Abstract
Stable isotope labeling by essential nutrients in cell culture (SILEC) was recently developed to generate isotopically labeled coenzyme A (CoA) and short-chain acyl-CoA thioesters. This was accomplished by modifying the widely used technique of stable isotope labeling by amino acids in cell culture to include [13C315N]-pantothenate (vitamin B5), a CoA precursor, instead of the isotopically labeled amino acids. The lack of a de novo pantothenate synthesis pathway allowed for efficient and near-complete labeling of the measured CoA species. This protocol provides a step-by-step approach for generating stable isotope-labeled short-chain acyl-CoA internal standards in mammalian and insect cells as well as instructions on how to use them in stable isotope dilution mass spectrometric-based analyses. Troubleshooting guidelines, as well as a list of unlabeled and labeled CoA species, are also included. This protocol represents a prototype for generating stable isotope internal standards from labeled essential nutrients such as pantothenate. The generation and use of SILEC standards takes approximately 2–3 weeks.
Introduction
High-performance liquid chromatography (HPLC) coupled with tandem mass spectrometry (MS/MS) provides a highly sensitive and specific platform to measure a broad range of analytes in a variety of complex biological matrices1. In addition, advancements in high-resolution MS2,3 and the development of more dynamic software platforms4–6 have provided scientists with the capability of measuring thousands of different analytes in a single run, making MS the optimal methodology for robust high-throughput metabolomic and proteomic analyses7,8. Nevertheless, appropriate internal standards are crucial for the success of any MS-based method, particularly when measuring endogenous metabolites in biological samples, because of matrix effects on analyte stability, extraction and ionization efficiency9–14. To account for these effects, an internal standard that faithfully reproduces the biological and chemical properties of the analyte of interest is needed15.
Stable isotope analogs represent the best internal standards for MS-based analyses because they have the same biological and physicochemical properties as the analyte of interest, but they can still be distinguished by a mass spectrometer15. Difficulty in chemically synthesizing stable isotope analogs of complex biological molecules has spurred the development of techniques to generate these standards in biological systems16–18. In the case of proteins, a method called stable isotope labeling by amino acids in cell culture (SILAC) has been developed19. In this method, cells are grown in the presence of a stable isotope-labeled essential amino acid such as lysine, with the unlabeled form specifically omitted. The inability of cells to synthesize lysine results in the exclusive uptake and incorporation of the labeled lysine into cellular proteins. After serial passages in this medium, greater than 99% of lysine residues are labeled. At this point, there are two possible modes by which these labeled cells can be used. In the ‘classical’ SILAC approach, an equal number of labeled and unlabeled cells are subjected to different stimuli, mixed, processed together and analyzed by MS17. Alternatively, the labeled cells can be lysed to produce a stable isotope-labeled proteome internal standard (SILAP)20, which can then be spiked into different experimental samples to normalize the samples for relative quantification21–23. The advantage of this approach is the ability to apply these standards to both in vitro or in vivo samples; it also avoids any biological perturbations introduced by the labeled amino acids.
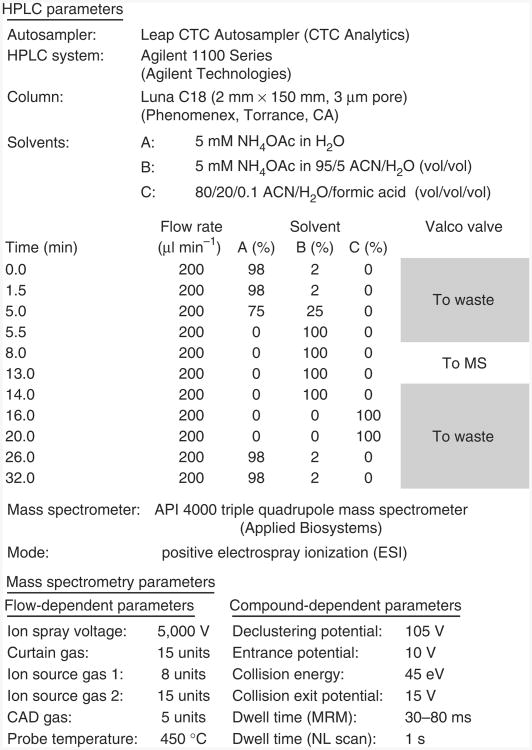
In addition to proteins, there are other endogenous molecules that have been difficult to chemically synthesize; consequently, labeled analogs for many of these compounds are not commercially available. One such compound is CoA, a ubiquitous fatty acyl carrier that has a crucial role in many metabolic and mitochondrial processes24–26. There are numerous CoA derivatives, including an acyl-CoA thioester for most individual fatty acids (Supplementary Table 1). MS/MS represents the optimal platform for developing CoA analysis. When subjected to collision-induced dissociation (CID), CoA derivatives show a characteristic fragmentation pattern (Fig. 1a), breaking at the ATP moiety, resulting in productions with a neutral loss of 507 atomic mass units (AMU)27,28. Typical parent protonated molecular ions observed for endogenous CoASH and its derivatives in a constant neutral loss/MS experiment are shown in Figure 1b. The use of this type of analysis not only facilitates LC– selected reaction monitoring (SRM)/MS method development, but also allows for more specific CoA discovery experiments. However, the lack of isotopically labeled CoA has limited the development of comprehensive stable isotope dilution mass spectral analysis of CoA compounds.
Figure 1.
LC-MS analysis of acyl-CoA thioesters. (a) Generalized CoA structure, showing the CID-induced neutral loss of the ATP moiety (m/z = 507). The substituent (R group) can include any thioester or derivative of CoA arising from endogenous or exogenous sources (see supplementary table 1 for a list of typical endogenous CoA species). (b) LC-constant neutral loss/MS scans (m/z = 507) of short-chain acyl-CoA species extracted from unlabeled Hepa 1c1c7 mouse hepatoma cells. The extraction was performed as described in the accompanying protocol using the specific LC-MS parameters provided in Figure 2. The precursor parent molecules of various short-chain acyl-CoA species are annotated in the spectrum. c.p.s., counts per second.
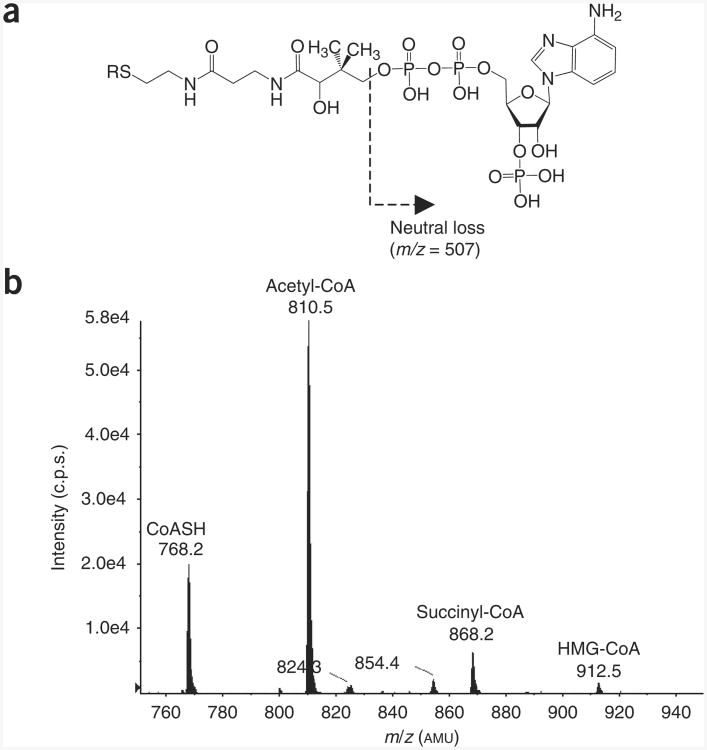
In this report, our focus is on generating stable isotope-labeled CoASH and short-chain acyl-CoA species, which would not only improve our ability to study metabolism29–33 but also aid in the diagnosis of various metabolic diseases34–38. The use of labeled standards is particularly important, as cellular thiols can be rapidly oxidized or can react with both intracellular and extracellular molecules39–41. The purpose of this protocol is to provide a practical guide to the generation of stable isotope-labeled short-chain acyl-CoA thioesters for use as internal standards in quantitative stable isotope dilution LC-MS assays. An in-depth discussion of how to conduct the LC-MS experiments has not been included because there is a substantial literature describing such methodology28,42–47. As a guideline, however, the specific LC-MS parameters used by our laboratory to analyze CoA derivatives have been provided (Fig. 2).
Figure 2.
LC-MS parameters used in our laboratory for the analysis of short-chain acyl-CoA thioesters.
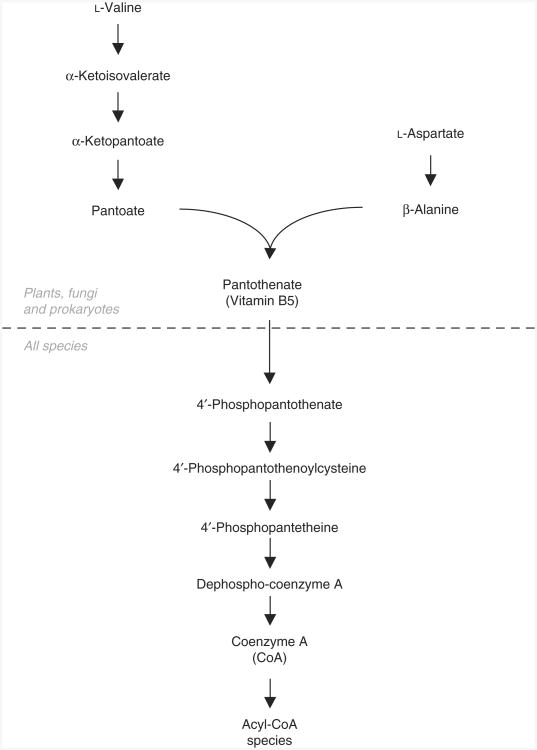
To biosynthetically generate labeled CoA standards, we recently developed SILEC, a method similar to SILAC, except that a labeled vitamin, pantothenate, is used instead of labeled amino acids18. Unlike plants, fungi and most prokaryotes, animals are not capable of de novo pantothenate synthesis (Fig. 3)48. As a result, pantothenate is a dietary requirement and a necessary cell culture medium supplement. After uptake, pantothenate is converted to CoA through five enzymatic steps24. By growing cells in the presence of [13C315N]-pantothenate, we have developed a method for biosynthetically generating stable isotope-labeled CoASH and acyl-CoA thioesters using a single culture. These standards can then be extracted and subsequently applied to cells, tissues or biological fluids before processing, and they can be used as internal standards for mass spectral analysis. To rigorously measure various short-chain acyl-CoA species using SILEC-labeled CoA standards, we have developed a stable isotope dilution LC-MS method (Fig. 2) on the basis of modifications of previously published procedures28,42–47.
Figure 3.
Pantothenate and coenzyme A biosynthesis. Plants, fungi and many prokaryotes are capable of de novo pantothenate synthesis from valine and aspartate, whereas animals require it as part of their diet.
Although this protocol focuses on the generation and utilization of short-chain acyl-CoA species, LC-MS analysis of medium- and long-chain acyl-CoA species reported in the literature27,45,49–53 could also be adapted to this SILEC approach and also used in the diagnosis of metabolic diseases54–56. Labeled CoASH that is generated by the SILEC method can also be used as a synthon to prepare isotope-labeled acyl-CoA species using conventional chemical synthesis methods57–60. A brief overview of the SILEC method, materials required, a step-by-step protocol and troubleshooting guidelines are provided below.
Overview
SILEC methodology can be used to label CoA in any cell system that does not contain a robust de novo pantothenate synthesis pathway, including mammalian and insect cell lines, both described here. Hepa 1c1c7 mouse hepatoma cells were used because of their rapid growth rates in SILEC medium, the abundance of short-chain acyl-CoA species of interest and relative ease in scalability. This method was also adapted to Drosophila S2 cells, which have even shorter doubling times and grow in suspension, facilitating larger-scale preparations of labeled standards.
Experimental design
Generation of labeled CoA
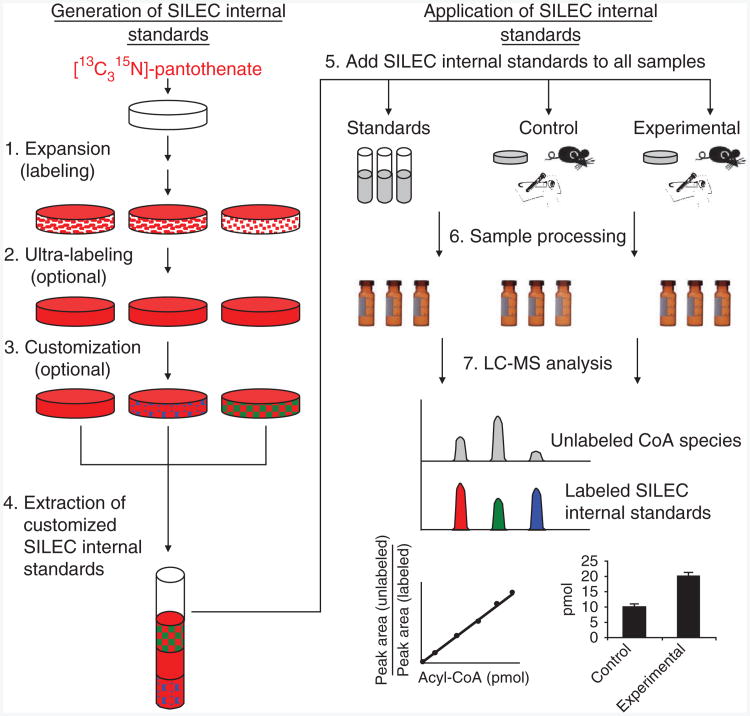
Biosynthetic generation of labeled CoA standards can be divided into four steps (Fig. 4). The objective of the first step is to label the majority of the intracellular CoA while still generating enough cellular material to accommodate the proposed experiment. To accomplish this, cells are passaged 3–5 times in medium containing 10% serum and 1 mg per liter [13C315N]-pantothenate, with unlabeled pantothenate specifically omitted. Unfortunately, undialyzed fetal bovine serum (uFBS) is often contaminated with numerous blood-borne hormones and nutrients, including pantothenate. Therefore, specialized serum such as dialyzed FBS (dFBS) or charcoal-dextran–stripped FBS (csFBS) must be used instead. In fact, we found that csFBS was optimal because it contained the least amount of residual unlabeled pantothenate18. By the third passage, approximately 98–99% of the CoA species in the cell are isotopically labeled. In the second step, the medium is replaced overnight with ultra-labeling medium containing a higher concentration of labeled pantothenate (2–3 mg per liter) and a lower concentration of serum (0–5%). This increases the concentration of labeled pantothenate in the medium while decreasing the level of the contaminating unlabeled pantothenate from the serum. Although serum-free culture could further decrease unlabeled pantothenate levels, such conditions can also affect growth, viability and metabolic characteristics of cells in culture61–63. Therefore, an optimal level of serum must be determined to balance the benefits of increased labeling while still maintaining cellular characteristics. For Hepa 1c1c7 cells, we found that using medium containing 3% csFBS and 3 mg per liter [13C315N]-pantothenate resulted in ≥99.5% labeling. At this point, the CoA species from a representative plate can be sampled to determine labeling efficiency and CoA profile using LC–neutral loss scan of m/z = 507. If the desired CoA species can be identified and measured, the third step is not necessary. However, if one or more of the labeled CoA species of interest are below a reproducibly quantifiable level, a customization step may be necessary. This can be accomplished in multiple ways. First, labeled cells can be supplemented with a particular fatty acid, which is then taken up by the labeled cells to generate a higher percentage of that fatty acyl-CoA. We have demonstrated this by adding propionate to generate propionyl-CoA, but can be extended to other lower-abundance CoA species, such as by adding β-hydroxybutyrate to generate β-hydroxybutyryl-CoA (Fig. 5), an important intermediate in fatty acid oxidation and ketogenesis64,65. In addition, cells can be subjected to different biological perturbations or toxicological insults to increase and decrease particular CoA species or to generate xenobiotic-CoA adducts such as menadione-CoA (Fig. 6). The CoA species in these cells can then be extracted and pooled together to generate a SILEC reference metabolome containing a more appropriate or more global CoA profile for the experiment. Alternatively, the cellular lysate can be hydrolyzed with a strong base, and the resulting [13C315N]-CoASH can be generated. This can be further purified and used to synthesize a particular CoA derivative57–60.
Figure 4.
General scheme for stable isotope labeling by essential nutrients in cell culture (SILEC). (1) Expansion: cells are serially passaged and expanded in the presence of labeled pantothenate. (2) Ultra-labeling: medium containing less serum and more labeled pantothenate is used to increase purity of labeled CoA. (3) Customization: labeled cells are subjected to different biological, pharmaceutical or toxicological exposures to modify the CoA derivative profile within the cells. (4) Extraction: cells are lysed and pooled together to generate a SILEC CoA internal standard mix with a more global CoA profile. (5) SILEC spike-in: equal amounts of SILEC internal standards are spiked into CoA standards, as well as in vitro, in vivo or into clinical experimental samples. (6) Sample processing: standards and samples are subjected to the same sample extraction procedures. (7) LC-MS analysis: CoA species are separated and analyzed by LC-SRM/MS. Co-eluting SILEC internal standards are used to confirm the identity and normalize for the different CoA species.
Figure 5.
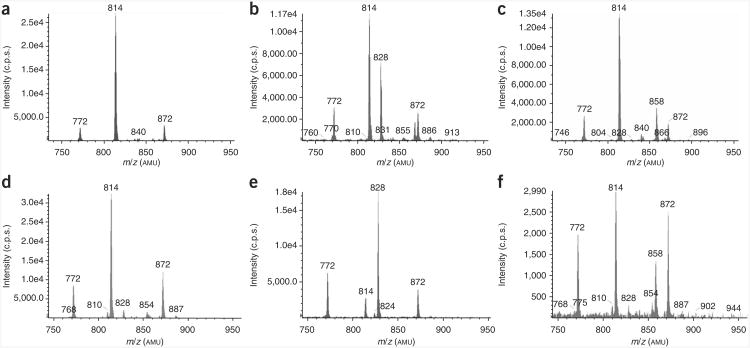
SILEC labeling and ‘customization’ of CoA metabolome. (a–c) LC–neutral loss/MS scans (m/z = 507) of acid-extracted acyl-CoA species in SILEC-labeled Hepa 1c1c7 cells that were untreated (a), treated with 10 mM propionate for 1 h (b) or treated with 10 mM β-hydroxybutyrate for 1 h (c). (d–f) LC–neutral loss/MS scans (m/z = 507) of acid-extracted acyl-CoA species in Drosophila S2 cells that were untreated (d), treated with 10 mM propionate for 1 h (e) or treated with 10 mM β-hydroxybutyrate for 1 h (f). The precursor ion masses of various labeled short-chain acyl-CoA species can be identified: [13C315N]-acetyl-CoA (m/z = 814), [13C315N]-succinyl-CoA (m/z = 872), [13C315N]-CoASH (m/z = 772), [13C315N]-propionyl-CoA (m/z = 828) and [13C315N]-β-hydroxybutyryl-CoA (m/z = 858). These ‘customized’ CoA extracts can be pooled together to generate a more comprehensive CoA profile. Extraction was performed as specified in the protocol (Fig. 2).
Figure 6.
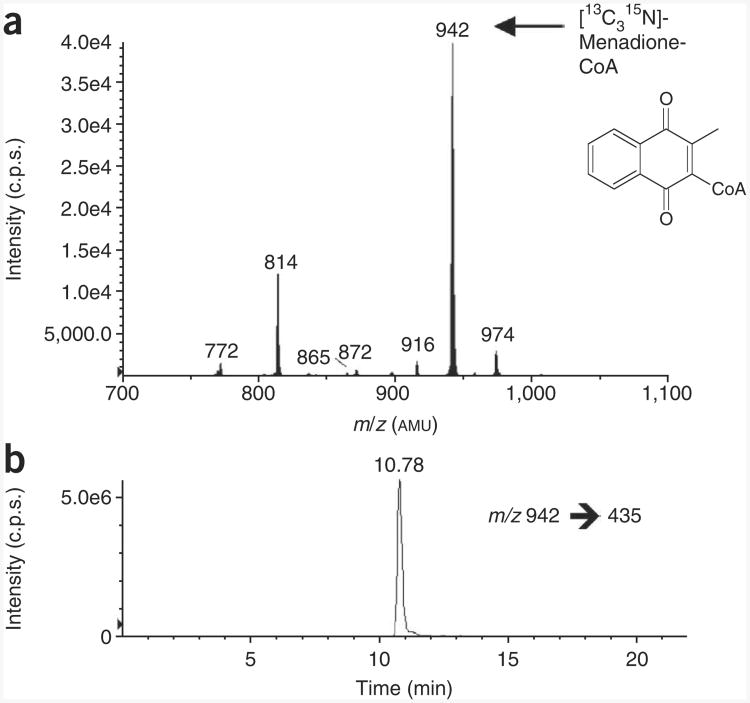
Biosynthetic generation of isotopically labeled menadione-CoA. (a) LC–neutral loss/MS scan (m/z = 507) of acid-extracted CoA species from Hepa 1c1c7 cells treated with 20 μM menadione for 1 h. (b) LC-SRM/MS chromatogram of [13C315N]-menadione-CoA (m/z = 942 → 435) derived from the same extract. Extraction and analysis were performed as specified. By spiking this extract into experimental samples, this isotopically labeled analyte can be used as an internal standard for quantifying unlabeled menadione-CoA.
Application of SILEC standards for LC-MS analysis
The cellular extracts containing the SILEC-labeled CoA internal standards can be applied to biological samples and used to normalize for the CoA species of interest. These biological samples can include cultured cells, tissue samples or clinical samples of obtainable cells such as fibroblasts, lymphocytes or platelets. In each case, a constant amount of SILEC standard is spiked into the samples at an early point during sample processing: to scraped cells for in vitro experiments, to snap-frozen or lyophilized tissues for in vivo experiments or into purified cells such as lymphocytes or platelets for clinical studies. In addition to spiking the samples, the same amounts of SILEC-labeled CoA internal standards are also spiked into known quantities of unlabeled CoA standards to generate a standard curve. Both biological samples and unlabeled CoA internal standards can then be extracted using solid-phase extraction (SPE) and analyzed by LC-MS. The peak area of each unlabeled standard is divided by the peak area of the corresponding SILEC internal standard analog to generate a standard curve of standards to peak area ratio. The same analysis is then performed on each sample, and the peak area ratio is then used to calculate the amount of each of the CoA species in the sample. These amounts are then compared between experimental groups. This method is comparable to the SILAP or SILAC ‘spike-in’ methods that have been described previously21,23. The strength of using a stable isotope dilution MS method is not only in the specificity of CoA elution, but also that the labeled compounds will have the same stability as the unlabeled analyte of interest. Therefore, there would be a parallel degradation of the labeled internal standard that does not affect the ratio of unlabeled analyte to labeled internal standard.
Additional considerations
Limitations of the SILEC approach are similar to those encountered with SILAC labeling in that a biological system is used to generate stable isotope standards17. Although numerous labeled CoA species can be generated in a single biological system, batch-to-batch variability persists even under well-controlled conditions.
If you are using a starting material other than pantothenate, such as folates, it is important to use medium and the appropriate serum that does not contain that essential nutrient. Therefore, an omitted medium along with a modified serum (such as csFBS or dFBS) is crucial for generating an isotopically pure standard. Finally, finding the appropriate cell line is also necessary for optimization. As this is largely empirical, many cell lines likely need to be tested to optimize scale-up and production. A time course should be established in order to determine the optimal time for harvesting that particular cell line.
Materials
Reagents
Cell line of choice. The protocol is written for Hepa 1c1c7 cells (ATCC, cat. no. CRL-2026) and Drosophila Schneider 2 cells (Invitrogen, cat. no. R690-07)
Medium
Mammalian cells: custom RPMI 1640 medium, pantothenate omitted (AthenaES). This is a special request item (cat. no. 0500-19)
Schneider's insect medium
Hank's buffered salt solution (HBSS)
Phosphate-buffered saline (PBS)
[13C315N]-pantothenate (vitamin B5-[13C615N2], Isosciences, cat. no. 5065)
csFBS (Gemini Biosciences, cat. no. 100-119)
Penicillin
Streptomycin
Glutamine
Trichloroacetic acid (TCA)
5-Sulfosalicyilic acid (SSA)
Potassium hydroxide (KOH)
Oasis HLB 1cc (30 mg) SPE columns (Waters, cat. no. WAT094225)
Conical glass centrifugation tubes, 10 ml
Nitrogen gas
Methanol
Ammonium acetate
Acetonitrile and water (mass spectrometry grade)
Formic acid
Propionic acid
β-Hydroxybutyric acid
Menadione
Equipment
Probe sonicator
Tissue culture dishes, 10 cm
Tissue culture flasks, 75 cm2
Cell scrapers
Microcentrifuge
Vacuum manifold
Nitrogen evaporator
Refrigerated autosampler
HPLC system with three pumps
Triple quadrupole mass spectrometer (API 4000 or instrument of similar sensitivity)
Computer with mass spectral computational software
Reagent Setup
Heat-inactivated csFBS
Thaw csFBS at 37 °C and heat-inactivate the complement for 30 min at 55 °C. Aliquot into 50-ml fractions and store at − 20 °C. The heat-inactivated csFBS can be used for up to 1 year when stored at − 20 °C.
[13C315N]-Pantothenate stocks
Prepare in double-distilled water (ddH2O) at 1 mg ml −1 and store at − 20 °C. The stocks are stable for 1 year if stored at − 20 °C
Unlabeled CoA standards
Prepare 5 mM stock solutions of acyl-CoA species in 5% (wt/vol) aqueous SSA. Store in 100-μl aliquots at − 80 °C. Thaw when needed and avoid repetitive freeze-thaw cycles. The aliquots are stable for 1 year when stored at − 80 °C.
SILEC expansion medium for mammalian cells
Prepare pantothenate-free medium with 10% (vol/vol) heat-inactivated csFBS, 2 mM l-glutamine, 100 IU ml −1 penicillin, 100 μg ml −1 streptomycin and 1 mg per liter [13C315N]-pantothenate.
SILEC expansion medium for insect cells (S2)
Prepare Schneider's medium with 10% (vol/vol) heat-inactivated csFBS, 2 mM l-glutamine, 100 IU ml −1 penicillin, 100 μg ml −1 streptomycin and 3 mg per liter [13C315N]-pantothenate.
SILEC ultra-labeling medium for mammalian cells
Prepare pantothenate-free medium with 3% (vol/vol) heat-inactivated csFBS, 2 mM l-glutamine, 100 IU ml −1 penicillin, 100 μg ml −1 streptomycin and 3 mg per liter [13C315N]-pantothenate.
SILEC ultra-labeling medium for insect cells (S2)
Prepare Schneider's medium with 3% (vol/vol) heat-inactivated csFBS, 2 mM l-glutamine, 100 IU ml −1 penicillin, 100g ml −1 streptomycin and 10 mg per liter [13C315N]-pantothenate.
Customization medium
Prepare HBSS medium supplemented with Ca2 +, Mg2 +, 1 g per liter d-glucose, 25 mM HEPES and the appropriate modifying compound (fatty acid, xenobiotic and so on) as shown in the table below.
All media should be filtered and can be stored at 4 °C for up to 3 months. Warm the media to 37 °C before cell treatment for mammalian cells and to room temperature (22–25 °C) for insect cells.
CoA extraction solution
Extraction solution is 10% (wt/vol) TCA in ddH2O. Chill it on ice before the experiment.
CoA elution solution
CoA elution solution is 25 mM ammonium acetate in methanol.
CoA resuspension solution
Resuspension solution is 5% (wt/vol) SSA in ddH2O.
CoA hydrolysis solution
If pure [13C315N]-CoASH is being generated, prepare 40% (wt/vol) KOH in ddH2O.
LC-MS solvents
Prepare, sonicate and degas HPLC solvents before LC-MS analysis. Use solvents of mass spectrometry grade (Fisher Optima-grade solvents are used for our analysis). Solvent A is 5 mM ammonium acetate in water. Solvent B is 95:5 ACN:H2O and 5 mM ammonium acetate in water. Solvent C is 80:20:0.1 ACN:H2O:formic acid.
Procedure
Generating labeled CoA standards
Thaw frozen cells directly into the SILEC expansion medium.
For Hepa 1c1c7 (adherent) cells, allow cells to reach confluence (3-4 d), wash two times with PBS, trypsinize and split 1:5 into SILEC expansion medium. For S2 (suspension) cells, when cells have reached ∼1 × 107 cells per ml, split into a new flask containing SILEC expansion medium at a concentration of 2 × 106 cells.
-
Repeat Step 2, passaging cells in SILEC expansion medium three more times. At this point, CoA species should be greater than 98% labeled. If cell growth or morphological characteristics are considerably affected, see TROUBLESHOOTING.
■Pause Point Harvest and freeze down one plate of labeled cells in SILEC expansion medium containing 10% (vol/vol) DMSO. These cells can be thawed to seed future SILEC expansions. ?Troubleshooting
After the fourth passage, allow cells to reach confluence or 1 × 107 cells per ml. Proceed to Step 6 if you do not want the ultimate incorporation of stable isotope label. Alternatively, perform Step 5.
(Optional) Wash cells with 10 ml of PBS, replace with SILEC ultra-labeling medium and incubate for 24 h. Perform this step if you want the highest level of stable isotope labeling, and then continue to Step 6. To produce purified [13C315N]-CoASH, continue to Step 27.
Testing of SILEC labeling
6. Short-chain acyl-CoA acid extraction (Steps 6-9). Harvest one plate of cells by scraping gently into the medium and briefly spin down at 1,000g. The remaining plates should be kept until the first plate has been tested (Steps 7-20).
7. Resuspend cells in 1 ml of ice-cold extraction solution (10% (wt/vol) TCA).
8. Sonicate cells for 30 s on ice at a rate of 1 pulse per s.
9. Spin the sonicated cell lysate at 15,000g for 5 min at 4 °C to precipitate the protein pellet.
10. SPE (Steps 10-16). Using a vacuum manifold, condition Oasis HLB SPE columns with 1 ml of methanol.
11. Equilibrate columns with 1 ml of water.
12. Load acid-extracted lysate (supernatant) onto the columns.
13. Wash the columns with 1 ml of water.
14. Elute CoA compounds into fresh tubes by applying three subsequent applications of 0.5 ml of elution solution.
15. Dry the samples under nitrogen.
16. Resuspend in 50 μl of 5% (wt/vol) SSA.
17. LC-MS analysis (Steps 17-20). Using the LC-MS specifications in Figure 2, perform a CID neutral loss scan of 507 AMU, scanning a parent mass range of m/z = 750–1,200 AMU. Sample injections of 10 μl should be used for analysis.
18. Characterizing the CoA SILEC Profile. Generate a m/z list from the neutral-loss scan and compare them against a m/z list of CoA species (supplementary table 1); prepare a SRM experiment for each of these parent ions and their respective productions. The singly charged protonated molecule should be used for the parent CoA, whereas the associated product ion of 507 AMU lower than the parent protonated molecule should be used as the SRM product ion. For example, acetyl-CoA has a parent protonated molecule of m/z = 810.1 and a production at m/z = 303.1, whereas [13C315N]-acetyl-CoA has a parent protonated molecule of m/z = 814.1 and a product ion at m/z = 307.1.
19. Determining labeling efficacy. Reinject and analyze the sample using the LC-SRM/MS method, monitoring both the labeled and unlabeled channels to determine the amount of unlabeled CoA remains for each acyl-CoA.
-
20. Confirmation of acyl-CoA species. To confirm the identity of the labeled CoA species, spike extracted SILEC internal standards with a mixture of the CoA thioesters of interest (1 pmol each) and repeat the LC-SRM/MS analysis. The labeled and unlabeled compounds should co-elute. This is particularly important when multiple peaks arise or if there are multiple isobaric CoA species with similar parent and productions.
▲ Critical step If there are certain CoA species you are interested in that are not represented in the mixture, further steps may need to be taken (see Generating customized SILEC standards, Steps 23–36). In the case of incomplete labeling, systematic troubleshooting may be necessary (see TROUBLESHOOTING). ?Troubleshooting
Harvesting
21. If labeling is complete (> 99% labeling) and CoA profile is satisfactory, perform CoA extraction for all plates (Steps 6–9).
-
22. Pool together acid-soluble SILEC extracts, label the batch and freeze at − 80 °C in 10-ml aliquots.
■ Pause Point The pooled acid-soluble extracts contain the SILEC internal standards that will later be spiked into cell or tissue samples. These standards can be stored for up to 1 year at − 80 °C, but they should be spiked into a new standard curve for each analysis. Avoid repetitive freeze-thaw cycles.
Generating customized SILEC standards
23. (Continued from Step 21) Wash half of the plates twice with PBS.
24. For each of the washed plates, replace with 10 ml of customization medium (see REAGENT SETUP) and incubate for 1 h.
25. Test labeling using Steps 17−20.
26. After incubation, harvest and extract CoA from all plates (Steps 6-9) and pool extracts together. This pooled extract should contain a SILEC CoA profile with increased levels of the species of interest.
Producing purified [3c315n]-CoASH
27. (Continued from Step 5) After overnight incubation in ultra-labeling medium, perform Steps 6−9 for all plates and pool together acid-soluble lysates, which contain CoASH and short-chain acyl-CoAs.
28. Adjust the pH of the lysate to ∼13 with 40% (wt/vol) KOH. Add 500 μl of KOH to 5 ml of TCA extract and incubate at room temperature for 1 h.
29. Adjust the pH of the lysate to 6 with formic acid.
30. Purify the labeled CoA using SPE extraction (Steps 10–16).
31. Dry down the hydrolyzed extract, which contains purified [13C315N]-CoASH. This product can now be used to chemically synthesize a particular CoA derivative57–60.
Using SILEC standards for CoA analysis
-
32. Thaw an adequate amount of previously generated acid-extracted SILEC CoA internal standard mixture. Generally, 1 ml of internal standard mixture is needed for every five samples. After thawing, vortex to ensure that the sample is mixed thoroughly and keep it on ice.
▲ Critical step It is important to determine the amount of labeled SILEC CoA standard needed before the experiment. If multiple batches are necessary for a single experiment, these batches must first be pooled together before application to samples/standard curve. Owing to the biosynthetic nature of SILEC generation, there can be substantial batch-to-batch variability; similarly, using different batches of labeled internal standards in different samples of the same experiment can lead to erroneous measurements.
33. To prepare standard curve samples, thaw and mix equal concentrations of unlabeled CoA standards of interest, creating a master mix, and perform twofold serial dilutions, from 1 μM down to ∼10 nM, in extraction solution (10% (wt/vol) TCA). Mix 50 μl of each dilution with 0.75 ml of extraction solution and 0.2 ml of thawed SILEC CoA internal standard extract (for a final volume of 1 ml) and vortex. This will generate a standard curve from approximately 100 fmol to 10 pmol of each CoA standard on the column.
34. To prepare experimental samples, add 0.8 ml of ice-cold TCA along with 0.2 ml of the same SILEC internal standard mixture to harvested cell pellets, snap-frozen or lyophilized tissues, or purified extracted cells (platelets, lymphocytes, fibroblasts and so on.)
35. For cell samples, sonicate for 30 s on ice in a pulsatile manner. For tissue samples, homogenize according to standard protocols. In either case, perform the same processing step for the standard curve samples as well.
36. Spin processed acid-extracted lysate at 15,000g for 5 min at 4 °C to precipitate the protein pellet.
37. Perform SPE extraction of CoA species as previously described (Steps 10–16).
-
38. Resuspend the samples in 50 μl of 5% (wt/vol) SSA.
■ Pause Point The samples and standards can be immediately analyzed or frozen at − 80 °C for future analysis. As the SILEC CoA internal standards are included, effects on sample stability should be reflected in the internal standards as well.
39. For analysis, inject 10 μl from each sample or standard mix and perform LC-SRM/MS analysis using the method developed previously.
40. From the raw data, develop a standard curve for each of the CoA species, with the x axis as the known amount of injected standard and the y axis as the ratio of unlabeled to labeled integrated peak areas.
41. Use the regression line from this standard curve to calculate the amount of each CoA thioester in the samples. ?Troubleshooting
Perturbation of cell growth kinetics by SILEC medium (step 3)
Changes in cell medium components may affect a number of cell growth parameters, such as doubling times, cell viability, adherence and morphological characteristics. Although slightly slower cell growth is expected with SILEC medium, if there is severely deranged cell growth or morphological changes, medium components should be serially investigated.
Serum
Although serum is necessary for many cell lines, the lack of serum standardization and lot-to-lot variability makes it very challenging to reproducibly scale up many biosynthetically generated compounds. Charcoal stripping is commonly used to standardize serum and csFBS is the optimal serum for our purposes, as it markedly reduces unlabeled pantothenate in the serum, which will contaminate the labeled CoA pool. In addition to removing pantothenate, however, charcoal stripping also removes a number of other serum components including hormones, growth factors and other B vitamins, which can affect cell growth66,67. One method to overcome this is by either increasing the amount of csFBS or using dFBS during the expansion stage, which may be enough to overcome these effects. Although dFBS would slightly increase the level of unlabeled pantothenate, it still has considerably less pantothenate than undialyzed serum. This may improve cell growth during the expansion phase. If it is found, however, that dFBS is necessary during the ultra-labeling step, the concentration should be substantially reduced and the concentration of pantothenate should be increased to lower the percentage of unlabeled pantothenate in the serum. If both csFBS and dFBS are prohibitive to cell growth, and uFBS is necessary, a different cell line should be considered, as the high level of residual pantothenate in uFBS would be difficult to overcome by simply adding more labeled compound.
Other medium components
Commercial services are available for the production of specialty medium such as pantothenate-omitted RPMI 1640 medium, which is used in our protocol. However, should you choose to prepare your own SILEC medium, ensure that all components are dissolved and that your cells show similar growth characteristics when this medium is supplemented with standard FBS and the same concentration of pantothenate. As certain medium components can also degrade over time, prepared medium should be kept at − 20 °C for long-term storage. Finally, as with any cell culture system, contamination with mycoplasma may result in decreased cell growth. Therefore, thaw fresh cells or perform simple tests for mycoplasma to eliminate this possibility.
Incomplete labeling (step 20)
There are three underlying causes for incomplete labeling: (i) contaminating unlabeled pantothenate in the medium, (ii) unlabeled pantothenate in the [13C315N]-pantothenate stock and (iii) insuffcient turnover of pantothenate or CoA pool within the cell line.
First, determine whether there is contaminating unlabeled pantothenate in the medium, as this is the most likely cause of the incomplete labeling. For mammalian cell culture, if the pantothenate-omitted medium was commercially prepared, verify that pantothenate is not in the formulation. If this medium was prepared in-house, verify that the base nutrient mix you are using does not contain any pantothenate. For insect cell culture, Schneider's medium contains yeast extract, which contains a variable (although marked) amount of different B vitamins, including pantothenate. For this reason, a higher concentration of labeled compound is needed for insect SILEC preparation.
If there is incomplete labeling in these cells, it could very well be coming from increased amounts of pantothenate in the yeast extract (yeastolate). To overcome this, the ultra-labeling step can be performed overnight with HBSS medium containing 4 mg per liter labeled pantothenate, which should increase labeling to greater than 99%. However, it is more likely that the unlabeled pantothenate is originating from the csFBS. Although there should be < 0.1 mg per liter of pantothenate remaining in the csFBS18 (resulting in an effective 0.01 mg per liter in medium containing 10% serum), it is possible that a particular lot may have more, or that a higher concentration of serum is needed for a particular cell line. Adding additional labeled pantothenate to the medium should overcome this problem. If this is a newly occurring problem, check the lot of the csFBS, as lot-to-lot variability can exist for serum. If a particular lot of csFBS has worked in the past, it may be worth ordering several bottles of this for future use. The serum can be frozen and stored at −20 °C for long-term storage.
The second possibility is that the stock itself is contaminated with unlabeled pantothenate. This can be assessed by performing mass spectral analysis of the labeled stock. To do this, reconstitute a small amount of the labeled pantothenate stock and using flow-infused syringe injection and SRM/MS analysis. The relative ratio of labeled to unlabeled pantothenate represents the maximal purity that the CoA can be labeled. If this is not acceptable, another source of labeled pantothenate should be sought. For our experiments, we found that the isotopic purity of the stock was ≥99.5%.
Finally, the last problem that can arise, particularly with suspension culture, is that a certain amount of the unlabeled compound is not turning over and remains in the cellular pool because of cell death or senescence. This is more of a problem with SILAC protocols because different proteins turn over at various frequencies, and salvage pathways of particular amino acids may cause these unlabeled species to continue to propagate in culture. In addition, unlabeled albumin may also be taken up by cells. For CoA species, this turnover is greater and more than 50% of the CoA can turn over in the cell in 24 h (ref. 18), although different CoA species may not turn over as rapidly. To overcome this problem, simply passage the cells two or three more times in the expansion medium. If the cells are being split into 1:3, < 0.5% of the original cell mass will remain after five passages. Although this may add an additional week to the expansion phase, it would replace at least 99.5% of the original cell mass and, likewise, of the original unlabeled CoA.
Problems in sample preparation and LC-Ms
Solid-phase extraction
Oasis HLB columns have been found to be optimal for short-chain acyl-CoA extraction, although other methods such as liquid-liquid extraction could be used as well18,47. However, when the experiment involves more complex biological matrix such as in tissue samples, or if a larger amount of biological material is being used (to measure lower abundance analytes), a greater matrix effect may be observed. In this case, repeating the SPE washing step once or twice may help. In addition, using a molecular weight cutoff filter before SPE extraction may also reduce the matrix effect, although this will increase processing time. The use of methanol containing concentrated ammonia may improve elution47.
HPLC
Although acyl-CoA analysis is generally robust, repeated injections of cell and tissue extracts can lead to buildup of biological materials on the column, which can lead to distortions in peak shape27. For this reason, we have added solvent C in a column wash step. If peak distortion is occurring, increasing the volume of the wash step, as well as slowing down the ramp time for washing, can improve column performance, although this will also increase run times. In addition, lowering the pH of this wash can also improve the ruggedness of the method.
Mass spectrometry
In the case of inadequate sensitivity, use acetonitrile instead of methanol in the mobile phase whenever possible, and decrease formic acid concentration during MS analysis. Moreover, if LC-SRM/MS analysis is increased to include a large number of species, a proper number of analytical points may not be achieved across each peak and may lead to incorrect quantification, even with internal standards. To overcome this, SRM dwell time should be reduced so as to increase the number of points across the peak. For example, if eight CoA species along with their internal standards are to be measured (16 analytes in total), using dwell times of 80–100 ms may be acceptable. However, if the number of analyzed CoA species is increased to 20–30 species (40–60 total), shorter dwell times (20–30 ms) may be necessary to achieve enough points across the peak. In addition, dividing up the analysis into shorter segments can allow for longer dwell times.
● Timing
Typical times used in our laboratory for the various steps are as follows:
SILEC labeling—Expansion: 12–16 d
SILEC labeling—Ultra-labeling: 1 d
SILEC labeling—Testing of labeling: 3 h
SILEC labeling—Customization: 1–12 h
SILEC labeling—Harvesting and extraction: 30 min
Application of internal standards—Sample preparation: ∼1 h
Application of internal standards—Analysis: 30 min per sample, 1 d per experiment (total time: 15–20 d)
Anticipated Results
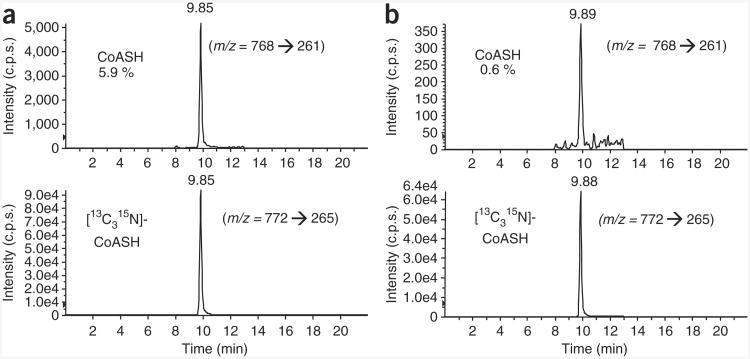
The SILEC approach presented in this study can be used to generate labeled short-chain acyl-CoA MS standards to improve specificity and precision in mass spectral analysis. As previously mentioned, a crucial aspect of the biosynthetic generation of labeled standards is minimizing the unlabeled fraction. Typical LC-SRM/MS chromatograms of reduced CoA (CoASH) harvested from [13C315N]-pantothenate-labeled cells are provided in Figure 7. Figure 7a shows an example of insufficient labeling, with > 5% of the CoASH unlabeled, whereas Figure 7b depicts CoASH from a SILEC batch with more complete labeling (< 1% unlabeled). In addition, cumulative spectra from a LC-constant neutral loss/MS scan analysis of CoA (m/z = 507) are presented throughout this report to illustrate the labeling and customization of various CoA reference metabolomes. Unlabeled cells are shown in Figure 1b, whereas examples of SILEC-labeled CoA reference metabolomes customized with fatty acids or xenobiotics are shown in Figures 5 and 6a, respectively. Furthermore, a representative chromatogram of menadione-CoA is presented in Figure 6b. It should be noted that using exogenous biological and toxicological stimuli to change the CoA profile in a given cell type is largely empirical and needs to be optimized for different cell systems and conditions. Before scaling up, optimization can be performed in unlabeled cells, although it should be confirmed in labeled cells as well.
Figure 7.
LC-SRM/MS chromatograms of CoASH harvested from [13C315N]-pantothenate-labeled cells. Both unlabeled CoASH (m/z = 768 → 261) and [13C315N]-CoASH (m/z = 772 → 265) chromatograms are presented to illustrate the labeling efficiency. (a) Suboptimal labeling: the amount of unlabeled CoASH was 5.7%. (b) Near-complete labeling: the amount of unlabeled CoASH was 0.5%. Extraction and analysis were performed as specified in the protocol.
Supplementary Material
| Desired acyl-CoA-thioester | Modifying compound (concentration) |
|---|---|
| Propionyl-CoA | Propionic acid (10 mM) |
| β-Hydroxybutyryl-CoA | β-Hydroxybutyric acid (10 mM) |
| Menadione-CoA | Menadione (20 μM) |
Acknowledgments
The support of US National Institutes of Health grants U01ES016004, P30ES013508 and 5T32HL007439 is gratefully acknowledged.
Footnotes
Note: Supplementary information is available via the HTML version of this article.
Author Contributions: Both authors wrote the paper and designed the experiments. S.S.B. conducted the actual experiments.
Competing Financial Interests: The authors declare no competing financial interests.
Reprints and permissions information is available online at http://www.nature.com/reprints/index.html.
References
- 1.Hsieh Y. HPLC-MS/MS in drug metabolism and pharmacokinetic screening. Expert Opin Drug Metab Toxicol. 2008;4:93–101. doi: 10.1517/17425255.4.1.93. [DOI] [PubMed] [Google Scholar]
- 2.Brown SC, Kruppa G, Dasseux JL. Metabolomics applications of FT-ICR mass spectrometry. Mass Spectrom Rev. 2005;24:223–231. doi: 10.1002/mas.20011. [DOI] [PubMed] [Google Scholar]
- 3.Hu QZ, et al. The Orbitrap: a new mass spectrometer. J Mass Spectrom. 2005;40:430–443. doi: 10.1002/jms.856. [DOI] [PubMed] [Google Scholar]
- 4.Whalen K, Gobey J, Janiszewski J. A centralized approach to tandem mass spectrometry method development for high-throughput ADME screening. Rapid Commun Mass Spectrom. 2006;20:1497–1503. doi: 10.1002/rcm.2469. [DOI] [PubMed] [Google Scholar]
- 5.Taylor CF, et al. A systematic approach to modeling, capturing, and disseminating proteomics experimental data. Nat Biotechnol. 2003;21:247–254. doi: 10.1038/nbt0303-247. [DOI] [PubMed] [Google Scholar]
- 6.Cox J, et al. A practical guide to the MaxQuant computational platform for SILAC-based quantitative proteomics. Nat Protoc. 2009;4:698–705. doi: 10.1038/nprot.2009.36. [DOI] [PubMed] [Google Scholar]
- 7.Madalinski G, et al. Direct introduction of biological samples into a LTQ-Orbitrap hybrid mass spectrometer as a tool for fast metabolome analysis. Anal Chem. 2008;80:3291–3303. doi: 10.1021/ac7024915. [DOI] [PubMed] [Google Scholar]
- 8.Wu CC, Yates JR., III The application of mass spectrometry to membrane proteomics. Nat Biotechnol. 2003;21:262–267. doi: 10.1038/nbt0303-262. [DOI] [PubMed] [Google Scholar]
- 9.Jemal M. High-throughput quantitative bioanalysis by LC/MS/MS. Biomed Chromatogr. 2000;14:422–429. doi: 10.1002/1099-0801(200010)14:6<422::AID-BMC25>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 10.Remane D, Wissenbach DK, Meyer MR, Maurer HH. Systematic investigation of ion suppression and enhancement effects of fourteen stable-isotope-labeled internal standards by their native analogues using atmospheric-pressure chemical ionization and electrospray ionization and the relevance for multi-analyte liquid chromatographic/mass spectrometric procedures. Rapid Commun Mass Spectrom. 2010;24:859–867. doi: 10.1002/rcm.4459. [DOI] [PubMed] [Google Scholar]
- 11.Prakash C, Shaffer CL, Nedderman A. Analytical strategies for identifying drug metabolites. Mass Spectrom Rev. 2007;26:340–369. doi: 10.1002/mas.20128. [DOI] [PubMed] [Google Scholar]
- 12.Bonfglio R, King RC, Olah TV, Merkle K. The effects of sample preparation methods on the variability of the electrospray ionization response for model drug compounds. Rapid Commun Mass Spectrom. 1999;13:1175–1185. doi: 10.1002/(SICI)1097-0231(19990630)13:12<1175::AID-RCM639>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 13.King R, Bonfglio R, Fernandez-Metzler C, Miller-Stein C, Olah T. Mechanistic investigation of ionization suppression in electrospray ionization. J Am Soc Mass Spectrom. 2000;11:942–950. doi: 10.1016/S1044-0305(00)00163-X. [DOI] [PubMed] [Google Scholar]
- 14.Matuszewski BK. Standard line slopes as a measure of a relative matrix effect in quantitative HPLC-MS bioanalysis. J Chromatogr B. 2006;830:293–300. doi: 10.1016/j.jchromb.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 15.Ciccimaro E, Blair IA. Stable-isotope dilution LC-MS for quantitative biomarker analysis. Bioanalysis. 2010;2:311–341. doi: 10.4155/bio.09.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bennett BD, Yuan J, Kimball EH, Rabinowitz JD. Absolute quantitation of intracellular metabolite concentrations by an isotope ratio-based approach. Nat Protoc. 2008;3:1299–1311. doi: 10.1038/nprot.2008.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ong SE, Mann M. A practical recipe for stable isotope labeling by amino acids in cell culture (SILAC) Nat Protoc. 2006;1:2650–2660. doi: 10.1038/nprot.2006.427. [DOI] [PubMed] [Google Scholar]
- 18.Basu SS, Mesaros C, Gelhaus SL, Blair IA. Stable isotope labeling by essential nutirents in cell culture for the preparation of labeled coenzyme A and its thioesters. Anal Chem. 2011;83:1363–1369. doi: 10.1021/ac1027353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ong SE, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 20.Yan Y, Weaver VM, Blair IA. Analysis of protein expression during oxidative stress in breast epithelial cells using a stable isotope labeled proteome internal standard. J Proteome Res. 2005;4:2007–2014. doi: 10.1021/pr050175d. [DOI] [PubMed] [Google Scholar]
- 21.Shah SJ, Yu KH, Sangar V, Parry SI, Blair IA. Identification and quantification of preterm birth biomarkers in human cervicovaginal fuid by liquid chromatography/tandem mass spectrometry. J Proteome Res. 2009;8:2407–2417. doi: 10.1021/pr8010342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rangiah K, et al. Differential secreted proteome approach in murine model for candidate biomarker discovery in colon cancer. J Proteome Res. 2009;8:5153–5164. doi: 10.1021/pr900518v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Geiger T, et al. Use of stable isotope labeling by amino acids in cell culture as a spike-in standard in quantitative proteomics. Nat Protoc. 2011;6:147–157. doi: 10.1038/nprot.2010.192. [DOI] [PubMed] [Google Scholar]
- 24.Brass EP. Overview of coenzyme A metabolism and its role in cellular toxicity. Chem Biol Interact. 1994;90:203–214. doi: 10.1016/0009-2797(94)90010-8. [DOI] [PubMed] [Google Scholar]
- 25.Robishaw JD, Neely JR. Coenzyme A metabolism. Am J Physiol. 1985;248:E1–E9. doi: 10.1152/ajpendo.1985.248.1.E1. [DOI] [PubMed] [Google Scholar]
- 26.Li LO, Klett EL, Coleman RA. Acyl-CoA synthesis, lipid metabolism and lipotoxicity. Biochim Biophys Acta. 2010;1801:246–251. doi: 10.1016/j.bbalip.2009.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Magnes C, Sinner FM, Regittnig W, Pieber TR. LC/MS/MS method for quantitative determination of long-chain fatty acyl-CoAs. Anal Chem. 2005;77:2889–2894. doi: 10.1021/ac048314i. [DOI] [PubMed] [Google Scholar]
- 28.Gao L, et al. Simultaneous quantification of malonyl-CoA and several other short-chain acyl-CoAs in animal tissues by ion-pairing reversed-phase HPLC/MS. J Chromatogr B. 2007;853:303–313. doi: 10.1016/j.jchromb.2007.03.029. [DOI] [PubMed] [Google Scholar]
- 29.MacDonald MJ, Smith AD, III, Hasan NM, Sabat G, Fahien LA. Feasibility of pathways for transfer of acyl groups from mitochondria to the cytosol to form short chain acyl-CoAs in the pancreatic beta cell. J Biol Chem. 2007;282:30596–30606. doi: 10.1074/jbc.M702732200. [DOI] [PubMed] [Google Scholar]
- 30.MacDonald MJ. Synergistic potent insulin release by combinations of weak secretagogues in pancreatic islets and INS-1 cells. J Biol Chem. 2007;282:6043–6052. doi: 10.1074/jbc.M606652200. [DOI] [PubMed] [Google Scholar]
- 31.Lowe DM, Tubbs PK. Succinylation and inactivation of 3-hydroxy-3-methylglutaryl-CoA synthase by succinyl-CoA and its possible relevance to the control of ketogenesis. Biochem J. 1985;232:37–42. doi: 10.1042/bj2320037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Donovan DJ, et al. CoASH and CoASSG levels in lungs of hyperoxic rats as potential biomarkers of intramitochondrial oxidant stresses. Pediatr Res. 2002;51:346–353. doi: 10.1203/00006450-200203000-00014. [DOI] [PubMed] [Google Scholar]
- 33.Wong YL, Smith CV, McMicken HW, Rogers LK, Welty SE. Mitochondrial thiol status in the liver is altered by exposure to hyperoxia. Toxicol Lett. 2001;123:179–193. doi: 10.1016/s0378-4274(01)00397-6. [DOI] [PubMed] [Google Scholar]
- 34.Feliz B, Witt DR, Harris BT. Propionic acidemia: a neuropathology case report and review of prior cases. Arch Pathol Lab Med. 2003;127:e325–e328. doi: 10.5858/2003-127-e325-PAANCR. [DOI] [PubMed] [Google Scholar]
- 35.Mitchell GA, et al. Hereditary and acquired diseases of acyl-coenzyme A metabolism. Mol Genet Metab. 2008;94:4–15. doi: 10.1016/j.ymgme.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 36.Van Hove JL, et al. D, L-3-hydroxybutyrate treatment of multiple acyl-CoA dehydrogenase deficiency (MADD) Lancet. 2003;361:1433–1435. doi: 10.1016/S0140-6736(03)13105-4. [DOI] [PubMed] [Google Scholar]
- 37.van Grunsven EG, et al. Peroxisomal D-hydroxyacyl-CoA dehydrogenase defciency: resolution of the enzyme defect and its molecular basis in bifunctional protein deficiency. Proc Natl Acad Sci USA. 1998;95:2128–2133. doi: 10.1073/pnas.95.5.2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Maldegem BT, Wanders RJ, Wijburg FA. Clinical aspects of short-chain acyl-CoA dehydrogenase deficiency. J Inherit Metab Dis. 2010;33:507–511. doi: 10.1007/s10545-010-9080-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Steghens JP, Flourie F, Arab K, Collombel C. Fast liquid chromatography-mass spectrometry glutathione measurement in whole blood: micromolar GSSG is a sample preparation artifact. J Chromatogr B Analyt Technol Biomed Life Sci. 2003;798:343–349. doi: 10.1016/j.jchromb.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 40.Blair IA. Endogenous glutathione adducts. Curr Drug Metab. 2006;7:853–872. doi: 10.2174/138920006779010601. [DOI] [PubMed] [Google Scholar]
- 41.Zhu P, Oe T, Blair IA. Determination of cellular redox status by stable isotope dilution liquid chromatography/mass spectrometry analysis of glutathione and glutathione disulfide. Rapid Commun Mass Spectrom. 2008;22:432–440. doi: 10.1002/rcm.3380. [DOI] [PubMed] [Google Scholar]
- 42.Hayashi O, Satoh K. Determination of acetyl-CoA and malonyl-CoA in germinating rice seeds using the LC-MS/MS technique. Biosci Biotechnol Biochem. 2006;70:2676–2681. doi: 10.1271/bbb.60269. [DOI] [PubMed] [Google Scholar]
- 43.Perera MA, Choi SY, Wurtele ES, Nikolau BJ. Quantitative analysis of short-chain acyl-coenzymeAs in plant tissues by LC-MS-MS electrospray ionization method. J Chromatogr B. 2009;877:482–488. doi: 10.1016/j.jchromb.2008.12.053. [DOI] [PubMed] [Google Scholar]
- 44.Dalluge JJ, et al. Separation and identification of organic acid-coenzyme A thioesters using liquid chromatography/electrospray ionization-mass spectrometry. Anal Bioanal Chem. 2002;374:835–840. doi: 10.1007/s00216-002-1554-x. [DOI] [PubMed] [Google Scholar]
- 45.Magnes C, et al. Validated comprehensive analytical method for quantification of coenzyme A activated compounds in biological tissues by online solid-phase extraction LC/MS/MS. Anal Chem. 2008;80:5736–5742. doi: 10.1021/ac800031u. [DOI] [PubMed] [Google Scholar]
- 46.Minkler PE, Kerner J, Kasumov T, Parland W, Hoppel CL. Quantification of malonyl-coenzyme A in tissue specimens by high-performance liquid chromatography/mass spectrometry. Anal Biochem. 2006;352:24–32. doi: 10.1016/j.ab.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 47.Park JW, Jung WS, Park SR, Park BC, Yoon YJ. Analysis of intracellular short organic acid-coenzyme A esters from actinomycetes using liquid chromatography-electrospray ionization-mass spectrometry. J Mass Spectrom. 2007;42:1136–1147. doi: 10.1002/jms.1240. [DOI] [PubMed] [Google Scholar]
- 48.Leonardi R, Zhang YM, Rock CO, Jackowski S, Coenzyme A back in action. Prog Lipid Res. 2005;44:125–153. doi: 10.1016/j.plipres.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 49.Kasuya F, Oti Y, Tatsuki T, Igarashi K. Analysis of medium-chain acyl-coenzyme A esters in mouse tissues by liquid chromatography-electrospray ionization mass spectrometry. Anal Biochem. 2004;325:196–205. doi: 10.1016/j.ab.2003.10.043. [DOI] [PubMed] [Google Scholar]
- 50.Minkler PE, Kerner J, Ingalls ST, Hoppel CL. Novel isolation procedure for short-, medium-, and long-chain acyl-coenzyme A esters from tissue. Anal Biochem. 2008;376:275–276. doi: 10.1016/j.ab.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang GF, et al. Catabolism of 4-hydroxyacids and 4-hydroxynonenal via 4-hydroxy-4-phosphoacyl-CoAs. J Biol Chem. 2009;284:33521–33534. doi: 10.1074/jbc.M109.055665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haynes CA, et al. Quantitation of fatty acyl-coenzyme As in mammalian cells by liquid chromatography-electrospray ionization tandem mass spectrometry. J Lipid Res. 2008;49:1113–1125. doi: 10.1194/jlr.D800001-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mauriala T, Herzig KH, Heinonen M, Idziak J, Auriola S. Determination of long-chain fatty acid acyl-coenzyme A compounds using liquid chromatography-electrospray ionization tandem mass spectrometry. J Chromatogr B. 2004;808:263–268. doi: 10.1016/j.jchromb.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 54.Bennett MJ, Hale DE. Medium chain acyl-coenzyme A dehydrogenase deficiency. N J Med. 1992;89:675–678. [PubMed] [Google Scholar]
- 55.Boneh A, et al. VLCAD deficiency: pitfalls in newborn screening and confirmation of diagnosis by mutation analysis. Mol Genet Metab. 2006;88:166–170. doi: 10.1016/j.ymgme.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 56.Wood JC, et al. Diagnosis of very long chain acyl-dehydrogenase deficiency from an infant's newborn screening card. Pediatrics. 2001;108:E19. doi: 10.1542/peds.108.1.e19. [DOI] [PubMed] [Google Scholar]
- 57.Chang SH, Wilken DR. Identity of a bovine liver nucleotide peptide with the unsymmetrical disulfied of coenzyme A and glutathione. J Biol Chem. 1965;240:3136–3139. [PubMed] [Google Scholar]
- 58.Kawaguchi A, Yoshimura T, Okuda S. A new method for the preparation of acyl-CoA thioesters. J Biochem. 1981;89:337–339. doi: 10.1093/oxfordjournals.jbchem.a133207. [DOI] [PubMed] [Google Scholar]
- 59.Olsen J, Bjornsdottir I, Tjornelund J, Honore HS. Chemical reactivity of the naproxen acyl glucuronide and the naproxen coenzyme A thioester towards bionucleophiles. J Pharm Biomed Anal. 2002;29:7–15. doi: 10.1016/s0731-7085(02)00026-2. [DOI] [PubMed] [Google Scholar]
- 60.van Wyk M, Strauss E. One-pot preparation of coenzyme A analogues via an improved chemo-enzymatic synthesis of pre-CoA thioester synthons. Chem Commun. 2007:398–400. doi: 10.1039/b613527g. [DOI] [PubMed] [Google Scholar]
- 61.Cooper S. Reappraisal of serum starvation, the restriction point, G0, and G1 phase arrest points. FASEB J. 2003;17:333–340. doi: 10.1096/fj.02-0352rev. [DOI] [PubMed] [Google Scholar]
- 62.Hasan NM, Adams GE, Joiner MC. Effect of serum starvation on expression and phosphorylation of PKC-alpha and p53 in V79 cells: implications for cell death. Int J Cancer. 1999;80:400–405. doi: 10.1002/(sici)1097-0215(19990129)80:3<400::aid-ijc11>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 63.Shin JS, et al. Serum starvation induces G1 arrest through suppression of Skp2-CDK2 and CDK4 in SK-OV-3 cells. Int J Oncol. 2008;32:435–439. [PubMed] [Google Scholar]
- 64.Cornille E, et al. Enhancement of L-3-hydroxybutyryl-CoA dehydrogenase activity and circulating ketone body levels by pantethine. Relevance to dopaminergic injury. BMC Neurosci. 2010;11:51. doi: 10.1186/1471-2202-11-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.MacDonald MJ, et al. Acetoacetate and beta-hydroxybutyrate in combination with other metabolites release insulin from INS-1 cells and provide clues about pathways in insulin secretion. Am J Physiol Cell Physiol. 2008;294:C442–C450. doi: 10.1152/ajpcell.00368.2007. [DOI] [PubMed] [Google Scholar]
- 66.Iwasaki K, et al. Effects of antiprogestins on the rate of proliferation of breast cancer cells. Mol Cell Biochem. 1999;198:141–149. doi: 10.1023/a:1006945813508. [DOI] [PubMed] [Google Scholar]
- 67.Cao Z, et al. Effects of resin or charcoal treatment on fetal bovine serum and bovine calf serum. Endocr Res. 2009;34:101–108. doi: 10.3109/07435800903204082. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.