Abstract
Clinical trial eligibility criteria define fine-grained characteristics of research volunteers for various disease trials and hence are a promising data source for disease profiling. This paper explores the feasibility of using disease-specific common eligibility features (CEFs) for representing diseases and understanding their relatedness. We extracted disease-specific CEFs from eligibility criteria on ClinicalTrials.gov for three illustrative categories – cancers, mental disorders and chronic diseases – each including seven diseases. We then constructed disease-specific CEF networks to assess the degree of overlap among the diseases. Using these automatically derived networks, we observed several findings that were confirmed in medicine. For example, we highlighted connections among schizophrenia, epilepsy and depression. We also identified a link between Crohn’s disease and arthritis. These observations confirm the value of using clinical trial eligibility criteria for identifying disease relatedness. We further discuss the implications of CEFs for standardizing clinical trial eligibility criteria through reuse.
Keywords: Medical informatics, clinical trials, data mining, feature discovery, information storage and retrieval
Introduction
Randomized Controlled Trials (RCTs) provide the strongest evidence for medical practice. As the central registry for RCT studies conducted in the United States of America, ClinicalTrials.gov gives public access to rich clinical trial summaries. As of December 2012, there were over 130,000 trials for about 5,000 diseases on the ClinicalTrials.gov repository, with a few thousand trials for each common disease [1]. Online clinical trials have been extensively used to accelerate trial recruitment and identify research gaps [2, 3].
Eligibility criteria are an important section of RCT summaries. They precisely specify the characteristics and medical conditions that make a person appropriate (i.e., eligible) or inappropriate (i.e., ineligible) for participation in a research study. According to informal observations, RCTs on the same or related diseases often include similar eligibility criteria. Anecdotally, when designing a new RCT, some clinical investigators often identify an existing RCT on the same disease, copy and paste the eligibility criteria section, and then adapt them for the new study.
Therefore, this study contains two hypotheses: (1) disease-specific Common Eligibility Features (CEFs), i.e., multi-word meaningful patterns, can be identified from multiple studies on the same disease and used to profile the disease; and (2) we can understand disease-relatedness by comparing disease profiles based on CEFs. Bhavani et al. constructed a network of medication interventions extracted from depression trials from ClinicalTrials.gov [4]. Using only the structured intervention field and one disease, i.e., depression, their study led us to envision the potential for using network analysis methods to understand disease relatedness.
Aiming to analyze multiple diseases simultaneously using scalable methods on information-rich, free-text eligibility criteria, we propose to profile each disease as a set of CEFs that can be automatically mined from each disease’s corresponding free-text eligibility criteria and to construct a disease-CEF association network to assess disease relatedness.
This study proves the feasibility of using free-text eligibility criteria to probe disease relatedness. At the same time, it also complements existing eligibility criteria standardization efforts, such as the “common core eligibility criteria” defined by the Clinical Data Interchange Standards Consortium (CDISC), by defining disease-specific CEFs [5].
Materials and Methods
Identifying common diseases and disease categories
For illustration purpose, we selected common diseases from the following three disease categories: mental disorders, cancers, and chronic diseases. To this aim, we relied on the classification provided by the Centers for Disease Control and Prevention and the National Institute of Neurological Disorders and Stroke. Mental disorders included epilepsy, Huntington’s disease, Parkinson’s disease, stroke, Amyloid Lateral Sclerosis (ALS), schizophrenia, and depression; cancers included breast cancer, prostate cancer, lung cancer, colorectal cancer, liver cancer, pancreatic cancer, and skin cancer; chronic diseases included diabetes, Crohn’s disease, chronic kidney disease (CKD), arthritis, obesity, atherosclerosis, and hypertension.
Extracting disease-specific common eligibility features
The free-text eligibility criteria are complex, full of temporal constraints and disease-associated traits. Automatically parsing eligibility criteria is a prerequisite for identifying disease-specific CEFs. In the context of this paper, a CEF is a multiword meaningful textual pattern in the free-text eligibility criteria that frequently appears within the clinical trials associated with a specific disease (e.g., computer tomography scan, negative pregnancy test, active malignancy).
Figure 1 shows the main steps in the proposed automated CEF extraction process. First, given a target disease, all the clinical trials associated with it were retrieved in XML format using a query by condition provided in the advanced search form on ClinicalTrials.gov [1]. For each disease, we considered only the trials labeled with one single condition, discarding those trials studying multiple morbidities. Trials were included regardless of trial phase.
Figure 1.
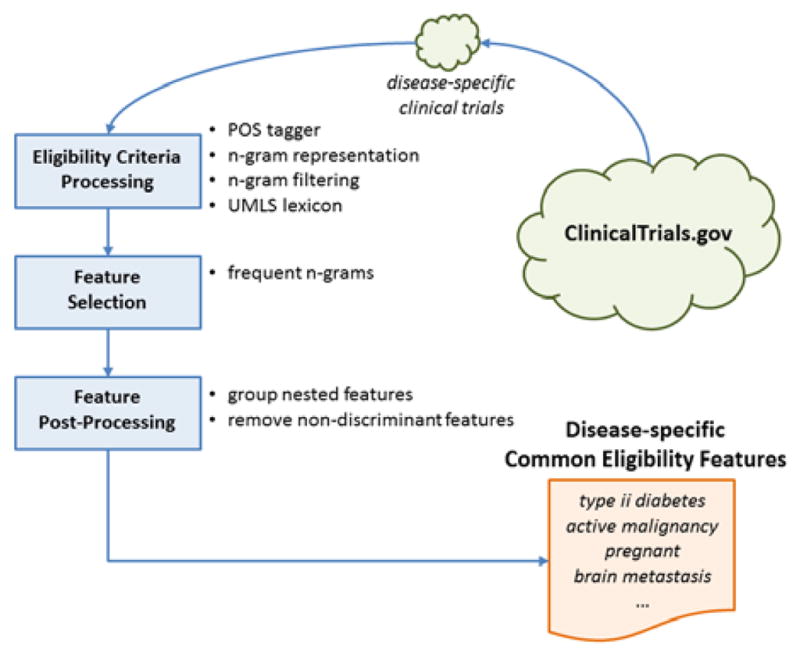
Block diagram for disease-specific CEF extraction
We used basic text processing techniques to parse the free-text eligibility criteria [6]. In particular, each criterion was annotated with a part-of-speech (POS) tagger to identify the grammar role of each word. The text was then processed to remove special characters and punctuation and to construct all possible n-grams (i.e., continuous sub-sequences of n words). N-grams composed of only English stop words or irrelevant grammar structures (e.g., adverbs, verbs, adjectives) were removed. We hypothesized that a meaningful CEF would be composed of at least one noun so that all the n-grams without a noun were automatically discarded.
Last, each n-gram was matched against the Unified Medical Languages System (UMLS) Metathesaurus and retained only if it contained at least one UMLS-recognizable term. For example, malignancy within the past 5 years was considered as a valid n-gram because at least one term (e.g., malignancy) is present in the UMLS lexicon. Each n-gram term, (e.g., x, y, z), found in the UMLS lexicon was also mapped to its normalized UMLS form, (e.g., x is the normalized term for y), in order to reduce the sparseness of the semantic concepts. If term y is found then it is considered as an appearance of term x (i.e., the normalized term). Therefore, each trial associated with the focus disease was summarized by a set of UMLS normalized n-grams representing the relevant concepts contained in the eligibility criteria.
We retained the n-grams appearing in at least 5% of the disease-associated trials as the CEFs1. This list of CEFs was further processed to discard features mostly appearing as substrings of longer features or sharing a similar semantic meaning (i.e., C-value analysis [6]) as well as features appearing too frequently (i.e., tf-idf analysis [7]). The remaining features represented the final list of CEFs associated to a particular disease, i.e., the disease profile.
Constructing disease-CEF network and UMLS analysis
We constructed a network of 621 CEFs associated with at least one of the 21 common diseases. Then we constructed disease-CEF networks for each disease-category. All networks were visualized using Cytoscape’s Biolayout [8].
We also analyzed the distribution of UMLS semantic types over CEFs shared by all possible subsets of disease combinations with varying sizes (i.e., 2, 3…7) respectively per category. We then ranked the most frequently occurring UMLS semantic types per disease category and report results for the top five types.
Results
Disease-specific clinical trials
We independently retrieved all clinical trials associated with each disease in the three disease categories from ClinicalTrials.gov, excluding trials studying multiple morbidities. Thus, we included a total of 13,905 cancer trials, 11,845 chronic disease trials and 5,027 mental disorder trials. The number of registered trials varied per disease, ranging from 4,223 trials for diabetes to 56 trials for Huntington’s disease.
The overall disease-CEF network
First, we analyzed the number of CEFs shared by at least two diseases within each disease category. At most, a CEF can be shared by seven diseases per category. Figure 2 shows the distribution of shared CEFs. Each bar refers to the number of CEFs per category shared by 2, 3…7 diseases, respectively. As it can be seen, there are 113 distinct CEFs shared by any two cancers. For example, progesterone is shared by breast and skin cancers, whereas pelvis is shared between colorectal and pancreatic cancers. Overall, the cancer category contained the largest number of shared CEFs with 23 features shared by all seven cancers.
Figure 2.
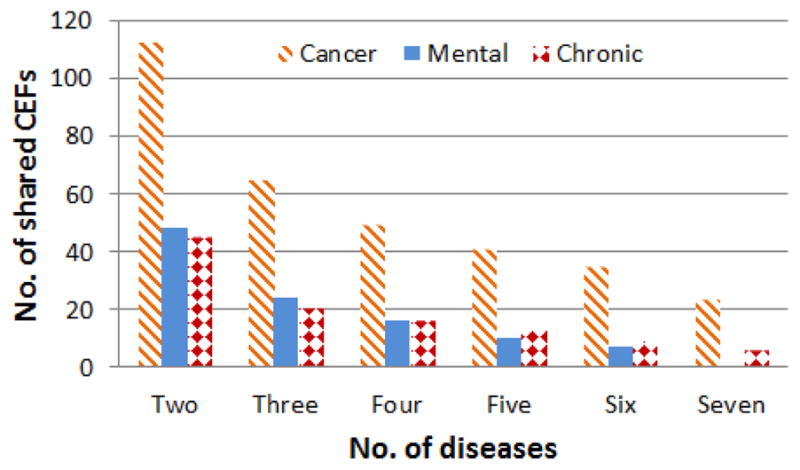
The distribution of CEFs shared within each category by the number of diseases in that category.
Each category includes at most seven diseases
Figure 3 shows the global disease-CEF network. Notice that CEFs successfully clustered similar diseases (i.e., diseases belonging to the same category) together. Cancers are clustered more tightly confirming that they share a greater number of CEFs than other disease categories. Examples of CEFs shared among all seven cancers include: effective contraception, MRI, brain metastasis, radio therapy, hypersensitivity, immunotherapy, karnofsky, and uncontrolled hypertension. In contrast, features shared among the chronic diseases were more general, e.g., pregnancy test, cancer, allergy, alcohol, pregnant and medical condition.
Figure 3.
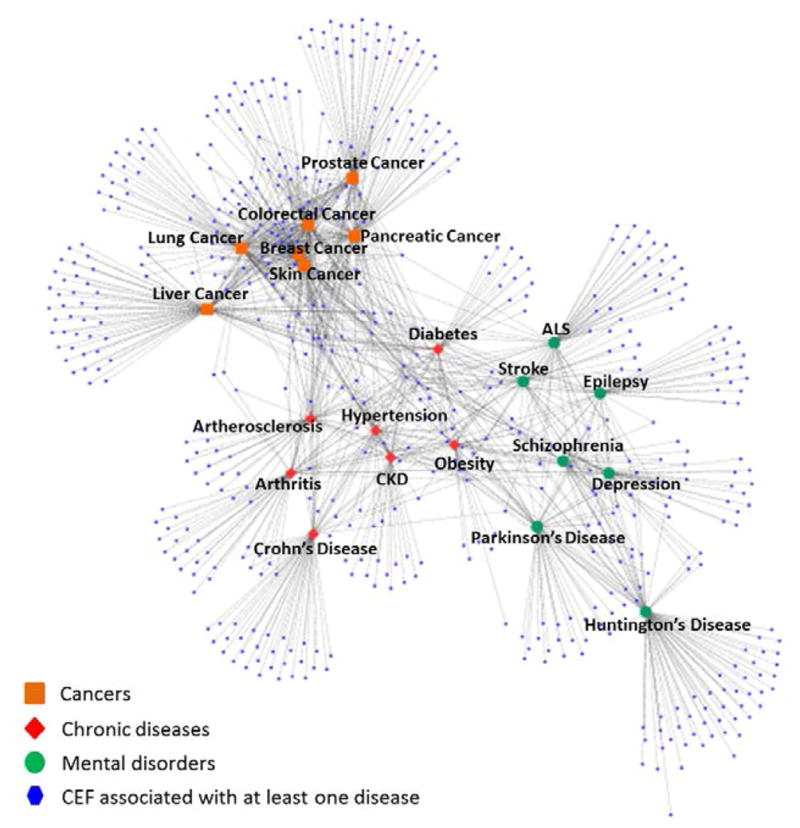
Disease-CEF network for 21 common diseases
Category-specific Disease-CEF networks
For a detailed analysis of each disease category, we visualized each category network – cancers, mental disorders and chronic diseases. First, Figure 4 illustrates the cancer network. The central cluster in the middle of the network represents the shared CEFs across the cancers. The distance between any two diseases in the network is proportional to the number of shared CEFs between the two diseases. Consequently, skin and breast cancers are closely related cancers sharing several features exclusively, such as estrogen, tamoxifen, trastuzumab, and progesterone. Lung cancer is also related to both skin and breast cancers (Figure 4 – it is the next closest disease to the skin, breast cancer cluster) sharing several features with them, e.g., adenocarcinoma and basal or squamous cell.
Figure 4.
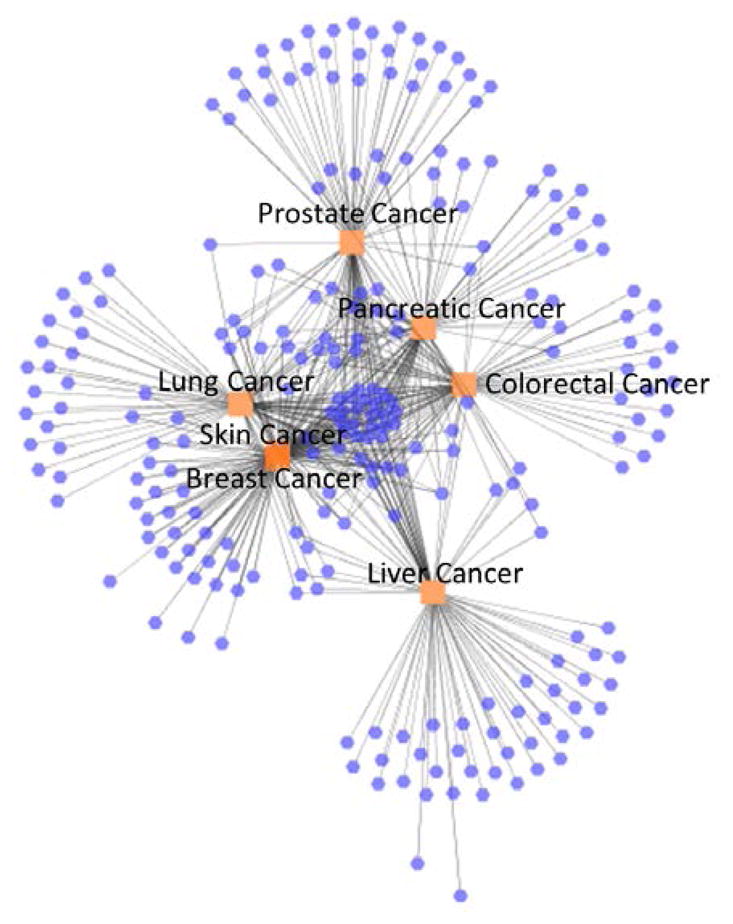
Cancer network
Second, Figure 5 illustrates the mental disorders network. In this case the center of the network is sparser than in Figure 4 because there are fewer shared CEFs. Interestingly, depression is located in the center of the network (in between schizophrenia and epilepsy) meaning that depression has fewer unique features associated (i.e., most of the CEFs associated with depression trials are also associated with trials for other mental disorders). Among the seven mental disorders considered in this study, Huntington’s disease is the most dissimilar, given its larger distance from other mental disorders.
Figure 5.
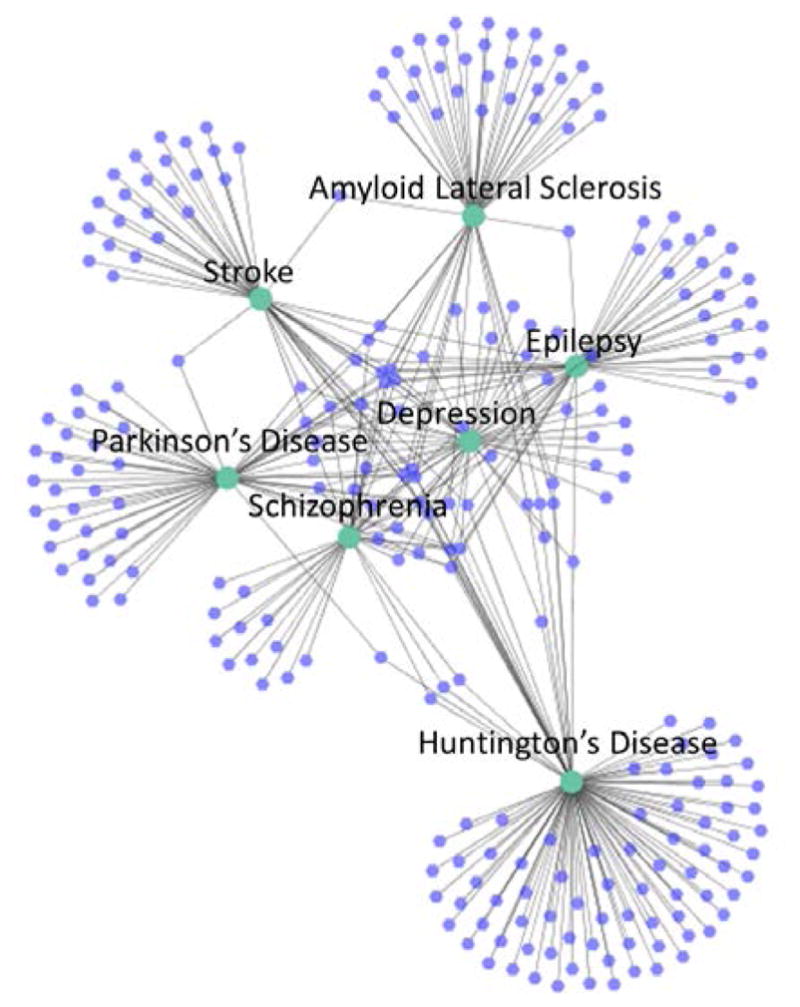
Mental disorder network
As shown in Figure 5, and to a lesser extent Figure 3, depression shares many of its CEFs with either epilepsy or schizophrenia. In fact out of the 25 depression CEFs shared with at least one other mental condition, only five are not shared with either epilepsy or schizophrenia. This means that depression CEFs overlap heavily with these two diseases. For the sake of completeness, we also measured the percent of disease-specific CEFs shared between each mental disorder, which is depicted in Figure 6.
Figure 6.
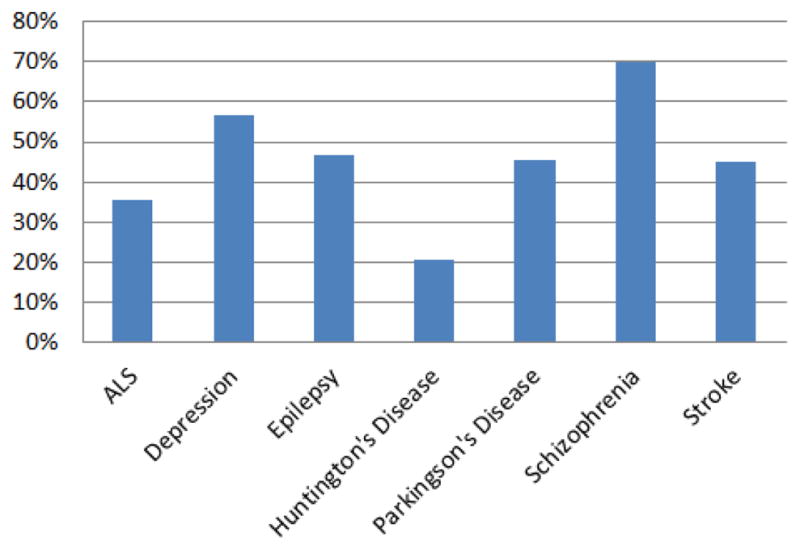
Disease-specific CEFs shared among mental disorders
We found that schizophrenia shared 70% of its features with other mental disorders followed by depression with 56.82%. This means that, proportionally, schizophrenia studies contained the least amount of schizophrenia-specific CEFs within the mental disorders category. Only 12 schizophrenia CEFs were not associated with other mental disorders, namely: clozapine, diagnosis of schizophrenia or schizoaffective, dsm-iv diagnosis of schizophrenia, hepatitis, history of neuroleptic malignant, iq, olanzapine, paliperidone, risperidone, axis diagnosis I, behavior, and depot antipsychotic.
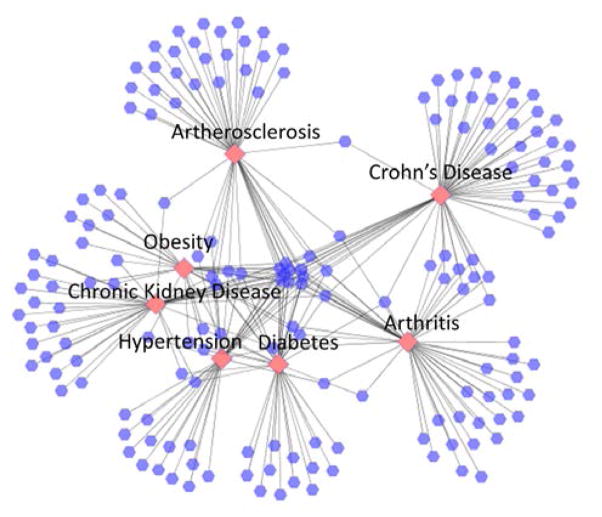
Last, the chronic disease network is shown in Figure 7. Obesity shares many CEFs with other chronic diseases, which accounts for its less distinctive position in the network. Trials for obesity, CKD, hypertension and diabetes are more closely related; conversely, arthritis and Crohn’s disease are both separated from the main hub of chronic diseases. However, a number of CEFs are shared between Crohn’s disease and arthritis (see the small cluster of nodes between them in Figure 7), e.g., TB, prednisone, methotrexate, adalimumab, CRP, infliximab.
Figure 7.
Chronic disease network
UMLS semantic type analysis
In this analysis, each CEF related to at least two diseases per category was mapped to the UMLS lexicon. Table 1 shows the top five most frequent UMLS semantic types per disease category ranked from most to least frequent. Overall, two semantic types appeared more frequently in all disease categories: “pharmacologic substance” and “disease or syndrome”. Therefore, those topics are commonly associated with a given disease regardless of disease category. Moreover, not surprisingly, some UMLS semantic types only appeared frequently among certain disease categories. For instance, “neoplastic process” occurred frequently in cancers only while “mental or behavioral dysfunction” and “pathologic function” occurred frequently only in mental disorders and chronic diseases, respectively. This indicates that some UMLS semantic types are biologically related to the disease category while others are common across all disease categories.
Table 1.
Top 5 UMLS semantic types per category; N is the number of unique CEFs per category associated with a given semantic type
| Cancers | Mental Disorders | Chronic Diseases |
|---|---|---|
| Unmapped2 (N=26) | Pharmacologic Substance (N=8) | Pharmacologic Substance (N=10) |
| Neoplastic Process (N=18) | Disease or syndrome (N=6) | Disease or syndrome (N=9) |
| Pharmacologic Substance (N=13) | Finding (N=6) | Unmapped (N=6) |
| Disease or syndrome (N=10) | Diagnostic procedure (N=5) | Pathologic function (N=5) |
| Therapeutic or preventive procedure (N=9) | Mental or behavioral dysfunction (N=4) | Therapeutic or preventive procedure (N=3) |
‘Unmapped’ means that the feature was not mapped to a UMLS semantic type. An example is diagnostic and statistical manual of mental disorder.
Discussion
Identifying relationships between diseases using CEFs
Using our disease-CEF network, we observed that biologically related diseases were often clustered together, indicating that related diseases often contain similar eligibility criteria features. For example in the cancer network, we found that skin and breast cancer were clustered tightly to each other and to lung cancer. Studies show that breast cancer and malignant melanoma (i.e., a form of aggressive skin cancer) are associated with each other [9] and that breast cancer survivors are more likely to develop melanoma than cancer-free controls [10]. Furthermore, breast and lung cancers are often studied together in research studies because of the close connection between the two diseases [11]. Based on our findings, we posit that the relatedness of diseases observed in our disease-CEF network may allude to the biological and clinical similarity between the diseases and their manifestations.
A closer look at the chronic disease-CEF network shows that all 7 chronic diseases shared only a few eligibility features. One of them was alcohol, which is most likely due to alcohol abuse being a common comorbidity among many chronic diseases. Compared to the cancer category, the number of shared CEFs across all chronic disorders was small. This suggests that clinical trials for chronic diseases could be mimicking the highly diverse etiologies and treatments for chronic conditions. We observed Crohn’s disease and arthritis to be separated from the main hub of chronic diseases. However, they shared a mutual subset of features. The features they shared were principally related to inflammation; for example, CRP (i.e., C-reactive protein) is part of the immune response, whereas other features referred to medications that regulate the immune response (i.e., prednisone, methotrexate, adalimumab, infliximab). In addition, our finding of a connection between Crohn’s disease and arthritis is largely supported in the literature, partially due to their shared treatment options; but also because inflammatory bowel disease (Crohn’s is a type of inflammatory bowel disease) and arthritis are associated with each other [12].
We also found a large overlap between the disease-specific CEFs for depression, schizophrenia and epilepsy. In fact, 80% of shared depression CEFs were also associated with schizophrenia and epilepsy indicating that these three diseases may be inter-connected. While Schmitz et al. failed to find a biological mechanism supporting a connection between the three diseases, they concluded that both neurological and sociological variables appear to link the diseases together [13]. This explains the inter-connectedness between the three diseases observed in our disease-CEF network because eligibility criteria contain both neurological and sociological variables (e.g., dsm-iv, willingness to consent).
Implications for eligibility criteria standards development
We developed a method for extracting CEFs from free-text clinical trial eligibility criteria that could be useful in standardization efforts. Standardization of criteria would enhance automatic electronic eligibility determination methods for clinical trials. We demonstrate that using CEFs is effective at stratifying common diseases into their respective categories (i.e., cancers, mental disorders, chronic diseases) demonstrating its further utility for studying disease relatedness.
Our method also complements the standardization efforts of CDISC, which is currently developing standards for a set of “common core eligibility criteria” that occur across trials regardless of disease type [5]. We focus on disease-specific CEFs, whereas CDISC identifies disease-neutral core eligibility criteria. We propose using our method in conjunction with theirs to achieve the common goal of standards-based eligibility criteria. Identifying CEFs shared across diseases within a disease category could also benefit protocol authors specializing within a certain disease area (e.g., oncology, psychiatry).
Limitations and future work
Using ClinicalTrials.gov as our sole data source presents two possible limitations: (1) trials deposited there may contain disease indexing errors; and (2) eligibility criteria may be incomplete or condensed for some studies, though these issues are common to any electronic data source in use for medical knowledge discovery and should not prevent methodology development. Despite these issues, we successfully utilized eligibility criteria from ClinicalTrials.gov to study disease relatedness and used medical literature to validate our findings. Also, we did not study pediatric diseases, e.g., autism, cystic fibrosis. Rather, we limited ourselves to primarily adult disorders. In the future, we will expand our research to cover all diseases and construct a comprehensive disease network.
Conclusion
We extracted disease-specific common eligibility features (CEFs) from ClinicalTrials.gov and used them to construct disease-CEF networks for 21 diseases and observed several interesting findings regarding relatedness between diseases. Some observed relationships were generally confirmed in the medical literature. For instance, we found a connection between schizophrenia, epilepsy and depression; similarly, we identified a link between Crohn’s disease and arthritis, which is related to their mutual role in the inflammatory response.
Acknowledgments
This research was supported by R01LM009886 from the National Library of Medicine, grant R01 HQ 1R01HS019853-01 from AHRQ and grant UL1 TR000040 from the National Center for Advancing Translational Sciences.
Footnotes
The 5% threshold was obtained after preliminary experiments. In fact, we detected that such value generally led to a good compromise between number of common features and noise reduction.
This article is published online with Open Access by IOS Press and distributed under the terms of the Creative Commons Attribution Non-Commercial License.
References
- 1.NIH. [Accessed in Nov 2012];ClinicalTrials.gov. http://www.clinicaltrials.gov.
- 2.Califf R, Zarin DA, Kramer JM, Sherman RE, Aberle LH, Tasneem A. Characteristics of clinical trials registered in clinicaltrials. gov, 2007–2010. JAMA. 2012;307(17):1838–47. doi: 10.1001/jama.2012.3424. [DOI] [PubMed] [Google Scholar]
- 3.Research Match. [Accessed in Dec. 2012]; https://www.researchmatch.org/
- 4.Bhavnani S, Carini S, Ross J, Sim I. Network Analysis of Clinical Trials on Depression: Implications for Comparative Effectiveness Research. AMIA Annu Symp Proc. 2010;2010:51–5. [PMC free article] [PubMed] [Google Scholar]
- 5.Kush R, Bain L. [Accessed in November, 2012];CDISC SHARP Teleconference. 2010 http://informatics.mayo.edu/sharp/images/4/42/CDISC_IHEs_RPE_A_SHARP_Solution.ppt.
- 6.Frantzi K, Ananiadou S, Mima H. Automatic recognition of multi-word terms: the C-value/NC-value method. Int J Digit Libr. 2000;3(2):115. [Google Scholar]
- 7.Salton G, Buckley C. Term-weighting approaches in automatic text retrieval. Inform Process Manag. 1988;24(5):513–24. [Google Scholar]
- 8.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ho WL, Comber H, Hill AD, Murphy GM. Malignant melanoma and breast carcinoma: a bidirectional correlation. Ir J Med Sci. 2011;180(4):901–3. doi: 10.1007/s11845-009-0297-5. [DOI] [PubMed] [Google Scholar]
- 10.Wassberg C, Thörn M, Yuen J, Hakulinen T, Ringborg U. Cancer risk in patients with earlier diagnosis of cutaneous melanoma In situ. Int J Cancer. 1999;83(3):314–7. doi: 10.1002/(sici)1097-0215(19991029)83:3<314::aid-ijc5>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 11.Watanabe J, Shimada T, Gillam EM, Ikuta T, Suemasu K, Higashi Y, Gotoh O, Kawajiri K. Association of CYP1B1 genetic polymorphism with incidence to breast and lung cancer. Pharmacogenetics. 2000;10(1):25–33. doi: 10.1097/00008571-200002000-00004. [DOI] [PubMed] [Google Scholar]
- 12.Lindsley CB, Schaller JG. Arthritis associated with inflammatory bowel disease in children. J Pediatr. 1974;84(1):16–20. doi: 10.1016/s0022-3476(74)80547-0. [DOI] [PubMed] [Google Scholar]
- 13.Schmitz EB, Robertson MM, Trimble MR. Depression and schizophrenia in epilepsy: social and biological risk factors. Epilepsy Research. 1999;35(1):59–68. doi: 10.1016/s0920-1211(98)00129-6. [DOI] [PubMed] [Google Scholar]