Abstract
Genomic structural variation (SV) can be thought of on a continuum from a single base pair insertion/deletion (INDEL) to large megabase-scale rearrangements involving insertions, deletions, duplications, inversions or translocations of whole chromosomes or chromosome arms. These variants can occur in coding or non-coding DNA, they can be inherited or arise sporadically in the germline or somatic cells. Many of these events are segregating in the population and can be considered common alleles while others are new alleles and thus rare events. All species studied to date harbor structural variants and these may be benign, contributing to phenotypes such as sensory perception and immunity, or pathogenic resulting in genomic disorders including DiGeorge/velocardiofacial, Smith-Margenis, Williams-Beuren and Prader-Willi syndromes. As structural variants are identified, validated and their significance, origin and prevalence elucidated it is of critical importance that this data be collected and collated in a way that can be easily accessed and analyzed.
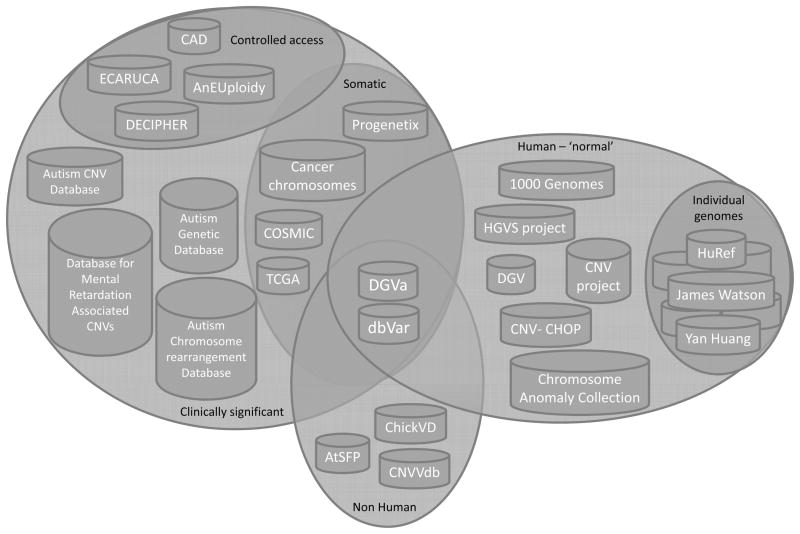
This chapter will describe current structural variation online resources (see Figure 1 and Table 1), highlight the challenges in capturing, storing and displaying SV data, and discuss how dbVar and DGVa, the genomic structural variation databases developed at NCBI and EBI respectively, were designed to address these issues.
Keywords: CNV, INDEL, SV, dbVar, DGVa, DGV
1. Introduction
In 1991, Charcot-Marie Tooth (CMT) disease was the first autosomal dominant disease associated with a gene dosage effect due to an inherited DNA rearrangement (1). It is widely accepted that copy number variants (CNVs) account for a number of genomic disorders including DiGeorge/velocardiofacial, Smith-Margenis, Williams-Beuren and Prader-Willi syndromes and number of genomic disorders is increasing including a learning disability phenotype associated with a 17q21.31 microdeletion (2, 3) and most recently with autism spectrum disorder associated with a 16p11.2 microdeletion (4).
In addition, evidence for copy number variation in disease resistance and susceptibility in humans is accumulating with publications on CCL3L1 and susceptibility to HIV/AIDS (5), FCGR3B and risk of systemic lupus erythematosus (6) and several independent studies correlating copy number of the beta defensin genes with predisposition to Crohn’s disease (7), risk of psoriasis (8) and sporadic prostate cancer (9).
Many genes that are found to be CNV (both in humans and in mouse) are involved in environmental response, for example sensory perception (olfactory receptors) and immunity (defensins) (10–15).
Although single nucleotide polymorphisms (SNPs) were initially thought to contribute the majority of human genomic variation (16, 17) it is now recognized that structural variation represents a significant, and at present poorly understood, contribution to an individual’s genetic diversity. It is only within the past 6 years, aided by the development of technologies such as high-throughput sequencing, paired-end mapping (PEM) and array comparative genome hybridization (aCGH), that the extent of structural variation in phenotypically normal individuals has been investigated.
As advances in technology are making it easier, faster and cheaper to sequence and analyze the genomes both within and between individuals of many species, numerous SV resources are being made available to access and analyze the data. Although not an exhaustive list, a number of these current resources are described below.
2. Resources
2.1 ‘Normal’ structural variation
Variants found in individuals that are healthy, or who have not been phenotyped, are often referred to as ‘normal’, or common variants. This is not to say that they are not of phenotypic consequence, merely that they have no known association with any disease phenotype. Several resources provide access to common structural variation data online and are described below.
2.1.1 Individual genomes
At present there are three purely de novo human genome assemblies, the GRCh37 reference sequence (18), Celera and HuRef (Venter) (19), although many of the read in the Celera assembly came from Craig Venter. The alignment of these assemblies to each other provides large scale and can be viewed in NCBI MapViewer (20). In addition, an increasing number of individual genomes, including James Watson (21) and Yan Huang (22), have been sequenced using next-generation technology (21–25). Both James Watson’s and Yan Huang’s genome sequences were aligned to NCBI36 (the previous version of GRCh37). The resulting SVs are available for download and can be viewed via the James Watson Genome Browser (26) and the Yan Huang GBrowser (27). Recently, de novo scaffolds of an Asian and African genome were integrated with NCBI36 to uncover move structural variation (PMID: 19997067)
The Copy Number Variation (CNV) Project
The Genome Structural Variation Consortium CNV Project (26) is a collaboration between groups at the Wellcome Trust Sanger Institute (Cambridge, UK), Harvard Medical School (Boston, USA) and the Hospital for Sick Children (Toronto, Canada). The CNV project provides data from the analysis of copy number variation in the 270 Phase I and II HapMap samples using aCGH with a genome-wide Whole Genome TilePath (WGTP) array consisting of ~27,000 bacterial artificial chromosome (BAC) clones using a single male reference, Coriell individual NA10851 (27). The project also conducted a CNV discovery project to identify common CNVs greater than 500 bp in size using a set of NimbleGen arrays consisting of ~42 million probes. They analyzed 40 females with European or African ancestry, against the same single male reference sample, Coriell individual NA10851. The SV data is available for download from the CNV Project website (26) which provides links to view the data as tracks in the UCSC (28) or Ensembl (29) genome browsers.
2.1.2 Human Genome Structural Variation (HGVS) Project
The Human Genome Structural Variation project (30) based at the University of Washington (Seattle, USA) aims to characterize structural variation at the sequence level. The project involves sequencing the ends of fosmids and BACs from multiple individuals and aligning them to NCBI35. The database currently contains results from an initial analysis of eight individuals (31, 32). The data, including validate SV sites and novel sequence not in NCBI35, are available for download and a link provides access to this data as tracks in the UCSC genome browser (30).
2.1.3 1000 Genomes
The 1000 Genomes project (33) is an international research consortium formed to create the most detailed and medically useful picture to date of human genetic variation from the sequencing and analysis of 1000 individuals. The project is expected to release high quality, validated structural variation data early 2010.
2.1.4 The Copy Number Variation project at the Children’s Hospital of Philadelphia (CHOP)
The CNV project at CHOP (34) represents an effort to identify all frequent copy number variations that exist in the human genome. Ongoing research uses the Illumina HumanHap 550 BeadChip to generate genotype data which is analyzed for CNVs using Illumina’s BeadStudio software in combination with in-house CNV detection methodologies. The database currently contains CNVs identified in 2,026 healthy children (35). This data is available for download in NCB35 coordinates and can be viewed in a genome browser (34).
2.1.6 Chromosome Anomaly Collection
The Chromosome Anomaly Collection (36) contains cases of unbalanced chromosome abnormalities (UBCAs) without phenotypic effect that have been directly transmitted from parents to children. The Collection also includes the cytogenetically visible euchromatic variants that can now be regarded as part of the continuum of copy number variation in the human genome. Cytogenetic data is represented on ideograms and provided in tables on the website.
2.1.7 Non-human structural variation
A number of studies have used aCGH to investigate the copy number of genes in other species, such as mouse (12, 37), rat (38) and macaque (39), and between human and other primate species including chimpanzee, bonobo, gorilla, orangutan and macaque in an attempt to define lineage-specific genes that may aid in understanding genome evolution (40–44).
As the sequence quality of non-human genomes increases a number of species-specific and interspecies databases are emerging including:
AtSFP (45), the Salk Institute Genome Analysis Laboratory (SIGnAL) Arabidopsis Single Feature Polymorphism database and genome browser (46).
ChickVD (47), the Beijing Genomics Institute Chicken Variation Database, which so far contains ~2.8 million non-redundant SNPs and 0.3 million indels. The data is available for download and can be viewed in a genome browser (48).
CNVVdb (49), the Taipei Genomics Research Center at Academia Sinica Copy Number Variations across Vertebrate genomes database that identifies potential inter-species CNVs by finding duplicated regions within a genome (paralogues) and between different genomes (orthologues) from pairwise sequence alignments between 16 vertebrate species (50).
2.1.7 Database of Genomic Variants (DGV)
The Database of Genomic Variants (51) is to date the most comprehensive database for the deposition, retrieval and visualization of phenotypically normal human structural variation. The database is continuously updated and curated with new data from peer reviewed research studies. Generally regions >3Mb are excluded and those 100bp-1kb are displayed in an indel track. Currently, original SV data from 35 publications is available to download. In addition these variants are mapped to NCBI35 and NCBI36 using the UCSC liftover tool (52) where necessary. This data can be downloaded and also viewed in a genome browser.
2.2 Clinically significant structural variation
Although only a small fraction of structural variants have been experimentally proven to be causative of a disease there are many variants that have been identified in individuals with a disease phenotype. As many of these variants are rare, only by collating this data and comparing with variants in ‘normal’ individuals can we begin to elucidate the significance of these variants and their relationship to disease. Several resources provide access, although often controlled-access, to clinically significant structural variation data online and are described below.
2.2.1 AnEUploidy Project
The goal of the AnEUploidy Project (53) is to understand the molecular mechanisms of gene dosage imbalance (aneuploidy) in human health and includes the identification and characterization of novel microaneuploidy syndromes and the establishment of a catalogue of copy number variants (CNVs) and segmental duplications (SDs) in Europeans. Access to clinical data is under controlled access.
2.2.2 Chromosome Abnormality Database (CAD)
CAD (54) is an online collection of both constitutional and acquired abnormal karyotypes reported by UK Regional cytogenetics centers and holds over 150,000 records collected from all UK NHS laboratories. Access to clinical data is under controlled access.
2.2.3 ECARUCA
ECARUCA (55), the European Cytogeneticists Association Register of Unbalanced Chromosome Aberrations, is a database which collects and provides cytogenetic and clinical information on rare chromosomal disorders, including microdeletions. The Register contains over 4500 cases with more than 5500 aberrations and links are provided to view all cases smaller than 30 Mb on NCBI35 in the UCSC or Ensembl genome browsers. Access to clinical data is under controlled access.
2.2.4 DECIPHER
DECIPHER (56), the DatabasE of Chromosomal Imbalance and Phenotype in Humans using Ensembl Resources, is a database of submicroscopic chromosomal imbalance that includes clinical information about chromosomal microdeletions/duplications/insertions, translocations and inversions. Coordinates are mapped to NCBI36 and access to non-consented clinical data is under controlled access.
2.2.5 Mental Retardation
The Database for Mental Retardation Associated CNVs (57) is a publicly accessible test database from the University of Tartu, Estonia, that gathers information about CNVs and related diseases.
2.2.6 Autism
There are several structural variation databases for autism spectrum disorder:
The Autism Chromosome Rearrangement Database (58) is a collection of hand curated breakpoints and other genomic features, related to autism, taken from publicly available literature, databases and unpublished data. This data can be viewed in NCB36 coordinates in a genome browser.
The Autism CNV Database (59) provides CNV data that was obtained using the Affymetrix GeneChip Mapping 10K 2.0 (60) and Affymetrix Whole Genome mapping 10K and 500K microarrays (61). This data can be viewed in NCB35 coordinates in a genome browser.
2.3 The Autism Genetic Database (62) is a comprehensive database for autism susceptibility gene-CNVs integrated with known non-coding RNAs and fragile sites and viewable in a genome browser. Cancer
It has been known for many years that somatic changes in cancer cells include gross copy number changes and structural alterations. As such several projects and databases provide access to structural variation data specifically found in cancer and are described below.
2.3.1 The Cancer Genome Project
The Wellcome Trust Sanger Institute (WTSI) Cancer Genome Project (63) is using the human genome sequence and high throughput mutation detection techniques to identify somatically acquired sequence variants/mutations and hence identify genes critical in the development of human cancers. The results from this work are collated and stored in the Catalogue of Somatic Mutations in Cancer, COSMIC (64), which also contains somatic mutation data published in the scientific literature.
2.3.2 The Cancer Genome Atlas (TCGA)
The National Cancer Institute (NCI) and the National Human Genome Research Institute (NHGRI) launched The Cancer Genome Atlas (TCGA) (65) program to create a comprehensive atlas of the genomic changes involved in more than 20 common types of cancer. The TCGA Data Portal (66) provides a platform for researchers to search, download, and analyze data sets generated by TCGA.
2.3.3 Progenetix
The University of Zurich Progenetix (67) database collects genomic CNV information from comparative genomic hybridization (CGH) experiments of individual cancer and leukemia cases, published in peer reviewed journals (68).
2.3.4 Cancer Chromosomes
Three databases, the NCI/NCBI SKY/M-FISH & CGH Database, the NCI Mitelman Database of Chromosome Aberrations in Cancer, and the NCI Recurrent Aberrations in Cancer, are now integrated into the NCBI Entrez system as Cancer Chromosomes (69, 70).
3. Limitations
As described above there are numerous resources for accessing and viewing structural variation. However, when interpreting data, or collating data from different databases, there are several limitations that should be taken into consideration and are described below.
3.1 Choice of technology
Currently, no single technology can accurately identify all structural variants in a sample. When comparing SV data derived from different methodologies, even if used on the same sample or samples, it is likely that the reported SVs will differ due to the resolution, genome coverage and variant type detectable by the chosen method. While the decreasing costs of sequencing will eventually make it feasible for labs to routinely sequence and genotype whole genomes the cost is still prohibitive enough to make aCGH and PEM the preferred methods of choice for SV detection in many labs.
aCGH generally uses two samples to determine relative gains and losses between the two. Using BAC aCGH, with a low resolution of ~1 Mb, means the identified region may contain several smaller CNVs and the extent of the CNV is often overestimated. Oligonucleotide arrays, with 45–85 bp probes, offer higher resolution than BAC arrays but the exact breakpoints of the structural variants can still not be accurately determined. Resolution of oligo arrays is in the order of 1 probe every 1–5 kb for whole genome arrays and greater than 1 probe every 50 bp for custom arrays. Oligo SNP arrays have an average resolution of 1 probe every 1–5 kb and can also be used to discover segregating deletion variants evident from genotyping patterns of null genotypes, Mendelian inconsistencies and Hardy-Weinberg disequilibrium (10, 71). However, the detection is limited to areas of the genome containing SNPs, while many regions of known structural variation are sparsely covered by SNPs, and many of the oligo arrays only contain probes to single copy regions of the genome. This is being improved by the new microarrays from, for example, NimbleGen (72) and Illumina (73) that contain >2 million probes and include coverage of novel CNV regions such as segmental duplications, megasatellites, and other unstable regions of the genome. However, array based methods are always limited to data known sequence, typically the reference assembly, and don’t provide for the identification of novel sequences.
Paired-end mapping (PEM) uses the end sequences of BACs, fosmids, and most recently 3 kb DNA fragments from next-generation sequencing technologies (74), to compare to a reference genome. The advantage of this methodology over aCGH is that it not only allows identification of insertions and deletion but also allows detection of balanced translocations and inversions, small indels from the end sequence alignments and novel insert sequences not present in the reference genome, all of which are not amenable to aCGH. However, like aCGH, PEM does not provide exact breakpoint resolution of the structural variant but if the clones are available from a genomic library the inserts can be fully sequenced and the nature of the structural variant determined.
3.2 Choice of SV detection algorithm
In addition to the different technologies available for identifying structural variants, many of which are used in parallel for validation, interpretation of the results and detection of a region as structurally variant is open to analysis, and reanalysis, using a whole suite of different software programs and algorithms. Continually being developed, these programs include BreakPtr (75), CNAG (76), CNVfinder (77), dCHIP (78), GEMCA (79), PennCNV (80) and VariationHunter (81). Although often optimized for a particular methodology or technique, many of these algorithms can be used on the same datasets to help achieve the most accurate and reproducible consensus set of SVs. Many of the array datasets are available via GEO (82) or ArrayExpress (83) and many sequences generated through paired-end sequencing or whole genome sequencing are available via the Trace (84) or Short Read Archive (SRA) (85). These resources provide a great opportunity to allow the data available from online databases to be reanalyzed and reinterpreted using different parameters of the original algorithm or using an additional or novel algorithm.
3.3 Choice of reference genome
Structural variants are generally defined as a region on a given reference assembly eg NCBI35 or GRCh37. However, the reference genome against which the SVs were identified may be different. aCGH identifies regions CNV between two different samples but the locations are placed on a reference assembly by virtue of the coordinates of the probes used on the arrays whereas PEM aligns sequences directly to the reference assembly. Hence, a ‘Loss’ displayed on a genome browser from an aCGH study is not necessarily the same as a ‘Deletion’ displayed on the same reference assembly in the genome browser from a PEM study. Indeed, an aCGH ‘Loss’ may even be the same as a PEM ‘Duplication’. Another major limitation of PEM, and other sequence analysis reliant on the reference assembly for comparison, is that a common loss or gain in the sample could simply reflect the presence of a minor allele in the reference. Therefore, the choice of reference genome should be taken into consideration when collating data across studies, even if reported on the same reference assembly.
3.4 Coordinate remapping
A major limitation with SVs being reported on a particular genome assembly eg NCBI35 is that they are often not transposable to a new or different assembly eg GRCh37 due to their complicated genomic structure (21, 31, 86). UCSC provides a liftover tool (52) to allow carryover of coordinates to a new assembly but essentially 10–15% of mappings are lost. In order to compare variants from different studies they all need to be reliable mapped to the same genome assembly and effort is underway at NCBI to develop a robust method for remapping.
3.5 Nomenclature
While the field of structural variation is still relatively new, and the methods and analyses are continuously changing, there is still a need for controlled vocabularies to facilitate searching and access of data. For example, depending on the technology, detection algorithm or the resource the data was submitted to, the same CNV could be described as a gain, duplication or amplification. In order to compare variants from different studies it would be of great benefit to the field if, for example, the methodologies and variant types could be defined as a controlled vocabulary and used throughout all the SV online resources and peer-reviewed publications.
3.6 Patient consent
Many clinical studies are under controlled access due to patient confidentiality. Unfortunately this means there is a wealth of structural variant data stored in controlled access databases such as dbGaP (87) and EGA (88) that cannot be incorporated into the public databases. As the significance of many of these SVs cannot be determined until compared with other studies there is a need to deposit de-identified and/or summary information from these controlled access database into the public domain.
4. dbVar and DGVa
As described in the previous sections, there are numerous structural variation databases but also numerous limitations. To address these issues NCBI and EBI (in collaboration with the Database of Genomic Variants) have recently launched the databases of genomic structural variation dbVar (89) and DGVa (90) respectively with the aims to:
Accession and track individual objects by providing study and variant accessions
Provide access to raw datasets for reanalysis (via links to eg GEO (82), ArrayExpress (83), Trace Archive (84) and SRA (85))
Represent both normal and clinically relevant data
Use controlled vocabularies where possible to facilitate searching
Represent data not on a sequenced genome assembly
Provide robust remapping
Provide resolution/confidence values to access quality of the data
Provide validation data
Store genotyping information to distinguish homozygous vs heterozygous variants
Store sample information to distinguish germline vs somatic variants
Provide summary data for controlled access data in dbGaP and EGA
Display data for species other than human
dbVar and DGVa are accepting submissions of structural variant data and will continue to develop to meet the needs of the community as the technology and analysis methods evolve.
Figure 1.
Online structural variation resources
Table 1.
Online resources for structural genomic variation
Acknowledgments
This research was supported [in part] by the Intramural Research Program of the NIH, National Library of Medicine.
References
- 1.Lupski JR, de Oca-Luna RM, Slaugenhaupt S, Pentao L, Guzzetta V, Trask BJ, Saucedo-Cardenas O, Barker DF, Killian JM, Garcia CA, Chakravarti A, Patel PI. DNA duplication associated with Charcot-Marie-Tooth disease type 1A. Cell. 1991;66:219–232. doi: 10.1016/0092-8674(91)90613-4. [DOI] [PubMed] [Google Scholar]
- 2.Koolen DA, Vissers LE, Pfundt R, de Leeuw N, Knight SJ, Regan R, Kooy RF, Reyniers E, Romano C, Fichera M, Schinzel A, Baumer A, Anderlid BM, Schoumans J, Knoers NV, van Kessel AG, Sistermans EA, Veltman JA, Brunner HG, de Vries BB. A new chromosome 17q21.31 microdeletion syndrome associated with a common inversion polymorphism. Nat Genet. 2006;38:999–1001. doi: 10.1038/ng1853. [DOI] [PubMed] [Google Scholar]
- 3.Shaw-Smith C, Pittman AM, Willatt L, Martin H, Rickman L, Gribble S, Curley R, Cumming S, Dunn C, Kalaitzopoulos D, Porter K, Prigmore E, Krepischi-Santos AC, Varela MC, Koiffmann CP, Lees AJ, Rosenberg C, Firth HV, de Silva R, Carter NP. Microdeletion encompassing MAPT at chromosome 17q21.3 is associated with developmental delay and learning disability. Nat Genet. 2006;38:1032–1037. doi: 10.1038/ng1858. [DOI] [PubMed] [Google Scholar]
- 4.Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, Fossdal R, Saemundsen E, Stefansson H, Ferreira MA, Green T, Platt OS, Ruderfer DM, Walsh CA, Altshuler D, Chakravarti A, Tanzi RE, Stefansson K, Santangelo SL, Gusella JF, Sklar P, Wu BL, Daly MJ. Association between Microdeletion and Microduplication at 16p11.2 and Autism. N Engl J Med. 2008 doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 5.Gonzalez E, Kulkarni H, Bolivar H, Mangano A, Sanchez R, Catano G, Nibbs RJ, Freedman BI, Quinones MP, Bamshad MJ, Murthy KK, Rovin BH, Bradley W, Clark RA, Anderson SA, O’Connell RJ, Agan BK, Ahuja SS, Bologna R, Sen L, Dolan MJ, Ahuja SK. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science. 2005;307:1434–1440. doi: 10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- 6.Fanciulli M, Norsworthy PJ, Petretto E, Dong R, Harper L, Kamesh L, Heward JM, Gough SC, de Smith A, Blakemore AI, Froguel P, Owen CJ, Pearce SH, Teixeira L, Guillevin L, Graham DS, Pusey CD, Cook HT, Vyse TJ, Aitman TJ. FCGR3B copy number variation is associated with susceptibility to systemic, but not organ-specific, autoimmunity. Nat Genet. 2007;39:721–723. doi: 10.1038/ng2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fellermann K, Stange DE, Schaeffeler E, Schmalzl H, Wehkamp J, Bevins CL, Reinisch W, Teml A, Schwab M, Lichter P, Radlwimmer B, Stange EF. A chromosome 8 gene-cluster polymorphism with low human beta-defensin 2 gene copy number predisposes to Crohn disease of the colon. Am J Hum Genet. 2006;79:439–448. doi: 10.1086/505915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hollox EJ, Huffmeier U, Zeeuwen PL, Palla R, Lascorz J, Rodijk-Olthuis D, van de Kerkhof PC, Traupe H, de Jongh G, den Heijer M, Reis A, Armour JA, Schalkwijk J. Psoriasis is associated with increased beta-defensin genomic copy number. Nat Genet. 2008;40:23–25. doi: 10.1038/ng.2007.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huse K, Taudien S, Groth M, Rosenstiel P, Szafranski K, Hiller M, Hampe J, Junker K, Schubert J, Schreiber S, Birkenmeier G, Krawczak M, Platzer M. Genetic Variants of the Copy Number Polymorphic beta-Defensin Locus Are Associated with Sporadic Prostate Cancer. Tumour Biol. 2008;29:83–92. doi: 10.1159/000135688. [DOI] [PubMed] [Google Scholar]
- 10.Conrad DF, Andrews TD, Carter NP, Hurles ME, Pritchard JK. A high-resolution survey of deletion polymorphism in the human genome. Nat Genet. 2006;38:75–81. doi: 10.1038/ng1697. [DOI] [PubMed] [Google Scholar]
- 11.Cooper GM, Nickerson DA, Eichler EE. Mutational and selective effects on copy-number variants in the human genome. Nat Genet. 2007;39:S22–29. doi: 10.1038/ng2054. [DOI] [PubMed] [Google Scholar]
- 12.Cutler G, Marshall LA, Chin N, Baribault H, Kassner PD. Significant gene content variation characterizes the genomes of inbred mouse strains. Genome research. 2007;17:1743–1754. doi: 10.1101/gr.6754607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graubert TA, Cahan P, Edwin D, Selzer RR, Richmond TA, Eis PS, Shannon WD, Li X, McLeod HL, Cheverud JM, Ley TJ. A high-resolution map of segmental DNA copy number variation in the mouse genome. PLoS Genet. 2007;3:e3. doi: 10.1371/journal.pgen.0030003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nguyen DQ, Webber C, Ponting CP. Bias of selection on human copy-number variants. PLoS Genet. 2006;2:e20. doi: 10.1371/journal.pgen.0020020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wong KK, de Leeuw RJ, Dosanjh NS, Kimm LR, Cheng Z, Horsman DE, MacAulay C, Ng RT, Brown CJ, Eichler EE, Lam WL. A comprehensive analysis of common copy-number variations in the human genome. Am J Hum Genet. 2007;80:91–104. doi: 10.1086/510560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 17.Sachidanandam R, Weissman D, Schmidt SC, Kakol JM, Stein LD, Marth G, Sherry S, Mullikin JC, Mortimore BJ, Willey DL, Hunt SE, Cole CG, Coggill PC, Rice CM, Ning Z, Rogers J, Bentley DR, Kwok PY, Mardis ER, Yeh RT, Schultz B, Cook L, Davenport R, Dante M, Fulton L, Hillier L, Waterston RH, McPherson JD, Gilman B, Schaffner S, Van Etten WJ, Reich D, Higgins J, Daly MJ, Blumenstiel B, Baldwin J, Stange-Thomann N, Zody MC, Linton L, Lander ES, Altshuler D. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature. 2001;409:928–933. doi: 10.1038/35057149. [DOI] [PubMed] [Google Scholar]
- 18.NCBI . http://www.ncbi.nlm.nih.gov/
- 19.HuRef Project. http://huref.jcvi.org/index.html.
- 20.MapViewer. http://www.ncbi.nlm.nih.gov/projects/mapview/
- 21.Wheeler DA, Srinivasan M, Egholm M, Shen Y, Chen L, McGuire A, He W, Chen YJ, Makhijani V, Roth GT, Gomes X, Tartaro K, Niazi F, Turcotte CL, Irzyk GP, Lupski JR, Chinault C, Song XZ, Liu Y, Yuan Y, Nazareth L, Qin X, Muzny DM, Margulies M, Weinstock GM, Gibbs RA, Rothberg JM. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- 22.Wang J, Wang W, Li R, Li Y, Tian G, Goodman L, Fan W, Zhang J, Li J, Zhang J, Guo Y, Feng B, Li H, Lu Y, Fang X, Liang H, Du Z, Li D, Zhao Y, Hu Y, Yang Z, Zheng H, Hellmann I, Inouye M, Pool J, Yi X, Zhao J, Duan J, Zhou Y, Qin J, Ma L, Li G, Yang Z, Zhang G, Yang B, Yu C, Liang F, Li W, Li S, Li D, Ni P, Ruan J, Li Q, Zhu H, Liu D, Lu Z, Li N, Guo G, Zhang J, Ye J, Fang L, Hao Q, Chen Q, Liang Y, Su Y, San A, Ping C, Yang S, Chen F, Li L, Zhou K, Zheng H, Ren Y, Yang L, Gao Y, Yang G, Li Z, Feng X, Kristiansen K, Wong GK, Nielsen R, Durbin R, Bolund L, Zhang X, Li S, Yang H, Wang J. The diploid genome sequence of an Asian individual. Nature. 2008;456:60–65. doi: 10.1038/nature07484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ahn SM, Kim TH, Lee S, Kim D, Ghang H, Kim DS, Kim BC, Kim SY, Kim WY, Kim C, Park D, Lee YS, Kim S, Reja R, Jho S, Kim CG, Cha JY, Kim KH, Lee B, Bhak J, Kim SJ. The first Korean genome sequence and analysis: full genome sequencing for a socio-ethnic group. Genome research. 2009;19:1622–1629. doi: 10.1101/gr.092197.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, Boutell JM, Bryant J, Carter RJ, Keira Cheetham R, Cox AJ, Ellis DJ, Flatbush MR, Gormley NA, Humphray SJ, Irving LJ, Karbelashvili MS, Kirk SM, Li H, Liu X, Maisinger KS, Murray LJ, Obradovic B, Ost T, Parkinson ML, Pratt MR, Rasolonjatovo IM, Reed MT, Rigatti R, Rodighiero C, Ross MT, Sabot A, Sankar SV, Scally A, Schroth GP, Smith ME, Smith VP, Spiridou A, Torrance PE, Tzonev SS, Vermaas EH, Walter K, Wu X, Zhang L, Alam MD, Anastasi C, Aniebo IC, Bailey DM, Bancarz IR, Banerjee S, Barbour SG, Baybayan PA, Benoit VA, Benson KF, Bevis C, Black PJ, Boodhun A, Brennan JS, Bridgham JA, Brown RC, Brown AA, Buermann DH, Bundu AA, Burrows JC, Carter NP, Castillo N, Chiara ECM, Chang S, Neil Cooley R, Crake NR, Dada OO, Diakoumakos KD, Dominguez-Fernandez B, Earnshaw DJ, Egbujor UC, Elmore DW, Etchin SS, Ewan MR, Fedurco M, Fraser LJ, Fuentes Fajardo KV, Scott Furey W, George D, Gietzen KJ, Goddard CP, Golda GS, Granieri PA, Green DE, Gustafson DL, Hansen NF, Harnish K, Haudenschild CD, Heyer NI, Hims MM, Ho JT, Horgan AM, Hoschler K, Hurwitz S, Ivanov DV, Johnson MQ, James T, Huw Jones TA, Kang GD, Kerelska TH, Kersey AD, Khrebtukova I, Kindwall AP, Kingsbury Z, Kokko-Gonzales PI, Kumar A, Laurent MA, Lawley CT, Lee SE, Lee X, Liao AK, Loch JA, Lok M, Luo S, Mammen RM, Martin JW, McCauley PG, McNitt P, Mehta P, Moon KW, Mullens JW, Newington T, Ning Z, Ling Ng B, Novo SM, O’Neill MJ, Osborne MA, Osnowski A, Ostadan O, Paraschos LL, Pickering L, Pike AC, Pike AC, Chris Pinkard D, Pliskin DP, Podhasky J, Quijano VJ, Raczy C, Rae VH, Rawlings SR, Chiva Rodriguez A, Roe PM, Rogers J, Rogert Bacigalupo MC, Romanov N, Romieu A, Roth RK, Rourke NJ, Ruediger ST, Rusman E, Sanches-Kuiper RM, Schenker MR, Seoane JM, Shaw RJ, Shiver MK, Short SW, Sizto NL, Sluis JP, Smith MA, Ernest Sohna Sohna J, Spence EJ, Stevens K, Sutton N, Szajkowski L, Tregidgo CL, Turcatti G, Vandevondele S, Verhovsky Y, Virk SM, Wakelin S, Walcott GC, Wang J, Worsley GJ, Yan J, Yau L, Zuerlein M, Rogers J, Mullikin JC, Hurles ME, McCooke NJ, West JS, Oaks FL, Lundberg PL, Klenerman D, Durbin R, Smith AJ. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim JI, Ju YS, Park H, Kim S, Lee S, Yi JH, Mudge J, Miller NA, Hong D, Bell CJ, Kim HS, Chung IS, Lee WC, Lee JS, Seo SH, Yun JY, Woo HN, Lee H, Suh D, Lee S, Kim HJ, Yavartanoo M, Kwak M, Zheng Y, Lee MK, Park H, Kim JY, Gokcumen O, Mills RE, Zaranek AW, Thakuria J, Wu X, Kim RW, Huntley JJ, Luo S, Schroth GP, Wu TD, Kim H, Yang KS, Park WY, Kim H, Church GM, Lee C, Kingsmore SF, Seo JS. A highly annotated whole-genome sequence of a Korean individual. Nature. 2009;460:1011–1015. doi: 10.1038/nature08211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.The Copy Number Variation (CNV) Project. http://www.sanger.ac.uk/humgen/cnv/
- 27.Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, Cho EK, Dallaire S, Freeman JL, Gonzalez JR, Gratacos M, Huang J, Kalaitzopoulos D, Komura D, MacDonald JR, Marshall CR, Mei R, Montgomery L, Nishimura K, Okamura K, Shen F, Somerville MJ, Tchinda J, Valsesia A, Woodwark C, Yang F, Zhang J, Zerjal T, Zhang J, Armengol L, Conrad DF, Estivill X, Tyler-Smith C, Carter NP, Aburatani H, Lee C, Jones KW, Scherer SW, Hurles ME. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.UCSC Genome Browser. http://genome.ucsc.edu/
- 29.Ensembl Genome Browser. http://www.ensembl.org/
- 30.Human Genome Structural Variation Project. http://hgsv.washington.edu/
- 31.Kidd JM, Cooper GM, Donahue WF, Hayden HS, Sampas N, Graves T, Hansen N, Teague B, Alkan C, Antonacci F, Haugen E, Zerr T, Yamada NA, Tsang P, Newman TL, Tuzun E, Cheng Z, Ebling HM, Tusneem N, David R, Gillett W, Phelps KA, Weaver M, Saranga D, Brand A, Tao W, Gustafson E, McKernan K, Chen L, Malig M, Smith JD, Korn JM, McCarroll SA, Altshuler DA, Peiffer DA, Dorschner M, Stamatoyannopoulos J, Schwartz D, Nickerson DA, Mullikin JC, Wilson RK, Bruhn L, Olson MV, Kaul R, Smith DR, Eichler EE. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tuzun E, Sharp AJ, Bailey JA, Kaul R, Morrison VA, Pertz LM, Haugen E, Hayden H, Albertson D, Pinkel D, Olson MV, Eichler EE. Fine-scale structural variation of the human genome. Nat Genet. 2005;37:727–732. doi: 10.1038/ng1562. [DOI] [PubMed] [Google Scholar]
- 33.1000 Genomes Project. http://www.1000genomes.org.
- 34.The Copy Number Variation project at the Children’s Hospital of Philadelphia (CHOP) http://cnv.chop.edu/
- 35.Shaikh TH, Gai X, Perin JC, Glessner JT, Xie H, Murphy K, O’Hara R, Casalunovo T, Conlin LK, D’Arcy M, Frackelton EC, Geiger EA, Haldeman-Englert C, Imielinski M, Kim CE, Medne L, Annaiah K, Bradfield JP, Dabaghyan E, Eckert A, Onyiah CC, Ostapenko S, Otieno FG, Santa E, Shaner JL, Skraban R, Smith RM, Elia J, Goldmuntz E, Spinner NB, Zackai EH, Chiavacci RM, Grundmeier R, Rappaport EF, Grant SF, White PS, Hakonarson H. High-resolution mapping and analysis of copy number variations in the human genome: a data resource for clinical and research applications. Genome research. 2009;19:1682–1690. doi: 10.1101/gr.083501.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chromosome Anomaly Collection. http://www.ngrl.org.uk/Wessex/collection/index.htm.
- 37.Snijders AM, Nowak NJ, Huey B, Fridlyand J, Law S, Conroy J, Tokuyasu T, Demir K, Chiu R, Mao JH, Jain AN, Jones SJ, Balmain A, Pinkel D, Albertson DG. Mapping segmental and sequence variations among laboratory mice using BAC array CGH. Genome research. 2005;15:302–311. doi: 10.1101/gr.2902505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guryev V, Saar K, Adamovic T, Verheul M, van Heesch SA, Cook S, Pravenec M, Aitman T, Jacob H, Shull JD, Hubner N, Cuppen E. Distribution and functional impact of DNA copy number variation in the rat. Nat Genet. 2008;40:538–545. doi: 10.1038/ng.141. [DOI] [PubMed] [Google Scholar]
- 39.Lee AS, Gutierrez-Arcelus M, Perry GH, Vallender EJ, Johnson WE, Miller GM, Korbel JO, Lee C. Analysis of copy number variation in the rhesus macaque genome identifies candidate loci for evolutionary and human disease studies. Hum Mol Genet. 2008 doi: 10.1093/hmg/ddn002. [DOI] [PubMed] [Google Scholar]
- 40.Fortna A, Kim Y, MacLaren E, Marshall K, Hahn G, Meltesen L, Brenton M, Hink R, Burgers S, Hernandez-Boussard T, Karimpour-Fard A, Glueck D, McGavran L, Berry R, Pollack J, Sikela JM. Lineage-specific gene duplication and loss in human and great ape evolution. PLoS Biol. 2004;2:E207. doi: 10.1371/journal.pbio.0020207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goidts V, Armengol L, Schempp W, Conroy J, Nowak N, Muller S, Cooper DN, Estivill X, Enard W, Szamalek JM, Hameister H, Kehrer-Sawatzki H. Identification of large-scale human-specific copy number differences by inter-species array comparative genomic hybridization. Hum Genet. 2006;119:185–198. doi: 10.1007/s00439-005-0130-9. [DOI] [PubMed] [Google Scholar]
- 42.Locke DP, Segraves R, Carbone L, Archidiacono N, Albertson DG, Pinkel D, Eichler EE. Large-scale variation among human and great ape genomes determined by array comparative genomic hybridization. Genome research. 2003;13:347–357. doi: 10.1101/gr.1003303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perry GH, Tchinda J, McGrath SD, Zhang J, Picker SR, Caceres AM, Iafrate AJ, Tyler-Smith C, Scherer SW, Eichler EE, Stone AC, Lee C. Hotspots for copy number variation in chimpanzees and humans. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:8006–8011. doi: 10.1073/pnas.0602318103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilson GM, Flibotte S, Missirlis PI, Marra MA, Jones S, Thornton K, Clark AG, Holt RA. Identification by full-coverage array CGH of human DNA copy number increases relative to chimpanzee and gorilla. Genome research. 2006;16:173–181. doi: 10.1101/gr.4456006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.AtSFP - The SIGnAL Arabidopsis SNP, Deletion and SFP Database. http://signal.salk.edu/cgi-bin/AtSFP.
- 46.Borevitz JO, Hazen SP, Michael TP, Morris GP, Baxter IR, Hu TT, Chen H, Werner JD, Nordborg M, Salt DE, Kay SA, Chory J, Weigel D, Jones JD, Ecker JR. Genome-wide patterns of single-feature polymorphism in Arabidopsis thaliana. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:12057–12062. doi: 10.1073/pnas.0705323104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang J, He X, Ruan J, Dai M, Chen J, Zhang Y, Hu Y, Ye C, Li S, Cong L, Fang L, Liu B, Li S, Wang J, Burt DW, Wong GK, Yu J, Yang H, Wang J. ChickVD: a sequence variation database for the chicken genome. Nucleic acids research. 2005;33:D438–441. doi: 10.1093/nar/gki092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chicken Variation Database (ChickVD) . http://chicken.genomics.org.cn/
- 49.Copy Number Variations across Vertebrate genomes. doi: 10.1093/bioinformatics/btp166. http://cnvvdb.genomics.sinica.edu.tw/ [DOI] [PMC free article] [PubMed]
- 50.Chen FC, Chen YZ, Chuang TJ. CNVVdb: a database of copy number variations across vertebrate genomes. Bioinformatics (Oxford, England) 2009;25:1419–1421. doi: 10.1093/bioinformatics/btp166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Database of Genomic Variants. http://projects.tcag.ca/variation/
- 52.UCSC liftover tool. http://genome.ucsc.edu/cgi-bin/hgLiftOver.
- 53.AnEUploidy Project. http://www.aneuploidy.eu/
- 54.CAD (Chromosome Abnormality Database) http://www.ukcad.org.uk./cocoon/ukcad/
- 55.ECARUCA (The European Cytogeneticists Association Register of Unbalanced Chromosome Aberrations) doi: 10.1016/j.ejmg.2013.06.010. http://www.ecaruca.net/ [DOI] [PubMed]
- 56.DECIPHER (DatabasE of Chromosomal Imbalance and Phenotype in Humans using Ensembl Resources) doi: 10.1016/j.ajhg.2009.03.010. https://decipher.sanger.ac.uk/ [DOI] [PMC free article] [PubMed]
- 57.Database of Mental Retardation Associated CNVs. http://bioinfo.ut.ee/dbcard/
- 58.The Autism Chromosome Rearrangement Database. http://projects.tcag.ca/autism/
- 59.Autism CNV Database. http://projects.tcag.ca/autism_500k/
- 60.Szatmari P, Paterson AD, Zwaigenbaum L, Roberts W, Brian J, Liu XQ, Vincent JB, Skaug JL, Thompson AP, Senman L, Feuk L, Qian C, Bryson SE, Jones MB, Marshall CR, Scherer SW, Vieland VJ, Bartlett C, Mangin LV, Goedken R, Segre A, Pericak-Vance MA, Cuccaro ML, Gilbert JR, Wright HH, Abramson RK, Betancur C, Bourgeron T, Gillberg C, Leboyer M, Buxbaum JD, Davis KL, Hollander E, Silverman JM, Hallmayer J, Lotspeich L, Sutcliffe JS, Haines JL, Folstein SE, Piven J, Wassink TH, Sheffield V, Geschwind DH, Bucan M, Brown WT, Cantor RM, Constantino JN, Gilliam TC, Herbert M, Lajonchere C, Ledbetter DH, Lese-Martin C, Miller J, Nelson S, Samango-Sprouse CA, Spence S, State M, Tanzi RE, Coon H, Dawson G, Devlin B, Estes A, Flodman P, Klei L, McMahon WM, Minshew N, Munson J, Korvatska E, Rodier PM, Schellenberg GD, Smith M, Spence MA, Stodgell C, Tepper PG, Wijsman EM, Yu CE, Roge B, Mantoulan C, Wittemeyer K, Poustka A, Felder B, Klauck SM, Schuster C, Poustka F, Bolte S, Feineis-Matthews S, Herbrecht E, Schmotzer G, Tsiantis J, Papanikolaou K, Maestrini E, Bacchelli E, Blasi F, Carone S, Toma C, Van Engeland H, de Jonge M, Kemner C, Koop F, Langemeijer M, Hijmans C, Staal WG, Baird G, Bolton PF, Rutter ML, Weisblatt E, Green J, Aldred C, Wilkinson JA, Pickles A, Le Couteur A, Berney T, McConachie H, Bailey AJ, Francis K, Honeyman G, Hutchinson A, Parr JR, Wallace S, Monaco AP, Barnby G, Kobayashi K, Lamb JA, Sousa I, Sykes N, Cook EH, Guter SJ, Leventhal BL, Salt J, Lord C, Corsello C, Hus V, Weeks DE, Volkmar F, Tauber M, Fombonne E, Shih A, Meyer KJ. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet. 2007;39:319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, Skaug J, Shago M, Moessner R, Pinto D, Ren Y, Thiruvahindrapduram B, Fiebig A, Schreiber S, Friedman J, Ketelaars CE, Vos YJ, Ficicioglu C, Kirkpatrick S, Nicolson R, Sloman L, Summers A, Gibbons CA, Teebi A, Chitayat D, Weksberg R, Thompson A, Vardy C, Crosbie V, Luscombe S, Baatjes R, Zwaigenbaum L, Roberts W, Fernandez B, Szatmari P, Scherer SW. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Autism Genetic Database. http://wren.bcf.ku.edu/
- 63.The Cancer Genome Project. http://www.sanger.ac.uk/genetics/CGP/
- 64.COSMIC. http://www.sanger.ac.uk/genetics/CGP/cosmic/
- 65.The Cancer Genome Atlas. http://cancergenome.nih.gov/
- 66.TCGA Data Portal. http://tcga-data.nci.nih.gov/tcga/homepage.htm.
- 67.Progenetix. www.progenetix.net/
- 68.Baudis M, Cleary ML. Progenetix. net: an online repository for molecular cytogenetic aberration data. Bioinformatics (Oxford, England) 2001;17:1228–1229. doi: 10.1093/bioinformatics/17.12.1228. [DOI] [PubMed] [Google Scholar]
- 69.Cancer Chromosomes. http://www.ncbi.nlm.nih.gov/cancerchromosomes.
- 70.Knutsen T, Gobu V, Knaus R, Padilla-Nash H, Augustus M, Strausberg RL, Kirsch IR, Sirotkin K, Ried T. The interactive online SKY/M-FISH & CGH database and the Entrez cancer chromosomes search database: linkage of chromosomal aberrations with the genome sequence. Genes, chromosomes & cancer. 2005;44:52–64. doi: 10.1002/gcc.20224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McCarroll SA, Hadnott TN, Perry GH, Sabeti PC, Zody MC, Barrett JC, Dallaire S, Gabriel SB, Lee C, Daly MJ, Altshuler DM. Common deletion polymorphisms in the human genome. Nat Genet. 2006;38:86–92. doi: 10.1038/ng1696. [DOI] [PubMed] [Google Scholar]
- 72.NimbleGen. http://www.nimblegen.com.
- 73.Illumina. http://www.illumina.com/
- 74.Korbel JO, Urban AE, Affourtit JP, Godwin B, Grubert F, Simons JF, Kim PM, Palejev D, Carriero NJ, Du L, Taillon BE, Chen Z, Tanzer A, Saunders AC, Chi J, Yang F, Carter NP, Hurles ME, Weissman SM, Harkins TT, Gerstein MB, Egholm M, Snyder M. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318:420–426. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.BreakPtr. http://breakptr.gersteinlab.org.
- 76.CNAG. http://www.genome.umin.jp/CNAGtop2.html.
- 77.Fiegler H, Redon R, Andrews D, Scott C, Andrews R, Carder C, Clark R, Dovey O, Ellis P, Feuk L, French L, Hunt P, Kalaitzopoulos D, Larkin J, Montgomery L, Perry GH, Plumb BW, Porter K, Rigby RE, Rigler D, Valsesia A, Langford C, Humphray SJ, Scherer SW, Lee C, Hurles ME, Carter NP. Accurate and reliable high-throughput detection of copy number variation in the human genome. Genome research. 2006;16:1566–1574. doi: 10.1101/gr.5630906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.dChip. http://biosun1.harvard.edu/complab/dchip/
- 79.Komura D, Shen F, Ishikawa S, Fitch KR, Chen W, Zhang J, Liu G, Ihara S, Nakamura H, Hurles ME, Lee C, Scherer SW, Jones KW, Shapero MH, Huang J, Aburatani H. Genome-wide detection of human copy number variations using high-density DNA oligonucleotide arrays. Genome research. 2006;16:1575–1584. doi: 10.1101/gr.5629106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.PennCNV. http://www.neurogenome.org/cnv/penncnv/
- 81.VariationHunter. http://compbio.cs.sfu.ca/strvar.htm.
- 82.GEO (Gene Expression Omnibus) http://www.ncbi.nlm.nih.gov/geo/
- 83.ArrayExpress. http://www.ebi.ac.uk/microarray-as/aer/?#ae-main[0]
- 84.Trace Archive. http://www.ncbi.nlm.nih.gov/Traces/trace.cgi?
- 85.Short Read Archive (SRA) http://www.ncbi.nlm.nih.gov/Traces/sra/sra.cgi?cmd=show&f=concepts&m=doc&s=concepts.
- 86.Levy S, Sutton G, Ng PC, Feuk L, Halpern AL, Walenz BP, Axelrod N, Huang J, Kirkness EF, Denisov G, Lin Y, MacDonald JR, Pang AW, Shago M, Stockwell TB, Tsiamouri A, Bafna V, Bansal V, Kravitz SA, Busam DA, Beeson KY, McIntosh TC, Remington KA, Abril JF, Gill J, Borman J, Rogers YH, Frazier ME, Scherer SW, Strausberg RL, Venter JC. The diploid genome sequence of an individual human. PLoS Biol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.dbGaP. http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gap.
- 88.European Genome-phenome Archive (EGA) http://www.ebi.ac.uk/ega/
- 89.dbVar. http://www.ncbi.nlm.nih.gov/dbvar/
- 90.DGVa