Abstract
Human rhinovirus (HRV) is the most common viral etiology in acute exacerbations of asthma. However, the exact mechanisms underlying HRV infection in allergic airways are poorly understood. IL-13 increases interleukin-1 receptor associated kinase M (IRAK-M) and subsequently inhibits airway innate immunity against bacteria. However, the role of IRAK-M in lung HRV infection remains unclear. Here, we provide the first evidence that IRAK-M over-expression promotes lung epithelial HRV-16 replication and autophagy, but inhibits HRV-16-induced IFN-β and IFN-λ1 expression. Inhibiting autophagy reduces HRV-16 replication. Exogenous IFN-β and IFN-λ1 inhibit autophagy and HRV-16 replication. Our data indicate the enhancing effect of IRAK-M on epithelial HRV-16 infection, which is partly through the autophagic pathway. Impaired anti-viral interferon production may serve as a direct or an indirect (e.g., autophagy) mechanism of enhanced HRV-16 infection by IRAK-M over-expression. Targeting autophagic pathway or administrating anti-viral interferons may prevent or attenuate viral (e.g., HRV-16) infections in allergic airways.
Keywords: Human rhinovirus, IRAK-M, viral replication, autophagy, airway epithelial cells
Introduction
Human rhinovirus (HRV) is the most common viral etiology in acute exacerbations of chronic respiratory diseases such as asthma, chronic obstructive pulmonary diseases, and cystic fibrosis (Contoli et al 2009, Kim and Gern 2012, Tan 2005, Wat et al 2008). HRV infection often leads to more severe and longer duration of lower respiratory tract symptoms in patients than in healthy individuals, significantly increases healthcare costs and negatively impacts the quality of life of patients and their families (van Elden et al 2008). Previous studies have shown that experimental infection with HRV-16 (a major group of HRV) in allergic asthma patients causes airway hyperresponsiveness compared with normal control subjects (Bardin et al 1994, Fraenkel et al 1995, Gern et al 1997, Gern and Busse 1999). However, the exact mechanisms underlying HRV-induced disease exacerbations are poorly understood.
Airway epithelial cells represent the primary site of respiratory microbial infections including HRV (Mosser et al 2005, Papadopoulos et al 2000). The degree of HRV replication in epithelial cells strongly influences the severity of respiratory symptoms during HRV infection. However, how host factors affect the course of HRV infection in airway epithelial cells remains unclear. In this study, we seek to explore the role of interleukin-1 receptor associated kinase M (IRAK-M) in HRV infection. IRAK-M, also known as IRAK-3, negatively regulates toll-like receptor (TLR)-mediated inflammatory responses to bacteria (Kobayashi et al 2002, Rosati and Martin 2002). We previously demonstrated epithelial IRAK-M up-regulation in asthmatic airways. Moreover, we found that IL-13, a Th2 cytokine prominent in asthma, increases airway epithelial IRAK-M and subsequently inhibits innate immunity against bacterial infections (Wu et al 2012). Although recent studies have shown that hepatitis C virus and influenza virus up-regulate IRAK-M in human antigen presenting cells and in mice (Chung et al 2010, Seki et al 2010), IRAK-M function in viral infections has not been investigated. Virus-induced production of type I (e.g., IFN-β) and III (e.g., IFN-λ1) interferons provides a critical first-line host defense against viruses (Sykes et al 2012, Vareille et al 2012). Deficiency of HRV-induced IFN-β and IFN-λ1 has been reported in asthmatic lungs, including bronchial epithelial cells (Baraldo et al 2012, Contoli et al 2006, Sykes et al 2012, Wark et al 2005). However, whether IRAK-M up-regulation in asthmatic airway epithelium regulates anti-viral responses is unknown.
Autophagy is an essential homeostatic pathway by which cells degrade damaged or obsolete organelles and proteins through the lysosomal machinery (Glick et al 2010, Xie and Klionsky 2007). Recent studies suggest autophagy is a novel mechanism by which the host defends against viral infection. Notably, autophagy recently has shown to exert both anti- and pro-viral functions. A recent study in HeLa cells demonstrated that autophagy promotes replication of HRV-2 (a minor group of HRV) (Klein and Jackson 2011). Likewise, poliovirus exploits the autophagy machinery to favor its replication (Jackson et al 2005, Taylor et al 2009, Taylor and Jackson 2009). Interestingly, poliovirus and HRV are both RNA viruses within the family of Picornaviridae. So far, the role of authophagy in the major group of HRV (e.g., HRV-16) infection and its regulation by IRAK-M have not been elucidated.
Central to the autophagy biogenesis is the formation of a double membrane-bound vesicle known as autophagosome, which undergoes several microtubule-dependent maturation events, including fusion with endosomes, multilamellar vesicles and lysosomes (Cesen et al 2012, Mizushima et al 2002). Beclin 1 plays an essential role in the initiation of autophagy (Cao and Klionsky 2007, Liang et al 1999). This process is mainly through promoting the activity of class III phosphoinositide 3-kinase (PI 3-kinase) Vps34 (Zeng et al 2006). LC3 is pivotal to the formation of autophagosomes and has two forms: LC3 I and LC3 II (Kabeya et al 2000). LC3 II is generated by the conjugation of LC3 I with phosphatidylethanolamine (PE) and is localized in the autophagosome membrane. Conversion of LC3 I into LC3 II is considered as a marker of autophagy (Klionsky et al 2012, Tang et al 2013).
In this study, we hypothesized that IRAK-M promotes lung HRV-16 infection via the autophagic pathway. To test our hypothesis, we first examined if IRAK-M over-expression in human lung epithelial cells promotes HRV-16 infection. Second, we knocked down beclin 1 in lung epithelial cells by using RNA interference to confirm the role of autophagy in IRAK-M-mediated HRV-16 replication. Third, we investigated whether IRAK-M promotes HRV-16 replication through inhibiting IFN-P and IFN-λ1 production in lung epithelial cells. Lastly, we determined the direct effects of exogenous IFN-β and IFN-λ1 on lung epithelial autophagy and HRV-16 infection.
Results
IRAK-M over-expression promotes HRV-16 replication
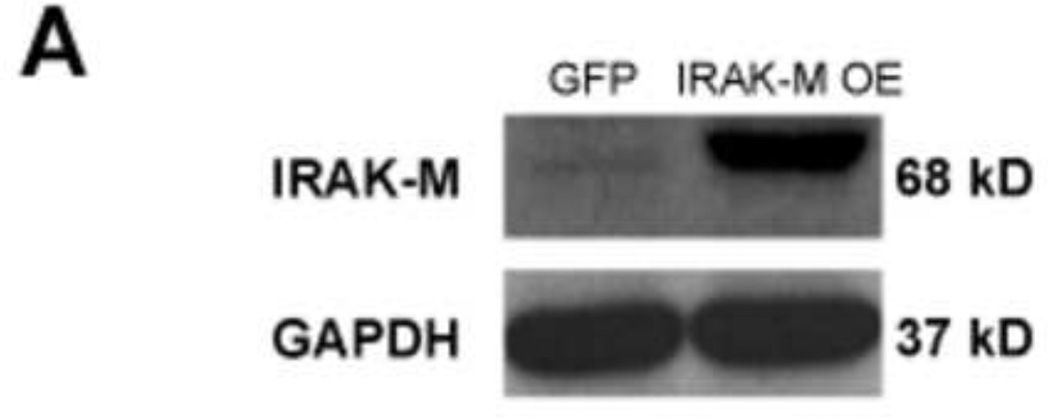
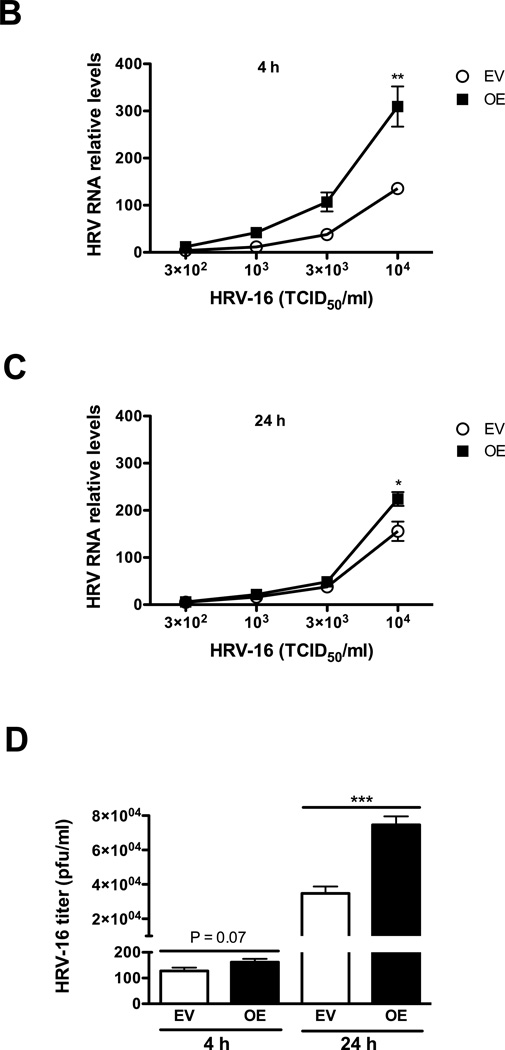
The IRAK-M over-expressing human lung epithelial NCI-H292 cell line (IRAK-M OE) and empty vector NCI-H292 cell line (EV) were established as previously described (Wu et al 2012). HRV-16 dose response and time course studies were performed in IRAK-M OE and EV NCI-H292 cells with various doses of HRV-16 (3 × 102, 103, 3 × 103 and 104 TCID50/well) or sterile phosphate-buffered saline (PBS) for 4 and 24 h to examine IRAK-M expression, viral RNA levels, and viral particles in cell supernatants. At the time of HRV infection, both cell lines reached the similar confluence (about 80%). No significant difference in cell growth (density) was observed during HRV infection period. Increased IRAK-M protein expression was confirmed in IRAK-M OE NCI-H292 cells (Fig. 1A). IRAK-M OE versus EV NCI-H292 cells consistently increased HRV-16 RNA levels at both 4 h (Fig. 1B) and 24 h (Fig. 1C) in a dose-dependent manner, particularly at 104 TCID50/well where a greater percentage of cells was infected. To support HRV RNA data, we measured infectious viral particles released into the supernatants from cells infected with HRV-16 at 104 TCID50/well by plaque assay. Like the RNA data, IRAK-M OE NCI-H292 cells had a higher viral titer starting at 4 h and reached a statistical significance at 24 h (Fig. 1D). Hence, 104 TCID50/well was chosen as the HRV-16 infection dose for all other experiments in this study.
Fig. 1. IRAK-M over-expression promotes HRV-16 replication in human lung epithelial cell line.
IRAK-M over-expressing (OE) and control (empty vector, EV) NCI-H292 cells were infected with various doses of HRV-16 or PBS for 4 and 24 h as described in Materials and methods. Cellular IRAK-M protein at the baseline of 4 h (A), HRV RNA levels at 4 h (B) and 24 h (C) post infection were determined by Western blot and quantitative RT-PCR, respectively. The released infectious viral particles (D) in supernatants of cells with HRV-16 infection at 104 TCID50/well were determined by plaque assay. Data are expressed as means ± SEM (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001.
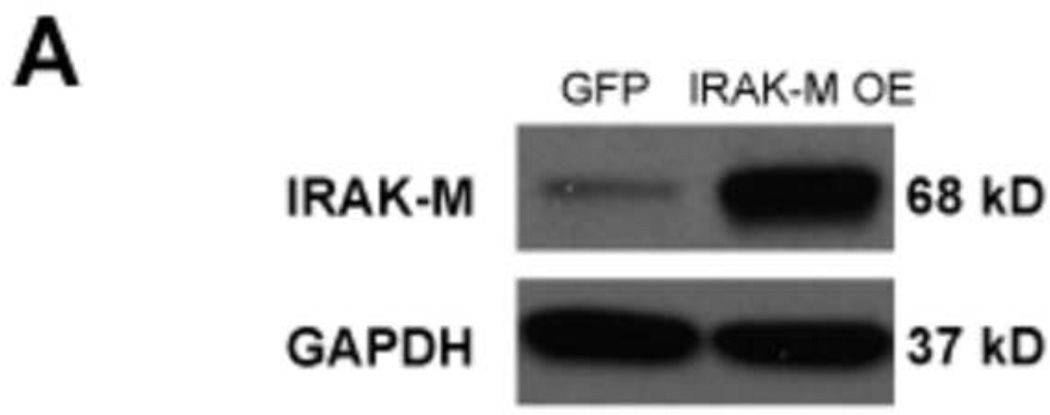
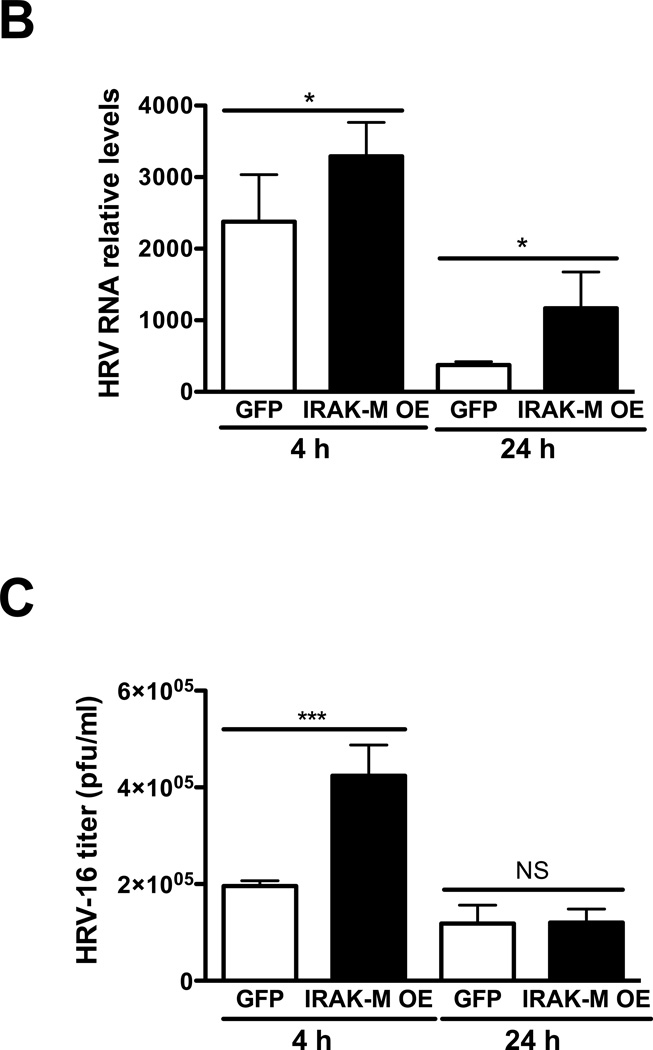
To validate our findings in NCI-H292 cell lines, IRAK-M over-expression was performed in submerged human primary airway epithelial cells by using lentivirus-mediated human IRAK-M cDNA transduction. These cells were then seeded in transwells, and cultured at air-liquid interface (ALI) to induce cell mucociliary differentiation. After 12 days of ALI culture, cells with or without IRAK-M over-expression were treated at the apical surface with HRV-16 or PBS as described previously (Chen et al 2006) to examine IRAK-M expression, viral RNA levels, and viral particles in apical supernatants. We confirmed over-expression of IRAK-M protein in human primary airway epithelial cells transduced with lentiviruses containing IRAK-M cDNA (Fig. 2A). In line with the NCI-H292 cell line data, IRAK-M over-expression in primary cells also increased HRV-16 RNA levels at both 4 and 24 h of infection (Fig. 2B). However, a significant increase in viral particles was observed only in IRAK-M over-expressing primary cell supernatants at 4 h post viral infection (Fig. 2C). This may be explained by the fact that primary cells need a longer (24 h versus 2 h for NCI-H292 cells) inoculation with HRV-16 to establish a successful viral infection.
Fig. 2. IRAK-M over-expression promotes HRV-16 replication in human primary airway epithelial cells.
Normal human tracheobronchial epithelial cells were transduced with lentiviruses containing either pLentilox 3.7-GFP (GFP) or pLentilox 3.7-IRAK-M (IRAK-M OE) and then exposed to HRV-16 at 104 TCID50/well or PBS for 4 and 24 h as described in Materials and methods. Cellular IRAK-M protein at the baseline of 4 h (A), HRV RNA levels (B), and apical supernatant infectious viral particles (C) were determined by Western blot, quantitative RT-PCR and plaque assay, respectively. Data are expressed as means ± SEM (n = 4). *P < 0.05, ***P < 0.001, NS = Not Significant.
Collectively, IRAK-M over-expression significantly enhanced HRV-16 replication in human lung epithelial cells.
IRAK-M over-expression enhances autophagy during HRV-16 infection
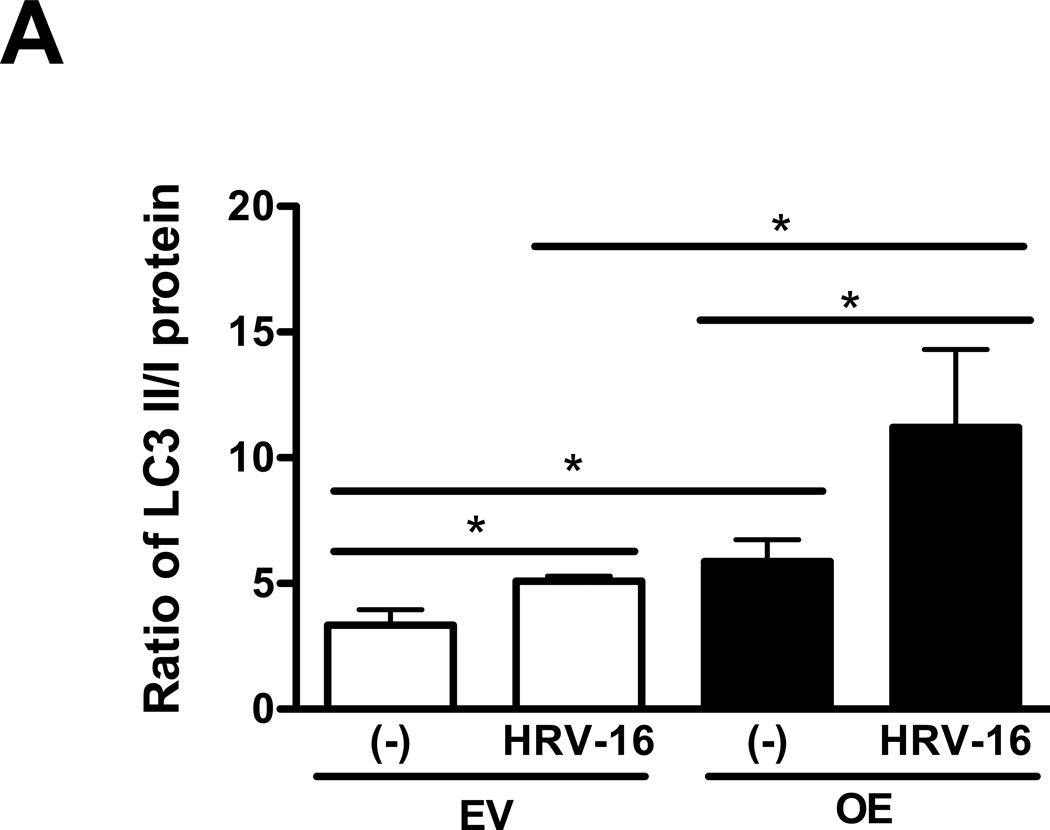
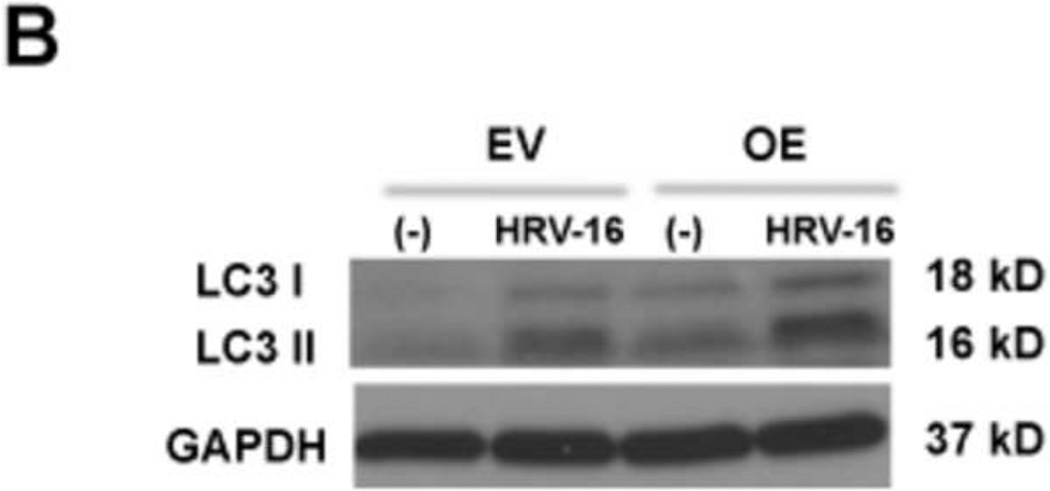
To determine the effect of IRAK-M on the autophagic pathway, LC3 I and LC3 II were evaluated in IRAK-M OE and EV NCI-H292 cells with or without HRV-16 infection for 4 h. As shown in Fig. 3, HRV-16 infection markedly increased the ratio of LC3 II/LC3 I protein (an indicator of autophagosome formation) in both IRAK-M OE and EV NCI-H292 cells. Notably, the LC3 II/LC3 I ratio at the baseline or following HRV-16 infection was significantly greater in IRAK-M OE NCI-H292 cells than that in EV NCI-H292 cells.
Fig. 3. IRAK-M over-expression enhances autophagy during HRV-16 infection.
IRAK-M over-expressing (OE) and control (empty vector, EV) NCI-H292 cells were infected with HRV-16 at 104 TCID50/well or PBS for 4 h as described in Materials and methods. Protein levels of LC3 I and LC3 II were measured by Western blot and are expressed as means ± SEM (A) (n = 3, *P < 0.05). A representative Western blot picture of LC3 I and LC3 II (B) was shown from 3 independent experiments.
Inhibition of the autophagic pathway reduces HRV-16 replication
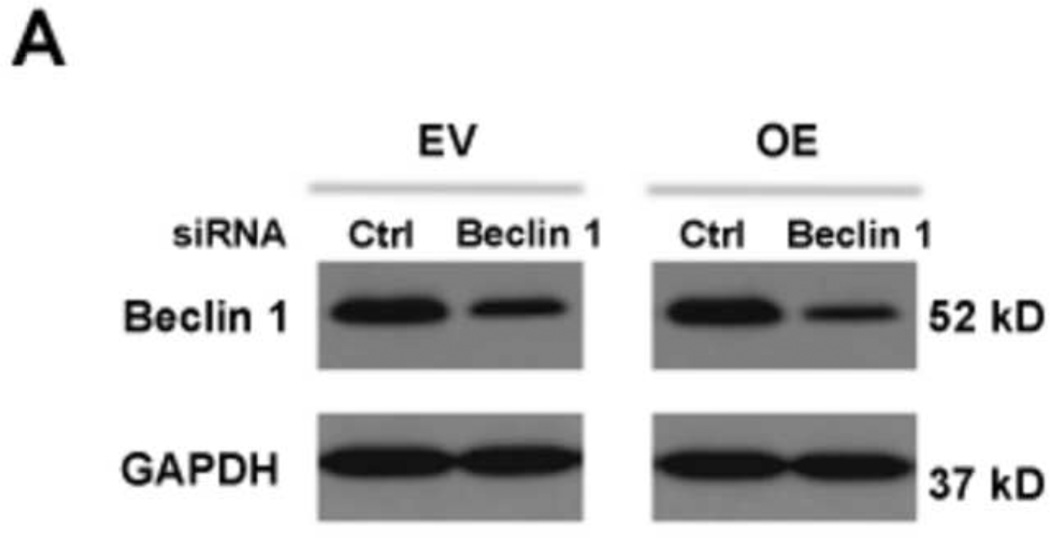
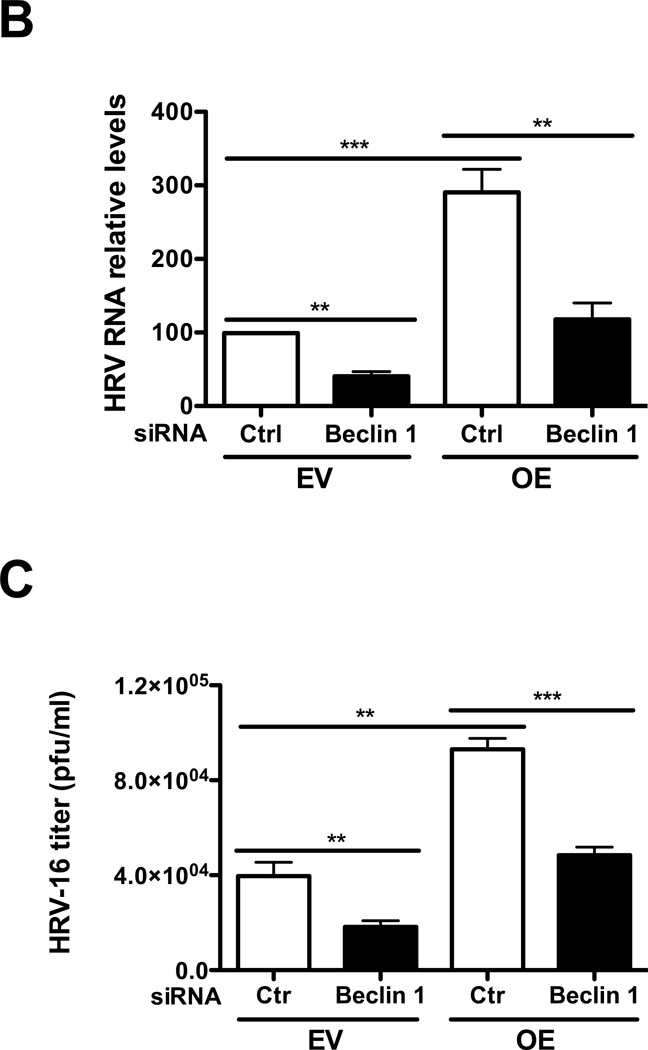
To explore whether the autophagic pathway is required for HRV-16 replication, we knocked down beclin 1 (a key initiator of autophagy biogenesis) by using target-specific RNA interference in IRAK-M OE and EV NCI-H292 cells. We then infected these cells with HRV-16 or PBS for 24 h to examine whether beclin 1 knockdown altered viral RNA levels and viral particles in cell supernatants. First, Western blot analysis confirmed the baseline beclin 1 protein reduction by beclin 1 siRNA in both IRAK-M OE and EV NCI-H292 cells (Fig. 4A). Following HRV-16 infection, in the absence of beclin 1 knockdown (control siRNA), IRAK-M OE versus EV NCI-H292 cells significantly increased viral RNA levels (Fig. 4B). Successful beclin 1 knockdown decreased HRV-16 RNA levels (Fig. 4B) and the release of infectious viral particles (Fig. 4C) in both NCI-H292 cell lines, particularly in IRAK-M OE NCI-H292 cells. These results suggest that autophagy may facilitate HRV-16 replication in the context of IRAK-M in human lung epithelial cells.
Fig. 4. Inhibition of the autophagic pathway reduces HRV-16 replication.
IRAK-M over-expressing (OE) and control (empty vector, EV) NCI-H292 cells were transfected with beclin 1 siRNA or control siRNA and then infected with HRV-16 at 10 TCID50/well or PBS for 24 h as described in Materials and methods. A representative Western blot picture of beclin 1 protein after PBS treatment (A) was shown from 3 independent experiments. HRV RNA levels (B) were examined by quantitative RT-PCR and the released infectious viral particles in cell supernatants (C) were determined by plaque assay. Data are expressed as means ± SEM (n = 3). **P < 0.01, ***P < 0.001.
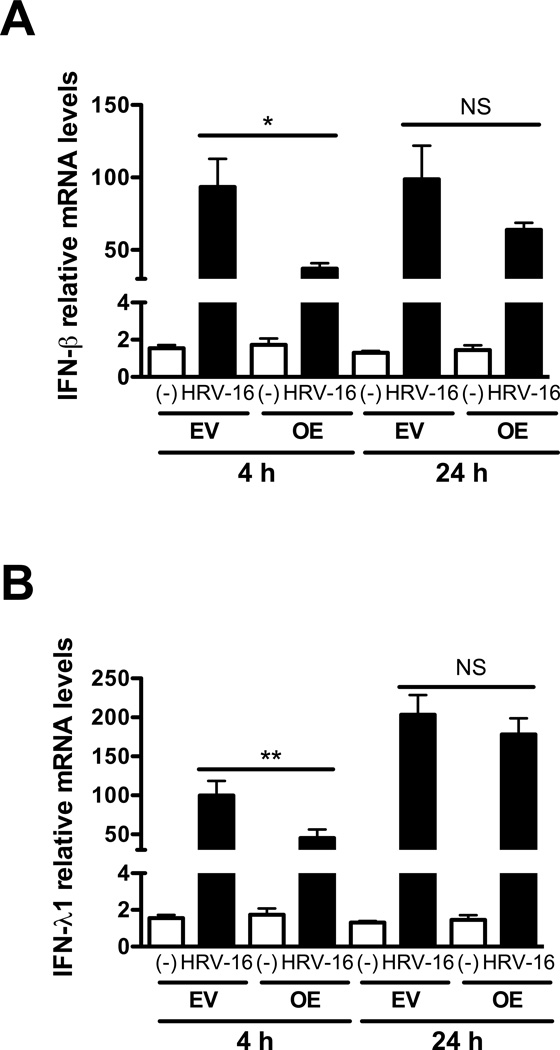
IRAK-M over-expression inhibits HRV-16-induced IFN-β and IFN-λ1
As IRAK-M over-expression increases HRV replication, we examined whether IRAK-M serves as a negative regulator of type I and III interferon production in HRV16-infected cells. IRAK-M OE and EV NCI-H292 cells, or well-differentiated human primary airway epithelial cells with or without IRAK-M over-expression, were infected with HRV-16 or PBS for 4 and 24 h after which we examined IFN-β and IFN-λ1 mRNA levels. After 4 h of HRV-16 infection, IRAK-M OE versus EV NCI-H292 cells significantly decreased IFN-β (Fig. 5A) and IFN-λ1 (Fig. 5B), which was maintained through 24 h. Likewise, IRAK-M over-expression in well-differentiated human primary epithelial cells resulted in lower mRNA expression of IFN-β (50% reduction) and IFN-λ1 (54% reduction) at 24 h post HRV-16 infection. Taken together, our data suggest that increasing IRAK-M expression may promote HRV-16 infection by impairing production of type I and III interferons in the infected human lung epithelial cells.
Fig. 5. IRAK-M over-expression inhibits HRV-16-induced IFN-β and IFN-λ1.
IRAK-M over-expressing (OE) and control (empty vector, EV) NCI-H292 cells were infected with HRV-16 at 104 TCID50/well or PBS for 4 and 24 h as described in Materials and methods. IFN-β (A) and IFN-λ1 (B) mRNA levels were examined by quantitative RT-PCR. Data are expressed as means ± SEM (n = 3). *P < 0.05, **P < 0.01, NS = Not Significant.
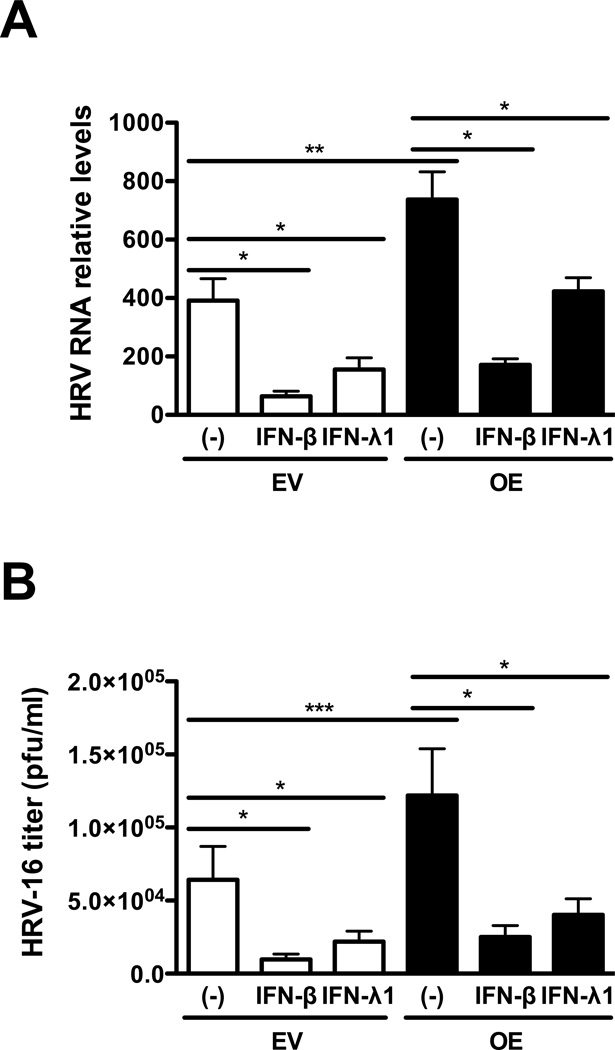
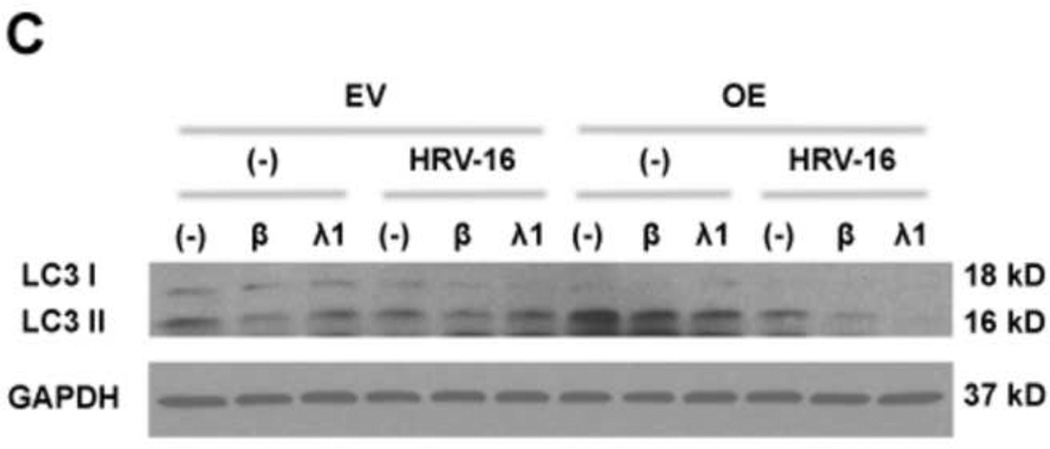
Exogenous IFN-β and IFN-λ1 inhibit autophagy and HRV-16 replication
Having shown that IRAK-M over-expression increases autophagy and HRV-16 replication coupled with decreased production of anti-viral interferons, we next tested whether impaired IFN-β and IFN-λ1 expression in IRAK-M over-expressing cells is responsible for the increased autophagy and HRV replication. IRAK-M OE and EV NCI-H292 cells were infected with HRV-16 or PBS in the absence or presence of IFN-β or IFN-λ1 for 24 h. Exogenous IFN-β and IFN-λ1 significantly protected lung epithelial cells from HRV-16 infection by reducing the levels of viral RNA (Fig. 6A) and the released infectious viral particles (Fig. 6B) in IRAK-M OE and EV NCI-H292 cells, which is associated with decreased LC II/LC I ratio and thus formation of autophagosomes (Fig. 6C).
Fig. 6. Exogenous IFN-β and IFN-λ1 inhibit autophagy and HRV-16 replication.
IRAK-M over-expressing (OE) and control (empty vector, EV) NCI-H292 cells were infected with HRV-16 at 104 TCID50/well or PBS in the absence or presence of IFN-β (3 ng/ml) and IFN-λ1 (10 ng/ml) for 24 h as described in Materials and methods. HRV RNA levels (A) were examined by quantitative RT-PCR and the released infectious viral particles in cell supernatants (B) were determined by plaque assay. Data are expressed as means ± SEM (n = 3). *P < 0.05, **P < 0.01. A representative Western blot picture of LC3 I and LC3 II (C) was shown from 2 independent experiments.
Discussion
This is the first study to reveal the function of IRAK-M in HRV infection in human lung epithelial cells. We discovered that IRAK-M over-expression significantly increases HRV-16 replication and the autophagic pathway. By silencing beclin 1 gene expression, we clearly demonstrated a critical role of autophagy in IRAK-M-mediated HRV-16 replication. Furthermore, IRAK-M was shown to down-regulate anti-viral interferon production in HRV-16-infected human lung epithelial cells. Exogenous IFN-β and IFN-λ1 was shown to restore the anti-viral mechanisms possibly by deceasing autophagy during HRV-16 infection.
Despite overwhelming evidence linking HRV infections with exacerbations of asthma, the mechanisms of HRV-induced disease exacerbations are poorly understood. Impaired IFN-β and IFN-λ1 induction in HRV-infected asthmatic airway epithelial cells and alveolar macrophages may explain the increased viral infection during acute exacerbations (Baraldo et al 2012, Contoli et al 2006, Sykes et al 2012, Wark et al 2005). However, the underlying impaired anti-viral mechanisms have not been well elucidated. Our previous work indicates that Th2 cytokine-induced airway epithelial IRAK-M up-regulation impairs host defense against bacterial infections (Wu et al 2012). Our findings in this study have advanced our understanding of IRAK-M function in viral infections. For the first time, we demonstrated that IRAK-M over-expression in lung epithelial cells increases HRV-16 replication, which may be explained by hindered host anti-viral defense – inhibition of HRV-16-induced IFN-β and IFN-λ1 expression. Therefore, IRAK-M may serve as a key mediator in promoting respiratory viral (e.g., HRV-16) infections in allergic lungs.
The autophagic pathway has been proposed as a novel mechanism against a variety of pathogens, but its role in viral infection may depend on specific viral strains and host cell types. In some studies, autophagy has been shown to facilitate sensing of intracellular viral elements by pattern-recognition receptors that trigger the secretion of anti-viral factors like type I interferon (Campbell and Spector 2012, Lee et al 2007). However, increasing numbers of recent studies suggest that the autophagic machinery appears to block the production of type I interferon, thereby promoting replication of certain RNA viruses (e.g., vesicular stomatitis virus, herpesvirus and hepatitis C virus) (Jounai et al 2007, Ke and Chen 2011, Takeshita et al 2008). Our current study has revealed that HRV-16 exploits the autophagic pathway to facilitate its replication in the context of IRAK-M. First, beclin 1 knockdown significantly attenuated the proviral role of IRAK-M. Second, the ratio of LC3 II/LC3 I protein (indication of autophagosome formation) at the baseline or following HRV-16 infection is significantly greater in IRAK-M over-expressing human lung epithelial cells. This increased LC3 II/I ratio may be due to increased conversion of LC3 I to LC3 II and/or decreased autophagosome fusion with lysosomes resulting in less LC3 II degradation. Thus, besides inducing autophagy, HRV-16 may impair the clearance of autophagosomes to promote their replication in IRAK-M over-expressing human lung epithelial cells. These potential mechanisms deserve future studies. Third, although we used the LC3 II/LC3 I ratio to monitor autophagic activity, we realize that LC3 may not fully indicate autophagic flux during HRV-16 infection. The adaptor protein p62 binds to LC3 and ubiquitinated substrates to form a complex, which is incorporated into the autophagosomes and subsequently degraded in autolysosomes. Thus, decreased p62 is suggested to be more indicative of autophagy flux (activity) (Bjorkoy et al 2005, Pankiv et al 2007, Tung et al 2010). In a preliminary study, we examined p62 expression in IRAK-M OE and EV NCI-H292 cells with or without HRV-16 infection for 4 h. We found that p62 protein levels at the baseline or following HRV-16 were lower in NCI-H292 IRAK-M OE cells than those in EV cells (data not shown). Intriguingly, HRV-16 infection reduced p62 level in EV cells, but not in IRAK-M OE cells. We speculate that while LC3 levels may change quickly, clearance of autophagy substrates may take a longer time, especially when autophagy levels were significantly higher in IRAK-M-over-expressing cells. We will test this hypothesis in our future study to improve our understating about the role of IRAK-M in regulating autophagy flux.
The interplay between autophagy and anti-viral interferons during viral infections is still unclear. IFN-γ is reported to induce autophagy in THP-1 cells and T cells (Dutta et al 2012, Fabri et al 2011). Although autophagy is involved in production of type I interferon (e.g., IFN-α) in response to RNA viruses (e.g., hepatitis C virus and HIV-1) (Henault et al 2012, Zhou et al 2012), the role of type I and III interferons in regulating autophagy remains unknown. Our findings clearly reveal that type I and III interferons inhibit HRV-16-induced autophagy in the context of IRAK-M in human lung epithelial cells. Therefore, targeting autophagic pathways or administrating type I and III interferons in IRAK-M over-expressing allergic airways may provide an effective therapy to reduce autophagy and subsequent HRV-16 replication.
As our data are derived using an in vitro cell culture model, important influences on viral infection and autophagy may be absent, including local and circulating factors and the influence of cells beneath the basement membrane. Future studies will need to consider in vivo animal models to further dissect the interplays of the components in the IRAK-M/autophagy/interferon axis. For example, the use IRAK-M and beclin 1 deficient mice may be helpful to reveal the in vivo functions of these two molecules during HRV infection in the context of allergic inflammation or Th2 cytokine exposure. Moreover, the molecular mechanisms by which type I and III interferons regulate the autophagic pathway warrant further study.
In summary, our findings indicate that IRAK-M promotes lung HRV-16 infection, which is in part through the autophagic pathway. Impaired anti-viral interferon production may serve as a direct or an indirect (e.g., autophagy) mechanism to enhance HRV-16 infection in IRAK-M over-expressing cells. A better understanding of the autophagic pathway in HRV infection may lead to novel interventions to attenuate viral (i.e., HRV-16) infections during acute exacerbations of asthma and other chronic lung diseases.
Materials and Methods
Preparation of HRV-16
HRV-16 (American Type Culture Collection, Manassas, VA) were propagated in H1-Hela cells (CRL-1958, ATCC), and purified as described previously (Hao et al 2012). Viral stocks were titrated by infecting H1-HeLa monolayers with serially diluted HRV-16 to assess cytopathic effect, and expressed by 50% tissue culture infective doses per ml (TCID50/ml) (Newcomb et al 2008).
HRV-16 infection in a human lung epithelial cell line stably over-expressing human IRAK-M
The IRAK-M over-expressing (OE) human lung epithelial cell line or control (empty vector, EV) cell line was established as previously described (Wu et al 2012). In brief, human IRAK-M cDNA was obtained from Open Biosystems (Huntsville, Ala), and cloned into a mammalian expression plasmid by PCR amplification. Human lung mucoepidermoid carcinoma derived NCI-H292 cells (clone CRL-1848, ATCC) were transfected with the IRAK-M expression vector or an empty vector (control), and selected by G418 (800 µg/ml, Invitrogen Life Technologies Inc., Carlsbad, CA) in RPMI1640 with 10% FBS to generate the stable cell lines. The cells were then maintained in the presence of G418 (400 µg/ml) until experiments.
To establish the HRV-16 infection model in NCI-H292 cells, cells were seeded at 5 × 105/well in 12-well cell culture plates and starved overnight in serum-free X-VIVO™ 10 medium (Lonza, Walkersville, MD). Thereafter, cells were infected with HRV-16 at various doses (3 × 102, 103, 3 × 103 and 104 TCID50/well) or sterile PBS as a mock infection. Two hours later, cells were washed three times in sterile RPMI1640 medium (no FBS) to remove unattached viruses and then cultured in X-VIVO™ 10 medium for additional 4 and 24 h. Cells and supernatants were processed to quantify viral load by quantitative RT-PCR and/or plaque assay. The IRAK-M protein at the baseline and auophagic pathway (e.g., LC3 I and LC3 II) in samples at 4 h of treatments was examined by Western blot. IFN-β and IFN-λ1 mRNA was measured by quantitative RT-PCR. The 4 and 24 h time points were chosen based on our preliminary time-course (4, 24 and 48 h) experiments by infecting IRAK-M OE and EV NCI-H292 cells with HRV-16 at the dose of 104 TCID50/well. We found that HRV-16 levels in IRAK-M OE versus EV NCI-H292 cells were increased at 4 h, and maintained at 24 h, but not at 48 h.
To test the effects of exogenous anti-viral interferon on HRV-16 replication and the autophagic pathway, cells were treated with recombinant human IFN-β (3 ng/ml) and IFN-λ1 (10 ng/ml) for 2 h prior to HRV-16 infection at 104 TCID50/well or PBS. After 24 h, samples were processed to examine viral RNA levels by quantitative RT-PCR, released viral particles in cell supernatants by plaque assay, and the auophagic pathway (LC3 I and LC3 II expression) by Western blot. IFN-β and IFN-λ1 concentrations were chosen based on previous publications in human lung epithelial cell cultures (Spurrell et al 2005, Suh et al 2007, Yoshikawa et al 2010) and our preliminary IFN dose (1, 3, and 10 ng/ml) and time course (4 and 24 h) optimization experiments in which 3 ng/ml of IFN-β and 10 ng/ml of IFN-λ1 demonstrated the maximal inhibitory effect on HRV-16 replication at 24 h.
IRAK-M over-expression in normal human primary airway epithelial cells
Lentivirus-mediated IRAK-M over-expression was performed in normal human primary airway epithelial cells by modifying previously described method (White et al 2009). In brief, human IRAK-M full-length cDNA was cloned into a lentiviral vector pLentilox (pLL) 3.7 by removing the GFP gene to generate pLL3.7-IRAK-M vector. The pLL3.7 vector expressing GFP (pLL3.7-GFP) was used as control. pLL3.7-GFP or pLL3.7-IRAK-M (4 µg) was co-transfected with psPAX2 (4 µg) and envelope protein vector pHCMV-G (4 µg) into 293FT packaging cells using the Effectene™ Transfection Reagent (Qiagen, Valencia, CA). Forty-eight hours after transfection, cell supernatants containing virus were harvested by centrifugation at 500 g for 5 min to remove cell debris and then used for infecting epithelial cells.
Normal human tracheobronchial epithelial cells were obtained from the tracheas and bronchi of de-identified organ donors whose lungs are not suitable for transplantation as previously described (Wu et al 2012). Epithelial cells at passage 1 were cultured in collagen-coated 60 mm tissue culture dishes containing bronchial epithelial cell growth medium (BEGM) with supplements (Lonza, Walkersville, MD) at 37°C, 5% CO2 until 80% confluence. Epithelial cells were then passed into 6-well culture plates at 1 × 105 cells/well and transduced with lentiviruses containing pLL3.7-GFP (GFP) or pLL3.7-IRAK-M (IRAK-M OE) by centrifugation of 2,000 rpm at room temperature (25°C) for 1 h. Ninety-six hours after the transduction, cells were transferred onto collagen-coated 12-well transwell plates at 4 × 104 cells/well (Corning Inc., Corning, NY) in DMEM/BEBM (1:1) with supplements (Lonza, Walkersville, MD). Cells were cultured in an immersed condition for 10 days when cells reached 100% confluence and were shifted to air-liquid interface (ALI) culture by reducing the apical surface medium volume to 50 µl. After 12 days of ALI culture, cells were well differentiated and incubated at the apical surface with 100 µl of PBS (control) or HRV-16 at 104 TCID50/well for 24 h to establish a successful viral infection (Chen et al 2006) as well-differentiated epithelial cells are difficult to be infected due to tight junctions and a mucus layer over the apical surface. Cells were washed three times to remove unattached viruses and then cultured for additional 4 and 24 h. Samples were processed to examine IRAK-M expression by Western blot, viral RNA levels by quantitative RT-PCR, and viral particles in cell supernatants by plaque assay.
Beclin 1 gene knockdown in a human lung epithelial cell line stably over-expressing human IRAK-M
To examine if the autophagic pathway promotes HRV replication, we knocked down beclin 1 in IRAK-M over-expressing (OE) or control (EV) NCI-H292 cells. Briefly, cells were seeded at 2.5 × 105/well in 12-well cell culture plates and transfected with beclin 1 siRNA or control siRNA using siRNA transfection medium and siRNA transfection reagent (Santa Cruz Biotechnology Inc., Santa Cruz, CA) according to manufacture’s instructions. Forty-eight hours after the transfection, cells were infected with HRV-16 at 104 TCID50/well or PBS for 24 h. Samples were processed to examine beclin 1 knockdown by Western blot, viral RNA levels by quantitative RT-PCR, and released viral particles in cell supernatants by plaque assay.
Viral plaque assay
Viral titer in cell culture supernatants was determined by plaque assay as previously described with modifications (Fiala 1968, Halfpap and Cooney 1983). Briefly, H1-HeLa cell monolayers in six-well plates were inoculated with 0.5 ml of serially diluted cell culture supernatants for 2 h at 37°C, 5% CO2. After removing the inoculum, cells were overlaid with 3 ml of minimum essential medium (Gibco®, Life Technologies, Grand Island, NY) containing 4% FBS, 1% Penicillin-streptomycin, 0.5% non-essential amino acid (NEAA) (HyClone, Thermo Scientific, Logan, UT), 60 µg/ml DEAE-Dextran hydrochloride (Sigma-Aldrich, St. Louis, MO), 30 mM MgCl2 and 0.5% SeaKem® LE Agarose (Lonza). After incubating for 3 days at 37°C, 5% CO2, monolayers of cells were stained with thiazolyl blue tetrazolium bromide (MTT, Sigma-Aldrich) to count the plaques.
Western blot analysis
Equal amounts of protein samples were separated on 7.5 % (for IRAK-M) or 15% (for LC3) SDS–PAGE, transferred onto nitrocellulose membranes, and probed with rabbit anti-human IRAK-M (Millipore, Billerica, MA), rabbit anti-LC3 (Sigma-Aldrich), or mouse anti-GAPDH (Santa Cruz Biotechnology Inc.). Blots were then incubated with appropriate HRP-linked secondary antibodies and Pierce® ECL Western blotting substrate. Densitometry was performed using the NIH Image-J software. The ratios of LC3 II/LC3 I protein were used to indicate the formation of autophagosomes.
Quantitative real-time RT-PCR
Taqman quantitative real-time RT-PCR was used to detect HRV RNA (Mosser et al 2005) and human IFN-β and IFN-λ1 mRNA (Wang et al 2009) as previously described. The specific primers and probes are: HRV (forward: 5′-CCT CCG GCC CCT GAA T-3′; reverse: 5′-GGT CCC ATC CCG CAA TT-3′, probe: 5′-CTA ACC TTA AAC CTG CAG CCA-3′), IFN-β (forward: 5′-CTT ACA GGT TAC CTC CGA AAC TGA A-3′; reverse: 5′-TTG AAG AAT GCT TGA AGC AAT TGT-3′, probe: 5′-ATC TCC TAG CCT GTG CCT CTG GGA CTG-3′); IFN-λ1 (forward: 5′-GGG AAC CTG TGT CTG AGA ACG T-3′; reverse: 5′-GAG TAG GGC TCA GCG CAT AAA TA-3′; probe: 5′-CTG AGT CCA CCT GAC ACC CCA CAC C-3′). Housekeeping gene GAPDH was evaluated as an internal positive control. The comparative cycle of threshold (∆∆Ct) method was used to demonstrate the relative levels of target genes.
Statistical analysis
Data are presented as means ± SEM. One-way analysis of variance (ANOVA) was used for multiple comparisons, and a Tukey’s post hoc test was applied where appropriate. Student’s t test was used when only two groups were compared. A p value < 0.05 was considered significant.
IRAK-M over-expression promotes rhinovirus infection in lung epithelial cells.
IRAK-M over-expression enhances autophagy during rhinovirus infection.
Inhibition of autophagy reduces rhinovirus infection.
Type I and III interferons are reduced in IRAK-M over-expressing epithelial cells.
Type I and III interferons inhibit autophagy and rhinovirus infection.
Acknowledgments
This study was financially supported by the National Institutes of Health R01 AI070175 and R01 HL088264. We thank Dr. Jieru Wang for advising the optimization of the HRV plaque assay protocol.
Abbreviations
- ALI culture
Air-liquid interface culture
- HRV
Human rhinovirus
- IRAK-M
Interleukin-1 receptor associated kinase M
- IFN-β
Interferon-β
- IFN-λ1
Interferon-λ1
- PE
phosphatidylethanolamine
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baraldo S, Contoli M, Bazzan E, Turato G, Padovani A, Marku B, et al. Deficient antiviral immune responses in childhood: distinct roles of atopy and asthma. J Allergy Clin Immunol. 2012;130:1307–1314. doi: 10.1016/j.jaci.2012.08.005. [DOI] [PubMed] [Google Scholar]
- Bardin PG, Fraenkel DJ, Sanderson G, Dorward M, Lau LC, Johnston SL, et al. Amplified rhinovirus colds in atopic subjects. Clin Exp Allergy. 1994;24:457–464. doi: 10.1111/j.1365-2222.1994.tb00934.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell GR, Spector SA. Autophagy induction by vitamin D inhibits both Mycobacterium tuberculosis and human immunodeficiency virus type 1. Autophagy. 2012;8:1523–1525. doi: 10.4161/auto.21154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Klionsky DJ. Physiological functions of Atg6/Beclin 1: a unique autophagy-related protein. Cell Res. 2007;17:839–849. doi: 10.1038/cr.2007.78. [DOI] [PubMed] [Google Scholar]
- Cesen MH, Pegan K, Spes A, Turk B. Lysosomal pathways to cell death and their therapeutic applications. Exp Cell Res. 2012;318:1245–1251. doi: 10.1016/j.yexcr.2012.03.005. [DOI] [PubMed] [Google Scholar]
- Chen Y, Hamati E, Lee PK, Lee WM, Wachi S, Schnurr D, et al. Rhinovirus induces airway epithelial gene expression through double-stranded RNA and IFN-dependent pathways. Am J Respir Cell Mol Biol. 2006;34:192–203. doi: 10.1165/rcmb.2004-0417OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H, Watanabe T, Kudo M, Chiba T. Hepatitis C virus core protein induces homotolerance and cross-tolerance to Toll-like receptor ligands by activation of Toll-like receptor 2. J Infect Dis. 2010;202:853–861. doi: 10.1086/655812. [DOI] [PubMed] [Google Scholar]
- Contoli M, Message SD, Laza-Stanca V, Edwards MR, Wark PA, Bartlett NW, et al. Role of deficient type III interferon-lambda production in asthma exacerbations. Nat Med. 2006;12:1023–1026. doi: 10.1038/nm1462. [DOI] [PubMed] [Google Scholar]
- Contoli M, Marku B, Conti V, Saturni S, Caramori G, Papi A. Viral infections in exacerbations of asthma and chronic obstructive pulmonary disease. Minerva Med. 2009;100:467–478. [PubMed] [Google Scholar]
- Dutta RK, Kathania M, Raje M, Majumdar S. IL-6 inhibits IFN-gamma induced autophagy in Mycobacterium tuberculosis H37Rv infected macrophages. Int J Biochem Cell Biol. 2012;44:942–954. doi: 10.1016/j.biocel.2012.02.021. [DOI] [PubMed] [Google Scholar]
- Fabri M, Stenger S, Shin DM, Yuk JM, Liu PT, Realegeno S, et al. Vitamin D is required for IFN-gamma-mediated antimicrobial activity of human macrophages. Sci Transl Med. 2011;3:104ra102. doi: 10.1126/scitranslmed.3003045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiala M. Plaque formation by 55 rhinovirus serotypes. Appl Microbiol. 1968;16:1445–1450. doi: 10.1128/am.16.10.1445-1450.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraenkel DJ, Bardin PG, Sanderson G, Lampe F, Johnston SL, Holgate ST. Lower airways inflammation during rhinovirus colds in normal and in asthmatic subjects. Am J Respir Crit Care Med. 1995;151:879–886. doi: 10.1164/ajrccm/151.3_Pt_1.879. [DOI] [PubMed] [Google Scholar]
- Gern JE, Calhoun W, Swenson C, Shen G, Busse WW. Rhinovirus infection preferentially increases lower airway responsiveness in allergic subjects. Am J Respir Crit Care Med. 1997;155:1872–1876. doi: 10.1164/ajrccm.155.6.9196088. [DOI] [PubMed] [Google Scholar]
- Gern JE, Busse WW. Association of rhinovirus infections with asthma. Clin Microbiol Rev. 1999;12:9–18. doi: 10.1128/cmr.12.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick D, Barth S, Macleod KF. Autophagy: cellular and molecular mechanisms. J Pathol. 2010;221:3–12. doi: 10.1002/path.2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfpap LM, Cooney MK. Isolation of rhinovirus intertypes related to either rhinoviruses 12 and 78 or 36 and 58. Infect Immun. 1983;40:213–218. doi: 10.1128/iai.40.1.213-218.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao W, Bernard K, Patel N, Ulbrandt N, Feng H, Svabek C, et al. Infection and propagation of human rhinovirus C in human airway epithelial cells. J Virol. 2012;86:13524–13532. doi: 10.1128/JVI.02094-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henault J, Martinez J, Riggs JM, Tian J, Mehta P, Clarke L, et al. Noncanonical autophagy is required for type I interferon secretion in response to DNA-immune complexes. Immunity. 2012;37:986–997. doi: 10.1016/j.immuni.2012.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson WT, Giddings TH, Jr., Taylor MP, Mulinyawe S, Rabinovitch M, Kopito RR, et al. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 2005;3:e156. doi: 10.1371/journal.pbio.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jounai N, Takeshita F, Kobiyama K, Sawano A, Miyawaki A, Xin KQ, et al. The Atg5 Atg12 conjugate associates with innate antiviral immune responses. Proc Natl Acad Sci U S A. 2007;104:14050–14055. doi: 10.1073/pnas.0704014104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke PY, Chen SS. Activation of the unfolded protein response and autophagy after hepatitis C virus infection suppresses innate antiviral immunity in vitro. J Clin Invest. 2011;121:37–56. doi: 10.1172/JCI41474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WK, Gern JE. Updates in the relationship between human rhinovirus and asthma. Allergy Asthma Immunol Res. 2012;4:116–121. doi: 10.4168/aair.2012.4.3.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein KA, Jackson WT. Human rhinovirus 2 induces the autophagic pathway and replicates more efficiently in autophagic cells. J Virol. 2011;85:9651–9654. doi: 10.1128/JVI.00316-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Hernandez LD, Galan JE, Janeway CA, Jr., Medzhitov R, Flavell RA. IRAK-M is a negative regulator of Toll-like receptor signaling. Cell. 2002;110:191–202. doi: 10.1016/s0092-8674(02)00827-9. [DOI] [PubMed] [Google Scholar]
- Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science. 2007;315:1398–1401. doi: 10.1126/science.1136880. [DOI] [PubMed] [Google Scholar]
- Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Ohsumi Y, Yoshimori T. Autophagosome formation in mammalian cells. Cell Struct Funct. 2002;27:421–429. doi: 10.1247/csf.27.421. [DOI] [PubMed] [Google Scholar]
- Mosser AG, Vrtis R, Burchell L, Lee WM, Dick CR, Weisshaar E, et al. Quantitative and qualitative analysis of rhinovirus infection in bronchial tissues. Am J Respir Crit Care Med. 2005;171:645–651. doi: 10.1164/rccm.200407-970OC. [DOI] [PubMed] [Google Scholar]
- Newcomb DC, Sajjan US, Nagarkar DR, Wang Q, Nanua S, Zhou Y, et al. Human rhinovirus 1B exposure induces phosphatidylinositol 3-kinase-dependent airway inflammation in mice. Am J Respir Crit Care Med. 2008;177:1111–1121. doi: 10.1164/rccm.200708-1243OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- Papadopoulos NG, Bates PJ, Bardin PG, Papi A, Leir SH, Fraenkel DJ, et al. Rhinoviruses infect the lower airways. J Infect Dis. 2000;181:1875–1884. doi: 10.1086/315513. [DOI] [PubMed] [Google Scholar]
- Rosati O, Martin MU. Identification and characterization of murine IRAK-M. Biochem Biophys Res Commun. 2002;293:1472–1477. doi: 10.1016/S0006-291X(02)00411-4. [DOI] [PubMed] [Google Scholar]
- Sarkar S, Korolchuk V, Renna M, Winslow A, Rubinsztein DC. Methodological considerations for assessing autophagy modulators: a study with calcium phosphate precipitates. Autophagy. 2009;5:307–313. doi: 10.4161/auto.5.3.7664. [DOI] [PubMed] [Google Scholar]
- Seki M, Kohno S, Newstead MW, Zeng X, Bhan U, Lukacs NW, et al. Critical role of IL-1 receptor-associated kinase-M in regulating chemokine-dependent deleterious inflammation in murine influenza pneumonia. J Immunol. 2010;184:1410–1418. doi: 10.4049/jimmunol.0901709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurrell JC, Wiehler S, Zaheer RS, Sanders SP, Proud D. Human airway epithelial cells produce IP-10 (CXCL10) in vitro and in vivo upon rhinovirus infection. Am J Physiol Lung Cell Mol Physiol. 2005;289:L85–L95. doi: 10.1152/ajplung.00397.2004. [DOI] [PubMed] [Google Scholar]
- Suh HS, Zhao ML, Rivieccio M, Choi S, Connolly E, Zhao Y, et al. Astrocyte indoleamine 2,3-dioxygenase is induced by the TLR3 ligand poly(I:C): mechanism of induction and role in antiviral response. J Virol. 2007;81:9838–9850. doi: 10.1128/JVI.00792-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykes A, Edwards MR, Macintyre J, del Rosario A, Bakhsoliani E, Trujillo-Torralbo MB, et al. Rhinovirus 16-induced IFN-alpha and IFN-beta are deficient in bronchoalveolar lavage cells in asthmatic patients. J Allergy Clin Immunol. 2012;129:1506–1514. doi: 10.1016/j.jaci.2012.03.044. e1506. [DOI] [PubMed] [Google Scholar]
- Takeshita F, Kobiyama K, Miyawaki A, Jounai N, Okuda K. The non-canonical role of Atg family members as suppressors of innate antiviral immune signaling. Autophagy. 2008;4:67–69. doi: 10.4161/auto.5055. [DOI] [PubMed] [Google Scholar]
- Tan WC. Viruses in asthma exacerbations. Curr Opin Pulm Med. 2005;11:21–26. doi: 10.1097/01.mcp.0000146781.11092.0d. [DOI] [PubMed] [Google Scholar]
- Tang SW, Chen CY, Klase Z, Zane L, Jeang KT. The cellular autophagy pathway modulates human T-cell leukemia virus type 1 replication. J Virol. 2013;87:1699–1707. doi: 10.1128/JVI.02147-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MP, Burgon TB, Kirkegaard K, Jackson WT. Role of microtubules in extracellular release of poliovirus. J Virol. 2009;83:6599–6609. doi: 10.1128/JVI.01819-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MP, Jackson WT. Viruses and arrested autophagosome development. Autophagy. 2009;5:870–871. doi: 10.4161/auto.9046. [DOI] [PubMed] [Google Scholar]
- Tung YT, Hsu WM, Lee H, Huang WP, Liao YF. The evolutionarily conserved interaction between LC3 and p62 selectively mediates autophagy-dependent degradation of mutant huntingtin. Cell Mol Neurobiol. 2010;30:795–806. doi: 10.1007/s10571-010-9507-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Elden LJ, Sachs AP, van Loon AM, Haarman M, van de Vijver DA, Kimman TG, et al. Enhanced severity of virus associated lower respiratory tract disease in asthma patients may not be associated with delayed viral clearance and increased viral load in the upper respiratory tract. J Clin Virol. 2008;41:116–121. doi: 10.1016/j.jcv.2007.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vareille M, Kieninger E, Alves MP, Kopf BS, Moller A, Geiser T, et al. Impaired type I and type III interferon induction and rhinovirus control in human cystic fibrosis airway epithelial cells. Thorax. 2012;67:517–525. doi: 10.1136/thoraxjnl-2011-200405. [DOI] [PubMed] [Google Scholar]
- Wang J, Oberley-Deegan R, Wang S, Nikrad M, Funk CJ, Hartshorn KL, et al. Differentiated human alveolar type II cells secrete antiviral IL-29 (IFN-lambda 1) in response to influenza A infection. J Immunol. 2009;182:1296–1304. doi: 10.4049/jimmunol.182.3.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wark PA, Johnston SL, Bucchieri F, Powell R, Puddicombe S, Laza-Stanca V, et al. Asthmatic bronchial epithelial cells have a deficient innate immune response to infection with rhinovirus. J Exp Med. 2005;201:937–947. doi: 10.1084/jem.20041901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wat D, Gelder C, Hibbitts S, Cafferty F, Bowler I, Pierrepoint M, et al. The role of respiratory viruses in cystic fibrosis. J Cyst Fibros. 2008;7:320–328. doi: 10.1016/j.jcf.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White SR, Martin LD, Abe MK, Marroquin BA, Stern R, Fu X. Insulin receptor substrate-1/2 mediates IL-4-induced migration of human airway epithelial cells. Am J Physiol Lung Cell Mol Physiol. 2009;297:L164–L173. doi: 10.1152/ajplung.90453.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Jiang D, Smith S, Thaikoottathil J, Martin RJ, Bowler RP, et al. IL-13 dampens human airway epithelial innate immunity through induction of IL-1 receptor-associated kinase M. J Allergy Clin Immunol. 2012;129:825–833. doi: 10.1016/j.jaci.2011.10.043. e822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- Yoshikawa T, Hill TE, Yoshikawa N, Popov VL, Galindo CL, Garner HR, et al. Dynamic innate immune responses of human bronchial epithelial cells to severe acute respiratory syndrome-associated coronavirus infection. PLoS One. 2010;5:e8729. doi: 10.1371/journal.pone.0008729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X, Overmeyer JH, Maltese WA. Functional specificity of the mammalian Beclin-Vps34 PI 3-kinase complex in macroautophagy versus endocytosis and lysosomal enzyme trafficking. J Cell Sci. 2006;119:259–270. doi: 10.1242/jcs.02735. [DOI] [PubMed] [Google Scholar]
- Zhou D, Kang KH, Spector SA. Production of interferon alpha by human immunodeficiency virus type 1 in human plasmacytoid dendritic cells is dependent on induction of autophagy. J Infect Dis. 2012;205:1258–1267. doi: 10.1093/infdis/jis187. [DOI] [PMC free article] [PubMed] [Google Scholar]