Abstract
Despite the therapeutic promise of the sub-nanomolar affinity cannabinoid CB2 antagonist, N-[(1S)-endo-1,3,3-trimethylbicyclo[2.2.1]heptan2-yl]-5-(4-chloro-3-methylphenyl)-1-[(4-methylphenyl)methyl]-1H-pyrazole-3-carboxamide (SR144528, 1), little is known about its binding site interactions and no primary interaction site for 1 at CB2 has been identified. We report here the results of Glide docking studies in our cannabinoid CB2 inactive state model that were then tested via compound synthesis, binding and functional assays. Our results show that the amide functional group of 1 is critical to its CB2 affinity and efficacy and that aromatic stacking interactions in the TMH5/6 aromatic cluster of CB2 are also important. Molecular modifications that increased the positive electrostatic potential in the region between the fenchyl and aromatic rings led to more efficacious compounds. This result is consistent with the EC-3 loop negatively charged amino acid, D275 (identified via Glide docking studies) acting as the primary interaction site for 1 and its analogs.
INTRODUCTION
The Cannabinoid CB2 receptor is highly expressed throughout the immune system1, 2 and has been described in the CNS under both pathological3 and physiological conditions.4 This quite specific localization, as well as, the fact that CB2 knock-out mice fail to respond to the immunomodulatory effects of classical cannabinoids,5 suggest that CB2 receptor ligands have potential therapeutic applications as immunomodulators for the treatment of inflammation and allergy. Several papers report the role of the CB2 receptor in modulating leukocyte migration,6–9 activation,10 and antigen processing.11 Additional applications could arise from studies on bone physiology, as blockade of CB2 has been reported to protect ovariectomized mice from bone loss.12
5-(4-Chloro-3-methyl-phenyl)-1-(4-methyl-benzyl)-1H-pyrazole-3-carboxylic acid (1,3,3-trimethyl-bicyclo[2.2.1]hept-2-yl)-amide (SR144528; 1), the first reported CB2 antagonist displays sub-nanomolar affinity for both the rat spleen and cloned human CB2 receptors (Ki = 0.60±0.13nM) and a 700-fold lower affinity for both the rat brain and cloned human CB1 receptors.13 CB2 receptor-transfected Chinese hamster cells exhibit high constitutive activity. This activity can be blocked by 1, working as an inverse agonist.14 Additional CB2 inverse agonists/antagonists include JTE-907,15 AM630, 16 SCH336 17, 18 and quinoline-3-carboxamide antagonists 19 (for a review see 20).
Given the therapeutic promise of 1 and other CB2 antagonists, it is surprising that the binding site for antagonists like 1 has not been more fully explored. We report here that Glide docking studies in our refined CB2 inactive state model suggest that 1 binds in the transmembrane helix (TMH) 1-2-3-5-6-7 region of CB2 with hydrogen bonding occurring between the amide hydrogen and EC-3 loop residue, D275 and no hydrogen bonding interaction for the central ring pyrazole N-1 nitrogen. To test this binding mode, we present the synthesis and evaluation of two sets of analogs. The first set was designed to test the importance of the hydrogen bonding of 1 at CB2 and the second set was designed to test the steric properties of the binding pocket. We show here that the amide functional group is crucial for the binding affinity and ability of 1 to block CB2 signaling, but that the N-1 pyrazole nitrogen is not important. We also show that the fenchyl ring positions the ligand amide functional group of 1 for productive receptor interaction. In addition, we show that shortening the N-2 benzyl substituent on the pyrazole ring of 1 to phenyl affects binding to the aromatic cluster in the TMH5/6 region, as well as, a loss in hydrogen bonding that results in a decrease in CB2 affinity and loss of antagonist activity at a concentration of 1 µM.
RESULTS
Modeling
The design of analogs presented here was based on docking studies of 1 in our CB2 receptor inactive state model that has been refined in an explicit lipid bilayer environment.21
Conformer analysis
The global minimum energy conformer of 1 has the amide co-planar with the pyrazole ring (N1-C2-C3-N4 = −2.02°) and the plane defined by the widest dimension of the fenchyl ring is almost perpendicular to the amide group (C3-N4-C5-H6 = 12.15°). The chloromethylphenyl ring is out of plane with the pyrazole (N2’-C3’’-C4’’-C5’’ = 57.27°). The methylbenzyl ring is attached to the pyrazole with a methylene group and its axis is perpendicular to the pyrazole plane (N1-N2’-C3’-C4’ = 90.34° and N2’-C3’-C4’-C5’ = −37.39°).
The global minimum energy conformer of 2 has the trans-ethylene group co-planar with the pyrazole ring (N1-C2-C3-C4 = −178.69°) and the plane defined by the widest dimension of the fenchyl ring is almost perpendicular to the trans-ethylene group (C3-N4-C5-H6 = −4.24°). The chloromethylphenyl ring is out of plane with the pyrazole (N2’-C3’’-C4’’-C5’’ = 56.19°). The methylbenzyl ring is attached to the pyrazole with a methylene group and its axis is perpendicular to the pyrazole plane (N1-N2’-C3’-C4’ = 89.36° and N2’-C3’-C4’-C5’ = −39.66°). Figure S1(A) illustrates an overlay of 2 with 1 which shows that the transethylene substituted analog, 2, has the same overall conformation as 1.
The global minimum energy conformer of 3 has the amide co-planar with the pyrrole ring (C1-C2-C3-N4 = 173.88°) and the plane defined by the widest dimension of the fenchyl ring is out of plane compared to the amide group (C3-N4-C5-H6 = −30.81°). The chloromethylphenyl ring is out of plane with the pyrrole (N2’-C3’’-C4’’-C5’’ = −55.03°). The methylbenzyl ring is attached to the pyrrole with a methylene group and its axis is perpendicular to the pyrrole plane (C1-N2’-C3’-C4’ = −86.39° and N2’-C3’-C4’-C5’ = 10.61°).
The global minimum energy conformer of 4 has the trans-ethylene group co-planar with the pyrrole ring (C1-C2-C3-C4 = −179.84°) and the plane defined by the widest dimension of the fenchyl ring is almost perpendicular to the trans-ethylene group (C3-C4-C5-H6 = −4.00°). The chloromethylphenyl ring is out of plane with the pyrrole (N2’-C3’’-C4’’-C5’’ = 51.73°). The methylbenzyl ring is attached to the pyrrole with a methylene group and its axis is perpendicular to the pyrrole plane (C1-N2’-C3’-C4’ = 80.52° and N2’-C3’-C4’-C5’ = 3.85°). Figure S1(B) illustrates an overlay of 4 with 1 which shows that the transethylene substituted analog, 4, has the same overall conformation as 1.
The structure of 5 is almost identical to that of 1. The only difference is that 1 has a fenchyl amide group attached to the pyrazole and 5 has a bornyl amide group. The global minimum energy conformer of 5 has the amide planar with the pyrazole ring (N1-C2-C3-N4 = − 0.16°) and the plane defined by the wide dimension of the bornyl ring is at an angle to the amide group (C3-N4-C5-H6 = 19.85°). The chloromethylphenyl ring is out of plane with the pyrazole (N2’-C3’’-C4’’-C5’’ = 57.01°). The methylbenzyl ring is attached to the pyrazole with a methylene group and its axis is perpendicular to the pyrazole plane (N1-N2’-C3’-C4’ = 90.41° and N2’-C3’-C4’-C5’ = −36.42°).
The global minimum energy conformer of 6 has the amide co-planar with the pyrazole ring (N1-C2-C3-N4 = −1.15°) and the plane defined by the widest dimension of the fenchyl ring is almost perpendicular to the amide group (C3-N4-C5-H6 = 12.37°). The chloromethylphenyl and the methylbenzyl rings are out of plane with the pyrazole (N2’-C3’’-C4’’-C5’’ = −133.25° and N1-N2’-C3’-C4’ = 53.42°).
Molecular Electrostatic Potential Maps for 1-6
Figure 1 illustrates the molecular electrostatic potential maps (ranges in kJ/mol given next to each ligand) of the docked conformations of 1–6 at CB2 R. These ligands have very similar shapes exposing the nitrogen of the pyrazole, or the CH that replaced it in 3 and 4, and the NH of the amide group, or the CH of the trans-ethylene group in 2 and 4, on the same face. The NH of the amide is an electropositive hot spot (blue) for 1 and analogs 3, 5 and 6. Compound 3 which has a pyrrole central ring (postion 1 has a carbon instead of a nitrogen) instead of a pyrazole, has also an electropositive region (blue) where the C-1 carbon is located that is broader than that seen for 1. Compounds 2 and 4 do not have an amide group, but rather a trans-ethylene moiety. Compound 2 has a weak electonegative region (yellow/orange) corresponding to the pyrazole nitrogen and the carbon C-4 of the trans-ethylene group is almost neutral. Compound 4, which has a pyrrole ring instead of a pyrazole, shows an electropositive region (blue) where the C-1 carbon is located. Also, the C-4 carbon of the trans-ethylene group appears to be very mildly electropositive (cyan). The range of the electrostatic potential has been normalized to be the same for all of the analogs and is given in kJ/mol.
Figure 1.
The molecular electrostatic potential maps of the docked conformations of compounds 1–6 are illustrated here. The electrostatic potential scale (in kJ/mol) is provided as a color scale. This scale is from blue (most electropositive) to red (most electronegative).
Ligand Docking
Glide docking studies of 1 in our CB2 inactive state model suggested that 1 spans the CB2 binding pocket and uses the EC-3 loop residue, D275 as its primary interaction site. These results are consistent with CB2 mutation and modeling studies (see Discussion section).
Figure 2 illustrates the final compound 1/CB2 R complex (see Table S1). The primary interaction is a hydrogen bond formed between the amide NH of 1 and the amino acid D275. The hydrogen bond heteroatom distance (N—O) and hydrogen bond N—H– –O) angle are 3.11Å and 161° respectively. Compound 1 forms a number of aromatic stacking interactions at CB2. The chloromethylphenyl ring forms an offset parallel stack with W6.48(258) with ring centroid to ring centroid distance of 4.53Å and also forms a T-stack with W5.43(194) with ring centroid to ring centroid distance of 5.97Å and the angle between the planes at 90°. Additionally, the methylbenzyl ring forms a tilted-T aromatic stack with W5.43(194) also. The ring centroid to ring centroid distance is 6.02Å and the angle between the ring planes is 48°. The conformational energy expense for 1 was 4.11 kcal/mol and the total pairwise interaction energy for 1 with CB2 is −71.70 kcal/mol (see Table S1). The major contributions to this interaction (greater than −5.00 kcal/mol) come from the hydrogen bonding interaction with D275, the favorable electrostatic and Van der Waals interactions with K3.28(109), the aromatic stacking interaction with W6.48(258), and the Van der Waals interaction with F2.57(87). Van der Waals interactions with the amino acids V3.32(113), M6.55(265), L6.54(264), M7.40(286) and V6.51(261) and the aromatic stacking interaction with W5.43(194) contribute between −5.00 and −3.00 kcal/mol to the interaction energy. The amino acids S7.39(285), S6.58(268), L6.52(262), F2.61(91), Q276, and I3.29(110) also contribute smaller amounts.
Figure 2.
This figure illustrates the final Compound 1/CB2 R complex. The view is from the lipid bilayer, with TMH6 and TMH7 closest to the viewer. The EC-3 loop residue, D2.75 (cyan) has been used as the primary interaction site for the most electropositive region of 1 (amide hydrogen). Residues displayed in orange are those for which the ligand interaction energy exceeded −3.50 kcal/mol (i.e., Int E<−3.50 kcal/mol). Additional interactions are present in the docked complex. Details of all interactions are provided in Tables S1 in Supporting Information.
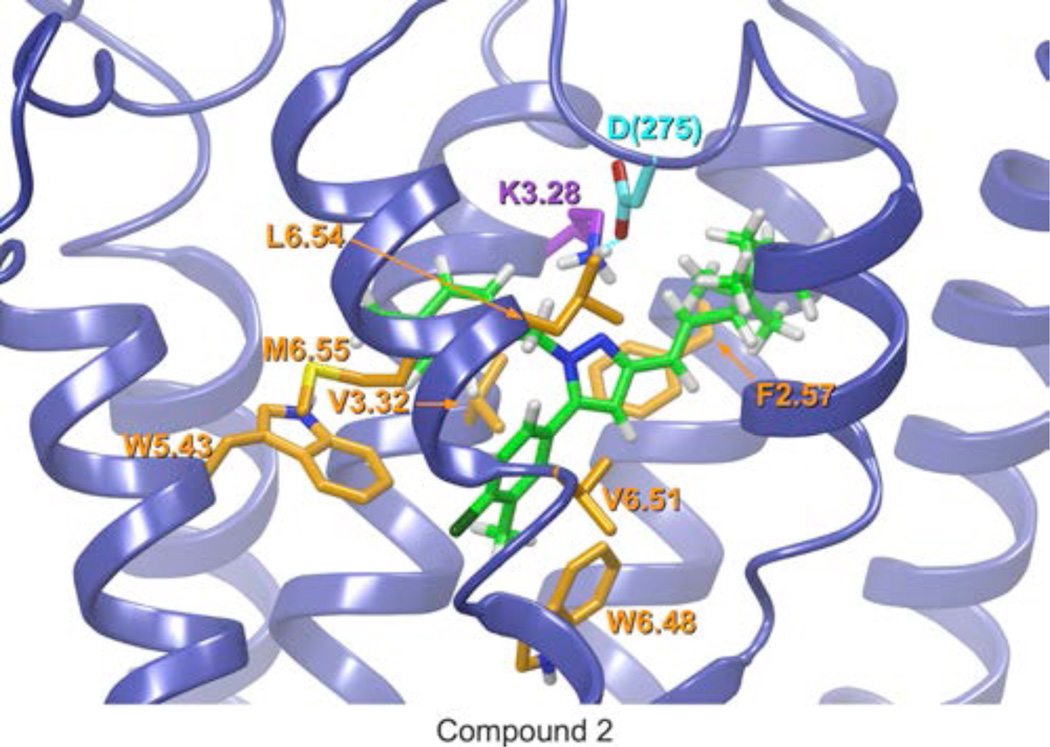
Figure 3 illustrates the final docked Compound 2/CB2 R complex (see Table S2). In Compound 2, the amide group has been replaced with a trans-ethylene group to mimic the geometry of the amide group in 1. Like 1, Glide docking studies reveal that 2 forms a number of aromatic stacking interactions. The chloromethylphenyl ring forms a offset parallel aromatic stack with W6.48(258). The ring centroid to centroid distance is 4.53Å and 5.74Å for the 6-membered and 5-membered rings of the tryptophan respectively. Additionally, the chloromethylphenyl ring forms an aromatic T-stack with W5.43(194). The ring centroid to centroid distance is 5.79Å (6-membered ring) and an angle of 84° between ring planes. Finally, the methylbenzyl ring forms a T-stack with W5.43(194) (ring centroid to centroid distance of 5.89Å and 6.32Å for the 6- and 5-membered rings respectively and an angle of 53° between ring planes). The conformational energy expense for 2 was 4.71 kcal/mol and the total interaction energy for 2 with CB2 is −63.96 kcal/mol. The highest interaction energy contributors (greater than −5.00 kcal/mol) for this complex are electrostatic and Van der Waals interactions with K3.28(109), aromatic stacking interactions with W6.48(258) and Van der Waals interactions with V3.32(113). M6.55(265), L6.54(264), F2.57(87), V6.51(261), and M7.40(286) contribute moderate Van der Waals and W5.43(194) contributes moderate aromatic energies of interaction between −5.00 and −3.00 kcal/mol. D275, S6.58(268), S7.39(285), Q276, F2.61(91), and I3.29(110) contribute smaller amounts of Van der Waals interaction energy.
Figure 3.
This figure illustrates the final Compound 2/CB2 R complex. In Compound 2, the amide group has been replaced with a trans-ethylene group to mimic the geometry of amide group in 1. The view is from the lipid bilayer, with TMH6 and TMH7 closest to the viewer. Glide docking studies reveal that 2 has highest electrostatic and Van der Waals interactions with K3.28(109) (magenta). Residues displayed in orange are those for which the ligand interaction energy exceeded −3.50 kcal/mol (i.e., Int E<−3.50 kcal/mol). Additional interactions are present in the docked complex. Details of all interactions are provided in Tables S2 in Supporting Information.
Figure 4 illustrates the final Compound 3/CB2 R complex (see Table S3). Compound 3 has a pyrrole central ring instead of a pyrazole so it has extra steric bulk where the N is substituted with a C-H moiety. The amide group of 3 forms a hydrogen bond with D275 with heteroatom to heteroatom distance (N—O) of 3.20Å and an angle (N—H– –O) of 158°. Compound 3 forms very similar aromatic stacking interactions to those formed by 1, 2 and 5 (see below). The chloromethylphenyl ring forms a parallel stack with W6.48(258). The ring centroid to centroid distances with the 6- and 5-membered rings of the W6.48(258) are 4.53Å and 5.74Å respectively. Also, the chloromethylphenyl ring forms a T-stack with W5.43(194); the ring centroid to centroid distance is 5.67Å and the angle between the ring planes is 84°. Lastly, the methylbenzyl ring of 3 has an aromatic T-stack interaction with W5.43(194); the ring centroid to centroid distance for the 6- and 5-membered rings are 5.89Å and 6.32Å respectively and the angle between the ring planes is 53°. The conformational energy expense for 3 was 3.79 kcal/mol and the total interaction energy between 3 and CB2 is −70.98 kcal/mol. D275, which forms a hydrogen bond with 3, is the single highest contributor of interaction energy followed by aromatic stacking contribution of W6.48(258) and the Van der Waals interactions with F2.57(87). The residues V3.32(113), M6.55(265), L6.54(264), M7.40(286), and V6.51(261) contribute significant amount of Van der Waals interaction energy along with the aromatic stacking interaction contributed by W5.43(194). Finally, the residues S6.58(268), S7.39(285), L6.52(262), F2.61(91), Q276 and I3.29(110) contribute to the interaction energy to a lesser extent.
Figure 4.
This figure illustrates the final Compound 3/CB2 R complex. Compound 3 has a pyrrole central ring instead of a pyrazole so it has extra steric bulk where the N is substituted with CH moiety. The view is from the lipid bilayer, with TMH6 and TMH7 closest to the viewer. Here the amide group of 3 forms a hydrogen bond with D275 (orange). Residues displayed in orange are those for which the ligand interaction energy exceeded −3.50 kcal/mol (i.e., Int E<−3.50 kcal/mol). Additional interactions are present in the docked complex. Details of all interactions are provided in Tables S3 in Supporting Information.
Figure 5 illustrates the final Compound 4/CB2 R complex (see Table S4). Compound 4 has no hydrogen bonding capability because it lacks both the amide group and has a pyrrole instead of a pyrazole central ring. However, 4 has all the aromatic stacking interactions found for 1, 2, 3 and 5 (see below). The chloromethylphenyl ring forms a parallel aromatic stack with W6.48(258); the ring centroid to ring centroid distance from the 6- and 5-membered rings of the W6.48(258) are 4.33Å and 5.51Å respectively. In addition, the chloromethylphenyl ring of 4 forms a T-stack with W5.43(194). The ring centroid to ring centroid distance is 5.77Å (6-membered ring) and the angle between the planes is 82°. Finally, the methylbenzyl ring forms an aromatic T-stack with W5.43(194) with ring centroid to centroid distance of 5.84Å and 6.29Å (6- and 5-membered ring respectively) and an angle of 55° between the ring planes. The conformational energy expense for 4 was 3.91 kcal/mol and the total energy of interaction between 4 and CB2 is −64.30 kcal/mol. The major contributors to the energy of interaction are the electrostatic and Van der Waals interaction with D275, the aromatic stacking interaction with W6.48(258) and the Van der Waals interaction with V3.32(113). The amino acids M6.55(265), L6.54(264), V6.51(261), F2.57(87), and M7.40(286) have significant Van der Waals energy contributions, as well as the aromatic stacking interaction with W5.43(194). Lastly, S6.58(268), L6.52(262), S7.39(285), Q276 and F2.61(91) have small but important Van der Waals energy contributions.
Figure 5.
This figure illustrates the final Compound 4/CB2 R complex. The view is from the lipid bilayer, with TMH6 and TMH7 closest to the viewer. Compound 4 has no hydrogen bonding capability because it lacks both the amide group and has a pyrrole instead of a pyrazole central ring. However, 4 has all the aromatic stacking interactions found for 1, 2, 3 and 5. Residues displayed in orange are those for which the ligand interaction energy exceeded −3.50 kcal/mol (i.e., Int E<−3.50 kcal/mol). Additional interactions are present in the docked complex. Details of all interactions are provided in Tables S4 in Supporting Information.
Figure 6 illustrates the final Compound 5/CB2 R complex (see Table S5). The main interaction energy contributor in the complex is D275 which forms a hydrogen bond with the amide group of 5. The hydrogen bond distance (N—O) and hydrogen bond (N—H– –O) angle are 3.26Å and 163° respectively. The aromatic rings of 5 are positioned in a very similar orientation as those in 1. The Compound 5 chloromethylphenyl ring forms an offset parallel aromatic stack with W6.48(258) with a ring centroid to ring centroid distance of 4.39Å with the 6-member ring and 5.61Å with the 5-member ring. The chloromethylphenyl group is also forming an aromatic T-stack with W5.43(194) (6-member ring) with a ring centroid to ring centroid distance of 5.67Å and a ring plane to plane angle of 84°. Finally, the methylbenzyl ring forms an aromatic T-stack interaction with W5.43(194). The ring centroid to centroid distance is 5.94Å with the 6-member ring and 6.51Å with the 5-member ring of the W5.43(194) and the ring plane to ring plane angle is 48°. The conformational energy expense for 5 was 3.58 kcal/mol and the total pairwise interaction energy for 5 with CB2 R was found to be −71.00 kcal/mol. The major interaction energy contributors to the complex are the electrostatic interaction with D275, the aromatic stacking interaction with W6.48(258), and the Van der Waals interactions with F2.57(87) and V3.32(113). The amino acids M6.55(265), L6.54(264), W5.43(194), S7.39(285), V6.51(261), and K3.28(109) also contribute a significant amount of Van der Waals interaction energy with M7.40(286), S6.58(268), F2.61(91), Q276, A7.36(282), L6.52(262) and I3.29(110) contributing to a lesser degree.
Figure 6.
This figure illustrates the final Compound 5/CB2 R complex. The view is from the lipid bilayer, with TMH6 and TMH7 closest to the viewer. Compound 5 has a bornyl ring substituted for the fenchyl ring of 1. The main interaction energy contributor in the complex is D275 which forms a hydrogen bond with the amide group of 5. Residues displayed in orange are those for which the ligand interaction energy exceeded −3.50 kcal/mol (i.e., Int E<−3.50 kcal/mol). Additional interactions are present in the docked complex. Details of all interactions are provided in Tables S5 in Supporting Information.
Figure 7 illustrates the final Compound 6/CB2 R complex (see Table S6). The lack of the methyl extension of the methyl phenyl ring causes a repositioning of 6 in the binding pocket such that the amide group of 6 is close enough to D275 to have favorable electrostatic interaction with it, but not close enough to form a hydrogen bond (N—O distance of 3.83Å). Compound 6 forms primarily aromatic stacking interactions with CB2. The central pyrazole ring forms an offset aromatic T-stack with F2.57(87). The ring centroid to ring centroid distance is 4.57Å and the angle between the ring planes is 39°. The chloromethylphenyl ring forms an offset parallel aromatic stack with W6.48(258) like 1 and the other analogs. The ring centroid to centroid distance is 4.66 Å and 5.95 Å with the 6- and 5-membered ring respectively. The conformational energy expense for 6 was 0.35 kcal/mol and the total interaction energy for the Compound 6/CB2 complex is −66.57 kcal/mol. The major contributors to the interaction energy with CB2 are the aromatic stacking interaction with F2.57(87), the electrostatic interaction with D275, aromatic stacking interaction with W6.48(258) and electrostatic and van der Waals interaction with K3.28(109) and M7.40(286). The residues V6.51(261), L6.54(264), M6.55(265), S7.39(285) and V3.32(113) have significant energy contributions. Finally, the amino acids F2.61(91) and Q276 add a small amount of interaction energy to the complex.
Figure 7.
This figure illustrates the final Compound 6/CB2 R complex. The view is from the lipid bilayer, with TMH6 and TMH7 closest to the viewer. The lack of the methyl extension of the methyl phenyl ring causes a repositioning of 6 in the binding pocket such that the amide group of 6 is close enough to D275 to have favorable electrostatic interaction with it, but not close enough to form a hydrogen bond. Residues displayed in orange are those for which the ligand interaction energy exceeded −3.50 kcal/mol (i.e., Int E<−3.50 kcal/mol). Additional interactions are present in the docked complex. Details of all interactions are provided in Tables S6 in Supporting Information.
Molecular Dynamics
Energy minimization of a complex provides the lowest energy structure for the complex, but this cannot provide dynamic information about this complex at biological temperature. Molecular dynamics (MD) simulations provide a snapshot of how the complex structure may fluctuate when kinetic energy is provided to the system. The receptor-ligand complexes presented in the previous section, therefore, were further studied using molecular dynamics (MD), with trajectory lengths of 22.5ns each. These trajectories were for the ligand/receptor complex itself in the absence of explicit lipid (see Methods section). The stability of the receptor (including sidechains) and ligand were monitored via RMSD plots. These plots (see Supporting Information, Figures S-3 and S-4) indicate that the RMSDs of receptor and ligand were stable over the 22.5 ns trajectories for compounds 1 to 5. For compound 6, the ligand showed a change in RMSD early in the trajectory and then leveled off, while the RMSD of the receptor remained stable. The origin of this change seen for Compound 6 will be discussed below. During the MD studies, we also monitored the position and interactions of each ligand in complex with CB2 for the occurrence of hydrogen bonds between each ligand and CB2 binding pocket residues. Only two residues have the potential for hydrogen bonding in the binding pocket: (1) The EC-3 residue, D(275), could serve as a hydrogen bond acceptor; and (2) K3.28 could potentially act as a hydrogen bond donor. Because of their proximity, D(275) and K3.28 can also form a salt bridge with each other and be unavailable for ligand hydrogen bonding. Results from these studies are summarized in Table 1. These results shed further light on each of the complexes discussed above. Compound 1 exhibited hydrogen bonding for 93.7% of the simulation. This hydrogen bond was between D(275) and the amide hydrogen of 1. During those periods when the hydrogen bond is not formed with 1, D(275) hydrogen bonds with K3.28. Although K3.28 is near the pyrazole nitrogen during part of the simulation, it cannot hydrogen bond with this nitrogen due to the poor geometry for a Nsp3-H…Nsp2 hydrogen bond.
Table 1.
Ki values of Compound 1 and Analogs for hCB2 vs [3H]-CP 55,940 Compared to Calculated Interaction Energies for Compound/CB2 Complexes
| Compound | Ki (nM) | SEM (n=3) | Int E (kcal/mol) | D(275 ) to Ligand Amide NH Hydrogen Bond (%)* |
|---|---|---|---|---|
| 1 | 2.23 | 0.31 | −71.70 | 93.7 |
| 2 | 527 | 126 | −63.96 | NA |
| 3 | 15.3 | 2.7 | −70.98 | 67.2 |
| 4 | 1220 | 38.1 | −64.30 | NA |
| 5 | 29.1 | 7.2 | −71.00 | 67.3 |
| 6 | 94.2 | 9.5 | −66.57 | 1.0 |
Percentage of time during 22.5 ns MD simulation that each ligand was hydrogen bonded to D(275). No hydrogen bonds with other residues were observed during these trajectories.
In Compound 2, a transethylene replaces the amide group. This region of 2, therefore, is incapable of hydrogen bonding. Compound 2 does retain the pyrazole nitrogen. However, K3.28 interacts with D(275) during the entire simulation and is never available for other hydrogen bonding interactions. This is likely due to the fact that the ligand sits lower in the binding pocket than 1 because there is no D(275)-amide hydrogen bond to hold the ligand closer to the EC-3 loop.
Compound 3 retains the amide functional group, but has no pyrazole nitrogen. In the removal of the pyrazole nitrogen, it was replaced with a C-H. The hydrogen of the C-H is pointed towards the K3.28/D(275) pair and therefore causes the ligand to sit slightly lower in the binding pocket to avoid steric overlap. The increased distance of the amide hydrogen from D(275) results in a decreased percentage of hydrogen bonding over the length of the trajectory (67.2% compared to 93.7% for 1). This decrease in hydrogen bonding correlates well with the increased Ki of 3.
Compound 4 has the amide functionality replaced by a transethylene group and the pyrazole nitrogen replaced by a C-H. This compound is incapable of forming hydrogen bonds. It’s significantly lowered affinity (1220nM) is consistent with this.
In Compound 5, the fenchyl ring is replaced with a bornyl ring. The protruding methyl groups of the bornyl ring cause the ligand to shift in the pocket in two directions: (1) towards TMH2/TMH3 and (2) towards TMH5. This shift displaces the ligand away from D(275) and results in the lowered incidence of hydrogen bond formation with this residue over the length of the trajectory (67.3% vs. 93.7% for 1). Although K3.28 comes near the pyrazole nitrogen during part of the simulation, it cannot hydrogen bond with this nitrogen due to the poor geometry for a Nsp3-H…Nsp2 hydrogen bond. The reduced incidence of the D(275)/amide N-H hydrogen bond for 5 is consistent with its increase in Ki. relative to 1.
RMSD plots and Torsion Angle traces for 6 during the MD trajectory indicate that 6 adjusts the position of the fenchyl ring via rotation of its ω4 torsion angle by 90 ° early in the simulation (~4ns), with this angle maintained at this new value for the rest of the simulation (See Figures S-3 and S-4). Compound 6 is missing the methylene spacer in the phenyl substituent. The loss of this methylene group places the phenyl ring in a position in which the ligand must move slightly (~1 Å) towards TMH1. More importantly, the phenyl ring’s proximity to the amide group restricts the accessibility of D(275) for the amide hydrogen. This is consistent with the lowered percentage of hydrogen bonds formed (1.0% compared to 93.7% for 1) and with its large increase in Ki. relative to 1.
Synthesis
5-(4-Chloro-3-methylphenyl)-1-(4-methyl-benzyl)-3-[(E)-2-endo-(1,3,3-trimethyl-bicyclo[2.2.1]hept-2-yl)-vinyl]-1H-pyrazole, 2
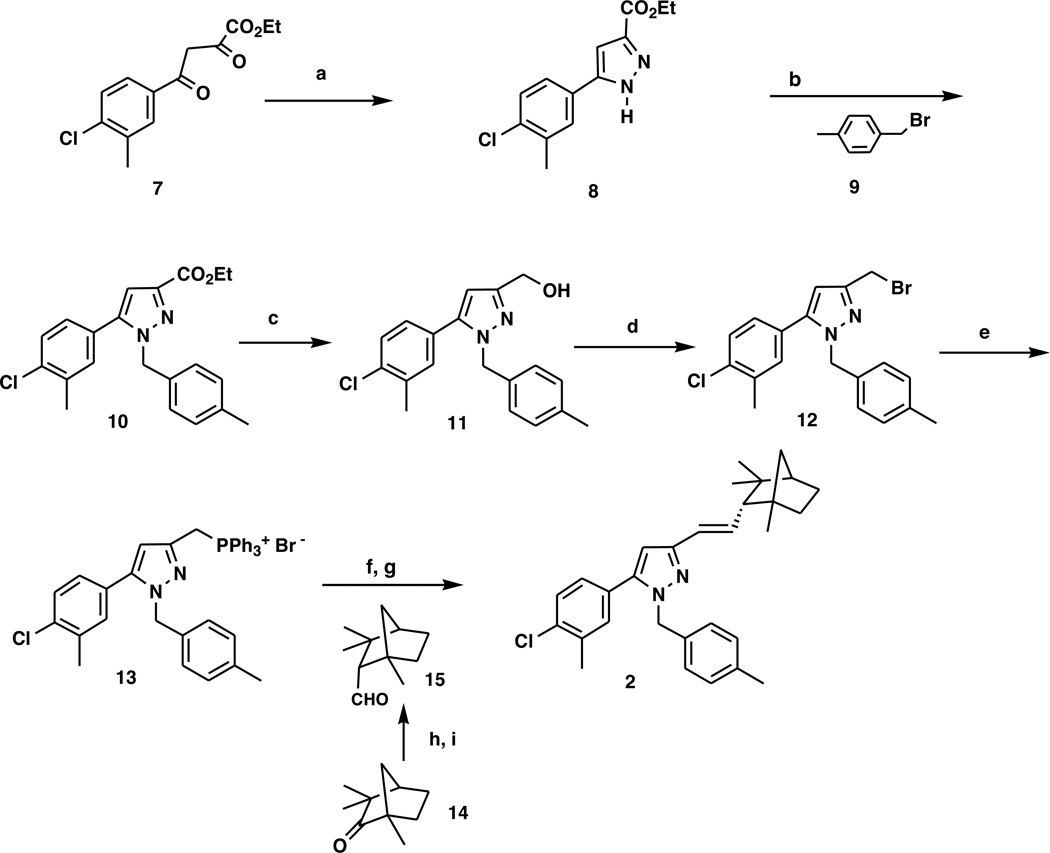
The target 2 replaces the fenchyl amide of the CB2 antagonist 1 with a trans vinyl fenchyl moiety that mimics the transamide conformation of the latter but without heteroatoms in that linkage, thus precluding H-bonding. Its synthesis started with cyclization of butanoate 7 25 with hydrazine provided a 49% chromatographed yield of the pyrazole 8. Benzylation of 8 with 4-methylbenzyl bromide (9) provided a 52% chromatographed yield of the 1,5 disubstituted pyrazole ester 10 25, 26. The 1,5-substitution was determined by HMBC NMR spectroscopy showing a correlation of the pyrazole C-5 carbon with the benzyl protons at N-1. The 1,3-isomer was not observed chromatographically or spectroscopically. Reduction of the ester 10 moiety to the benzylic alcohol 11 in 85% chromatographed yield followed by conversion of the alcohol to the benzylic bromide 12 with PBr3 in 97% chromatographed yield. Treatment of 12 with triphenylphosphine formed the phosphonium salt 13 in 89% yield. Deprotonation of the latter with n-Butyllithium afforded the corresponding ylide, which was coupled with endo-fenchylcarboxaldehyde (15) 27 in a Wittig olefination to yield the target compound 2 (see Chart 2).
Chart 2.
a) H2NNH2, HOAc ; b) NaH; c) LiAlH4; d) PBr3; e) Ph3P; f) nBuLi; g) 15; h) Ph2POCH2OCH3, LDA; i) Cl3C-CO2H, (85:15 endo:exo);
The structure of 2 was determined by 1H NMR, CMR, MS, and 2D-NMR. The preferred E-olefin from stabilized ylides 28 was confirmed by the trans coupling constant of 15.5 Hz of the vinyl protons and the absence of a ROESY interaction for the vinyl protons while a COSY interaction was observed, consistent with a trans geometry. Further, a second olefin isomer was obtained which did exhibit a ROESY interaction of the vinyl protons indicative of the cis geometry and further validating the trans assignment based on the absence of an interaction. The E-olefin geometry is in agreement with the previously synthesized vinylcyclohexyl analog of SR141716, 29 which was prepared via the same chemistry.
The endo configuration of the fenchyl group follows from the known endo configuration of the precursor aldehyde and the observed ROESY interaction of the fenchyl C2-H with the apical syn C7-H, which is only possible with the exo C2-H (hence an endo vinyl group). Additionally, the exo C2-H is identified by its observed interaction with two exo methyls (at C1 and C3); if the C2-H were endo, it would interact with only one methyl (endo at C3). The 1,5-substituted pyrazole assignment was determined by HMBC NMR spectroscopy showing a correlation of the pyrazole C-5 carbon with the benzyl protons at N-1. Target 2 derives from the intermediate 10 with similarly proven geometry. The 1,3-isomer was not observed chromatographically or spectroscopically.
5-(4-Chloro-3-methyl-phenyl)-1-(4-methyl-benzyl)-1H-pyrrole-3-carboxylic acid (1,3,3-trimethyl-bicyclo[2.2.1]hept-2-yl)-amide, 3
Compound 3 is a pyrrole analog of 1 wherein the N-2 pyrazole nitrogen of 1 was replaced with carbon. The pyrrole intermediate 22 was prepared by the cycloaddition reaction between ethyl acrylate 21 and an α-tosylbenzylisocyanide 20. 30, 31 Thus, 4-chloro-3-methylbenzaldehyde 16 was condensed with p-toluenesulfinic acid (17) and formamide (18) in the presence of formic acid to afford the corresponding N-(α-tosyl-4-chloro-3-methylbenzyl)formamide 19 as outlined in Chart 3. 30, 32 Dehydration of 19 afforded the substituted tosybenzylisocyanide 20. A 3 + 2 cycloaddition of 20 with ethyl acrylate 21, initiated with sodium hydride, yielded the 3,5-disubstituted pyrrole ester 22 in close parallel with the unsubstituted phenyl analog. 30 Benzylating the pyrrole nitrogen 33 with 4-methylbenzyl bromide 9 yielded the intermediate 23 common to both 3 and 4 syntheses.
Chart 3.
a) HCO2H; b) POCl3; c) NaH; d) ethyl acrylate, 21; e) 9; f) KOH, aq MeOH; g) SOCl2; h) (IS)-endo-fenchylamine, 24
Continuing to follow the pyrazole synthesis of 5, saponification, activation of the acid as the acid chloride with thionyl chloride, and treatment with endo-fenchylamine 24 yielded 3. The structure of 3 is supported by mass spectrometry and by 1H NMR of 22, 23 and 3 which exhibit the pyrrole aromatic hydrogens with a small coupling constant (d, J = 1.7 Hz) establishing the 2,4-substitution pattern, in agreement with similarly substituted pyrroles 34 and the mechanism for this reaction. 30 The endo-configuration of the fenchylamine establishes the endo-configuration of the corresponding amide 3.
5-(4-Chloro-3-methylphenyl)-1-[(4-methylphenyl)methyl]-3-[(E)-2-[(1S,2S,4R)-1,3,3-trimethylbicyclo[2.2.1]heptan-2-yl]ethenyl]-1H-pyrrole, 4
The 3-trans-vinyl fenchyl pyrrole analog 4 replaces both the N-2 pyrazole nitrogen and the amide heteroatoms of 1 with carbons or hydrogen. While the vinyl group maintains the preferred trans conformation of the amide, it lacks the hydrogen bonding capability of the amide O and N. The synthetic approach with which we sought to transform the pyrrole ester 23 to the target vinylfenchyl compound 4 examined the same Wittig chemistry that was successful with the pyrazole analog 10 in the synthesis of 2. Thus, the ester 23 was reduced with LiAlH4 to the alcohol 25 (Chart 4). Bromination of 25 with carbon tetrabromide and triphenylphosphine 35 or with PBr3, however, failed to afford the benzylic bromide 26 (Table G), affording instead extensive degradation suggesting an instability of 26. The 4-methyl benzyl N-substituent was changed to an electron withdrawing p-toluenesulfonyl (tosyl) protecting group by treating the pyrrole 22 with tosyl chloride and NaH to obtain 27 (80%). The latter ester was reduced with LiAlH4 to provide the alcohol 28 (78%). Successful conversion of the alcohol to the bromide 29 with PBr3 (90%) was in marked contrast to the results with 25. The bromide was carried onto the triphenylphosphonium salt 30 (85%). Treatment of 30 with nBuLi, potassium t-butoxide, or KOH/18-crown-6 ether for deprotonation and endo-2-fenchanecarboxaldehyde 15 27 did not yield 31 nor give an olefin, even though deprotonation with nBuLi was confirmed by quenching with MeOD.
Chart 4.
a) LiAlH4, THF; b) CBr4, Ph3P; c) Dess-Martin periodinane, CH2Cl2, r.t.; d) G6, nBuLi, THF; e) MgBr2•Et2O, dioxane, 100 C; f) Ph2POCH2OCH3, LDA; g) Cl3C-CO2H, (85:15 endo:exo); h) (MeO)2P(O)H, LiHMDSi, THF, −78 C; i) S=C-im2, THF, 50 C; j) Bu3SnH, AIBN
Returning to the N-(4-methylbenzyl) pyrrole 25, the ylide and carbonyl roles of the pyrrole and fenchyl intermediates in the olefination were reversed. Thus, the pyrrole 25 was oxidized to the aldehyde 32 with the Dess-Martin periodinane reagent in 38% yield. Correspondingly, endo-2-fenchanecarboxaldehyde 15 was converted to the Horner-Wadsworth-Emmons ylide precursor 34 in three steps: 1) LiHMDS promoted addition of dimethylphosphite to the aldehyde 15 affording 33 (83%); 36 2) acylation of the resulting carbinol with thiocarbonyl diimidazole to yield the corresponding thiocarbonylimidazolide (56%); and 3) reduction of the latter with tri-n-butyl tin hydride mediated by AIBN to give dimethylphosphonate 34 (51%). 37 Treatment of 34 with n-butyllithium and the aldehyde 32 gave the adduct 35, which partly eliminated to the olefin 4. Treatment of the residual 35 with MgBr2 and heating was expected to provide the corresponding phosphate monoester 38 but gave instead the target olefin 4. The combined 4 containing fractions were purified chromatographically (2.7%). The small scale for these reactions likely contributed to the low yields in the olefination sequence.
The purified and tested material was a single compound (HPLC, 1H NMR). The mass spectrum showed a positive ion of m/e 458 for the M + 1 ion. The 1H NMR spectrum supported the structure given for 4 with special note that the trans-olefin geometry is consistent with the vinyl proton-coupling constant of 15 Hz.
5-(4-Chloro-3-methylphenyl)-1-[(4-methylphenyl)methyl]-N-[(1S,2R,4S)-1,7,7-trimethylbicyclo[2.2.1]heptan-2-yl]-1H-pyrazole-3-carboxamide, 5
The target compound 5 is a 2-(−)-endo-bornyl amide analog of the CB2 antagonist (1) fenchyl amide. Its synthesis is an amalgam of that reported by Sanofi 26 and our approach developed for tritiated 1 25 (see Chart 5). Saponification of the ester 10 (prepared as described for compound 2) gave the corresponding acid in 85% yield, which was converted to the acid chloride and treated with (−)-endo-bornyl-2-amine39 to yield the endo-bornylamide 5 in 34% yield after chromatography and two recrystallizations. The 98% pure (HPLC) product was characterized by 1H NMR and MS. The (−)-endo-bornyl-2-amine 38 was determined to be 95% endo by 1H NMR spectroscopic observation of the resonances of the exo and endo proton alpha to the nitrogen. 39
Chart 5.
a) KOH, aq MeOH; b) SOCl2; c) 38; d) H2NOH; e) Na0
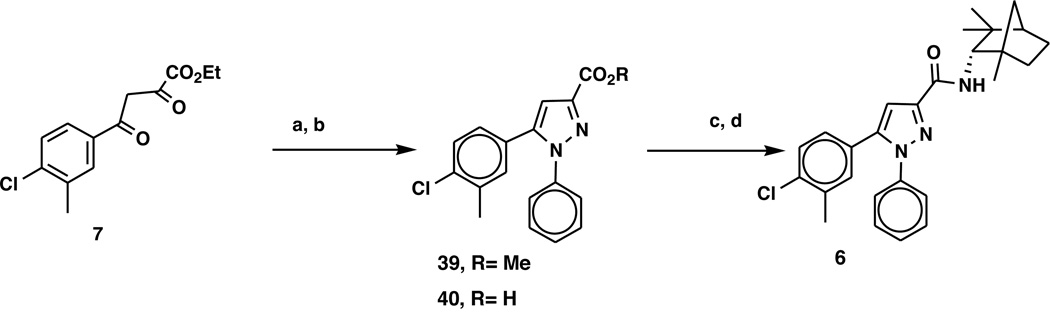
5-(4-Chloro-3-methylphenyl)-1-phenyl-N-{1,3,3-trimethylbicyclo[2.2.1]heptan-2-yl}-1H-pyrazole-3-carboxamide, 6
The target 6 deletes the 4-methyl and the benzyl methylene from the 4-methylbenzyl moiety of 1. Chart 6 illustrates the synthesis of 6. Cyclization and concomitant transesterification of the ethyl ester 7 with phenylhydrazine hydrochloride in methanol provided a 65% yield of the 1,5 disubstituted pyrazole methyl ester 40. While ROESY results for 40 were inconclusive regarding direct confirmation of 1,5-configuration due to insufficient separation of resonances, the 1,5 diaryl substitution was assigned on the basis of chemical shift of the single pyrazole Proton40 and the precedented strong preference for condensation of phenylhydrazines with 2,4-diketobutanoic acid esters to yield 1,5-diaryl-pyrazoles. 40–42 Saponification of 40 provided a 90% yield of the acid 41 that was converted to its acid chloride and which was then treated with (−)-fenchylamine 24 to yield the target 6 (62%) in >99% purity (HPLC) after chromatography. The structure of the compound was confirmed by 1H NMR, CMR, and mass spectrometry.
Chart 6.
a) Phenylhydrazine HCl, MeOH, b) NaOH, EtOH, c) SOCl2; d) (IS)-endo-fenchylamine, 24
Radioligand Binding Evaluation
Table 1 provides a summary of the binding affinities for compounds 1–6 vs. [3H]CP55940. This table indicates that the loss of the amide functionality in 1 (analog 2) results in a 236-fold loss of affinity compared to 1. In contrast, replacement of the pyrazole nitrogen with carbon (C-H; analog 3) results in only a 7-fold drop in affinity. Replacing both the amide group and the pyrazole nitrogen (analog 4) results in a 547-fold loss in CB2 affinity. Replacement of the the N-2 benzyl substituent with a phenyl group (analog 5) results in a 13-fold drop in affinity compared to 1, while replacing the fenchyl ring with the bornyl ring (analog 6) results in a 42-fold decrease in affinity. Values for the Interaction Energy (Int E) calculated for each compound/CB2 complex (See Tables S-1 to Tables S-6) are also provided in Table 1. As can be seen in Table 1, these values follow the same trend as the Ki values for each compound.
Effect of 1 and Analogs on CP-55,940-Induced Inhibition of cAMP Accumulation
In the current study, we investigated the antagonistic activities of 1 (CB2 selective antagonist) and five analogs (2–6). Table 2 summarizes the effect that 1 and each of the analogs (2–6) had on the potency of CP-55,940 in inhibiting cAMP accumulation. CP-55,940 concentration-dependently inhibited forskolin-stimulated cAMP accumulation in HEK293 cells stably expressing CB2 with an EC50 value of 4.21 ± 1.16 nM. 1 µM of 1 was found to antagonize the effects of CP-55,940. In the presence of 1, the EC50 value of CP-55,940 was significantly increased to 23.31 ± 5.94 nM. In the presence of 1 µM of 3, the EC50 value for CP-55,940 was found to be 42.62 ± 4.17 nM, which is significantly greater than both the EC50 values of CP-55,940 in the presence of vehicle or in the presence of 1. In the presence of 1 µM of 5, the EC50 value for CP-55,940 was found to be 17.19 ± 1.30 nM, which is not significantly different than the EC50 value for CP-55,940 in the presence 1. In the presence of 1 µM of 2, 4 and 6, the EC50 values for CP-55,940 were found to be 5.32 ± 1.760 nM, 11.56 ± 1.10 nM and 10.09 ± 1.89 nM,, respectively, which is significantly lower than the EC50 value for CP-55,940 in the presence of 1, and not significantly different than the EC50 value for CP-55,940 in the presence of vehicle.
Table 2.
Effect of Compound 1 and Analogs on CP-55,940-induced Inhibition of cAMP Accumulation
| Compound | CP-55,940 EC50 (Mean ± SEM), nM |
|---|---|
| Vehicle | 4.21 ± 1.16 |
| 1 | 23.31 ± 5.94* |
| 2 | 5.32 ± 1.76 |
| 3 | 42.62 ± 4.17* |
| 4 | 11.56 ± 1.10 |
| 5 | 17.19 ± 1.30* |
| 6 | 10.09 ± 1.89 |
Significantly different (P < 0.05) compared with vehicle. Values represent means ± S.E.M. of at least three experiments each performed in duplicate.
DISCUSSION
Compound 1 is a large ligand that modeling studies predict to span the entire CB2 binding pocket with fenchyl ring near TMH1/2/7, amide functionality near TMH3/7 and aromatic moieties near TMH3/5/6. The CB2 mutation literature has focused primarily on important functional residues, such as the DRY motif43, 44 in TMH3, W4.50,45 the ionic lock residue D6.30 in TMH6,46 and the NPXXY motif in TMH7.47 A few mutations have been reported that address CB2 binding pocket residues important for the binding of 1. These binding pocket studies are discussed below in the context of the hydrogen bonding, hydrophobic and aromatic stacking capabilities of the ligands reported here (1–6).
Primary Interaction Site at CB2
Glide docking studies reported here suggested that the EC-3 loop residue, D275 is the primary interaction site for 1 at CB2. In CB1, K3.28 has been shown to be the key ligand interaction site for endocannabinoids, classical and non-classical cannabinoid agonists,22, 48 as well as the CB1 antagonist, SR141716A.23 However, the K3.28A mutation in CB2 was found to have no effect on the binding or signaling of a panel of cannabinoid agonists (1 was not evaluated in this study).24 This suggests that some other residue must be the primary site of interaction for many CB2 ligands. In our microsecond timescale simulations of 2-AG entry and activation of CB2, the headgroup of 2-AG establishes a long standing interaction with EC-3 loop residue, D275.21 In our CB2 models (inactive and activated states), K3.28 is engaged in a salt bridge with this EC-3 loop residue. The interaction between D275 and K3.28 makes K3.28 less available for direct ligand binding. Thus, mutation studies suggest that K3.28 in CB2 may not be an important residue for ligand binding, while modeling and docking studies instead point to D275 as an important residue for ligand binding in both the inactive and activated states of CB2. If a negatively charged residue is the primary interaction site for 1 at CB2, then there should exist a concentration of positive electrostatic potential on 1 that would promote receptor recognition. The electrostatic potential map of 1 (see Figure 1) shows a concentrated region of positive electrostatic potential localized immediately adjacent to the fenchyl ring on the face of the molecule illustrated in Figure 1. This region corresponds to the amide hydrogen of 1.
To test the importance of this electropositive region in 1, we designed Compounds 2–4. In Compound 2, we replaced the amide functionality with an isosteric trans-ethylene group which preserves the molecular geometry of 1 (see Figure S1(A)), but eliminates the region of positive electrostatic potential associated with the amide hydrogen on the face of the molecule illustrated in Figure 1. (see 2). This substitution removes any possibility of the amide functionality contributing to hydrogen bonding with the receptor (either from the amide hydrogen or the carboxamide oxygen). We found that this change resulted in a 236-fold loss of binding affinity for 2 compared to 1 and the inability of Compound 2 (at a 1 µM concentration) to antagonize CP55940 (see Table 1).
In Compound 3, we replaced the pyrazole nitrogen (N-1) in the central ring of 1 with a carbon (C–H). This substitution eliminates any hydrogen bonding potential for the pyrazole (now a pyrrole ring) with the receptor. This change resulted in a modest change in Ki from 2.23nM for 1 to 15.3nM for 3 (see Table 1). Interestingly, the EC50 of CP55940 in the presence of 3 was significantly greater (P < 0.05) than the EC50 value of CP-55,940 in the presence of 1 (see Table 2), indicating 3 is a more potent CB2 antagonist than 1. Comparison of the electrostatic potential map of compound 3 with that of 1 (see Figure 1) indicates that the replacement of the pyrazole nitrogen with a C-H creates a broad positively charged electrostatic potential region (blue) between the fenchyl ring and the aromatic system of 1 on the molecular face displayed for 3 in Figure 1. This region should be more attractive to negatively charged amino acids, such as D275 in the EC-3 loop. This result is consistent with our Glide docking study, as the interaction energy of 3 with D275 was found to be −15.84 kcal/mol, while that of 1 with D275 was −9.73 kcal/mol (See Supporting Information, Tables S1 and S3). However, our MD study showed that the hydrogen of the C-H is pointed towards the K3.28/D(275) pair and therefore causes the ligand to sit slightly lower in the binding pocket to avoid steric overlap. The increased distance of the amide hydrogen from D(275) results in a decreased percentage of hydrogen bonding over the length of the trajectory (67.2% compared to 93.7% for 1). Thus two opposing effects, larger positive MEP for interaction balanced by the lower position of the ligand in the binding pocket (caused by C-H introduction) and concomitant decrease in hydrogen bonding, likely result in a Ki that is higher (6.8-fold) than the Ki of 1.
In Compound 4, we combined the modifications seen in 2 and 3 into the same molecule, i.e. replacement of the amide group with a trans-ethylene group and replacement of the pyrazole N-1 nitrogen with C-H. The electrostatic potential map of 4 shows that the region between the fenchyl ring and the aromatic system on the face of this molecule illustrated in Figure 1 is essentially neutral. This change is very consistent with the severely reduced affinity of 4 for CB2 (547-fold loss in CB2 affinity, see Table 1) and the failure of 4 (at a 1 µM concentration) to antagonize the CP55940 effects in the cAMP assay (see Table 2).
Importance of Hydrophobic Interactions in the TMH2/7 Region
Compound 5 showed a 13-fold loss in CB2 affinity, but an EC50 value for CP-55,940 in the presence of 5 which is not significantly different from the EC50 value for CP-55,940 in the presence SR 144528 (see Table 2). Compound 5 was designed to probe if the binding pocket could accommodate other rings comparable to the fenchyl ring of 1 in bulk, but varying in the placement of this bulk in the binding pocket. In 5, the fenchyl ring was replaced with a bornyl ring (see Chart 1 for compound drawings). It is clear in Figure 1, that while the fenchyl ring of 1 is relatively symmetrical about the plane of the amide group, the bornyl ring of 5 is not. This causes 5 to place more bulk towards TMH7 when docked and energy minimized in the CB2 model (see Figure 6). Docking studies suggested that this ligand can still interact with both W5.43 and W6.48 (as well as with D275) and the ligand produces an electrostatic potential surface quite similar to that of 1 as might be expected (see Figure 1). However, MD simulations show that the protruding methyl groups of the bornyl ring cause the ligand to shift in the pocket in two directions: (1) towards TMH2/TMH3 and (2) towards TMH5. This shift displaces the ligand away from D(275) and results in the lowered incidence of hydrogen bond formation with this residue over the length of the trajectory (67.3% vs. 93.7% for 1). The reduced incidence of the D(275)/ amide N-H hydrogen bond for 5 correlates with its increase in Ki.
Chart 1.
Importance of Aromatic Binding Pocket Residues
The docking studies reported here suggest that 1 occupies a significant amount of the binding pocket, with its aromatic rings interacting within the CB2 aromatic cluster residues on TMH5–6 which include Y5.39, W5.43 and W6.48. Early chimera studies of CB1/CB2 conducted at Sanofi showed that the TMH4/EC-2/TMH5 region of CB2 were critical for the binding of 1.49 The Xie lab found that a W5.43(194)Y mutation produced a decrease in the binding of 1, while W5.43A/F mutations resulted in no detectable binding of all ligands, including 1 (suggesting a misfolded protein).50 The key change in Compound 6, was the elimination of the methylene linker to the paramethylphenyl ring (C3’ in drawing of 1, Chart 1). This changes the orientation of the methyl phenyl ring which then results in loss of aromatic stacking interaction with W5.43. Compound 6 has a 42-fold loss in CB2 affinity and shows no antagonism of CP55940’s cAMP effect at a concentration of 1 µM (see Table 2). This result suggests that aromatic stacking interaction with W5.43 may be important and therefore that one significance of the methylene linker may be that it positions 1 for a productive aromatic stacking interaction with W5.43.
Ligand Moieties That Are Important to the Antagonism of Compound 1
Results for compounds 2, 4 and 6 reported here point to the amide group of 1 as a critical functional group for its CB2 affinity. Compounds 2 and 4 contain the transethylene substitution for the amide group, while 6 retains an amide functional group, but access to the amide group is severely restricted by the paramethylphenyl ring (C3’ in drawing of 1, Chart 1) in this analog. These results suggest that hydrogen bonding is a critical interaction for the formation of the 1/CB2 complex. These same three compounds (2, 4 and 6) were found to lack antagonist properties in the cAMP assay, while compounds 1, 3 and 5 (which possess the amide functionality and can form a hydrogen bond with D(275)) were found to antagonize the cAMP effects of CP55940. Admittedly, 2 and 4 have poor affinity for CB2, however, 6 has an affinity below 100 nM. It is tempting to speculate that this hydrogen bonding potential may be the key feature of these ligands that favors the inactive state of CB2 and renders 1,3 and 5 antagonists. A similar parallel has been found for the CB1 receptor and SR141716A. Here amide interaction with K3.28 was found to be key to the inverse agonism of SR141716A. 29
Other Interaction Sites for 1 Proposed in the Literature
Gouldson and co-workers proposed that S4.53 and S4.57 hydrogen bonded either to the carbonyl oxygen of the amide group of 1 or to the pyrazole nitorgen (N-1) (or both). They found that S4.53(161)A and S4.57(165)A mutations abolished the binding of 1.51 Although these investigators suggested that each of these effects were likely due to loss of hydrogen bonding with 1, this is likely not the case. In our model of the CB2 receptor, both S4.53 and S4.57 are in the TMH4-TMH3 interface and do not face into the binding pocket. However, mutations of serines can have profound effects on ligand binding pockets even when the serines are not facing into the binding pocket. Serines and threonines have the capacity to bend alpha helices via hydrogen bonding back to their own helix backbone by assuming the χ1 rotametric state of g−.52 In our previous CB1 S7.39A mutation study, we have shown the steric consequences of the loss of the serine to the conformation of a helix. In this case, the mutation resulted in the loss of binding of CP55940 and reduction in binding of HU210.53 In unpublished CB2 studies, we have shown that each of the single point mutations (S4.53A or S4.57A) performed by Gouldson bends TMH4 into the binding pocket. It is therefore very likely that the S4.53(161)A and S4.57(165)A mutations change the conformation of TMH4, sterically blocking the binding of 1. It should be noted, however, that the S4.53A or S4.57A mutations performed by Gouldson do suggest that 1 binds near TMH4, as is suggested by our modeling.
CONCLUSIONS
Results for compounds 2, 4 and 6 reported here point to the amide group of 1 as a critical functional group for its CB2 affinity and antagonist efficacy, suggesting that hydrogen bonding is a critical interaction for the formation of the 1/CB2 complex. Because interaction with the EC-3 loop residue, D(275), yielded the largest single contribution to the interaction energy for 1, the design of other CB2 antagonists that can substitute for the amide group, another group that can generate a more strongly positive electrostatic potential in this molecular region should yield higher affinity antagonists. It should also be noted that although modeling studies reported here suggest that an important hydrogen bond is between the amide hydrogen of 1 and D(275), results for the transethylene substituted compounds (2 and 4) indicate only that the amide group is important for CB2 interaction. These results do not show what part of the amide functional group is critical (carboxamide oxygen or amide hydrogen). To assess directly which part of the amide group is the interaction site will require mutant cycle studies29 that are planned for the future.
EXPERIMENTAL SECTION
Synthesis
Unless otherwise noted, all materials were obtained from commercial suppliers and used without further purification. Anhydrous solvents were obtained from Aldrich and used directly. All reactions involving air- or moisture-sensitive reagents were performed under a nitrogen atmosphere. Analytical thin-layer chromatography (TLC) was carried out on plates precoated with silica gel GHLF (250 µM thickness). TLC visualization was accomplished with a UV lamp or a solution of phosphomolybdic acid in EtOH. Silica gel chromatography was performed using RediSep prepacked silica gel cartridges. . HPLC analyses were performed using a Waters Emperor chromatography system comprised of a 1525 Binary Pump, 2487 Dual l Absorbance Detector, and a 717 plus Autosampler using a C-18 reverse phase NovaPak column (4m; 8 mm × 10 cm; 254 nm; 2 mL/min). Gas chromatography was done on a Hewlett Packard 5890 Series II gas chromatograph with a DB-17 column at 90 oC. 1H and 13C NMR spectra were run on a Bruker Avance 300 MHz or a Varian Unity Inova 500 MHz NMR spectrometer. Mass spectra (MS) were run on a Perkin-Elmer Sciex API 150 EX mass spectrometer outfitted with APCI (atmospheric pressure chemical ionization) or ESI (turbospray) sources in positive or negative modes.
Ethyl 5-(4-chloro-3-methylphenyl)-1H-pyrazole-3-Carboxylate (8)
7 (1.32 g, 0.0049 mol), hydrazine hydrate (64–65%) (0.36 mL, 0.0048 mol), and EtOH (absolute) (20 mL) were heated at reflux with stirring under nitrogen for 2 h. The reaction mixture was concentrated to give a pale yellow solid. Acetic acid (10 mL) was added to the solid, and the stirred mixture was heated overnight at reflux under nitrogen. The reaction mixture was concentrated, and the resulting oil was partitioned between CH2Cl2 and saturated NaHCO3 with frequent venting. The layers were separated, and the aqueous phase was extracted with CH2Cl2 (2X). The combined organic extracts were washed with brine, dried (MgSO4), filtered, and concentrated to give a yellow-tan solid that was purified by flash chromatography over silica (24 g) with a hexane– EtOAc gradient (0 – 60% EtOAc) to give 0.632 g (49%) of 8 as an off-white solid. TLC (SiO2, hexane–EtOAc (4:1), UV) = 0.15. 1H NMR (300 MHz, DMSO-d6): δ 14.03 (br s, 1H), 7.88 (m, J = 1H), 7.70 (dd, J = 1.6, 8.3 Hz, 1H), 7.48 (d, J = 8.3 Hz, 1H), 7.28 (s, 1H), 4.32 (q, J = 7.1 Hz, 2H), 2.38 (s, 3H), 1.32 (t, J = 7.1 Hz, 3H). MS (ES+) 265 (M + H+), 287 (M + Na+), 551 (2M + Na+).
Ethyl-5-(4-chloro-3-methylphenyl)-1-[(4-methylphenyl)methyl]-1H-pyrazole-3-carboxylate (10)
Under anhydrous conditions, 8 (0.62 g, 0.00234 mol) and NaH (60% oil dispersion) (0.31 g, 0.00775 mol) in anhydrous toluene (25 mL) were stirred at 60 °C under nitrogen for 30 min. The oil bath was removed, and the reaction mixture was allowed to stir at room temperature for 10 min. 4-Methylbenzyl bromide (0.91 g, 0.0049 mol) in anhydrous toluene (3 mL) was added at ambient temperature, and the reaction mixture was heated at reflux under nitrogen for 16 h. The reaction mixture was cooled in an ice-water bath, quenched with saturated ammonium chloride (30 mL), transferred to a separatory funnel, and extracted with EtOAc (2X). The combined extracts were washed with brine, dried (MgSO4), filtered, and concentrated to give a yellow oil. Purification by flash chromatography over silica (40 g) with a hexane–EtOAc gradient (0 – 60% EtOAc) gave 0.45 g (52%) of 10 as a yellow oil. An additional 0.175 g of impure product was also obtained. 1H NMR (300 MHz, DMSO-d6): δ 7.50 (d, J = 8.3 Hz, 1H), 7.46 (m, 1H), 7.29 (dd, J = 1.9, 8.3 Hz, 1H), 7.11 (d, J = 7.9 Hz, 2H), 6.94 (s, 1H), 6.88 (d, J = 8.0 Hz, 2H), 5.42 (s, 2H), 4.29 (q, J = 7.1 Hz, 2H), 2.33 (s, 3H), 2.24 (s, 3H), 1.30 (t, J = 7.1 Hz, 3H). 13C NMR (500 MHz, CDCl3): d 162.64, 144.47, 143.37, 137.76, 136.80, 135.62, 133.72, 131.76, 129.55, 129.54, 128.30, 127.78, 127.03, 109.60, 61.23, 54.24, 21.30, 20.22, 14.65. MS (ES+) 369 (M + H+), 391 (M + Na+).
5-(4-Chloro-3-methylphenyl)-1-[(4-methylphenyl)methyl]-1H-pyrazole-3-carboxylic acid (10a)
To a stirred solution of 10 (0.238 g, 0.000645 mol) in EtOH (5 mL) and THF (5 mL) was added 1 N NaOH (3 mL, 3 mmol). The reaction mixture was stirred at 80 °C under nitrogen for 2.5 h. The THF and EtOH were removed in vacuo, and the aqueous mixture was acidified to pH ~1 with 1N HCl and extracted with CH2Cl2. The organic phase was separated, washed with water followed by brine, dried (MgSO4), filtered, and concentrated to give 0.188 g (85%) of 10a as an off-white solid. 1H NMR (300 MHz, CD3OD): δ 7.43 (d, J = 8.2 Hz, 1H), 7.25 (s, 1H), 7.18 (dm, J = 8.2 Hz, 1H), 7.11 (d, J = 7.8 Hz, 2H), 6.91 (d, J = 8.0 Hz, 2H), 6.88 (s, 1H), 5.40 (s, 2H), 2.36 (s, 3H), 2.30 (s, 3H). MS (ES-) 339 (M - H+).
[5-(4-Chloro-3-methylphenyl)-1-[(4-methylphenyl)methyl]-1H-pyrazole-3-yl]methanol (11)
To a stirred mixture of LiAlH4 (113 mg, 2.98 mmol) and ~10 mL dry Et2O under dry nitrogen and cooled to 0 °C, ethyl 5-(4-chloro-3-methylphenyl)-1-[(4-methylphenyl)methyl]-1H-pyrazole-3-carboxylate (1.62 g, 0.00439 mol) in 30 mL Et2O was added vial cannula. The reaction was warmed to room temperature and stirred for 1 hour at which point another 28 mg (0.74 mmol) LiAlH4 was added. The reaction was complete 10 minutes later by TLC (SiO2, EtOAc–hexanes (2:3), UV). The reaction was worked up by adding 140 µL H2O, 140 µL 15% NaOH (aq) and then 420 µL H2O followed by MgSO4. The solids were filtered off and the filtrate was evaporated to yield 1.45 g of impure material that was chromatographed on 60 g SiO2 eluting with 20% acetone in hexanes. Collecting 20 mL fractions, the product was in fractions 15–26 which were recrystallized in acetone–hexane to form white spiked crystals, 1.22 g (0.00373 mol, 85% TY).
3-(Bromomethyl)-[5-(4-Chloro-3-methylphenyl)-1-[(4-methylphenyl)methyl]-1H-pyrazole (12)
11 (339 mg, 1.04 mmol) and PBr3 in CH2Cl2 (1 mL of 1.0 M) were stirred at room temperature for 3 h, at which point TLC (SiO2, EtOAc–hexanes (2:3), UV) revealed no starting material. The reaction was worked up by partitioning between CH2Cl2 and NaHCO3 (aq), drying the organics over Na2SO4 and evaporating to yield 485 mg of yellow oil. Chromatography on 1.5 g of SiO2 eluting with 20 mL of EtOAc–hexanes (2:3) resulted in 394 mg (1.01 mmol, 97% TY) of the title compound as a clear oil.
[5-(4-Chloro-3-methylphenyl)-1-[(4-methylphenyl)methyl]-1H-pyrazole-3-yl]methyl-triphenylphosphonium Bromide, 13
To solid triphenlyphosphine (231 mg, 0.882 mmol) in a 5 mL reactivial with stir bar capped under N2 was added a solution of 12 (394 mg, 1.01 mmol) in 4 mL dry toluene. The capped reaction was heated to 90 °C for 24 h. The resulting mixture was refrigerated for two days,and the resulting solid was filtered and washed with hexanes and dried in vacuo to yield 514 mg (89%) of white solid product.
(1S,2S,4R)-1,3,3-trimethylbicyclo[2.2.1]heptane-2-carbaldehyde, (endo-Fenchaldehyde) (15)
To a 1-L three-neck round bottom flask equipped with a stir bar, thermometer, powder addition funnel, rubber septum, and nitrogen inlet was added lithium diisopropylamide (2 M in heptane–THF–ethyl benzene) (40 mL, 80 mmol) followed by anhydrous THF (190 mL) at room temperature under dry nitrogen. The stirred solution was cooled between −70 and −80 °C with a dry ice–acetone bath, and the (methoxymethyl)diphenylphosphine oxide (19.5 g, 0.079 mol) was added portionwise over 33 min. The powder addition funnel was rinsed with THF (5 mL). The dry ice–acetone bath was replaced with an ice-water–NaCl bath, and the reaction mixture was allowed to warm to between −9 and −12 °C and maintained at that temperature for 20 min. The reaction mixture was then cooled to between −70 and −80 °C, and a solution of fenchone 14 (4 mL, 3.8 g, 0.025 mol) in anhydrous THF (14 mL) was added dropwise. The reaction mixture was stirred for 3 h and allowed to slowly warm to room temperature overnight. After 45 h, the red-orange suspension was cooled to between −70 and −80 °C and water (100 mL) was added dropwise. The dry ice– acetone bath was removed, and the reaction mixture was stirred at room temperature for ~1 h. The reaction mixture was extracted with EtOAc (2 × 250 mL). The organic phase was washed with brine, dried (MgSO4), filtered, and concentrated to give the crude product. Diethyl ether was added, and the suspension was filtered. The filtrate was concentrated and purified by flash chromatography over SiO2 (120 g) with a hexane–EtOAc gradient (0 – 30% EtOAc) to give 3.7 g (82%) of 2-(methoxymethylidene)-1,3,3-trimethylbicyclo[2.2.1]heptane as a pale yellow liquid mixture of 2E- and 2Z-isomers. 1H NMR (olefin H 5.57 ppm, OMe 3.48 ppm); (olefin’ H 5.59 ppm, OMe 3.46 ppm).
To a rapidly stirred solution of these isomers (0.51 g, 0.0028 mol) in CHCl3 (17 mL) was added a solution of trichloroacetic acid (1.82 g, 0.0111 mol) in water (10 mL) at room temperature. The reaction mixture was stirred vigorously at room temperature for 3 h and transferred to a separatory funnel with CH2Cl2, and the layers were separated. The organic phase was washed with saturated NaHCO3 (2X), dried (MgSO4), filtered, and concentrated to give 0.38 g (82%) of 15 as a colorless oil. 1H NMR indicates that 15 is an 87:13 mixture of endo:exo diastereomers. 1H NMR (300 MHz, CDCl3); δ: major isomer 9.85 (d, J = 3.5 Hz, 1H), 1.87 (dd, J = 2.5 Hz, 4.0 Hz, 1H), 1.82 (m, 3H), 1.56 (m, 2H), 1.43–1.23 (m, 2H), 1.16 (s, 3H), 1.11 (s, 3H), 1.09 (s, 3H); minor isomer: 9.60 (d, J = 6.5 Hz, 1H, minor isomer), 1.10 (s, 3H, minor isomer), 1.07 (s, 3H, minor isomer), 1.05 (s, 3H, minor isomer).
[5-(4-Chloro-3-methylphenyl)-1-(4-methylphenyl)methyl]-3-[(E)-2-[(1S,2S,4R)-1,3,3-trimethylbicyclo[2.2.1]heptan-2-yl]ethenyl]-1H-pyrazole, 2
To 13 (257 mg, 0.394 mmol) in 10 mL anhydrous THF (some insolubles), cooled to –72 °C, was added 1.0 M freshly titrated BuLi (0.38 mL, 0.38 mmol) dropwise to achieve an orange mixture. To the ylide was added fenchyl aldehyde 14 (196 mg in 2.5 mL Et2O, 1.18 mmol), and the reaction was allowed to warm to room temperature. The reaction turned a clear yellow after ~30 min. TLC (SiO2, Acetone– hexanes (1:9), UV) showed the olefin product(s) and a by-product which was the corresponding pyrazole-3-methyl by-product that derived from hydrolysis of the ylide from 13. Stirring overnight at room temperature showed no significant change by TLC. The reaction mixture was worked up by evaporating the THF and partitioning the remaining residue between CH2Cl2 and H2O (aq. pH = 12). Adjusted to pH 6 with 1 N HCl (aq). Removed the organic phase and extracted the aqueous layer with CH2Cl2. The combined organics were washed with brine, dried (Na2SO4) and evaporated to yield 370 mg of crude yellow resin. Chromatography of the resin on 20 g SiO2 eluting with Et2O–hexanes (8:92) collecting 8 mL fractions provided the product in fractions 12–15, 37 mg (20%). TLC (SiO2; 10% acetone-hexanes; UV) Rf (trans) = 0.48; Rf (cis) = 0.44.
TRANS ENDO
1H NMR (CDCl3, 500 MHz) d 7.32–7.34 (d, J = 5.1 Hz, 1 H, Ar-H), 7.17 (s, 1 H, 2’H), 7.06– 7.10 (m, 2 H, Ar-H), 6.97–6.98 (d, J = 4.8 Hz, 2 H, Ar-H), 6.43 (s, 1 H, pyr-H), 6.38–6.41 (d, J = 9.3 Hz, 1 H, olefin a to pyr), 6.20–6.25 (dd, J = 9.3 Hz, .021 ppm, 1 H, olefin b to pyr), 5.22 (s, 2 H, benz CH2), 2.37 (s, 3 H, 3’Me), 2.33 (s, 3 H, benz-Me), 1.88–1.90 (d, J = 6.3 Hz, 1 H, fench-2), 1.77 (m, 1 H, fench-4), 1.65–1.75 (m, 1 H, exo fench-5), 1.59–1.61 (d, J = .019 ppm, 1 H, fench-7 syn), 1.54–1.57 (m, 1 H, exo fench-6), 1.44–1.52 (m, 1 H, endo fench-5), 1.23–1.25 (d, J = 6.0 Hz, 1 H, fench-7 anti), 1.07–1.14 (m, 1H, endo fench-6), 1.03 (s, 3 H, bridgehead Me), 1.01 (s, 3 H, exo gem Me), 0.90 (s, 3 H, endo gem Me). 13C NMR (CDCl3, 500 MHz) d 150.7 (pyr-3), 143.8 (pyr-5), 137.1, 136.4, 134.8, 134.6, 131.4 (2’), 130.7 (olefin b pyr), 129.3, 129.2, 127.3, 126.7, 124.6 (olefin a pyr), 102.8 (pyr-4), 61.5 (fench-2), 52.9 (benzyl), 49.5 (fench-3), 48.5 (fench-4), 44.9 (fench-7), 40.8 (fench-1), 32.0 (exo Me), 28.0 (fench-6), 26.4 (fench-5), 22.7 (endo Me), 21.1 (benz-Me), 20.6 (bridge Me), 20.0 (3’Me).
Pertinent 2D interaction:
Fenchyl syn 7H w/fenchyl 2H ROESY; therefore exo.
Olefins have COSY interaction, not ROESY.
Fractions 7–11 were rechromatographed on 3 g SiO2 eluting with 5% Et2O / 95% petroleum ether. Fractions 10–14 yielded 9 mg (5%) of the cis-isomer.
CIS ENDO
1H NMR (CDCl3, 300 MHz) d 7.32–7.35 (k, J = 8.15 Hz, 1 H, Ar-H), 7.14 (s, 1 H, Ar-H), 7.04– 7.10 (m, 3 H, Ar-H), 6.97–6.99 (d, J = 8.03 Hz, 2 H, Ar-H), 6.50–6.54 (d, J = 11.9 Hz, 1 H, olefin a to pyr), 6.39 (s, 1 H, pyr-H), 5.66–5.74 (dd, J = 11.8, 11.8 Hz, 1 H, olefin b to pyr), 2.87–2.91 (d, J = 11.6 Hz, 1 H, fench-2), 2.35 (s, 3 H, Ar-Me), 2.31 (s, 3 H, Ar-Me), 1.68–1.72 m, 2 H, fench-4), 1.61–1.64 (d, J = 8.6 Hz, 1 H, fench-7 syn), 1.29–1.46 (m, 3 H, fench 5 & 6), 1.20–1.25 (m, 1 H, fench-7 anti), 1.04 (s, 3 H, gem Me exo), 1.00 (s, 3 H, bridgehead Me), 0.89– 0.92 (m, 1 H, fench 5 or 6), 0.88 (s, 3 H, gem Me endo). 13C NMR (CDCl3, 500 MHz) d 149.3, 143.4, 137.4, 136.6, 135.0, 134.9, 132.2, 131.7, 129.52, 129.49, 129.46, 127.8, 123.4, 106.5, 54.5, 53.2, 50.4, 49.2, 45.4, 41.4, 32.6, 27.9, 26.6, 22.7, 21.3, 21.0, 20.3.
Pertinent 2D interaction:
Fenchyl syn 7H w/ fenchyl 2H ROESY; therefore exo.
Olefins exhibit ROESY interaction; therefore cis.
3-Methyl-5-(4-chloro-3-methylphenyl)-1-(4-methylphenyl)methyl]-1H-pyrazole (62 mg, 50%) was also isolated. 1H NMR (300 MHz, CD3Cl): δ 7.30 (m, 1H), 7.14 (m, 1H), 7.04 (m, 3H), 6.93 (m, 2H), 6.10 (m, 1H), 5.20 (d, J= 2 Hz, 2 H), 2.33 (s, 3H), 2.31 (s, 3H), 2.30 (s, 3H).
N-[(4-Chloro-3-methyl-phenyl)-(toluene-4-sulfonyl)-methyl]-formamide (19)
To a solution of sodium p-toluenesulfinate tetrahydrate (24.97 g, 0.0998 mol) in water (125 mL) was added tert-butylmethyl ether (125 mL) followed by the dropwise addition of 12 N HCl (8.2 mL) over 5 min. The solution was stirred for 20 min, transferred to a separatory funnel, and the aqueous layer was removed. The organic solution was transferred to a round bottom flask followed by toluene (125 mL). The mixture was reduced to ~¼ volume in vacuo at bath temperature between 35–40 °C. Heptane (40 mL) was added to the turbid mixture, and the suspension was filtered. The filtered solid was washed with heptane (50 mL) and dried under vacuum at room temperature to give 13.07 g (84%) of p-toluenesulfinic acid 17 as a white solid.32
A stirred solution of 4-chloro-3-methylbenzaldehyde 16 (7.05 g, 0.0456 mol), acetonitrile (30 mL), toluene (30 mL), formamide 18 (4.4 mL, 0.111 mol), and chlorotrimethylsilane (6.2 mL, 0.0489 mol) was heated at 50 °C under nitrogen for 5 h. The reaction mixture was allowed to cool at room temperature and p-toluenesulfinic acid (10.2 g, 0.0653 mol) was added followed by acetonitrile (8 mL) and toluene (8 mL). The suspension was heated at 50 °C under nitrogen for 5 h and allowed to stand overnight at room temperature. To the suspension was added tert-butylmethyl ether (30 mL). The mixture was stirred for 5 min and water (130 mL) was added. The stirred two-phase mixture was cooled in an ice-water bath and stirring was continued for 1 h. The thick white suspension was filtered with a tert-butylmethyl ether rinse (2 × 15 mL). The solid was dried under vacuum at 60 °C to give 13.33 g (87%) of crude 19 as an off-white solid. The 1H NMR of 19 indicates that it exists as a mixture of rotamers at 25 °C in DMSO-d6 as noted for an analogous compound described in the above cited reference. The 1H NMR data is reported for the major rotamer. 1H NMR (300 MHz, DMSO-d6): d 9.76 (d, J = 11.2 Hz, 1H), 7.94 (m, 1H), 7.74 (d, J = 8.3 Hz, 2H), 7.55 (m, 1H), 7.46 (m, 4H), 6.39 (d, J = 10.6 Hz, 1H), 2.42 (s, 3H), 2.34 (s, 3H).
[(4-Chloro-3-methyl-phenyl)-(toluene-4-sulfonyl)-methyl]-isocyanide (20)
To a stirred solution of 19 (10.75 g, 0.0318 mol) in anhydrous tetrahydrofuran (70 mL) was added phosphorus V oxychloride (6 mL, 0.0644 mol) dropwise at room temperature under nitrogen. The solution was stirred for 5 min and cooled in an ice-water bath. To the cold stirred solution was added triethylamine (27 mL, 0.194 mol) slowly dropwise over a period of 30 min maintaining the temperature between 3– −8 °C. The reaction mixture was stirred with cooling for 7 min, and ethyl acetate (50 mL) was added followed by water (50 mL). The two-phase mixture was transferred to a separatory funnel, and the aqueous phase was removed. The organic phase was washed with water (2 × 50 mL), followed by saturated sodium bicarbonate (2 × 50 mL), and finally brine (20 mL). The organic solution was dried (MgSO4), filtered, and concentrated under vacuum at a bath temperature of 35–40 °C to give the crude product as a dark red-orange viscous oil (9.95 g). The crude F5 was used directly without further purification. In an earlier run of this reaction the crude product was crystallized from isopropanol to provide an authentic sample for which 1H NMR data is provided. 1H NMR (300 MHz, CDCl3): d 7.67 (d, J = 8.3 Hz, 2H), 7.37 (m, 3H), 7.22 (m, 1H), 7.09 (dd, J = 2.0, 8.3 Hz, 1H), 5.53 (s, 1H), 2.49 (s, 3H), 2.38 (s, 3H). [Safety Note: As noted in the above cited reference, similar isocyanides have been found to be thermally unstable at temperatures above 80 °C. Therefore, to ensure a margin of safety, avoid heating the isocyanide above 35–40 °C.]
5-(4-Chloro-3-methyl-phenyl)-1H-pyrrole-3-carboxylic acid ethyl ester (22)
A stirred suspension of sodium hydride (60% oil dispersion) (2.65 g, 0.0662 mol) in anhydrous diethyl ether (100 mL) was cooled under nitrogen in an ice-water bath. A solution of crude 20 (9.86 g) and ethyl acrylate (2.9 mL, 0.0266 mol) in anhydrous dimethylsulfoxide (100 mL) and anhydrous diethyl ether (200 mL) was added dropwise at a moderately fast rate to the stirred cold sodium hydride suspension over a period of 75 min while maintaining the temperature of the reaction mixture between 0–10 °C. The ice-water bath was removed, and the reaction mixture was stirred for 1 h, again cooled in an ice-water bath, and quenched by the very slow dropwise addition of water and gradually increasing the addition rate when the evolution of hydrogen ceased (500 mL). The quenched reaction mixture was transferred to a separatory funnel, and the aqueous layer was removed. The organic phase was washed with water (500 mL) followed by brine. The combined aqueous solutions used to quench the reaction and wash the crude product, as well as the brine wash, were back extracted with ethyl acetate. The organic solutions were combined, dried (MgSO4), filtered, and concentrated to give the crude product which was purified by flash chromatography over silica (120 g) eluting with a hexanes–ethyl acetate gradient (0 – 50% EtOAc) to give 1.4 g (17% over two steps) of 22 as a red-brown solid. 1H NMR (300 MHz, DMSO-d6): d 11.98 (s, 1H), 7.72 (s, 1H), 7.53 (m, 2H), 7.40 (d, J = 8.3 Hz, 1H), 6.91 (m, 1H), 4.20 (q, J = 7.1 Hz, 2H), 2.35 (s, 3H), 1.27 (t, J = 7.1 Hz, 3H). MS (ES-) 262(M - H+).
5-(4-Chloro-3-methyl-phenyl)-1-(4-methyl-benzyl)-1H-pyrrole-3-carboxylic acid ethyl ester (23)
To a stirred ice-water cooled suspension of sodium hydride (60% dispersion in oil) (0.294 g, 0.00735 mol) in anhydrous N,N-dimethylformamide (12 mL) was added dropwise a solution of 22 (1.21 g, 0.00461 mol) in anhydrous N,N-dimethylformamide (24 mL) under nitrogen. The cold reaction mixture was stirred for 15 min, and a solution of 4-methylbenzyl bromide (1.1 g, 0.00594 mol) in anhydrous N,N-dimethylformamide (12 mL) was added at a moderate rate. The ice-water bath was removed, and the reaction mixture was stirred under nitrogen for 2 h, cooled in an ice-water bath, and water (50 mL) was added slowly dropwise. To the quenched reaction mixture was added ethyl acetate (150 mL), and the two-phase mixture was transferred to a separatory funnel. The organic phase was separated, washed with water (100 mL) followed by brine, dried (MgSO4), filtered, and concentrated to give the crude product which was purified by flash chromatography over silica (40 g) with a hexanes–ethyl acetate gradient (0 – 30% EtOAc) to give 1.17 g (69%) of 23 as a gold-yellow oil. 1H NMR (300 MHz, CDCl3): δ 7.36 (d, J = 1.7 Hz, 1H), 7.30 (d, J = 8.2 Hz, 1H), 7.15 (m, 1H), 7.12 (d, J = 8.0 Hz, 2H), 7.05 (dd, J = 2.1, 8.2 Hz, 1H), 6.90 (d, J = 8.0 Hz, 2H), 6.64 (d, J = 1.7 Hz, 1H), 5.04 (s, 2H), 4.28 (q, J = 7.1 Hz, 2H), 2.33 (s, 6H), 1.31 (t, J = 7.1 Hz, 3H). MS (ES+) 368 (M + H+), 390 (M + Na+).
5-(4-Chloro-3-methyl-phenyl)-1-(4-methyl-benzyl)-1H-pyrrole-3-carboxylic acid (1,3,3-trimethyl-bicyclo[2.2.1]hept-2-yl)-amide (3)
To a stirred solution of 23 (0.099 g, 0.00027 mol) in ethanol (3 mL) was added sodium hydroxide (2 N) (0.55 mL, 0.00110 mol). The reaction mixture was heated at 60 °C under nitrogen for 8 h, allowed to stand at room temperature for 4 days, and heated at 60 °C under nitrogen for 90 min. The ethanol was removed in vacuo, and water (6 mL) was added. The aqueous suspension was washed with diethyl ether (6 mL), and the layers were separated. Hydrochloric acid (1 N, ~0.8 mL) was added to the aqueous phase, and the viscous mixture was extracted with ethyl acetate (12 mL). The aqueous phase was separated and acidified to pH 2 (indicator paper) using 1 N HCl. The ethyl acetate solution was washed with the acidic aqueous solution, and the two phases were separated. The EtOAc solution was dried (MgSO4), filtered, and concentrated to give 0.069 g (75%) of the acid analog of the 23 ester as an off-white solid. 1H NMR (300 MHz, CD3OD) δ 7.48 (d, J = 1.8 Hz, 1H), 7.32 (d, J = 8.2 Hz, 1H), 7.18 (s, 1H), 7.10 (m, 3H), 6.86, (d, J = 8.0, 2H), 6.57 (d, J = 1.9 Hz, 1H), 5.13 (s, 2H), 2.31 (s, 3H), 2.29 (s, 3H). MS ES(−) 338 (M - H+).
Thionyl chloride (0.03 mL, 0.41 mmol) was added dropwise to a stirred mixture of the above acid (0.069 g, 0.00020 mol) and anhydrous toluene (4 mL) at room temperature under nitrogen and then heated at reflux under nitrogen for 90 min. The reaction mixture was concentrated in vacuo, and the crude acid chloride was dissolved in anhydrous dichloromethane (5 mL). To the acid chloride solution was added a solution of (−)-fenchylamine 24 (0.052 g, 0.00034 mol) in anhydrous dichloromethane (1 mL) followed by the dropwise addition of triethylamine (0.084 mL, 0.00060 mol) at room temperature under nitrogen. After 1 h, the reaction mixture was diluted with dichloromethane (4 mL) and washed with saturated sodium carbonate (5 mL). The layers were separated, and the aqueous phase was extracted with dichloromethane (5 mL). The organic extracts were combined, dried (MgSO4), filtered, and concentrated to give the crude product, which was purified by flash chromatography over silica (4 g) with a hexanes–ethyl acetate gradient (0 – 50% EtOAc) to give 0.049 g (52%) of 3 as an off-white amorphous solid. Less pure fractions were also obtained (0.032 g, 34%) as an off-white amorphous solid. 1H NMR (300 MHz, CDCl3): δ 7.31 (m, 2), 7.17 (m, 1), 7.11 (d, J = 7.9 Hz, 2H), 7.06 (dd, J = 2.2, 8.2 Hz, 1H), 6.92 (d, J = 8.0 Hz, 2H), 6.38 (d, J = 2 Hz, 1H), 5.78 (br d, J = 9.4 Hz, 1H), 5.03 (s, 2H), 3.81 (dd, J = 1.6, 9.5 Hz, 1H), 2.34 (s, 3H), 2.33 (s, 3H), 1.72 (m, 3H), 1.48 (m, 1H), 1.25 (m, 3H), 1.16 (s, 3H), 1.09 (s, 3H), 0.84 (s, 3H). 13C NMR (300 MHz, CDCl3): δ 164.87, 137.55, 136.22, 134.51, 134.24, 133.99, 131.68, 130.76, 129.50, 129.09, 127.68, 126.76, 125.35, 120.03, 106.76, 62.82, 51.05, 48.56, 48.17, 42.65, 39.36, 30.83, 27.45, 25.99, 21.22, 21.04, 20.01, 19.65. MS (EI) 474 (M+).
[5-(4-Chloro-3-methylphenyl)-1-[(4-methylphenyl)methyl]-1H-pyrrol-3-yl]methanol (25)
To an ice-water bath cooled solution of LiAlH4 (1 M in THF, 1.6 mL, 1.6 mmol) was added dropwise under nitrogen a solution of 23 (0.292 g, 0.00071 mol) in anhydrous THF (3 mL). The ice-water bath was removed, and the yellow solution was stirred at room temperature for 4.5 h. The reaction mixture was cooled in an ice-water bath, and quenched by the sequential addition of water (0.06 mL), 15% aqueous sodium hydroxide (0.06 mL), and water (0.16 mL). The suspension was filtered through a pad of Celite, and the pad was washed with EtOAc. The filtrate was concentrated to give 0.25 g (>100%) of crude 25 as a yellow oil. The crude product was used directly without further purification. 1H NMR (300 MHz, CDCl3): δ 7.28 (m, 1H), 7.11 (m, 4H), 6.92 (d, J = 7.8 Hz, 2H), 6.72 (s, 1H), 6.26 (m, 1H), 5.02 (s, 2H), 4.57 (d, J = 3.4 Hz, 2H), 2.33 (s, 6H).
5-(4-Chloro-3-methylphenyl)-1-[(4-methylphenyl)methyl]-1H-pyrrole-3-carbaldehyde (32)
To a stirred solution of 25 (0.431 g, 0.0013 mol) in CH2Cl2 (15 mL) was added portionwise Dess-Martin Periodinane (0.69 g, 0.0016 mol) at room temperature. The dark purple reaction mixture was stirred at room temperature under nitrogen for 3.5 h. EtOAc (40 mL) was added followed by a solution of sodium thiosulfate pentahydrate (4.7 g) in saturated sodium bicarbonate (6 mL), and the reaction mixture was stirred vigorously for 10 min. The aqueous phase was separated, and the organic phase was washed with saturated sodium bicarbonate followed by brine, dried (MgSO4), filtered, and concentrated to give a red-orange oil which was purified by flash chromatography over SiO2 (24 g) with a hexane–EtOAc gradient (0 – 50% EtOAc) to give 0.25 g (60%) of 32 as an orange oil. 1H NMR (300 MHz, CDCl3): δ 9.75 (s, 1H), 7.34 (m, 2H), 7.17 (m, 1H), 7.14 (d, J = 8.0, 2H), 7.07 (dd, J = 2.3, 8.2 Hz, 1H), 6.92 (d, J = 8.0 Hz, 2H), 6.67 (d, J = 1.8 Hz, 1H), 5.06 (s, 2H), 2.35 (s, 3H), 2.34 (s, 3H).
Dimethyl {hydroxy[(1S,2S,4R)-1,3,3-trimethylbicyclo[2.2.1]heptan-2-yl]methyl}phosphonate (33)
To a stirred solution of dimethylphosphite (0.25 mL, 2.73 mmol) in anhydrous THF (12 mL) cooled in a dry ice–acetone bath was added LiHMDS (1 M in THF) (2.8 mL, 0.0028 mol) dropwise under nitrogen. After 15 min, a solution of 15 (0.38 g, 0.0023 mol) in anhydrous THF (20 mL) was added, and stirring was continued for 1 h. Saturated aqueous ammonium chloride (10 mL) and Et2O (48 mL) were added, and the reaction mixture was stirred with cooling for 15 min and then at room temperature for 15 min. The mixture was extracted with Et2O. The organic extracts were combined, dried (MgSO4), filtered, and concentrated to give a yellow oil, which was purified by flash chromatography over SiO2 (12 g) eluting with CH2Cl2 followed by methanol. The product was dissolved in CH2Cl2, filtered, and concentrated to give 0.53 g (83%) of 33 as a yellow oil. 1H NMR (300 MHz, CDCl3): δ 4.15 (m, 1H), 3.83 (m, 6H), 2.01–0.99 (m, 18H). MS (ES+) 277 (M + H+), 299 (M + Na+), 575(2M+Na+).
Dimethyl {[(1S,2S,4R)-1,3,3-trimethylbicyclo[2.2.1]heptan-2-yl]methyl}phosphonate (34)
To a stirred solution of 33 (0.405 g, 0.00147 mol) in anhydrous THF (10 mL) was added 1,1’-thiocarbonyldiimidazole (0.57 g, 0.0032 mol) at room temperature. The reaction mixture was stirred at room temperature under nitrogen overnight, heated at 50 °C for 6 h, and allowed to stand overnight at room temperature. The reaction mixture was concentrated to give an oil which was partially purified by flash chromatography over SiO2 (24 g) with a hexane–EtOAc gradient (0 – 100% EtOAc) to give 0.32 g of a pale yellow oil. While 1H NMR and TLC (SiO2, EtOAc) clearly show the corresponding thiocarbonylimidazolide was impure, MS analysis indicates the desired product is present [(ES+) 387 (M + H+)]. The impure thiocarbamate (0.32 g) was dissolved in anhydrous toluene (12 mL) and then treated with azobisisobutyrylnitrile (0.01 g, 0.0006 mol) and tributyltin hydride (0.27 mL, 1.00 mmol) at room temperature. The reaction mixture was heated at reflux under nitrogen for 3 h and allowed to stand at room temperature overnight. The reaction mixture was concentrated, and the crude product was purified by flash chromatography over SiO2 (12 g) with EtOAc to give 0.111 g (29% over two steps) of 34 as a colorless oil. 1H NMR (300 MHz, CDCl3): δ 3.76 (d, J = 2.3 Hz, 3H), 3.72 (d, J = 2.3 Hz, 3H), 1.83–1.62 (m, 3H), 1.51–1.07 (m, 6H), 1.05 (s, 3H), 1.03 (s, 3H), 0.99 (m, 1H), 0.91 (s, 3H). MS (ES+) 261 (M + H+), 283 (M + Na+).
Dimethyl {2-[5-(4-chloro-3-methylphenyl)-1-[(4-methylphenyl)methyl]-1H-pyrrol-3-yl]-2-hydroxy-1-[(1S,2S,4R)-1,3,3-trimethylbicyclo[2.2.1]heptan-2-yl]ethyl}phosphonate (35)
To a dry ice/acetone cooled stirred solution of 34 (0.052 g, 0.00020 mol) in anhydrous THF (0.60 mL) was added dropwise n-butyllithium (2.2 M in hexane) (0.09 mL, 0.20 mmol) under nitrogen. After ~15 min, a solution of 32 (0.065 g, 0.00020 mol) in anhydrous THF (0.25 mL) was added dropwise and stirring was continued for 1 h. The reaction was quenched by the dropwise addition of saturated aqueous NH4Cl (~0.75 mL). The cold bath was removed, and the reaction mixture was partitioned between EtOAc and water. The organic phase was separated, dried (MgSO4), filtered, and concentrated to give the crude product, which was purified by flash chromatography over SiO2 (12 g) with a hexane–EtOAc gradient (0 – 100% EtOAc) to give 0.0227 g (20%) of 35 as a gold-yellow oil. Interestingly, 7.4 mg of impure 4 was also obtained. 1H NMR (35): 1H NMR (300 MHz, CDCl3): δ 7.27 (d, J = 8.0 Hz, 1H), 7.13 (s, 1H), 7.09 (d, J = 7.8 Hz, 2H), 7.02 (d, J = 7.7 Hz, 1H), 6.91 (d, J = 7.9 Hz, 2H), 6.78 (s, 1H), 6.30 (s, 1H), 5.03 (m, 4H), 3.72 (d, J = 10.8 Hz, 3H), 3.32 (d, J = 11.1 Hz, 3H), 2.51 (m, 1H), 2.32 (s, 6H), 1.67 (m, 6H), 1.43 (m, 1H), 1.15 (s, 3H), 1.11 (s, 3H), 1.06 (s, 3H). MS (ES+) 606 (M + Na+).
5-(4-Chloro-3-methylphenyl)-1-[(4-methylphenyl)methyl]-3-[(E)-2-[(1S,2S,4R)-1,3,3-trimethylbicyclo[2.2.1]heptan-2-yl]ethenyl]-1H-pyrrole, 4
To a 1-dram glass vial containing a solution of 35 (0.023 g, 0.000039 mol) in anhydrous 1,4-dioxane (0.5 mL) was added magnesium bromide ethyl etherate (0.0104 g, 0.000040 mol). The stirred reaction mixture was heated in the sealed vial at 100 °C. After 1 h, anhydrous 1,4-dioxane (0.10 mL) was added to the reaction mixture, and heating was resumed at 100 °C. After 3 h, the oil bath was removed, and the reaction mixture was allowed to stand at room temperature for 3 days. Heating was resumed at 100 °C for 18.5 h. The reaction mixture was partitioned between EtOAc and water. The layers were separated, and the organic phase was washed with brine, dried (MgSO4), filtered, and concentrated to give 18 mg of a gold-yellow oil. The crude product was dissolved in CH2Cl2, and the solution was filtered through a plug of SiO2 and eluted with CH2Cl2. The eluate was concentrated to give 10 mg of impure product,which was combined with the impure alkene obtained during the synthesis of 35. Flash chromatography over SiO2 (4 g) with hexane gave 2.6 mg (2.7% over two steps) of 4 as an amorphous solid. 1H NMR (300 MHz, CD3OD): δ 7.28 (d, J = 8.2 Hz, 1H), 7.19 (m, 1H), 7.08 (m, 3H), 6.86(d, J = 8.0 Hz, 2H), 6.76 (d, J = 1.9 Hz, 1H), 6.31(d, J = 1.9 Hz, 1H), 6.16 (d, J = 15 Hz, 1H), 5.91 (dd, J = 10.1, 15.5 Hz, 1H), 5.04 (s, 2H), 2.30 (s, 3H), 2.28 (s, 3H), 1.77 (m, 3H), 1.51 (m, 3H), 1.25 (m, 1H), 1.09 (m, 1H), 0.98 (s, 6H), 0.87 (s, 3H). MS (APCI+) 458 (M + H+).
N-{1,7,7-Trimethylbicyclo[2.2.1]heptan-2-ylidene}hydroxylamine (37)
This compound was prepared as reported.54 A mixture of (1S)-(-)-camphor 36 (3.0 g, 0.0197 mol), hydroxylamine hydrochloride (2.1 g, 0.0302 mol) and water (11 mL) was heated at 80 °C under nitrogen. To the stirred suspension was added MeOH (16 mL) via a syringe followed by a solution of NaOAc (3.9 g, 0.0475 mol) in water (8 mL). The reaction mixture was heated at reflux under nitrogen for 9 h, the methanol was removed in vacuo, and the aqueous suspension was filtered. The filtered solid was washed with water, recrystallized from 95% EtOH, and dried to give 2.33 g (71%) of 37 as a white solid.
(-)-endo-Bornan-2-amine, (38)
This compound was prepared as reported [3,19] with modification. A solution of 37 (1.04 g, 0.0062 mmol) in absolute EtOH (16 mL) was heated at 80 °C with stirring under nitrogen while sodium (1.1 g, 0.0478 mol) was carefully added portionwise over 1 h. The reaction mixture was heated for another 2 h, cooled in an ice-water bath, and treated carefully with water (16 mL). The aqueous ethanol mixture was partially concentrated to ~16 mL, and the aqueous mixture was extracted with CH2Cl2. The organic phase was separated, washed with brine, dried (MgSO4), filtered, and concentrated to give a white pasty solid (0.854 g). The crude amine was dissolved in 2N HCl, washed with diethyl ether (2 × 10 mL), and the acidic aqueous solution was concentrated to give the crude hydrochloride salt which was recrystallized twice from 2N HCl (6 mL and 1 mL, respectively) to give ~ 0.187 g of a white solid. The hydrochloride salt (0.18 g) was partitioned between 1N NaOH (2 mL) and dichloromethane (10 mL). The organic phase was separated, and the aqueous phase was extracted twice with dichloromethane (5 mL and 6 mL, respectively). The organic extracts were combined, dried (MgSO4), filtered, and concentrated to give 0.098 g (10%) of 38 as a sticky white solid. 1H NMR (CDCl3) indicates that 38 is ~95:5 endo:exo. The filtrates from the above recrystallizations were independently concentrated to give 0.537 g of the amine hydrochloride salt.]
5-(4-Chloro-3-methylphenyl)-1-[(4-methylphenyl)methyl]-N-[(1S,2R,4S)-1,7,7-trimethylbicyclo[2.2.1]heptan-2-yl]-1H-pyrazole-3-carboxamide (5)
To a stirred suspension of 10a (0.082 g, 0.00024 mol) in anhydrous toluene (5 mL) was added thionyl chloride (0.039 mL, 0.00053 mol) dropwise at room temperature under nitrogen. The reaction mixture was heated at reflux for 90 min, concentrated, and toluene (3 mL) was added to the resulting oil. The toluene was removed in vacuo and replaced with CH2Cl2 (5 mL). To the crude acid chloride solution was added triethylamine (0.10 mL, 0.72 mmol) followed by a solution of 38 (0.042 g, 0.00028 mol) in CH2Cl2 (1 mL). After stirring at room temperature under nitrogen for 2 h the reaction mixture was concentrated, and the crude product was purified by flash chromatography over silica (12 g) with a hexanes–EtOAc gradient (0 – 40% EtOAc) to give the desired product as an off-white amorphous solid (0.085 g). TLC (SiO2, hexanes–EtOAc (4:1) as well as CH2Cl2, UV) showed a single UV-absorbing spot. However, HPLC [C-18 reverse phase NovaPak column; 4 µ; 8 mm × 10 cm; 254 nm; 2 mL/min; CH3CN–H2O (90:10)] and 1H NMR show one or more impurities were present. The product was approximately 94.8% pure by HPLC. Two recrystallizations from MeOH (1 mL) gave 0.039 g (34%) of 5 as a white solid (98.3% pure by HPLC). 1H NMR (300 MHz, CDCl3): δ 7.34 (d (J = 8.1 Hz, 1H), 7.12 (m, 3H), 7.04 (dd, J = 2.0, 8.2 Hz, 1H), 6.96 (m, 3H), 6.84 (s, 1H), 5.29 (s, 2H), 4.44 (m, 1H), 2.40 (m, 1H), 2.34 (s, 6H), 1.61–1.83 (m, 3H), 1.24–1.50 (m, 3H), 1.01 (s, 3H), 0.91 (s, 3H), 0.89 (s, 3H). MS ES(+) 476 (M + H+), 498 (M + Na+).
Methyl 5-(4-chloro-3-methylphenyl)-1-phenyl-1H-pyrazole-3-carboxylate (39)
7 (0.498 g, 0.00185 mol), phenylhydrazine hydrochloride (0.305 g, 0.0021 mol), and MeOH (6 mL) were combined, and the reaction mixture was heated in a sealed tube at 80 °C for 16 h. The reaction mixture was allowed to cool at room temperature and concentrated to give a solid. Dichloromethane and methanol were added to the crude product, and the turbid mixture was filtered. The filtrate was partially concentrated in vacuo and purified by flash chromatography over silica (24 g) using a hexane–ethyl acetate gradient (0 – 40% EtOAc) to give 0.393 g (65%) of the methyl ester as a gold-yellow oil. 1H NMR (300 MHz, CDCl3): δ 7.35 (m, 5H), 7.25 (m, 1H), 7.14 (d, J = 1.8 Hz, 1H), 7.04 (s, 1H), 6.91 (dd, J = 1.9, 8.1 Hz, 1H), 3.97 (s, 3H), 2.31 (s, 3H). MS ES(+) 327 (M + H+), 349 (M + Na+).
5-(4-Chloro-3-methylphenyl)-1-phenyl-1H-pyrazole-3-carboxylic acid (40)
To a stirred solution of 39 (0.37 g, 0.00113 mol) in THF (8 mL) and MeOH (8 mL) was added 1N NaOH (5 mL, 5 mmol). The reaction mixture was heated at 80 °C under nitrogen for 2.5 h and then allowed to stand at room temperature overnight. The THF and MeOH were removed in vacuo. The aqueous mixture was acidified to pH 1 (indicator strip) with 1N HCl and extracted with CH2Cl2. The organic extract was separated, dried (MgSO4), filtered, and concentrated to give 0.318 g (90%) of 40 as a pale tan solid. 1H NMR (300 MHz, CD3OD): δ 7.47 (m, 3H), 7.36 (m, 2H), 7.32 (d, J = 8.3 Hz, 1H), 7.23 (m, 1H), 7.08 (s, 1H), 7.03 (dd, J = 1.7, 8.1 Hz, 1H), 2.31 (s, 3H). MS (ES-) 311 (M - H+).
5-(4-Chloro-3-methylphenyl)-1-phenyl-N-{1,3,3-trimethylbicyclo[2.2.1]heptan-2-yl}-1H-pyrazole-3-carboxamide (6)
To a stirred suspension of 40 (0.092 g, 0.00029 mol) in anhydrous toluene (6 mL) was added dropwise SOCl2 (0.044 mL, 0.00060 mol) at room temperature under nitrogen. The reaction mixture was heated at reflux under nitrogen for 90 min and then concentrated. The crude acid chloride was dissolved in CH2Cl2 (5 mL) and treated dropwise with a solution of (-)-fenchylamine 24 (0.054 g, 0.00035 mol) in CH2Cl2 (1 mL) followed by Et3N (0.12 mL, 0.00086 mol) at room temperature under nitrogen. After stirring at room temperature for 1 h, the reaction mixture was partitioned between CH2Cl2 and saturated NaHCO3. The organic phase was separated, dried (MgSO4), filtered, and concentrated to give the crude amide which was purified by flash chromatography over silica (12 g) with a hexane–ethyl acetate gradient (0 – 50% EtOAc) to give a solid which was dried at 65 °C under vacuum to give 0.081 g (62%) of 6 as an off-white amorphous solid. 1H NMR (300 MHz, CDCl3): δ 7.39 (m, 3H), 7.33 (m, 2H), 7.25 (m, 1H), 7.12 (s, 1H), 7.07 (m, 1H) 7.01 (s, 1H), 6.90 (dm, J = 8.4 Hz, 1H), 3.84 (d, J = 9.9 Hz, 1H), 2.31 (s, 3H), 1.75 (m, 3H), 1.46 (m, 2H), 1.26 (d, J = 9.8 Hz, 2H), 1.18 (s, 3H), 1.12 (s, 3H), 0.87 (s, 3H). 13C NMR (300 MHz, CDCl3): δ 162.30, 147.48, 143.86, 139.56, 136.50, 134.88, 131.14, 129.20, 129.03, 128.43, 128.20, 127.35, 125.48, 108.11, 63.14, 48.64, 48.21, 42.75, 39.54, 30.96, 27.37, 26.01, 21.30, 20.00, 19.68. MS (ES+) 448 (M + H+), 470 (M + Na+).
Ligand Binding Assay
Test compounds were evaluated in competitive binding assays vs. [3H]CP-55,940, utilizing membranes prepared from cells transfected with human CB2 receptors (Perkin Elmer LAS, Boston, MA). Dilutions of the compounds were prepared in assay buffer consisting of 50 mM Tris-HCl, pH 7.4, 1 mM EDTA, 3 mM MgCl2, and 0.5% (w/v) bovine serum albumin (BSA) at concentrations ranging from approximately 0.05 nM to 100 uM. The assays were conducted in a total volume of 0.5 mL in 1.2 mL polypropylene test tubes. Reaction mixtures (in triplicate) consisted of test compound, 0.72 nM [3H]CP-55,940, and 80 pM receptor preparation, all in assay buffer. Unlabeled CP-55,940 for determination of nonspecific binding was at a final concentration of 10 µM. After mixing by vortex, the reaction mixtures were incubated at 30°C for 1 h. When the incubation period was complete, the reaction was terminated by vacuum filtration of the reaction mixture. Separation of bound and free radioactivity was accomplished by filtration though a 96-well shallow GF/C filter plate (Perkin Elmer LAS) and a 96-manifold cell harvester (Brandel, Gaithersburg, MD) prepared by priming approximately 1 L of cold 50 mM Tris-HCl, pH 7.4, containing 0.1% (w/v) BSA wash buffer through the harvester. Reaction tubes were then rinsed with approximately 3 mL of wash buffer each. The filter plate was removed from the harvester and allowed to dry thoroughly. Approximately 50 µL of MicroScinät 20 liquid scintillation cocktail (Packard, Meriden, CT) was added to each plate well, which was then sealed and counted in a liquid scintillation counter.
Measurements of cAMP Accumulation
Materials
Dulbecco’s Modified Eagles’s Medium (DMEM), penicillin/streptomycin, L-glutamine, trypsin, and geneticin were purchased from Mediatech (Manassas, VA). Fetal bovine serum was obtained from Atlanta Biologicals (Lawrenceville, GA). Glass tubes used for cAMP accumulation assays were obtained from Kimble Chase (Vineland, NJ). These tubes were silanized by exposure to dichlorodimethylsilane (Sigma-Aldrich, St. Louis, MO) vapor for 3 h under vacuum. 384-well, round bottom, low volume white plates were purchased from Grenier Bio One (Monroe, NC). The cell-based HTRF cAMP HiRange assay kits were purchased from CisBio International (Bedford, MA). Forskolin was obtained from Sigma (St. Louis, MO). Ro 20–1724 was purchased from Enzo Life Sciences (Farmingdale, NY).
Cell Transfection and Culture
Human Embryonic Kidney 293 (HEK293) cells (purchased from ATCC, Manassas, VA) were maintained in DMEM containing 10% fetal bovine serum, 2 mM glutamine, 100 units/ml penicillin, and 100 mg/ml streptomycin in a humidified atmosphere consisting of 5% CO2 at 37°C. Expression plasmid containing CB2 receptor was stably transfected into HEK293 cells using lipofectamine, according to manufacturer’s instructions. Stably transfected cells were selected in culture medium containing 800 mg/ml geneticin and maintained in growth medium containing 400 mg/ml of geneticin (G418) until needed for experiments.
Cell-based HTRF cAMP assay
Cellular cAMP levels were measured using reagents supplied by Cisbio International (HTRF HiRange cAMP kit). Cultured cells were washed twice with phosphate-buffered saline (8.1 mM NaH2PO4, 1.5 mM KH2PO4, 138 mM NaCl, and 2.7 mM KCl, pH 7.2), and then dissociated in phosphate-buffered saline containing 1 mM EDTA. Dissociated cells were collected by centrifugation for 5 min at 2000g. The cells were resuspended in cell buffer (DMEM plus 0.2 % fatty acid free bovine serum albumin) and centrifuged a second time at 2000g for 5 min at 4°C. Subsequently, the cells were resuspended in an appropriate final volume of cell buffer plus the phosphodiesterase inhibitor Ro 20–1724 (2 mM).
CP-55940 was diluted in drug buffer (DMEM plus 2.5 % fatty acid free bovine serum albumin and 2 mM forskolin) and added to the assay plate at 5 ml per well. Cells were pretreated for 20 minutes with vehicle, or antagonists 1, 2, 3, 4, 5, 6. Subsequently, 5000 cells were added at 5ml per well into 384-well, round bottom, low volume white plates (Grenier Bio One, Monroe, NC). Following incubation of cells with CP-55940 for 7 minutes at room temperature, d2-conjugated cAMP and Europium cryptate-conjugated anti-cAMP antibody were added to the assay plate at 5 ml per well. After 2 hour incubation at room temperature, the plate was read on a TECAN GENious Pro microplate reader with excitation at 337 nm and emissions at 665 nm and 620 nm.
Data Analysis
For ligand binding assays, the amount of radiolabel specifically bound in the absence of competing compounds was calculated by subtracting nonspecific binding from total binding. The percentage of this specific binding was then calculated for the amount of radiolabel bound in the presence of various concentrations of the competing compound. Data were analyzed using GraphPad Prism© (GraphPad Software, Inc., San Diego, CA), which fit the displacement data to a one-site binding model using a goodness-of-fit quantification based on sum-of-squares and calculated the Ki for each competing compound.
For cAMP accumulation assays, data analyses were performed based on the ratio of fluorescence intensity of each well at 620 nm and 665 nm. Data are expressed as delta F%, which is defined as [(standard or sample ratio – ratio of the negative control) / ratio of the negative control] × 100. The standard curves were generated by plotting delta F% versus cAMP concentrations using non-linear least squares fit (Prism software, GraphPad, San Diego, CA). Unknowns are determined from the standard curve as nanomolar concentrations of cAMP. The data from the cAMP assays were expressed as percentage of forskolin-stimulated cAMP accumulation. The sigmoidal concentration-response equations were used (via GraphPad Prism) to determine EC50 and Emax values of the tested compounds
Modeling
Model Development
The model of the CB2 receptor employed here is based on the crystal structure of the Class A GPCR, rhodopsin in the dark state.55 The rhodopsin structure was chosen as the template because no modifications were made to its structure in order to crystallize it. In addition, rhodopsin shares some unusual sequence motifs with the cannabinoid receptors, including the GWNC motif at the extracellular (EC) end of TMH4 that forms an aromatic stacking interaction with Y5.39. This interaction affects the EC positions of TMH3-4-5 and is therefore important for CB2. We calculated low free energy conformations for any TMH with an important sequence divergence from rhodopsin. This would be TMH2 (GG helix distorting motif in Rho vs. no Pro or GG in CB2) and TMH5 (Pro at 5.50 in Rho vs.no Pro at 5.50 in CB2). The resultant model has been tested by substituted cysteine accessibility studies to identify residues that face into the CB2 binding pocket; 56, 57 by mutation to identify key ligand interactions sites;24, 56–58 and, by covalent labeling of CB2 59 to identify the method of ligand entry into the CB2 binding pocket.
The CB2 inactive state receptor model used in docking and MD studies here was pre-equilibrated in a stearoyl-docosahexaenoylphosphatidylcholine (SDPC) bilayer for 300ns to allow it to adjust to a lipid environment. 21 Figure S-2 in Supporting Information presents the RMSD plot for this simulation. Here it is clear that the CB2 TMH bundle stabilized early (~50 ns) into this trajectory. For ligand docking and MD studies of ligand binding pocket described below, only the resultant equilibrated CB2 model was used (i.e., no explicit lipid was present). These studies were performed at a dielectric of 1 to mimic the lipid non-polar environment. This substitution of a low dielectric for explicit lipids was made because Glide, for example, treats the receptor protein and ligand only. Subsequent energy minimizations of the receptor/ligand complexes restrain helix backbone phi and psi angles, allowing for some flexibility only in the final stage of minimization. Susequent MD simulations of the Glide generated receptor/ligand complexes using the dynamics module of Macromodel 9.1 (see below) apply restraints to helix backbones to preserve the global fold of the receptor, while allowing all amino acid sidechains and all torsion angles of the ligand to vary. As is clear in Figure S-3 in Supporting Information, the receptor model (including amino acid sidechains) maintains a low RMSD throughout the 22.5 ns simulations.
Conformer analysis of 1 and its analogs
Complete conformational analyses of 1 and its analogs were performed using ab initio Hartree-Fock calculations at the 6-31G* level, within the Spartan molecular modeling program (Wavefunction, Inc., Irvine, CA). HF 6-31G* 6-fold conformer searches were performed for all the rotatable bonds (Compounds 1, 3 and 5: C2-C3, N4-C5, N2’-C3’, C3’-C4’, and C3’’-C4’’ bonds; Compound 6: C2-C3, N4-C5, N2’-C3’, and C3’’-C4’’ bonds; Compounds 2 and 4: C2-C3, C4-C5, N2’-C3’, and C3’’-C4’’ bonds). In each conformer search, local energy minima were identified by rotation of a subject torsion angle through 360 in 60 increments (6-fold search), followed by HF 6-31G* energy minimization of each rotamer generated. To calculate the difference in energy between the global minimum energy conformer of each compound and its final docked conformation, the rotatable bonds in the global minimum energy conformer were driven to the their corresponding value in the final docked conformation and the single point energy of the resultant structure was calculated at the HF 6-31G* level. This energy difference for each compound is provided in the Results section and in Supporting Information (see Conformational Energy listing for each compound).
Electrostatic potential map calculation
The electrostatic potential density surface of the ligands was calculated using Spartan ‘08 (Wavefunction, Inc., Irvine, CA). The electrostatic potential energy was calculated using the Hartree-Fock method at the 6-31G* energy level of theory and was mapped on the 0.002 isodensity surface of each ligand. The electron-rich regions are colored red and the electron-poor regions are colored blue.
Docking of ligands
The lowest free energy conformer of 1 was docked in our inactive state model of the CB2 receptor using the automatic docking program Glide (Schrodinger Inc.). Glide was used to generate a grid based on the centroid of select residues in the binding pocket. The box size was set to the default value of 30×30×30 Angstroms, with the inner box size was set to 10×10×10 Angstroms. This default box size encompasses the entire CB2 binding pocket both in width and depth. Standard precision (SP) and flexible docking with ring sampling were selected for the docking setup. The torsions of the amide and bonds connecting it to the bicyclic ring were constrained to prevent the generation of high energy conformations during the automatic docking. Docking of the global minima of compounds 2–5 began from the Glide identified docking position of 1 followed by energy minimization. The analog 6 which lacks the “elbow” methylene of 1 and has a phenyl ring attached on the nitrogen (N-2’) of the pyrazole ring was docked using Glide in the defined binding pocket of 1 using the same protocol as described above.
Receptor-ligand complex minimization
The receptor-ligand complexes were minimized using the OPLS2005 all atom force field in Macromodel 9.1 (Schrodinger Inc.). An 8.0-Å nonbonded cutoff (updated every 10 steps), a 20.0-Å electrostatic cutoff, and a 4.0-Å hydrogen bond cutoff were used in each stage of the calculation. The first stage consisted of 20000 steps of conjugate gradient minimization using a distance-dependent dielectric coefficient of 1. No harmonic constraints were placed on the side chains, but there were constraints applied to hold the backbone phi/psi torsion angles of the helices (1000 kcal/mol force constant). During the second stage, backbone torsional constraints were reduced to 50 kcal/mol, and the minimization allowed to converge to a gradient of 0.01 kcal/mol·A2. To relax the loops, a 1000 step Polak-Ribier conjugate gradient minimization of the loop regions was performed. The loop and termini regions were unrestrained while the transmembrane regions were not allowed to move during this final minimization. An 8.0 Å extended nonbonded cutoff (updated every 10 steps), 20.0 Å electrostatic cutoff, and 4.0 Å hydrogen bond cutoff were used in this calculation, and the generalized Born/surface area (GB/SA) continuum solvation model for water available in Macromodel was employed.
Receptor-ligand molecular dynamics
The receptor-ligand complexes were further studied using molecular dynamics. The docking poses for each compound were subjected to the following minimization/dynamics protocol. For each pose the OPLS2005 force field was utilized with a distance dependent dielectric (coefficient of 1 to match the docking studies). The extended non-bonded treatment was employed, as in the minimization procedure discussed above. The dynamics module of Macromodel 9.1 was invoked, using stochastic dynamics at 300K with the use of SHAKE constraints for bonds to hydrogen allowing a 1.5fs timestep. Each system was first minimized for 500 steps with restraints on all the heavy atoms, using a large force constant of 4184.kJ/mol. The molecular dynamics was then initialized to 300K and an initial 100ps of molecular dynamics was run. Subsequently these restraints were slowly released for the side chain heavy atoms (4184kJ/mol to 0.05kJ/mol halving in each step for a total of 16 steps), at each step 150ps of dynamics was performed. Finally, for each complex, a 22.5ns molecular dynamics simulations was conducted. Since the goal of these simulations was to explore the dynamic behavior of each ligand in the binding pocket, in these MD simulations only the amino acid side chains and the ligand were free to move. Hydrogen bonding analysis for D275 to the amide moiety in 1 and its analogs was performed in VMD 60 using a 3.5 Å heavy atom to heavy atom cutoff and an angle of 60°. The results were averaged over the 22.5ns trajectory.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by NIDA grants DA003934 and DA021358 to PHR.
ABBREVIATIONS USED
- CB2
cannabinoid receptor sub-type 2
- CNS
central nervous system
- T-stack
aromatic ring T stacking interaction
- MD
molecular dynamics
- TMH
transmembrane helix
- DRY motif
ASP-ARG-TYR sequence motif on transmembrane helix 3
- NPXXY motif
ASN-PRO-X-X-TYR amino acid sequence motif on transmembrane helix 7, where X can be any amino acid
- EC
extracellular
- EC-n
extracellular loop n
- χ1
the torsion angle formed by N backbone-C-alpha backbone-C beta sidechain-C gamma-sidechain
- g−
torsion angle value of +60°
Footnotes
SUPPORTING INFORMATION AVAILABLE: Additional figures and ligand/receptor interaction energy tables. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Galiegue S, Mary S, Marchand J, Dussossoy D, Carriere D, Carayon P, Bouaboula M, Shire D, Le Fur G, Casellas P. Expression of central and peripheral cannabinoid receptors in human immune tissues and leukocyte subpopulations. Eur J Biochem. 1995;232:54–61. doi: 10.1111/j.1432-1033.1995.tb20780.x. [DOI] [PubMed] [Google Scholar]
- 2.Howlett AC, Barth F, Bonner TI, Cabral G, Casellas P, Devane WA, Felder CC, Herkenham M, Mackie K, Martin BR, Mechoulam R, Pertwee RG. International Union of Pharmacology. XXVII. Classification of cannabinoid receptors. Pharmacol Rev. 2002;54:161–202. doi: 10.1124/pr.54.2.161. [DOI] [PubMed] [Google Scholar]
- 3.Benito C, Nunez E, Tolon RM, Carrier EJ, Rabano A, Hillard CJ, Romero J. Cannabinoid CB2 receptors and fatty acid amide hydrolase are selectively overexpressed in neuritic plaque-associated glia in Alzheimer’s disease brains. J Neurosci. 2003;23:11136–11141. doi: 10.1523/JNEUROSCI.23-35-11136.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van Sickle MD, Duncan M, Kingsley PJ, Mouihate A, Urbani P, Mackie K, Stella N, Makriyannis A, Piomelli D, Davison JS, Marnett LJ, Di Marzo V, Pittman QJ, Patel KD, Sharkey KA. Identification and functional characterization of brainstem cannabinoid CB2 receptors. Science. 2005;310:329–332. doi: 10.1126/science.1115740. [DOI] [PubMed] [Google Scholar]
- 5.Buckley NE, McCoy KL, Mezey E, Bonner T, Zimmer A, Felder CC, Glass M. Immunomodulation by cannabinoids is absent in mice deficient for the cannabinoid CB(2) receptor. Eur J Pharmacol. 2000;396:141–149. doi: 10.1016/s0014-2999(00)00211-9. [DOI] [PubMed] [Google Scholar]
- 6.Massi P, Fuzio D, Vigano D, Sacerdote P, Parolaro D. Relative involvement of cannabinoid CB(1) and CB(2) receptors in the Delta(9)-tetrahydrocannabinol-induced inhibition of natural killer activity. Eur J Pharmacol. 2000;387:343–347. doi: 10.1016/s0014-2999(99)00860-2. [DOI] [PubMed] [Google Scholar]
- 7.Jorda MA, Verbakel SE, Valk PJ, Vankan-Berkhoudt YV, Maccarrone M, Finazzi-Agro A, Lowenberg B, Delwel R. Hematopoietic cells expressing the peripheral cannabinoid receptor migrate in response to the endocannabinoid 2-arachidonoylglycerol. Blood. 2002;99:2786–2793. doi: 10.1182/blood.v99.8.2786. [DOI] [PubMed] [Google Scholar]
- 8.Kishimoto S, Gokoh M, Oka S, Muramatsu M, Kajiwara T, Waku K, Sugiura T. 2-arachidonoylglycerol induces the migration of HL-60 cells differentiated into macrophage-like cells and human peripheral blood monocytes through the cannabinoid CB2 receptor-dependent mechanism. J Biol Chem. 2003;278:24469–24475. doi: 10.1074/jbc.M301359200. [DOI] [PubMed] [Google Scholar]
- 9.Franklin A, Stella N. Arachidonylcyclopropylamide increases microglial cell migration through cannabinoid CB2 and abnormal-cannabidiol-sensitive receptors. Eur J Pharmacol. 2003;474:195–198. doi: 10.1016/s0014-2999(03)02074-0. [DOI] [PubMed] [Google Scholar]
- 10.Kishimoto S, Kobayashi Y, Oka S, Gokoh M, Waku K, Sugiura T. 2-Arachidonoylglycerol, an endogenous cannabinoid receptor ligand, induces accelerated production of chemokines in HL-60 cells. J Biochem (Tokyo) 2004;135:517–524. doi: 10.1093/jb/mvh063. [DOI] [PubMed] [Google Scholar]
- 11.McCoy KL, Matveyeva M, Carlisle SJ, Cabral GA. Cannabinoid inhibition of the processing of intact lysozyme by macrophages: evidence for CB2 receptor participation. J Pharmacol Exp Ther. 1999;289:1620–1625. [PubMed] [Google Scholar]
- 12.Idris AI, van‘t Hof RJ, Greig IR, Ridge SA, Baker D, Ross RA, Ralston SH. Regulation of bone mass, bone loss and osteoclast activity by cannabinoid receptors. Nat Med. 2005;11:774–779. doi: 10.1038/nm1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rinaldi-Carmona M, Barth F, Millan J, Derocq JM, Casellas P, Congy C, Oustric D, Sarran M, Bouaboula M, Calandra B, Portier M, Shire D, Breliere JC, Le Fur GL. SR 144528, the first potent and selective antagonist of the CB2 cannabinoid receptor. J Pharmacol Exp Ther. 1998;284:644–650. [PubMed] [Google Scholar]
- 14.Bouaboula M, Desnoyer N, Carayon P, Combes T, Casellas P. Gi protein modulation induced by a selective inverse agonist for the peripheral cannabinoid receptor CB2: implication for intracellular signalization cross-regulation. Mol Pharmacol. 1999;55:473–480. [PubMed] [Google Scholar]
- 15.Iwamura H, Suzuki H, Ueda Y, Kaya T, Inaba T. In vitro and in vivo pharmacological characterization of JTE-907, a novel selective ligand for cannabinoid CB2 receptor. J Pharmacol Exp Ther. 2001;296:420–425. [PubMed] [Google Scholar]
- 16.Ross RA, Brockie HC, Stevenson LA, Murphy VL, Templeton F, Makriyannis A, Pertwee RG. Agonist-inverse agonist characterization at CB1 and CB2 cannabinoid receptors of L759633, L759656, and AM630. Br J Pharmacol. 1999;126:665–672. doi: 10.1038/sj.bjp.0702351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lavey BJ, Kozlowski JA, Hipkin RW, Gonsiorek W, Lundell DJ, Piwinski JJ, Narula S, Lunn CA. Triaryl bis-sulfones as a new class of cannabinoid CB2 receptor inhibitors: identification of a lead and initial SAR studies. Bioorg Med Chem Lett. 2005;15:783–786. doi: 10.1016/j.bmcl.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 18.Shankar BB, Lavey BJ, Zhou G, Spitler JA, Tong L, Rizvi R, Yang DY, Wolin R, Kozlowski JA, Shih NY, Wu J, Hipkin RW, Gonsiorek W, Lunn CA. Triaryl bis-sulfones as cannabinoid-2 receptor ligands: SAR studies. Bioorg Med Chem Lett. 2005;15:4417–4420. doi: 10.1016/j.bmcl.2005.07.023. [DOI] [PubMed] [Google Scholar]
- 19.Pasquini S, Ligresti A, Mugnaini C, Semeraro T, Cicione L, De Rosa M, Guida F, Luongo L, De Chiaro M, Cascio MG, Bolognini D, Marini P, Pertwee R, Maione S, Di Marzo V, Corelli F. Investigations on the 4-quinolone-3-carboxylic acid motif. 3. Synthesis, structure-affinity relationships, and pharmacological characterization of 6-substituted 4-quinolone-3-carboxamides as highly selective cannabinoid-2 receptor ligands. J Med Chem. 2010;53:5915–5928. doi: 10.1021/jm100123x. [DOI] [PubMed] [Google Scholar]
- 20.Lunn CA. Updating the chemistry and biology of cannabinoid CB2 receptor-specific inverse agonists. Curr Top Med Chem. 2010;10:768–778. doi: 10.2174/156802610791164193. [DOI] [PubMed] [Google Scholar]
- 21.Hurst DP, Grossfield A, Lynch DL, Feller S, Romo TD, Gawrisch K, Pitman MC, Reggio PH. A lipid pathway for ligand binding is necessary for a cannabinoid G protein-coupled receptor. J Biol Chem. 2010;285:17954–17964. doi: 10.1074/jbc.M109.041590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song ZH, Bonner TI. A lysine residue of the cannabinoid receptor is critical for receptor recognition by several agonists but not WIN55212-2. Mol Pharmacol. 1996;49:891–896. [PubMed] [Google Scholar]
- 23.Hurst DP, Lynch DL, Barnett-Norris J, Hyatt SM, Seltzman HH, Zhong M, Song ZH, Nie J, Lewis D, Reggio PH. N-(piperidin-1-yl)-5-(4-chlorophenyl)-1-(2,4-dichlorophenyl)-4-methyl-1H-p yrazole-3-carboxamide (SR141716A) interaction with LYS 3.28(192) is crucial for its inverse agonism at the cannabinoid CB1 receptor. Mol Pharmacol. 2002;62:1274–1287. doi: 10.1124/mol.62.6.1274. [DOI] [PubMed] [Google Scholar]
- 24.Tao Q, McAllister SD, Andreassi J, Nowell KW, Cabral GA, Hurst DP, Bachtel K, Ekman MC, Reggio PH, Abood ME. Role of a conserved lysine residue in the peripheral cannabinoid receptor (CB2): evidence for subtype specificity. Mol Pharmacol. 1999;55:605–613. [PubMed] [Google Scholar]
- 25.Seltzman HH, Foster MC, Wyrick CD, Burgess JP, Carroll FI. Tritiation of the cannabinoid receptor antagonist SR144528 involving lithium aluminum tritide reduction; assessment of the kinetic isotope effect by 3H-NMR. Journal of Labelled Compounds and Radiopharmaceuticals. 2005;48:589–596. [Google Scholar]
- 26.Barth F, Casellas P, Millan J, Qustric D, Rinaldi M, Sarran M. 3-Pyrazolecarboxamide Derivaties Having Cannabinoid Receptor Affinity. Patent WO9721682. 1997 [Google Scholar]
- 27.Corey EJ, Tius MA. 1-Diphenylphosphono-1-Methoxymethyllithium, A Useful Reagent for the Synthesis of Aldehydes from Hindered Ketones. Tet. Lett. 1980;21:3535–3538. [Google Scholar]
- 28.Vedejs E, Peterson MJ. Topics in Stereochemistry. Vol. 21. John Wiley & Sons; 1994. Stereochemistry and Mechanism in the Wittig Reaction; pp. 1–157. [Google Scholar]
- 29.Hurst D, Lynch D, Barnett-Norris J, Hyatt S, Seltzman H, Zhong M, Song ZH NJ, Lewis D, Reggio P. N-(piperidin-1-yl)-5-(4-chlorophenyl)-1-(2,4-dichlorophenyl)-4-methyl-1H-pyrazole-3-carboxamide (SR141716A) interaction with LYS 3.28(192) is crucial for its inverse agonism at the cannabinoid CB1 receptor. Mol Pharmacol. 2002;62:1274–1287. doi: 10.1124/mol.62.6.1274. [DOI] [PubMed] [Google Scholar]
- 30.Di Santo R, Costi R, Massa S, Artico M. 4-Substituted 2-Phenyl- and 2-Phenyl-3-Aryl Pyrroles by Reaction of Tosyl Benzyl Isocyanide (TosBIC) with Michael Acceptors. Synth. Commun. 1995;25:795–802. [Google Scholar]
- 31.van Leusen AM, Wildeman J, Oldenziel OH. Base-Induced Cycloaddition of Sulfonylmethyl isocyanides to C,N Double Bonds. Synthesis of 1,5-Disubstituted and 1,4,5-Trisubstituted Imidazoles from Aldimines and Imidoyl Chlorides. Journal of Organic Chemistry. 1977;42:1153–1159. [Google Scholar]
- 32.Sisko J, Mellinger M, Sheldrake PW, Baine NH. α-TOSYLBENZYL ISOCYANIDE. Organic Synthesis. 2000;77:198–205. [Google Scholar]
- 33.Tarzia G, Duranti A, Tontini A, Piersanti G, Mor M, Rivara S, Plazzi PV, Park C, Kathuria S, Piomelli D. Design, Synthesis, and Structure-Activity Relationships of Alkylcarbamic Acid Aryl Esters, a New Class of Fatty Acid Amide Hydrolase Inhibitors. J. Med. Chem. 2003;46:2352–2360. doi: 10.1021/jm021119g. [DOI] [PubMed] [Google Scholar]
- 34.Padwa A, Haffmanns G, Tomas M. Generation of Azomethine Ylides via the Desilylation Reaction of Immonium Salts. Journal of Organic Chemistry. 1984;49:3314–3322. [Google Scholar]
- 35.Kocienski PJ, Cernigliaro G, Feldstein G. A Synthesis of (+/−)-Methyl n-Tetradeca-trans-2,4,5-trienoate, an Allenic Ester Produced by the Male Dried Bean Beetle Acanthoscelides obtectus (Say) J. Org. Chem. 1977;42:353–355. [Google Scholar]
- 36.Haiduch J, Nam G, Ju Kim E, Frohlich R, Hanoverb JA, Kirka KL. A convenient synthesis of the C-1-phosphonate analogue of UDP-GlcNAc and its evaluation as an inhibitor of O-linked GlcNAc transferase (OGT) Carbohydrate Research. 2008;343:189–195. doi: 10.1016/j.carres.2007.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chevrier C, Le Nouën D, Defoin A, Tarnus C. Synthesis of Amino-L-Lyxose Phosphonates as Fucosyl-Phosphate Mimics. European Journal of Organic Chemistry. 2006:2384–2392. [Google Scholar]
- 38.Reichwein JF, Pagenkopf BL. A New Horner-Wadsworth-Emmons Type Coupling Reaction between Nonstabilized â-Hydroxy Phosphonates and Aldehydes or Ketones. Journal of the American Chemical Society. 2003;125:1821–1824. doi: 10.1021/ja027658s. [DOI] [PubMed] [Google Scholar]
- 39.Carman RM, Greenfield K. L. Aust. J. Chem. 1984;37:1785–1790. [Google Scholar]
- 40.Murray WV, Wachter MP. A Simple Regioelective Synthesis of Ethyl 1,5-Diarylpyrazole-3-carboxylates. J. Heterocycl. Chem. 1989;26:1389–1392. [Google Scholar]
- 41.Seltzman HH, Carroll FI, Burgess JP, Wyrick CD, Burch DF. Tritiation of SR141716 by metallation-iodination-reduction: tritium-proton nOe study. J. Label. Comp. Radiopharm. 2002;44:1–12. [Google Scholar]
- 42.Talley JJ, Penning TD, Collins PW, Rogier JDJ, Malecha JW, Miyashiro JM, Bertenshaw SR, Khanna IK, Graneto MJ, Rogers RS, Carter JS, Docter SH, Yu SS. Substituted Pyrazolyl Benzenesulfonamides for the Treatment of Inflammation. 1998 Jun 2;5,760,068 1998. [Google Scholar]
- 43.Feng W, Song ZH. Effects of D3.49A, R3.50A, and A6.34E mutations on ligand binding and activation of the cannabinoid-2 (CB2) receptor. Biochem Pharmacol. 2003;65:1077–1085. doi: 10.1016/s0006-2952(03)00005-4. [DOI] [PubMed] [Google Scholar]
- 44.Rhee MH, Nevo I, Levy R, Vogel Z. Role of the highly conserved Asp-Arg-Tyr motif in signal transduction of the CB2 cannabinoid receptor. FEBS Lett. 2000;466:300–304. doi: 10.1016/s0014-5793(00)01094-2. [DOI] [PubMed] [Google Scholar]
- 45.Rhee MH, Nevo I, Bayewitch ML, Zagoory O, Vogel Z. Functional role of tryptophan residues in the fourth transmembrane domain of the CB(2) cannabinoid receptor. J Neurochem. 2000;75:2485–2491. doi: 10.1046/j.1471-4159.2000.0752485.x. [DOI] [PubMed] [Google Scholar]
- 46.Nebane NM, Kellie B, Song ZH. The effects of charge-neutralizing mutation D6.30N on the functions of CB1 and CB2 cannabinoid receptors. FEBS Lett. 2006;580:5392–5398. doi: 10.1016/j.febslet.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 47.Feng W, Song ZH. Functional roles of the tyrosine within the NP(X)(n)Y motif and the cysteines in the C-terminal juxtamembrane region of the CB2 cannabinoid receptor. FEBS Lett. 2001;501:166–170. doi: 10.1016/s0014-5793(01)02642-4. [DOI] [PubMed] [Google Scholar]
- 48.Chin C, Lucas-Lenard J, Abadji V, Kendall DA. Ligand Binding and Modulation of Cyclic AMP Levels Depend on the Chemical Nature of Residue 192 of the Human Cannabinoid Receptor 1. J. Neurochem. 1998;70:366–373. doi: 10.1046/j.1471-4159.1998.70010366.x. [DOI] [PubMed] [Google Scholar]
- 49.Shire D, Calandra B, Bouaboula M, Barth F, Rinaldi-Carmona M, Casellas P, Ferrara P. Cannabinoid receptor interactions with the antagonists SR 141716A and SR 144528. Life Sci. 1999;65:627–635. doi: 10.1016/s0024-3205(99)00285-4. [DOI] [PubMed] [Google Scholar]
- 50.Zhang Y, Xie Z, Wang L, Schreiter B, Lazo JS, Gertsch J, Xie XQ. Mutagenesis and computer modeling studies of a GPCR conserved residue W5.43(194) in ligand recognition and signal transduction for CB2 receptor. Int Immunopharmacol. 2011;11:1303–1310. doi: 10.1016/j.intimp.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gouldson P, Calandra B, Legoux P, Kerneis A, Rinaldi-Carmona M, Barth F, Le Fur G, Ferrara P, Shire D. Mutational analysis and molecular modelling of the antagonist SR 144528 binding site on the human cannabinoid CB(2) receptor. Eur J Pharmacol. 2000;401:17–25. doi: 10.1016/s0014-2999(00)00439-8. [DOI] [PubMed] [Google Scholar]
- 52.Ballesteros JA, Deupi X, Olivella M, Haaksma EE, Pardo L. Serine and threonine residues bend alpha-helices in the chi(1) = g(-) conformation. Biophys J. 2000;79:2754–2760. doi: 10.1016/S0006-3495(00)76514-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kapur A, Hurst DP, Fleischer D, Whitnell R, Thakur GA, Makriyannis A, Reggio PH, Abood ME. Mutation Studies of Ser7.39 and Ser2.60 in the Human CB1 Cannabinoid Receptor: Evidence for a Serine-Induced Bend in CB1 Transmembrane Helix 7. Mol Pharmacol. 2007;71:1512–1524. doi: 10.1124/mol.107.034645. [DOI] [PubMed] [Google Scholar]
- 54.Paquette LA, Doehner RF., Jr. Synthesis of Optically Active Triazolinediones and Examination of their Utility for Inducing Asymmetry in Diels-Alder Cycloaddition Reactions. J. Org. Chem. 1980;45:5105–5113. [Google Scholar]
- 55.Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 56.Zhang R, Hurst DP, Barnett-Norris J, Reggio PH, Song ZH. Cysteine 2.59(89) in the second transmembrane domain of human CB2 receptor is accessible within the ligand binding crevice: evidence for possible CB2 deviation from a rhodopsin template. Mol Pharmacol. 2005;68:69–83. doi: 10.1124/mol.104.007823. [DOI] [PubMed] [Google Scholar]
- 57.Nebane NM, Hurst DP, Carrasquer CA, Qiao Z, Reggio PH, Song ZH. Residues accessible in the binding-site crevice of transmembrane helix 6 of the CB2 cannabinoid receptor. Biochemistry. 2008;47:13811–13821. doi: 10.1021/bi8007802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Song ZH, Slowey CA, Hurst DP, Reggio PH. The difference between the CB(1) and CB(2) cannabinoid receptors at position 5.46 is crucial for the selectivity of WIN55212-2 for CB(2) Mol Pharmacol. 1999;56:834–840. [PubMed] [Google Scholar]
- 59.Pei Y, Mercier RW, Anday JK, Thakur GA, Zvonok AM, Hurst D, Reggio PH, Janero DR, Makriyannis A. Ligand-binding architecture of human CB2 cannabinoid receptor: evidence for receptor subtype-specific binding motif and modeling GPCR activation. Chem Biol. 2008;15:1207–1219. doi: 10.1016/j.chembiol.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.