Abstract
Peroxisome proliferator-activated receptor (PPAR) α is a transcription factor that regulates genes involved in fatty acid catabolism. Here we provide evidence that PPARα is constitutively expressed in nuclei of hippocampal neurons and surprisingly controls calcium influx and the expression of various plasticity-related genes via direct transcriptional regulation of CREB. Accordingly, Ppara-null, but not Pparb-null, mice are deficient in CREB and memory-associated genes, and have decreased spatial learning and memory. While shRNA knockdown of PPARα in the hippocampus suppressed CREB and NR2A rendering wild type animals markedly poor in consolidating spatial memory, introduction of PPARα to the hippocampus of Ppara-null mice increased hippocampal CREB and NR2A and improved spatial learning and memory. These results together with detailed analyses of CREB, NR2A and spatial learning and memory in bone marrow chimeric animals lacking PPARα in the CNS describe a novel mechanism for transcriptional control of Creb and associated plasticity genes by PPARα.
INTRODUCTION
Memory loss is a major issue in gerontology and thus it would be of great value to preserve memory throughout the life. Although problems with memory become increasingly common as people age, in some individuals, memories last a long time, even a life time. On the other hand, some people experience milder to substantial memory problems even at an earlier age. Among many factors that control memory, cAMP response element-binding (CREB) has been shown to be an integral part in the formation of variety of complex forms of memory, including spatial and social learning (Abel and Kandel, 1998; Yin et al., 1995). Therefore, understanding molecular mechanisms by which CREB and memory are regulated is an important area of subject. Peroxisome proliferator-activated receptors (PPARs) are a group of three transcription factors (PPARα, PPARβ/δ and PPARγ) that are involved in control of lipid homeostasis; PPARγ controls lipid uptake and storage while PPARα controls fatty acid catabolism. PPARα activates the expression of genes responsible for fatty acid transport and oxidation notably in the liver, heart and kidney, and plays the most important role in the hepatic catabolism of fatty acids (Keller et al., 1993; Kersten et al., 2000; Marcus et al., 1993). Accordingly, PPARα is highly expressed in liver, the major lipid-metabolizing organ (Chakravarthy et al., 2009; Schoonjans et al., 1996).
Here we demonstrate that PPARα is expressed in the hippocampus and that PPARα, but not PPARβ and PPARγ, modulates the expression of various plasticity-related molecules and their functions via direct transcriptional control of the master regulator CREB. Furthermore, from analysis of Ppara-null and bone marrow chimeric mice, and lentiviral manipulation of PPARα expression in the adult hippocampus, we establish a direct role of PPARα in the regulation of CREB and NR2A and the formation of hippocampal memory.
RESULTS
Expression and Distribution of PPAR in Hippocampus
We examined the distribution of PPARα in different regions of hippocampus. Consistent to previous reports (Braissant et al., 1996; Moreno et al., 2004), PPARα was seen to be present in the hippocampus as our immunohistochemical analyses revealed that PPARα protein was localized in CA1, CA2, CA3, and DG subfields of the hippocampus of wild-type, but not Ppara-null mice (Fig. S1A). Although we have observed the presence of PPARα in rodent hippocampus, primate hippocampus may not contain PPARα because humans and other primates have considerably lower levels of PPARα in liver than rodents that is responsible in part for species differences in response to peroxisome proliferators (Tugwood et al., 1998). However, as evident from supplementary figure 1B, PPARα was also localized in all different subfields of hippocampus of rhesus monkey, suggesting that both rodents and primates have PPARα in hippocampus. Similarly, PPARα mRNA was also detected in the hippocampus of wild-type (WT) mice, but not Ppara-null mice (Fig. S1C). On the other hand, both WT and Ppara-null hippocampi expressed equivalent levels of PPARβ and PPARγ mRNAs (Fig. S1C), suggesting that the deficiency of PPARα does not affect other PPARs.
The role of PPARα in synaptic function
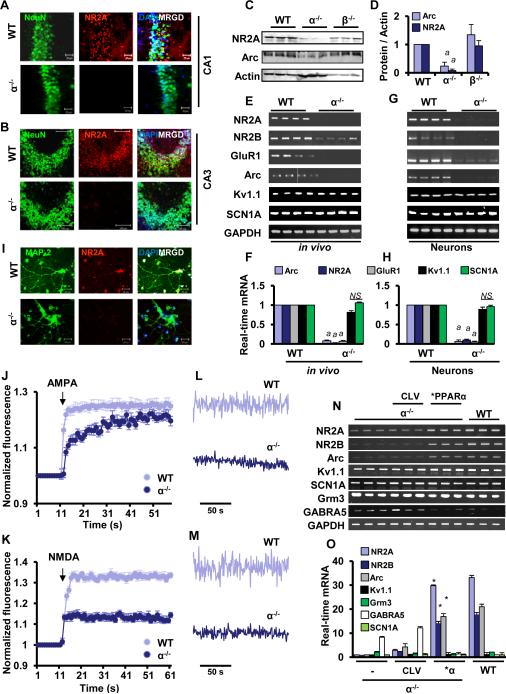
Because hippocampus is associated with the formation of long-term memory, next we investigated the role of PPARα in calcium oscillation and the regulation of various hippocampal plasticity-related genes. NMDA receptor subunit NR-2A (Sakimura et al., 1995), NR-2B (Sakimura et al., 1995), AMPA-receptor subunit GluR1 (Lee et al., 2003) and AMPA-receptor associated activity-related cytoskeletal protein, Arc (Tzingounis and Nicoll, 2006), are some of the major plasticity-related genes in the hippocampus. As evident from immunofluorescence analysis (Fig. 1A-B), the expression of NR2A was markedly higher in the hippocampus of WT, but not Ppara-null mice. This is also supported by immunoblot data on NR2A and Arc (Fig. 1C-D). Moreover, mRNA expression analysis of hippocampus (Fig. 1E-F) demonstrates that hippocampal NR2A, NR2B, GluR1, and Arc mRNAs were higher in WT mice than Ppara-null mice. However, the mRNA expression of voltage-gated ion channel genes including Kv1.1 and Scn1a was found to be same in both WT and Ppara-null hippocampus (Fig. 1E-F). It is interesting because recent genome-wide survey has identified Scn1a as an important molecule for short-term memory (Papassotiropoulos et al., 2011).
Figure 1. PPARα Regulates the Expression of Plasticity-related Molecules in Hippocampus.
Paracoronal cryosections from CA1(A) and CA3(B) regions of hippocampus in 6-8 weeks old male WT and Ppara-null (KO) mice were double-immunolabeled with NeuN (green) and NR2A (red). (C) Hippocampi of adult WT (n=3), α KO (n=3), and β KO (n=3) mice were immunobloted with NR2A and Arc. (D) The levels of NR2A and Arc proteins were normalized with β Actin immunoblot analysis. (ap<0.001 vs. WT) RT-PCR (E) and real-time PCR (F) analyses of different plasticity-related molecules were performed in hippocampi of four 6-8 wks old WT and KO mice (marked as in vivo) (ap<0.001 vs. WT). The mRNA expression of different plasticity-related molecules in E18 hippocampal neurons (marked as in vitro) was carried out by RT-PCR (G) and real-time PCR (H) (ap<0.001 vs. WT). (I) The cultured hippocampal neurons (WT and KO) were also immunostained with NR2A (red), MAP2 (green) and DAPI (blue). Results represent three separate experiments. Normalized fluorescence intensity of fluo-4 (Ca2+ indicator) in WT and KO neurons in presence of AMPA (J) and NMDA (K). Calcium oscillation in WT, KO, and PPARα-transduced neurons under influence of AMPA (L) and NMDA (M). α KO hippocampal neurons were infected with lentivirions containing either PPARα over-expression construct (*PPARα) or empty vector (CLV). After 48 h of infection, RT-PCR (N) and real-time PCR (O) analyses of NR2A, NR2B, Arc, Kv1.1, Scn1a, Grm3, and GABRA5 in WT, KO, CLV-, and *PPARα-infected α KO hippocampal neurons. Results are analyzed from fetal neurons of either three or four different pregnant WT and KO mice (*p<0.0001 vs. KO).
Similar to hippocampal tissues, cultured Ppara-null hippocampal neurons were also observed to express significantly low level of plasticity-related genes (Fig. 1G-H) and proteins (Fig.1I) as compared to WT hippocampal neurons. These results were specific as PPARα knockdown did not alter the expression of Kv1.1 and Scn1a in hippocampal neurons (Fig. 1G-H). Interestingly, the expression of NR2A, NR2B, GluR1, and Arc was not compensated by PPARβ and PPARγ, which are present at the normal level in the hippocampus of Ppara-null mice, indicating that PPARα, but neither PPARβ nor PPARγ, is critically required for the expression of these plasticity-related genes. Since calcium oscillation through metabotropic receptors has been implicated in synaptic plasticity, next we monitored calcium oscillation in cultured WT and Ppara-null hippocampal neurons in the presence of AMPA (Fig.1J &L) and NMDA (Fig. 1K & M). Interestingly, both AMPA and NMDA elicited a stronger calcium influx and a larger amplitude oscillation in WT neurons than Ppara-null hippocampal neurons, suggesting that PPARα plays a direct role in controlling the synaptic plasticity in hippocampal neurons.
Re-instating PPARα upregulates the expression of plasticity-related genes and restores calcium oscillation in Ppara-null hippocampal neurons
Results described above suggest that PPARα plays an important role in calcium oscillation and the transcription of plasticity-related genes. However, many different factors regulate plasticity and it is not clear if PPARα alone is sufficient to control calcium current and the transcription of various plasticity-associated genes. To test this hypothesis, we supplemented PPARα in Ppara-null hippocampal neurons via lentiviral transduction. Interestingly, lentiviral over-expression of PPARα, but not an empty vector, in mouse Ppara-null hippocampal neurons significantly upregulated the mRNA expression of plasticity-related genes such as NR2A, NR2B, Arc, and GluR1 (Fig. 1N-O). This effect was specific as lentiviral manipulation of PPARα had no effect on the expression of ion channel genes (Kv1.1 and Scn1a) and Grm3, a metabotropic glutamate receptor subunit, in hippocampal neurons (Fig. 1N-O). Furthermore, in contrast to the upregulation of plasticity-related genes, PPARα over-expression suppressed the expression of GABRA5, receptor subunit of γ amino butyric acid and a molecule that is involved in long-term depression (Martin et al., 2013), in Ppara-null hippocampal neurons (Fig. 1N-O).
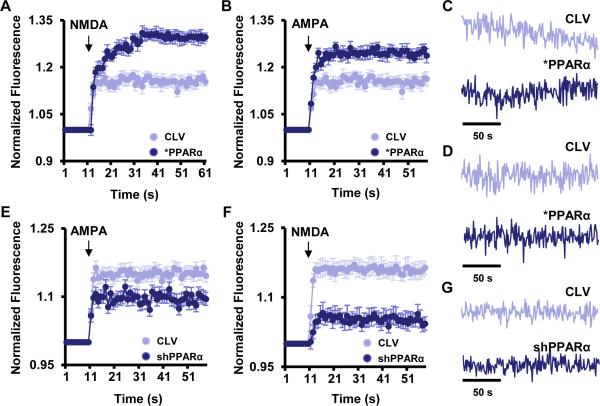
Next, we investigated if restoration of PPARα normalizes calcium influx in Ppara-null hippocampal neurons. Interestingly, lentiviral reinstallation of PPARα in Ppara-null hippocampal neurons elicited increased calcium influx (Fig. 2A-B) and larger oscillatory amplitude (Fig. 2C-D) in the presence of NMDA (Fig 2A & 2C) and AMPA (Fig. 2B & 2D) as compared to lentivector-transduced Ppara-null neurons. Conversely, lentiviral knockdown of PPARα by lenti-shPPARα also inhibited the calcium influx (Fig 2E-2F) and signaling amplitude (Fig. 2G) through ionotropic calcium channels in cultured WT neurons. Together, these results indicate that PPARα alone is sufficient to control the expression of plasticity-related genes and their functions in hippocampal neurons without disturbing the expression of other essential genes associated with voltage gated ion conductance.
Figure 2. Effect of PPARα on NMDA- and AMPA-induced calcium oscillation in cultured hippocampal neurons.
E18 hippocampal neurons were infected with lentivirions containing either PPARα over-expression construct (*PPARα) or empty vector (CLV). After 48 h of infection, neurons were analyzed for Fluo-4 labeled calcium influx in the presence of 50 μM NMDA (A) and AMPA (B). Oscillatory amplitude of calcium signaling after the treatment of NMDA (C) and AMPA (D) was shown in CLV- and *PPARα- transduced neurons. Normalized fluo-4 fluorescence value that represents AMPA- (E) and NMDA- (F) dependent calcium influx in lenti-shControl (CLV) and lenti-shPPARα-(shPPARα) infected hippocampal neurons. Arrow indicates the application of AMPA and NMDA into the culture. (G) The calcium oscillation in CLV- and lenti-shPPARα–infected neurons. Results represent three independent experiments.
Involvement of PPARα in the transcription of CREB
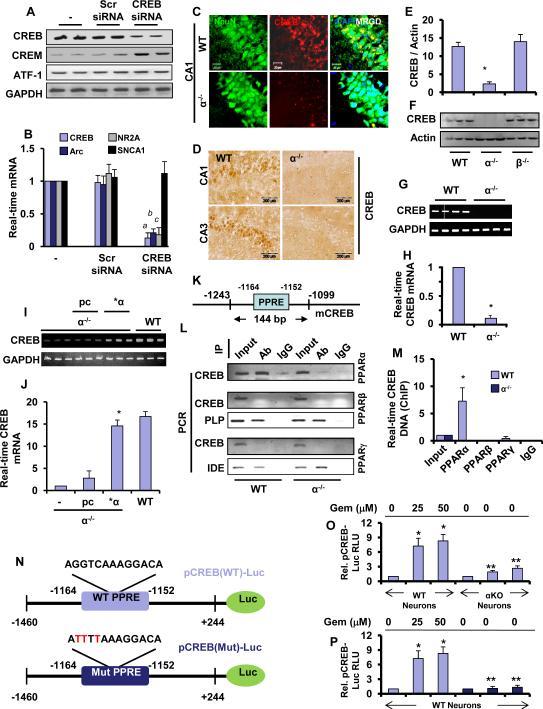
Next, we investigated mechanisms by which PPARα controls the expression of plasticity-related genes. Interestingly, the promoters of all these plasticity-associated genes do not contain any PPAR-responsive elements (PPRE), suggesting that PPARα does not directly regulate the expression of these genes. On the other hand, these promoters harbor multiple cAMP response elements (CRE) (Guzowski and McGaugh, 1997; Impey et al., 1998; Lee et al., 2003; Yin et al., 1995). Therefore, we examined the role of CREB in the expression of these plasticity-related genes using CREB siRNA. Knockdown of CREB was specific as CREB siRNA suppressed the expression of only CREB, but not other CREB-related molecules such as, CREM and ATF-1 (Fig. 3A). Consistently, siRNA knockdown of CREB (Fig. 3A) inhibited the expression of NR2A and Arc mRNAs (Fig. 3B) in hippocampal neurons. Again, this effect was specific as siRNA knockdown of CREB did not suppress the expression of short-term memory-associated molecule Scn1a (Fig. 3A-B).
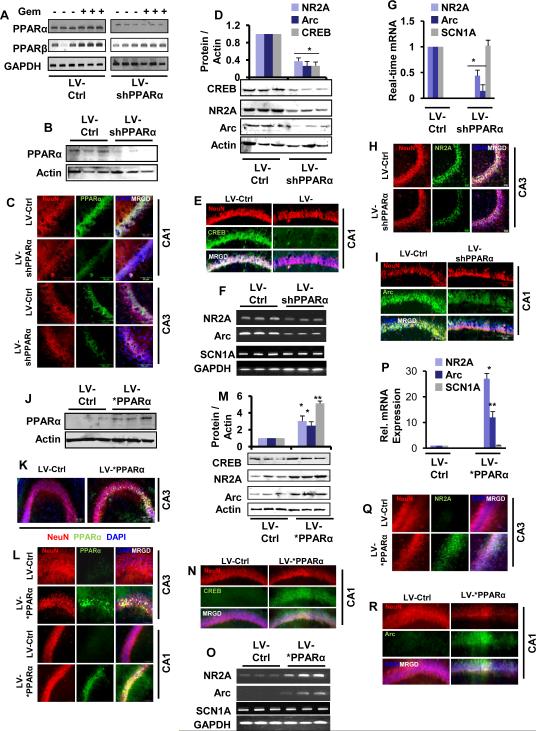
Figure 3. PPARα Regulates the Expression of CREB.
(A) The efficiency of CREB siRNA for knocking down the expression of CREB and other Creb family genes (CREM and ATF-1) was analyzed in cultured hippocampal neurons by RT-PCR. (B) Hippocampal neurons were transfected with CREB siRNA and scrambled siRNA and after 24 h of transfection, cells were analyzed for the mRNA expression of Arc, NR2A and Scn1a by quantitative real-time PCR. CREB was also run to verify siRNA knockdown. Results are mean ± SD of three independent experiments. a p < 0.0001 vs cont siRNA for CREB; b p < 0.0001 vs cont siRNA for Arc, and c p < 0.0001 vs cont siRNA for NR2A. (C) Immunofluorescence analysis of CREB in PPARα WT and KO hippocampus. CA1 regions of 6-8 weeks old mice were stained with NeuN (green), CREB (red) and DAPI (blue). (D) DAB-immunostaining of WT and KO hippocampus. Both CA1 and CA3 hippocampal regions were stained with biotinylated antibody against mouse CREB. (E) Hippocampal tissue of 6-8 wks old WT (n=3), Ppara-null (α KO) (n=3) and Pparb-null (β KO) (n=3) mice were immunoblotted for CREB. (F) Relative density of CREB was analyzed by image J software. Results are ± SD of three independent experiments. The mRNA expression of CREB in the hippocampus of four 6-8 wks old WT and KO mice was carried out by RT-PCR (G) and real-time PCR (H). PPARα KO hippocampal neurons were transfected with 0.25 μg either human PPARα cDNA or an empty vector (pcDNA3). Twenty-four h after transfection, the mRNAs encoding Creb was analyzed by RT-PCR (I) and quantitative real-time PCR (J). *p<0.0001 vs. empty vector pcDNA3. (K) The map of mouse Creb promoter region that harbors one 12 bp long consensus PPRE (position −1152 to −1164). In situ ChIP for PPARα, PPARβ and PPARγ followed by semi-quantitative (L) and quantitative PCR (M) analyses of Creb promoter were performed in the hippocampus of WT and KO mice. Transcriptional activities of PPARβ and PPARγ was further validated by amplifying the promoters of their known target genes PLP (proteolipid protein) and IDE (insulin degrading enzyme), respectively. Results represent three separate analyses. (N) MAP of wild-type and mutated Creb promoter constructs. (O) WT hippocampal neurons were transfected with pCREB-Luc (light blue) and pCREB(mut)-Luc (deep blue). Ppara-null (KO) hippocampal neurons were transfected with pCREB-Luc (gray). After 24 h after transfection, cells were stimulated with gemfibrozil for 4 h followed by measuring luciferase activity. Results are mean ± SD of three independent experiments. *p<0.001 vs control; **p<0.001 vs respective treatment of WT neurons.
Upon analysis of the creb gene promoter by MatInspector promoter analysis tool (Genomatix software GmbH), we found one consensus PPRE in the distal region (−1164 to −1152) of the Creb promoter (Fig. 3K). Therefore, the expression of CREB in hippocampus of WT and Ppara-null mice was analyzed. As evident from our immunofluorescence (Fig. 3C), DAB-staining (Fig. 3D), immunoblot (Fig. 3E-F), and mRNA analysis (Fig. 3G-H), the expression of CREB was significantly higher in both WT and Pparb-null hippocampi than Ppara-null hippocampus. Moreover, the reinstatement of PPARα in Ppara-null hippocampal neurons significantly restored the expression of CREB mRNA (Fig. 3I-J), suggesting that PPARα may play an important role in the expression of CREB in mouse hippocampal neurons. Next, we investigated mechanism by which PPARα could control the transcription of CREB. We first cloned the mouse CREB promoter and then performed site-directed mutagenesis to mutate the PPRE (Fig. 3N). As evident from figure 3O, gemfibrozil, an agonist of PPARα (Roy and Pahan, 2009), markedly induced CREB promoter-driven luciferase activity in hippocampal neurons isolated from WT, but not Ppara-null, mice. However, in WT hippocampal neurons, gemfibrozil remained unable to induce the luciferase activity driven by a CREB promoter in which PPRE was mutated (Fig. 3P).
CREB is also present in non-neuronal cells and accordingly, we also found that PPARα was present in nuclei of astrocytes (Fig. S2A). Therefore, we investigated if CREB was also regulated in astrocytes the same way as in neurons by PPARα. Increasing doses of different PPARα agonists such as gemfibrozil (known as Lopid® in the pharmacy), fenofibrate (known as Tricor® in the pharmacy) and WY14643, a synthetic agonist of PPAR-α, induced the activation of PPRE-driven luciferase activity (Fig. S2B) and CREB promoter-driven luciferase activity (Fig. S2C) in astrocytes isolated from WT, but not PPAR-α (−/−), mice. These results demonstrate an essential role of PPARα in the activation of CREB promoter in neurons as well as non-neuronal cells.
Recruitment of PPARα to the Creb Promoter In Vivo in the Hippocampus
To further support the transcriptional regulation of Creb by PPARα, we performed in situ chromatin immunoprecipitation (ChIP) analysis, in which the recruitment of PPARα to the Creb promoter (Fig. 3L) was monitored in vivo in the hippocampus. We were able to amplify a 144 bp fragment encompassing the PPRE of the Creb promoter (Fig. 3K) in hippocampus isolated from wild-type, but not Ppara-null mice (Fig. 3L; upper panel). On the other hand, no amplification product of the Creb promoter was observed in any of the immunoprecipitates obtained with PPARβ (Fig. 3L; middle panel), PPARγ (Fig. 3L; lower panel) or control IgG. This effect was confirmed by real-time PCR as well (Fig. 3M). To understand whether PPARβ and PPARγ are functionally active in hippocampus or not, we examined the recruitment of PPARβ and PPARγ to promoters of proteolipid protein (PLP) (Jana et al., 2012) and insulin degrading enzyme (IDE) (Du et al., 2009), respectively. For details about the location of PPRE in the promoters of PLP and IDE, please see Figure S3A-B. While we observed the recruitment of PPARβ to the PLP promoter (Fig. 3L; middle panel), PPARγ was recruited to IDE promoter (Fig. 3L; bottom panel). PPARα knockdown had no effect on the recruitment of either PPARβ to the PLP promoter or PPARγ to the IDE promoter (Fig. 3L). Together, these data suggest that although PPARβ and PPARγ are transcriptionally active in vivo in the hippocampus, only PPARα, but neither PPARβ nor PPARγ, is recruited to the PPRE of the Creb promoter.
Does PPARα Control the Expression of Plasticity-related Genes and Calcium Influx in Hippocampal Neurons via CREB?
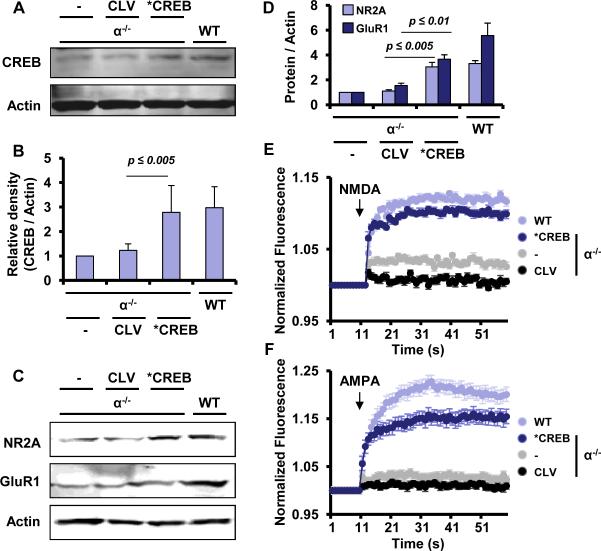
Since we are hypothesizing that PPARα controls the expression of plasticity-related genes by regulating the transcription of CREB, next, we examined if CREB over-expression increased the expression of NR2A and GluR1 and normalized calcium influx in Ppara-null hippocampal neurons. Interestingly, lentiviral over-expression of CREB (Fig. 4A-B) significantly stimulated the expression of NR2A and GluR1 in Ppara-null neurons (Fig. 4C-D). Accordingly, lenti-CREB, but not lenti-control, stimulated NMDA- (Fig. 4E) and AMPA- (Fig.4F) dependent calcium influx in Ppara-null neurons. These results suggest that PPARα regulates plasticity-related molecules and their functions in hippocampal neurons via CREB.
Figure 4. Lentiviral over-expression of CREB induces the expression of plasticity-related molecules and restores calcium influx in Ppara-null hippocampal neurons.
(A) E18 Ppara-null hippocampal neurons were transduced with control (CLV) and CREB (*CREB) lentivirus at a MOI of 10. After 48 h, the expression of CREB protein was monitored in untransduced, CLV, and CREB-transduced neurons by Western blot. (B) Densitometric analyses of CREB expression. Results are mean ± SD of three independent results. ap< 0.005 vs. CLV. (C) Immunoblot analyses of NR2A and GluR1 in untransduced, CLV, and CREB lentiviral-transduced Ppara-null hippocampal neurons. (D) Densitometric analysis of NR2A and GluR1. Results are mean ± SD of three different experiments. ap< 0.005 and bp<0.01 vs. NR2A and GluR1, respectively in CLV-transduced neurons. Calcium influx in WT, Ppar-null, CLV, and CREB- transduced Ppara-null neurons was assayed in the presence of (E) NMDA and (F) AMPA. Results are mean ±SD of three independent experiments.
A Direct Role for PPARα in the Expression of Hippocampal Plasticity-related Genes in Adult Mouse Hippocampus
Here we used lentiviral-mediated manipulations of PPARα in vivo in the hippocampus. At first, we checked the specificity of lenti-shPPARα. As evident from Figure 5A, gemfibrozil treatment led to the stimulation of both PPARα and PPARβ in hippocampal neurons. However, lenti-shPPARα suppressed the upregulation of PPARα, but not PPARβ, in hippocampal neurons (Fig. 5A). These results were specific as lenti-control shRNA did not exhibit any effect (Fig. 5A). Next, EGFP constructed lentiviral (Lenti-GFP) particles were infused bilaterally in the pyramidal layer of CA2 region and in the subgranular layer (SGL) of dentate gyrus (DG) (Fig. S4A-B) of 6-8 weeks old adult mice. Three weeks after the infusion, we observed a marked distribution of EGFP in the entire pyramidal (CA1, CA2 and CA3) and DG region (Fig. S4C-D). Although, the distribution was observed both in dividing and non-dividing population, neurons were the major cell types that expressed EGFP (Fig. S4E-F). Accordingly, the bilateral infusion of lentiviral PPARα shRNA (Fig S4G) significantly inhibited the expression of PPARα, but not PPARβ, in different regions of hippocampus (Fig. 5B-C). The immunoblot (Fig. 5D) and immunofluorescence (Fig. 5E) analyses indicate strong inhibition of CREB in lenti-shPPARα- as compared to lenti-shcontrol-infused hippocampus. Consistently, we observed strong inhibition in the expression of NR2A and Arc in lenti-shPpara-, but not in lenti-shcontrol-transduced hippocampus (Fig. 5D, F-I). This effect was specific as lentiviral knockdown of PPARα did not modulate the short-term memory-related molecule Scn1a in vivo in the hippocampus (Fig. 5F-G).
Figure 5. Lentiviral manipulation of PPARα alters the Expression of CREB and plasticity-associated molecules in the Hippocampus of Adult Mice.
(A) Mouse primary hippocampal neurons were infected with lenti-control and lenti-PPARα shRNA in the presence or absence of 10 μM gemfibrozil. The specificity of lenti-shPPAR-α was monitored by RT-PCR of PPARα and PPARβ. Twelve weeks after the injection of lentiparticles, hippocampal regions of control (n=6) and shPPARα transduced (n=6) animals were analyzed for the expression of PPARα by immunoblot (n=3) (B) and immunofluorescence (n=3) analyses (C). (D) Immunoblot analyses and densitometric analyses (upper panel) of CREB, NR2A and Arc were performed in hippocampus (n=3) after 12 weeks of lenti-control and lenti-shPPAR-α transduction. Immunofluorescence analysis of CREB in CA1 (E). RT-PCR (F) and real-time PCR (G) were performed to analyze the mRNA expression of NR2A, Arc and Scn1a in the hippocampus of lenti-control and lenti-shPPARα-transduced animals. Results are mean ± SEM of 3 mice per group. *p<0.001 vs Lenti-control. Immunofluorescence analyses of NR2A (H) and Arc (I) were monitored in lenti-control (n = 3) and lenti-shPPARα-transduced (n = 3) hippocampus. Six to eight wks old male C57BL6/J animals (n=6 per group) received vector (Lenti-control) and PPARα expressing lentiviral particles (Lenti-*PPARα) in the hippocampus. Twelve weeks after the injection, hippocampal regions of control (n=3) and lenti-*PPARα (n=3) animals were analyzed for the expression of PPARα by immunoblot (J) and immunofluorescence (K and L) analyses. (M) Immunoblot analyses and densitometric analyses (upper panel) of CREB, NR2A and Arc were performed in hippocampus. Immunofluorescence analysis of CREB in CA1 (N). RT-PCR (O) and real-time PCR (P) were performed to analyze the mRNA expression of NR2A, Arc and Scn1a in the hippocampus. Results are mean ± SEM of 3 mice per group. Results are mean ± SEM of 3 mice per group. *p<0.05 and **p<0.01 vs Lenti-control. Immunofluorescence analyses of hippocampal neurons for NR2A and Arc were performed after 12 weeks of lenti-control and lenti-*PPARα infection.
Next, we examined if exogenously introduced PPARα (lenti-*Ppara) (Fig. S4H) could regulate hippocampal genes in Ppara-null mouse brain hippocampus. Bilateral infusion of lenti-*Ppara, but not the lenti-vector, significantly restored the expression of PPARα in the hippocampus (Fig. 5J-L) and subsequently stimulated the expression of CREB protein at 12 week of lentiviral injection (Fig. 5M). Accordingly, we also observed increased mRNA and protein expression of NR2A and Arc in different parts of the hippocampus of Ppara-null mice after lenti-*Ppara injection (Fig. 5M-R). On the other hand, lenti-*Ppara remained unable to alter the mRNA expression of Scn1a (Fig. 5O-P).
The Role of PPARα in the Process of Learning and Memory
The hippocampus regulates the generation of long term memory (Kesner and Connor, 1972) and spatial learning (Morris et al., 1986). First, we investigated if there was in fact any change in the expression of hippocampal plasticity-related genes during memory consolidation (Fig. S5A-B4). Consistent to a previous report (Cavallaro et al., 2002), we observed that the expression of Arc was significantly high after one hour (Fig. S5C1 & S5D1) whereas the levels both NR2A (Fig. S5C2 & S5D2) and GluR1 (Fig. S5C3 & S5D3) genes were stimulated at 24 hours post probe trial. Interestingly, there was no temporal change of gene expression in cage control and poor performers (Fig. S5). These results suggest that memory consolidation upregulates the expression of plasticity-related genes. Therefore, we analyzed the performances of both WT (C57/BL6J) and Ppara-null mice before and after of hippocampal Ppara gene manipulation in different learning and memory experimental paradigms. There was no significant difference in body weight between WT and Ppara-null mice (Table S1) after lentiviral infusion, nullifying its metabolic interference in the test.
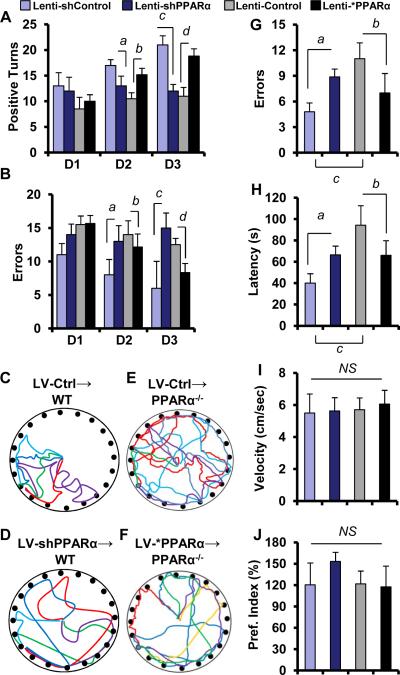
First, we performed T maze to determine whether there was any difference in spatial memory between lenti-shcontrol and lenti-shPPAR animals. On day 3, 4 and 5, we performed appetitive motivated tasks (Deacon, 2006). Interestingly, lenti-shPpara-transduced animals exhibited less number of positive turns in day 2 and day 3[F2, 6 = 7.93; p<0.05(=0.018)] and committed more errors in day 2 and day3 [F2, 6 = 4.118; p<0.01(=0.002)] than lenti-shControl-transduced animals in T maze apparatus (Fig. 6A-B) as analyzed by 2-way ANOVA with Tukey HSD posthoc test. On the other hand lenti-*Ppara, but not lenti-vector-transduced Ppara-null animals, significantly improved the hippocampus dependent memory performances as the lenti-*Ppara animals exhibited more positive turns [F2, 6 = 28.24; p<0.01(=0.0044)] and less errors [F2, 6 = 32.77; p<0.01(=0.0039)] than lenti-vector-transduced animals in T maze (Fig. 6A-B). Next, we studied the performance of these groups in the Barnes circular maze test, a hippocampus-dependent cognitive task that requires spatial reference memory. Interestingly, lenti-shPpara-transduced animals exhibited more number of errors [F2, 6 = 31.23; p<0.01(=0.008)] and more latency [F2, 6 = 28.614; p<0.05(=0.014)] (Fig. 6C-D, 6G-H) whereas lenti-*Ppara animals exhibited less number of errors [F2, 6 = 41.23; p<0.01(=0.0054)] and less latency [F2, 6 = 71.11; p<0.01(=0.00114)] (Fig. 6E-F, 6G-H) in Barnes maze. In all these experiments, lenti-shcontrol wild type group always remained better performer than lenti-control Ppara-null group in terms of making successful turns in T maze or taking less time in reaching target in Barnes maze. This difference in Barnes maze performance among different groups was not due to any change in velocity of mice, which remained almost same (Fig. 6I). Furthermore, we also did not find any significant difference in novel object recognition, an indicator of short-term memory, among different groups (Fig. 6J). Together, these results suggest an essential role of hippocampal PPARα in controlling the long-term, but not short-term, memory.
Figure 6. Barnes maze and T maze analyses of lenti-control, lenti-shRNA, and lenti-*PPAR-α animals.
In T maze analyses lenti-shControl (n=5), lenti-shPPARα (n=5), lenti-control (n=5), and lenti-*PPARα (n=5) groups of animals were analyzed for (A) number of positive turns and (B) number of errors. Lenti-shPPARα mice committed less number of positive turns (2-way repeated measures ANOVA;ap < 0.05 vs. Lenti-shControl at day 2 and cp < 0.01 vs. Lenti-shControl at day 3) and more number of errors (ap < 0.01 vs. Lenti-shControl at day 2 and cp < 0.001 vs. Lenti-shControl at day 3) in an appetitive T maze- based task when compared with lenti-control mice. On the other hand Lenti-*PPAR-α mice committed significantly more number of positive turns (bp < 0.05 vs. Lenti-Control at day 2 and dp < 0.01 vs. Lenti-Control at day 3) and less number of errors (bp < 0.05 vs. Lenti-Control at day 2 and dp < 0.01 vs. Lenti-Control at day 3) compared with lenti-control mice . Track plots of (C) lenti-shControl (n=5), (D) lenti-shPPARα (n=5) (E) lenti-control (n=5), and (F) lenti-*PPARα (n=5) transduced animals on an appetitive Barnes maze conditioning task. Animals (n=5) were analyzed for the (G) number of errors in Barnes maze. ap < 0.01 vs. Lenti-shControl; bp < 0.01 vs. Lenti-control; and cp < 0.001 vs. Lenti-shControl. (H) Latency, which is total time taken before entering into the target hole in Barnes maze was also analyzed. ap < 0.05 vs. Lenti-shcontrol; bp < 0.01 vs. Lenti-control; and cp < 0.001 vs. Lenti-shControl. Results are mean ±SD of three independent results. I represents the velocity of animals on Barnes maze apparatus. (J) Novel object recognition test was performed to monitor the short-term memory. Results are mean ± SEM of five mice per group. NS = No significance.
The Involvement of CNS PPARα, but not Peripheral PPARα, in the Regulation of Learning and Memory
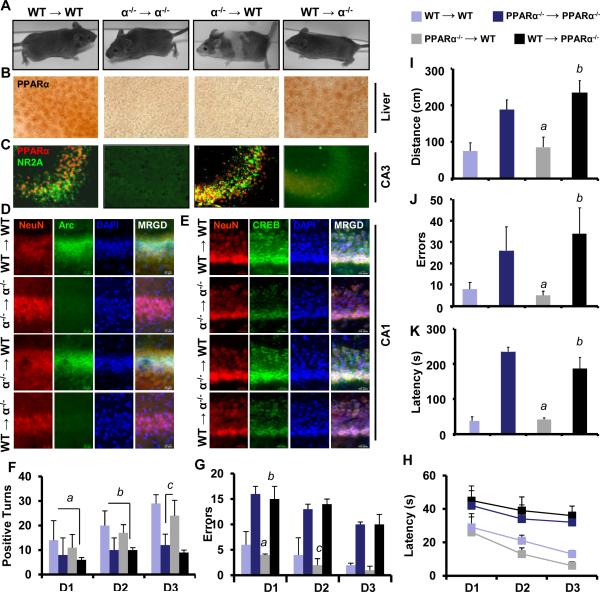
Since PPARα is present in the periphery (e.g. liver) as well as in the CNS, to understand the role of CNS PPARα in memory and learning, we generated bone marrow chimera mice. Eight weeks after bone marrow cell reconstitution, all KO→WT mice developed striped skin color phenotype, whereas all other groups of animals did not show any change in skin color (Fig. 7A). Moreover, chimera establishment did not affect the metabolic activities of animals as both WT→KO and KO→WT animals gained similar body weight (Table S2). We confirmed the establishment of chimeras by immunohistochemical analysis of PPARα in liver and brain hippocampus (Fig. 7B). As expected, hippocampi of WT→KO and KO→KO mice were devoid of PPARα (Fig. 7C). Consistent with the level of PPARα, both KO→WT and WT→WT mice expressed similar level of NR2A (Fig. 7C) and Arc (Fig. 7D) in hippocampus, suggesting that peripheral disruption of PPARα in KO →WT chimeric mice does not affect the expression of plasticity-associated molecules in hippocampus. Similarly, the level of CREB was significantly higher in the hippocampus of both KO→WT and WT→WT mice compared to that of WT→KO and KO→KO animals (Fig. 7E). Next we analyzed behavioral performances by T maze and Barnes maze. In T maze analyses, we observed very similar performances between WT→WT and KO→WT chimeric mice in terms of making number of positive turns (Fig. 7F) [ F2,6= 1.27, p>0.05], errors (Fig. 7G) [ F2,6 = 1.24, p>0.05], and latency (Fig. 7H) [ F2,6 = 1.07, p>0.05]. On the other hand, both KO→KO and WT→KO chimeric animals displayed similar, but significantly less number of positive turns, more errors and took longer time before making correct turns than WT →WT and KO→WT chimeric mice. In KO→WT mice, the improvement is consistent from day1 to day2 [t= 12.112, p<0.01] and from day2 to day3 [t = 26.342, p <0.005]. In contrast, WT→KO chimeric mice did not improve much as evident from the paired t-test with [t = 0.112, p >0.1 (n = 5)] from day 1 to day 2 and with [t = 0.448, p >0.1 (n = 5)] from day 2 to day 3. Similarly in Barnes maze analyses, KO→WT chimeric mice displayed significantly better performances (Fig. 7I-K) in travelling total distances [t = 12.371, p <0.05(=0.0192)], making errors [t = 37.561, p<0.05(=0.0217)] and taking time [t = 23.371, p <0.05(=0.0122)] than WT→KO chimeric mice. Together, CNS, but not peripheral, PPARα controls plasticity-related molecules and hippocampal learning and memory.
Figure 7. Generation of Bone Marrow Chimeric Mice and Analyzing Their Performances in T-maze and Barnes Maze Spatial Learning Tasks.
(A) Four weeks old male WT (C57/BL6J) and Ppara-null (KO) animals were used to generate bone marrow chimera mice. The establishment of chimeras was confirmed by PPARα staining in liver (B) and hippocampal (C) sections. Hippocampal sections stained with PPARα (green) were also stained with NR2A (red). Hippocampal sections of all four groups of chimeric animals were immunostained with NeuN (red) /Arc (green) (D) and NeuN (red)/CREB (green) (E). In a T maze, number of positive attempts made to discover the target (F) ( a,bp < 0.05 vs. WT→WT; cp < 0.01 vs. KO→KO ), number of errors (G) ( ap < 0.01 vs. KO→KO; bp < 0.001 vs. WT→WT; and cp < 0.001 vs. KO→KO) and the mean latency (H) are plotted as a function of number of days. In Barnes maze analysis, WT→WT, KO→ KO, WT→KO, and KO→WT chimeric mice were monitored for the total distance traveled (I) (ap < 0.001 vs. KO→KO; bp < 0.001 vs. WT→WT), the total number of errors made (J) (ap < 0.001 vs. KO→KO; bp < 0.001 vs. WT→WT) (), and the latency or total time taken (K) (ap < 0.001 vs. KO→KO; bp < 0.001 vs. WT→WT)before entering into the target hole (n=5 per group). Results are mean ±SEM of three separate experiments.
DISCUSSION
Although hippocampus does not metabolize fat (Perera et al.; Seyfried and Mukherjee, 2005; Yue and Lam) here we demonstrate that PPARα, a major regulator of hepatic fatty acid metabolism, is also expressed in hippocampus. Interestingly, despite the presence of normal levels of PPARβ and PPARγ, either Ppara-null hippocampus or Ppara-null hippocampal neurons have deficient calcium influx, lower expression of various plasticity-related molecules (NR2A, NR2B, GluR1, and Arc) and decreased spatial learning and memory as compared to wild-type mice. Moreover, shRNA knock down of PPARα in hippocampal neurons and adult hippocampus decreased calcium influx, reduced the expression of various hippocampal plasticity-related molecules and thereby depleted hippocampus-dependent memory and learning. On the other hand, lentiviral expression of PPARα in Ppara-null hippocampal neurons and the hippocampus of Ppara-null mice improved calcium current, restored the expression of various plasticity-related genes and recovered memory and learning. Recent genome-wide behavioral genetics approach combined with functional brain imaging studies has identified an important role of the α subunit of the voltage-gated sodium channel, type I (protein: Nav1. 1; gene: Scn1a) in human short-term memory (Papassotiropoulos et al., 2011). However, PPARα remained unable to regulate Scn1a either in cultured hippocampal neurons or in vivo in the hippocampus. By MatInspector analysis, we also did not find any CRE or PPRE in the promoter of Scn1a and siRNA knockdown of CREB also had no effect on Scn1a. Accordingly, PPARα manipulations in the hippocampus did not modulate short-term memory, thus indicating that PPARα regulates long-term, but not short-term, memory.
While investigating the underlying mechanism, we found no PPREs in the promoters of Nr2a, Nr2b, Glur1, and Arc genes. On the other hand, one conserved PPRE was noted in the promoter of Creb, encoding CREB, the master regulator of memory and learning. Several lines of evidence clearly highlight a direct role for PPARα in the transcriptional regulation of Creb. First, immunofluorescence and immunoblot analyses revealed that expression of CREB was significantly lower in Ppara-null hippocampus as compared to wild-type hippocampus. Second, PPARα knockdown inhibited the level of CREB and reduced calcium oscillation in cultured WT hippocampal neurons. Third, lentiviral over-expression of PPARα in Ppara-null hippocampal neurons stimulated the expression of CREB and restored calcium current. Fourth, in situ ChIP assays demonstrated that although PPARβ and PPARγ were active in the hippocampus and recruited to their respective target genes, only PPARα, but not PPARβ and PPARγ, were recruited to the Creb promoter in vivo in the hippocampus. Fifth, gemfibrozil, an agonist of PPARα, activated the Creb promoter in wild-type, but not Ppara-null, neurons. Sixth, gemfibrozil remained unable to activate mutated Creb promoter in which the conserved PPRE is mutated by site-directed mutagenesis. Finally, while several CREs are present in the promoters of plasticity-related genes (NR2A, Arc etc) and CREB is required for the transcription of these genes, we did not find any CRE in the promoter of Scn1a and siRNA knockdown of CREB also did not inhibit the mRNA expression of snc1a. Accordingly, over-expression of PPARα in Ppara-null hippocampal neurons induced the expression of CREB and CREB-dependent plasticity related molecules (NR2A and Arc), but not CREB-independent short-term memory-related gene (Scn1a). Taken together, these results show that PPARα directly controls CREB and thereby regulates CREB-associated hippocampal functions.
Although, we have proposed a unique role of hippocampal PPARα in controlling memory, the involvement of peripheral PPARα in this regard has been reported recently (Campolongo et al., 2009). PPARα in the gut generates a noradrenergic transmission in the basolateral amygdale, which facilitates the retention of spatial memory, and this particular autonomic neurotransmission is absent in mice lacking expression of PPARα thus rendering them poor consolidators of spatial memory. In contrast, we demonstrated that PPARα is constitutively present directly in the hippocampus and particularly within the nuclei of hippocampal neurons. Interestingly, lentiviral knockdown of PPARα in the adult hippocampus strongly inhibited the spatial memory and learning process, whereas, implementing PPARα in the hippocampus of Ppara-null animals incurred significant recovery, suggesting a direct role for hippocampal PPARα in the formation of memory. Furthermore, bone marrow chimeric mice with peripherally ablated PPARα have intact hippocampal NR2A and the ability of generating spatial memory, whereas, the ablation of PPARα in the CNS lowers hippocampal NR2A expression and makes animals markedly poor in consolidating spatial memory. Therefore, although the peripheral PPARα regulates noradrenergic neurotransmission to amygdala via vagal innervations in response to N-oleoylethanolamide (Campolongo et al., 2009) this pathway is not the only mechanism involved in PPARα-mediated regulation of learning and memory. This could be an indirect mechanism as peripheral PPARα should not regulate the hippocampal master regulator CREB at the transcriptional level. Therefore, our study highlights that the major and direct effect on spatial memory comes from hippocampal PPARα and the absence of this transcription factor in the hippocampus abrogates the learning and memory acquisition process via inhibition of Creb transcription and subsequent suppression of different memory-related genes.
EXPERIMENTAL PROCEDURES
Isolation of Mouse Hippocampal Neurons and Human Fetal Neurons
Hippocampal neurons were isolated from fetuses (E18) of pregnant female Ppara-null and strain-matched wild-type littermate mice as described by us (Jana et al., 2007; Saha et al., 2009) with some modifications. Briefly, dissection and isolation procedures were performed in an ice-cold, sucrose buffer solution (sucrose 0.32 M, Tris 0.025 M; pH 7.4) (Gorini et al., 1999). The skin and the skull were carefully removed from the brain by scissors followed by peeling off the meninges by a pair of fine tweezers. Next, a fine incision was made in the middle line around the circle of Willis and medial temporal lobe was opened up. Hippocampus was isolated as a thin slice of tissue located near the cortical edge of medial temporal lobe. Hippocampal tissues isolated from all fetal pups (n>10) were combined together and homogenized with 1 ml of Trypsin for 5 minutes at 37°C followed by neutralization of trypsin. The single cell suspension of hippocampal tissue was plated in the poly-D-lysine pre-coated 75 mm flask. Five min after plating, the supernatants were carefully removed and replaced with complete neurobasal media. The next day, 10 μM AraC was added to remove glial contamination in the neuronal culture. The pure cultures of hippocampal neurons were allowed to differentiate fully for 9-10 days before treatment (Jana et al., 2007; Saha et al., 2009; Saha and Pahan, 2007).
Calcium Influx Measurement
Cultured hippocampal neurons were loaded with Fluo4-fluorescence conjugated calcium buffer (Invitrogen Molecular Probes, Cat# F10471, F10472, F10473) and incubated at 37°C for 60 mins following manufacture's protocol. After that, fluorescence excitation and emission spectra were recorded in a Perkin–Elmer Victor X2 Luminescence Spectrometer in the presence of NMDA (50 μM) and AMPA (50 μM). The recording was performed with 300 repeats at 0.1ms intervals.
Cloning of Creb Promoter and Site-Directed Mutagenesis
Mouse genomic DNA isolated from mouse BV-2 microglial cells were used as template in PCR. The 5′ flanking sequence of mouse Creb (−1460/+244) gene was cloned by PCR. Primers were designed from gene bank sequences. Creb: sense: 5′-aga tct (BglII) CTG CCT CCT CCT CCT GCT CCT CTT-3′; antisense: 5′-acg cgt (MluI) GGC TCA GAT GAC TCC TGC ACA GGA-3′. The sense primer was tagged with BglII restriction site while the antisense primer was tagged with MluI. The PCR was performed using Advantage-2 PCR kit (Clontech) and resulting fragments were gel purified and ligated into the PGEM-TEasy vector (Promega). These fragments were further subcloned into the PGL-3 Enhancer vector and verified by sequencing in the automated sequencer of the University of Nebraska at Lincoln Biotechnology Center.
Site-directed mutagenesis was done by using the site directed mutagenesis kit (Stratagene, USA). Two primers in opposite orientation were used to amplify the mutated plasmid in a single PCR reaction. The PCR product was precipitated with ethanol and then phosphorylated by T4 kinase. The phosphorylated fragment was self-ligated by T4 DNA ligase and digested with restriction enzyme DpnI to eliminate the non-mutated template. The mutated plasmid was cloned and amplified in Escherichia coli (DH5-α strain) competent cells.
In situ ChIP
Recruitment of PPARα to the Creb promoter in vivo in the hippocampus of wild-type and Ppara null mice was examined by in situ ChIP analysis. Mice (n=4 in each group) were perfused with 4% paraformaldehyde at a speed of 3ml/hr for 30 mins. After fixation in formaldehyde, brains were kept in 4% paraformaldehyde for overnight and dorsomedial telencephalon was dissected out, washed with PBS and then homogenized in Tris-EDTA buffer (pH 7.6). The homogenates were kept in 500 μL lysis buffer at 52°C for overnight until tissue fragments were dissolved completely. After that, the genomic DNA was isolated (Singh et al., 1998) and sonicated followed by immunoprecipitation with antibodies against different PPAR isoforms (α, β and γ) and control IgG according to standard protocol as described by us (Jana et al., 2007). Immunoprecipitated DNA was analyzed by PCR to quantify the PPREs in Creb using following primers:
Creb promoter primers
Sense 5’...TGG AAC TCG GAA AGG AGC TTA AGG A...3’; Anti-sense 5’...CGA GAA CCC ACG TGG TAG AAA GGA GAG AA...3’.
Lenti-PPARα Plasmid Construction and Generation of Lentivirus
PPARα cDNA was sub-cloned into a pLenti-III expression plasmid under the control of a CMV promoter. High-titer lentivirus generation and purification were performed in Applied Biological Materials, Inc.
Lentiviral Administration of PPARα Over-expressing Construct and PPARα shRNA in the Adult Mouse Hippocampus
For stereotaxic injection of lentivirus overexpressing GFP, PPARα, and shPPARα, experimental procedures were followed according to the guidelines of Laboratory Animal Manual of the National Institute of Health Guide to the Care and Use of Animals, which were approved by the Rush University Animal Care Committee. 6 to 8-week-old male C57BL6/J mice (Jackson Laboratories, Inc) or Ppara-null mice were anesthetized with pentobarbital (60 mg/kg). An injection cannula (26.5 gauges) was applied stereotaxically into the hippocampus (Anterioposterior, −2.54 mm from bregma; Mediolateral, 2.4 mm; Dorsoventral, 1.30 mm and 1.80 mm). The infusion was performed bilaterally in two different places of hippocampus at a rate of 0.2 ml/min and wound healing and recovery were monitored after the injection was done. Three months after injection, animals were monitored for the hippocampus dependent memory tasks for another week. In another case, animals were anesthetized and perfused with PBS followed by 4% paraformaldehyde. Brains were post-fixed with 4% paraformaldehyde, cryoprotected in 30% sucrose, and processed for immunohistochemistry. Forty-mm coronal sections of hippocampus were immunostained with a 1:1000 dilution of rabbit polyclonal anti-PPARα, anti-NR2A, anti-CREB, and anti-Arc (Abcam) antibodies followed by visualization under Olympus BX41 fluorescence microscope.
Bone Marrow Transplantation and Establishment of Chimera
Both wild-type (C57/BL6J) and Ppara-null recipient animals (4 wks old) were given γ-ray irradiation in a security-restricted γ cell chamber. The whole body of mouse was irradiated at 9 GY units for 13 min with a rate of 0.70 GY/minute. In order to minimize the stress, animals were irradiated twice for 6.5 minutes at an interval of 4-5 hours. Immediate after irradiation, animals were sent to a sterile room in an autoclaved cage with antibiotics-mixed food chow and water. The next day, bone marrow was isolated from femur and tibia bones of donor animals and was suspended in RPMI complete media. After RBC lysis and centrifugation, cells were resuspended in HBSS, counted, and then injected (106 cells/mouse) into recipient mice via tail vein. In order to establish chimeric animals, bone marrow cells from wild-type mice were transferred to wild-type and Ppara-null recipient mice and cells from Ppara-null mice were transferred to Ppara-null and wild-type recipient mice. Animals were maintained in sterile conditions until the chimera were fully established. After 10 weeks, establishment of the chimera was examined by immunofluorescence analysis of PPARα in liver and hippocampus.
Statistical Analysis
All values are expressed as the mean ± SD. Differences among means were analyzed using one- or two-way ANOVA with time or genotype as the independent factors. Differences in behavioral measures were examined by independent one-way or repeated-measures ANOVAs using SPSS. Homogeneity of variance between test groups was examined using Levene's test. Post-hoc analyses of between-subjects effects were conducted using Scheffe's, Tukey's or Games-Howell tests, where appropriate. p < 0.05 was considered statistically significant.
Supplementary Material
HIGHLIGHTS.
Transcriptional regulation of CREB by hippocampal PPARα
Involvement of PPARα in synaptic function
Regulation of long-term, but not short-term, memory by PPARα
Independence of hippocampal memory apparatus from peripheral PPARα
Acknowledgements
This study was supported by grants from Alzheimer's Association (IIRG-12-241179) and NIH (AT6681).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdominal fat boosts later dementia risk. The Harvard mental health letter / from Harvard Medical School. 2008;25:7. [PubMed] [Google Scholar]
- Abel T, Kandel E. Positive and negative regulatory mechanisms that mediate long-term memory storage. Brain Res Brain Res Rev. 1998;26:360–378. doi: 10.1016/s0165-0173(97)00050-7. [DOI] [PubMed] [Google Scholar]
- Braissant O, Foufelle F, Scotto C, Dauca M, Wahli W. Differential expression of peroxisome proliferator-activated receptors (PPARs): tissue distribution of PPAR-alpha, -beta, and -gamma in the adult rat. Endocrinology. 1996;137:354–366. doi: 10.1210/endo.137.1.8536636. [DOI] [PubMed] [Google Scholar]
- Campolongo P, Roozendaal B, Trezza V, Cuomo V, Astarita G, Fu J, McGaugh JL, Piomelli D. Fat-induced satiety factor oleoylethanolamide enhances memory consolidation. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:8027–8031. doi: 10.1073/pnas.0903038106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavallaro S, D'Agata V, Manickam P, Dufour F, Alkon DL. Memory-specific temporal profiles of gene expression in the hippocampus. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:16279–16284. doi: 10.1073/pnas.242597199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarthy MV, Lodhi IJ, Yin L, Malapaka RR, Xu HE, Turk J, Semenkovich CF. Identification of a physiologically relevant endogenous ligand for PPARalpha in liver. Cell. 2009;138:476–488. doi: 10.1016/j.cell.2009.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deacon RM. Appetitive position discrimination in the T-maze. Nature protocols. 2006;1:13–15. doi: 10.1038/nprot.2006.3. [DOI] [PubMed] [Google Scholar]
- Du J, Zhang L, Liu S, Zhang C, Huang X, Li J, Zhao N, Wang Z. PPARgamma transcriptionally regulates the expression of insulin-degrading enzyme in primary neurons. Biochem Biophys Res Commun. 2009;383:485–490. doi: 10.1016/j.bbrc.2009.04.047. [DOI] [PubMed] [Google Scholar]
- Guzowski JF, McGaugh JL. Antisense oligodeoxynucleotide-mediated disruption of hippocampal cAMP response element binding protein levels impairs consolidation of memory for water maze training. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:2693–2698. doi: 10.1073/pnas.94.6.2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Impey S, Smith DM, Obrietan K, Donahue R, Wade C, Storm DR. Stimulation of cAMP response element (CRE)-mediated transcription during contextual learning. Nature neuroscience. 1998;1:595–601. doi: 10.1038/2830. [DOI] [PubMed] [Google Scholar]
- Jana A, Pahan K. Fibrillar amyloid-beta peptides kill human primary neurons via NADPH oxidase-mediated activation of neutral sphingomyelinase. Implications for Alzheimer's disease. The Journal of biological chemistry. 2004a;279:51451–51459. doi: 10.1074/jbc.M404635200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jana A, Pahan K. Human immunodeficiency virus type 1 gp120 induces apoptosis in human primary neurons through redox-regulated activation of neutral sphingomyelinase. J Neurosci. 2004b;24:9531–9540. doi: 10.1523/JNEUROSCI.3085-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jana M, Jana A, Pal U, Pahan K. A simplified method for isolating highly purified neurons, oligodendrocytes, astrocytes, and microglia from the same human fetal brain tissue. Neurochemical research. 2007;32:2015–2022. doi: 10.1007/s11064-007-9340-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jana M, Mondal S, Gonzalez FJ, Pahan K. Gemfibrozil, a lipid-lowering drug, increases myelin genes in human oligodendrocytes via peroxisome proliferator-activated receptor-beta. J Biol Chem. 2012;287:34134–34148. doi: 10.1074/jbc.M112.398552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller H, Dreyer C, Medin J, Mahfoudi A, Ozato K, Wahli W. Fatty acids and retinoids control lipid metabolism through activation of peroxisome proliferator-activated receptor-retinoid X receptor heterodimers. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:2160–2164. doi: 10.1073/pnas.90.6.2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersten S, Desvergne B, Wahli W. Roles of PPARs in health and disease. Nature. 2000;405:421–424. doi: 10.1038/35013000. [DOI] [PubMed] [Google Scholar]
- Kesner RP, Connor HS. Independence of short- and long-term memory: a neural system analysis. Science (New York. NY. 1972;176:432–434. doi: 10.1126/science.176.4033.432. [DOI] [PubMed] [Google Scholar]
- Lee HK, Takamiya K, Han JS, Man H, Kim CH, Rumbaugh G, Yu S, Ding L, He C, Petralia RS, et al. Phosphorylation of the AMPA receptor GluR1 subunit is required for synaptic plasticity and retention of spatial memory. Cell. 2003;112:631–643. doi: 10.1016/s0092-8674(03)00122-3. [DOI] [PubMed] [Google Scholar]
- Marcus SL, Miyata KS, Zhang B, Subramani S, Rachubinski RA, Capone JP. Diverse peroxisome proliferator-activated receptors bind to the peroxisome proliferator-responsive elements of the rat hydratase/dehydrogenase and fatty acyl-CoA oxidase genes but differentially induce expression. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:5723–5727. doi: 10.1073/pnas.90.12.5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson MP. Pathways towards and away from Alzheimer's disease. Nature. 2004;430:631–639. doi: 10.1038/nature02621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S, Farioli-Vecchioli S, Ceru MP. Immunolocalization of peroxisome proliferator-activated receptors and retinoid X receptors in the adult rat CNS. Neuroscience. 2004;123:131–145. doi: 10.1016/j.neuroscience.2003.08.064. [DOI] [PubMed] [Google Scholar]
- Morris RG, Anderson E, Lynch GS, Baudry M. Selective impairment of learning and blockade of long-term potentiation by an N-methyl-D-aspartate receptor antagonist, AP5. Nature. 1986;319:774–776. doi: 10.1038/319774a0. [DOI] [PubMed] [Google Scholar]
- Papassotiropoulos A, Henke K, Stefanova E, Aerni A, Muller A, Demougin P, Vogler C, Sigmund JC, Gschwind L, Huynh KD, et al. A genome-wide survey of human short-term memory. Mol Psychiatry. 2011;16:184–192. doi: 10.1038/mp.2009.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera TD, Lu D, Thirumangalakudi L, Smith EL, Yaretskiy A, Rosenblum LA, Kral JG, Coplan JD. Correlations between hippocampal neurogenesis and metabolic indices in adult nonhuman primates. Neural Plast. 2011:1–6. doi: 10.1155/2011/875307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Pahan K. Gemfibrozil, stretching arms beyond lipid lowering. Immunopharmacology and immunotoxicology. 2009;31:339–351. doi: 10.1080/08923970902785253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha RN, Ghosh A, Palencia CA, Fung YK, Dudek SM, Pahan K. TNF-alpha preconditioning protects neurons via neuron-specific up-regulation of CREB-binding protein. J Immunol. 2009;183:2068–2078. doi: 10.4049/jimmunol.0801892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakimura K, Kutsuwada T, Ito I, Manabe T, Takayama C, Kushiya E, Yagi T, Aizawa S, Inoue Y, Sugiyama H, et al. Reduced hippocampal LTP and spatial learning in mice lacking NMDA receptor epsilon 1 subunit. Nature. 1995;373:151–155. doi: 10.1038/373151a0. [DOI] [PubMed] [Google Scholar]
- Schoonjans K, Peinado-Onsurbe J, Lefebvre AM, Heyman RA, Briggs M, Deeb S, Staels B, Auwerx J. PPARalpha and PPARgamma activators direct a distinct tissue-specific transcriptional response via a PPRE in the lipoprotein lipase gene. The EMBO journal. 1996;15:5336–5348. [PMC free article] [PubMed] [Google Scholar]
- Seyfried TN, Mukherjee P. Targeting energy metabolism in brain cancer: review and hypothesis. Nutr Metab (Lond) 2005;2:30. doi: 10.1186/1743-7075-2-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh I, Pahan K, Khan M, Singh AK. Cytokine-mediated induction of ceramide production is redox-sensitive. Implications to proinflammatory cytokine-mediated apoptosis in demyelinating diseases. The Journal of biological chemistry. 1998;273:20354–20362. doi: 10.1074/jbc.273.32.20354. [DOI] [PubMed] [Google Scholar]
- Tugwood JD, Holden PR, James NH, Prince RA, Roberts RA. A peroxisome proliferator-activated receptor-alpha (PPARalpha) cDNA cloned from guinea-pig liver encodes a protein with similar properties to the mouse PPARalpha: implications for species differences in responses to peroxisome proliferators. Archives of toxicology. 1998;72:169–177. doi: 10.1007/s002040050483. [DOI] [PubMed] [Google Scholar]
- Tzingounis AV, Nicoll RA. Arc/Arg3.1: linking gene expression to synaptic plasticity and memory. Neuron. 2006;52:403–407. doi: 10.1016/j.neuron.2006.10.016. [DOI] [PubMed] [Google Scholar]
- Yin JC, Del Vecchio M, Zhou H, Tully T. CREB as a memory modulator: induced expression of a dCREB2 activator isoform enhances long-term memory in Drosophila. Cell. 1995;81:107–115. doi: 10.1016/0092-8674(95)90375-5. [DOI] [PubMed] [Google Scholar]
- Yue JT, Lam TK. Lipid sensing and insulin resistance in the brain. Cell Metab. 15:646–655. doi: 10.1016/j.cmet.2012.01.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.