Abstract
T-box transcription factors are critical developmental regulators in all multi-cellular organisms, and altered T-box factor activity is associated with a variety of human congenital diseases and cancers. Despite the biological significance of T-box factors, their mechanism of action is not well understood. Here we examine whether SUMOylation affects the function of the C. elegans Tbx2 sub-family T-box factor TBX-2. We have previously shown that TBX-2 interacts with the E2 SUMO-conjugating enzyme UBC-9, and that loss of TBX-2 or UBC-9 produces identical defects in ABa-derived pharyngeal muscle development. We now show that TBX-2 is SUMOylated in mammalian cell assays, and that both UBC-9 interaction and SUMOylation depends on two SUMO consensus sites located in the T-box DNA binding domain and near the TBX-2 C-terminus, respectively. In co-transfection assays, a TBX-2:GAL4 fusion protein represses expression of a 5xGal4:tk:luciferase construct. However, this activity does not require SUMOylation, indicating SUMO is not generally required for TBX-2 repressor activity. In C. elegans, reducing SUMOylation enhances the phenotype of a temperature-sensitive tbx-2 mutant and results in ectopic expression of a gene normally repressed by TBX-2, demonstrating that SUMOylation is important for TBX-2 function in vivo. Finally, we show mammalian orthologs of TBX-2, Tbx2, and Tbx3, can also be SUMOylated, suggesting SUMOylation may be a conserved mechanism controlling T-box factor activity.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-013-1336-y) contains supplementary material, which is available to authorized users.
Keywords: T-box, SUMOylation, C. elegans, TBX-2, Genetic enhancer
Introduction
T-box proteins are a family of transcription factors found in all multicellular animals where they play important roles in the development of a variety of tissues [1, 2]. The defining feature of this family is the conserved T-box DNA-binding domain, and T-box factors are grouped into distinct sub-families based on sequence conservation within this domain. In many cases, the level of T-box factor activity is crucial to normal function. For example, reduced expression of the human Tbx2 sub-family genes TBX3, TBX4, and TBX5 resulting from loss of one functional allele results in ulnar-mammary syndrome, small patella syndrome, and Holt-Oram syndrome, respectively [3–7]. In contrast, over-expression of the Tbx2-subfamily genes TBX2 and TBX3 is found in a number of human cancers [8]. Despite their developmental and clinical importance, relatively little is known about the mechanism by which T-box factors function.
We are interested in the role that SUMOylation plays in T-box factor activity. SUMOylation is the covalent and reversible post-translational attachment of the small ubiquitin-like modifier peptide (SUMO) to specific lysine residues in target proteins [9, 10], and it has been implicated in diverse processes, including modifying function, nuclear localization, and sub-nuclear localization of transcriptional regulators [11]. SUMOylation of transcription factors is typically associated with repression [12], but it has also been implicated in transcriptional activation by some factors [13, 14]. The SUMO conjugation pathway is analogous to the ubiquitination pathway and involves an E1-activating enzyme (Aos1/Uba2) and an E2 conjugating enzyme (Ubc9) sufficient for specific SUMO attachment in vitro [15, 16]. In addition, a variety of E3 ligases have been identified that promote SUMO transfer from E2 to specific substrates in vivo. Ubc9 recognizes the ΨKX(D/E) SUMO consensus site (where Ψ is a large hydrophobic amino acid and K is the residue attached to SUMO) [17, 18], and many SUMOylation substrates have been identified by their interaction with Ubc9 in yeast two-hybrid screens [19]. SUMOylation also occurs at non-consensus sites, and non-covalent SUMO/substrate or E3 ligase/substrate interactions are involved in directing SUMOylation at these sites [9].
We hypothesize that function of the C. elegans T-box factor TBX-2 depends on SUMOylation [20]. TBX-2 is the sole C. elegans member of the Tbx2 subfamily and is necessary for formation of anterior pharyngeal muscles. In yeast two-hybrid assays, TBX-2 interacts with the E2 SUMO conjugating enzyme UBC-9, and loss of UBC-9 produces pharyngeal phenotypes identical to those resulting from tbx-2 loss-of-function. In addition, sub-nuclear localization of a TBX-2::GFP fusion protein is altered when SUMOylation is reduced.
Here, we ask if TBX-2 is SUMOylated and whether SUMOylation affects TBX-2 activity in vivo. We first used the two-hybrid assay to map interaction sites between TBX-2 and UBC-9 and found two SUMO consensus sites in TBX-2 that mediate interaction with UBC-9. One of these sites is located near the TBX-2 C-terminus, while the other is located in a highly conserved region of the T-box DNA binding domain. We next showed that TBX-2 is SUMOylated in mammalian cell assays, and that TBX-2 SUMOylation depends on both of these UBC-9 interaction sites. We then examined TBX-2 transcriptional activity and found that in mammalian cells a TBX-2-GAL4 DNA-binding domain (GAL4-DBD) fusion protein represses expression of a GAL4-responsive reporter, but surprisingly this repression did not require SUMOylation. To determine whether SUMOylation is important for TBX-2 activity in vivo, we asked if tbx-2 and ubc-9 interact genetically. We found that reduction of SUMOylation enhances the effect of a tbx-2 hypomorphic mutant on embryonic viability and pharyngeal muscle development, and that repression of a downstream target of TBX-2 depends on SUMOylation. Finally, we examined SUMOylation of two mammalian orthologs of TBX-2 and found that human TBX2 and mouse Tbx3 can also be SUMOylated. We suggest SUMOylation is a common mechanism regulating activity of T-box transcription factors.
Materials and methods
Nematode handling, transformation, and strains
C. elegans were grown under standard conditions [21]. Germ line transformation was performed using standard techniques with pRF4 containing rol-6(su1006) as a dominant marker for transformation [22]. The following strains were used in these studies: OK0660 tbx-2(bx59) obtained by outcrossing from EM207 tbx-2(bx59); him-5(e1490); OK0666 cuEx553[D2096.6::gfp]; OK0692 tbx-2(bx59); cuEx553[D2096.6::gfp]; OK0741 tbx-2(ok529)/dpy-17(e164) unc-32(e189); cuEx553[D2096.6::gfp].
Genotyping tbx-2(bx59) mutants
tbx-2(bx59) is a G→A substitution located at position 24,597 of the cosmid F21H11 (accession FO081200) (K. Chow, pers. comm.) and disrupts a BstCI restriction enzyme site. Animals were genotyped by single worm PCR [23] using primers PO931 [AGTTTGACACCGATTTTCTCG] and PO932 [GTGATGATGGATCTTGTTCCG] followed by digestion with BstCI and gel electrophoresis.
General methods for nucleic acid manipulations and plasmid construction
Standard methods were used to manipulate plasmid DNAs and oligonucleotides [24], and all plasmid sequences are available from the authors. For yeast two-hybrid assays, the LKIE and VKKE SUMOylation sites were separately mutated using the Stratagene QuikChange II Kit in the tbx-2 bait plasmid pOK187.01 containing the full-length tbx-2 orf [20] to generate pOK222.01 and pOK222.06, respectively. The LKIE/VKKE→AAAA double mutant was constructed by ligation of fragments pOK222.01 and pOK226.06 to create the plasmid pOK225.02. Plasmids for expressing TBX-2 (pOK241.05), TBX-2LKIE→AAAA (pOK241.10), TBX-2VKKE→AAAA (pOK241.13), and TBX-2LKIE/VKKE→AAAA (pOK241.17) were constructed by inserting the PCR amplified tbx-2 orf from the two-hybrid vectors into pCDNA3.1 using TOPO cloning (Invitrogen). Plasmids for expressing TBX-2K231R (pOK263.01), TBX-2K400R (pOK244.18), and the TBX-22KR double mutant (pOK261.03) were made by site-directed mutagenesis of pOK241.05 using the Stratagene QuikChange II Kit. The plasmid encoding HA::SUMO-1 (pcDNA3 HA SUMO-1, pOK251.01) was a gift from Jorge A. Iñiguez-Lluhí (University of Michigan), and it was mutated using the Stratagene QuikChange II Kit to encode HA::SUMO-1(∆GG) (pOK263.05). cDNA clones for human TBX2 (IMAGE:6339405) and mouse Tbx3 (IMAGE:30547736) were purchased from Open Biosystems and inserted into pCDNA3.1 using TOPO cloning to make pOK246.01 and pOK245.01.
Plasmids for expressing TBX-2:GAL4 (pOK253.01) and TBX-2LKIE/VKKE→AAAA:GAL4 (pOK253.04) for co-transfection assays were made by cloning the amplified tbx-2 orf from pOK241.05 and 241.17, respectively, into pcDNA HA:GAL4(1-100) (provided by Jorge A. Iñiguez-Lluhí, University of Michigan). For mock transfections, the HA:GAL4 fragment was removed from pcDNA HA:GAL4(1–100) (pOK293.03). The 5xGAL4:tk:luc reporter was a gift from Elizaveta Benevolenskaya, University of Illinois at Chicago.
Yeast two-hybrid assays
Yeast 2-hybrid assays were carried out in L40 yeast containing HIS3 and lacZ reporters regulated by LexA binding sites with the ubc-9 prey plasmid pOK193.11 in the pACT vector and tbx-2 bait plasmids pOK187.01 (wild-type tbx-2), pOK222.01 (tbx-2 LKIE→AAAA), pOK222.06 (tbx-2 VKKE→AAAA), or pOK225.02 (tbx-2 LKIE/VKKE→AAAA) in the pLexA-NLS vector as previously described [20]. ß-galactosidase expression in yeast was quantified in at least three assays as previously described [25].
RNAi analyses
Feeding RNAi was performed as previously described [26] using plasmids obtained from Geneservice containing genomic fragments of ubc-9 or smo-1 cloned into L4440 [27]. To assess enhancement of the tbx-2(bx59) mutant phenotype, N2 or OK0660 [tbx-2(bx59)] L4 hermaphrodites raised at 16 °C were transferred to plates seeded with RNAi feeding E. coli or OP50 and incubated at 25 °C for 24 h. These animals were transferred to fresh feeding plates at 25 °C and allowed to lay eggs for 4 h. Progeny embryos were transferred to fresh feeding plates and counted. Larvae and terminally arrested embryos were counted 24 h later to assay embryonic lethality, or examined by DIC microscopy after hatching to assess the pharyngeal phenotype.
To examine D2096.6::gfp expression, OK0666 [cuEx553] L4 hermaphrodites were transferred to plates seeded with ubc-9 RNAi feeding E. coli or OP50 and grown 20 h at 20 °C. These animals were transferred to fresh feeding plates, and GFP expression was examined in progeny embryos and larvae.
SUMOylation and co-transfection assays
COS-1 cells were maintained in D-MEM with 10 % FBS, 10 mM HEPES, and 1× Antibiotic–Antimycotic (Invitrogen). For SUMOylation assays, ~2 × 106 cells were seeded into 10-cm plates 24 h prior to transfection. Plates were transfected with plasmids expressing wild-type or mutant TBX-2 (10 μg), HA-SUMO-1 or HA-SUMO-1(ΔGG) (10 μg), and peGFP-N3 (4 μg; Clontech) using Lipofectamine 2000 in OPTI-MEM following the manufacturer’s instructions (Invitrogen). After 48 h, COS-1 cells were harvested in PBS, lysed in 0.75 ml lysis buffer [8 M urea, 0.5 M NaCl, 45 mM Na2HPO4, 5 mM NaH2PO4,10 mM imidazole, 10 mM NEM (pH 8.0)], sonicated, and incubated with 50 μl Ni–NTA magnetic beads (Qiagen). Beads were washed twice with 1 ml wash [8 M urea, 0.4 M NaCl, 17.6 mM Na2HPO4, 32.4 mM NaH2PO4, 10 mM imidazole, 10 mM NEM (pH 6.75)] on a MagnaRack (Invitrogen). Protein was eluted in 50 μl (250 mM imidazole, 5 % SDS, 0.15 M Tris pH6.7, 30 % glycerol, 0.72 M ßME), resolved by SDS-PAGE, and blotted. Proteins were detected using anti-V5 (Invitrogen) or anti-HA (Covance), HRP-conjugated secondary antibody (Goat anti-mouse, Millipore), and ECL Plus (GE Healthcare) detection reagent. Chemiluminescence was recorded using ECL hyperfilm (GE Healthcare) or recorded and quantified using a STORM 860 Molecular Imager and ImageQuant software (Molecular Dynamics).
For co-transfection assays, 2 × 105 COS-1 cells were seeded to wells of a 24-well plate 24 h prior to transfection. Wells were transfected with plasmids expressing TBX-2 (25–500 ng), 5xGAL4:tk:luc (280 ng), and CMV ßgal (20 ng) using Lipofectamine 2000 in OPTI-MEM following the manufacturer’s instructions (Invitrogen) and harvested. Luciferase and ß-gal were measured in triplicate samples using Steady Glo Luciferase system (Promega) and a Clarity Luminescence Micro-plate reader (BIO-TEK), and ßgal activity was measured with a Genesys 10 UV spectrophotometer (Thermo-Fisher) [28].
Microarray and data analysis
Mixed stage populations of N2 and OK0660 [tbx-2(bx59)] animals grown at 25 °C were treated with bleach/sodium hypochlorite to isolate embryos [21]. Aliquots of embryos were examined to verify comparable age distributions, and RNA was isolated using TRIzol (Invitrogen) and further purified using RNeasy Kit (Qiagen) following manufacturers’ protocols.
Total RNA from two independent populations of N2 embryos and three independent tbx-2(bx59) embryos were labeled and hybridized to Affymetrix C. elegans Genome GeneChips by the UIC Core Genomic Facility (CGF). The microarray data was analyzed using the R statistical programming language, using the Bioconductor suite of tools [29], and the Affy package. Normalization to correct for chip-to-chip variation was done using the Robust Multiarray Averaging (RMA) method of microarray normalization [30]. Microarray results were pre-filtered using the genefilter function (25 % of the probes have a measured intensity of at least 100 on the original scale and the coefficient of variation is between 0.7 and 10 on the original scale) [31]. The limma package [32] was used to calculate differential expression using the limma linear model fit, eBayes smoothing of standard errors, and Benjamini-Hochberg (BH) multiple test correction with a false discovery rate of 5 % [33]. Probes were matched to genes using the Affymetrix-to-WormBase ID table for WS210 (http//:www.wormbase.org). Probes mapping to more than one gene were discarded. When one or more probes mapping to a gene were differentially expressed, that gene was considered to be differentially expressed. One GeneChip hybridized with tbx-2(bx59) RNA exhibited high variation in the spiked in control probes (TBXa) compared to the other samples, and data from this chip was not included in our analysis.
Microscopy
Worms were visualized using a Zeiss Axioskop microscope equipped for DIC and fluorescence microscopy, and images were captured using an Axiocam MRm camera and AxioVision software.
Results
TBX-2 interacts with UBC-9 via two SUMO consensus sites
We previously showed using yeast two-hybrid assays that TBX-2 specifically interacts with the E2 SUMO-conjugating enzyme UBC-9 [20], and we used this assay to identify sites in TBX-2 mediating this interaction. TBX-2 contains several sites matching the SUMO consensus site ΨKX(D/E) (Supplementary Table 1) [17, 18]. The two highest-scoring matches to this consensus are an LK231IE sequence located near the C-terminus of the T-box DNA binding domain and a VK400KE sequence located near the TBX-2 C-terminus (Fig. 1a). LKIE is located in a conserved region of the T-box, and a SUMO consensus site is found at this position in many T-box factors, including all members of the Tbx2 sub-family [34]. VKKE is located in a region that is not highly conserved among T-box factors, although high scoring SUMO consensus sites are found near the C-terminus of TBX-2 proteins from C. elegans, C. briggsae, and C. remanei, suggesting this site may be functionally conserved (Fig. 1b). We mutated each of these two sites in C. elegans TBX-2 to all alanines either in single mutants (LKIE→AAAA or VKKE→AAAA) or in a double mutant (LKIE/VKKE→AAAA) and tested whether these mutants affected the ability of a TBX-2 bait to interact with UBC-9 prey. Interactions were scored in plate assays for histidine prototrophy and ß-galactosidase (ß-gal) expression, and the level of interaction was quantified by measuring ß-gal activity.
Fig. 1.
TBX-2 interacts with UBC-9. a Schematic diagram of the TBX-2 protein (Accession CCD69847) indicating the location of the T-box DNA binding domain (black) and the positions of the LK231IE and VK400KE SUMO consensus sites. b T-Coffee alignment of the C-terminus of TBX-2 proteins from C. elegans (CeTBX-2), C. briggsae (CbTBX-2; WormBase ID CBP05056), and C. remanei (CrTBX-2; WormBase ID RP21057) [62]. High-scoring SUMO consensus sites are indicated in grey, and identical residues are marked with asterisks. c Quantification of ß-galactosidase activity in yeast expressing the indicated TBX-2 protein or the empty pLexA as bait and UBC-9 prey in replicate samples from three independent experiments (n = 7). Differences between mutants and wild-type TBX-2 or different mutants (bracket) are statistically significant at *p < 0.05 or **p < 0.005. Error bars indicate the standard error of the mean
We found that UBC-9 interaction with TBX-2 was affected by mutations affecting both the LKIE and VKKE sites. UBC-9 interacted with both the TBX-2LKIE→AAAA and TBX-2VKKE→AAAA single mutants in plate assays, but this interaction was reduced to 63 and 22 % of the levels observed for wild-type TBX-2, respectively (Fig. 1b). In comparison, UBC-9 failed to interact with the TBX-2LKIE/VKKE→AAAA double mutant in plate assays, and the ß-gal activity was close to that obtained using an empty bait plasmid (Fig. 1b, Supplementary Figure 1). As a control, we found that wild-type and all of the mutant TBX-2 proteins retained the ability to interact with an unrelated protein UNC-37 in yeast two-hybrid assays, indicating the mutant proteins were expressed (Supplementary Figure 1). Thus, both the LKIE and VKKE sites can interact with UBC-9. Because mutating both of these sites reduces interaction to near background, we believe they are the primary sites in TBX-2 that mediate this interaction. We have not tested other potential TBX-2 SUMOylation sites for interaction with UBC-9 in two-hybrid assays.
TBX-2 can be SUMOylated in mammalian cell assays
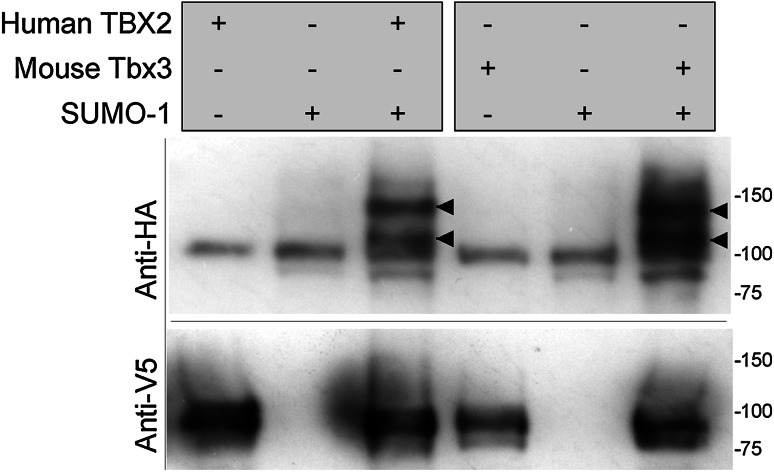
To determine if TBX-2 can be SUMOylated, we co-expressed full-length TBX-2 and human SUMO-1 in COS-1 cells. TBX-2 was fused to poly-histidine and pulled down using Ni2+-beads under denaturing conditions, while TBX-2 and SUMO-1 were tagged with V5 and HA epitope tags, respectively, for detection on Western blots. A SUMO-1∆GG mutant lacking the C-terminal Gly–Gly motif required for conjugation to target lysine residues was used as a control to demonstrate SUMO conjugation.
When co-expressed with SUMO-1, wild-type TBX-2 formed several more slowly migrating bands detectable with both anti-HA and anti-V5 (Fig. 2a). These bands likely represent mono- and multi-SUMOylated TBX-2, although proteins SUMOylated at different sites can also migrate at different positions due to the branched nature of the SUMOylated protein [35]. In comparison, no SUMOylated TBX-2 was detected when co-expressed with SUMO-1∆GG. As for many SUMOylated proteins, we found only a fraction of TBX-2 (~10 %) is SUMOylated in these assays. Similar results were obtained when TBX-2 was co-expressed with human SUMO-2 or SUMO-3 (Supplementary Figure 2).
Fig. 2.
SUMOylation of TBX-2 is mediated via two SUMO consensus sites. Western blots of Ni–NTA pulled-down wild-type and mutant TBX-2/V5/HIS probed to detect TBX-2 (bottom) or SUMO-1 (top). Combinations of proteins (grey boxes) were expressed in COS-1 cells. a Wild-type TBX-2 and mutants with SUMOylation sites converted to all alanines. b Wild-type TBX-2 and mutants with SUMO acceptor lysines converted to arginines. The position of the fastest migrating SUMOylated form of TBX-2 is indicated (bar and circle) and the position of molecular weight markers are indicated in kDa (bars). Signal in the lower panel in A was detected using a STORM Molecular Imager and clearly demonstrates more slowly migrating TBX-2 bands when co-transfected with SUMO-1. TBX-2/V5/HIS is ~52 kDa, and HA-SUMO-1 is ~13 kDa. The fastest migrating SUMOylated form migrates somewhat slower than predicted by its molecular weight, which is a common feature of SUMOylated proteins
We next asked how mutations in the UBC-9 interaction sites affected TBX-2 SUMOylation. SUMOylation of TBX-2LKIE→AAAA and TBX-2VKKE→AAAA were reduced to approximately 30 and 70 % of the level of wild-type TBX-2, respectively, while SUMOylation of the TBX-2LKIE/VKKE→AAAA double mutant was further reduced to a level comparable to background (Fig. 2a, lanes 2 and 9). These results indicate that TBX-2 can be SUMOylated, and that the LKIE and VKKE sites for UBC-9 interaction are required for TBX-2 SUMOylation.
We next mutated the SUMO-conjugated lysine residues in the LKIE and VKKE sites to arginine, which is a conservative substitution that cannot be conjugated to SUMO, and we examined SUMOylation in COS-1 cells (Fig. 2b). The TBX-2K231R mutant affecting LKIE exhibited reduction of the more slowly migrating SUMOylated products, but these products were not eliminated, while the fastest migrating band appeared unaffected. In comparison, overall SUMOylation of the TBX-2K400R mutant affecting VKKE was strongly reduced, and the fast migrating form of SUMOylated TBX-2 was nearly completely eliminated. SUMOylation of the TBX-22KR double mutant containing K231R and K400R was similar to that of the TBX-2K400R single mutant, but this mutant was still SUMOylated above background levels (compare Fig. 2b, lanes 2 and 9). Because both of these conservative mutations affect the pattern of TBX-2 SUMOylation, we believe lysine residues in both the LKIE and VKKE SUMO consensus sites are SUMOylated.
TBX-2 is a transcriptional repressor in mammalian cells
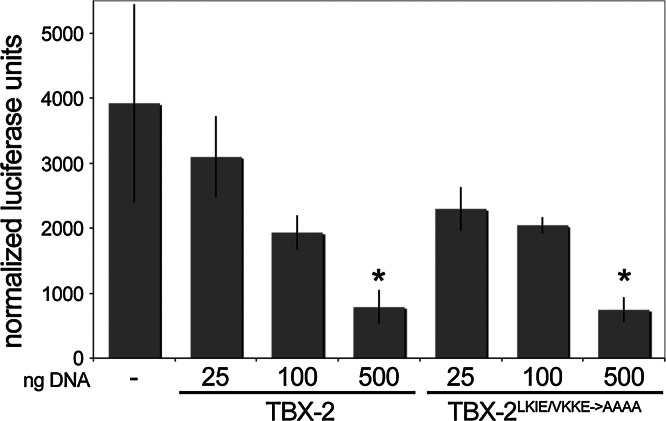
C. elegans TBX-2 is most closely related to the mammalian T-box repressors Tbx2 and Tbx3 [20]. We wanted to ask if TBX-2 functions similarly to repress transcription and, if so, whether this activity depends on SUMOylation. Because mutations affecting the LKIE SUMOylation would likely affect DNA binding, we asked if TBX-2 fused to the heterologous GAL4 DNA binding domain (TBX-2:GAL4) could repress expression of the 5xGAL4:tk:luc reporter. This reporter contains five copies of the GAL4 binding site upstream of thymidine kinase promoter:luciferase reporter, and TBX-2:GAL4 repressed expression of this reporter up to fivefold (Fig. 3). SUMOylation is most often associated with transcriptional repression, and we expected that mutation of the LKIE and VKKE SUMO sites would reduce this repressor activity. However, we found that the TBX-2LKIE/VKKE→AAAA double mutant repressed 5xGAL4:tk:luc similarly to wild-type TBX-2. Co-expressing SUMO-1 did not affect repression of 5xGAL4:tk:luc with either wild-type or mutant TBX-2 (Supplementary Figure 3). Thus, SUMOylation is not required for TBX-2:GAL4 repressor activity in COS-1 cells.
Fig. 3.
Dose-dependent transcriptional repression by TBX-2:GAL4. Relative luciferase activity in experiments co-transfecting increasing amounts of wild-type and mutant TBX-2:GAL4 with the 5xGAL4:tk:luc reporter. Data shown is the average of three assays and is representative of multiple independent experiments. Error bars indicate standard deviation. Statistically significant differences from control transfections are marked (*p < 0.05)
TBX-2 function is SUMO-dependent in C. elegans
mab-22(bx59) is a temperature-sensitive mutant that exhibits defects in male tail ray formation and partially penetrant larval lethality. bx59 has recently been identified as a missense mutation in tbx-2 (King Chow, pers. comm.), and we subsequently refer to this mutation as tbx-2(bx59). We examined the viability and pharyngeal morphology of tbx-2(bx59) mutants produced from hermaphrodites shifted to the non-permissive temperature (25 °C) at the L4 stage. Forty-four % of these animals arrested at the L1 stage (n = 64) with variable pharyngeal abnormalities (Fig. 4a–c). These phenotypes are similar to those observed in tbx-2(RNAi) animals and are not as severe as those observed in tbx-2 null mutants [20, 36], and we conclude that tbx-2(bx59) is a hypomorphic allele.
Fig. 4.
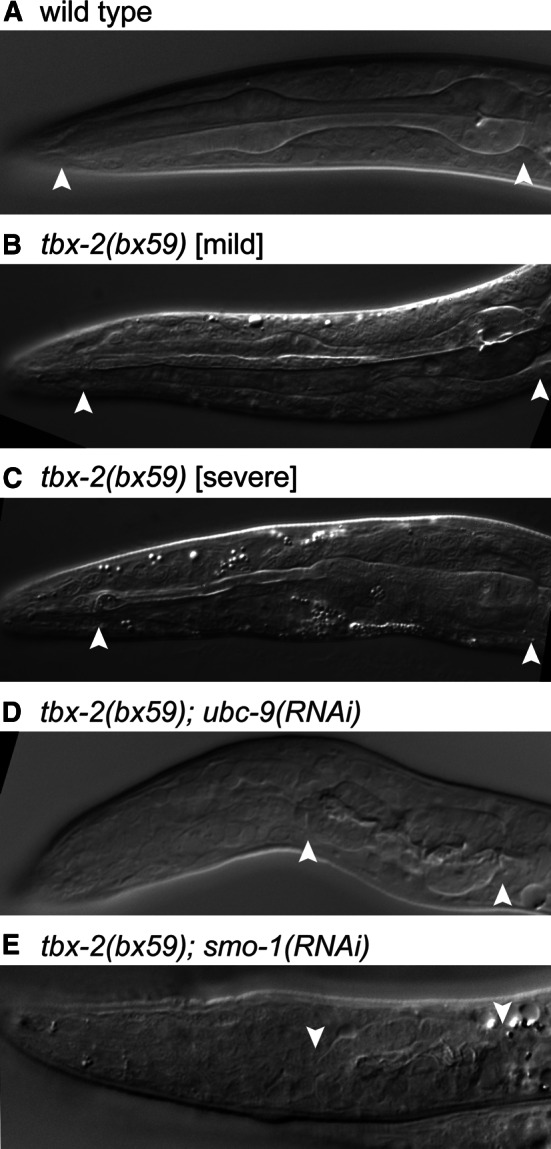
Pharyngeal defects in tbx-2(bx59) mutant are enhanced by reduced SUMOylation DIC micrographs of the pharynx of L1 larvae of the indicated genotypes raised at the non-permissive temperature (25 °C). a Wild-type N2. b tbx-2(bx59) exhibiting a mild pharyngeal defect. c tbx-2(bx59) exhibiting a more severe pharyngeal defect. tbx-2(bx59); ubc-9(RNAi) (d) and tbx-2(bx59); smo-1(RNAi) (e) L1 larvae exhibiting very severe defects resembling those of tbx-2 null mutants. Arrowheads mark the extent of pharyngeal tissue. Anterior is left. The frequency of these phenotypes is indicated in Table 1
We hypothesize that TBX-2 function depends on SUMOylation. To test this hypothesis, we asked if inhibiting SUMOylation by reducing UBC-9 or the SUMO protein SMO-1 by RNAi could enhance the phenotype of tbx-2(bx59) mutants. Using the RNAi-feeding method, we found that ubc-9(RNAi) produced a relatively low frequency of embryonic arrest in a wild-type background (Table 1) [26]. In comparison, tbx-2(bx59); ubc-9(RNAi) double mutants exhibited a synergistic increase in the frequency of arrested embryos (Table 1). tbx-2(bx59); smo-1(RNAi) double mutants also exhibited an increased frequency of embryonic lethality compared to each single mutant; however, the smo-1(RNAi) lethality alone was higher making it difficult to determine if the double mutant lethality was more than additive (Table 1).
Table 1.
Reduction of SUMOylation enhances tbx-2(bx59) embryonic lethality and pharyngeal defects
| Genotypea | % embryonic arrest (n) | Pharyngeal phenotypes in hatched L1s (percentage of total hatched animals) | |||
|---|---|---|---|---|---|
| Severe Tbx-2 pharynx | Mild Tbx-2 pharynx | Wild-type pharynx | n | ||
| tbx-2(bx59) | 7 (120) | 19 | 56 | 25 | 54 |
| ubc-9(RNAi) | 19 (186) | 7 | 6 | 87 | 69 |
| tbx-2(bx59); ubc-9(RNAi) | 78 (200) | 70 | 30 | 0 | 47 |
| smo-1(RNAi) | 65 (347) | 22 | 39 | 39 | 49 |
| tbx-2(bx59); smo-1(RNAi) | 75 (359) | 69 | 26 | 5 | 64 |
aL4 animals raised at 16 °C were shifted to 25 °C, and defects were scored in the F1 progeny
Many of the tbx-2(bx59) mutants that hatch grow to adulthood, but nearly all of the tbx-2(bx59); ubc-9(RNAi) and tbx-2(bx59); smo-1(RNAi) arrested as L1 larvae. We examined newly hatched larvae to determine if this enhanced L1 arrest results from pharyngeal defects. We found that both tbx-2(bx59); ubc-9(RNAi) and tbx-2(bx59); smo-1(RNAi) double mutants exhibited a synergistic increase in the frequency of animals with a severe anterior pharyngeal defect compared to the single mutants (Table 1; Fig. 4). Together, these results strongly suggest that SUMOylation is necessary for TBX-2 function for anterior pharyngeal development.
TBX-2 and SUMOylation are required for repression of D2096.6 gene expression
To identify genes downstream of TBX-2, we used microarrays to compare mRNA levels in populations of wild-type and tbx-2(bx59) embryos grown at 25 °C. We found 1,276 protein coding genes that are differentially expressed in tbx-2(bx59) (BH corrected p ≤ 0.05) (Supplementary Table 2). A total of 1,030 of these genes (80.7 %) are upregulated in tbxc-2(bx59), consistent with our hypothesis that TBX-2 functions as a transcriptional repressor.
We focused on the gene D2096.6, which had previously been shown to be specifically expressed in the pharyngeal muscles, marginal cells, and epithelial cells under control of the FoxA-family transcription factor PHA-4 [37, 38]. We observed an approximately 1.8-fold increase in D2096.6 expression in tbx-2(bx59) mutants in our microarray (BH corrected p = 0.03). While several candidate T-box binding sites are located upstream of D2096.6, our preliminary characterization of this promoter suggests it is indirectly regulated by TBX-2.
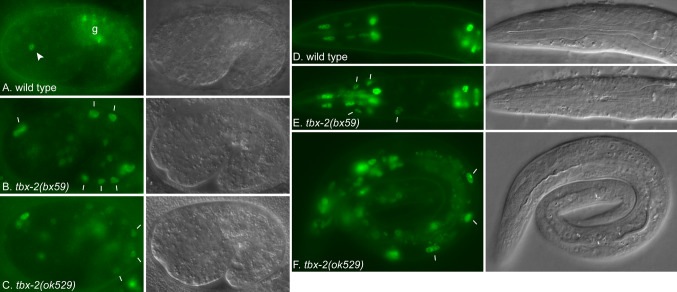
To determine how TBX-2 regulates D2096.6 expression, we compared expression of a D2096.6::gfp reporter in wild-type and tbx-2 mutants. Consistent with previous studies [37], we observed that a D2096.6::gfp reporter was expressed in wild-type embryos specifically in the pharynx at beginning approximately at the bean stage when the pharyngeal primordium forms. Expression was typically observed in one to two cells in the pharynx in one and one-half fold embryos (Fig. 5), and no expression was observed outside the pharynx. The number of GFP-expressing cells increased and animals hatched as L1s with GFP expression in pharyngeal muscles, marginal cells, and epithelial cells (Fig. 5d). In comparison, in tbx-2(bx59) and tbx-2(ok529) embryos D2096.6::gfp was expressed in more cells in the pharynx, and expression was observed in many cells outside the pharynx, including body wall muscles and hypodermal cells (Fig. 5b, c; Table 2). Ectopic D2096.6::gfp expression continued into the L1 larval stage where it was observed in bodywall muscle, hypodermal, and gut cells (Fig. 5e, f). These results indicate TBX-2 is an upstream regulator that represses D2096.6 expression both temporally and spatially.
Fig. 5.
D2096.6::gfp is ectopically expressed in tbx-2 mutants. Fluorescence (left) and DIC (right) micrographs of 1.5-fold stage embryos (a–c) and L1 larvae (d–f) expressing D2096.6::gfp. a tbx-2(+); cuEx553[D2096.6::gfp] embryo containing a single GFP-expressing pharyngeal nucleus (arrowhead) and auto-fluorescent gut granules (g). b, c tbx-2(bx59); cuEx553 and tbx-2(ok529); cuEx553 embryos exhibiting widespread D2096.6::gfp expression outside the pharynx. d tbx-2(+); cuEx553 L1 with GFP expression in pharyngeal nuclei. e, f tbx-2(bx59); cuEx553 and tbx-2(ok529); cuEx553 L1 larvae. Representative bodywall muscle and hypodermal nuclei ectopically expressing D2096.6::gfp are marked (bars)
Table 2.
Ectopic expression of D2096.6::gfp
| Genotype | % animals with ectopic D2096.6::gfp expression (n) |
|---|---|
| cuEx553[D2096.6::gfp] | 5 (136) |
| tbx-2(bx59); cuEx553 a | 71 (55) |
| tbx-2(ok529); cuEx553 b | 20 (35) |
| ubc-9(RNAi); cuEx553 | 75 (48) |
aL4 animals raised at 16 °C were shifted to 25 °C, and defects were scored in the F1 progeny
bProgeny segregating from tbx-2(ok529)/dpy-17(e164) unc-32(e189) hermaphrodites were scored. Twenty-five % of these progeny are expected to be tbx-2(ok529) homozygotes
To ask if SUMOylation is necessary for TBX-2 function, we examined D2096.6 expression in animals where activity of UBC-9 was reduced using feeding-RNAi. The most severely affected ubc-9(RNAi) animals have a highly disorganized morphology that makes it difficult to identify specific tissues [20]. Therefore we characterized D2096.6::gfp expression in older embryos that had undergone morphogenesis and the surviving L1 larvae. ubc-9(RNAi) resulted in D2096.6::gfp expression in posterior body wall muscles in embryos in a pattern similar to that which we have observed in tbx-2(bx59) and tbx-2(ok529) embryos (Fig. 6a, b). In larvae, we observed expression in body wall muscles and hypodermal cells in the posterior of the worm similar to the expression pattern we see in tbx-2 mutants (Fig. 6 c, d). Thus, SUMO-dependent mechanisms repress D2096.6::gfp expression and the similarities in the pattern of ectopic expression in ubc-9(RNAi) and tbx-2 mutants strongly suggests TBX-2 function depends on SUMOylation.
Fig. 6.
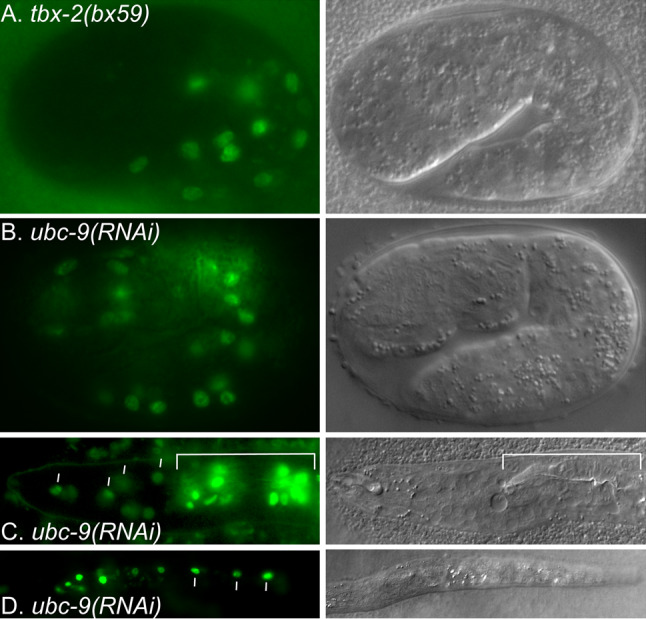
D2096.6::gfp is ectopically expressed in ubc-9(RNAi) animals. Fluorescence (left) and DIC (right) micrographs of a tbx-2(bx59); cuEx553 embryo (a) and a ubc-9(RNAi); cuEx553 embryo (b), and L1 larva (c, d) expressing D2096.6::gfp in bodywall muscle and hypodermal nuclei (bars). The white bracket indicates the partial pharynx in c
Mammalian Tbx2 subfamily members can be SUMOylated
To determine if SUMOylation is a conserved mechanism regulating T-box factor activity, we asked if mammalian orthologs of TBX-2 could be SUMOylated. Human TBX2 or mouse Tbx3 tagged with poly-histidine and V5 were co-expressed in COS-1 cells with or without HA:SUMO-1 and pulled down using Ni2+-beads under denaturing conditions similarly to C. elegans TBX-2. When co-expressed with SUMO-1, both TBX2 and Tbx3 formed two more slowly migrating SUMOylated bands (Fig. 7). This data indicates that other Tbx2-subfamily members can be SUMOylated, and we hypothesize that SUMOylation may be a common regulatory method of T-box factor activity.
Fig. 7.
SUMOylation of mammalian Tbx2 subfamily members Western blots of Ni–NTA pulled-down human TBX2/V5/HIS and mouse Tbx3/V5/HIS probed to detect TBX2 and Tbx3 (bottom) or SUMO-1 (top). Combinations of proteins expressed in COS-1 cells are indicated (grey boxes). SUMOylated forms of TBX2 and Tbx3 are marked with arrowheads, and MW weight markers are indicated (kDa). A cross-reacting background band was detected in all lanes using anti-HA
Discussion
Here we show that C. elegans TBX-2 and its mammalian orthologs human TBX2 and mouse Tbx3 can be SUMOylated, and that TBX-2 SUMOylation depends on two SUMO consensus sites that mediate interaction with the E2 SUMO conjugating enzyme UBC-9. We further demonstrate that C. elegans TBX-2 can function as a transcriptional repressor when fused to a heterologous DNA-binding domain; however, mutations that eliminate SUMOylation do not affect this repressor activity. Finally, we provide genetic evidence that SUMOylation is required for TBX-2 function in vivo by showing that reduction of SUMOylation enhances the phenotype of a hypomorphic tbx-2 mutant and phenocopies the loss of tbx-2 on expression of one gene that is downstream of TBX-2.
Two TBX-2 SUMO consensus sites interact with UBC-9 and mediate SUMOylation
TBX-2 contains two predicted high-scoring SUMO consensus sites. VKKE located near the C-terminus and LKIE located within the T-box DNA-binding domain. Each of theses sequences interacts with the E2 SUMO-conjugating enzyme UBC-9 in yeast two-hybrid assays, and they are the only sites that can mediate this interaction.
Our data strongly suggests that both of these sites are SUMOylated. We observed multiple SUMOylated forms of TBX-2 in COS-1 cells, and mutations affecting either SUMO site reduce the amount of SUMOylated TBX-2. However, there are differences in how specific mutations in these sites affect TBX-2 SUMOylation. For VKKE, mutation of the acceptor lysine (K400R) results in a more severe reduction in SUMOylation than mutating this site completely to alanines. In comparison, mutating the LKIE site to alanines results in a large decrease in SUMOylation whereas mutation of the acceptor lysine (K231R) has a moderate affect and preferentially affects the more slowly migrating SUMOylated forms of TBX-2. Mutations affecting only the acceptor lysine likely retain interaction with UBC-9 (Sampson et al. 2001), whereas mutations converting the SUMO site to alanines eliminate UBC-9 binding. This difference likely underlies the different effects we observed on TBX-2 SUMOylation when these sites are mutated.
T-boxes have a highly conserved structure when bound to DNA [39–42], and TBX-2 LKIE is located within the α3 helix that spans the DNA backbone. While SUMOylation can occur in α helices [43, 44], this is an unusual 2˚ structure for UBC-9 interaction and SUMOylation, as UBC-9 has been shown to bind SUMO consensus sites in extended loops [18, 45]. However, recent evidence indicates that some T-boxes have significant structural flexibility that might allow SUMOylation at LKIE. The Tbx20 T-box exists as a molten globule with an unstable tertiary structure allowing flexibility between 2° structural domains [46]. Likewise in TBX5, the 310-helix located just C terminal to the α3 helix is unstructured in the absence of DNA [42]. While the α3 helix remains structured when TBX5 is not bound to DNA, the LKIE site would be more accessible to bind UBC-9.
LKIE was also investigated as a potential SUMOylation site in the human T-box factor TBX22 [47]. Interestingly, mutation of this site eliminated TBX22 SUMOylation, however, this is believed to result indirectly from possible effects on DNA binding as was observed with several DNA-binding defective mutants. Biochemical analyses of SUMOylated T-box factors is necessary to explicitly determine if this site is SUMOylated in different proteins.
tbx-2 genetically interacts with ubc-9 and smo-1
tbx-2(bx59); ubc-9(RNAi) and tbx-2(bx59); smo-1(RNAi) animals exhibit penetrant embryonic arrest and enhanced pharyngeal defects, indicating that reduced SUMOylation affects TBX-2 activity in vivo. In particular, the enhanced pharyngeal phenotype of these animals resembles that of tbx-2 null mutants, strongly suggesting that tbx-2 and SUMOylation function in the same pathway to specify pharyngeal muscle fate [48]. These observations are consistent with the hypothesis that TBX-2 function is SUMO-dependent.
In comparison, we do not know why enhanced embryonic lethality was observed in tbx-2(bx59); ubc-9(RNAi) and tbx-2(bx59); smo-1(RNAi) animals. Neither tbx-2 null mutants nor tbx-2(RNAi) animals exhibit embryonic lethality [20], indicating lethality does not result from loss of zygotic tbx-2(bx59) activity. One possibility is that tbx-2(bx59) may have a partial gain-of-function character, and reducing SUMOylation deregulates this activity. tbx-2(bx59) mutates a conserved histidine residue within the dimerization domain of the T-box to a tyrosine (H145Y; accession CCD69847), and mutations affecting this domain in human TBX1 result in gain-of-function associated with some cases of DiGeorge and velocardiofacial syndromes [49]. Alternatively, decreased SUMOylation of a parentally provided, RNAi-resistant protein or mRNA tbx-2 gene product might be required for viability. Finally, decreased SUMOylation may affect SUMO-dependent activity of both TBX-2 and another factor with a partially redundant activity required for embryogenesis.
How might SUMOylation affect TBX-2 activity?
SUMOylation of transcription factors is usually associated with transcriptional repression, and it can promote recruitment of chromatin remodeling and histone modifying complexes [12, 50]. Indeed, SUMOylation of the C. elegans Ets-domain factor LIN-1 leads to interaction with MEP-1 and the NuRD chromatin repressor complex [51]. However, our results argue that SUMOylation affects TBX-2 function by a different mechanism. Unlike LIN-1, repressor activity of TBX-2:GAL4 fusion protein in mammalian cells is independent of SUMOylation, and in extensive yeast two-hybrid screens TBX-2 has not been observed to interact with MEP-1 [20].
SUMOylation at the LKIE site in the T-box would likely affect the ability of TBX-2 to bind sites in the genome and to interact with other factors binding TBX-2 regulated promoters. If this is the case, our assays for TBX-2:GAL4 repressor activity would be insensitive to SUMOylation, since they depend on a heterologous DNA-binding domain targeting a synthetic promoter. Indeed, SUMOylation has been shown to affect DNA-binding activity of specific transcription factors [52–54].
It is difficult to predict how SUMOylation at the VKKE site would affect TBX-2 activity. T-box proteins are poorly conserved outside of the DNA-binding domain, and the function of the TBX-2 C-terminus is unknown. Interestingly, our preliminary results indicate VKKE is located near an interaction site for the Groucho-family co-repressor UNC-37 (T. Crum and P. Okkema, unpublished). SUMOylation can regulate interaction with Groucho-family proteins [55, 56], and we hypothesize SUMOylation at VKKE has a similar function. Groucho-interaction motifs are enriched in T-box factors from C. elegans, Drosophila, and humans [57], and several T-box factors have been shown to interact with Groucho-family proteins [58–61]. SUMOylation may be a common mechanism for regulating T-box factor interaction with Groucho.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
The authors are indebted to Jorge A. Iñiguez-Lluhí, Elizaveta Benevolenskaya, Philip Stanier, Andy Fire, Jeb Gaudet, and Hans-Georg Simon for plasmids, strains, and advice. This project was supported by NIH grant 5R01GM82865 (P.G.O.), a UIC LASURI Award (A.V.P.), and State of Illinois funding to the Laboratory for Molecular Biology. Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). The authors gratefully acknowledge the Research Open Access Publishing (ROAAP) Fund of the University of Illinois at Chicago for financial support towards the open access publishing fee for this article.
References
- 1.Greulich F, Rudat C, Kispert A. Mechanisms of T-box gene function in the developing heart. Cardiovasc Res. 2011;91(2):212–222. doi: 10.1093/cvr/cvr112. [DOI] [PubMed] [Google Scholar]
- 2.Naiche LA, Harrelson Z, Kelly RG, Papaioannou VE. T-box genes in vertebrate development. Annu Rev Genet. 2005;39:219–239. doi: 10.1146/annurev.genet.39.073003.105925. [DOI] [PubMed] [Google Scholar]
- 3.Bamshad M, Lin RC, Law DJ, Watkins WC, Krakowiak PA, Moore ME, Franceschini P, Lala R, Holmes LB, Gebuhr TC, Bruneau BG, Schinzel A, Seidman JG, Seidman CE, Jorde LB. Mutations in human TBX3 alter limb, apocrine and genital development in ulnar-mammary syndrome. Nat Genet. 1997;16(3):311–315. doi: 10.1038/ng0797-311. [DOI] [PubMed] [Google Scholar]
- 4.Basson CT, Bachinsky DR, Lin RC, Levi T, Elkins JA, Soults J, Grayzel D, Kroumpouzou E, Traill TA, Leblanc-Straceski J, Renault B, Kucherlapati R, Seidman JG, Seidman CE. Mutations in human TBX5 cause limb and cardiac malformation in Holt-Oram syndrome. Nat Genet. 1997;15(1):30–35. doi: 10.1038/ng0197-30. [DOI] [PubMed] [Google Scholar]
- 5.Bongers EM, Duijf PH, van Beersum SE, Schoots J, Van Kampen A, Burckhardt A, Hamel BC, Losan F, Hoefsloot LH, Yntema HG, Knoers NV, van Bokhoven H. Mutations in the human TBX4 gene cause small patella syndrome. Am J Hum Genet. 2004;74(6):1239–1248. doi: 10.1086/421331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li QY, Newbury-Ecob RA, Terrett JA, Wilson DI, Curtis AR, Yi CH, Gebuhr T, Bullen PJ, Robson SC, Strachan T, Bonnet D, Lyonnet S, Young ID, Raeburn JA, Buckler AJ, Law DJ, Brook JD. Holt-Oram syndrome is caused by mutations in TBX5, a member of the Brachyury (T) gene family. Nat Genet. 1997;15(1):21–29. doi: 10.1038/ng0197-21. [DOI] [PubMed] [Google Scholar]
- 7.Packham EA, Brook JD. T-box genes in human disorders. Hum Mol Genet 12 Spec No. 2003;1:R37–44. doi: 10.1093/hmg/ddg077. [DOI] [PubMed] [Google Scholar]
- 8.Lu J, Li XP, Dong Q, Kung HF, He ML. TBX2 and TBX3: the special value for anticancer drug targets. Biochim Biophys Acta. 2010;1806(2):268–274. doi: 10.1016/j.bbcan.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gareau JR, Lima CD. The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat Rev Mol Cell Biol. 2010;11(12):861–871. doi: 10.1038/nrm3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilkinson KA, Henley JM. Mechanisms, regulation and consequences of protein SUMOylation. Biochem J. 2010;428(2):133–145. doi: 10.1042/BJ20100158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gill G. SUMO and ubiquitin in the nucleus: different functions, similar mechanisms? Genes Dev. 2004;18(17):2046–2059. doi: 10.1101/gad.1214604. [DOI] [PubMed] [Google Scholar]
- 12.Gill G. Something about SUMO inhibits transcription. Curr Opin Genet Dev. 2005;15(5):536–541. doi: 10.1016/j.gde.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 13.Wang J, Feng XH, Schwartz RJ. SUMO-1 modification activated GATA4-dependent cardiogenic gene activity. J Biol Chem. 2004;279(47):49091–49098. doi: 10.1074/jbc.M407494200. [DOI] [PubMed] [Google Scholar]
- 14.Wang J, Li A, Wang Z, Feng X, Olson EN, Schwartz RJ. Myocardin sumoylation transactivates cardiogenic genes in pluripotent 10T1/2 fibroblasts. Mol Cell Biol. 2007;27(2):622–632. doi: 10.1128/MCB.01160-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Desterro JM, Rodriguez MS, Kemp GD, Hay RT. Identification of the enzyme required for activation of the small ubiquitin-like protein SUMO-1. J Biol Chem. 1999;274(15):10618–10624. doi: 10.1074/jbc.274.15.10618. [DOI] [PubMed] [Google Scholar]
- 16.Okuma T, Honda R, Ichikawa G, Tsumagari N, Yasuda H. In vitro SUMO-1 modification requires two enzymatic steps, E1 and E2. Biochem Biophys Res Commun. 1999;254(3):693–698. doi: 10.1006/bbrc.1998.9995. [DOI] [PubMed] [Google Scholar]
- 17.Sampson DA, Wang M, Matunis MJ. The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J Biol Chem. 2001;276(24):21664–21669. doi: 10.1074/jbc.M100006200. [DOI] [PubMed] [Google Scholar]
- 18.Bernier-Villamor V, Sampson DA, Matunis MJ, Lima CD. Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell. 2002;108(3):345–356. doi: 10.1016/S0092-8674(02)00630-X. [DOI] [PubMed] [Google Scholar]
- 19.Johnson ES. Protein modification by SUMO. Annu Rev Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- 20.Roy Chowdhuri S, Crum T, Woollard A, Aslam S, Okkema PG. The T-box factor TBX-2 and the SUMO conjugating enzyme UBC-9 are required for ABa-derived pharyngeal muscle in C. elegans . Dev Biol. 2006;295(2):664–677. doi: 10.1016/j.ydbio.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 21.Lewis JA, Fleming JT. Basic culture methods. In: MiC Biology., editor. Caenorhabditis elegans: modern biological analysis of an organism. San Diego: Academic Press; 1995. pp. 4–30. [Google Scholar]
- 22.Mello C, Fire A. DNA transformation. In: Epstein HF, Shakes DC, editors. Caenorhabditis elegans: modern biological analysis of an organism. San Diego: Academic Press; 1995. pp. 451–482. [Google Scholar]
- 23.Beaster-Jones L, Okkema PG. DNA binding and in vivo function of C. elegans PEB-1 require a conserved FLYWCH motif. J Mol Biol. 2004;339(4):695–706. doi: 10.1016/j.jmb.2004.04.030. [DOI] [PubMed] [Google Scholar]
- 24.Ausubel FM (1990) Current protocols in molecular biology. Greene Pub. Associates and Wiley-Interscience: Wiley, New York
- 25.Amberg DC, Burke DJ, Strathern JN. Methods in yeast genetics: a cold spring harbor laboratory course manual. New York: Cold Spring Harbor Laboratory Press; 2005. [Google Scholar]
- 26.Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, Ahringer J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2001;2:RESEARCH0002. doi: 10.1186/gb-2000-2-1-research0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans . Gene. 2001;263(1–2):103–112. doi: 10.1016/S0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- 28.Rogers ED, Ramalie JR, McMurray EN, Schmidt JV. Localizing transcriptional regulatory elements at the mouse Dlk1 locus. PLoS ONE. 2012;7(5):e36483. doi: 10.1371/journal.pone.0036483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19(2):185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 31.Chiaretti S, Li X, Gentleman R, Vitale A, Vignetti M, Mandelli F, Ritz J, Foa R. Gene expression profile of adult T-cell acute lymphocytic leukemia identifies distinct subsets of patients with different response to therapy and survival. Blood. 2004;103(7):2771–2778. doi: 10.1182/blood-2003-09-3243. [DOI] [PubMed] [Google Scholar]
- 32.Smyth GK (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Statistical applications in genetics and molecular biology 3:Article3. doi:10.2202/1544-6115.1027 [DOI] [PubMed]
- 33.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc Ser B (Methodol) 1995;57(1):289–300. [Google Scholar]
- 34.Papaioannou VE. T-box genes in development: from hydra to humans. Int Rev Cytol. 2001;207:1–70. doi: 10.1016/S0074-7696(01)07002-4. [DOI] [PubMed] [Google Scholar]
- 35.Costa MW, Lee S, Furtado MB, Xin L, Sparrow DB, Martinez CG, Dunwoodie SL, Kurtenbach E, Mohun T, Rosenthal N, Harvey RP. Complex SUMO-1 regulation of cardiac transcription factor Nk2–5. PLoS ONE. 2011;6(9):e24812. doi: 10.1371/journal.pone.0024812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith PA, Mango SE. Role of T-box gene tbx-2 for anterior foregut muscle development in C. elegans . Dev Biol. 2007;302(1):25–39. doi: 10.1016/j.ydbio.2006.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gaudet J, Mango SE. Regulation of organogenesis by the Caenorhabditis elegans FoxA protein PHA-4. Science. 2002;295(5556):821–825. doi: 10.1126/science.1065175. [DOI] [PubMed] [Google Scholar]
- 38.Nakano S, Ellis RE, Horvitz HR. Otx-dependent expression of proneural bHLH genes establishes a neuronal bilateral asymmetry in C. elegans . Development. 2010;137(23):4017–4027. doi: 10.1242/dev.058834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coll M, Seidman JG, Muller CW. Structure of the DNA-bound T-box domain of human TBX3, a transcription factor responsible for ulnar-mammary syndrome. Structure (Camb) 2002;10(3):343–356. doi: 10.1016/S0969-2126(02)00722-0. [DOI] [PubMed] [Google Scholar]
- 40.El Omari K, De Mesmaeker J, Karia D, Ginn H, Bhattacharya S, Mancini EJ. Structure of the DNA-bound T-box domain of human TBX1, a transcription factor associated with the DiGeorge syndrome. Proteins. 2011 doi: 10.1002/prot.23208. [DOI] [PubMed] [Google Scholar]
- 41.Muller CW, Herrmann BG. Crystallographic structure of the T domain-DNA complex of the Brachyury transcription factor. Nature. 1997;389(6653):884–888. doi: 10.1038/39929. [DOI] [PubMed] [Google Scholar]
- 42.Stirnimann CU, Ptchelkine D, Grimm C, Muller CW. Structural basis of TBX5-DNA recognition: the T-box domain in its DNA-bound and -unbound form. J Mol Biol. 2010;400(1):71–81. doi: 10.1016/j.jmb.2010.04.052. [DOI] [PubMed] [Google Scholar]
- 43.Knipscheer P, Flotho A, Klug H, Olsen JV, van Dijk WJ, Fish A, Johnson ES, Mann M, Sixma TK, Pichler A. Ubc9 sumoylation regulates SUMO target discrimination. Mol Cell. 2008;31(3):371–382. doi: 10.1016/j.molcel.2008.05.022. [DOI] [PubMed] [Google Scholar]
- 44.Pichler A, Knipscheer P, Oberhofer E, van Dijk WJ, Korner R, Olsen JV, Jentsch S, Melchior F, Sixma TK. SUMO modification of the ubiquitin-conjugating enzyme E2–25K. Nat Struct Mol Biol. 2005;12(3):264–269. doi: 10.1038/nsmb903. [DOI] [PubMed] [Google Scholar]
- 45.Lin D, Tatham MH, Yu B, Kim S, Hay RT, Chen Y. Identification of a substrate recognition site on Ubc9. J Biol Chem. 2002;277(24):21740–21748. doi: 10.1074/jbc.M108418200. [DOI] [PubMed] [Google Scholar]
- 46.Macindoe I, Glockner L, Vukasin P, Stennard FA, Costa MW, Harvey RP, Mackay JP, Sunde M. Conformational stability and DNA binding specificity of the cardiac T-box transcription factor Tbx20. J Mol Biol. 2009;389(3):606–618. doi: 10.1016/j.jmb.2009.04.056. [DOI] [PubMed] [Google Scholar]
- 47.Andreou AM, Pauws E, Jones MC, Singh MK, Bussen M, Doudney K, Moore GE, Kispert A, Brosens JJ, Stanier P. TBX22 missense mutations found in patients with X-linked cleft palate affect DNA binding, sumoylation, and transcriptional repression. Am J Hum Genet. 2007;81(4):700–712. doi: 10.1086/521033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Herman RK, Yochem J (2005) Genetic enhancers. Worm Book:1-11. doi:10.1895/wormbook.1.27.1 [DOI] [PMC free article] [PubMed]
- 49.Zweier C, Sticht H, Aydin-Yaylagul I, Campbell CE, Rauch A. Human TBX1 missense mutations cause gain of function resulting in the same phenotype as 22q11.2 deletions. Am J Hum Genet. 2007;80(3):510–517. doi: 10.1086/511993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Garcia-Dominguez M, Reyes JC. SUMO association with repressor complexes, emerging routes for transcriptional control. Biochim Biophys Acta. 2009;1789(6–8):451–459. doi: 10.1016/j.bbagrm.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 51.Leight ER, Glossip D, Kornfeld K. Sumoylation of LIN-1 promotes transcriptional repression and inhibition of vulval cell fates. Development. 2005;132(5):1047–1056. doi: 10.1242/dev.01664. [DOI] [PubMed] [Google Scholar]
- 52.Anckar J, Sistonen L. Heat shock factor 1 as a coordinator of stress and developmental pathways. Adv Exp Med Biol. 2007;594:78–88. doi: 10.1007/978-0-387-39975-1_8. [DOI] [PubMed] [Google Scholar]
- 53.Campbell LA, Faivre EJ, Show MD, Ingraham JG, Flinders J, Gross JD, Ingraham HA. Decreased recognition of SUMO-sensitive target genes following modification of SF-1 (NR5A1) Mol Cell Biol. 2008;28(24):7476–7486. doi: 10.1128/MCB.00103-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hong Y, Rogers R, Matunis MJ, Mayhew CN, Goodson ML, Park-Sarge OK, Sarge KD. Regulation of heat shock transcription factor 1 by stress-induced SUMO-1 modification. J Biol Chem. 2001;276(43):40263–40267. doi: 10.1074/jbc.M104714200. [DOI] [PubMed] [Google Scholar]
- 55.Lee PC, Taylor-Jaffe KM, Nordin KM, Prasad MS, Lander RM, LaBonne C. SUMOylated SoxE factors recruit Grg4 and function as transcriptional repressors in the neural crest. J Cell Biol. 2012;198(5):799–813. doi: 10.1083/jcb.201204161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sung KS, Go YY, Ahn JH, Kim YH, Kim Y, Choi CY. Differential interactions of the homeodomain-interacting protein kinase 2 (HIPK2) by phosphorylation-dependent sumoylation. FEBS Lett. 2005;579(14):3001–3008. doi: 10.1016/j.febslet.2005.04.053. [DOI] [PubMed] [Google Scholar]
- 57.Copley RR. The EH1 motif in metazoan transcription factors. BMC Genomics. 2005;6:169. doi: 10.1186/1471-2164-6-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Farin HF, Bussen M, Schmidt MK, Singh MK, Schuster-Gossler K, Kispert A. Transcriptional repression by the T-box proteins Tbx18 and Tbx15 depends on Groucho corepressors. J Biol Chem. 2007;282(35):25748–25759. doi: 10.1074/jbc.M703724200. [DOI] [PubMed] [Google Scholar]
- 59.Formaz-Preston A, Ryu JR, Svendsen PC, Brook WJ. The Tbx20 homolog midline represses wingless in conjunction with Groucho during the maintenance of segment polarity. Dev Biol. 2012;369(2):319–329. doi: 10.1016/j.ydbio.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 60.Kawamura A, Koshida S, Takada S. Activator-to-repressor conversion of T-box transcription factors by the Ripply family of Groucho/TLE-associated mediators. Mol Cell Biol. 2008;28(10):3236–3244. doi: 10.1128/MCB.01754-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Miller RR, Okkema PG. The Caenorhabditis elegans T-Box factor MLS-1 requires Groucho co-repressor interaction for uterine muscle specification. PLoS Genet. 2011;7(8):e1002210. doi: 10.1371/journal.pgen.1002210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Notredame C, Higgins DG, Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302(1):205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.