Abstract
Sirtuins, which are class III NAD-dependent histone deacetylases (HDACs) that regulate a number of physiological processes, play important roles in the regulation of metabolism, aging, oncogenesis and cancer progression. More recently, a role for the sirtuins in the regulation of steroid hormone receptor signaling is emerging. In this mini-review, we will summarize current research into the regulation of estrogen, androgen, progesterone, mineralocorticoid and glucocorticoid signaling by sirtuins in cancer. Sirtuins can regulate steroid hormone signaling through a variety of molecular mechanisms, including acting as co-regulatory transcription factors, deacetylating histones in the promoters of genes with nuclear receptor binding sites, directly deacetylating steroid hormone nuclear receptors, and regulating pathways which modify steroid hormone receptors through phosphorylation. Furthermore, disruption of sirtuin activity may be an important step in the development of steroid hormone-refractory cancers.
Keywords: Sirtuin, steroid hormone receptor activity/regulation, SIRT1, endocrine cancer
Introduction
Steroid hormone receptors, such as the estrogen (ER) and androgen receptors (AR), as well as the progesterone (PR), glucocorticoid (GR), and mineralocorticoid receptors (MR) are part of a large nuclear receptor family of eukaryotic transcription factors (Aoyagi and Archer 2008; Mangelsdorf, et al. 1995; Tsai and Omalley 1994). Steroid hormone receptors play essential roles in numerous biological processes, such as homeostasis, metabolism, cell growth and development (Chawla, et al. 2001; Mangelsdorf et al. 1995; Tsai and Omalley 1994).
Initiation of steroid hormone receptor activity normally requires the binding of a specific ligand, such as estrogen in the case of the ER. The steroid hormone receptors then dimerize and in some cases undergo post-translational modifications, translocate to the nucleus, and bind to their cognate hormone-responsive elements in the promoter or other regulatory regions of their target genes. The promoter-bound steroid hormone receptor then recruits transcriptional co-regulatory proteins and ultimately RNA-polymerase II (RNA Pol II) and other components of the transcriptional machinery, eliciting a transcriptional response (Kinyamu and Archer 2004; Perissi and Rosenfeld 2005; Tsai and Omalley 1994).
Regulation of steroid hormone receptor signaling is complex. Disruption in the physiological functions of these receptors can lead to several types of malignancies such as breast cancer, leukemia and lymphoma, prostate cancer, ovarian cancer, and lung cancer among others (Gronemeyer, et al. 2004; Huang, et al. 2010; Miller and Langdon 1997).
Understanding of the steroid hormone-dependency of certain cancer cells and the mechanisms underlying their subsequent evolution to a hormone-refractory state is an important and challenging goal of molecular oncology. Abnormal function of the AR has been linked to both the pathogenesis and the progression of human prostate cancer (Edwards and Bartlett 2005; Fu, et al. 2006; Wang, et al. 2005), and the ER plays a similar role in pathogenesis and progression of breast cancer. In the healthy mammary glands, estrogen receptor-positive cells are largely quiescent. Estrogen stimulates these cells to produce growth factors that cause the growth of neighboring ER-negative cells through paracrine effects involving stromal-epithelial cell interaction. Estrogen receptor-positive breast cancer cells are stimulated to proliferate in response to estrogen. Estrogen not only stimulates growth in these cells, but also suppresses apoptosis. One early step in the neoplastic transformation of the breast epithelium is the autocrine mechanism becoming predominant (Aghmesheh, et al. 2005; Fuqua, et al. 2000; Song and Santen 2006; Streuli and Haslam 1998). Prostate cancer progression can evolve through a similar pattern, including autocrine production of androgens, and eventual activation of AR signaling in a ligand-independent fashion (Prins and Putz 2008).
Post-translational modification of the steroid hormone receptors can play a role in the evolution of steroid responsiveness and independence in these tumors, by regulating signaling in response to steroid hormones. These modifications include phosphorylation, ubiquitination, glycosylation and acetylation. For example, PRMT1-mediated acetylation of ERα is required for recruitment of the co-effector molecules SRC, PI3K and Fak (Le Romancer, et al. 2008). ERα can be acetylated by p300 on two lysine residues located in the hinge region of the molecule in vitro. Acetylation at these sites decreases the hormone sensitivity of the receptor (Margueron, et al. 2004; Wang, et al. 2001). A mutation at one of these acetylation sites, lysine 303, has been linked to premalignant lesions of the breast (Fuqua et al. 2000; Wang et al. 2001). It has not yet been determined whether acetylation affects ERα stability, intracellular localization, or its ability to interact with co-activators or co-repressors (Margueron et al. 2004).
Sirtuins
The sirtuins comprise a family of enzymes with increasing relevance to the regulation of steroid hormone receptor activity. The sirtuins are encoded by a family of genes that has remained highly conserved from archaebacteria to eukaryotes (Frye 1999, 2000). In yeast there are four other sirtuins in addition to Sir2p, whereas seven homologs have been identified in mammals, SIRTs 1–7 (Ford, et al. 2005; Frye 1999, 2000; Yamamoto, et al. 2007). Sirtuin activities are not limited to histone deacetylation; SIRT members can also deacetylate non-histone proteins and have diverse functions in multiple cellular compartments. Sirtuin (SIRT) 1, 6 and 7 localize to the nucleus, whereas SIRT3, 4 and 5 are mitochondrial, and SIRT2 is primarily cytoplasmic (Liu, et al. 2005; Michishita, et al. 2008; Michishita, et al. 2005). SIRT6 has been found to deacetylate histone H3, thereby regulating transcription and maintaining genomic stability (Kawahara, et al. 2009; Liu et al. 2005; Lombard, et al. 2008; Michishita et al. 2008; Michishita et al. 2005). SIRT7 regulates RNA polymerase I (RNA-Pol I)-mediated expression of ribosomal RNA genes. SIRT7 has also been linked to angiogenesis (Ford, et al. 2006; Potente, et al. 2007). SIRT2 is involved in cell cycle regulation, can deacetylate histone H4, and also acts as a tumor suppressor in certain gliomas (Dryden, et al. 2003; Hiratsuka, et al. 2003; Vaquero, et al. 2006). Mice carrying inactivating mutations of SIRT1, SIRT2 or SIRT3 develop cancers, predominantly in the mammary glands and the liver, accompanied by genomic instability and/or abnormal energy metabolism. SIRT2-deficient mice initially develop normally, but develop mammary tumors and hepatocellular carcinoma (HCC) as they age (Deng1, et al. 2011 Apr 2–6). The functions of SIRTs 3–5 are, as of yet, unknown. Substrates for these sirtuins include acetyl- coenzyme A synthetase 2, glutamate dehydrogenase and cytochrome C (Haigis and Guarente 2006; Haigis, et al. 2006; Hallows, et al. 2006; Hirschey, et al. 2010; Schlicker, et al. 2008; Schwer and Verdin 2008). Of all the sirtuins, SIRT1 remains the most intensively studied, and its actions on steroid hormone signaling will be the focus of this review (Table 1).
Table 1.
Summary of actions of SIRT1 on steroid hormone receptor activity.
| SIRT1 Regulation | Deacetylase Activity-dependent | Physiological Function of SIRT1 | |
|---|---|---|---|
| Estrogen Receptor | SIRT1 inhibits ligand-independent activation. | Yes | Represses transcription |
| Androgen Receptor | SIRT1 regulates AR ability to transform prostate cells. | Yes | Represses transcription |
| Progesterone Receptor | SIRT1 regulates nucleo-cytoplasmic shuttling of the receptor as well as the induction of slow versus rapid progesterone response genes. | Yes | Transcriptional regulator |
| Glucocorticoid Receptor | SIRT1 regulation protects cells from the stress response. | Yes | Cell metabolism and survival |
| Mineralocorticoid Receptor | SIRT1 regulates the tissue damage response. | No | Cell metabolism and survival |
SIRT1
SIRT1 is a nicotinamide adenosine dinucleotide (NADP+P)-dependent histone deacetylase (HDAC) that functions by deacetylating histone (H1, H3, and H4) and non-histone proteins. SIRT1 has been linked to control of longevity, gene silencing, cell cycle progression, apoptosis and energy homeostasis (Dali-Youcef, et al. 2007; Greiss and Gartner 2009; Haigis and Guarente 2006; Yamamoto et al. 2007; Yang, et al. 2006). SIRT1 also has functions relating to inflammation and neurodegeneration (Yamamoto et al. 2007). SIRT1 interacts with nuclear steroid hormone receptor co-activator proteins such as p300, PPARγ and PGC-α in the differentiation of muscle cells, adipogenesis, fat storage and metabolism in the liver (Fulco, et al. 2003; Picard, et al. 2004; Puigserver, et al. 2005; Rodgers, et al. 2005).
More recently, our understanding of the physiological control of SIRT1 activity in the cell and its consequences on steroid hormone receptor activity is enlarging. Endogenous SIRT1 activity in normal cells is regulated by the absolute levels of SIRT1 protein (which do not generally appear to change temporally among cells of the same type), and more importantly by the ability of SIRT1 to act as a cellular energy sensor controlled by the intracellular NAD/NADH ratio. Whether changes in the energy state of the cell thereby have an effect on the function of steroid hormone receptors, however, has not yet been explored. Furthermore, in tumor cells there exist a number of other possible levels of SIRT1 regulation, ranging from over-expression of the protein, which is common in tumor cells, to dramatic changes in sub-cellular localization and post-translational modifications of SIRT1 itself (Byles, et al. 2010), and aberrant control of SIRT1 gene transcription.
The transcription of SIRT1 is regulated at the SIRT1 promoter by FOXO3a and p53, among other transcription factors. Upon activation, cytoplasmic FOXO3a translocates to the nucleus and induces the removal of p53 from two binding sites present at the SIRT1 promoter (Nemoto, et al. 2004). E2F1 is a crucial positive regulator of SIRT1 transcription, with two E2F1 two binding sites within the SIRT1 promoter. In non-neoplastic cells, SIRT1 binds to and deacetylates E2F1, deactivating it and also decreasing transcription and expression of SIRT1, thus forming a negative-feedback-loop (Wang, et al. 2006).
SIRT1 also regulates its own expression through other mechanisms. SIRT1 can form a complex with, and deacetylate, the hypermethylated in cancer-1 (HIC-1) protein at residue L314, thereby activating the transcriptional repressor functions of HIC-1. HIC-1 then negatively regulates SIRT1 transcription (Stankovic-Valentin, et al. 2007). HIC-1 is of particular interest in the regulation of the estrogen-response because of its role in the response of breast cancer cells to estrogen-antagonists. In estrogen-antagonist sensitive cells, estrogen-antagonists induce HIC-1 expression via a c-Jun N-terminal kinase 1 (JNK1) -and prohibitin-mediated signaling pathways. It is therefore possible that when SIRT1 activity is inhibited, the repressive functions of HIC-1 are also impaired, but this has yet to be established. Investigation of the relationship between SIRT1 and HIC-1 in both estrogen-dependent and estrogen-independent breast cancer cells may elucidate important reciprocal roles for these molecules.
Additionally, SIRT1 is post-translationally modified by phosphorylation via CyclinB/Cdk1 (Sasaki, et al. 2008), and sumoylation (Kwon and Oft 2008). Phosphorylation of SIRT1 increases its deacetylase activity. Mutations at residues T530 and S540 which eliminate phosphorylation at those residues disturb normal cell cycle progression. Sumoylation also increases SIRT1 activity (Yang 2007). Whether there are post-translational modifications to SIRT1 in response to steroid hormone receptor activity, which in some cells promotes cell cycle progression and ultimately cyclin B activation, has not yet been studied.
SIRT1 and Cancer
SIRT1 has been linked to cancer in several ways, through its regulation of tumor cell apoptosis, senescence and the DNA damage response, all promoting tumor cell survival. SIRT1 associates with the tumor suppressor protein p53 (Bouras, et al. 2005; Haigis and Guarente 2006; Nemoto, et al. 2005; Rodgers et al. 2005; Yamamoto et al. 2007), and regulates protein levels of p53 through deacetylation of residue L382, which destabilizes p53, thereby promoting cell survival (Zhao, et al. 2008). Accordingly, over-expression of SIRT1 has been found in cancer cells to promote tumor cell survival (Ford et al. 2005; Ota, et al. 2006a; Ota, et al. 2006b). SIRT1 can deacetylate the DNA repair factor Ku70, which sequesters the pro-apoptotic protein BAX in the mitochondria, thus preventing apoptosis (Cohen, et al. 2004a). In addition, SIRT1 can deacetylate FOXO family proteins, which repress the transcription of Bim, a pro-apoptotic protein (Brunet, et al. 2004; Cohen et al. 2004a; Cohen, et al. 2004b; Luo, et al. 2001; Motta, et al. 2004; Yi and Luo 2010).
SIRT1 also regulates epigenetic mechanisms in the transformation and progress of cancer. SIRT1 localizes to the promoters of several aberrantly-silenced, densely-hypermethylated tumor suppressor genes (TSGs SFRP1 and SFRP2) in MCF-7 and MDA-231 breast cancer cell lines. Interestingly, SIRT1 does not localize to these same promoters in normal or tumor cell lines in which these genes are not hypermethylated. Inhibition of SIRT1 in breast and colon cancer cells causes increases in H4-K16 and H3-K9 acetylation at endogenous promoters and subsequent re-expression of silenced TSGs. Surprisingly, this reactivation occurs despite the fact the hypermethylated state of the promoter remains unchanged. These results indicate that SIRT1 may comprise a new form of epigenetic silencing that may link some of the epigenetic changes found in aging with those found in cancers. In the future, this action of SIRT1 may prove to be important for the therapeutic targeting and reactivation of these TSGs (Pruitt, et al. 2006).
SIRT1 activity increases the expression of drug resistance-promoting genes in drug-resistant cancer cell lines. Conversely, suppression of SIRT1 by siRNA decreases the drug-resistant phenotype (Chu, et al. 2005). These results indicate that SIRT1 may be involved in the transformation of cancer cells from drug-responsive to drug-refractory phenotypes, an ongoing clinical challenge in the treatment of many types of cancer. SIRT1 may play a role in determining drug-resistance or sensitivity, and thus, SIRT1 activity may prove to be important as a prognostic indicator of responses to chemotherapeutics.
In Sirt1-transgenic mice, Sirt1 is able to protect from aging-associated spontaneous cancer development, as well as from metabolic syndrome-associated liver cancer (Pfluger, et al. 2008; Serrano, 2011). This beneficial effect of Sirt1 appears due both to its impact on metabolism (Herranz, et al. 2010; Pfluger et al. 2008) and on genomic integrity (Pfluger et al. 2008). However, there are other experimental scenarios in which Sirt1 over-expression is able to accelerate tumorigenesis in certain tissues (tissues which also show higher Sirt1 expression when malignant), and these observations constitute the first in vivo demonstrations of a potentially oncogenic role for Sirt1 (Serrano, 2011).
The transformation of steroid hormone-responsive cancer cells from a responsive to a refractory state is an important step in the progression of breast and prostate cancers. For example, estrogen-dependent cancers are defined by the presence of the ER and the requirement of estrogen for cell survival and growth (Davidson 1992; Osborne 1998; Osborne and Schiff 2005; Osborne, et al. 2005; Skliris, et al. 2008). Estrogen-dependent cancers are considered treatable and are associated with a more favorable prognosis, although these initially estrogen-dependent tumors generally transform over time to estrogen-independence. Breast cancers which are ER-negative at diagnosis have lost ERα expression, most often due to gene silencing, and are capable of proliferating in the absence of estrogen. Estrogen-independent cancers are generally more aggressive and associated with a less favorable prognosis. These cancer phenotypes are the least treatable (Cleator, et al. 2007). Prostate cancers also inevitably undergo an evolution from hormone-responsive to hormone-independent (castration-resistant cancer, CRC), although loss of AR expression is rare (Bennett, et al. 2010; Risbridger, et al. 2010; Scher, et al. 2004).
Several lines of evidence suggest that SIRT1 activity may be required to prevent the transition of cancer cells from a steroid hormone-dependent to and independent state (Byles et al. 2010; Dai, et al. 2007; Dai, et al. 2008; Wang, et al. 2008). An understanding of SIRT1/steroid hormone receptor interactions may thus provide new approaches to the clinical problem of hormone-independence in breast and prostate tumors.
Regulation of Androgen Receptor Activity by SIRT1
Prostate cancer, like breast cancer, requires gonadal steroids for initiation and early development. Tumors that arise from both breast and prostate tissue are typically hormone-dependent and the underlying cellular biology of these two tumors is often strikingly similar. Prostate cancer cells are initially dependent upon androgen, and the tumor cells predictably progress to a hormone-refractory state within 18 months of the institution of endocrine-based therapies. The primary treatment modality for prostate cancer is androgen-ablation therapy, which utilizes anti-androgenic approaches such as the androgen-antagonists flutamide and casodex, suppression of the hypothalamic/gonadal axis, or castration (Risbridger et al. 2010).
The AR is a DNA-binding transcription factor that governs male sexual development and differentiation. The induction of AR activity is regulated by androgenic hormones, including dihydrotestosterone (DHT), which enhances co-activator (p300 and SRC, among others) association and reduces co-repressor protein (NCoR, HDAC, and Smad) association with the AR (Dai et al. 2008).
Deacetylation of the AR by SIRT1 inactivates its ability to transform prostate cells. SIRT1 binds to and deacetylates the AR at a conserved lysine motif, down-regulating its levels in the cell and repressing androgen-induced AR transcription (Popov, et al. 2007). Furthermore, a recent report found that SIRT1 regulatesDHT-responsive miRNAs, with putative binding sites withincrucial developmental genes, such as the homeobox gene Nkx3.1. Interestingly, this regulation appears to be AR-independent. This suggests that SIRT1 not only can regulate androgen signaling, which in turn effects DHT-dependent cellular proliferation and growth, but also theexpression of specific homeobox genes essential for normal prostate development (Powell, et al. 2009).
SIRT1 is required for androgen antagonist-mediated transcriptional repression and growth suppression of prostate cancer cells (Dai et al. 2007). Androgen-antagonists promote the physical association of SIRT1 with the AR and NCoR, and androgen antagonist-bound AR then recruits SIRT1 and NCoR to AR-responsive promoters, and deacetylates histone H3-K9 locally in the AR-responsive promoters. SIRT1 recruitment to these promoters is required for androgen antagonist-mediated transcriptional repression. SIRT1 down-regulation by pharmacological means or by siRNA increased the sensitivity of androgen-responsive genes to androgen stimulation, and enhanced the sensitivity of prostate cancer cell proliferation in response to androgens. This finding was the first demonstration of ligand-dependent recruitment of a class III HDAC into a co-repressor transcriptional complex, and of a necessary functional role for a class III HDAC as a transcriptional co-repressor in AR antagonist-induced transcriptional repression. While deacetylation at K632/K633 reduces the affinity of AR to androgen in vitro (Fu, et al. 2000), it has not yet been determined whether the proliferative effects observed following inhibition of SIRT1 activity are due in part to direct deacetylation of AR by SIRT1 in vivo.
This activity of SIRT1 on AR signaling suggests that SIRT1 itself may serve as a tumor suppressor gene product in prostate cancer cells -- loss of SIRT1 eliminates repression of AR-regulated gene activity and renders hormone-antagonist therapy ineffective. The role(s) of SIRT1 in prostate cancer may, therefore, be pleiotropic and may depend in part on its aberrant sub-cellular localization in cancer cells. SIRT1 is over-expressed in many prostate cancer cells, as well as many other steroid hormone-dependent or-independent cancer cell phenotypes. This over-expression is the result of SIRT1 expression in the cytoplasmic compartment, rather than being restricted to the nucleus. This predominant cytoplasmic localization of SIRT1 is regulated by elevated mitotic activity and PI3K/IGF-1R signaling in cancer cells, producing increased SIRT1 protein stability. SIRT1 is required for PI3K-mediated prostate cancer cell growth, and promotes prostate cancer cell survival (Byles et al. 2010). Conversely, a separate study employing HeLa cells suggests that enforced cytoplasmic expression of SIRT1, achieved through truncation of nuclear localization motifs, enhances sensitivity of the cells to apoptosis (Jin, et al. 2007). Interestingly, this pro-apoptotic activity of the truncated SIRT1 is not dependent upon its deacetylase activity.
Regulation of Estrogen Receptor Activity by SIRT1
A significant and increasingly important role for SIRT1 in the development and progression of breast cancer is emerging. SIRT1 transcription and activity is repressed by the deleted in breast cancer-1 (DBC-1) protein. In non-malignant cells, SIRT1 and DBC-1 expression are balanced. However, in many cancers, and particularly breast cancer, DBC-1 is dysregulated, while SIRT1 is highly up-regulated (Sung, et al. 2010; Zhao et al. 2008). A role for SIRT1 as a potential tumor promoter in breast cancer has therefore been proposed. Conversely, activation of SIRT1 elicits a more profound inhibitory effect on BRCA1 mutant cancer cells than on BRCA1-wild-type cancer cells both in vitro and in vivo, suggesting a tumor suppressor role for SIRT1 in BRCA1-mutant breast cancer development (Wang et al. 2008).
ERα and estrogen receptor-beta (ERβ) both belong to the steroid/nuclear superfamily of ligand-regulated transcription factors. ERα contains six domains, termed A–F, which can be divided into a hormone-independent activation function (AF-1) region, a DNA-binding domain (DBD), a hinge region which contains the three nuclear localization sequences (NLS) that mediate the translocation of the receptor from the cytoplasm to the nucleus, and a hormone-binding domain (HBD) on the C-terminus of the receptor. This latter region is responsible for the dimerization of ERα and hormone-dependent activation function. Binding of estrogen to the HBD leads to a series of conformational changes in the protein structure. The changes uncover areas on the external surface of ERα that are responsible for the binding of co-activator molecules (Carroll, et al. 2006; Song and Santen 2006).
One report has suggested that SIRT1 inhibition leads to inhibition of ER signaling (Yao, et al. 2010), implying that SIRT1 serves as an ERα co-activator. Repression of SIRT1 activity in that study led to lower estrogen-responsive gene activity and thus slower breast cancer cell growth. Those findings are inconsistent, however, with other reports in the literature. In a different experimental system, for example, exposure of breast cancer cells to resveratrol, a phytoestrogen and an activator of SIRT1 (albeit not a specific one) led to an inhibition of breast cancer cell growth and an up-regulation of SIRT1 mRNA and protein expression, suggesting that instead of being an ERα co-activator, SIRT1 actually serves as an ERα-repressor (Lin, et al. 2010). Our own work, using much more specific SIRT1 inhibitors such as sirtinol and splitomycin as well as anti-SIRT1 siRNA, has demonstrated that SIRT1 activity represses estrogen-independent ERα-regulated gene activity in breast cancer cells, represses PI3K/AKT-dependent ERα activation (as assayed by ERα phosphorylation at S118 and ERα nuclear translocation), and represses cell growth in the absence of estrogen, via an ERα-dependent mechanism.
One study found that human breast cancer tissues exhibited reduced levels of SIRT1, 2 and 3, suggesting a potential tumor suppressor role for these proteins in breast tissue (Deng, et al. 2011 Apr 2–6). Another recent report found that DBC-1 and SIRT1 were expressed in 71% (87/122) and 67% (82/122) of human breast carcinomas, respectively, and DBC-1 and SIRT1 expression was significantly associated with chemotherapeutic resistance, distant metastatic relapse in ER-positive tumors, and a shorter relapse-free survival rate in ER-negative tumors (Lee, et al. 2011). Collectively, these findings support the theory that SIRT1 generally serves a tumor suppressor function in certain types of ageing-associated cancers (including breast and prostate cancer) and in metabolic syndrome-associated cancers, through, in part, its effects on steroid hormone receptor signaling, and consequently that disruption of SIRT1 expression and activity has oncogenic potential (Herranz and Serrano 2010; Yi and Luo 2010).
These findings also highlight a potential role for SIRT1 in decreasing breast cancer cell dependency on estrogen. When considered together with the reports that SIRT1 activity increases the expression of drug-resistance genes, SIRT1 levels in tumors may represent a useful biomarker with regard to prognosis, or for tailoring patient-specific treatment regimens (Chu et al. 2005). Furthermore, pharmacological modulation of SIRT1 activity in breast tumors may prove useful in future breast cancer therapeutic strategies.
It is therefore important to note the gaps in our understanding of the role of SIRT1 in ERα signaling. The molecular mechanism directly linking SIRT1 activity and ER-dependent signaling has not yet been established. A direct enzymatic action of SIRT1 on ERα is not likely to be the mechanism, as acetylation or deacetylation of the estrogen receptor itself produces only modest effects, at best, on ERα activity (Cui, et al. 2004; Ma, et al. 2010; Popov et al. 2007). Furthermore, although there is clearly an effect of SIRT1 activation on PI3K regulation and subsequent downstream AKT-mediated activation/phosphorylation of ERα, the pathway linking SIRT1 and PI3K is not yet rigorously established. For example, any functional effects of acetylation on the PI3K pathway components p85, p110 or AKT have not been established.
SIRT1 has, however, been directly linked to one PI3K co-regulatory protein, phosphatase and tensin homolog (PTEN). PTEN is a lipid phosphatase that inhibits PI3K activation by specifically dephosphorylating the three position of phosphatidylinositols (PI3P, PI3,4P2, and PI3,4,5P3), which are the product of PI3K, thereby inhibiting PI3K signaling (Ikenoue, et al. 2008; Maehama and Dixon 1998; Medema, et al. 2000; Myers, et al. 1998; Stambolic, et al. 1998). SIRT1 deacetylates PTEN at residue L402, thereby inactivating PTEN and relieving its repression of PI3K signaling (Ikenoue et al. 2008). One would expect PTEN activity to increase in response to SIRT1 inhibition, thus decreasing PI3K signaling. However, our unpublished findings demonstrate an increase in PI3K activity in response to SIRT1 inhibition, making it unlikely that PTEN plays a role in SIRT1-mediated repression of estrogen-independent PI3K activity.
Regulation of Progesterone Receptor Activity by SIRT1
The Progesterone Receptor (PR) is an intracellular steroid hormone receptor that specifically binds progesterone (Misrahi, et al. 1987). The PR has two main isoforms, A and B (Gadkar-Sable, et al. 2005; Sachdeva, et al. 2005). The acetylation state of the PR has been linked to modest degrees of functional modulation. Acetylation of the PR regulates the nuclear-cytoplasmic shuttling of the receptor as well as the transcriptional activation of “slow” versus “rapid” progesterone-responsive genes. For instance, constitutive-acetylation-mimic mutations at the PR hinge region display delayed nuclear entry upon progesterone binding compared to wild-type, whereas acetylation-defective mutants induce more rapid induction of c-MYC gene expression (c-MYC is a proto-typical “rapid” progesterone-responsive gene), when compared to wild type or constitutive-acetylation-mimic mutants (Daniel, et al. 2010). These results indicate that acetylation of the hinge region of the progesterone receptor regulates the kinetics of PR nucleo-cytoplasmic transport, and thereby subsequent transcriptional activity.
Additionally, c-MYC binds to the SIRT1 promoter, increasing SIRT1 expression, while at the same time, SIRT1 deacetylates c-MYC, decreasing its stability and compromising the transformative ability of c-MYC. Thus, the c-MYC/SIRT interaction comprises a negative-feedback loop. This not only suggests that SIRT1 has a role(s) in PR “rapid-response” regulation, but also solidifies the notion that SIRT1 acts as a tumor suppressor (Yuan, et al. 2009). It will be therefore of great interest to further investigate the role(s) SIRT1 expression and activity has in the ability c-MYC to transform PR-expressing cells.
It has yet to be determined whether SIRT1 plays a major role in the de-acetylation of the progesterone receptor itself. When breast cancer cells were treated with nicotinamide (NAM), a SIRT1 inhibitor, progesterone-dependent transcription decreases, suggesting that SIRT1 activity is necessary for PR-mediated transcription. However, when more specific suppression of SIRT1 activity by knockdown with siRNA was employed, the reduction of PR-dependent signaling in response to NAM treatment was shown to be independent of SIRT1. The authors suggested that NAM inhibits the coordination of basal transcription machinery assembly after chromatin remodeling of the progesterone-responsive promoter by a SIRT1-independent mechanism (Aoyagi and Archer 2008). As NAM is a very non-specific SIRT1 inhibitor, it is likely that any actions on PR-mediated transcription as a result of SIRT1 inhibition by NAM are masked by the off-target effects of NAM. In order to conclusively determine whether SIRT1 has a role in regulating basal PR-dependent gene activity, more selective inhibitors of SIRT1 should be employed.
SIRT1 Regulation of the Glucocorticoid and Mineralocorticoid Receptors
The glucocorticoid receptor (GR) is widely expressed and is pleiotropic in its functions, depending in part on when in development, and in what tissue, it is expressed (Tsai and Omalley 1994). The interaction between SIRT1 activity and GR function is best characterized in myocyte metabolism.
In skeletal muscle cells, glucocorticoids play a major role in the response of skeletal muscle to catabolic conditions such as sepsis, severe injury, or burn. Glucocorticoids are important mediators of muscle-wasting and mitochondrial dysfunction in such conditions (Hasselgren 1999). Glucocorticoids activate transcription of uncoupling protein-3 (UCP3), a mitochondrial membrane transporter that protects muscle cells from an overload of fatty acids and protects against excessive production of reactive oxygen species (Amat, et al. 2007; Brand and Esteves 2005).
Glucocorticoid receptor activity, induced by ligand binding, activates p300, a histone acetylase (HAT) and SIRT1 co-regulatory protein, as well as releasing class I and II HDACs, resulting in increased UPC3 gene transcription. Interestingly, SIRT1 deacetylase activity is required for the repression of UPC3 gene transcription. SIRT1 represses UPC3 gene transcription by inhibiting the interaction between GR and the co-activator, p300, at the promoter. Furthermore, the SIRT1 activator resveratrol is able to completely inhibit the induction of the UP3C gene upon treatment with glucocorticoids (Amat et al. 2007). (Due to the inherent lack of specificity of resveratrol, however, this result should be verified with other SIRT1 activators.) Collectively, it therefore appears that, in contrast to the roles of SIRT1 in ERα and AR signaling which in turn regulate proliferation and differentiation, the functional endpoint of SIRT1 and GR interactions is a metabolic one. SIRT1 activity in turn is controlled by the metabolic state of the cell, acting as an energy sensor controlled by the NAD/NADH ratio, in this case linking transcriptional regulation of metabolic genes to the stress response.
The Mineralocorticoid Receptor (MR) and its steroid hormone ligand, aldosterone, regulates electrolyte balance in the kidney (Nakano, et al. 2010). Aldosterone increases renal tubular Na+ absorption in large part by increasing transcription of the epithelial Na+ channel α-subunit (α-ENaC) expressed in the apical membrane of collecting duct principal cells in the kidney (Zhang, et al. 2009b).
SIRT1 has recently been identified as a modulator of the aldosterone signaling pathway (Zhang et al. 2009b). SIRT1 plays an essential role in the genesis of aldosterone-induced renal injury. This renal injury is the result of cellular senescence in renal tissue secondary to a MR/SIRT1-dependent signaling pathway involving the cyclin-dependent kinase inhibitor p21 and p53. The resulting renal damage is due to delayed repair of tubular cells (Nakano et al. 2010).
The physical interaction between SIRT1 and Disruptor of telomeric silencing-1 (Dot1), a histone-methyltransferase, results in global H3K79 hypermethylation in chromatin along the α-ENaC 5′-flanking region and inhibition of α-ENaC gene transcription. Interestingly, this effect appears to be mediated by an action of SIRT1 to support the distributive methyltransferase activity of Dot1 on H3K79 methylation. Surprisingly, however, the deacetylase activity of SIRT1 is not required for this action. SIRT1 also inhibits aldosterone-induced α-ENaC gene transcription via a pathway that is largely independent of MR (Zhang et al. 2009b). The authors assert that SIRT1 is therefore able to regulate proteins that do not serve as substrates for its deacetylase or ADP-ribosylase activities (Zhang et al. 2009b). Thus, in some cases, the actual physical interaction of SIRT1 with other proteins confers actions beyond its catalytic activity.
SIRT1 Regulation of Steroid Hormone Receptor FOXO Co-Regulatory Proteins, and the PI3K Activation Pathway
Many avenues of research have demonstrated that FOXO proteins are involved in the regulation of steroid hormone receptor signaling, and that many FOXO proteins are in turn regulated by SIRT1. FOXO proteins are members of the forkhead superfamily of proteins. Since the first member of this family of genes was discovered in Drosophila melanogaster, more than 100 structurally-related forkhead transcription factors have been identified. Forkhead proteins share a conserved 100-residue DNA binding domain called the forkhead (FKH) domain. Crystal structure analysis indicates that this domain contains three major α-helices and two large wing-like loops, leading to these genes also being called winged-helix transcription factors (Clark, et al. 1993; Huang and Tindall 2007; Lai, et al. 1993; Weigel, et al. 1989).
FOXO proteins are tightly-regulated transcription factors that are able to stimulate expression of proteins involved in cell cycle arrest or initiation of apoptosis (Lengyel, et al. 2007). FOXO proteins in turn are regulated by growth factors and cellular stress. Growth factors regulate FOXO proteins by threonine/serine phosphorylation and nuclear exclusion (Brownawell, et al. 1999; Kops, et al. 1999). Cellular stresses result in FOXO acetylation, deactivating FOXO activity (Brunet 2004; Brunet et al. 2004; Daitoku, et al. 2004; Huang and Tindall 2007; Motta et al. 2004). Whereas deacetylation of transcription factors is normally associated with a decrease in their activity, deacetylation of the FOXO proteins results in an increase in their activity. SIRT1 targets many FOXO proteins, including FOXO1 and FOXO3a (Brunet et al. 2004; Daitoku et al. 2004; Giannakou and Partridge 2004; Huang and Tindall 2007).
SIRT1, which is a sensor of cellular energy/stress levels, is a selective activator of FOXO signaling (Berdichevsky and Guarente 2006; Giannakou and Partridge 2004; Yeung, et al. 2004). Interplay between FOXOs and SIRT1 potentiates cellular resistance to oxidative stress and enhances cell-cycle arrest in response to stress conditions, which promotes cellular survival and longevity (Frescas, et al. 2005).
FOXO proteins interact with many of the steroid hormone receptors. For example, the amino terminal domain of FOXO3a (amino acids 1–300) binds to ERα, suggesting that FOXO3a could possibly repress ERα activity directly. When FOXO3a is over-expressed in estrogen-treated MCF-7 cells, expression of ER-regulated genes is decreased. FOXO3a interacts with, and regulates, FOXOM1, which has a binding site on the ERα promoter and can regulate ERα expression (Delpuech, et al. 2007; Madureira, et al. 2006). Furthermore, silencing of endogenous FOXO3a can convert non-tumorigenic, estrogen-dependent breast cancer cells into tumorigenic, estrogen-independent cells. Wild-type MCF-7 cells injected into the footpads of female athymic mice are not able to form tumors without being given supplemental estradiol. MCF-7 cells transduced by retroviruses expressing siRNA against human FOXO3a, however, were able to form and promote tumor growth without the addition of supplemental estradiol. This observation has been taken as evidence that FOXO3a serves as a tumor suppressor and that endogenous FOXO3a may normally prevent hormone-independent cell growth of breast cancer cells in vivo (Zou, et al. 2008). FOXO3a repression of estrogen-independent breast cancer cell proliferation parallels the effects of SIRT1 on repression of estrogen-independent breast cancer cell proliferation, engendering the hypothesis that SIRT1 and FOXO3a are both components of the same regulatory pathway governing ER-mediated signaling; both SIRT1 and FOXO3a together would be necessary to repress ligand-independent ERα activation, through the PI3K pathway (Table 2).
Table 2.
Similarities between SIRT1- and FOXO3a-mediated regulation of breast cancer cell proliferation suggests the potential interaction of the two pathways.
Similarities Between SIRT1- and FOXO3a-mediated Regulation of Breast Cancer Cell Proliferation
| SIRT1-Mediated Repression of Breast Cancer Cell Proliferation | FOXO3a-Mediated Repression of Breast Cancer Cell Proliferation |
|---|---|
| SIRT1 represses estrogen-regulated gene expression. | FOXO3a represses estrogen-regulated gene expression. |
| Repression of proliferation by SIRT1 is ERα-dependent. | FOXO3a binds to and represses ERα activity. |
| SIRT1 represses estrogen-independent PI3K/AKT activation. | FOXO3a represses PI3K/AKT activity. |
| SIRT1 represses estrogen-independent cell growth. | FOXO3a represses estrogen-independent cell growth. |
| SIRT1 expression is regulated by FOXO3a. | FOXO3a regulates SIRT1 expression. |
| SIRT1 regulates FOXO3a activity. | FOXO3a activity is regulated by SIRT1. |
The PI3K/AKT pathway is an important component of many of the steroid hormone signaling pathways, both upstream and downstream of the steroid receptor. PI3K/AKT activity is required for phosphorylation and activation of certain steroid hormone receptors (Ward and Weigel 2009). Subsequently, PI3K/AKT is activated as a consequence of steroid hormone receptor activation (Dillon, et al. 2007; Wang, et al. 2007), generating a positive-feedback loop. In the case of prostate cancer cells, the AR-signaling pathway and the PI3K/AKT pathway cross-talk through FOXO proteins, among other routes (Wang et al. 2007).
FOXO proteins and the PI3K/AKT signaling pathway play a similar role in the catabolic actions of the GR in the regulation of genes involved in myopathy (Schakman, et al. 2008). Muscle atrophy is an important consequence of many diseases. Glucocorticoids induce insulin resistance and subsequent activation of the ubiquitin-proteosome pathway. Elevated glucocorticoid production results in increased expression of atrogenes such as IRS1 and 2, AT-1 and UbC. FOXO3a mediates cross-talk between the PI3K/AKT, MEK/ERK, and GR signaling pathway (Zheng, et al. 2010).
Discussion
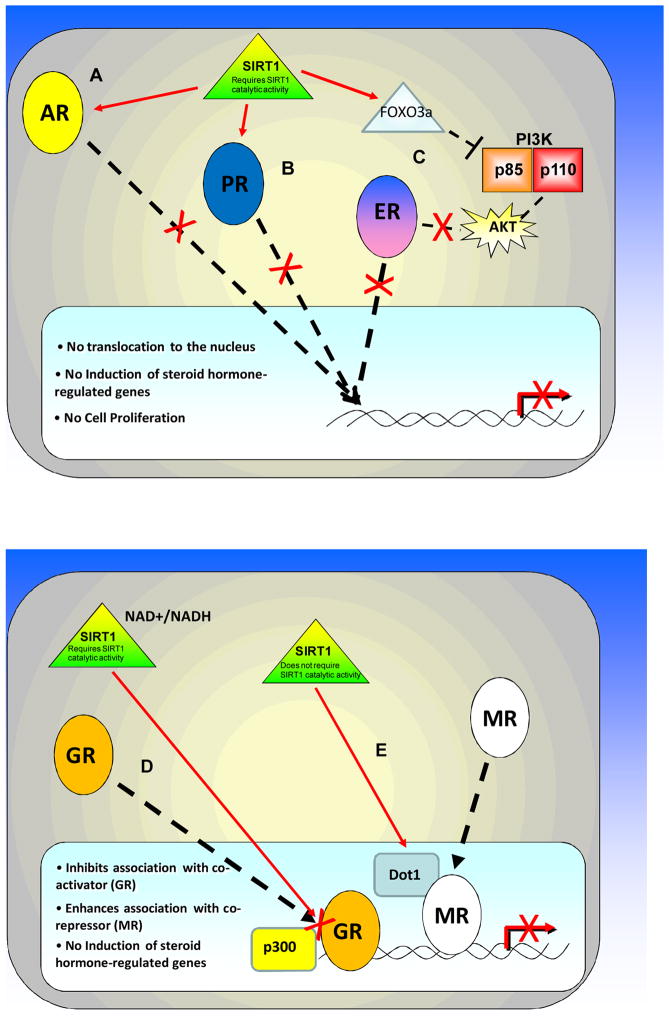
SIRT1 activity has been linked to steroid hormone receptor sensitivity to ligands, activation, and function. The literature to date suggests that SIRT1 in general functions to repress steroid hormone receptor activity. In the case of the ER, SIRT1 represses ligand-independent activation. In the case of AR and GR, SIRT1 regulates ligand-sensitivity and the subsequent transcriptional response to androgen and glucocorticoids respectively. In the case of the PR, it is likely that SIRT1 regulates the kinetics of nucleo-cytoplasmic shuttling and, consequently, “slow” versus “rapid” progesterone-responsive gene transcription. In the case of the MR, SIRT1 represses MR-induced gene transcription and can also cause cellular senescence and subsequent renal injury (Fig. 1).
Figure 1. Model of SIRT1 regulation of steroid hormone receptor activity.
SIRT1 regulates individual steroid hormone receptors via different mechanisms. A) SIRT1 regulates the AR through direct deacetylation, thereby inhibiting AR activation and translocation and the transcription of AR-dependent genes. B) SIRT1 inhibits the PR by regulating the nucleo-cytoplasmic translocation of the receptor, and thereby the downstream activation of PR-regulated genes. C) SIRT1 regulates ERα by inhibiting the PI3K/AKT (p85/p110) pathway, possibly through FOXO3a. This inhibits the translocation of ERα from the cytoplasm to the nucleus, thus decreasing ERα binding to DNA and ERα-dependent gene transcription. D) The energy-sensing capabilities of SIRT1 regulate its effect on GR signaling. The cellular NAD/NADH ratio regulates SIRT1-mediated deacetylation of the GR and thus its ability to interact with the co-activator p300. E) SIRT1 regulates the MR by binding DOT1, thereby enhancing DOT1-mediated repression of MR-regulated genes. SIRT1 regulation of MR is independent of SIRT1 deacetylase activity.
However, for the most part, direct deacetylation of steroid hormone receptors by SIRT1 has not been demonstrated, or the functional effects of such a direct interaction (receptor acetylation status) have been modest. In the case of the PR and ER, SIRT1 regulation of receptor activity appears independent of direct SIRT1 acetylation. Moreover, regulation of the MR activity by SIRT1 is independent of SIRT1 deacetylase activity. It is rather more likely the SIRT1 regulation of steroid hormone receptor activity is “indirect.” For at least one steroid hormone receptor, AR, SIRT1 appears to serve as a transcriptional cofactor. The AR, upon binding of an androgen antagonist, physically associates with SIRT1 and the co-repressor NCoR, and recruits these proteins to the promoter of AR-responsive genes, repressing their transcription.
SIRT1 appears to regulate the ER through a different indirect mechanism, via modulation of a PI3K-dependent pathway, thus activating AKT, which in turn phosphorylates and activates ERα. In this case, and potentially for other steroid hormone receptors, the FOXO proteins may provide a critical link between SIRT1 activity and signaling pathways which activate or inactivate steroid hormone receptors.
In prostate cancer cells, SIRT1 inhibition via selective inhibitors or siRNA produces an increase of FOXO1 expression and acetylation, resulting in a decrease in cell viability that is independent of p53 (Jung-Hynes and Ahmad 2009). FOXO proteins may thus provide an important or even necessary co-regulatory component linking steroid hormone receptor activity and SIRT1 activity.
A recent study reports that SIRT1 is expressed in 67% (82/122) of breast carcinomas investigated, and with concurrent expression of DBC-1, is significantly associated with distant metastatic relapse and a shorter relapse-free survival rate (Lee et al. 2011). Additionally, activation of SIRT1 elicits a more profound inhibitory effect on BRCA1 mutant cancer cells (women possessing genetic mutations in BCRA1 have an 80% risk of developing breast cancer in their lifetime) than on BRCA1-wild-type cancer cells both in vitro and in vivo (Wang et al. 2008). When considered together with the reports that SIRT1 activity increases the expression of drug-resistance genes (Chu et al. 2005), SIRT1 expression and modulation may have important clinical implications for breast cancer patients, and potentially other steroid hormone-dependent cancer patients as well.
Understanding the effects of physiological and pathological changes in SIRT1 activity on steroid receptor function in normal cells and tumor cells is therefore of potentially great importance. New methods of pharmacological or genetic modulation of SIRT1 activity are under active development for a number of potential applications. In particular, SIRT1 may prove to be an important target molecule for new therapies of diseases of steroid hormone signaling and endocrine-related cancers.
Acknowledgments
This work was supported by grants from the National Cancer Institute: R01-CA101992 (DVF) and CA101992S1 (RLM), and by the Karin Grunebaum Cancer Research Foundation (DVF).
References
- Aghmesheh M, Edwards L, Clarke CL, Byth K, Katzenellenbogen BS, Russell PJ, Friedlander M, Tucker KM, de Fazio A. Expression of steroid hormone receptors in BRCA1-associated ovarian carcinomas. Gynecologic Oncology. 2005;97:16–25. doi: 10.1016/j.ygyno.2004.12.030. [DOI] [PubMed] [Google Scholar]
- Amat R, Solanes G, Giralt M, Villarroya F. SIRT1 is involved in glucocorticoid-mediated control of uncoupling protein-3 gene transcription. Journal of Biological Chemistry. 2007;282:34066–34076. doi: 10.1074/jbc.M707114200. [DOI] [PubMed] [Google Scholar]
- Aoyagi S, Archer TK. Nicotinamide uncouples hormone-dependent chromatin remodeling from transcription complex assembly. Molecular and Cellular Biology. 2008;28:30–39. doi: 10.1128/MCB.01158-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett NC, Gardiner RA, Hooper JD, Johnson DW, Gobe GC. Molecular cell biology of androgen receptor signalling. International Journal of Biochemistry & Cell Biology. 2010;42:813–827. doi: 10.1016/j.biocel.2009.11.013. [DOI] [PubMed] [Google Scholar]
- Berdichevsky A, Guarente L. A stress response pathway involving sirtuins, forkheads and 14–3–3 proteins. Cell Cycle. 2006;5:2588–2591. doi: 10.4161/cc.5.22.3513. [DOI] [PubMed] [Google Scholar]
- Bouras T, Fu MF, Sauve AA, Wang F, Quong AA, Perkins ND, Hay RT, Gu W, Pestell RG. SIRT1 deacetylation and repression of p300 involves lysine residues 1020/1024 within the cell cycle regulatory domain 1. Journal of Biological Chemistry. 2005;280:10264–10276. doi: 10.1074/jbc.M408748200. [DOI] [PubMed] [Google Scholar]
- Brand MD, Esteves TC. Physiological functions of the mitochondrial uncoupling proteins UCP2 and UCP3. Cell Metabolism. 2005;2:85–93. doi: 10.1016/j.cmet.2005.06.002. [DOI] [PubMed] [Google Scholar]
- Brownawell AM, Kops G, Macara IG, Burgering BMT. Mechanism of nuclear transport of the forkhead transcription factor, AFX. Molecular Biology of the Cell. 1999;10:1648. doi: 10.1128/MCB.21.10.3534-3546.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet A. The multiple roles of FOXO transcription factors. M S-Medecine Sciences. 2004;20:856–859. doi: 10.1051/medsci/20042010856. [DOI] [PubMed] [Google Scholar]
- Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin YX, Tran H, Ross SE, Mostoslavsky R, Cohen HY, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- Byles V, Chmilewski LK, Wang J, Zhu L, Forman LW, Faller DV, Dai Y. Aberrant cytoplasm localization and protein stability of SIRT1 is regulated by PI3K/IGF-1R signaling in human cancer cells. Int J Biol Sci. 2010;6:599–612. doi: 10.7150/ijbs.6.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, et al. Genome-wide analysis of estrogen receptor binding sites. Nature Genetics. 2006;38:1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- Chawla A, Repa JJ, Evans RM, Mangelsdorf DJ. Nuclear receptors and lipid physiology: Opening the X-files. Science. 2001;294:1866–1870. doi: 10.1126/science.294.5548.1866. [DOI] [PubMed] [Google Scholar]
- Chu F, Chou PM, Zheng X, Mirkin BL, Rebbaa A. Control of multidrug resistance gene mdr1 and cancer resistance to chemotherapy by the longevity gene sirt1. Cancer Research. 2005;65:10183–10187. doi: 10.1158/0008-5472.CAN-05-2002. [DOI] [PubMed] [Google Scholar]
- Clark KL, Halay ED, Lai ES, Burley SK. CO-CRYSTAL STRUCTURE OF THE HNF-3/FORK HEAD DNA-RECOGNITION MOTIF RESEMBLES HISTONE-H5. Nature. 1993;364:412–420. doi: 10.1038/364412a0. [DOI] [PubMed] [Google Scholar]
- Cleator S, Heller W, Coombes RC. Triple-negative breast cancer: therapeutic options. Lancet Oncology. 2007;8:235–244. doi: 10.1016/S1470-2045(07)70074-8. [DOI] [PubMed] [Google Scholar]
- Cohen HY, Lavu S, Bitterman KJ, Hekking B, Imahiyerobo TA, Miller C, Frye R, Ploegh H, Kessler BM, Sinclair DA. Acetylation of the C terminus of Ku70 by CBP and PCAF controls Bax-mediated apoptosis. Molecular Cell. 2004a;13:627–638. doi: 10.1016/s1097-2765(04)00094-2. [DOI] [PubMed] [Google Scholar]
- Cohen HY, Miller C, Bitterman KJ, Wall NR, Hekking B, Kessler B, Howitz KT, Gorospe M, de Cabo R, Sinclair DA. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004b;305:390–392. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- Cui Y, Mao Z, Pestell R, Curran EM, Welshons WV, Fuqua SAW. Phosphorylation of estrogen receptor a blocks its acetylation and regulates estrogen sensitivity. Cancer Research. 2004;64:9199–9208. doi: 10.1158/0008-5472.CAN-04-2126. [DOI] [PubMed] [Google Scholar]
- Dai Y, Ngo D, Forman LW, Qin DC, Jacob J, Faller DV. Sirtuin 1 is required for antagonist-induced transcriptional repression of androgen-responsive genes by the androgen receptor. Molecular Endocrinology. 2007;21:1807–1821. doi: 10.1210/me.2006-0467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Y, Ngo D, Jacob J, Forman LW, Faller DV. Prohibitin and the SWI/SNF ATPase subunit BRG1 are required for effective androgen antagonist-mediated transcriptional repression of androgen receptor-regulated genes. Carcinogenesis. 2008;29:1725–1733. doi: 10.1093/carcin/bgn117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daitoku H, Hatta M, Matsuzaki H, Aratani S, Ohshima T, Miyagishi M, Nakajima T, Fukamizu A. Silent information regulator 2 potentiates Foxo1-mediated transcription through its deacetylase activity. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:10042–10047. doi: 10.1073/pnas.0400593101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dali-Youcef N, Lagouge M, Froelich S, Koehl C, Schoonjans K, Auwerx J. Sirtuins: The ‘magnificent seven’, function, metabolism and longevity. Annals of Medicine. 2007;39:335–345. doi: 10.1080/07853890701408194. [DOI] [PubMed] [Google Scholar]
- Daniel AR, Gaviglio AL, Czaplicki LM, Hillard CJ, Housa D, Lange CA. The Progesterone Receptor Hinge Region Regulates the Kinetics of Transcriptional Responses Through Acetylation, Phosphorylation, and Nuclear Retention. Molecular Endocrinology. 2010;24:2126–2138. doi: 10.1210/me.2010-0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson NE. Biology of breast cancer and its clinical implications. Current Opinion in Oncology. 1992;4:1003–1009. doi: 10.1097/00001622-199212000-00002. [DOI] [PubMed] [Google Scholar]
- Delpuech O, Griffiths B, East P, Essafi A, Lam EWF, Burgering B, Downward J, Schulze A. Induction of Mxi1-SR alpha by FOXO3a contributes to repression of Myc-dependent gene expression. Molecular and Cellular Biology. 2007;27:4917–4930. doi: 10.1128/MCB.01789-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng C, Kim1 Hyun-Seok, Vassilopoulos1 Athanassios, Wang1 Rui-Hong, Lahusen1 Tyler, Xiao2 Zhen, Xu1 Xiaoling, Li1 Cuiling, Veenstra2 Timothy D, Ji3 Junfang, et al. Knockout mouse models for sirtuins: Genome integrity, metabolism, and cancer [abstract]. Orlando, Florida. 2011; Proceedings of the 102nd Annual Meeting of the American Association for Cancer Research.Philadelphia (PA): AACR; 2011. Apr 2–6, [Google Scholar]
- Deng C, Kim H-S, Vassilopoulos A, Wang R-H, Lahusen T, Xiao Z, Xu X, Li C, Veenstra TD, Ji J, et al. Knockout mouse models for sirtuins: Genome integrity, metabolism, and cancer [abstract]. Orlando, Florida. 2011; Proceedings of the 102nd Annual Meeting of the American Association for Cancer Research.Philadelphia (PA): AACR; 2011. Apr 2–6, [Google Scholar]
- Dillon RL, White DE, Muller WJ. The phosphatidyl inositol 3-kinase signaling network: implications for human breast cancer. Oncogene. 2007;26:1338–1345. doi: 10.1038/sj.onc.1210202. [DOI] [PubMed] [Google Scholar]
- Dryden SC, Nahhas FA, Nowak JE, Goustin AS, Tainsky MA. Role for human SIRT2 NAD-dependent deacetylase activity in control of mitotic exit in the cell cycle. Molecular and Cellular Biology. 2003;23:3173–3185. doi: 10.1128/MCB.23.9.3173-3185.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards J, Bartlett JMS. The androgen receptor and signal-transduction pathways in hormone-refractory prostate cancer. Part 2: androgen-receptor cofactors and bypass pathways. Bju International. 2005;95:1327–1335. doi: 10.1111/j.1464-410X.2005.05527.x. [DOI] [PubMed] [Google Scholar]
- Ford E, Voit R, Liszt G, Magin C, GrumMt I, Guarente L. Mammalian Sir2 homolog SIRT7 is an activator of RNA polymerase I transcription. Genes & Development. 2006;20:1075–1080. doi: 10.1101/gad.1399706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford J, Jiang M, Milner J. Cancer-specific functions of SIRT1 enable human epithelial cancer cell growth and survival. Cancer Research. 2005;65:10457–10463. doi: 10.1158/0008-5472.CAN-05-1923. [DOI] [PubMed] [Google Scholar]
- Frescas D, Valenti L, Accili D. Nuclear trapping of the forkhead transcription factor FoxO1 via Sirt-dependent deacetylation promotes expression of glucogenetic genes. Journal of Biological Chemistry. 2005;280:20589–20595. doi: 10.1074/jbc.M412357200. [DOI] [PubMed] [Google Scholar]
- Frye RA. Characterization of five human cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins (sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochemical and Biophysical Research Communications. 1999;260:273–279. doi: 10.1006/bbrc.1999.0897. [DOI] [PubMed] [Google Scholar]
- Frye RA. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochemical and Biophysical Research Communications. 2000;273:793–798. doi: 10.1006/bbrc.2000.3000. [DOI] [PubMed] [Google Scholar]
- Fu MF, Liu MR, Sauve AA, Jiao XM, Zhang XP, Wu XF, Powell MJ, Yang TL, Gu W, Avantaggiati ML, et al. Hormonal control of androgen receptor function through SIRT1. Molecular and Cellular Biology. 2006;26:8122–8135. doi: 10.1128/MCB.00289-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu MF, Wang CG, Reutens AT, Wang J, Angeletti RH, Siconolfi-Baez L, Ogryzko V, Avantaggiati ML, Pestell RG. p300 and p300/cAMP-response element-binding protein-associated factor acetylate the androgen receptor at sites governing hormone-dependent transactivation. Journal of Biological Chemistry. 2000;275:20853–20860. doi: 10.1074/jbc.M000660200. [DOI] [PubMed] [Google Scholar]
- Fulco M, Schiltz RL, Iezzi S, King MT, Zhao P, Kashiwaya Y, Hoffman E, Veech RL, Sartorelli V. Sir2 regulates skeletal muscle differentiation as a potential sensor of the redox state. Molecular Cell. 2003;12:51–62. doi: 10.1016/s1097-2765(03)00226-0. [DOI] [PubMed] [Google Scholar]
- Fuqua SAW, Wiltschke C, Zhang QX, Borg A, Castles CG, Friedrichs WE, Hopp T, Hilsenbeck S, Mohsin S, O’Connell P, et al. A hypersensitive estrogen receptor-alpha mutation in premalignant breast lesions. Cancer Research. 2000;60:4026–4029. [PubMed] [Google Scholar]
- Gadkar-Sable S, Shah C, Rosario G, Sachdeva G, Puri C. Progesterone receptors: Various forms and functions in reproductive tissues. Frontiers in Bioscience. 2005;10:2118–2130. doi: 10.2741/1685. [DOI] [PubMed] [Google Scholar]
- Giannakou ME, Partridge L. The interaction between FOXO and SIRT1: tipping the balance towards survival. Trends in Cell Biology. 2004;14:408–412. doi: 10.1016/j.tcb.2004.07.006. [DOI] [PubMed] [Google Scholar]
- Greiss S, Gartner A. Sirtuin/Sir2 phylogeny, evolutionary considerations and structural conservation. Molecules and Cells. 2009;28:407–415. doi: 10.1007/s10059-009-0169-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronemeyer H, Gustafsson JA, Laudet V. Principles for modulation of the nuclear receptor superfamily. Nature Reviews Drug Discovery. 2004;3:950–964. doi: 10.1038/nrd1551. [DOI] [PubMed] [Google Scholar]
- Haigis MC, Guarente LP. Mammalian sirtuins - emerging roles in physiology, aging, and calorie restriction. Genes & Development. 2006;20:2913–2921. doi: 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- Haigis MC, Mostoslavsky R, Haigis KM, Fahie K, Christodoulou DC, Murphy AJ, Valenzuela DM, Yancopoulos GD, Karow M, Blander G, et al. SIRT4 inhibits glutamate dehydrogehase and opposes the effects of calorie restriction in pancreatic beta cells. Cell. 2006;126:941–954. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- Hallows WC, Lee S, Denu JM. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:10230–10235. doi: 10.1073/pnas.0604392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasselgren PO. Glucocorticoids and muscle catabolism. Curr Opin Clin Nutr Metab Care. 1999;2:201–205. doi: 10.1097/00075197-199905000-00002. [DOI] [PubMed] [Google Scholar]
- Herranz D, Munoz-Martin M, Canamero M, Mulero F, Martinez-Pastor B, Fernandez-Capetillo O, Serrano M. Sirt1 improves healthy ageing and protects from metabolic syndrome-associated cancer. Nature Communications. 2010:1. doi: 10.1038/ncomms1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herranz D, Serrano M. SIRT1: recent lessons from mouse models. Nature Reviews Cancer. 2010;10:819–823. doi: 10.1038/nrc2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsuka M, Inoue T, Toda T, Kimura N, Shirayoshi Y, Kamitani H, Watanabe T, Ohama E, Tahimic CGT, Kurimasa A, et al. Proteomics-based identification of differentially expressed genes in human gliomas: down-regulation of SIRT2 gene. Biochemical and Biophysical Research Communications. 2003;309:558–566. doi: 10.1016/j.bbrc.2003.08.029. [DOI] [PubMed] [Google Scholar]
- Hirschey MD, Shimazu T, Goetzman E, Jing E, Schwer B, Lombard DB, Grueter CA, Harris C, Biddinger S, Ilkayeva OR, et al. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. 2010;464:121–U137. doi: 10.1038/nature08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HJ, Tindall DJ. Dynamic FoxO transcription factors. Journal of Cell Science. 2007;120:2479–2487. doi: 10.1242/jcs.001222. [DOI] [PubMed] [Google Scholar]
- Huang PX, Chandra V, Rastinejad F. Structural Overview of the Nuclear Receptor Superfamily: Insights into Physiology and Therapeutics. Annual Review of Physiology. 2010;72:247–272. doi: 10.1146/annurev-physiol-021909-135917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikenoue T, Inoki K, Zhao B, Guan KL. PTEN acetylation modulates its interaction with PDZ domain. Cancer Research. 2008;68:6908–6912. doi: 10.1158/0008-5472.CAN-08-1107. [DOI] [PubMed] [Google Scholar]
- Jin QH, Yan TT, Ge XJ, Sun C, Shi XG, Zhai QW. Cytoplasm-localized SIRT1 enhances apoptosis. Journal of Cellular Physiology. 2007;213:88–97. doi: 10.1002/jcp.21091. [DOI] [PubMed] [Google Scholar]
- Jung-Hynes B, Ahmad N. Abstract #5092: Inhibition of Sirt1 by sirtinol causes p53-independent activation of FoxO1 in human prostate cancer cells. AACR Meeting Abstracts. 2009;2009:5092. [Google Scholar]
- Kawahara TLA, Michishita E, Adler AS, Damian M, Berber E, Lin M, McCord RA, Ongaigui KCL, Boxer LD, Chang HY, et al. SIRT6 Links Histone H3 Lysine 9 Deacetylation to NF-kappa B-Dependent Gene Expression and Organismal Life Span. Cell. 2009;136:62–74. doi: 10.1016/j.cell.2008.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinyamu HK, Archer TK. Modifying chromatin to permit steroid hormone receptor-dependent transcription. Biochimica Et Biophysica Acta-Gene Structure and Expression. 2004;1677:30–45. doi: 10.1016/j.bbaexp.2003.09.015. [DOI] [PubMed] [Google Scholar]
- Kops G, de Ruiter ND, De Vries-Smits AMM, Powell DR, Bos JL, Burgering BMT. Direct control of the Forkhead transcription factor AFX by protein kinase B. Nature. 1999;398:630–634. doi: 10.1038/19328. [DOI] [PubMed] [Google Scholar]
- Kwon HS, Oft M. The ups and downs of SIRT1. Trends in Biochemical Sciences. 2008;33:517–525. doi: 10.1016/j.tibs.2008.08.001. [DOI] [PubMed] [Google Scholar]
- Lai E, Clark KL, Burley SK, Darnell JE. HEPATOCYTE NUCLEAR FACTOR 3/FORK HEAD OR WINGED HELIX PROTEINS - A FAMILY OF TRANSCRIPTION FACTORS OF DIVERSE BIOLOGIC FUNCTION. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:10421–10423. doi: 10.1073/pnas.90.22.10421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Romancer M, Treilleux I, Leconte N, Robin-Lespinasse Y, Sentis S, Bouchekioua-Bouzaghou K, Goddard S, Gobert-Gosse S, Corbo L. Regulation of estrogen rapid signaling through arginine methylation by PRMT1. Molecular Cell. 2008;31:212–221. doi: 10.1016/j.molcel.2008.05.025. [DOI] [PubMed] [Google Scholar]
- Lee H, Kim KR, Noh SJ, Park HS, Kwon KS, Park BH, Jung SH, Youn HJ, Lee BK, Chung MJ, et al. Expression of DBC1 and SIRT1 is associated with poor prognosis for breast carcinoma. Human Pathology. 2011;42:204–213. doi: 10.1016/j.humpath.2010.05.023. [DOI] [PubMed] [Google Scholar]
- Lengyel F, Vertes Z, Kovacs KA, Kornyei JL, Sumegi B, Vertes M. Effect of estrogen and inhibition of phosphatidylinositol-3 kinase on Akt and FOXO1 in rat uterus. Steroids. 2007;72:422–428. doi: 10.1016/j.steroids.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Lin JN, Lin VCH, Rau KM, Shieh PC, Kuo DH, Shieh JC, Chen WJ, Tsai SC, Way TD. Resveratrol Modulates Tumor Cell Proliferation and Protein Translation via SIRT1-Dependent AMPK Activation. Journal of Agricultural and Food Chemistry. 2010;58:1584–1592. doi: 10.1021/jf9035782. [DOI] [PubMed] [Google Scholar]
- Liu SY, Michishita E, Park JY, Nunez N, Hursting SD, Barrett C, Horikawa I. Analysis of mammalian SIRT6, a homolog of the yeast Sir2 protein, during adipogenesis and calorie-restriction. Cancer Epidemiology Biomarkers & Prevention. 2005;14:2731S–2731S. [Google Scholar]
- Lombard DB, Schwer B, Alt FW, Mostoslavsky R. SIRT6 in DNA repair, metabolism and ageing. Journal of Internal Medicine. 2008;263:128–141. doi: 10.1111/j.1365-2796.2007.01902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo JY, Nikolaev AY, Imai S, Chen DL, Su F, Shiloh A, Guarente L, Gu W. Negative control of p53 by Sir2 alpha promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- Ma YX, Fan SJ, Hu CY, Meng QH, Fuqua SA, Pestell RG, Tomita YA, Rosen EM. BRCA1 Regulates Acetylation and Ubiquitination of Estrogen Receptor-alpha. Molecular Endocrinology. 2010;24:76–90. doi: 10.1210/me.2009-0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madureira PA, Varshochi R, Constantinidou D, Francis RE, Coombes RC, Yao KM, Lam EWF. The forkhead box M1 protein regulates the transcription of the estrogen receptor alpha in breast cancer cells. Journal of Biological Chemistry. 2006;281:25167–25176. doi: 10.1074/jbc.M603906200. [DOI] [PubMed] [Google Scholar]
- Maehama T, Dixon JE. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. Journal of Biological Chemistry. 1998;273:13375–13378. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, et al. THE NUCLEAR RECEPTOR SUPERFAMILY - THE 2ND DECADE. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Duong V, Castet A, Cavailles V. Histone deacetylase inhibition and estrogen signalling in human breast cancer cells. Biochemical Pharmacology. 2004;68:1239–1246. doi: 10.1016/j.bcp.2004.04.031. [DOI] [PubMed] [Google Scholar]
- Medema RH, Kops G, Bos JL, Burgering BMT. AFX-like Forkhead transcription factors mediate cell-cycle regulation by Ras and PKB through p27(kip1) Nature. 2000;404:782–787. doi: 10.1038/35008115. [DOI] [PubMed] [Google Scholar]
- Michishita E, McCord RA, Berber E, Kioi M, Padilla-Nash H, Damian M, Cheung P, Kusumoto R, Kawahara TLA, Barrett JC, et al. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008;452:492–U416. doi: 10.1038/nature06736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Molecular Biology of the Cell. 2005;16:4623–4635. doi: 10.1091/mbc.E05-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller WR, Langdon SP. Steroid hormones and cancer. 3. Observations from human subjects. European Journal of Surgical Oncology. 1997;23:163–177. doi: 10.1016/s0748-7983(97)80014-5. [DOI] [PubMed] [Google Scholar]
- Misrahi M, Atger M, Dauriol L, Loosfelt H, Meriel C, Fridlansky F, Guiochonmantel A, Galibert F, Milgrom E. COMPLETE AMINO-ACID-SEQUENCE OF THE HUMAN PROGESTERONE-RECEPTOR DEDUCED FROM CLONED CDNA. Biochemical and Biophysical Research Communications. 1987;143:740–748. doi: 10.1016/0006-291x(87)91416-1. [DOI] [PubMed] [Google Scholar]
- Motta MC, Divecha N, Lemieux M, Kamel C, Chen D, Gu W, Bultsma Y, McBurney M, Guarente L. Mammalian SIRT1 represses forkhead transcription factors. Cell. 2004;116:551–563. doi: 10.1016/s0092-8674(04)00126-6. [DOI] [PubMed] [Google Scholar]
- Myers MP, Pass I, Batty IH, Van der Kaay J, Stolarov JP, Hemmings BA, Wigler MH, Downes CP, Tonks NK. The lipid phosphatase activity of PTEN is critical for its tumor suppressor function. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:13513–13518. doi: 10.1073/pnas.95.23.13513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano D, Fan YY, Kohno M, Hitomi H, Felder R, Nishiyama A. Aldosterone induces renal senescence in proximal tubular cells via mineralocorticoid receptor/p21-dependent pathway. Endocrine Journal. 2010;57:S544–S544. [Google Scholar]
- Nemoto S, Fergusson MM, Finkel T. Nutrient availability regulates SIRT1 through a forkhead-dependent pathway. Science. 2004;306:2105–2108. doi: 10.1126/science.1101731. [DOI] [PubMed] [Google Scholar]
- Nemoto S, Fergusson MM, Finkel T. SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1 alpha. Journal of Biological Chemistry. 2005;280:16456–16460. doi: 10.1074/jbc.M501485200. [DOI] [PubMed] [Google Scholar]
- Osborne CK. Steroid hormone receptors in breast cancer management. Breast Cancer Research and Treatment. 1998;51:227–238. doi: 10.1023/a:1006132427948. [DOI] [PubMed] [Google Scholar]
- Osborne CK, Schiff R. Estrogen-receptor biology: Continuing progress and therapeutic implications. Journal of Clinical Oncology. 2005;23:1616–1622. doi: 10.1200/JCO.2005.10.036. [DOI] [PubMed] [Google Scholar]
- Osborne CK, Shou J, Massarweh S, Schiff R. Crosstalk between estrogen receptor and growth factor receptor pathways as a cause for endocrine therapy resistance in breast cancer. Clinical Cancer Research. 2005;11:865S–870S. [PubMed] [Google Scholar]
- Ota H, Kaneki M, Kobayashi T, Iijima K, Eto M, Kozaki K, Akishita M, Ouchi Y. Sirt1, a longevity gene, regulates senescence-like phenotype in human endothelial cells. Journal of Hypertension. 2006a;24:131–131. [Google Scholar]
- Ota H, Tokunaga E, Chang K, Hikasa M, Iijima K, Eto M, Kozaki K, Akishita M, Ouchi Y, Kaneki M. Sirt1 inhibitor, Sirtinol, induces senescence-like growth arrest with attenuated Ras-MAPK signaling in human cancer cells. Oncogene. 2006b;25:176–185. doi: 10.1038/sj.onc.1209049. [DOI] [PubMed] [Google Scholar]
- Perissi V, Rosenfeld MG. Controlling nuclear receptors: The circular logic of cofactor cycles. Nature Reviews Molecular Cell Biology. 2005;6:542–554. doi: 10.1038/nrm1680. [DOI] [PubMed] [Google Scholar]
- Pfluger PT, Herranz D, Velasco-Miguel S, Serrano M, Tschop MH. Sirt1 protects against high-fat diet-induced metabolic damage. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:9793–9798. doi: 10.1073/pnas.0802917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard F, Kurtev M, Chung NJ, Topark-Ngarm A, Senawong T, de Oliveira RM, Leid M, McBurney MW, Guarente L. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature. 2004;429:771–776. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popov VM, Wang CG, Shirley LA, Rosenberg A, Li SW, Nevalainen M, Fu MF, Pestell RG. The functional significance of nuclear receptor acetylation. Steroids. 2007;72:221–230. doi: 10.1016/j.steroids.2006.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potente M, Ghaeni L, Baldessari D, Mostoslavsky R, Rossig L, Dequiedt F, Haendeler J, Mione M, Dejana E, Alt FW, et al. Sirt1 controls endothelial angiogenic functions during vascular growth and maturation. Circulation. 2007;116:37–37. doi: 10.1101/gad.435107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell M, Yu Z, Casimiro M, Marampon F, Yeow W-S, Parlow AF, Cardiff R, Katiyar S, He X, McCue P, et al. Abstract #4126: Sirt1 is required for normal prostate gland development and androgen signaling in vivo. AACR Meeting Abstracts. 2009;2009:4126. [Google Scholar]
- Prins GS, Putz O. Molecular signaling pathways that regulate prostate gland development. Differentiation. 2008;76:641–659. doi: 10.1111/j.1432-0436.2008.00277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt K, Zinn RL, Ohm JE, McGarvey KM, Kang SHL, Watkins DN, Herman JG, Baylin SB. Inhibition of SIRT1 reactivates silenced cancer genes without loss of promoter DNA hypermethylation. Plos Genetics. 2006;2:344–352. doi: 10.1371/journal.pgen.0020040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigserver P, Rodgers J, Lerin C, Haas W, Gygi S, Spiegelman B. Nutrient control of glucose metabolism through PGC-1?/SIRT1 complex. Gerontologist. 2005;45:56–56. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- Risbridger GP, Davis ID, Birrell SN, Tilley WD. Breast and prostate cancer: more similar than different. Nature Reviews Cancer. 2010;10:205–212. doi: 10.1038/nrc2795. [DOI] [PubMed] [Google Scholar]
- Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1 alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- Sachdeva G, Gadkar S, Shah CA, Kholkute SD, Puri CP. Characterization of a critical region in the hormone binding domain of sperm progesterone receptor. International Journal of Andrology. 2005;28:120–124. doi: 10.1111/j.1365-2605.2005.00511.x. [DOI] [PubMed] [Google Scholar]
- Sasaki T, Maier B, Koclega KD, Chruszcz M, Gluba W, Stukenberg PT, Minor W, Scrable H. Phosphorylation Regulates SIRT1 Function. Plos One. 2008:3. doi: 10.1371/journal.pone.0004020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schakman O, Gilson H, Thissen JP. Mechanisms of glucocorticoid-induced myopathy. Journal of Endocrinology. 2008;197:1–10. doi: 10.1677/JOE-07-0606. [DOI] [PubMed] [Google Scholar]
- Scher HI, Buchanan G, Gerald W, Butler LM, Tilley WD. Targeting the androgen receptor: improving outcomes for castration-resistant prostate cancer. Endocrine-Related Cancer. 2004;11:459–476. doi: 10.1677/erc.1.00525. [DOI] [PubMed] [Google Scholar]
- Schlicker C, Gertz M, Papatheodorou P, Kachholz B, Becker CFW, Steegborn C. Substrates and regulation mechanisms for the human mitochondrial Sirtuins Sirt3 and Sirt5. Journal of Molecular Biology. 2008;382:790–801. doi: 10.1016/j.jmb.2008.07.048. [DOI] [PubMed] [Google Scholar]
- Schwer B, Verdin E. Conserved metabolic regulatory functions of sirtuins. Cell Metabolism. 2008;7:104–112. doi: 10.1016/j.cmet.2007.11.006. [DOI] [PubMed] [Google Scholar]
- Serrano M. Sirt1 transgenic and cancer models [abstract]. Orlando, Florida. Proceedings of the 102nd Annual Meeting of the American Association for Cancer Research.Philadelphia (PA): AACR; 2011. [Google Scholar]
- Skliris GP, Leygue E, Watson PH, Murphy LC. Estrogen receptor alpha negative breast cancer patients: Estrogen receptor beta as a therapeutic target. Journal of Steroid Biochemistry and Molecular Biology. 2008;109:1–10. doi: 10.1016/j.jsbmb.2007.12.010. [DOI] [PubMed] [Google Scholar]
- Song RXD, Santen RJ. Membrane initiated estrogen signaling in breast cancer. Biology of Reproduction. 2006;75:9–16. doi: 10.1095/biolreprod.105.050070. [DOI] [PubMed] [Google Scholar]
- Stambolic V, Suzuki A, de la Pompa JL, Brothers GM, Mirtsos C, Sasaki T, Ruland J, Penninger JM, Siderovski DP, Mak TW. Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell. 1998;95:29–39. doi: 10.1016/s0092-8674(00)81780-8. [DOI] [PubMed] [Google Scholar]
- Stankovic-Valentin N, Deltour S, Seeler J, Pinte S, Vergoten G, Guerardel C, Dejean A, Leprince D. An acetylation/deacetylation-SUMOylation switch through a phylogenetically conserved psi KXEP motif in the tumor suppressor HIC1 regulates transcriptional repression activity. Molecular and Cellular Biology. 2007;27:2661–2675. doi: 10.1128/MCB.01098-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streuli CH, Haslam SZ. Introduction - Control of mammary gland development and neoplasia by stromal-epithelial interactions and extracellular matrix. Journal of Mammary Gland Biology and Neoplasia. 1998;3:107–108. [Google Scholar]
- Sung JY, Kim R, Kim JE, Lee J. Balance between SIRT1 and DBC1 expression is lost in breast cancer. Cancer Science. 2010;101:1738–1744. doi: 10.1111/j.1349-7006.2010.01573.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MJ, Omalley BW. MOLECULAR MECHANISMS OF ACTION OF STEROID/THYROID RECEPTOR SUPERFAMILY MEMBERS. Annual Review of Biochemistry. 1994;63:451–486. doi: 10.1146/annurev.bi.63.070194.002315. [DOI] [PubMed] [Google Scholar]
- Vaquero A, Scher MB, Lee DH, Sutton A, Cheng HL, Alt FW, Serrano L, Sternglanz R, Reinberg D. SirT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Genes & Development. 2006;20:1256–1261. doi: 10.1101/gad.1412706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CG, Chen LH, Hou XH, Li ZY, Kabra N, Ma YH, Nemoto S, Finkel T, Gu W, Cress WD, et al. Interactions between E2F1 and SirT1 regulate apoptotic response to DNA damage. Nature Cell Biology. 2006;8:1025–U1109. doi: 10.1038/ncb1468. [DOI] [PubMed] [Google Scholar]
- Wang CG, Fu MF, Angeletti RH, Siconolfi-Baez L, Reutens AT, Albanese C, Lisanti MP, Katzenellenbogen BS, Kato S, Hopp T, et al. Direct acetylation of the estrogen receptor alpha hinge region by p300 regulates transactivation and hormone sensitivity. Journal of Biological Chemistry. 2001;276:18375–18383. doi: 10.1074/jbc.M100800200. [DOI] [PubMed] [Google Scholar]
- Wang L, Hsu CL, Chang CS. Androgen receptor corepressors: An overview. Prostate. 2005;63:117–130. doi: 10.1002/pros.20170. [DOI] [PubMed] [Google Scholar]
- Wang RH, Zheng Y, Kim HS, Xu XL, Cao L, Luhasen T, Lee MH, Xiao CY, Vassilopoulos A, Chen WP, et al. Interplay among BRCA1, SIRT1, and Survivin during BRCA1-Associated Tumorigenesis. Molecular Cell. 2008;32:11–20. doi: 10.1016/j.molcel.2008.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Kreisberg JI, Ghosh PM. Cross-talk between the androgen receptor and the phosphatidylinositol 3-kinase/akt pathway in prostate cancer. Current Cancer Drug Targets. 2007;7:591–604. doi: 10.2174/156800907781662248. [DOI] [PubMed] [Google Scholar]
- Ward RD, Weigel NL. Steroid receptor phosphorylation: Assigning function to site-specific phosphorylation. Biofactors. 2009;35:528–536. doi: 10.1002/biof.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Jurgens G, Kuttner F, Seifert E, Jackle H. THE HOMEOTIC GENE FORK HEAD ENCODES A NUCLEAR-PROTEIN AND IS EXPRESSED IN THE TERMINAL REGIONS OF THE DROSOPHILA EMBRYO. Cell. 1989;57:645–658. doi: 10.1016/0092-8674(89)90133-5. [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Schoonjans K, Auwerx J. Sirtuin functions in health and disease. Molecular Endocrinology. 2007;21:1745–1755. doi: 10.1210/me.2007-0079. [DOI] [PubMed] [Google Scholar]
- Yang SIRT1 sumoylation regulates its deacetylase activity and cellular response to genotoxic stress (vol 9, pg 1253, 2007) Nature Cell Biology. 2007;9:1442–1442. doi: 10.1038/ncb1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang TL, Fu MF, Pestell R, Sauve AA. SIRT1 and endocrine signaling. Trends in Endocrinology and Metabolism. 2006;17:186–191. doi: 10.1016/j.tem.2006.04.002. [DOI] [PubMed] [Google Scholar]
- Yao Y, Li HZ, Gu YS, Davidson NE, Zhou Q. Inhibition of SIRT1 deacetylase suppresses estrogen receptor signaling. Carcinogenesis. 2010;31:382–387. doi: 10.1093/carcin/bgp308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung F, Hoberg JE, Ramsey CS, Keller MD, Jones DR, Frye RA, Mayo MW. Modulation of NF-kappa B-dependent transcription and cell survival by the SIRT1 deacetylase. Embo Journal. 2004;23:2369–2380. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi JJ, Luo JY. SIRT1 and p53, effect on cancer, senescence and beyond. Biochimica Et Biophysica Acta-Proteins and Proteomics. 2010;1804:1684–1689. doi: 10.1016/j.bbapap.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Dykhouse K, Lou Z. Abstract #4441: c-Myc/SIRT1 feedback loop: A novel regulation of c-Myc induced cellular transformation. AACR Meeting Abstracts. 2009;2009:4441. [Google Scholar]
- Zhang DY, Li SY, Cruz P, Kone BC. Sirtuin 1 Functionally and Physically Interacts with Disruptor of Telomeric Silencing-1 to Regulate alpha-ENaC Transcription in Collecting Duct. Journal of Biological Chemistry. 2009b;284:20917–20926. doi: 10.1074/jbc.M109.020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao WH, Kruse JP, Tang Y, Jung SY, Qin J, Gu W. Negative regulation of the deacetylase SIRT1 by DBC1. Nature. 2008;451:587–U511. doi: 10.1038/nature06515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Ohkawa S, Li HY, Roberts-Wilson TK, Price SR. FOXO3a mediates signaling crosstalk that coordinates ubiquitin and atrogin-1/MAFbx expression during glucocorticoid-induced skeletal muscle atrophy. Faseb Journal. 2010;24:2660–2669. doi: 10.1096/fj.09-151480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou YY, Tsai WB, Cheng CJ, Hsu C, Chung YM, Li PC, Lin SH, Hu MCT. Forkhead box transcription factor FOXO3a suppresses estrogen-dependent breast cancer cell proliferation and tumorigenesis. Breast Cancer Research. 2008:10. doi: 10.1186/bcr1872. [DOI] [PMC free article] [PubMed] [Google Scholar]