Figures 6F). While Rab11 recruitment was independent of PI4P production by PI4KIIIβ (Figure S6D), it remains to be determined whether PI4KIIIβ plays a scaffold role. Regardless, enteroviral 3A proteins, by harnessing Rab11, target cholesterol to replication organelles and prevent its cycling back to the PM, and thereby contribute to the net increase in intracellular free cholesterol pools (Figure 5A). Furthermore together with PI4KIIIβ, 3A proteins facilitate the biogenesis of PI4P and cholesterol enriched replication organelle membranes.
Finally, acute treatment of cells with Ezetimibe, a highly specific inhibitor of the NPC1L1 cholesterol receptor (Chang and Chang 2008), blocked enteroviral replication (Figure 6G; Figure S6E). Since NPC1L1, traffics cholesterol via clathrin/AP2 mediated endocytosis to Rab11 recycling endosomes (Wang and Song 2012), this data provides further support for the viral exploitation of the CME pathway for the enrichment and delivery of cholesterol to replication organelles.
Discussion
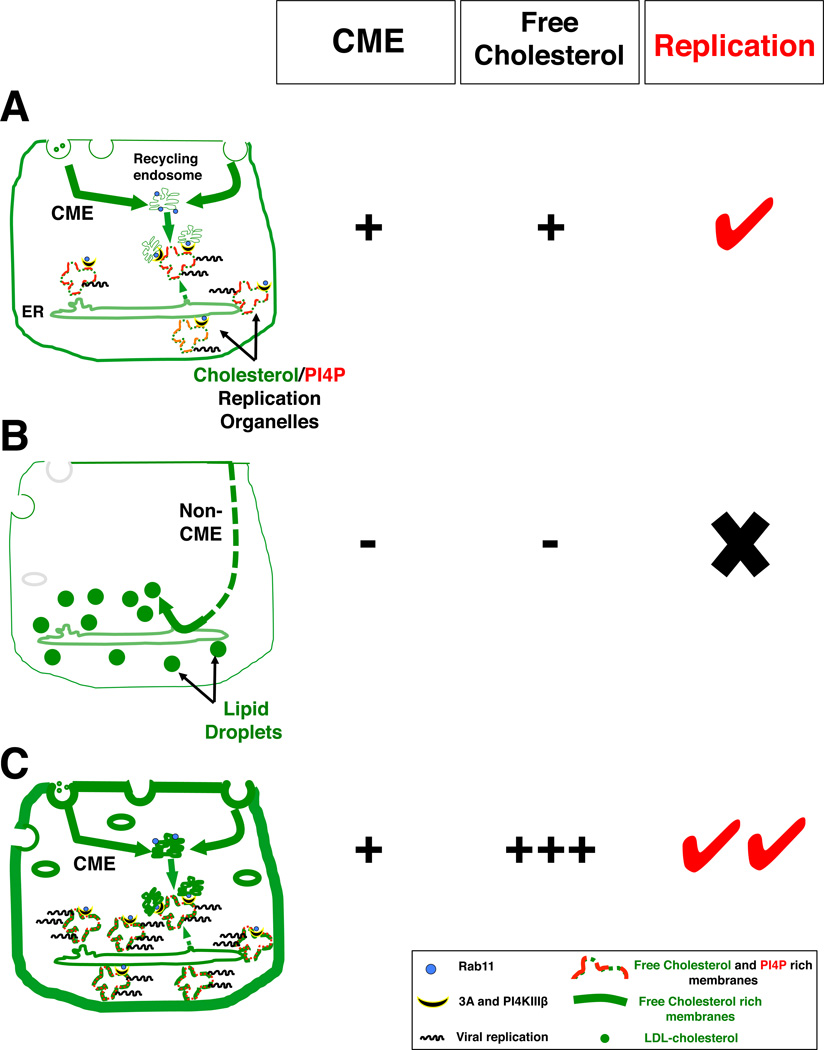
We have shown here that CME is harnessed by enteroviruses to enrich intracellular free cholesterol (i.e., increase LDL uptake; internalize PM cholesterol pools) and subsequently traffic cholesterol to replication organelles where cholesterol modulates proteolytic processing of viral 3CDpro and facilitates viral RNA synthesis. Furthermore we found that enteroviral replication can be stimulated in cells with high free cholesterol pools and functional CME pathways while replication is inhibited when CME is disrupted (Figure 7). In the latter, CME machinery is not only unavailable to traffic cholesterol to replication organelles but PM cholesterol pools are instead trafficked to lipid droplets for storage.
Figure 7. Cholesterol landscape and enteroviral replication.
(A) Upon infection, viral proteins (e.g. 2BC) modulate CME to enhance the net uptake of PM and extracellular cholesterol pools. Internalized cholesterol is pooled in Rab11 recycling endosomes and targeted to PI4P enriched replication organelles via protein-protein interactions among viral 3A, Rab11 and PI4KIIIβ proteins. Additionally some endocytosed cholesterol is transferred to the replication organelles indirectly, through the ER, as the organelles emerge from ER exit sites.
(B) Enteroviral replication is inhibited when CME is disrupted: cholesterol cannot be internalized/transported to replication organelles; PM free cholesterol pools are instead trafficked by alternate pathways to lipid droplets for storage.
(C) Enteroviral replication is stimulated in cells with functional CME and high free cholesterol pools at the PM and endosomal compartments (e.g. NPC or Cav1 or Cav2 depleted).
Based on our findings we propose the following model for the role CME in regulating enteroviral replication. Early in infection, there is a net increase in clathrin-mediated internalization of cholesterol (i.e., LDL-cholesterol, NPC1L1-cholesterol, PM free cholesterol). This is potentially modulated through the expression of newly synthesized viral 2BC proteins. A large fraction of internalized cholesterol pools is then transported to recycling endosomes while the remainder traffcis to the ER through alternative pathways, leading to a decrease in cholesterol biosynthesis. Furthermore the gradual absorption into the ER of cholesterol rich Golgi membranes, as a result of enteroviral 3A proteins interference with coatomer recruitment (Hsu et al., 2010; Wessels et al., 2006), may also potentiate this decrease in biosynthesis.
By peak replication times (Figure 7A), replication organelles will have emerged from ER exit sites, carrying cholesterol away from the ER and leading to the resumption of cholesterol biosynthesis. Unlike cholesterol biosynthesis, which proceeds throughout infection, cholesterol storage activities, through yet unknown mechanisms, are virally inhibited, which further enhances cellular free cholesterol pools. Meanwhile 3A proteins by recruiting Rab11 positive recycling endosomes to replication organelles along with PI4KIIIβ, enrich these organelles with both cholesterol and PI4P lipids, which then facilitates viral polyprotein processing and RNA synthesis. Targeting recycling endosomes to replication organelles also prevents endocytosed cholesterol pools, other lipids as well as plasma membrane proteins, such as LDL-receptor and MHC, from being recycled back to the cell surface. Preventing LDL-receptor recycling may explain the decrease observed in LDL-uptake at 4hrs post infection, while intracellular trapping of MHC may contribute to evasion of the immune system (Deitz et al., 2000; Cornell et al., 2006).
When cholesterol is abundant, cells esterify and store within lipid droplets some of their PM free cholesterol pools in order to maintain cholesterol homeostasis (Lange et al., 1993). Here we found that this process was enhanced when CME was disrupted (Figure 7B). While the mechanisms remain to be determined, CME perturbation may trigger free cholesterol to be trafficked by non-clathrin vesicular or entirely non-vesicular pathways such as direct exchange via ER-PM contact sites or ORP carriers (English and Voeltz 2013; Jansen et al., 2011). Alternatively, the normal recycling of free cholesterol pools back to the PM may be inhibited when CME is disrupted (van Dam and Stoorvogel 2002) and thus resulting in transfer of these pools to the ER for storage in lipid droplets. The physical proximity of the ER to the PM and endosomes and the increase in its esterification activity, suggest that free cholesterol is trafficked first to the ER prior to storage although some fraction of sterol may be directly trafficked to the lipid droplets.
We found that disrupting CME machinery had an opposite impact on enteroviral replication than disrupting caveolins. The former not only resulted in PM free cholesterol pools being routed for storage but also prevented enteroviruses from harnessing the CME machinery to traffic these pools to replication organelles. In contrast, in caveolin depleted cells, as well as NPC cells, the presence of functional CME machinery and abundant free cholesterol pools generated an ideal environment within which enteroviruses could replicate (Figure 7C). Notably for NPC cells intracellular cholesterol trafficking from the late endosomal stores to the PM occurs at a normal rate (Lange et al., 2002). Thus, the reduction in movement of cholesterol to the ER, a primary defect in NPC, may in fact promote the availability of sterol for the replication machinery from the PM.
For the majority of the siRNAs tested, their impact on CVB3 and PV replication were of similar magnitude and small differences observed were potentially a consequence of differences in replication kinetics, which may provide opportunity for cells to mount antiviral responses, which combined with CME loss, can result in stronger inhibition of the slower replicating virus. However the impact of depleting DAB2, an adaptor for LDL-receptor, was significantly greater on CVB3 then PV, suggesting a larger dependence of CVB3 on LDL to enhance free cholesterol.
Our data also revealed that by trafficking cholesterol to PI4P rich replication organelle membranes, enteroviruses might be able to regulate their levels of 3CDpro proteins. Cholesterol domains help partition and organize lipids and transmembrane proteins within membrane bilayers (Simons and Sampaio, 2011; Lippincott-Schwartz and Phair, 2010; Bretscher and Munro, 1993). Replication complex components 3CDpro, 3Cpro and 3Dpol all localize to PI4P enriched membranes and 3Dpol has PI4P lipid specific binding domains (Hsu et al., 2010). PI4P enriched membranes can be highly fluid (Zhendre et al., 2011) which may prevent viral proteins from assembling on them. Cholesterol can counter this fluidity and thereby facilitate both replication complex assembly and position 3CDpro in a specific conformation such that autocatalytic processing will be attenuated.
Our findings here may also have implications for understanding the pathogenesis of enteroviral infections. The cells of the human gastrointestinal tract serve as initial replication sites for many enteroviruses before dissemination to the rest of the body (Bopegamage et al., 2005; Iwasaki et al., 2002). These polarized cells are specialized for maximum absorption of dietary cholesterol and express high levels of NPC1L1 at their PM (Jia et al., 2011). Thus they would be ideal for enteroviral replication: high cholesterol absorption along with functional CME machinery, including Rab11 recycling endosomes through which both apical and basolateral PM cholesterol pools can be trafficked (Maxfield and Wustner 2002). Furthermore mice made hypercholesterolemic by diet develop infections with high enteroviral loads but whether this is due to compromised antiviral responses or enhanced replication remains to be investigated (Campbell et al., 1982).
Finally cholesterol is a highly abundant critical component of the central and peripheral nervous systems (Chang et al., 2010; Karasinska et al., 2011). In Alzheimer’s disease (AD) and Huntington’s disease (HD) disruptions of both CME and cholesterol homeostasis have been frequently reported, including a significant increase in the number of neuronal lipid droplets containing esterified cholesterol (Area-Gomez et al., 2012; Li and DiFiglia 2012; Martinez-Vicente et al., 2010; Chang et al., 2010; Cataldo et al., 2000). Huntingtin protein, the primary causative agent for HD, interacts with HIP1 and clathrin (Velier et al., 1998) and mutant Huntingtin expression alone can disrupt CME and cholesterol homeostasis (Trushina et al., 2006). Similarly, in AD, amyloid β proteins were shown to cause CME defects (Treusch et al., 2011) and enhanced cholesterol esterification and storage is a hallmark of familial AD (Andrea-Gomez et al., 2012; Chang et al., 2010). Indeed blocking cholesterol esterification alleviates AD symptoms and reduces amyloid plaque formation (Bryleva et al., 2010). Our findings here coupling the disruption of CME with accumulation of esterified cholesterol may provide insight and therapeutic strategies for these neurological conditions. At any rate, whenever CME components are perturbed, the latter’s impact on cholesterol homeostasis should be given consideration when interpreting experimental results.
In summary our results identify a wider role of host endocytic proteins in shaping the cellular cholesterol landscape and impacting the viral lifecycle beyond attachment, entry and export. These findings may provide new panviral therapeutic strategies for treating enteroviral infections including blocking cholesterol uptake or biosynthesis; stimulating cholesterol storage; and preventing cholesterol from being trafficked to replication organelles by disrupting the viral recruitment of Rab11 proteins.
Experimental Procedures
Confocal time-lapse imaging and immunofluorescence was performed as described (Hsu et al., 2010). All images were analyzed with Zeiss LSM or ImageJ software.
Super-resolution 3D-SIM imaging was performed on a Zeiss ELYRA S.1 system (Carl Zeiss, USA). Images were acquired with a Plan-Apochromat 63x/1.40 oil immersion objective and an Andor iXon 885 EMCCD camera. 15 images per plane (5 phases, 3 rotations) and 0.125 mm z-section of 3 mm height were required for generating super-resolution images. Raw images were reconstructed and processed to demonstrate structure with greater resolution by the ZEN 2011 microscope software (Carl Zeiss, USA).
Cell viability quantification Optimal plasmid expression times, siRNA and drug concentrations/incubation times that maximize cell viability were assessed by both quantifying cell number and by CellTiter-Glo cell viability assays (Promega Corp, WI). Plasmid concentration range and siRNA concentration range tested were 0.1µg/µl-1µg/µl and 25nM-100nM respectively.
Lipid Assays Lipid Loading: Top Fluor (BODIPY) cholesterol in complex with MβCD at a molar ratio 1/10 was applied to cells. BODIPY- LDL was loaded at 20µg/ml in FBS free medium for 20 min at 37°C. Lipid Staining: Nile Red and Filipin III were utilized at 0.5 µg/ml and 50 µg/ml respectively for 5 minutes.
Transfections All DNA transfections were performed with Fugene 6 reagent (Roche Applied Science, IN). All siRNA transfections were performed with Dharmafect 1 (Dharmacon, CO).
Replicon Assays were performed as described in Hsu et al., 2010. Capped Firefly luciferase mRNA containing poly-A tail was used for control of RNA transfection.
Chemical treatments and analysis Cells were incubated in Lovastatin (Enzo Life Sciences Inc., NY) (5–25µM) and mevalonate (Sigma, MO) (250µM) for 72hrs in media with 5% lipoprotein depleted serum (Milipore, MA). Cholesterol was depleted by incubating HeLa or NPC cells with 10mM MβCD for 1hr or 2hrs respectively at 37°C. Dynasore (Sigma), was used at 80µM; Ezetimibe (Santa Cruz Inc, CA) concentration range was 1–30µM; PIK93 (Knight et al., 2006)(Symansis, Auckland New Zealand) concentration range was 500nM-1µM.
Cell-free Translation and Replication Assays Cell-free Translation and Replication Assays were performed as described in Hsu et al., 2010.
Cholesterol quantification Free and esterified cholesterol was determined enzymatically using Amplex Red (Invitrogen). Samples were diluted to equal amount of protein.
Statistical Analysis Data were expressed and plotted as means ± standard error of the mean (SEM). Unpaired student’s t tests were used to compare the mean of control and experimental groups. The actual p value and sample size of each experimental group are provided in the respective figure legends.
Supplementary Material
Highlights.
Enteroviruses harness endocytosis to traffic cholesterol to replication organelles
Cholesterol regulates viral polyprotein processing and facilitates replication
Enteroviral replication is stimulated in cells with high cholesterol levels
Endocytosis block triggers cholesterol storage by host and inhibits replication
Acknowledgements
We thank Jennifer Lippincott-Schwartz, Ellie Ehrenfeld, Cathy Jackson, Sandy Simon, Grégoire Altan-Bonnet, Nan Gao, Radek Dobrowolski, Eckard Wimmer for critical reading of the manuscript; Ilya Raskin, Carolyn Ott, Elise Shumsky for technical support. Awards from NIH R01AI091985 and NSF MCB-0822058 supported NAB; NIH DK38389 and Ara Parseghian Medical Research Foundation supported JS; National Center for Research Resources RR-021120 supported JLD.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- Andino R, Rieckhof GE, Baltimore D. A functional ribonucleoprotein complex forms around the 5' end of poliovirus RNA. Cell. 1990;63:369–380. doi: 10.1016/0092-8674(90)90170-j. [DOI] [PubMed] [Google Scholar]
- Area-Gomez E, Del Carmen Lara Castillo M, Tambini MD, Guardia-Laguarta C, de Groof AJ, Madra M, Ikenouchi J, Umeda M, Bird TD, Sturley SL, Schon EA. Upregulated function of mitochondria-associated ER membranes in Alzheimer disease. EMBO J. 2012;31:4106–4123. doi: 10.1038/emboj.2012.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton DJ, Flanegan JB. Coupled translation and replication of poliovirus RNA in vitro: synthesis of functional 3D polymerase and infectious virus. J Virol. 1993;67:822–831. doi: 10.1128/jvi.67.2.822-831.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belov GA, Nair V, Hansen BT, Hoyt FH, Fischer ER, Ehrenfeld E. Complex dynamic development of poliovirus membranous replication complexes. J Virol. 2012;86:302–312. doi: 10.1128/JVI.05937-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bopegamage S, Kovacova J, Vargova A, Motusova J, Petrovicova A, Benkovicova M, Gomolcak P, Bakkers J, van Kuppeveld F, Melchers WJ, Galama JM. Coxsackie B virus infection of mice: inoculation by the oral route protects the pancreas from damage, but not from infection. J Gen Virol. 2005;86:3271–3280. doi: 10.1099/vir.0.81249-0. [DOI] [PubMed] [Google Scholar]
- Bretscher MS, Munro S. Cholesterol and the Golgi apparatus. Science. 1993;261:1280–1281. doi: 10.1126/science.8362242. [DOI] [PubMed] [Google Scholar]
- Brown MS, Goldstein JL. A receptor-mediated pathway for cholesterol homeostasis. Science. 1986;1232:34–47. doi: 10.1126/science.3513311. [DOI] [PubMed] [Google Scholar]
- Brown MS, Goldstein JL. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a free transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- Brown MS, Goldstein JL. Sterol regulatory element binding proteins (SREBPs): controllers of lipid synthesis and cellular uptake. Nutr Rev. 1998;56:S1–S3. doi: 10.1111/j.1753-4887.1998.tb01680.x. discussion S54–75. [DOI] [PubMed] [Google Scholar]
- Bryleva EY, Rogers MA, Chang CC, Buen F, Harris BT, Rousselet E, Seidah NG, Oddo S, LaFerla FM, Spencer TA, Hickey WF, Chang TY. ACAT1 gene ablation increases 24(S)-hydroxycholesterol content in the brain and ameliorates amyloid pathology in mice with AD. Proc Natl Acad Sci U S A. 2010;107:3081–3086. doi: 10.1073/pnas.0913828107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell AE, Loria RM, Madge GE, Kaplan AM. Dietary hepatic cholesterol elevation: effects on coxsackievirus B infection and inflammation. Infect Immun. 1982;37:307–317. doi: 10.1128/iai.37.1.307-317.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cataldo AM, Peterhoff CM, Troncoso JC, Gomez-Isla T, Hyman BT, Nixon RA. Endocytic pathway abnormalities precede amyloid beta deposition in sporadic Alzheimer's disease and Down syndrome: differential effects of APOE genotype and presenilin mutations. Am J Pathol. 2000;157:277–286. doi: 10.1016/s0002-9440(10)64538-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TY, Chang C. Ezetimibe blocks internalization of the NPC1L1/cholesterol complex. Cell Metab. 2008;7:469–471. doi: 10.1016/j.cmet.2008.05.001. [DOI] [PubMed] [Google Scholar]
- Chang TY, Chang CC, Bryleva E, Rogers MA, Murphy SR. Neuronal cholesterol esterification by ACAT1 in Alzheimer's disease. IUBMB Life. 2010;62:261–267. doi: 10.1002/iub.305. [DOI] [PubMed] [Google Scholar]
- Cohen AW, Razani B, Schubert W, Williams TM, Wang XB, Iyengar P, Brasaemle DL, Scherer PE, Lisanti MP. Role of caveolin-1 in the modulation of lipolysis and lipid droplet formation. Diabetes. 2004;53:1261–1270. doi: 10.2337/diabetes.53.5.1261. [DOI] [PubMed] [Google Scholar]
- Cornell CT, Semler BL. Subdomain specific functions of the RNA polymerase region of poliovirus 3CD polypeptide. Virology. 2002;298:200–13. doi: 10.1006/viro.2002.1481. [DOI] [PubMed] [Google Scholar]
- Cornell CT, Kiosses WB, Harkins S, Whitton JL. Inhibition of protein trafficking by coxsackievirus b3: multiple viral proteins target a single organelle. J Virol. 2006;80:6637–6647. doi: 10.1128/JVI.02572-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deitz SB, Dodd DA, Cooper S, Parham P, Kirkegaard K. MHC I-dependent antigen presentation is inhibited by poliovirus protein 3A. Proc Natl Acad Sci U S A. 2000;97:13790–13795. doi: 10.1073/pnas.250483097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Boon JA, Ahlquist P. Organelle-like membrane compartmentalization of positive-strand RNA virus replication factories. Annu Rev Microbiol. 2010;64:241–256. doi: 10.1146/annurev.micro.112408.134012. [DOI] [PubMed] [Google Scholar]
- Dutta D, Donaldson JG. Search for inhibitors of endocytosis: Intended specificity and unintended consequences. Cell Logist. 2012 Oct;1(24):203–208. doi: 10.4161/cl.23967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English AR, Voeltz GK. Endoplasmic reticulum structure and interconnections with other organelles. Cold Spring Harb Perspect Biol. 2013;5(4) doi: 10.1101/cshperspect.a013227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco D, Pathak HB, Cameron CE, Rombaut B, Wimmer E, Paul AV. Stimulation of poliovirus RNA synthesis and virus maturation in a HeLa cell-free in vitro translation-RNA replication system by viral protein 3CDpro. Virol J. 2005;2:86. doi: 10.1186/1743-422X-2-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Hoang A, Escher G, Parton RG, Krozowski Z, Sviridov D. Expression of caveolin-1 enhances cholesterol efflux in hepatic cells. J Biol Chem. 2004;279:14140–14146. doi: 10.1074/jbc.M311061200. [DOI] [PubMed] [Google Scholar]
- Greninger AL, Knudsen GM, Betegon M, Burlingame AL, Derisi JL. The 3A protein from multiple picornaviruses utilizes the Golgi Adaptor Protein ACBD3 to Recruit PI4KIIIbeta. J Virol. 2012;86:3605–3616. doi: 10.1128/JVI.06778-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson MG, Shao L, Carlton PM, Wang CJ, Golubovskaya IN, Cande WZ, Agard DA, Sedat JW. Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination. Biophys J. 2008;94:4957–4970. doi: 10.1529/biophysj.107.120345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtta-Vuori M, Uronen RL, Repakova J, Salonen E, Vattulainen I, Panula P, Li Z, Bittman R, Ikonen E. BODIPY-cholesterol: a new tool to visualize sterol trafficking in living cells and organisms. Traffic. 2008;9:1839–1849. doi: 10.1111/j.1600-0854.2008.00801.x. [DOI] [PubMed] [Google Scholar]
- Howes MT, Mayor S, Parton RG. Molecules, mechanisms, and cellular roles of clathrin-independent endocytosis. Current Opinion in Cell Biology. 2010;22:519–527. doi: 10.1016/j.ceb.2010.04.001. [DOI] [PubMed] [Google Scholar]
- Hsieh K, Lee YK, Londos C, Raaka BM, Dalen KT, Kimmel AR. Perilipin family members preferentially sequester to either triacylglycerol- or cholesteryl ester-specific intracellular lipid storage droplets. J Cell Sci. 2012 doi: 10.1242/jcs.104943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu NY, Ilnytska O, Belov G, Santiana M, Chen YH, Takvorian PM, Pau C, van der Schaar H, Kaushik-Basu N, Balla T, et al. Viral Reorganization of the Secretory Pathway Generates Distinct Organelles for RNA Replication. Cell. 2010;141:799–811. doi: 10.1016/j.cell.2010.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu TH, Spindler KR. Identifying host factors that regulate viral infection. PLoS Pathog. 2012;8:e1002772. doi: 10.1371/journal.ppat.1002772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikonen E. Cellular cholesterol trafficking and compartmentalization. Nat Rev Mol Cell Biol. 2008;9:125–138. doi: 10.1038/nrm2336. [DOI] [PubMed] [Google Scholar]
- Iwasaki A, Welker R, Mueller S, Linehan M, Nomoto A, Wimmer E. Immunofluorescence analysis of poliovirus receptor expression in Peyer's patches of humans, primates, and CD155 transgenic mice: implications for poliovirus infection. J Infect Dis. 2002;186:585–592. doi: 10.1086/342682. [DOI] [PubMed] [Google Scholar]
- Jansen M, Ohsaki Y, Rita Rega L, Bittman R, Olkkonen VM, Ikonen E. Role of ORPs in sterol transport from plasma membrane to ER and lipid droplets in mammalian cells. Traffic. 2011;12:218–231. doi: 10.1111/j.1600-0854.2010.01142.x. [DOI] [PubMed] [Google Scholar]
- Jia L, Betters JL, Yu L. Niemann-pick C1-like 1 (NPC1L1) protein in intestinal and hepatic cholesterol transport. Annu Rev Physiol. 2011;73:239–259. doi: 10.1146/annurev-physiol-012110-142233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasinska JM, Hayden MR. Cholesterol metabolism in Huntington disease. Nat Rev Neurol. 2011;7:561–572. doi: 10.1038/nrneurol.2011.132. [DOI] [PubMed] [Google Scholar]
- Knight ZA, Gonzalez B, Feldman ME, Zunder ER, Goldenberg DD, Williams O, Loewith R, Stokoe D, Balla A, Toth B, et al. A pharmacological map of the PI3-K family defines a role for p110alpha in insulin signaling. Cell. 2006;125:733–747. doi: 10.1016/j.cell.2006.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krukemyer JJ, Talbert RL. Lovastatin: a new cholesterol-lowering agent. Pharmacotherapy. 1987;7:198–210. doi: 10.1002/j.1875-9114.1987.tb03524.x. [DOI] [PubMed] [Google Scholar]
- Lange Y, Strebel F, Steck TL. Role of the plasma membrane in cholesterol esterification in rat hepatocytes. J Biol Chem. 1993;268:13838–13843. [PubMed] [Google Scholar]
- Lange Y, Ye J, Rigney M, Steck TL. Dynamics of lysosomal cholesterol in Niemann-Pick type C and normal human fibroblasts. J Lipid Res. 2002;43:198–204. [PubMed] [Google Scholar]
- Li X, DiFiglia M. The recycling endosome and its role in neurological disorders. Prog Neurobiol. 2012;97:127–141. doi: 10.1016/j.pneurobio.2011.10.002. [DOI] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Phair RD. Lipids and cholesterol as regulators of traffic in the endomembrane system. Annu Rev Biophys. 2010;39:559–578. doi: 10.1146/annurev.biophys.093008.131357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macia E, Ehrlich M, Massol R, Boucrot E, Brunner C, Kirchhausen T. Dynasore, a cell-permeable inhibitor of dynamin. Dev Cell. 2006;10:839–850. doi: 10.1016/j.devcel.2006.04.002. [DOI] [PubMed] [Google Scholar]
- Mackenzie JM, Khromykh AA, Parton RG. Cholesterol manipulation by West Nile virus perturbs the cellular immune response. Cell Host Microbe. 2007;2:229–239. doi: 10.1016/j.chom.2007.09.003. [DOI] [PubMed] [Google Scholar]
- Martinez-Vicente M, Talloczy Z, Wong E, Tang G, Koga H, Kaushik S, de Vries R, Arias E, Harris S, Sulzer D, Cuervo AM. Cargo recognition failure is responsible for inefficient autophagy in Huntington's disease. Nat Neurosci. 2010;13:567–576. doi: 10.1038/nn.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxfield FR, Wüstner D. Intracellular cholesterol transport. J Clin Invest. 2002;110:891–898. doi: 10.1172/JCI16500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCloskey MA, Poo MM. Rates of membrane-associated reactions: reduction of dimensionality revisited. J Cell Biol. 1986;102:88–96. doi: 10.1083/jcb.102.1.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer J, Schelhaas M, Helenius A. Virus entry by endocytosis. Annu Rev Biochem. 2010;79:803–833. doi: 10.1146/annurev-biochem-060208-104626. [DOI] [PubMed] [Google Scholar]
- Miller S, Krijnse-Locker J. Modification of intracellular membrane structures for virus replication. Nat Rev Microbiol. 2008;6:363–374. doi: 10.1038/nrmicro1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molla A, Harris KS, Paul AV, Shin SH, Mugavero J, Wimmer E. Stimulation of poliovirus proteinase 3Cpro-related proteolysis by the genome-linked protein VPg and its precursor 3AB. J Biol Chem. 1994;269:27015–27020. [PubMed] [Google Scholar]
- Nichols BJ, Kenworthy AK, Polishchuk RS, Lodge R, Roberts TH, Hirschberg K, Phair RD, Lippincott-Schwartz J. Rapid cycling of lipid raft markers between the cell surface and Golgi complex. Journal of Cell Biology. 2001;153:529–541. doi: 10.1083/jcb.153.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parton RG, Del Pozo MA. Caveolae as plasma membrane sensors, protectors and organizers. Nat Rev Mol Cell Biol. 2013;14:98–112. doi: 10.1038/nrm3512. [DOI] [PubMed] [Google Scholar]
- Paul AV, Yang CF, Jang SK, Kuhn RJ, Tada H, Nicklin M, Krausslich HG, Lee CK, Wimmer E. Molecular events leading to poliovirus genome replication. Cold Spring Harb Symp Quant Biol. 1987;52:343–352. doi: 10.1101/sqb.1987.052.01.039. [DOI] [PubMed] [Google Scholar]
- Prinz WA. A bridge to understanding lipid droplet growth. Dev Cell. 2013;24:335–336. doi: 10.1016/j.devcel.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum AI, Maxfield FR. Niemann-Pick type C disease: molecular mechanisms and potential therapeutic approaches. J Neurochem. 2011;116:789–795. doi: 10.1111/j.1471-4159.2010.06976.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe RK, Suszko JW, Pekosz A. Roles for the recycling endosome, Rab8, and Rab11 in hantavirus release from epithelial cells. Virology. 2008;382:239–249. doi: 10.1016/j.virol.2008.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki J, Ishikawa K, Arita M, Taniguchi K. ACBD3-mediated recruitment of PI4KB to picornavirus RNA replication sites. EMBO Journal. 2011;31:754–766. doi: 10.1038/emboj.2011.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons K, Sampaio JL. Membrane organization and lipid rafts. Cold Spring Harb Perspect Biol. 2011;3:a004697. doi: 10.1101/cshperspect.a004697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treusch S, Hamamichi S, Goodman JL, Matlack KE, Chung CY, Baru V, Shulman JM, Parrado A, Bevis BJ, Valastyan JS, Han H, Lindhagen-Persson M, Reiman EM, Evans DA, Bennett DA, Olofsson A, DeJager PL, Tanzi RE, Caldwell KA, Caldwell GA, Lindquist S. Functional links between Aβ toxicity, endocytic trafficking, and Alzheimer's disease risk factors in yeast. Science. 2011;334:1241–1245. doi: 10.1126/science.1213210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trushina E, Singh RD, Dyer RB, Cao S, Shah VH, Parton RG, Pagano RE, McMurray CT. Mutant huntingtin inhibits clathrin-independent endocytosis and causes accumulation of cholesterol in vitro and in vivo. Hum Mol Genet. 2006;15:3578–3591. doi: 10.1093/hmg/ddl434. [DOI] [PubMed] [Google Scholar]
- van Dam EM, Stoorvogel W. Dynamin-dependent transferrin receptor recycling by endosome-derived clathrin-coated vesicles. Mol Biol Cell. 2002b;13:169–182. doi: 10.1091/mbc.01-07-0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velier J, Kim M, Schwarz C, Kim TW, Sapp E, Chase K, Aronin N, DiFiglia M. Wild-type and mutant huntingtins function in vesicle trafficking in the secretory and endocytic pathways. Exp Neurol. 1998;152:34–40. doi: 10.1006/exnr.1998.6832. [DOI] [PubMed] [Google Scholar]
- Wang LJ, Song BL. Niemann-Pick C1-Like 1 and cholesterol uptake. Biochim Biophys Acta. 2012;1821:964–972. doi: 10.1016/j.bbalip.2012.03.004. [DOI] [PubMed] [Google Scholar]
- Warnock DE, Roberts C, Lutz MS, Blackburn WA, Young WW, Jr, Baenziger JU. Determination of plasma membrane lipid mass and composition in cultured Chinese hamster ovary cells using high gradient magnetic affinity chromatography. J Biol Chem. 1993;268(14):10145–10153. [PubMed] [Google Scholar]
- Wessels E, Duijsings D, Niu TK, Neumann S, Oorschot VM, de Lange F, Lanke KH, Klumperman J, Henke A, Jackson CL, et al. A viral protein that blocks Arf1-mediated COP-I assembly by inhibiting the guanine nucleotide exchange factor GBF1. Dev Cell. 2006;11:191–201. doi: 10.1016/j.devcel.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Zhendre V, Grélard A, Garnier-Lhomme M, Buchoux S, Larijani B, Dufourc EJ. Key role of polyphosphoinositides in dynamics of fusogenic nuclear membrane vesicles. PLoS One. 2011;6:e23859. doi: 10.1371/journal.pone.0023859. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.