Abstract
Quorum sensing (QS) is a mechanism of bacterial gene regulation in response to increases in population density. Perhaps most studied are QS pathways mediated by acylhomoserine lactones (AHLs) in Gram-negative bacteria. Production of small molecule QS signals, their accumulation within a diffusion-limited environment and their binding to a LuxR-type receptor trigger QS-controlled gene regulatory cascades. In Pseudomonas aeruginosa, for example, binding of AHLs to their cognate receptors (LasR, RhlR) controls production of virulence factors, pigments, antibiotics and other behaviors important for its interactions with eukaryotic hosts and other bacteria. We have previously shown that marine cyanobacteria produce QS-inhibitory molecules, including 8-epi-malyngamide C (1), malyngamide C (2) and malyngolide (3). Here we isolated a new small cyclopropane-containing fatty acid, lyngbyoic acid (4), as a major metabolite of the marine cyanobacterium, Lyngbya cf. majuscula, collected at various sites in Florida. We screened 4 against four reporters based on different AHL receptors (LuxR, AhyR, TraR and LasR) and found that 4 most strongly affected LasR. We also show that 4 reduces pyocyanin and elastase (LasB) both on the protein and transcript level in wild-type P. aeruginosa, and that 4 directly inhibits LasB enzymatic activity. Conversely, dodecanoic acid (9) increased pyocyanin and LasB, demonstrating that the fused cyclopropane “tag” is functionally relevant and potentially confers resistance to β-oxidation. Global transcriptional effects of 4 in some ways replicate the gene expression changes of P. aeruginosa during chronic lung infections of cystic fibrosis patients, with reduced lasR signaling, increased biofilm and expression of the virulence locus HSI-I. Compound 4 may therefore prove to be a useful tool in the study of P. aeruginosa adaption during such chronic infections.
Introduction
Quorum sensing (QS) is a mechanism by which bacteria regulate their behavior in response to increases in their population density. Within a diffusion-limited environment, the local concentration of small molecule cues increases with bacterial population, acting to upregulate virulence genes in opportune situations (i.e., when the population of bacteria is large enough to overwhelm host defenses).1 QS pathways, therefore, are an attractive target for antimicrobial defense. In the marine environment, QS also contributes to the formation of biofilms by bacteria, the first step in the process of colonization of abiotic surfaces (biofouling).2
In Gram-negative bacteria, acylhomoserine lactones (AHLs, Figure 1a) of varying alkyl chain lengths and oxidation states at C-3 are used in quorum signaling.3 AHLs with short side chains diffuse freely across cell membranes and can bind intracellularly with receptor proteins (R proteins), which typically act as transcriptional activators of target genes.4 One of these targets is generally the gene responsible for the synthesis of the signaling molecule itself. In this way QS is a positive feedback loop, and QS signaling molecules are sometimes referred to as “autoinducers”.5
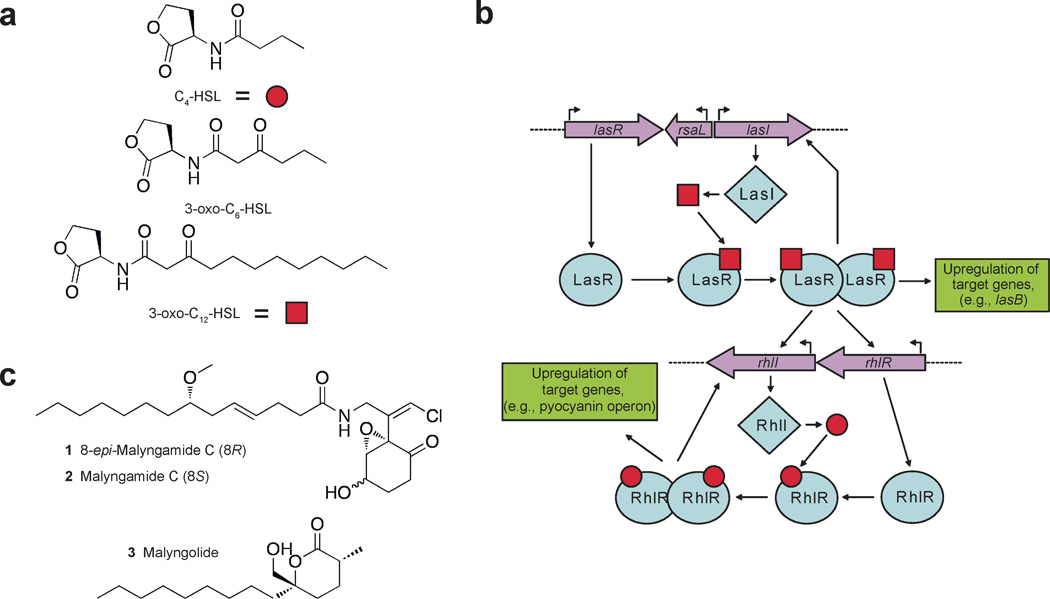
Figure 1.
a) Structures of three AHL signaling molecules used in Gram negative bacteria for quorum sensing, C4-HSL, 3-oxo-C6-HSL and 3-oxo-C12-HSL. b) Scheme showing the hierarchy of AHL pathways in P. aeruginosa. c) Three previously identified quorum sensing inhibitors, 8-epi-malyngamide C (1), malyngamide C (2) and malyngolide (3).
Bacterial infections and the increase in antibiotic-resistant pathogens are an ever escalating problem. Such infections complicate and prolong hospital stays and increase the cost to individuals and society.6 Antibiotics exert strong selective pressure on bacteria, and consequently, resistance is strongly associated with increased antibiotic use.7 Agents that are not bacteriocidal, but instead modulate harmful bacterial behavior, could perhaps exert less selective pressure for resistance. It has been suggested that this may be a strategy used by marine organisms that lack cellular immune systems,8 due to the fact that the ecologically relevant concentrations of natural antibiotics are sometimes in the sub-lethal range.
One prime target for QS-based therapy is the opportunistic pathogen Pseudomonas aeruginosa. This organism is a particular problem in cystic fibrosis, where it can persistently establish itself in chronic lung infections.9 Persistence is largely due to the formation of antibiotic-resistant biofilms, a phenotype which is modulated by QS. P. aeruginosa can also cause serious eye infections in wearers of contact lenses,9 with extensive tissue damage mediated by the QS-controlled proteolytic enzymes, including the elastase LasB.9
P. aeruginosa has multiple QS pathways, mediated through two AHLs (C4-HSL‡ and 3-oxo-C12-HSL)5 and a group of quinolone compounds (the Pseudomonas quinolone signaling pathway, PQS).10 The interplay of the two AHL pathways is shown in Figure 1b. One important feature of these is that the RhlR-RhlI system is subordinate to the LasR-LasI system,11 as expression of both RhlR and C4-HSL synthesis (via RhlI) is regulated by LasR/3-oxo-C12-HSL (Figure 1a). Therefore both C4-HSL and 3-oxo-C12-HSL are required for expression of RhlR target genes. The expression of LasR is not under the control of AHL-mediated signaling,12,13 and it therefore represents an upstream target for QS inhibition in P. aeruginosa. Interplay of AHL signaling with the quinolone pathway is complex. While on the one hand, quinolone signaling is thought to be dependent on LasR-LasI,10 this pathway has been shown to act independently under some circumstances.14
For some time, we have been involved in the search for novel and bioactive secondary metabolites. Such natural products have historically been a major source of pharmaceuticals, or have provided inspiration to medicinal chemists.15 Toxic compounds that are presumably used for chemical defense in competitive environments have found use as antibiotics or anticancer drugs.15,16 AHL signaling pathways are widely used among gram negative bacteria, and thus QS modulation by small molecules may be an effective strategy to alter competitor behavior in complex communities.17 Such small molecules could prove useful as drug candidates or tool compounds.
There is increasing evidence that various marine organisms can interfere with bacterial quorum sensing. Several halogenated furanones that inhibit AHL signaling have been isolated from the marine red alga Delisea pulchra.18 It has been shown that these compounds do not compete with AHLs but instead accelerate turnover of the LuxR protein.19 LuxR is the master regulator of three QS pathways in many luminescent Vibrio spp., and so furanones block all AHL signaling in these organisms.20 Recently, tumonoic acids E–H from the cyanobacterium Blennothrix cantharidosmum were shown to reduce luminescence in wild-type Vibrio harveyi, although the mechanism of action was not determined.21 In a screen of 284 extracts of marine organisms, 23% were found to exhibit quorum sensing antagonism in a LuxR-based reporter.22 We have recently identified three other compounds that inhibit AHL-mediated quorum sensing in reporter systems, 8-epi-malyngamide C (1),23 malyngamide C (2)23 and malyngolide (3)24 (Figure 1c). Therefore, it would seem that QS modulation may be a fairly widespread phenomenon amongst marine organisms for pathogen defense or for maintenance of bacterial symbionts. In addition, there is increasing evidence that interspecies crosstalk of quorum sensing is widespread,17 as are interactions between infection hosts and pathogenic QS pathways.25 From a biomedical perspective, natural QS modulators may prove useful in the treatment and prevention of infection and be complementary to natural antibiotics, which have long provided benefit in the treatment of infections (e.g., penicillins, macrolides and glycopeptides).
In the present work we describe the isolation and structure determination of a simple cyclopropane-containing fatty acid from a marine cyanobacterium, termed lyngbyoic acid (4, Figure 2). Because the compound was produced in relatively large amounts, and considering its small size, we hypothesized that it could be a signaling molecule. In view of the importance of QS to biofouling in the marine environment, we screened 4 against several AHL-responsive reporter constructs and found robust inhibitory activity against the 3-oxo-C12-HSL-responsive reporter pSB1075. Further investigations using pSB1075 and related control plasmids aimed to 1) determine the role of the AHL-binding site of LasR to the inhibitory activity, and 2) to determine preliminary structure-activity relationships using several compounds with structures related to 4. Subsequently, we investigated the inhibitory effect of 4 in the wild-type Gram-negative bacterium that expresses LasR, P. aeruginosa. Although this organism may not be an ecologically relevant target for the cyanobacterium, it is widespread in the environment,26 including the sea27 and it is a pathogen with an extremely broad host range, which includes protozoa, plants and humans.28
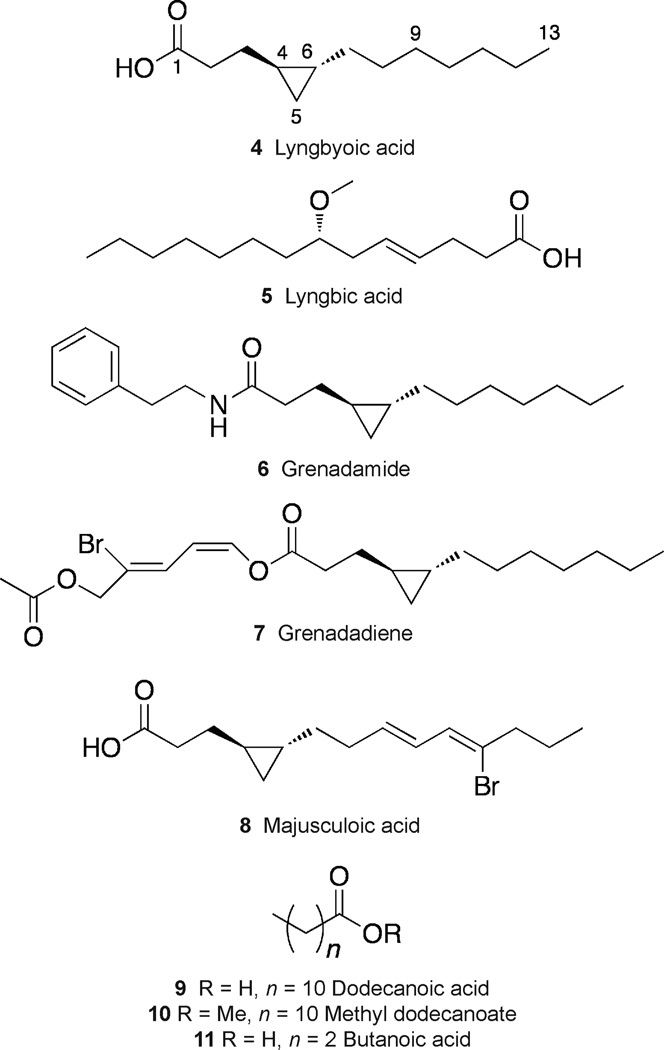
Figure 2.
Structures of lyngbyoic acid (4), lyngbic acid (5), grenadamide (6), grenadadiene (7), majusuloic acid (8), dodecanoic acid (9), methyl dodecanoate (10) and butanoic acid (11). Note that the depicted absolute configurations for 7 and 8 are arbitrary because only relative configurations have been determined.
Results and discussion
Isolation and structure determination
Samples of Lyngbya cf. majuscula were collected near Fort Pierce, Florida, in the Indian River Lagoon. Fractionation of the non-polar extract of this material yielded 4 (1.32% of extracted lipophilic material, Figure 2). Samples of Lyngbya cf. majuscula collected within the Dry Tortugas National Park, Florida, extracted and fractionated in a similar manner, afforded 4 (0.03% of extracted lipophilic material), malyngolide (3, Figure 1b) (0.007%) and lyngbic acid (5, Figure 2) (0.11%).
NMR data combined with a [M – H]− peak of 211.1702 in the HRESI/APCIMS of 4 suggested a molecular formula of C13H24O2. Perusal of the 1H NMR spectrum revealed the presence of a carboxylic acid exchangeable proton (broad peak at δH 10.18) and a cyclopropane ring (shielded signals at δH 0.45 and 0.21). Analysis of 1D and 2D NMR data allowed the construction of the planar structure (Table 1). The relative configuration of the cyclopropane ring was assigned trans, as the H-5 methylene protons are magnetically equivalent due to “pseudo C2ν symmetry”, as in grenadamide (6, Figure 2), grenadadiene (7)29 and majusculoic acid (8).30 The absolute configuration could be determined as (4R,6R) from the optical rotation, which was equal magnitude and opposite sign to that of the synthetic enantiomer.31 Although 4 is essentially the fatty acid side chain of 6 and 7 it has not been previously reported as the free acid. Compounds 6 and 7 were previously shown to be moderately cytotoxic to brine shrimp and human cancer cell lines, respectively, and 6 binds to cannabinoid receptors.29 Compound 8 was found to have some antifungal activity,30 but no quorum sensing-related activities for 6–8 have been reported.
Table 1.
NMR data for lyngbyoic acid (4) in CDCl3 (500 MHz)
| C/H No. | δC, mult.a | δH (J in Hz) | COSY | HMBCb |
|---|---|---|---|---|
| OH | 10.18, br | |||
| 1 | 180.7, s | |||
| 2 | 34.5, t | 2.42, t (7.5) | H-3a, H-3b | 1, 3, 4 |
| 3a | 29.6, t | 1.56, m | H2-2, H-4 | 1, 2, 4, 6 |
| 3b | 1.52, m | H2-2, H-4 | 1, 2, 4, 5, 6 | |
| 4 | 18.3, d | 0.45, m | H2-5, H-3a, H-3b | 5,c 7c |
| 5 | 12.0, t | 0.21, m (2H) | H-4, H-6 | 3, 4, 6, 7 |
| 6 | 19.2, d | 0.45, m | H2-5, H-7a, H-7b | 5,c 7c |
| 7a | 34.3, t | 1.21, m | H-6, H-7b, H2-8 | 4, 5, 6, 8 |
| 7b | 1.13, m | H-6, H-7a, H2-8 | 4, 5, 6, 8 | |
| 8 | 29.8, t | 1.33, m | H-7a, H-7b, H2-9 | 6, 7 |
| 9 | 29.7,d t | 1.25, m | H2-8 | 7 |
| 10 | 29.5,d t | 1.25, m | 12 | |
| 11 | 32.1, t | 1.25, m | 12 | |
| 12 | 22.2, t | 1.27, m | H2-11, H3-13 | 11, 13 |
| 13 | 14.0, q | 0.88, t (7.0) | H2-12 | 11, 12 |
Multiplicity is derived from APT and HSQC spectra.
Protons showing long-range correlation to indicated carbon.
It could not be distinguished which proton shows HMBC correlations to C-5 and C-7.
Assignment of these carbons is interchangeable.
Initial AHL quorum sensing reporter screening
Compound 4 was found in relatively large amounts, similar to the tumonoic acids.32 Natural quorum sensing inhibitors generally exhibit IC50s only in the micromolar range (including tumonoic acids,21 halogenated furanones18 and manoalides22), and consequently they have to be present in high concentrations in the native organism. Interestingly, in addition to the main collection that produced 4 only, in another collection of Lyngbya cf. majuscula from Dry Tortugas, we co-isolated 4 with two other QS inhibitors, malyngolide24 and lyngbic acid (3 and 5, respectively) in smaller amounts. With this in mind, and also with the consideration that 4 somewhat resembled some natural AHL disrupters, we screened it against three reporter plasmids in E. coli (pSB401, pSB536 and pSB1075, Figure 3, Table S1, Supporting Information).33 Each plasmid encodes different R-proteins (that respond to different AHLs) and contains its cognate binding site within the QS-regulated promoter, cloned upstream of a promoterless light-producing luxCDABE cassette. In each, the R-protein is under the control of its native promoter.33 Additionally, we tested 4 against an Agrobacterium tumefaciens lacZ-based reporter35 that responds to 3-oxo-C8-HSL.
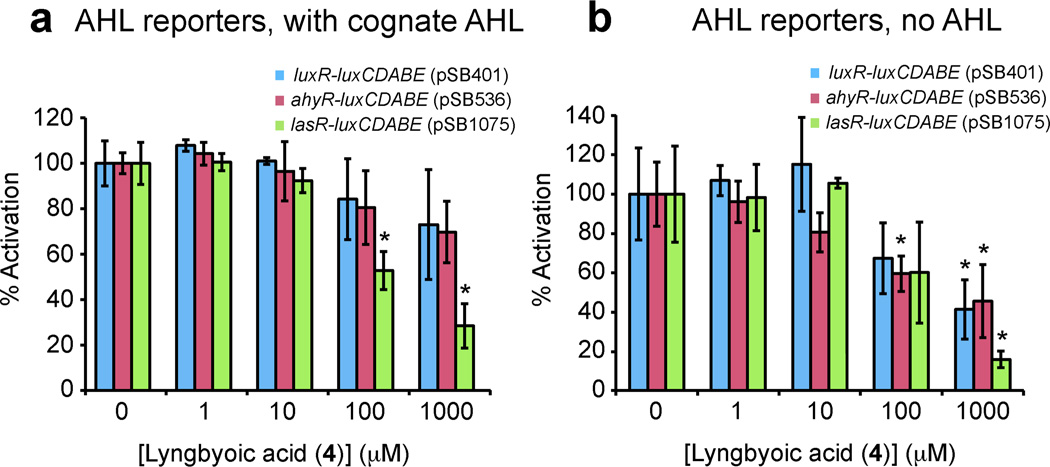
Figure 3.
Determination of inhibitory activity of compound 4 in luxCDABE reporter constructs expressed in E. coli (see Table S1, Supporting Information). (*) Indicates downregulation with statistical significance of P < 0.05 (t-test), compared with untreated controls, and error bars depict standard deviation. a) Effect of lyngbyoic acid (4) on luminescence of the reporter strains pSB401, pSB536 and pSB1075 in the presence of 3-oxo-C6-HSL, C4-HSL and 3-oxo-C12-HSL, respectively. Compound 4 and the appropriate cognate AHL were added to a 96-well plate and the solvent was allowed to dry off before 100 µL cultures of the appropriate reporter strain were added to each well. Plates were incubated for 6.5 h at 37 °C before fluorescence was measured. Results are expressed as % activation compared to control wells treated with cognate AHL alone (100%). Compound 4 was able to inhibit 3-oxo-C12-HSL mediated luminescence in pSB1075 and to a lesser extent in the other reporters. b) Effect of lyngbyoic acid (4) on background luminescence, in the absence of cognate AHL. The same protocol as for panel a) was used, except that cognate AHLs were not added to the plate. Results are expressed as % activation compared to untreated control (100). Compound 4 reduced background luminescence in all reporters.
Reporter strains were treated with compound 4 both in the presence and absence of the cognate AHL signaling molecule (Figure 3a and 3b), in order to detect antagonism or agonism of AHL signaling, respectively. Compound 4 antagonized 3-oxo-C12-HSL mediated light production through LasR (pSB1075) with an apparent IC50 of approximately 100 µM, and to a much lesser extent in the other two reporters (Figure 3a). Interestingly, 4 also reduced the baseline luminescence in all three reporters in the absence of cognate AHL (Figure 3b), perhaps indicating either an inverse-agonist type activity or an effect on expression of the R-protein, the luxCDABE cassette, or both. Compound 4 was not able to antagonize the production of blue pigment in the A. tumefaciens reporter in the presence of 3-oxo-C8-HSL (see Figure S1, Supporting Information).
Investigation of dependence on the LasR AHL-binding site
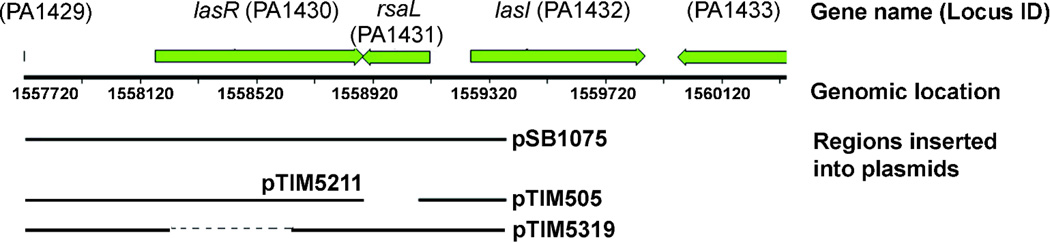
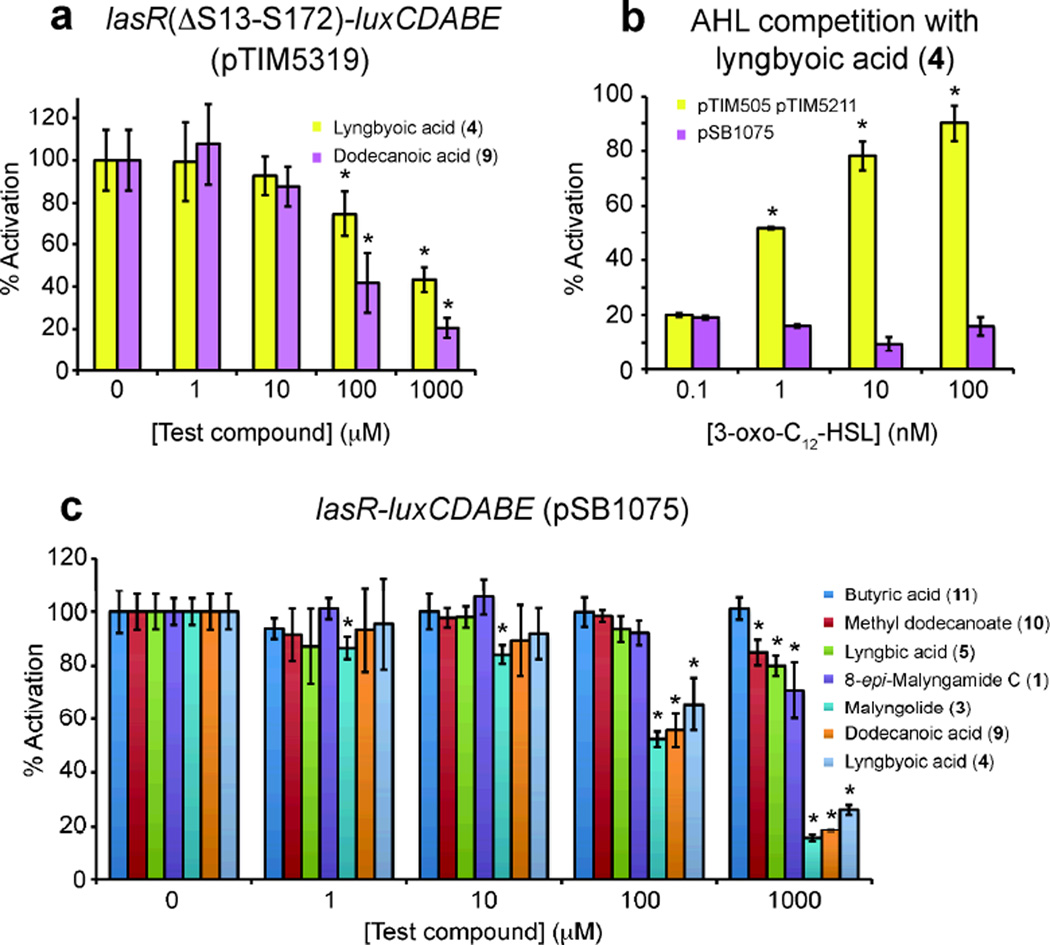
To test whether the AHL-binding site was required for inhibition, we used a reporter (pTIM5319) that lacks this domain, but in other respects is identical to pSB107534 (Figure 4). Compound 4 reduced baseline luminescence in this reporter (Figure 5a), indicating that neither the cognate AHL, nor the AHL-binding domain of the AHL receptor is required. Additionally, we found that by varying the concentration of 3-oxo-C12-HSL, 4 is not competitive with this ligand in pSB1075 (see Figure 5b). However, in a reporter that lacks the transcriptional repressor rsaL (pTIM505 pTIM5211), which in the PAO1 genome resides in the region between lasR and lasI, 3-oxo-C12-HSL is clearly able to compete with 4 (Figure 5b). Taken together, these results suggest that the effects of 4 are exerted both through the AHL-binding domain of LasR, and independently of it. The repressor rsaL is potentially implicated in the latter. To exclude a general effect on bacterial physiology or on the activity of the lux reporter cassette, we tested 4 on a reporter where the lux operon is under control of the constitutively active λ phage promoter (pTIM2442, Table S1, Supporting Information), and found no effect (Figure S2, Supporting Information).
Figure 4.
Schematic of selected reporter plasmids and controls used (see Table S1, Supporting Information, and text). Regions cloned upstream of the promoterless luxCDABE cassette are indicated by a thick line. pTIM5319 carries a truncated lasR in which the AHL binding pocket (S13-S172, indicated by a thin dashed line) was removed using primers GCGTGGCGATGGGCCGACAGTG and GCGTTCCAGCTCAAGAAAACCGTC; AAACCGGTGGTTCTGACCAGCCGG and CACTAACGTCCCAGCCTTTGCGCTC, as described in Rajamani et al., 2008.34
Figure 5.
Probing of the mechanism of inhibition by 4 by use of luxCDABE reporter constructs, and comparison of 4 with compounds of related structure. (*) Indicates downregulation with statistical significance of P < 0.05 (t-test), compared with untreated controls, and error bars depict standard deviation. a) Effect of lyngbyoic acid (4) and dodecanoic acid (9) on a reporter that lacks a functional AHL-binding domain (pTIM5319).34 Cultures of the reporter were grown in 96-well plates in the presence of varying concentrations of 4 or 9. Results are expressed as % activation compared to untreated control (set to 100%). b) 3-oxo-C12-HSL is able to compete with 4 (1 mM) in a reporter strain lacking rsaL (pTIM505 5211), but not in pSB1075. c) Effect of lyngbyoic acid (4) and other compounds on luminescence in pSB1075.
Investigation of related compounds in pSB1075
We compared the effect of 4 on pSB1075 to that of compounds of related structure, such as dodecanoic acid (9) and the cyanobacterial metabolite lyngbic acid (5, see Figures 2 and 5c), which was co-produced with 4 by one of the Lyngbya cf. majuscula samples we investigated. The most closely related dodecanoic acid (9) exerted an inhibitory effect of similar magnitude to 4, as did the previously identified quorum-sensing inhibitor malyngolide (3).24 Similar to 4, 9 was found to inhibit the AHL-binding deficient reporter pTIM5319 (Figure 5a), indicating that this site is non-essential for inhibition in both cases. We recently disclosed that 8-epi-malyngamide C (1) weakly inhibited pSB1075.23 It can now be seen that compound 4 is more potent than 1. Interestingly, both 1 and its free side chain, lyngbic acid (5), have similar potency. Considering that the fatty acids 4 and 9 inhibit stimulation of pSB1075 by 3-oxo-C12-HSL, it could be that the ring-opened form of malyngolide is the active species. The methyl ester of dodecanoic acid (10) had only a small inhibitory effect, and butyric acid (11) was completely inactive, indicating a preference for free acids and longer alkyl chains, respectively (Figure 5c).
Effects of lyngbyoic acid (4) and related compounds in wild-type P. aeruginosa and PAO-JP2
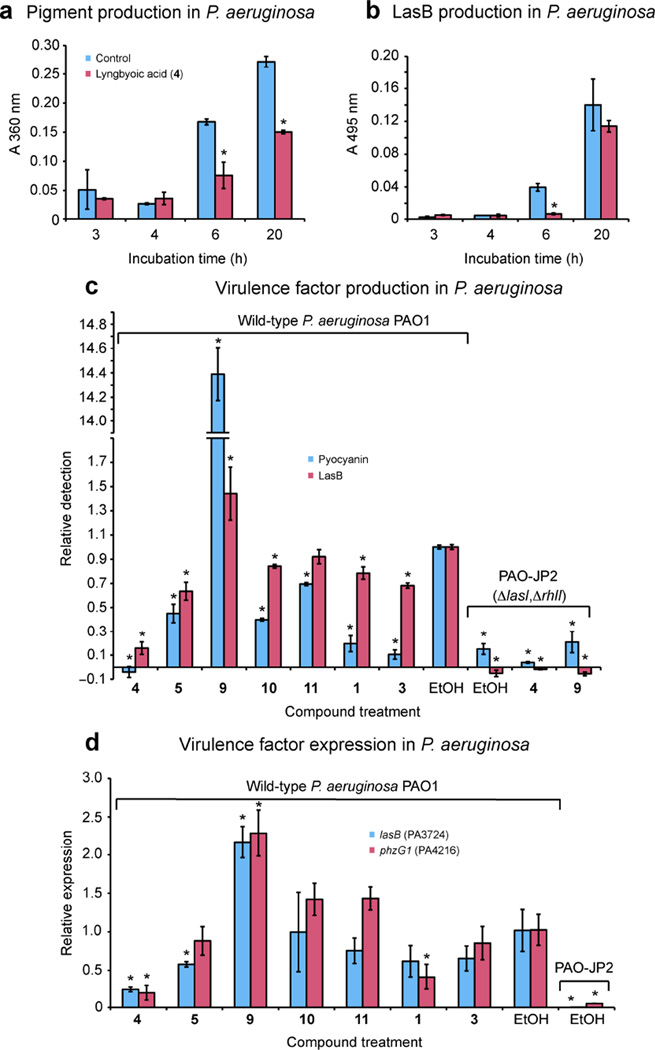
To assess whether 4 is able to inhibit a native quorum-sensing system, we treated wild-type P. aeruginosa cultures with 4 (1 mM) in a preliminary experiment measuring secreted pigments (assessed by UV absorption of culture supernatants at 360 nm), and LasB (assessed by degradation of elastin Congo red, see Figures 6a and 6b). It was observed that 4 was able to lower pigment production by the 6 h and 20 h timepoints, and that lower LasB activity levels were observed at 6 h only. Therefore the 6 h timepoint was used for subsequent experiments. To exclude an effect on the viability of PAO1, cultures were grown in the presence and absence of 1 mM 4. No effect on growth, as assessed by OD600, was observed (see Figure S3, Supporting Information). Because fatty acids can inhibit proteases,36 we tested compound 4 for direct inhibition of purified LasB. Indeed, we found that 4 inhibited LasB with a Ki of 5.4 µM (see Figure S4, Supporting Information). However, we did not detect 4 by LC-MS in the filtered supernatants. Therefore, elastase activity in supernatants is a true reflection of expression and there is no direct inhibition of secreted enzyme by 4. Since 4 is quite lipophilic it may be sequestered within cells, membranes, or membrane vesicles, or else it is degraded by cellular enzymes.
Figure 6.
Effect of lyngbyoic acid (4) and other compounds on wild-type P. aeruginosa and the ΔlasIΔrhlI mutant PAO-JP2. In all graphs (*) indicates downregulation with statistical significance of P < 0.05 (t-test) compared to untreated controls. a) Lyngbyoic acid (4) is able to reduce pigment production by PAO1. Cultures (500 µL containing 1 mM 4) were incubated at 37 °C with shaking for the appropriate time before they were spun down and the absorbance of the supernatant at 360 nm was measured. b) Lyngbyoic acid (4) is able to reduce LasB production in PAO1. A portion of supernatants (50 µL) was incubated for 5 h at 37 °C with shaking in the presence of elastin Congo red (200 µL of a 12.5 mg/mL suspension in 30 mM Tris-HCl, pH 7.2), then spun down and the absorbance at 495 nm was measured. c) Effect of lyngbyoic acid (4) and other compounds (1 mM) on pyocyanin and LasB production in PAO1 and PAO-JP2. Cultures (1 mL) were grown in the presence of test compounds or EtOH for 6 h at 37 °C with shaking, before being spun down. A portion (100 µL) of each supernatant was added to 900 µL 5 mg/mL elastin Congo red and incubated at 37 °C with shaking for 18 h, then the mixtures were spun down and the absorbance of the supernatant was measured at 495 nm to measure LasB activity. Another portion (500 µL) of culture supernatants was extracted with 500 µL CHCl3 and then back-extracted with 0.2 N HCl. The absorbance of this acidic aqueous layer was measured at 385 nm to quantify pyocyanin production. d) Effect of lyngbyoic acid (4) and other compounds on the gene expression of lasB and phzG1 as assessed by RT-qPCR. Cultures were grown under the same conditions as in c).
To assess potential differences between our reporter system and P. aeruginosa, we tested the complete set of compounds in PAO1 (see Figures 6c and 6d). Through extraction of pyocyanin from supernatants according to a published procedure,37 and measurement of LasB activity, it could be seen that compound 4 reduced both pyocyanin and LasB by the greatest extent. The most striking contrast with results in reporter assays came from the effects of dodecanoic acid (9) in PAO1. This compound greatly increased pyocyanin compared to control, and LasB to a lesser extent (see Figure 6c). Intriguingly, this effect was not replicated in the ΔlasIΔrhlI mutant PAO-JP2,38 indicating that it is dependent on either lasI or rhlI genes, or their downstream targets. Plausibly, dodecanoic acid could act as a substrate for β-oxidation pathways to produce the 3-oxo acid, which if bound to an acyl-carrier protein (ACP), is one of the substrates for LasI.39 This would suggest that the cyclopropane of 4 precludes β-oxidation at the 3-position and does not allow it to be utilized by LasI. We therefore describe 4 as “tagged”, as the cyclopropane may allow the compound to persist in both the producing cyanobacterium and target organisms, by avoiding metabolism through β-oxidation. Gene expression studies by RT-qPCR revealed the effects on virulence factors due to compound treatments was largely duplicated in the transcript levels of lasB (PA3724) and phzG1 (PA4216), a member of the pyocyanin biosynthetic operon (see Figure 6d).40
Global gene expression analysis of lyngbyoic acid (4)-treated P. aeruginosa
Some aspects of the reporter studies suggested that 4 may have effects on gene expression independent of AHL signaling. We investigated the effects of 4 on the transcriptome of PAO1 through microarray analysis, which revealed extensive gene expression changes (at ≥1.8-fold change with p < 0.01: 969 genes upregulated, 887 genes downregulated, see Scheme S1, Supporting Information). Importantly, comparison of microarray data revealed a high overlap with two landmark transcriptome studies of LasR-LasI and RhlR-RhlI controlled genes (Figure 7).12,13 This included downregulation of pyocyanin synthesis (phzM, phzA1–G1, phzS [PA4209–18], phzA2–G2 [PA1899–1905] and phzH [PA0051]), secreted enzymes (lasA [PA1871], lasB [PA3724], chiC [PA2300] and aprA [PA1249]) and rhamnolipid production (rhlA and B, PA3479 and PA3480). Also, the pqsABCDE and phnAB operons (PA0996–PA1002), responsible for synthesis of QS quinolone signal molecules,10 were significantly decreased (−3.4 to −11.9-fold). These operons have previously been shown to be under the control of the Las system,12,13 but treatment with 4 did not affect the expression level of lasR (PA1430), lasI (PA1432), rhlR (PA3477) or rhlI (PA3476). Interestingly, expression of hydrogen cyanide production genes (hcnA-C, PA2193–5) was unaffected, even though they were previously identified as QS-controlled,12,13 and were downregulated by a previously described QS inhibitor.41
Figure 7.
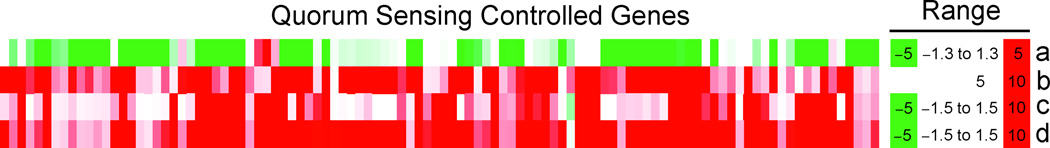
Comparison of lyngbyoic acid (4) induced changes in gene expression (a) with previous studies of quorum-sensing controlled genes, Wagner et al.13 (b) and Schuster et al.12 (c and d). a) Cultures of PAO1 were treated with 1 mM 4 for 6 h at 37 °C with shaking. b) PAO-JP2 treated with exogenous 3-oxo-C12-HSL and C4-HSL. c) PAO-MW1 treated with 3-oxo-C12-HSL. d) PAO-MW1 treated with 3-oxo-C12-HSL and C4-HSL.
In addition to effects on quorum sensing, lyngbyoic acid (4) reduced the expression of 36 genes previously identified as induced by iron-starvation (see Table S2, Supporting Information).42 For example the expression of some genes for the biosynthesis of pyoverdin43 (pvdA, D, I and J [PA2386, PA2399 and PA2400/2401, respectively]), a fluorescent siderophore, were reduced (−2.9 to −4.5-fold) along with the outer membrane pyoverdin receptor fvpA (PA2398, −2.2-fold) and the regulatory σ-factor pvdS (PA2426, −4.7-fold). Genes for the biosynthesis of pyochelin (pchABCDG, PA4228–4231, −2.6 to −3.5-fold), another siderophore, were also reduced. Effects on both iron-regulated and QS-regulated genes potentially implicate an involvement of the regulator vqsR.44,45 A vqsR mutant was shown to have decreased expression of genes related to quorum sensing, as well as pyoverdine and pyochelin.45
Transcript levels of some other regulators that have previously been implicated in quorum sensing were also affected in a complex manner. rpoS (PA3622),46 rsmA (PA0905)47 and qscR (PA1898)48 were all decreased (−4.3, −2.4 and −2.0-fold, respectively), whereas rpoN (PA4462),49 mvfR (PA1003)50 and pmpR (PA0964)51 were all increased (+2.2, +1.8 and +2.8-fold, respectively). The downstream QS effects of 4 could perhaps in part depend on alteration of the balance of positive and negative regulators, as well as posttranscriptional/posttranslational effects. It is possible that 4 similarly modulates transcriptional regulators in E. coli, potentially accounting for the inhibitory effects seen in reporter systems that were independent of the lasR AHL-binding site (see Figures 5a and 5b). The effects in both systems could be due to regulators conserved between the two species, such as the GasS/A system, which is found in many Gram-negative bacteria.52
The effect of 4 on biofilm genes was also complex. While some members of the psl operon (pslA and B [PA2231–2232] and pslN [PA2244]) were downregulated (−1.9 to −3.4-fold), the entire pelABCDEFG operon (PA3058–64) was upregulated (+2.0 to +7.2-fold). The pel operon is required for the synthesis of a glucose-rich matrix exopolysaccharide that is an important component of biofilms, and its expression has been shown to be dependent on lasI.53 Therefore, it would seem that in lyngbyoic acid (4)-treated cells, expression of pel is uncoupled from general QS effects, which are inhibited. Recently, it has been shown that pel genes are repressed by the tyrosine phosphatase TpbA (PA3885).54 Lyngbyoic acid (4) paradoxically increased the expression of tpbA by 3.3 fold, perhaps suggesting a posttranscriptional or direct effect on the protein.
Compound 4 also increased the expression of a type VI secretion virulence locus, HSI-I55 (PA0071–91, +1.8 to +5.7-fold). This locus expresses a secreted protein, Hcp1, along with its secretion apparatus. Hcp1 has been detected in CF patients that harbor chronic P. aeruginosa infections, and the expression of HSI-I is antagonistically regulated by RetS (repression) and LadS (activation). These two regulators also control exopolysaccharide production, are implicated in the control of virulence factor expression in acute (RetS) and chronic (LadS) infections,55 and modulate the activity of the GasS/A pathway, that controls expression of QS post-transcriptionally.56
We found that 4 caused extensive gene expression changes in P. aeruginosa. Similarly, using a more stringent cutoff (≥2.0-fold change) 437 genes were found by Son et al.57 to be induced in vivo using P. aeruginosa isolates from CF patients, including many involved in general metabolism. This was attributed to nutrient sources within CF lungs, including lipid surfactants (e.g., phosphatidylcholine) and amino acids. As in Son et al.,57 4 affected many genes involved in central metabolism, for example the operons of the fatty acid sensors PsrA58 and DesT59 were upregulated and downregulated, respectively (see Scheme S1, Supporting Information). In another example, several genes involved in choline degradation (PA3933, betA, betB, betI and betT1 [PA5372–5375]), which were all upregulated by 4 (+9.6 to +18.1-fold) and also previously shown to be upregulated by phosphatidylcholine.57 Importantly, although many of the gene expression changes could be as a result of involvement of 4 in central metabolism pathways, the quorum sensing effects are dependent on the cyclopropane moiety as evidenced by the QS-stimulatory effects seen after treatment with 9.
The characterization of 4 may have biomedical significance. After initial infection of cystic fibrosis patients, P. aeruginosa adapts to the CF lung environment, acquiring a phenotype characterized by reduced quorum sensing, overproduction of exopolysaccharide (mucoid phenotype), and reduced motility.60 This is often accompanied by a loss of lasR.60 Compound 4 mimics many of these effects, including a general inhibition of quorum sensing and expression of the virulence determinant HSI-I in wild-type P. aeruginosa. It may therefore prove a valuable tool compound for modeling the process of adaption in CF, perhaps by replicating the response of P. aeruginosa to certain fatty acids present in CF sputum. In P. aeruginosa, quorum sensing circuits are known to cross-talk with environmental pressures independent of cell density, for example stringent starvation61 and membrane fluidity.62 The observed differential physiological and molecular responses to structurally related simple fatty acids suggest a major role of fatty acids to switch-on or switch-off certain pathways and adjust to environmental conditions.
Experimental
General experimental procedures
Optical rotation was measured on a Perkin-Elmer 341 polarimeter. UV spectra were recorded on SpectraMax M5 (Molecular Devices). 1H, 13C, and 2D NMR spectra were recorded in CDCl3 on a Bruker Avance 500 MHz using residual solvent signals (δH 7.26, δC 77.0) as internal standards. HSQC experiments were optimized for 1JC,H = 145 Hz, and HMBC experiments were optimized for nJC,H = 8 Hz. HRMS data were obtained using an Agilent LC-TOF mass spectrometer equipped with an APCI/ESI multimode ion source detector. Luminescence assays and OD600 measurements were read on a Perkin-Elmer Victor3 microtiter plate reader. UV spectra of culture supernatants and elastase activity assays were read on a SpectraMax M5 (Molecular Devices). LC-MS data were obtained using a 3200 Q Trap LC/MS/MS system (Applied Biosystems). Real-time quantitative PCR experiments were carried out on an Applied Biosystems 7300 instrument. C4-HSL and 3-oxo-C6-HSL were obtained from Sigma Aldrich, 3-oxo-C12-HSL was supplied by Cayman Biochemicals.
Extraction and isolation
Samples of Lyngbya cf. majuscula were collected in the Indian River Lagoon (IRL), near Fort Pierce (27°26.668′ N, 80°18.095′ W) on June 23, 2006. This was a recollection of the sample designated IRL1. A voucher sample is maintained at the Smithsonian Marine Station, Fort Pierce, FL. The freeze-dried material was extracted with EtOAc–MeOH (1:1) to furnish a crude non-polar extract, which was subsequently partitioned between 80% aqueous MeOH and hexanes. The H2O–MeOH fraction was further partitioned between n-BuOH and H2O. The n-BuOH extract (1.69 g) was subjected to silica gel chromatography, eluting fractions with increasing proportions of i-PrOH in CH2Cl2. The fraction eluting with 6% i-PrOH in CH2Cl2 was further purified by semi-preparative reversed-phase HPLC (Phenomenex Ultracarb 5u ODS column, 250 × 10 mm, 2.0 mL/min; UV detection at 220 and 240 nm) using a MeOH−H2O linear gradient (60–100% MeOH over 50 minutes and then 100% MeOH for 20 min) to furnish compound 4, tR 49.0 min (42.4 mg). The yield was 1.32% of lipophilic material (the n-BuOH and hexanes fractions, excluding the H2O fraction).
Samples of Lyngbya cf. majuscula were collected at Garden Key, within the Dry Tortugas National Park, FL on April 22, 2007. A voucher sample is maintained at the Smithsonian Marine Station, Fort Pierce, FL (DRTO0000003). The freeze-dried material was extracted with EtOAc–MeOH (1:1) and then subjected to solvent-solvent partitioning as with the IRL material. The n-BuOH extract (669 mg) was subjected to silica gel chromatography, eluting fractions with increasing proportions of i-PrOH in CH2Cl2. The fraction eluting with 10% i-PrOH in CH2Cl2 showed evidence by 1H NMR of the presence of 3, 4 and 5, and so was further purified by semi-preparative reversed-phase HPLC (Phenomenex Synergi Hydro column, 250 × 10 mm, 2.0 mL/min; UV detection at 220 and 240 nm) using an ACN–0.1% HCOOH linear gradient (40–100% ACN over 20 min then 100% ACN for 30 min) to furnish compounds 3, tR 25.5 min (0.3 mg), 4, tR 26.5 (1.3 mg) and 5, tR 24.2 min (4.4 mg). Adjacent silica fractions also show the oxygenated methylene doublets (δH 3.71 and 3.47) of malyngolide (3) and the distinctive upfield cyclopropane (δH 0.45 and 0.21) signals of 4 and so the total yield of these compounds is expected to be greater.
Lyngbyoic acid (4): Colorless oil; [α]20D −15.5 (c 0.1, CHCl3); UV (EtOH) λmax (log ɛ) 202 (2.41), 230 (1.95), 260 (1.56); IR (film) νmax 3400–2400 (br), 3061, 2923, 2854, 1710, 1541, 1456, 1414, 1283, 1213, 1120, 1079, 1021, 936, 772, and 722 cm−1; 1H NMR, 13C NMR, APT, COSY, HSQC and HMBC data, see Table 1; HRESI/APCIMS m/z [M – H]− 211.1702 (calcd. for C13H23O2, 211.1698).
Bacterial strains and culture conditions
Bacterial strains and plasmids used in this study are listed in Table S1, Supporting Information. Reporter strains were grown overnight in Luria-Bertani (LB) medium at 37 °C with agitation as previously described.63 Briefly, overnight cultures were grown in the presence of the appropriate antibiotic (Table S1). The following day, cultures were diluted 100-fold with fresh LB and antibiotic, and incubated for 1 h, then diluted 100-fold again and incubated for a further 2 h. Cultures were diluted 10-fold with fresh LB and the appropriate antibiotic was added before cultures were used in assays. P. aeruginosa strain PAO1 was also grown using the same protocol, without added antibiotic.
luxCDABE-Based reporter assays
Test compounds and/or cognate AHL where appropriate were added to black 96-well plates, and the solvent was allowed to evaporate at room temperature. 100 µL cultures of the appropriate reporter were added to each well, and the plates were incubated at 37 °C in a humid environment for 6.5 h before their luminescence was recorded. For each assay, untreated wells (+/− AHL where appropriate) were used as controls. The final concentrations used were 10 µM (C4-HSL), 10 µM (3-oxo-C6-HSL) and 1 nM (3-oxo-C12-HSL), except for the 3-oxo-C12-HSL competition experiment (see Figure 5b). These concentrations corresponded to the experimentally determined IC50 of the AHLs against the relevant reporter strains under the same conditions as the assays.
A. tumefaciens reporter assay
The A. tumefaciens lacZ-based reporter was grown overnight in LB in the presence of gentamicin at 30 °C with shaking. The culture was diluted 100-fold then grown for a further 24 hours in M9 medium supplemented with sucrose (0.2% w/v), subcultured for 4 h and then mixed 1:1 with M9 sucrose containing 1.12% agar and immediately 100 µL of this mixture was added to each well of a 96-well plate containing compound 4 with or without 3-oxo-C8-HSL. The final concentration of 3-oxo-C8-HSL used was 1 nM, a concentration found to produce ~half maximal blue coloration by visual inspection. Plates were incubated in a humidified atmosphere at 30 °C and then visually inspected for blue coloration.
Pigment and elastase production in P. aeruginosa
Cultures (1 mL) of strain PAO1 were grown in 15 mm diameter glass tubes with shaking at 37 °C in the presence of 1 mM test compounds, added directly to the cultures as 10 µL of 100 mM stocks (in EtOH), for 6 h. Negative controls consisted of PAO1 cultures with 10 µL EtOH added, while positive controls were PAO-JP2 cultures with 10 µL EtOH added. To obtain an estimate of the pigment production, cultures were spun down and the absorbance of the supernatant at 360 nm was measured, corresponding to one of the UV maxima reported for pyocyanin.64 The supernatants were then passed through a 0.2 µm filter. Following the procedure of Müh et al.37 100 µL of each supernatant was added to 900 µL of a 5 mg/mL suspension of elastin Congo red (ECR, Elastin Products Company, Inc.).65 The mixtures were incubated in 15 mm plastic tubes at 37 °C with shaking for 18 h, at which point the reaction was stopped by addition of 100 µL 0.12 M EDTA. Soluble reaction product was quantified by UV absorption of the supernatants at 495 nm after centrifugation. Pyocyanin was quantified according to the procedure of Müh et al.,37 with some differences. A portion (500 µL) of the filtered culture supernatants was extracted with 500 µL CHCl3 in an Eppendorf tube. The CHCl3, which took on a visible blue color in cultures with high levels of pyocyanin, was transferred to a new tube and back-extracted with 150 µL of 0.2 N HCl. Under acidic conditions the UV spectrum of pyocyanin changes and solutions take on a visible red color in high-pyocyanin samples.64 A portion (100 µL) of the aqueous layer was transferred to a 384-well plate, and the absorbance at 385 nm was measured. The UV maximum at 385 nm has a higher ɛ than the maximum at 520 nm64 used elsewhere,37 and thus is more suitable for small-scale cultures.
LC-MS of PAO1 culture supernatants
A portion (1 µL) of supernatants from the elastase activity assay were subjected to an LC separation step followed by MS detection (Phenomenex Luna 5u C8 column, 4.6 × 50 mm, 0.5 mL/min; detection by ESIMS, MRM scan in negative mode) using an isocratic solvent system (90:10 MeOH–H2O, both with 0.1% HCOOH). Samples were compared to a standard solution of 4 at the expected concentration, and spiked control cultures. Compound 4 was detected in spiked control cultures containing non-denatured excreted LasB and other proteins, and thus specific and non-specific protein binding did not significantly affect detection. Authentic lyngbyoic acid (4) eluted at tR 6.3 min. The MS parameters were as follows: MRM ion pair 211→193, DP −66, EP −6.3, CEP −12, CE −25, CXP −3.4, CUR 30, CAD Medium, IS −4500, TEM 500, GS1 50, GS2 60.
In vitro inhibition of LasB
The in vitro inhibition of LasB (Elastin Products Company, Inc.) was assessed using BODIPY-FL casein (Invitrogen). 2 µL stock solutions of lyngbyoic acid (4) were added to a mixture of 1 µL LasB (10 µg/ml), 20 µL H2O and 77 µL assay buffer (10 mM Tris-HCl, pH 7.8), and incubated at 37 °C for 30 min. 100 µL BODIPY-FL was then added (10 µg/mL), and the reaction was followed by fluorescence (λex/λem 505/589 nm). EDTA (10 mM in H2O), a zinc chelating compound known to inhibit LasB, was used as a positive control. Similar results were seen using ECR as the substrate.
The Ki of lyngbyoic acid (4) was determined according to the protocol recommended by Copeland.66 First, the KM of the substrate BODIPY-FL casein was determined by measuring the slope of reactions in the presence of different substrate concentrations. Reaction mixtures consisted of 189 µL 10 mM Tris-HCl, pH 7.8, 1 µL of 10 µg/mL LasB, and 10 µL substrate solution. Initial slope was plotted against substrate concentration and the substrate concentration that gave half-maximal rate (the KM) was calculated by non-linear curve fitting in Graphpad to be 20 µg/mL. The IC50 of lyngbyoic acid (4) at this substrate concentration was then determined under the same conditions to be 4.3 µM. The Ki was then determined by running reactions in the presence of different substrate concentrations (10×, 5×, 2.5×, 1.25×, 0.625×, 0.3125× and 0.1563×KM) and different inhibitor concentrations (0, 1 µM, 3.16 µM and 10 µM). Best fit was obtained by assuming a noncompetitive inhibition model in Graphpad, and the Ki was calculated to be 5.4 µM.
Transciptome analysis
Cultures (1 mL) of PAO1 were grown either in the presence or absence of 1 mM 4 (added as 10 µL of a 100 mM stock solution in EtOH), for 6 h at 37 °C with shaking in 15 mm diameter glass tubes. Parallel cultures of each condition were grown alongside, and after ~5.5 h these were spun down and the UV absorbance of their supernatants was measured to confirm differential pyocyanin expression. RNA was stabilized in vivo by use of RNAprotect bacteria reagent (Qiagen) according to the manufacturer’s instructions. RNA was extracted using the RNeasy Kit (Qiagen) according to the manufacturer’s instructions.
RNA samples were quantified by UV absorbance (Nanodrop 8000, Thermo), and DNA contamination was quantified by RT-qPCR of the samples using a primer/probe set for rpsL (PA4268, see Table S3, Supporting Information). To reduce DNA contamination, Turbo DNA-free (Ambion) was used according to the manufacturer’s “stringent” treatment protocol. RNA quality was assessed using the Agilent 2100 Bioanalyzer (Agilent Technologies, Inc.) and samples were reverse transcribed, fragmented and labeled according to Affymetrix’s protocol for prokaryotic target preparation.67 Samples were hybridized with rotation at 50 °C for 16 h to the Affymetrix GeneChip® P. aeruginosa Genome Arrays. The arrays were washed and stained with the reagents supplied in GeneChip® Hybridization Wash and Stain Kit (Affymetrix, Inc.) on an Affymetrix Fluidics Statino 450, and scanned with a GeneChip® 7G Scanner (Affymetrix, Inc.). Statistical tests were performed using Bioconductor statistical software (http://www.bioconductor.org) and the R program (R: A language and environment for statistical computing).68 Raw data were normalized by Robust Multichip Analysis (RMA) approach.69 Then the probe set’s detection call was estimated using the Wilcoxon signed rank-based algorithm. Probe sets that were absent in all the study samples were removed from further analysis. A linear modeling approach and empirical Bayes statistics as implemented in the limma package70 in the R software were employed for differential expression analysis. The p-values obtained were controlled for multiple testing (FDR, false discovery rate) using the Benjamini-Hochberg method.71 Differentially expressed genes were then ranked by their p-values, and genes with p-value less than 0.01 and with greater than or equal to 1.8-fold change were considered as differentially expressed genes at a statistically significant level.
The microarray data were validated by realtime quantitative PCR using probes for lasB, phzG1, retS, fadA5 and lasR (vide infra). Comparison of real-time PCR and microarray data is shown in Table S4, Supporting Information. The global transcriptome data have been deposited in NCBI’s Gene Expression Omnibus72 and are accessible through GEO Series accession number GSE22999.
Reverse transcription and real-time quantitative PCR
RNA for use in RT-qPCR experiments was extracted and treated with DNase as for the GeneChip® samples. Samples were reverse-transcribed using Superscript II reverse transcriptase (Invitrogen) and random primers (Invitrogen). TaqMan primers/probes were custom designed by Applied Biosystems, using FAM as the fluorescent reporter and NFQ as the quencher. The sequences of primers and probes used is shown in Table S3, Supporting Information, and in all experiments the housekeeping gene rpoD (PA0576) was used as the endogenous control, as previously it has been found to have very stable expression, suitable for its use as a control in RT-qPCR experiments.73 Additionally, this gene was found to not be affected by lyngbyoic acid (4) in the microarray experiment. Real-time PCR was performed by using 12.5 µL of TaqMan 2× gene expression master mix (Applied Biosystems), 1.25 µL of 20× TaqMan gene expression assay mix (see Table S3, Supporting Information), 0.5 µL of cDNA and 10.75 µL of sterile water, in a total volume of 25 µL per well reaction in a 96-well plate (Applied Biosystems). The thermocycler program consisted of 2 min at 50 °C, 10 min at 95 °C, and 40 cycles of 95 ° C for 15 s and 60 °C for 1 min. Each assay was carried out in triplicate.
Conclusions
We have described a new cyclopropane-containing fatty acid that we termed lyngbyoic acid (4). This compound was found to inhibit the response of LasR-based QS reporter plasmids to 3-oxo-C12-HSL. The AHL-binding site of LasR was not essential to this effect, but competition experiments indicated that 4 likely has a dual mechanism, acting both through the AHL-binding site and independently of it. Comparison of 4 with related compounds revealed a structure–activity relationship. While dodecanoic acid (9) had a similar potency in pSB1075 compared to 4, either esterification (10) or shortening of the alkyl chain (11) reduced activity.
In an organism that possesses native quorum sensing circuitry, P. aeruginosa, we found 4 to reduce downstream pigment and elastase production. This was reflected by reduced expression of genes required for the biosynthesis of the pigment pyocyanin and the elastase LasB. Additionally, we found that 4 is able to directly inhibit purified LasB. Strikingly, despite minimal structural differences, dodecanoic acid (9) had opposite effects in P. aeruginosa, increasing pyocyanin and LasB on the transcript level. This indicates the functional relevance of the “tag” in 4.
Global gene expression analysis revealed that 4 downregulates the majority of genes previously identified as controlled by quorum sensing. In addition, differential expression of known QS regulators was noted, as well as complex effects on biofilm genes. Compound 4 also had various effects on central metabolism and upregulated the CF virulence locus HSI-I.
Supplementary Material
Acknowledgements
This research was supported by the National Institutes of Health, NIGMS grant P41GM086210 (VP and HL) and the University of Florida College of Pharmacy. MT’s contributions were supported by Florida Sea Grant # R/LR-MB-27 NA060AR4170014. We thank J. R. Rocca (UF) for assistance with NMR data acquisition and K. Arthur and C. Ross for help in collecting the cyanobacterium (Fort Pierce material). We thank Florida Institute of Oceanography for supporting use of R/V Bellows, and the National Park Service for granting permission to collect within Dry Tortugas National Park. We thank the crew of R/V Bellows, K. Arthur, F. Gurgel, S. Matthew, R. Ritson-Williams, K. Taori, and R. Wang for help in collecting L. cf. majuscula at Dry Tortugas. We also wish to thank D. and M. Littler for their helpful comments on cyanobacterial taxonomy, J. Li for assistance with the bioinformatics, and Y. Zhang for helpful discussions on sample preparation for microarray analysis. This is contribution #838 of the Smithsonian Marine Station at Fort Pierce.
Footnotes
Electronic Supplementary Information (ESI) available: Figures S1–S4, Scheme S1, Tables S1–S4, Supplementary References and NMR Spectra for 4. See DOI: 10.1039/b000000x/
Abbreviations used: CAD, Collisionally Activated Decomposition; CE, Collission Energy; COSY, Correlation SpectroscopY; CUR, CURtain gas; CXP, Collision-cell eXit Potential; DP, Declustering Potential; EP, Entrance Potential; ESIMS, ElectroSpray Ionization Mass Spectrometry; GS1, Gas 1; GS2, Gas 2; HMBC, Heteronuclear Multiple-Bond Correlation spectroscopy; HMQC, Heteronuclear Multiple-Quantum Correlation spectroscopy; HRESI/APCIMS, High Resolution Electrospray Ionization/Atmospheric Pressure Chemical Ionization Mass Spectrometry (dual probe); HRMS, High Resolution Mass Spectrometry; HSL, HomoSerine Lactone; HSQC, Heteronuclear Single-Quantum Correlation spectroscopy; IS, IonSpray voltage; MRM, Multiple Reaction Monitoring; ROESY, Rotating frame nuclear Overhauser Effect SpectroscopY; RT-qPCR, Real-time quantitative Polymerase Chain Reaction after Reverse Transcription; TEM, TEMperature
Notes and references
- 1.Pomianek ME, Semmelhack MF. ACS Chem. Biol. 2007;2:293–295. doi: 10.1021/cb700098c. [DOI] [PubMed] [Google Scholar]
- 2.Dobretsov S, Teplitski M, Paul V. Biofouling. 2009;25:413–427. doi: 10.1080/08927010902853516. [DOI] [PubMed] [Google Scholar]
- 3.McDougald D, Rice SA, Kjelleberg S. Anal. Bioanal. Chem. 2007;387:445–453. doi: 10.1007/s00216-006-0761-2. [DOI] [PubMed] [Google Scholar]
- 4.Nasser W, Reverchon S. Anal. Bioanal. Chem. 2007;387:381–390. doi: 10.1007/s00216-006-0702-0. [DOI] [PubMed] [Google Scholar]
- 5.Suga H, Smith KM. Curr. Opin. Chem. Biol. 2003;7:586–591. doi: 10.1016/j.cbpa.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 6. [Accessed: December 1st 2010];Centers for Disease Control and Prevention, A Public Health Action Plan to Combat Antimicrobial Resistance, Part I: Domestic Issues. Available: http://www.cdc.gov/drugresistance/actionplan/aractionplan.pdf.
- 7.Heinemann JA, Ankenbauer RG, Amábile-Cuevas CF. Drug Discov. Today. 2000;5:195–204. doi: 10.1016/s1359-6446(00)01483-5. [DOI] [PubMed] [Google Scholar]; Čižman M. Int. J. Antimicrob. Agents. 2003;21:297–307. doi: 10.1016/s0924-8579(02)00394-1. [DOI] [PubMed] [Google Scholar]
- 8.Engel S, Jensen PR, Fenical W. J. Chem. Ecol. 2002;28:1971–1985. doi: 10.1023/a:1020793726898. [DOI] [PubMed] [Google Scholar]
- 9.Willcox MDP, Zhu H, Conibear TCR, Hume EBH, Givskov M, Kjelleberg S, Rice SA. Microbiol. 2008;154:2184–2194. doi: 10.1099/mic.0.2008/019281-0. [DOI] [PubMed] [Google Scholar]
- 10.Diggle SP, Cornelis P, Williams P, Cámara M. Int. J. Med. Microbiol. 2006;296:83–91. doi: 10.1016/j.ijmm.2006.01.038. [DOI] [PubMed] [Google Scholar]
- 11.Schuster M, Greenberg EP. Int. J. Med. Microbiol. 2006;296:73–81. doi: 10.1016/j.ijmm.2006.01.036. [DOI] [PubMed] [Google Scholar]
- 12.Schuster M, Lostroh CP, Ogi T, Greenberg EP. J. Bacteriol. 2003;185:2066–2079. doi: 10.1128/JB.185.7.2066-2079.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wagner VE, Bushnell D, Passador L, Brooks AI, Iglewski BH. J. Bacteriol. 2003;185:2080–2095. doi: 10.1128/JB.185.7.2080-2095.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diggle SP, Winzer K, Chhabra SR, Worrall KE, Cámara M, Williams P. Mol. Microbiol. 2003;50:29–43. doi: 10.1046/j.1365-2958.2003.03672.x. [DOI] [PubMed] [Google Scholar]
- 15.Newman DJ, Cragg GM. J. Nat. Prod. 2007;70:461–477. doi: 10.1021/np068054v. [DOI] [PubMed] [Google Scholar]
- 16.Cragg GM, Grothaus PG, Newman DJ. Chem. Rev. 2009;109:3012–3043. doi: 10.1021/cr900019j. [DOI] [PubMed] [Google Scholar]
- 17.Ryan RP, Dow JM. Microbiol. 2008;154:1845–1858. doi: 10.1099/mic.0.2008/017871-0. [DOI] [PubMed] [Google Scholar]
- 18.Givskov M, de Nys R, Manefield M, Gram L, Maximilien R, Eberl L, Molin S, Steinberg PD, Kjelleberg S. J. Bacteriol. 1996;178:6618–6622. doi: 10.1128/jb.178.22.6618-6622.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koch B, Liljefors T, Persson T, Nielsen J, Kjelleberg S, Givskov M. Microbiol. 2005;151:3589–3602. doi: 10.1099/mic.0.27954-0. [DOI] [PubMed] [Google Scholar]
- 20.Defoirdt T, Miyamoto CM, Wood TK, Meighen EA, Sorgeloos P, Verstraete W, Bossier P. Environ. Microbiol. 2007;9:2486–2495. doi: 10.1111/j.1462-2920.2007.01367.x. [DOI] [PubMed] [Google Scholar]
- 21.Clark BR, Engene N, Teasdale ME, Rowley DC, Matainaho T, Valeriote FA, Gerwick WH. J. Nat. Prod. 2008;71:1530–1537. doi: 10.1021/np800088a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Skindersoe ME, Ettinger-Epstein P, Rasmussen TB, Bjarnsholt T, de Nys R, Givskov M. Mar. Biotechnol. 2008;10:56–63. doi: 10.1007/s10126-007-9036-y. [DOI] [PubMed] [Google Scholar]
- 23.Kwan JC, Teplitski M, Gunasekera SP, Paul VJ, Luesch H. J. Nat. Prod. 2010;73:463–466. doi: 10.1021/np900614n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dobretsov S, Teplitski M, Alagely A, Gunasekera SP, Paul VJ. Env. Microbiol. Rep. 2010;2:739–744. doi: 10.1111/j.1758-2229.2010.00169.x. [DOI] [PubMed] [Google Scholar]
- 25.Lowery CA, Dickerson TJ, Janda KD. Chem. Soc. Rev. 2008;37:1337–1346. doi: 10.1039/b702781h. [DOI] [PubMed] [Google Scholar]
- 26.Mena KD, Gerba CP. Rev. Environ. Contam. Toxicol. 2009;201:71–115. doi: 10.1007/978-1-4419-0032-6_3. [DOI] [PubMed] [Google Scholar]
- 27.Kimata N, Nishino T, Suzuki S, Kogure K. Microb. Ecol. 2004;47:41–47. doi: 10.1007/s00248-003-1032-9. [DOI] [PubMed] [Google Scholar]
- 28.He J, Baldini RL, Déziel E, Saucier M, Zhang Q, Liberati NT, Lee D, Urbach J, Goodman HM, Rahme LG. Proc. Natl. Acad. Sci. U. S. A. 2004;101:2530–2535. doi: 10.1073/pnas.0304622101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sitachitta N, Gerwick WH. J. Nat. Prod. 1998;61:681–684. doi: 10.1021/np970576a. [DOI] [PubMed] [Google Scholar]
- 30.MacMillan JB, Molinski TF. J. Nat. Prod. 2005;68:604–606. doi: 10.1021/np049596k. [DOI] [PubMed] [Google Scholar]
- 31.The S,S isomer of 4 was prepared as an intermediate in the synthesis of the enantiomer of genadamide. [α]24D + 14.8 (c 1.05, CHCl3), in: Al Dulayymi JR, Baird MS, Jones, K K. Tetrahedron. 2004;60:341–345.
- 32.Harrigan GG, Luesch H, Yoshida WY, Moore RE, Nagle DG, Biggs J, Park PU, Paul VJ. J. Nat. Prod. 1999;62:464–467. doi: 10.1021/np980460u. [DOI] [PubMed] [Google Scholar]
- 33.Winson MK, Swift S, Fish L, Throup JP, Jørgensen F, Chhabra SR, Bycroft BW, Williams P, Stewart GSAB. FEMS Microbiol. Lett. 1998;163:185–192. doi: 10.1111/j.1574-6968.1998.tb13044.x. [DOI] [PubMed] [Google Scholar]
- 34.Rajamani S, Bauer WD, Robinson JB, Farrow JM, Pesci EC, Teplitski M, Gao M, Sayre RT, Phillips DA. Mol. Plant-Microbe Interact. 2008;21:1184–1192. doi: 10.1094/MPMI-21-9-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shaw PD, Ping G, Daly SL, Cha C, Cronan JE, Rinehart KL, Farrand SK. Proc. Natl. Acad. Sci. U. S. A. 1997;94:6036–6041. doi: 10.1073/pnas.94.12.6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rennert B, Melzig MF. Planta Med. 2002;68:767–769. doi: 10.1055/s-2002-34411. [DOI] [PubMed] [Google Scholar]
- 37.Müh U, Schuster M, Heim R, Singh A, Olson ER, Greenberg EP. Antimicrob. Agents Chemother. 2006;50:3674–3679. doi: 10.1128/AAC.00665-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pesci EC, Pearson JP, Seed PC, Iglewski BH. J. Bacteriol. 1997;179:3127–3132. doi: 10.1128/jb.179.10.3127-3132.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gould TA, Schweizer HP, Churchill MEA. Mol. Microbiol. 2004;53:1135–1146. doi: 10.1111/j.1365-2958.2004.04211.x. [DOI] [PubMed] [Google Scholar]; Pappas KM, Weingart CL, Winans SC. Mol. Microbiol. 2004;53:755–769. doi: 10.1111/j.1365-2958.2004.04212.x. [DOI] [PubMed] [Google Scholar]
- 40.Mavrodi DV, Bonsall RF, Delaney SM, Soule MJ, Phillips G, Thomashow LS. J. Bacteriol. 2001;183:6454–6465. doi: 10.1128/JB.183.21.6454-6465.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hentzer M, Wu H, Andersen JB, Riedel K, Rasmussen TB, Bagge N, Kumar N, Schembri MA, Song Z, Kristoffersen P, Manefield M, Costerton JW, Molin S, Eberl L, Steinberg P, Kjelleberg S, Høiby N, Givskov M. EMBO J. 2003;22:3803–3815. doi: 10.1093/emboj/cdg366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ochsner UA, Wilderman PJ, Vasil AI, Vasil ML. Mol. Microbiol. 2002;45:1277–1287. doi: 10.1046/j.1365-2958.2002.03084.x. [DOI] [PubMed] [Google Scholar]
- 43.Ravel J, Cornelis P. Trends Microbiol. 2003;11:195–200. doi: 10.1016/s0966-842x(03)00076-3. [DOI] [PubMed] [Google Scholar]
- 44.Cornelis P, Aendekerk S. Microbiol. 2004;150:752–756. doi: 10.1099/mic.0.27086-0. [DOI] [PubMed] [Google Scholar]
- 45.Juhas M, Wiehlmann L, Huber B, Jordan D, Lauber J, Salunkhe P, Limpert AS, von Götz F, Steinmetz I, Eberl L, Tümmler B. Microbiol. 2004;150:831–841. doi: 10.1099/mic.0.26906-0. [DOI] [PubMed] [Google Scholar]
- 46.Schuster M, Hawkins AC, Harwood CS, Greenberg EP. Mol. Microbiol. 2004;51:973–985. doi: 10.1046/j.1365-2958.2003.03886.x. [DOI] [PubMed] [Google Scholar]
- 47.Brencic A, Lory S. Mol. Microbiol. 2009;72:612–632. doi: 10.1111/j.1365-2958.2009.06670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lequette Y, Lee J-H, Ledgham F, Lazdunski A, Greenberg EP. J. Bacteriol. 2006;188:3365–3370. doi: 10.1128/JB.188.9.3365-3370.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heurlier K, Dénervaud V, Pessi G, Reimmann C, Haas D. J. Bacteriol. 2003;185:2227–2235. doi: 10.1128/JB.185.7.2227-2235.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Déziel E, Gopalan S, Tampakaki AP, Lépine F, Padfield KE, Saucier M, Xiao G, Rahme LG. Mol. Microbiol. 2005;55:998–1014. doi: 10.1111/j.1365-2958.2004.04448.x. [DOI] [PubMed] [Google Scholar]
- 51.Liang H, Li L, Dong Z, Surette MG, Duan K. J. Bacteriol. 2008;190:6217–6227. doi: 10.1128/JB.00428-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Heeb S, Haas D. Mol. Plant-Microbe Interact. 2001;14:1351–1363. doi: 10.1094/MPMI.2001.14.12.1351. [DOI] [PubMed] [Google Scholar]
- 53.Sakuragi Y, Kolter R. J. Bacteriol. 2007;189:5383–5386. doi: 10.1128/JB.00137-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ueda A, Wood TK. PLoS Pathog. 2009;5:e1000483. doi: 10.1371/journal.ppat.1000483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mougous JD, Cuff ME, Raunser S, Shen A, Zhou M, Gifford CA, Goodman AL, Joachimiak G, Ordoñez CL, Lory S, Walz T, Joachimiak A, Mekalanos JJ. Science. 2006;312:1526–1530. doi: 10.1126/science.1128393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brencic A, McFarland KA, McManus HR, Castang S, Mogno I, Dove SL, Lory S. Mol. Microbiol. 2009;73:434–445. doi: 10.1111/j.1365-2958.2009.06782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Son MS, Matthews WJ, Kang Y, Nguyen DT, Hoang TT. Infect. Immun. 2007;75:5313–5324. doi: 10.1128/IAI.01807-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kang Y, Nguyen DT, Son MS, Hoang TT. Microbiol. 2008;154:1584–1598. doi: 10.1099/mic.0.2008/018135-0. [DOI] [PubMed] [Google Scholar]
- 59.Zhu K, Choi K-H, Schweizer HP, Rock CO, Zhang Y-M. Mol. Microbiol. 2006;60:260–273. doi: 10.1111/j.1365-2958.2006.05088.x. [DOI] [PubMed] [Google Scholar]
- 60.D'Argenio DA, Wu M, Hoffman LR, Kulasekara HD, Déziel E, Smith EE, Nguyen H, Ernst RK, Freeman TJL, Spencer DH, Brittnacher M, Hayden HS, Selgrade S, Klausen M, Goodlett DR, Burns JL, Ramsey BW, Miller SI. Mol. Microbiol. 2007;64:512–533. doi: 10.1111/j.1365-2958.2007.05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van Delden C, Comte R, Bally M. J. Bacteriol. 2001;183:5376–5384. doi: 10.1128/JB.183.18.5376-5384.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Baysse C, Cullinane M, Dénervaud V, Burrowes E, Dow JM, Morrisey JP, Tam L, Trevors JT, O'Gara F. Microbiol. 2005;151:2529–2542. doi: 10.1099/mic.0.28185-0. [DOI] [PubMed] [Google Scholar]
- 63.Gao M, Teplitski M, Robinson JB, Bauer WD. Mol. Plant-Microbe Interact. 2000;16:827–834. doi: 10.1094/MPMI.2003.16.9.827. [DOI] [PubMed] [Google Scholar]; Teplitski M, Robinson JB, Bauer WD. Mol. Plant-Microbe Interact. 2000;13:637–648. doi: 10.1094/MPMI.2000.13.6.637. [DOI] [PubMed] [Google Scholar]
- 64.Watson D, MacDermot J, Wilson R, Cole PJ, Taylor GW. Eur. J. Biochem. 1986;159:309–313. doi: 10.1111/j.1432-1033.1986.tb09869.x. [DOI] [PubMed] [Google Scholar]
- 65.Note that for preliminary experiments (see Figure 6b), different assay conditions were employed to quantify LasB activity because smaller scale cultures (500 µL) were used. A portion of supernatants (50 µL) was incubated for 5 h at 37 °C with shaking in the presence of elastin Congo red (200 µL of a 12.5 mg/mL suspension in 30 mM Tris-HCl, pH 7.2), after which the reaction was spun down and the absorbance at 495 nm measured
- 66.Copeland RA. Evaluation of Enzyme Inhibitors In Drug Discovery: A Guide for Medicinal Chemists and Pharmacologists. Hoboken: John Wiley & Sons; 2005. pp. 48–81. [PubMed] [Google Scholar]; Copeland RA. Evaluation of Enzyme Inhibitors In Drug Discovery: A Guide for Medicinal Chemists and Pharmacologists. Hoboken: John Wiley & Sons; 2005. pp. 111–140. [PubMed] [Google Scholar]
- 67. [Accessed: December 1st 2010];Affymetrix, GeneChip Expression Analysis Technical Manual. Available: http://media.affymetrix.com/support/downloads/manuals/expression_analysis_technical_manual.pdf.
- 68.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JYH, Zhang J. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bolstad BM, Irizarry RA, Åstrand M, Speed TP. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 70.Smyth GK. Stat. Appl. Genet. Mol. Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article 3. [DOI] [PubMed] [Google Scholar]
- 71.Benjamini Y, Hochberg Y. J. Royal Stat. Soc. B. 1995;57:289–300. [Google Scholar]
- 72.Edgar R, Domrachev M, Lash AE. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Savli H, Karadenizli A, Kolayli F, Gundes S, Ozbek U, Vahaboglu H. J. Med. Microbiol. 2003;52:403–408. doi: 10.1099/jmm.0.05132-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.