Abstract
Recently, the enhanced resolution and sensitivity offered by chemoselective isotope tags have enabled new and enhanced methods for detecting hundreds of quantifiable metabolites in biofluids using nuclear magnetic resonance (NMR) spectroscopy or mass spectrometry. However, the inability to effectively detect the same metabolites using both complementary analytical techniques has hindered the correlation of data derived from the two powerful platforms and thereby the maximization of their combined strengths for applications such as biomarker discovery of the identification of unknown metabolites. With the goal of alleviating this bottleneck, we describe a smart isotope tag, 15N-cholamine, which possesses two important properties: an NMR sensitive isotope, and a permanent charge for MS sensitivity. Using this tag, we demonstrate the detection of carboxyl group containing metabolites in both human serum and urine. By combining the individual strengths of the 15N label and permanent charge, the smart isotope tag facilitates effective detection of the carboxyl-containing metabolome by both analytical methods. This study demonstrates a unique approach to exploit the combined strength of MS and NMR in the field of metabolomics.
Keywords: Metabolomics, nuclear magnetic resonance spectroscopy, mass spectrometry, smart isotope tag, cholamine, carboxylic acids
Introduction
The metabolomics field has witnessed exponential growth over the last decade due to its capabilities for systems biology research and potential applications in numerous disciplines including biomedicine, toxicology, food and nutrition, drug development and environmental science.1-5 Commonly used analytical techniques such as nuclear magnetic resonance (NMR) spectroscopy and/or mass spectrometry (MS) have evolved in response to the high demand for resolving the complexity of biological mixtures and identifying the large pool of quantifiable metabolites. However, despite numerous advances, the biological complexity still often outweighs the capabilities of these advanced analytical methods; no single technique currently is capable of detecting all metabolites in a single experiment. Each analytical method is sensitive to certain classes of metabolites, and depending on the nature of the metabolites of interest, generally one or sometimes a combination of NMR or MS techniques are used to profile as many metabolites as possible and thereby derive the biological meaning. A major hurdle of such an approach is that the metabolite data obtained from NMR and LC-MS or GC-MS methods for the same or similar samples are often not directly comparable. The inability to compare and correlate data from different analytical techniques for the same or similar samples is a significant challenge that prevents drawing meaningful conclusions from the vast amount of metabolite data existing in the literature and exploiting the combined strength of NMR and MS for unknown metabolite identification. The main contributing factors for this bottleneck are the limited NMR sensitivity, complex spectral signatures and variable MS ionization efficiency or suppression.
The use of chemo-selective tags provides an avenue to improve the sensitivity of metabolite detection by both NMR and MS methods. For example, the sensitivity of MS detection is shown to be enhanced by three orders of magnitude or more by tagging metabolites with a chemo-selective tags containing a permanent charge.6-10 Because of the permanent charge, the tagged metabolites are effectively detected with high sensitivity and better quantitative accuracy, irrespective of the pH or nature of the solvents used to separate metabolites before detection by MS. Separately, based on differential dansylation using 12C/13C-dansyl chloride, absolute or relative quantitation of amine and phenol containing metabolites has been achieved with a sensitivity enhancement of three orders of magnitude.11,12 Similarly, NMR-sensitive isotope based chemo-selective tags have been shown to detect many quantifiable metabolites with high sensitivity and resolution by NMR.13-17 Using 15N-ethanolamine as the tag, for example, over a hundred carboxyl-containing metabolites have been detected by 1H-15N two-dimensional NMR with high resolution and sensitivity.13 However, while metabolites can be detected with high sensitivity by both MS and NMR separately using chemoselective tags, the inability to compare and correlate the data from the two methods is a major bottleneck in the metabolomics field.
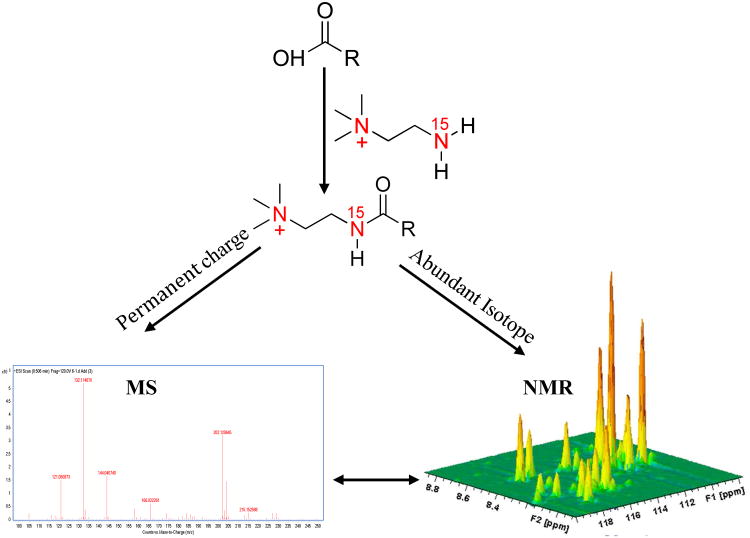
The ability to more easily detect the same metabolites by both NMR and MS methods would offer new avenues to compare data between MS and NMR platforms and to exploit the combined strength of the complimentary methods. Towards this goal, we introduce a new “smart isotope tag” approach, using 15N-cholamine in this case, which possesses the characteristics of high NMR sensitivity and resolution through its isotope enrichment and high MS sensitivity through its permanent positive charge (see schematic Figure 1). The tag combines the strengths of individual chemo-selective tags, demonstrated previously and separately for NMR and MS detection,6, 13 and offers news avenues to exploit the combined strength of these powerful and complementary techniques for areas such as metabolite profiling and unknown metabolite identification.
Figure 1.
Schematic figure illustrating the “smart isotope tag” approach used to detect the same metabolites using NMR and MS with high sensitivity. Tagging carboxyl-containing metabolites with 15N-cholamine enables their detection by both NMR and MS with high sensitivity.
Experimental
Chemicals and Biofluids
A total of 48 carboxyl-containing metabolite standards (Table 1), (2-bromoethyl)trimethylammonium bromide, dimethylformamide (DMF), methanol, acetonitrile, isopropanol, acetone, hydrochloric acid (HCl), sodium hydroxide (NaOH) (all from Sigma-Aldrich, St. Louis, MO), 4-(4,6-dimethoxy[1.3.5]triazin-2-yl)-4-methylmorpholinium chloride (DMTMM) (Acros Organic, Pittsburgh, PA), 15N-phthalimide potassium and deuterium oxide (Cambridge Isotope Laboratories, Andover, MA) were used without further purification. Human serum samples were obtained from Innovative Research, Inc. (Novi, MI) and urine from healthy volunteers, in accordance with the Internal Review Board at Purdue University. Deionized (DI) water was from in-house Synergy Ultrapure Water System from Millipore (Billerica, MA)
Table I.
1H and 15N NMR chemical shifts for 15N-cholamine tagged carboxyl-containing metabolites that were measured with reference to smart tagged formic acid.
| Label | Name | 1H (ppm) | 15N (ppm) | Label | Name | 1H (ppm) | 15N (ppm) |
|---|---|---|---|---|---|---|---|
| 1 | Cis-Aconitic acid | 8.5 | 118.24 | 23 | 2-hydroxyisobutyric acid | 7.95 | 117.51 |
| 8.14 | 121.47 | 24 | DL-Isocitric acid | 8.40 | 117.15 | ||
| 8.06 | 119.49 | 8.11 | 120.77 | ||||
| 8.07 | 120.21 | 25 | Isoleucine | 8.37 | 118.19 | ||
| 8.23 | 116.00 | 26 | Isovaleric acid | 8.07 | 121.92 | ||
| 8.14 | 120.81 | 27 | α-Ketoglutaric acid | 8.69 | 116.34 | ||
| 2 | Adipic acid | 8.05 | 120.57 | 8.63 | 111.84 | ||
| 3 | DL-Alanine | 8.30 | 114.39 | 28 | Lactic acid | 8.23 | 114.18 |
| 4 | 4-minobenzoic acid | 8.25 | 111.35 | 8.49 | 114.45 | ||
| 5 | Arginine | 8.34 | 115.96 | 29 | Leucine | 8.34 | 115.74 |
| 6 | Asparagine | 8.31 | 116.03 | 30 | Lysine | 8.33 | 115.88 |
| 7 | Aspartic acid | 8.15 | 120.01 | 31 | Maleic acid | 8.39 | 120.39 |
| 8.38 | 115.27 | 32 | Malic acid | 8.28 | 122.83 | ||
| 8.31 | 115.6 | 8.29 | 115.14 | ||||
| 8.08 | 122.15 | ||||||
| 8.16 | 121.35 | 33 | Malonic acid | 8.19 | 121.44 | ||
| 8 | Betaine | 8.55 | 122.69 | 34 | Methionine | 8.36 | 116.08 |
| 9 | Citric acid | 8.20 | 121.46 | 35 | Oxalic acid | 8.47 | 117.13 |
| 8.07 | 123.95 | 36 | Oxaloacetic acid | 8.35 | 112.67 | ||
| 7.87 | 121.88 | 8.63 | 111.40 | ||||
| 10 | Cysteine | 8.35 | 115.93 | 37 | L-phenylalanine | 8.21 | 118.85 |
| 11 | Cystine | 8.5 | 115.22 | 38 | L-proline | 8.35 | 115.58 |
| 12 | Formic acid | 8.05 | 123.93 | 39 | Propionic acid | 7.95 | 118.85 |
| 13 | Fumaric acid | 8.42 | 122.68 | 40 | Pyroglutamic acid | 8.29 | 115.88 |
| 8.56 | 124.24 | 41 | Pyruvic acid | 8.63 | 111.39 | ||
| 14 | Glucuronic acid | 8.38 | 119.54 | 8.35 | 112.72 | ||
| 15 | Glutamic acid | 8.28 | 115.99 | 42 | Serine | 8.17 | 117.63 |
| 8.05 | 120.42 | 43 | Succinic acid | 7.96 | 119.64 | ||
| 8.01 | 119.16 | ||||||
| 16 | Glutamine | 8.35 | 115.90 | 44 | Succinyl-COA | 8.03 | 119.17 |
| 8.11 | 119.67 | ||||||
| 17 | Glycine | 8.2 | 115.45 | 45 | L-threonine | 8.34 | 117.79 |
| 18 | Glycolic acid | 8.22 | 114.97 | 46 | L-tryptophan | 7.98 | 119.37 |
| 8.37 | 115.19 | 47 | Tyrosine | 8.27 | 118.05 | ||
| 19 | Hippuric acid | 8.2 | 115.62 | 48 | Valine | 8.38 | 118.20 |
| 20 | Histidine | 8.36 | 116.60 | ||||
| 21 | 3-Hydroxybutyric acid | 8.07 | 122.20 | ||||
| 22 | 4-Hydroxy-L-proline | 8.5 | 115.89 | ||||
| 8.36 | 117.62 |
Design and Synthesis of the smart isotope tag- 15N-Cholamine
Synthesis of 15N-cholamine involved a two-step reaction and followed the Gabriel synthesis procedure with modifications as described below to yield the pure product.18,19 The first step involved reacting potassium 15N-phthalimide with (2-bromoethyl)trimethylammonium bromide in DMF to obtain the 15N substituted phthalimide intermediate (Scheme 1). The second step involved alkaline and acid hydrolyses of the 15N substituted phthalimide to yield the smart isotope tag, 15N-cholaimne (Scheme 2).
Scheme 1.
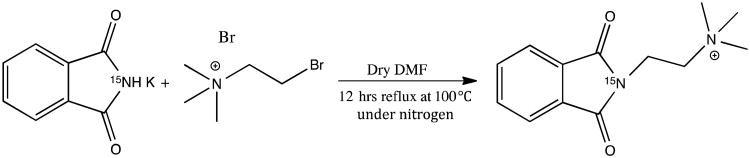
Synthesis of 15N substituted phthalimide.
Scheme 2.
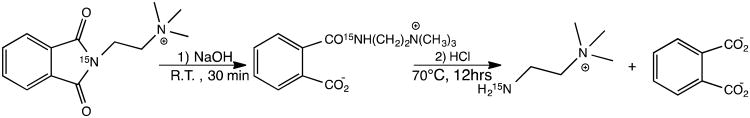
Alkaline and acid hydrolyses of the 15N substituted phthalimide to yield 15N-cholamine.
Briefly, for the synthesis of 15N substituted phthalimide (Scheme 1), (2-bromoethyl)trimethylammonium bromide (9.5 mmol, 2.35 g) was mixed with 15N-phthalimide potassium (10 mmol, 1.86 g) in a 250 mL round bottom flask and dry DMF (100 mL) was added under nitrogen gas. The mixture was then refluxed at 100 °C with stirring for 12 h. The supernatant from the reaction mixture was separated and the solvent was removed using a rotary evaporator.18 The resulting crude residue was washed thrice using acetonitrile (5 mL each time), twice with acetone (2 mL each time) followed by washing again once with acetonitrile (3 mL) to obtain the pure 15N-substituted phthalimide. 1H NMR spectra in D2O at each step were monitored to assess the purity of the intermediate product. For the synthesis of 15N-cholamine, in the second step, the 15N-substituted phthalimide (538 mg) (Scheme 1) was dissolved in DI water (24 mL); 1 N NaOH (2.69 mL) was added to the solution and the mixture was left at room temperature with stirring for 30 min to complete the alkaline hydrolysis (Scheme 2).19 Subsequently, 12 N HCl (1.8 mL) was added to the solution, the temperature was raised to 70°C and left for 12 h with stirring to complete the acid hydrolysis (Scheme 2).19 The solvent was then removed using a rotary evaporator. The resulting crude residue was washed thrice with acetonitrile (4 mL each time) followed by washing thrice with 25:75 water/acetone mixture (2 mL each time) to yield the pure product, 15N-cholamine. 1H NMR spectra in D2O at each step were monitored to assess the purity of the final product.
Tagging metabolites using the smart isotope tag - 15N-cholamine
15N-cholamine (5mg, 50 μmol) was added to 500 μL sample in an eppendorf tube, the pH of the mixture was adjusted to 7.0 with 1 M HCl or NaOH. 21 mg DMTMM was added to initiate the reaction.13,20,21 The mixture was stirred at room temperature for 4 h to complete the reaction. The general reaction for tagging metabolites with the smart isotope tag is shown in Scheme 3. To maintain 15N amide protonation, the pH was adjusted to 5.0 by adding 1 M HCl or 1 M NaOH, and the solution volume was adjusted to 580 μL by adding DI water and 30 μL of D2O prior to NMR detection. Serum was deproteinized using methanol prior to metabolite tagging and urine was used with no pretreatment.13
Scheme 3.
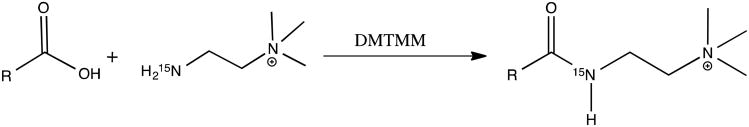
General reaction for tagging carboxyl-containing metabolites with the smart isotope tag 15N-cholamine.
NMR spectroscopy
For each sample, 550 μl was mixed with 30 μl D2O and placed in a 5 mm NMR tube. NMR experiments were performed on a Bruker DRX 500 MHz or Avance III 800 spectrometer equipped with a room temperature or cryoprobe probe, respectively, suitable for 1H inverse detection with Z-gradients at 298 K. A one pulse sequence with or without solvent signal suppression using presaturation was used for 1H 1D NMR experiments. The sensitivity-enhanced 1H-15N 2D HSQC experiments employed an INEPT transfer delay of 6 ms corresponding to the JNH of 90 Hz. Spectral widths for the 1H and 15N dimensions were approximately 8 kHz and 3 kHz, respectively. 128 free induction decays of 1,024 data points each were collected in the indirect (t1) dimension with 1 or 4 transients per increment. Nitrogen decoupling during the direct acquisition (t2 dimension) was achieved with the GARP (Globally Optimized Alternating-Phase Rectangular Pulses) sequence. The resulting 2D data were zero-filled to 1,024 points in the t1 dimension after forward linear prediction to 256 or 512 points. A 45° shifted sine-bell window function was applied to both dimensions before Fourier transformation. Chemical shifts were referenced to the 1H signal of TSP for the 1D spectra or the derivatized formic acid signal (1H: 8.05 ppm; 15N: 123.93 ppm) in the HSQC spectra. Bruker Topspin versions 3.0 or 3.1 software packages were used for NMR data acquisition or processing.
Mass spectrometry
LC-MS and LC-MS/MS experiments were performed using an Agilent 1200 SL-LC system coupled online with an Agilent 6520 Q-TOF mass spectrometer (Agilent Technologies, Santa Clara, CA). The sample (8 μL) was injected onto an Agilent Poroshell 120 EC-C18 column (30×50 mm, 2.7-micron), which was heated to 50 °C. The flow rate was 0.5 mL/min. Mobile phase A was 5 mM ammonium acetate in water, and mobile phase B was 0.1% water in ACN. The mobile phase composition was initially kept isocratic at 3% B for 1 min, then increased to 90% B over 4 min; after another 4 min at 90% B, the mobile phase composition returned to 3% B. Electrospray ionization (ESI) was used in positive mode, and the voltage was 3.5 kV. The mass analyzer was scanned over a range of 50–1000 m/z. The collision energy for auto LC-MS/MS experiments was fixed at 10 V, targeting pre-selected compounds.
Results and Discussion
The smart isotope tag, 15N-cholamine, designed, developed and used in this stu`dy retains all the characteristics of the 15N-ethanolamine tag including the solubility of the tagged metabolites in aqueous media, large one-bond J-coupling between 1H and 15N of ∼90 Hz for efficient polarization transfer between 1H and 15N nuclei, and wide chemical shift dispersion for different metabolites in the resulting 2D NMR spectra.13 In addition, and importantly, 15N-cholamine possesses a permanent positive charge, which enables efficient positive mode detection of the same carboxyl-containing metabolites by MS, irrespective of the pH or solvent conditions of the eluting media, commonly used for chromatographic separation before detection by MS.6
Synthesis of 15N-cholamine involved a two-step reaction and followed the Gabriel synthesis procedure with suitable modifications to yield the pure product.18, 19 As seen in the 1H NMR spectrum (Supplementary Figure S1), the pure intermediate compound, 15N substituted phthalimide, was obtained after the first step of the synthesis. Hydrolysis of this compound yielded the 15N-cholamine in pure form as can be ascertained from its 1H NMR spectrum (Supplementary Figure S2; peaks at 3.16; 3.48; 3.64 ppm). The accurate MS and MS/MS spectra for 15N-cholamine, shown in Supplementary Figure S3, help further verify the identity and purity of the synthesized smart isotope tag (m/z= 104.120).
The compound was then used to tag 48 metabolites that were selected for their prominence as carboxyl-containing metabolites in biofluids that represent a variety of metabolic pathways. The general reaction for tagging metabolites with the smart isotope tag is shown in Scheme 3. To maintain 15N amide protonation, the pH was adjusted to 5.0 by adding 1 M HCl or 1 M NaOH, and the solution volume was adjusted to 580 μL by adding DI water and 30 μL of D2O prior to NMR detection. Serum was deproteinized using methanol prior to metabolite tagging and urine was used with no pretreatment.13
The 1H and 15N chemical shift data derived from the 2D NMR experiments, after tagging with 15N cholamine, are shown in Table I. Because the 15N-cholamine and the previously used 15N-ethanolamine differ only in their terminal group, the tagging efficiency, reproducibility and chemical shift values for metabolites with 15N-cholamine tag were similar to those obtained using the 15N-ethanolamine tag.13
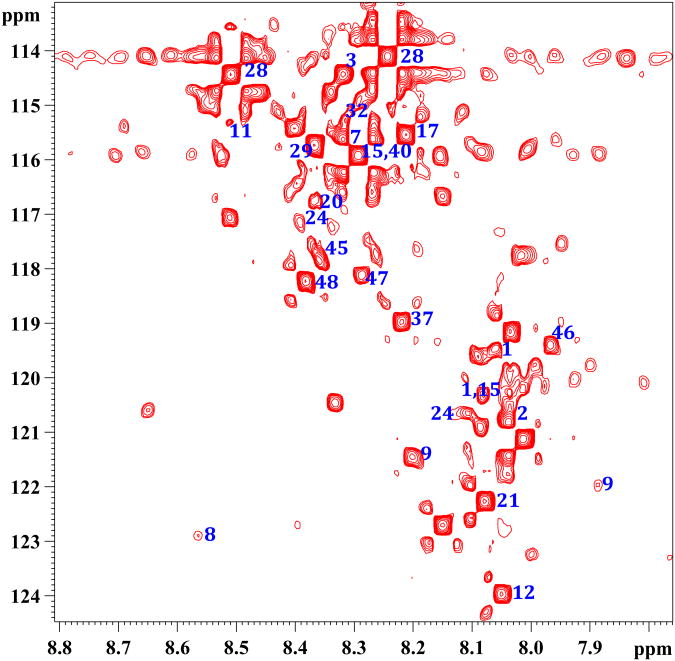
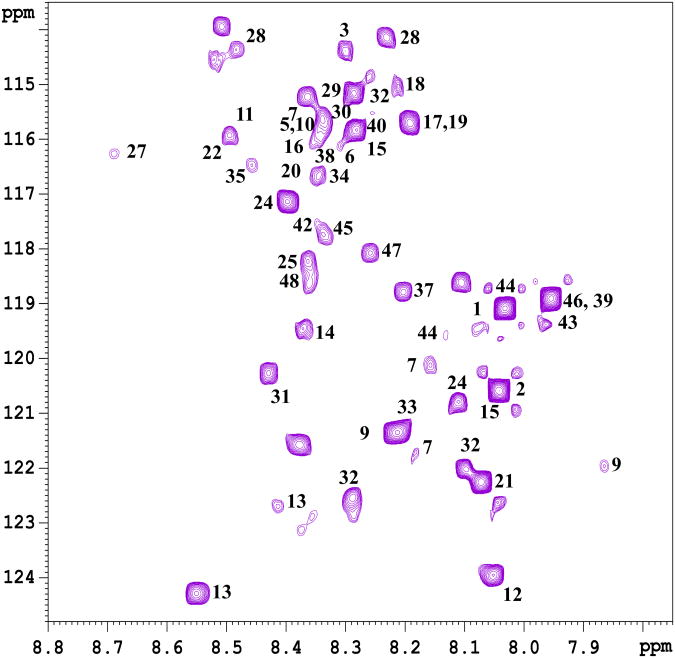
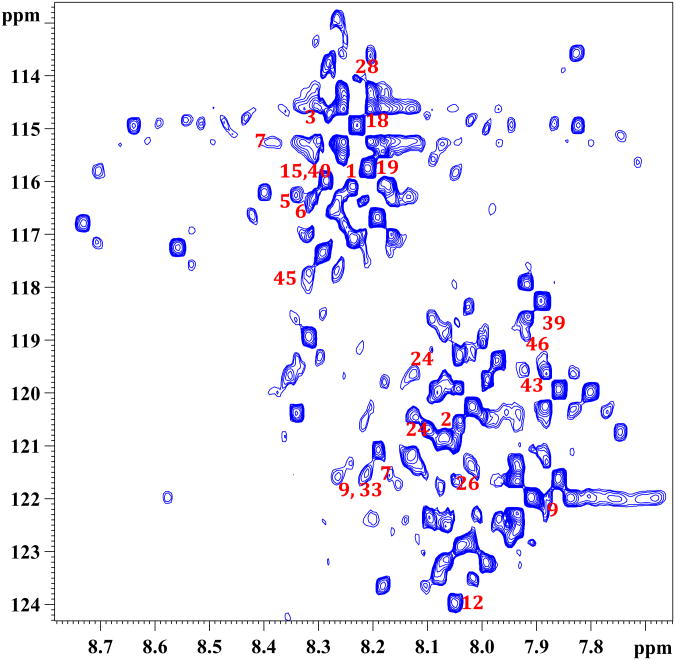
Importantly, as anticipated based on the 15N-ethanolimane tagging approach shown earlier in our laboratory,13 the 15N-cholamine tagging of metabolites in human serum provided a rich NMR spectrum due to the large number of carboxyl-containing metabolites normally present in blood (Figure 2a). The low natural abundance of 15N (0.37%) ensures that none of the nitrogen containing metabolites interferes with the detection of carboxyl-metabolites through 15N-cholamine tag. Each peak in the spectrum corresponds to different metabolite from the carboxylic acid class. However, metabolites, which contain more than one carboxyl group, provide additional peaks depending on the number of carboxyl groups and molecular symmetry. In addition, metabolites such as lactate, which possess α-hydroxyl groups, show more than one peak for the same metabolite as we described earlier using the 15N-ethanolmaine tag.13 Some of the peaks assigned based on the chemical shift values for the standard compounds are marked with the corresponding number shown in Table I and Figure 2b. Similarly, tagging of metabolites in human urine with 15N-cholamine also enabled the detection of peaks corresponding to well over a hundred carboxylic acid group containing metabolites (Figure 3). Peaks tentatively assigned based on the values for the standard compounds are marked by their numbers shown in Table I and Figure 2b.
Figure 2.
Figure 2a. A portion of the 1H-15N HSQC spectrum of human serum tagged with 15N-cholamine. 1: aconitic acid; 2: adipic acid; 3: alanine; 7: aspartic acid; 8: betaine; 9: citric acid; 11: cystine; 12: formic acid; 15: glutamic acid; 17: glycine; 20; histidine; 21: 3-hydroxybutyric acid; 24: isocitric acid; 28: lactic acid; 29: leucine; 32: malic acid; 37: phenylalanine; 40: pyroglutamic acid; 45: threonine; 46: tryptophan; 47: tyrosine; 48: valine.
Figure 2b. A portion of the 1H-15N HSQC spectrum of a mixture of standard compounds at various concentrations obtained after tagging with 15N-cholamine. The peak numbers correspond to the compounds shown in Table I.
Figure 3.
A portion of the 1H-15N HSQC spectrum of human urine tagged with 15N-cholamine. 1: aconitic acid; 2: adipic acid; 3: alanine; 5: arginine; 6: asparagine; 7: aspartic acid; 9: citric acid; 12: formic acid; 15: glutamic acid; 18: glycolic acid; 19: hippuric acid; 24: isocitric acid; 28: lactic acid; 33: malonic acid; 39: propionic acid; 40: pyroglutamic acid; 43: succinic acid; 45: threonine; 46: tryptophan.
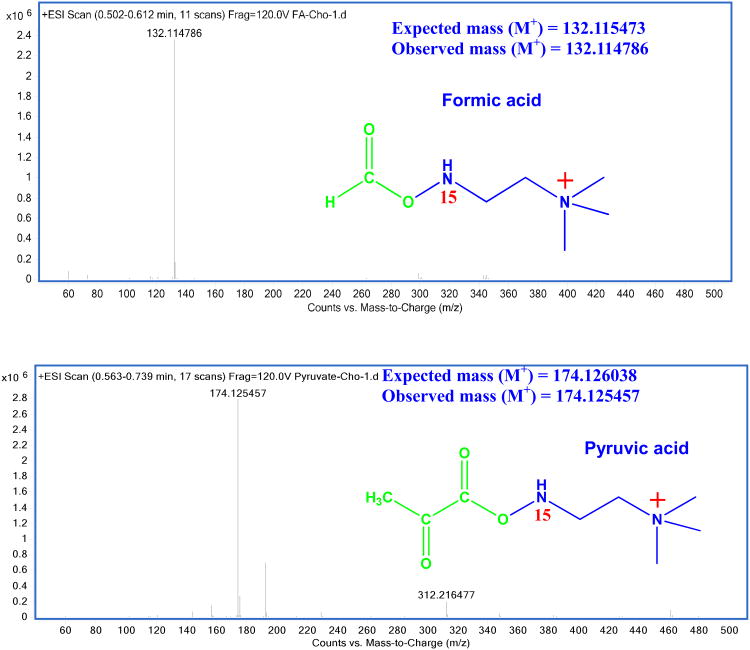
The 15N-cholamine tagging of metabolites in aqueous media enabled a sensitivity enhancement of up to three orders of magnitude or more in the MS detection of carboxyl metabolites. The derivatized metabolites could be detected easily in positive ion mode as compared to the same metabolites detected in negative ion mode without the tag. For example, the sensitivity for pyruvic acid detected in positive ion mode after 15N-cholamine tagging was enhanced by a factor of about 1500 when compared to that detected for the same metabolite without the 15N-cholamine tag, in negative ion mode. Figure 4 shows typical mass spectra for formic acid and pyruvic acid after tagging with 15N-cholamine. The enhancement in sensitivity is primarily due to the high ionization efficiency imparted by the permanent positive charge of the 15N-cholamine and is in agreement with results by Smith and co-workers for fatty acid analysis using the heavy and light forms of cholamine.6 In that study, reactions of metabolites with cholamine were made in organic solution in contrast to the aqueous media used here. The 15N-cholamine derivatized serum samples were then analyzed by LC-MS. As anticipated, due to the presence of the permanent positive charge, tagged metabolites could be readily detected in positive ion mode with high sensitivity. Sensitivity enhancement by a factor of up to nearly 3000 could be achieved for tagged acids. The extracted ion chromatograms for a few typical carboxylic acids detected in serum with 15N-cholamine tag are shown in the Supplementary Figure S4.
Figure 4.
Typical LC-QTOF-MS spectra for formic acid and pyruvic acid obtained after tagging with the smart isotope tag, 15N-cholamine. The permanent charge on the tagged metabolites enables their sensitive detection; the observed peak is from the intact tagged metabolite.
One potential issue is the effect on chromatographic retention time caused by the addition of the cholamine tag. However, separation of the tagged metabolites using HILIC columns offers opportunity to effectively separate before detection using MS. For example, the results of separation of a mixture of standard carboxylic and amino acids performed using a HILIC column, without attempting to optimize chromatography conditions, indicate that 15N-cholamine tagged metabolites can be separated effectively (Supplementary Figure S5). More broadly, we can contemplate the use of dual purpose smart tags for other NMR-MS combinations. For GC-MS, the addition of a charged species will likely cause problems related to reduced volatility; however, a different tag, such as 13C or even 29Si labeled silyl-type tags can be contemplated.22 Another alternative is the use of smart tags for capillary electrophoresis (CE) coupled to MS, which is increasingly of interest in metabolomics.23 In fact, positively charged derivatization agents (based on pyridinum containing compounds) have been demonstrated for the use of metabolite profiling of carboxylic acids by CE-MS.24 Thus, the potential for the use of smart tags such as cholamine for CE-MS and NMR is quite promising.
In conclusion, we have developed a smart isotope tag, 15N-cholamine, which possesses dual characteristics for metabolite profiling in complex biological mixtures using the powerful analytical techniques of NMR and MS. By combining the individual strengths of the 15N label and permanent charge, the smart isotope tag facilitates detection of carboxyl-containing metabolome by both NMR and LC-MS techniques with high sensitivity. Detection of the same metabolites by both NMR and MS (Supplementary Figure S6), effectively opens unique opportunities for identification of unknown metabolites and direct comparison of metabolite data from the two powerful analytical platforms.
Supplementary Material
Acknowledgments
This work was supported by a grant from the National Institutes of Health (2R01GM085291). We thank the reviewer who suggested we think more broadly about possible applications of smart tags.
Footnotes
Financial Disclosure: DR is an officer at Matrix-Bio.
References
- 1.Gowda GAN, Zhang S, Gu H, Asiago V, Shanaiah N, Raftery D. Expert Rev Diagn. 2008;8(5):617–33. doi: 10.1586/14737159.8.5.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shintu L, Banudin R, Navratil V, Prot JM, Pontoizeau C, Defernez M, Blaise BJ, Domange C, PÉRY AR, Toulhoat P. Anal Chem. 2012;84:1840–48. doi: 10.1021/ac2011075. [DOI] [PubMed] [Google Scholar]
- 3.Wishart DS. Trends in Food Sci Technol. 2008;19:482–93. [Google Scholar]
- 4.Lindon JC, Holmes E, Nicholson JK. Pharm Res. 2006;23:1075–88. doi: 10.1007/s11095-006-0025-z. [DOI] [PubMed] [Google Scholar]
- 5.Veldhoen N, Ikonomou MG, Helbing CC. Ecotoxicol Environ Saf. 2012;76:23–38. doi: 10.1016/j.ecoenv.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 6.Lamos SM, Shortreed MR, Frey BL, Belshaw PJ, Smith LM. Anal Chem. 2007;79:5143–9. doi: 10.1021/ac062416m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang WC, Adamec J, Regnier FE. Anal Chem. 2007;79:5150–7. doi: 10.1021/ac070311t. [DOI] [PubMed] [Google Scholar]
- 8.Yang WC, Sedlak M, Regnier FE, Mosier N, Ho N, Adamec J. Anal Chem. 2008;80:9508–16. doi: 10.1021/ac801693c. [DOI] [PubMed] [Google Scholar]
- 9.Yang WC, Regnier FE, Silva D, Adamec J. J Chromatogr B. 2008;870:233–40. doi: 10.1016/j.jchromb.2008.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang WC, Regnier FE, Jiang Q, Adamec J. J Chromatogr A. 2010;1217:667–75. doi: 10.1016/j.chroma.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo K, Li L. Anal Chem. 2009;81:3919–3932. doi: 10.1021/ac900166a. [DOI] [PubMed] [Google Scholar]
- 12.Wu Y, Li L. Anal Chem. 2013 Mar 29; doi: 10.1021/ac400330z. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 13.Ye T, Mo H, Shanaiah N, Gowda GAN, Zhang S, Raftery D. Anal Chem. 2009;81(12):4882–8. doi: 10.1021/ac900539y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ye T, Zhang S, Mo H, Tayyari F, Nagana Gowda GA, Raftery D. Anal Chem. 2010;82(6):2303–9. doi: 10.1021/ac9024818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shanaiah N, Desilva MA, Nagana Gowda GA, Raftery MA, Hainline BE, Raftery D. Proc Natl Acad Sci U S A. 2007;104:11540–11544. doi: 10.1073/pnas.0704449104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeSilva MA, Shanaiah N, Gowda GAN, Rosa-Perez K, Hanson BA, Raftery D. Magn Reson Chem. 2009;47:S74–S80. doi: 10.1002/mrc.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gowda GAN, Tayyari F, Ye T, Suryani Y, Wei S, Shanaiah N, Raftery D. Anal Chem. 2010;82:8983–90. doi: 10.1021/ac101938w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iida K, Ohtaka K, Kajiwara M. J Label Compd Radiopharm. 2007;50:251. [Google Scholar]
- 19.Niyaz Khan M. J Org Chem. 1996;61(23):8063–8. doi: 10.1021/jo961172a. [DOI] [PubMed] [Google Scholar]
- 20.Kunishima M, Kawachi C, Monta J, Terao K, Iwasaki F, Tani S. Tetrahedron. 1999;55:13159–70. [Google Scholar]
- 21.Kunishima M, Kawachi C, Hioki K, Terao R, Tani S. Tetrahedron. 2001;57:1551–8. [Google Scholar]
- 22.Schraml J. Prog NMR Spect. 1990;22:289–348. [Google Scholar]
- 23.Ramautar R, Somsen GW, de Jong GJ. Electrophoresis. 2013;34(1):86–98. doi: 10.1002/elps.201200390. [DOI] [PubMed] [Google Scholar]
- 24.Yang WC, Regnier FE, Adamec J. Electrophoresis. 2008;29(22):4549–60. doi: 10.1002/elps.200800156. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.