Abstract
The galanin 3 receptor (GalR3) belongs to the large G protein–coupled receptor (GPCR) family of proteins. GalR3 and two other closely related receptors, GalR1 and GalR2, together with their endogenous ligand galanin, are involved in a variety of physiological and pathophysiological processes. GalR3 in particular has been strongly implicated in addiction and mood-related disorders such as anxiety and depression. It has been the target of many drug discovery programs within the pharmaceutical industry, but despite the significant resources and effort devoted to discovery of galanin receptor subtype selective small molecule modulators, there have been very few reports for the discovery of such molecules. GalR3 has proven difficult to enable in cell-based functional assays due to its apparent poor cell surface expression in recombinant systems. Here, we describe the generation of a modified GalR3 that facilitates its cell surface expression while maintaining wild-type receptor pharmacology. The modified GalR3 has been used to develop a high-throughput screening–compatible, cell-based, cAMP biosensor assay to detect selective small molecule modulators of GalR3. The performance of the assay has been validated by challenging it against a test library of small molecules with known pharmacological activities (LOPAC; Sigma Aldrich). This approach will enable identification of GalR3 selective modulators (chemical probes) that will facilitate dissection of the biological role(s) that GalR3 plays in normal physiological processes as well as in disease states.
Introduction
Galanin is a neuropeptide of 29 amino acids (30 in human) that is derived from a 123–amino acid galanin precursor.1 It is widely expressed in the central nervous system (CNS)2–4 as well as in peripheral tissues including the gastrointestinal tract4 and pancreas.5 Galanin exerts its physiological actions through at least three galanin receptor subtypes, denoted GalR1, GalR2, and GalR3, that belong to the G protein–coupled receptor (GPCR) superfamily.6,7 Galanin together with its cognate receptors is involved in a broad range of physiological processes including cognition and memory,8 sensory/pain processing,9–11 feeding behavior,12,13 and neuronal survival and regeneration.14 Consequently, the galanin system is also involved in pathological conditions such as depression,15,16 Alzheimer's disease,17 diabetes,18 and drug addiction.19–21 In the brain, the galanin receptors have widespread, partially overlapping distribution as shown by radioligand binding3,22 and in situ hybridization studies.23,24 GalR3 is the least abundantly expressed of the galanin receptor subtypes and is most densely expressed in the hypothalamus,25 a brain region that regulates appetite and feeding.26 GalR3 has also been implicated in mood-related disorders such as anxiety and depression,27 alcoholism,26,28 and addiction.19,29 Studies suggest the rewarding effects of alcohol may be mediated by hippocampal GalR3,26 and single nucleotide polymorphisms in the GalR3 gene have been linked to an increased risk of alcoholism.26 Thus, development of selective GalR3 modulators may represent a novel pharmacological treatment for addiction and mood disorders.
While the signaling properties of GalR3 are poorly understood, the receptor has been found to stimulate inward K+ currents in a pertussis toxin–sensitive manner when co-expressed with GIRKs in Xenopus oocytes,30 suggesting that, like GalR1, GalR3 can couple to the Gi class of G proteins. Unfortunately, due to reasons addressed herein, GalR3 has until now proven difficult to enable in cell-based functional assays. As a result, the only available galanin receptor agonists are either of the peptide type such as galanin itself, a nonselective ligand acting as a full agonist at all three receptors, or nonpeptide type with relatively low affinity (micromolar) such as galnon and galmic, which are also nonselective.31,32 To date, there are only two “scaffolds” or “chemotypes” of GalR3-selective small molecule modulators described in the literature, the first of these is an indolone scaffold that the GalR3 antagonist SNAP37889 is derived from.27 This compound has served as a useful tool for probing GalR3 function in vivo and has shown antidepressant-like activity in several animal models27; however, it suffers from low solubility and poor oral bioavailability. The second series of GalR3 small molecule modulators appears exclusively in the patent literature and is based on a series of pyrimidine structures.33,34 These compounds also behave as antagonists and are only moderately active against GalR3, but they have shown efficacy in animal models of depression. With much room for improvement, these are currently the best-in-class GalR3 antagonists. Here we describe the development and optimization of a high-throughput screening (HTS)–compatible cell-based functional assay to identify GalR3 selective ligands for use as molecular probes to interrogate GalR3 receptor function in normal physiological processes and disease states.
Materials and Methods
Cell Culture and Transfection
All media and serum were purchased from Gibco, and plasticware from Falcon. HEK293 cells expressing the cAMP nucleotide-gated (CNG) channel, namely HEK293-CNG, (purchased from BD Biosciences, now available from Codex Biosystems) and stable cell lines derived from these cells were cultured in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum (FBS). Chinese hamster ovarian (CHO) cells were cultured in F12 media containing 10% FBS. Transfections were performed using Fugene HD (Roche) according to manufacturer's instructions. Selection antibiotics for maintenance of HEK293-CNG-GPCR stable cell lines were G418 (Thermo-Fisher) to maintain expression of the CNG channel, and hygromycin B (Invitrogen) to maintain GPCR expression, both 300 μg/mL. All cells were maintained in a humidified incubator at 37°C, 5% CO2 and passaged by splitting at a ratio of 1:10 such that confluency did not exceed 80%.
cDNA constructs
HA-GalR1 (pcDNA3.1) was purchased from UMR cDNA Resource Center; HA-GalR3 (pReciever-M06) was purchased from GeneCopoeia Inc. The GalR3/1ctR chimera was created by in-frame fusion of HA-GalR3 (residues 1–299) and the carboxy terminus of HA-GalR1 (residues 299–350). GalR3 1–299 was cut from the HA-GalR3 construct using Not1 and BsrB1 restriction endonucleases while GalR1 299–350 was amplified from HA-GalR1 by polymerase chain reaction (PCR) using forward primer 5′-ccg ctc att tat gca ttt ctc tct ga-3′ and reverse primer 5′-tca cac atg agt aca att ggt tga ctc gag-3′. A three-way ligation reaction was performed with the two fragments in the presence of pcDNA3.1 expression vector to generate the GalR3/1ctR chimera.
CNG cAMP biosensor assay.
The CNG cAMP biosensor assay is described in Table 1.
Table 1.
cAMP Nucleotide-Gated Channel cAMP Biosensor Assay
| Step | Parameter | Value | Description |
|---|---|---|---|
| 1 | Plate cells | 20 μL | 10,000 cells per well |
| 2 | Incubation | 18 h | 37°C, 5% CO2 |
| 3 | Dye load | 20 μL | ACT:One membrane potential dye |
| 4 | Incubation | 1 h | Room temperature, dark |
| 5 | Compound (antagonist) addition | 5 μL | From 10× compound plate |
| 6 | Incubation | 1 h | Room temperature, dark |
| 7 | Agonist challenge | 5 μL | Fluid transfer performed in FLIPR |
| 8 | Read fluorescence at time=0 | T0 | Use 510–545/565–625 bandpass FLIPR filter |
| 9 | Incubation | 30 min | Room temperature in FLIPR chamber |
| 10 | Read fluorescence at time=30 | T30 | Use 510–545/565–625 bandpass FLIPR filter |
Step Notes
1. BD Biosciences black clear bottom poly-d-lysine coated 384-well plate.
4. Generate compound plate during this step.
5. Final DMSO concentration in assay plate=0.7%. Made in PBS.
6. Generate agonist plate during this step.
7. Challenge with EC90 porcine galanin (American Peptide) for antagonist mode, or CRC of porcine galanin for control, all in the presence of EC90 of forskolin (MP Biochemicals), (10 μM in 10× plate; 1 μM final concentration), and PDE inhibitor (Sigma Aldrich) (250 μM in 10× plate; 25 μM final concentration), made in PBS, final DMSO concentration ∼0.7%.
10. The T30/T0 ratio is used to graph the concentration response of test compounds.
FLIPR, fluorescent imaging plate reader; DMSO, dimethyl sulfoxide; PBS, phosphate-buffered saline; EC90, 90% of maximal response; CRC, concentration response curve; PDE, phosphodiesterase.
Indirect Immunofluorescence
Forty-eight hours after transient transfection with HA-GalR3 or HA-GalR1, HEK293 cells were either fixed in 4% paraformaldehyde (PFA) only (Sigma Aldrich) or fixed and permeabilized in 4% paraformaldehyde and 0.3% Triton X-100 (Thermo Scientific) made up in phosphate-buffered saline (PBS) for 20 min. After 10-min quench in 0.1 M glycine (Sigma Aldrich)/PBS, cells were washed once with PBS and then blocked in 5% goat serum (Invitrogen)/PBS for 30 min. Primary antibody incubations were performed at a dilution of 1:300 in 5% goat serum/PBS for 1 h at room temperature. After five washes with 5% goat serum/PBS, secondary antibody incubations were also performed in 5% goat serum/PBS for 1 h at room temperature at a dilution of 1:700. After three more washes with 5% goat serum/PBS and three washes with PBS, cells were mounted using prolong gold (Invitrogen), sealed, and imaged by confocal microscopy.
Impedance Measurement
Impedance measurements were performed using the Cellkey system (Molecular Devices). This technology is based on applying an electrical current to cells within a microplate format and measuring changes in impedance. Receptor activation leads to changes in cell morphology, cell adherence and/or cell–cell interaction. These changes individually or collectively affect the flow of extracellular and transcellular current, leading to a decrease or an increase in impedance depending on the G protein–coupling preference of the receptor being activated.35 HEK293-CNG cells expressing GalR1, GalR3, or Gal3/1ctR were seeded at a density of 40,000 cells per well into Cellkey microplates (MDS SCIEX) and cultured overnight. The following day, cells were placed in the Cellkey instrument. Using on-board microfluidics, cells were washed three times with assay buffer (Hank's balanced salt solution containing 20 mM HEPES, pH 7.4 and 0.1% fatty acid free BSA [Sigma]) and allowed to equilibrate at room temperature for 30 min. After a 90-s read to establish a baseline, porcine galanin was added, and the changes in cellular impedance were measured over a 1000-s period. Individual values were exported at time (t)=600 s and CRCs were determined by nonlinear regression analysis using GraphPad Prism 5.0 software (GraphPad Software).
Results
GalR3 Expresses Poorly at the Plasma Membrane in Recombinant Systems
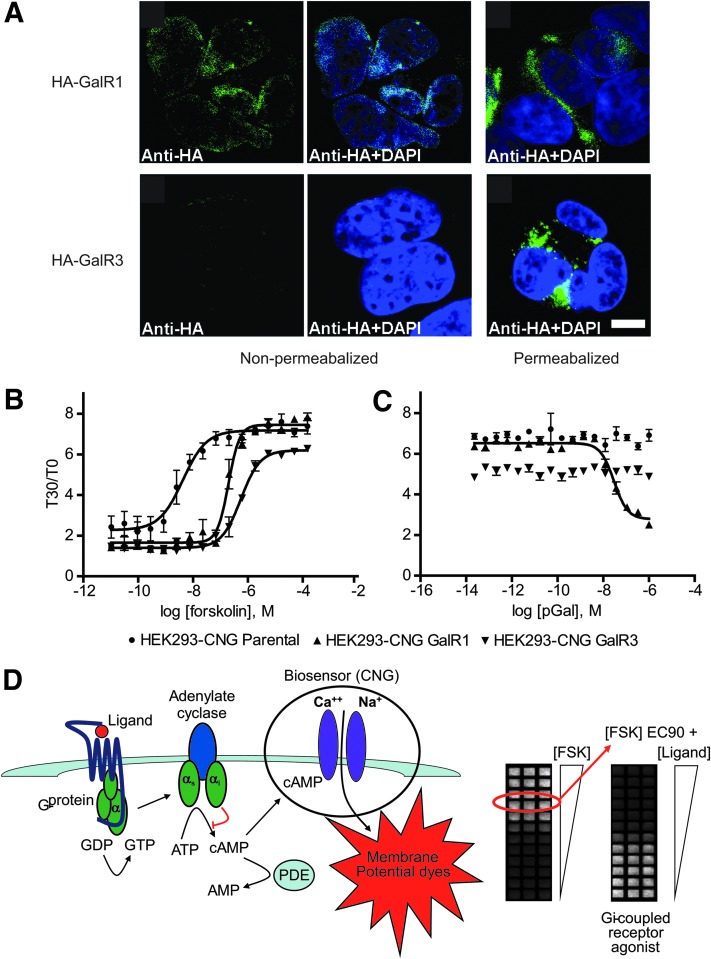
The fact that GalR3 has proven difficult to enable in cell-based functional assays is likely due to its poor expression at the plasma membrane in recombinant systems. Indirect immunofluorescence using an anti-HA antibody showed that HA-tagged GalR1 transiently transfected into HEK293 cells can traffic efficiently to the plasma membrane allowing antibody detection in both permeabilized and nonpermeabilized cells (Fig. 1A, top panel). In contrast, transient transfection of equivalent levels of HA-tagged GalR3 resulted in a predominantly intracellular staining pattern that could only be visualized if the cells were permeabilized (Fig. 1A, bottom right panel). Thus, GalR3 but not GalR1 fails to traffic efficiently to the plasma membrane. Similar data were observed in CHO and PC12 cells (data not shown).
Fig. 1.
Galanin 3 receptor (GalR3) expresses poorly at the plasma membrane in recombinant systems. (A) Forty-eight hours after transient transfection with HA-GalR3 or HA-GalR1, HEK293 cells were fixed and permeabilized (or not) as described in Materials and Methods. Fixed cells were incubated with anti-HA primary antibody (rat) (Roche) and then with anti-rat Alexafluor 588 secondary antibody (Invitrogen) before imaging by confocal microscopy. Scale bar=10 μm. (B) HEK293-CNG parental cells (●) or HEK293-CNG cells stably transfected with Gal1R (▲) or Gal3R (▼) were plated at 14,000 cells per well in a 384-well FLIPR plate. After 18-h incubation at 37°C, 5% CO2, cells were loaded with ACT:One membrane potential dye for 2 h at room temperature. cAMP production in response to increasing concentrations of forskolin was determined according to ACT:One manufacturer's instructions and as described in detail in Table 1. Forskolin EC90s were extrapolated from the concentration response curves for each cell line. (C) Cells were plated and dye-loaded as in (B) before treatment with an EC90 of forskolin together with increasing concentrations of porcine galanin. cAMP levels were determined as in (B). The data presented are means±SEM of triplicate wells (n=3). (D) cAMP biosensor assay principle. The cAMP biosensor assay utilizes a stable cell line expressing a modified cyclic nucleotide gated channel (CNG) acting as a biosensor for cAMP. Changes in cAMP can be measured with a combination of membrane potential dyes and a FLIPR. Gi-coupled GPCRs (e.g., GalR3) promote exchange of guanosine diphosphate (GDP) for guanosine triphosphate (GTP) in Gi α-subunits. Activated Gi acts to inhibit cAMP production at the effector adenylate cyclase, thus Gi-coupled GPCR activity is determined by inhibition of forskolin-stimulated cAMP production. Phosphodiesterases (PDEs), which catalyze the conversion of cAMP into adenosine monophosphate (AMP), are inhibited to maximize the assay window. GPCR, G protein–coupled receptor.
Consistent with its failure to traffic efficiently to the plasma membrane, GalR3 also failed to signal effectively in recombinant systems. We established a 384-well plate format cell-based functional assay to monitor galanin receptor activity using the ACT:One (Codex Biosystems) live cell cAMP technology. This assay utilizes a stable HEK-293 cell line co-expressing the receptor of interest and a modified cyclic nucleotide gated channel (CNG) acting as a biosensor for cAMP. Changes in cAMP are measured with a combination of membrane potential dyes and a fluorescent imaging plate reader (FLIPR); see Table 1 for detailed protocol and Figure 1D for a schematic illustrating the technology. To monitor the activity of Gi-coupled receptors such as the GalR3 or GalR1, a concentration of forskolin (a direct activator of adenylate cyclases) that produces 90% of its maximal response (EC90) is used to stimulate cAMP. Gi-coupled receptor agonist activity is then measured as the ability to inhibit the EC90 forskolin-stimulated cAMP response. HEK293-CNG parental cell line and HEK293-CNG cell lines stably expressing GalR1 or GalR3 were treated with increasing concentrations of forskolin for 30 min (Fig. 1B). In all cases a robust concentration-dependent cAMP response was observed in the cAMP biosensor assay and 1 μM was determined to be a suitable EC90 forskolin challenge for both the GalR1 and GalR3 expressing cell lines (Fig. 1B). Next we treated the cells with increasing concentrations of porcine galanin in the presence of 1 μM forskolin to monitor galanin-induced inhibition of the forskolin-stimulated cAMP response (Fig. 1C). The HEK293-CNG-GalR1 cells responded with an EC50 of 29 nM. However, we observed no response in the HEK293-CNG cells stably expressing wild-type (wt)GalR3 (Fig. 1C), consistent with its poor surface expression as observed in Figure 1A. Thus, GalR3, unlike GalR1, expresses poorly at the plasma membrane and fails to signal via Gi in recombinant systems.
Generation of a Gal3/1ctR Chimera to Enable Screening for Small Molecule Probes of GalR3
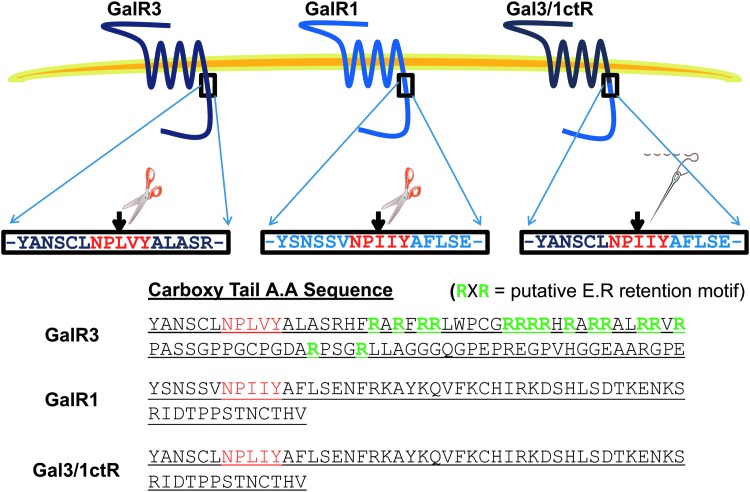
While the transmembrane regions of GalR1 and GalR3 are strongly conserved (42% identical), the carboxy tail region of the two receptors is weakly conserved (11% identical). We therefore reasoned that motifs contained within the carboxy tail of GalR3 but lacking in GalR1 might be responsible for its observed intracellular retention. On examination of the amino acid sequence of the GalR3 carboxy tail, we identified multiple overlapping putative endoplasmic reticulum (ER) retention motifs (RXR; Fig. 2). These motifs have been shown to regulate trafficking of a number of different GPCRs to the plasma membrane, including the GABAB receptor.36 Given that these motifs are absent from GalR1 (Fig. 2), we hypothesized that substitution of the carboxy tail of GalR3 with that of GalR1 to generate a modified GalR3, namely Gal3/1ctR, would rescue cell surface expression sufficiently to enable a cell-based functional assay to monitor Gal3/1ctR activity.
Fig. 2.
Generation of a Gal3/1ctR chimera. The Gal3/1ctR chimera, for which “1ctR” refers to the carboxy tail of GalR1, was generated by in-frame fusion of wild-type GalR3 (residues 1–299) with the carboxy tail of GalR1 (residues 299–350) as described in Materials and Methods. Removal of the arginine-rich putative endoplasmic reticulum (ER)-retention motifs (highlighted in green) found in the carboxy tail of wild-type GalR3 was proposed to enable plasma membrane expression of the receptor. The highly conserved characteristic NPxxY motif found in the seventh transmembrane domain of most GPCRs is highlighted in red.
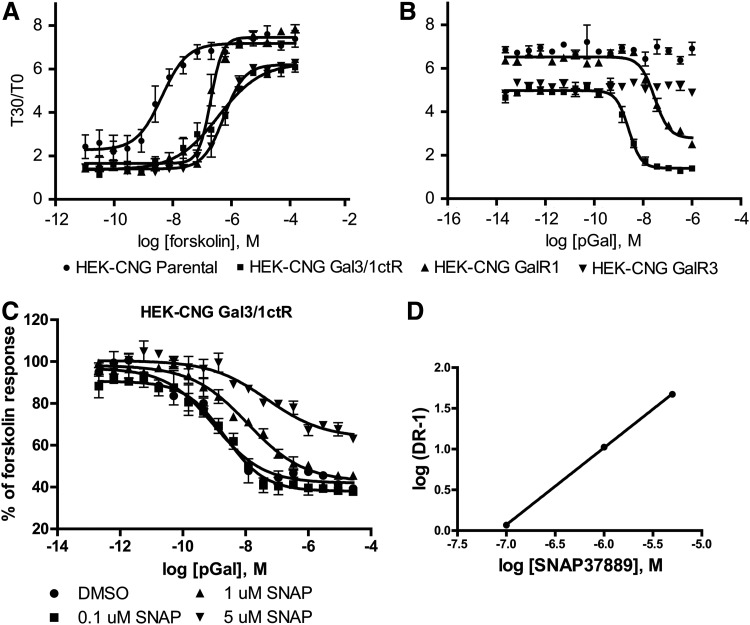
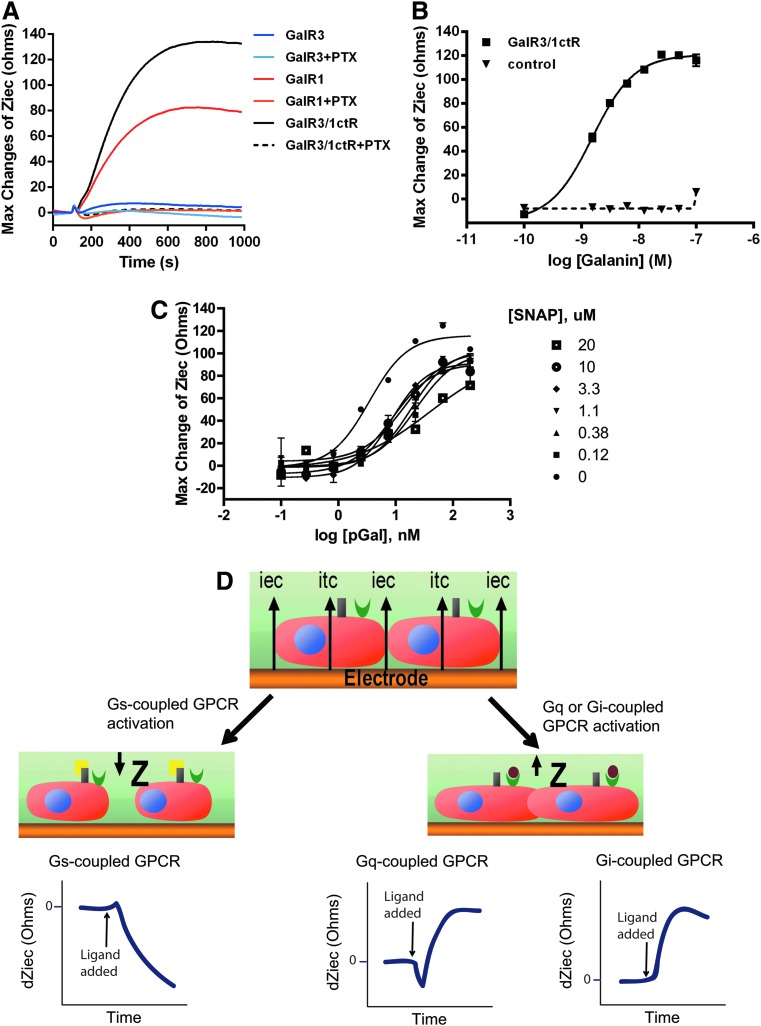
We generated the Gal3/1ctR construct as described in Figure 2 and introduced it into the HEK293-CNG cell line. HEK293-CNG-Gal3/1ctR cells responded robustly to forskolin in the cAMP biosensor assay and, as for wtGalR1 and wtGalR3, 1 μM was determined to be a suitable EC90 forskolin challenge (Fig. 3A). Next we treated HEK293-CNG-Gal3/1ctR cells with increasing concentrations of porcine galanin in the presence of 1 μM forskolin and observed a galanin response with a signal-to-background ratio (S:B) equivalent to that seen in HEK293-CNG cells stably expressing GalR1 (Fig. 3B, concentration response curves for wtGalR1 and wtGalR3 from Fig. 1 are reproduced here for comparison). The HEK293-CNG-Gal3/1ctR cells responded with an EC50 of 3 nM, while the HEK293-CNG-GalR1 cells responded with an EC50 of 29 nM, consistent with literature values.31 Then, using a label-free bioimpedance-based technology as described in Materials and Methods and illustrated in Figure 4D, we evaluated the effect of porcine galanin on HEK293-CNG-Gal3/1ctR cells, as well as on cells stably expressing wtGalR3, or the wtGalR1. In this label-free assay, stimulation of cells expressing either the Gal3/1ctR or the GalR1 with porcine galanin (50 nM) induced an increase in impedance that was consistent with the prototypical response profile for Gi-coupled receptors (Fig. 4A).35 In addition, the porcine galanin-induced increase in impedance was fully inhibited when cells were treated with the Gi inhibitor pertussis toxin (Fig. 4A), confirming that wtGalR1 and the Gal3/1ctR signal through Gi protein such that the pharmacology of the Gal3/1ctR is consistent with that of wtGalR3. In good agreement with our previous data, addition of porcine galanin had no effect on cells expressing wtGalR3 (Fig. 4A). Importantly, porcine galanin induced a concentration-dependent increase in cellular impedance in the HEK293-CNG-Gal3/1ctR cells with an EC50 value of 1.5 nM (Fig. 4B) that correlates well with the EC50 value we previously determined using the cAMP biosensor assay. We then investigated the effect of increasing concentrations of the GalR3-selective antagonist SNAP37889 on porcine galanin CRC in the HEK293-CNG-Gal3/1ctR cells using the cAMP biosensor assay and the bioimpedance-based assay. Despite its low potency and solubility, SNAP37889 causes a concentration-dependent rightward shift in the concentration response curve to galanin in both the cAMP biosensor assay (Fig. 3C) and the bioimpedance-based assay (Fig. 4C), further demonstrating GalR3 specificity. Schild regression analysis of galanin EC50 values in the presence of increasing concentrations of SNAP37889 obtained in the cAMP biosensor assay (Fig. 3C) afforded a line with slope close to unity (0.95±0.01, Fig. 3D). Constraining the slope to 1 afforded a pKb estimate of 7.08 (Kb=83 nM) in good agreement with the only previously published Kb value for SNAP37889 at wtGalR3 (Kb=29 nM).27 Together these data indicate that the HEK293-CNG-Gal3/1ctR cell line can be used to monitor small molecule modulation of GalR3 activity and as such enable the development of an HTS-compatible cell-based functional assay.
Fig. 3.
cAMP biosensor assay development. (A) HEK293-CNG parental cells (●) or HEK293-CNG cells stably transfected with the Gal3-1R chimera (■), Gal1R (▲), or Gal3R (▼) were plated at 14,000 cells per well in a 384-well FLIPR plate. After 18 h incubation at 37°C, 5% CO2, cells were loaded with ACT:One membrane potential dye for 2 h at room temperature. cAMP production in response to increasing concentrations of forskolin was determined according to ACT:One manufacturer's instructions and as described in detail in Table 1. Forskolin EC90s were extrapolated from the concentration response curves for each cell line. (B) Cells were plated and dye-loaded as in (A) before treatment with an EC90 of forskolin together with increasing concentrations of porcine galanin. cAMP levels were determined as in (B). (C) Cells were plated and dye-loaded as in (B) before treatment with an EC90 of forskolin together with increasing concentrations of porcine galanin in the presence of the indicated concentrations of SNAP37889 (SNAP). The data presented are means±SEM of triplicate wells (n=3). (D) Schild regression of the mean EC50 values from (C) affords a slope of 0.95±0.01 and, when constrained to a unit slope, a pKb value of 7.08 (Kb=83 nM).
Fig. 4.
Cellular dielectric spectroscopy. (A) Time-dependent cellular dielectric spectroscopy (CDS) profiles induced by porcine galanin in HEK293-CNG cells expressing either GalR3, GalR1, or the Gal3/1ctR chimera. After 90 s of baseline read, 50 nM of porcine galanin was added and responses were collected for 1000 s. Plotted are individual data points collected every 2 s, each trace is a representative of three separate experiments. Pretreatment with pertussis toxin (PTX; 0.1 μg/mL) inhibited galanin-induced increase in cellular impedance observed in cells expressing the GalR1 and the Gal3/1ctR chimera. (B) HEK293-CNG-Gal3/1ctR cells (■) were treated with a range of porcine galanin concentrations and impedance values were exported at t=600 s. CRCs yielded an EC50 value of 1.5 nM. No response was observed in HEK293-CNG cells (control, ▼) (C) HEK293-CNG-Gal3/1ctR cells were treated with a range of porcine galanin concentrations in the presence of the indicated concentrations of SNAP37889 (SNAP) and impedance values were exported at t=600 s. Addition of SNAP37889 resulted in a concentration-dependent right shift of galanin CRC. In (B) and (C), data presented are means±SEM of triplicate wells (n=3). (D) Principle of Cellkey system. A voltage is applied at a number of frequencies. At low frequencies the voltage induces extracellular currents (iec) that pass around the cells, whereas at high frequency they induce transcellular current (itc). Changes in impedance are reported kinetically for each well allowing time-dependent traces in response to ligand to be plotted, such as in (A). Activation of a Gq-coupled receptor or a Gi-coupled receptor leads to an increase in actin polymerization and will cause cells to move closer to the electrode and to one another generating an increase in impedance. In contrast, activation of a Gs-coupled receptor results in actin depolymerization and will cause cells to move away from the electrode and to one another generating a decrease in impedance. Thus, the time-dependent response profiles are indicative of Gi, Gq, or Gs coupling. Alternatively, impedance at a single time point can be exported from each well and plotted against a range of ligand concentrations to generate dose response curves as in (B) and (C).
Assay Optimization and Pilot Screen
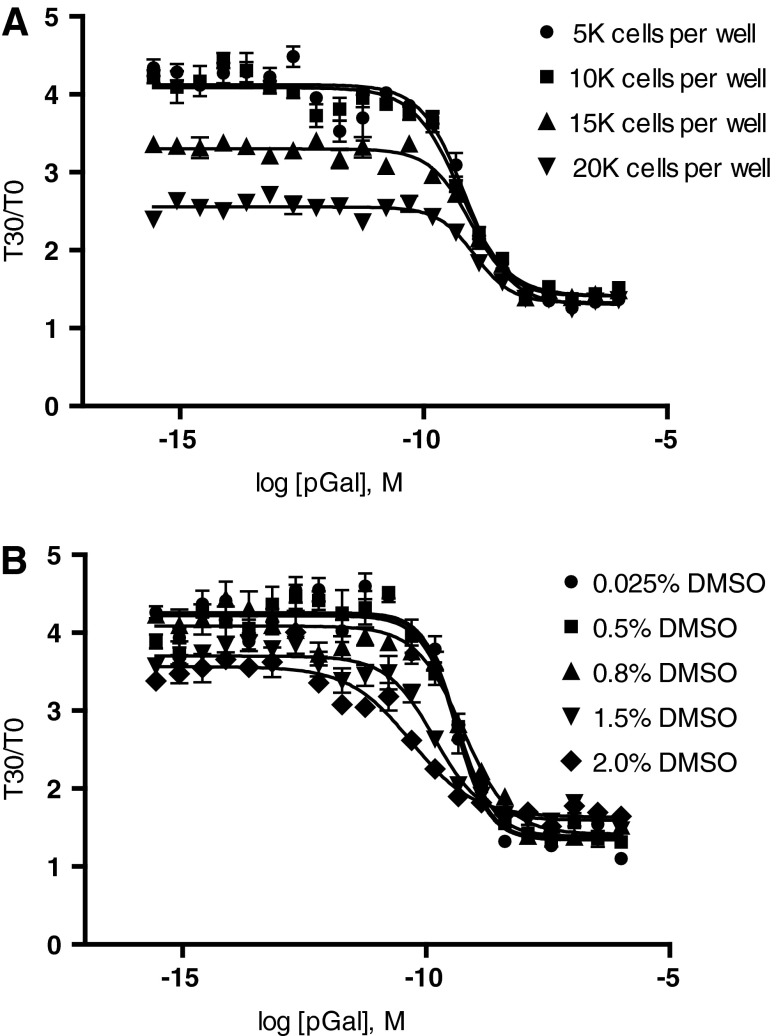
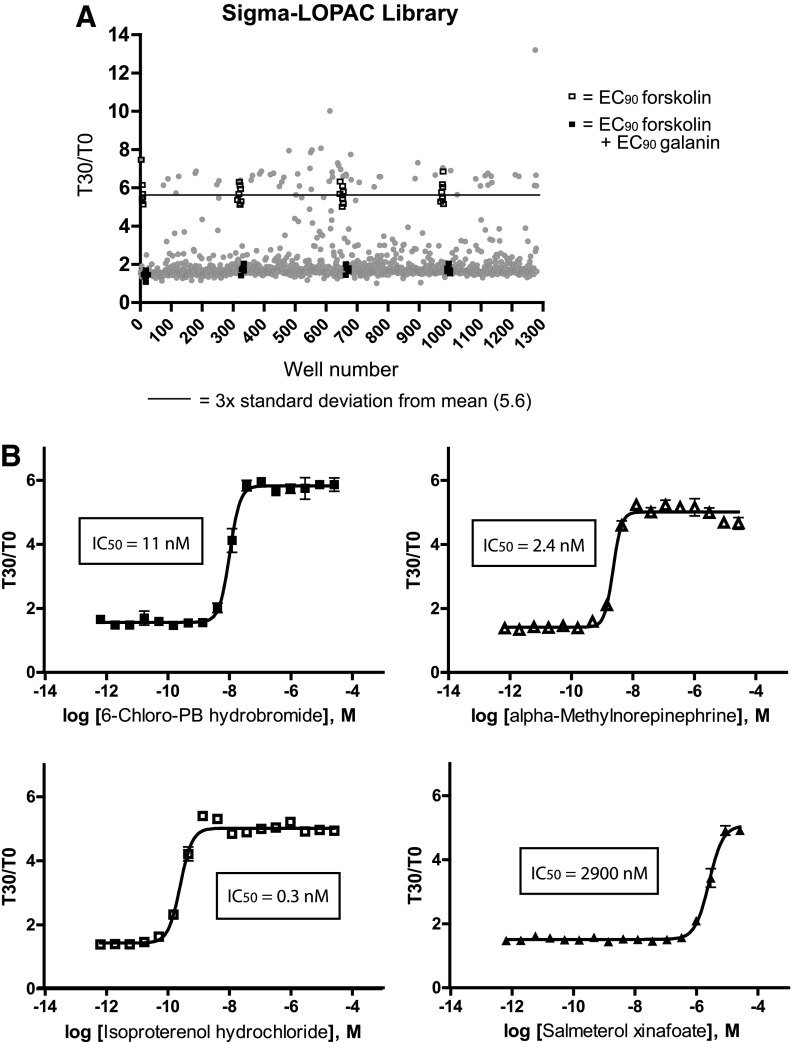
Initially we plated cells at a density of 14,000 cells/well per manufacturer's instructions. As seen in Figure 3B, this yielded a S:B of 2.8. Although this is suitable for HTS, for assay optimization purposes various cell densities were investigated (5,000, 10,000, 15,000, and 20,000 cells/well), and a cell density of 10,000 cells/well was identified as the optimal cell density based on the S:B and resulting pharmacology, allowing us to reduce the number of cells used without compromising the assay window. Plating at 10,000 cells/well resulted in a S:B of 2.9 and a galanin EC50 of 1 nM (Fig. 5A). These parameters were used in subsequent assay optimization. Most compound libraries are plated in 100% dimethyl sulfoxide (DMSO), therefore we next tested the DMSO tolerance of the cells in the cAMP biosensor assay. Cells were incubated with 1 μM forskolin and increasing concentrations of porcine galanin in the presence of varying concentrations of DMSO up to 2% (Fig. 5B). The Gal3/1ctR cAMP assay performance, both in terms of S:B and galanin EC50 values, was not significantly affected by DMSO concentrations up to 2% (Fig. 5B). Final DMSO concentration used in the HTS assay was 0.7%. Finally, to determine the performance of our optimized Gal3/1ctR assay in terms of robustness (Z′) under HTS conditions, the assay was screened against LOPAC (Library of Pharmacologically Active Compounds; Sigma Aldrich). Briefly, ∼1280 compounds were analyzed at a single concentration of 10 μM (0.7% DMSO) and challenged with an EC90 of forskolin together with an EC90 of porcine galanin in order to identify Gal3/1ctR antagonists (Fig. 6A). A scatterplot of all compound activities as well as the high (EC90 forskolin alone) and low (EC90 forskolin+EC90 galanin) controls is shown in Figure 6, demonstrating Z values >0.7 for the entire pilot screen. This is indicative of an excellent assay window. We found that the Gal3/1ctR antagonist assay was reproducible, robust, and suitable for HTS using the ACT:One cAMP biosensor technology. The primary screen hit rate was ∼5.4% (Fig. 6A, Table 2), with 68 compounds that gave greater than three times the standard deviation from the mean of all compounds tested. It is important to note that a high number of hits are expected from LOPAC screens due to the high proportion of pharmacologically active compounds in the LOPAC itself. Not surprisingly, the majority of the top hits were modulators of the cAMP pathway. Concentration response curves for four of the top “hits” and their IC50 values are shown in Figure 6B. While the majority of the pharmacologically active compounds identified as hits likely increased cAMP by acting as agonists at endogenous Gs-coupled GPCRs, the LOPAC screen serves to demonstrate that the assay is reproducible with Z values >0.7 and S:B suitable for HTS. The Gal3/1ctR HTS-compatible assay was submitted to the Molecular Libraries Screening Center Network (MLSCN) HTS resource via the National Institutes of Health (NIH) fast track mechanism and assigned to the Scripps Research Institute Molecular Screening Center (SRIMSC), one of the national comprehensive MLSCN centers. The results of the primary HTS (https://pubchem.ncbi.nlm.nih.gov/assay/assay.cgi?aid=652245) and counterscreen against HEK293-CNG parental cells (https://pubchem.ncbi.nlm.nih.gov/assay/assay.cgi?aid=652268) can be viewed in PubChem. As reported in PubChem, the hit rate in the primary screen was 0.44% with an average Z value of 0.76±0.06 and S:B of 3.84±0.37. The elimination rate by counterscreen was 10%.
Fig. 5.
Assay optimization. (A) HEK293-CNG cells stably expressing the Gal3/1ctR chimera were plated at 5000 (●), 10,000 (■), 15,000 (▲), or 20,000 (▼) cells per well in a 384-well plate. After 18-h incubation at 37°C, 5% CO2, cells were loaded with ACT:One membrane potential dye for 2 h at room temperature. cAMP levels were determined in response to increasing concentrations of porcine galanin in the presence of an EC90 of forskolin (1 μM) according to ACT:One manufacturer's instructions and as described in detail in Table 1. The optimal cell density was determined to be 10,000 cells per well. At this density signal-to-background ratio (S:B) was 2.9. (B) HEK293-CNG cells stably expressing the Gal3/1ctR chimera were plated at 10,000 cells per well and cAMP levels at increasing concentrations of porcine galanin were determined as in (A) in the presence of an EC90 of forskolin (1 μM) and in the presence of 0.025% (●), 0.5% (■), 0.8% (▲), 1.5% (▼), or 2% (♦) DMSO. The Gal3/1ctR cAMP assay performance, both in terms of S:B and galanin EC50 values, was not significantly affected by DMSO concentrations up to 2%. Final DMSO concentration used in the high-throughput screening (HTS) assay was 0.7%. The data presented are means±SEM of triplicate wells (n=3).
Fig. 6.
Pilot screen. (A) The LOPAC screening set (Library of Pharmacologically Active Compounds; Sigma Aldrich) containing ∼1280 compounds was tested at 10 μM against the Gal3/1ctR chimera in the cAMP biosensor assay under conditions as optimized in Figure 5 (10,000 cells per well), in the presence of an EC90 of porcine galanin (antagonist mode). Compounds that displayed activity greater than three times the standard deviation from the mean of all the compounds tested were designated as hits (see Table 2). The S:B was 3.4 if signal was generated with an EC90 of forskolin alone and baseline using an EC90 of forskolin together with an EC90 of galanin. An average Z score of 0.7 was calculated from eight wells of EC90 forskolin alone (signal, open squares) and eight wells of EC90 forskolin+EC90 galanin (baseline, closed squares) on each plate (total n=32) and the primary hit rate was ∼5.4%. (B) Four of the top 10 hits were challenged at the indicated concentration ranges with an EC90 of forskolin together with an EC90 of galanin. The data presented in (B) are means±SEM of triplicate wells (n=3).
Table 2.
Top 20 Hits From LOPAC Screen Described in Figure 6
| Compound name | Cat no. | Sigma class | Description |
|---|---|---|---|
| Salmeterol xinafoate | S 5068 | Adrenoceptor | Beta2 adrenoceptor agonist |
| GW1929 | G 5668 | Transcription | PPAR-gamma agonist |
| (−)-Alpha-methylnorepinephrine | D 5290 | Adrenoceptor | Adrenoceptor agonist; vasoconstrictor; antihypertensive |
| cis-(Z)-Flupenthixol dihydrochloride | F-114 | Dopamine | Dopamine receptor antagonist; antipsychotic |
| HE-NECA | H 8034 | Adenosine | A2 adenosine receptor agonist |
| Felodipine | F 9677 | Ca2+ channel | L-type calcium channel blocker |
| (−)-Isoproterenol hydrochloride | I 6504 | Adrenoceptor | Beta-adrenoceptor agonist; increases cytosolic cAMP |
| N-phenylanthranilic acid | 144509 | Cl− channel | Cl− channel blocker |
| R(−)-N6(2-phenylisopropyl)adenosine | P 4532 | Adenosine | A1 adenosine receptor agonist |
| 5′-N-ethylcarboxamidoadenosine | E 2387 | Adenosine | Adenosine A1 and A2 receptor agonist |
| R(−)-isoproterenol (+)-bitartrate | I 2760 | Adrenoceptor | Agonist at beta adrenoceptors; bronchodilator. |
| Ritodrine hydrochloride | R 0758 | Adrenoceptor | Beta2-adrenoceptor agonist; relaxes uterine muscle contractions |
| Formoterol | F 9552 | Adrenoceptor | Beta2-adrenoceptor agonist |
| Isotharine mesylate | I 3639 | Adrenoceptor | Beta-adrenoceptor agonist; bronchodilator |
| (±)-6-Chloro-PB hydrobromide | S-143 | Dopamine | D1 dopamine receptor agonist |
| 2-Chloroadenosine triphosphate tetrasodium | C-145 | P2 receptor | P2Y receptor agonist |
| 5′-(N-cyclopropyl)carboxamidoadenosine | C 6042 | Adenosine | Potent A2 adenosine receptor agonist |
| Dihydrexidine hydrochloride | D 5814 | Dopamine | D1 dopamine receptor agonist |
| 5′-N-methyl carboxamidoadenosine | A-024 | Adenosine | Adenosine receptor agonist with selectivity for A2 over A1 |
| Fenoterol hydrobromide | F 1016 | Adrenoceptor | Beta2-adrenoceptor agonist; bronchodilator |
LOPAC, Library of Pharmacologically Active Compounds; PPAR, peroxisome proliferator activated receptor.
Discussion
There is considerable interest in developing GalR3 selective small molecule modulators within the pharmaceutical industry as novel therapeutics to treat mood and addiction disorders, as indicated by the number of patent applications focused on GalR3 antagonists appearing in the patent literature. For example, the indolone series of compounds has seen 17 U.S. and WO patent applications since 2002 (e.g., U.S. patents 6936607, 7465750).33,34 Despite this, there is a lack of potent and selective antagonists available, with those that do currently exist in the literature, including SNAP37889, suffering low solubility and poor oral bioavailability. The difficulties in screening against GalR3 are likely due to its poor surface expression and thus lack of functionality in recombinant systems (Fig. 1). Our findings strongly suggest that this is due to its predominantly intracellular expression in recombinant systems. On examining the carboxy-tail amino acid sequence of the GalR3 we identified multiple overlapping putative ER retention signals that most likely mediate GalR3 intracellular expression. These motifs are basic in nature and comprised of RXR amino acid motifs in which X can be any amino acid.37 We hypothesized that if the carboxy-terminal RXR motifs are functioning as ER retention signals in the wtGalR3, then removal or replacement of these motifs with the carboxy tail of a receptor lacking such motifs would facilitate cell surface expression of GalR3. We investigated this possibility by generating a receptor chimera in which the carboxy tail of the GalR3 was replaced by the carboxy tail of a related receptor, namely GalR1, that does not contain any known ER retention signals and demonstrates good cell surface expression in recombinant systems.
Many GPCRs contain various carboxy-terminal motifs known to be involved in their forward trafficking.37 Thus, the tail-swapping approach described herein may provide a general approach for enabling HTS of GPCRs that traffic poorly to the plasma membrane. A tail-swapping approach has been used previously to increase the affinity of certain GPCRs for β-arrestin in order to enable β-arrestin recruitment assays.38 These studies showed that receptor pharmacology with regards to G protein coupling was not altered, further validating the tail-swapping strategy for enabling high throughput screening of poor surface-expressing GPCRs.
The limitations in enabling a cell-based functional assay for the screening of large compound collections may have contributed to the lack of success in identifying novel lead GalR3 antagonists. Both the indolone and pyrimidine scaffolds were identified using a traditional radioligand binding competition assay and cell membranes generated from mammalian cultured cells heterologously expressing the GalR3. This methodology has a strong bias towards detecting orthosteric (endogenous ligand binding site) ligands. In contrast, the cell-based functional assay developed here has the potential to detect a wider spectrum of compounds including allosteric modulators because the only prerequisite is that the compounds perturb receptor activity irrespective of where they bind. Allosteric modulators offer several advantages over orthosteric ligands including the potential for greater selectivity, novel modes of efficacy, and perhaps a reduced potential for unwanted side effects. Therefore, we are likely to find novel GalR3 pharmacological tools that possess a wider spectrum of pharmacological activities and greater structural diversity than those that currently exist in the literature.
In conclusion, we have shown that the tail-swapping approach (Fig. 2) facilitates cell-surface expression of GalR3/1ctR and enables a high-throughput compatible, cell-based functional assay for the identification of GalR3 small molecule modulators. Our data indicate that the ACT:One cAMP biosensor technology provides a robust and reproducible assay for the detection of Gal3/1ctR activity in a productive screening set-up as indicated by the LOPAC screening results (Fig. 6). We and others have previously demonstrated success using the HEK293-CNG cAMP biosensor approach to develop HTS-compatible assays for monitoring Gi- and Gs-coupled receptors (NPY-Y2 and TSH, respectively).39,40 Our NPY-Y2 receptor screening campaign led to the identification of novel NPY-Y2 receptor small molecule antagonists that are currently being pursued as lead compounds for the treatment of alcoholism.41 Furthermore, using the optimized and validated Gal3/1ctR cAMP assay that we have described, an HTS campaign against the MLPCN 300K compound collection available through the Scripps Molecular Libraries' Screening Center (one of the national comprehensive MLSCN centers) has been completed (AID=652245), with counterscreening against the HEK293-CNG parental cell line (AID=652268). Our current follow-up studies include counterscreening using the HEK293-CNG-GalR1 cell line to enable us to assess selectivity of any potential lead compounds.
In addition, we are currently investigating the mechanisms that regulate wtGalR3 trafficking and function. A detailed understanding of the mechanisms that control the trafficking and signaling of GalR3 may lead to novel insights into its function in vivo and will complement our drug discovery efforts.
Abbreviations
- cAMP
cyclic adenosine monophosphate
- CHO
Chinese hamster ovary
- CNG
cAMP nucleotide-gated channel
- CNS
central nervous system
- CRC
concentration response curve
- DMSO
dimethyl sulfoxide
- EC50
50% of maximal response
- EC90
90% of maximal response
- ER
endoplasmic reticulum
- FBS
fetal bovine serum
- FLIPR
fluorescent imaging plate reader
- GalR1
galanin receptor 1
- GalR2
galanin receptor 2
- GalR3
galanin receptor 3
- GIRK
G protein–coupled inwardly rectifying potassium channel
- GPCR
G protein–coupled receptor
- HEK
human embryonic kidney
- HTS
high-throughput screening
- LOPAC
Library of Pharmacologically Active Compounds
- MLSCN
Molecular Libraries Screening Center Network
- PBS
phosphate-buffered saline
- PCR
polymerase chain reaction
- PDE
phosphodiesterase
- PFA
paraformaldehyde
- S:B
signal-to-background ratio
- wt
wild-type
Acknowledgments
We thank Darinka Obradovich for technical assistance. This work was supported by a grant from the National Institutes of Health (NS067631 awarded to P. McDonald).
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Tatemoto K. Rokaeus A. Jornvall H. McDonald TJ. Mutt V. Galanin—a novel biologically active peptide from porcine intestine. FEBS Lett. 1983;164:124–128. doi: 10.1016/0014-5793(83)80033-7. [DOI] [PubMed] [Google Scholar]
- 2.Skofitsch G. Jacobowitz DM. Galanin-like immunoreactivity in capsaicin sensitive sensory neurons and ganglia. Brain Res Bull. 1985;15:191–195. doi: 10.1016/0361-9230(85)90135-2. [DOI] [PubMed] [Google Scholar]
- 3.Melander T. Kohler C. Nilsson S, et al. Autoradiographic quantitation and anatomical mapping of 125I-galanin binding sites in the rat central nervous system. J Chem Neuroanat. 1988;1:213–233. [PubMed] [Google Scholar]
- 4.Rokaeus A. Melander T. Hokfelt T, et al. A galanin-like peptide in the central nervous system and intestine of the rat. Neurosci Lett. 1984;47:161–166. doi: 10.1016/0304-3940(84)90423-3. [DOI] [PubMed] [Google Scholar]
- 5.McDonald TJ. Brooks BD. Rokaeus A. Tinner B. Staines WA. Pancreatic galanin: molecular forms and anatomical locations. Pancreas. 1992;7:624–635. [PubMed] [Google Scholar]
- 6.Iismaa TP. Shine J. Galanin and galanin receptors. Results Probl Cell Differ. 1999;26:257–291. doi: 10.1007/978-3-540-49421-8_12. [DOI] [PubMed] [Google Scholar]
- 7.Branchek TA. Smith KE. Gerald C. Walker MW. Galanin receptor subtypes. Trends Pharmacol Sci. 2000;21:109–117. doi: 10.1016/s0165-6147(00)01446-2. [DOI] [PubMed] [Google Scholar]
- 8.Sundstrom E. Archer T. Melander T. Hokfelt T. Galanin impairs acquisition but not retrieval of spatial memory in rats studied in the Morris swim maze. Neurosci Lett. 1988;88:331–335. doi: 10.1016/0304-3940(88)90233-9. [DOI] [PubMed] [Google Scholar]
- 9.Hobson SA. Holmes FE. Kerr NC. Pope RJ. Wynick D. Mice deficient for galanin receptor 2 have decreased neurite outgrowth from adult sensory neurons and impaired pain-like behaviour. J Neurochem. 2006;99:1000–1010. doi: 10.1111/j.1471-4159.2006.04143.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jimenez-Andrade JM. Zhou S. Yamani A. Valencia de Ita S. Castaneda-Hernandez G. Carlton SM. Mechanism by which peripheral galanin increases acute inflammatory pain. Brain Res. 2005;1056:113–117. doi: 10.1016/j.brainres.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 11.Liu HX. Hokfelt T. The participation of galanin in pain processing at the spinal level. Trends Pharmacol Sci. 2002;23:468–474. doi: 10.1016/s0165-6147(02)02074-6. [DOI] [PubMed] [Google Scholar]
- 12.Kyrkouli SE. Stanley BG. Leibowitz SF. Galanin: stimulation of feeding induced by medial hypothalamic injection of this novel peptide. Eur J Pharmacol. 1986;122:159–160. doi: 10.1016/0014-2999(86)90175-5. [DOI] [PubMed] [Google Scholar]
- 13.Zorrilla EP. Brennan M. Sabino V. Lu X. Bartfai T. Galanin type 1 receptor knockout mice show altered responses to high-fat diet and glucose challenge. Physiol Behav. 2007;91:479–485. doi: 10.1016/j.physbeh.2006.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Theodorsson A. Holm L. Theodorsson E. Hypothermia-induced increase in galanin concentrations and ischemic neuroprotection in the rat brain. Neuropeptides. 2008;42:79–87. doi: 10.1016/j.npep.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 15.Kuteeva E. Hokfelt T. Wardi T. Ogren SO. Galanin, galanin receptor subtypes and depression-like behaviour. Cell Mol Life Sci. 2008;65:1854–1863. doi: 10.1007/s00018-008-8160-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holmes A. Kinney JW. Wrenn CC, et al. Galanin GAL-R1 receptor null mutant mice display increased anxiety-like behavior specific to the elevated plus-maze. Neuropsychopharmacology. 2003;28:1031–1044. doi: 10.1038/sj.npp.1300164. [DOI] [PubMed] [Google Scholar]
- 17.McMillan PJ. Peskind E. Raskind MA. Leverenz JB. Increased galanin receptor occupancy in Alzheimer's disease. Neurobiol Aging. 2004;25:1309–1314. doi: 10.1016/j.neurobiolaging.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 18.Adeghate E. Ponery AS. Large reduction in the number of galanin-immunoreactive cells in pancreatic islets of diabetic rats. J Neuroendocrinol. 2001;13:706–710. doi: 10.1046/j.1365-2826.2001.00682.x. [DOI] [PubMed] [Google Scholar]
- 19.Picciotto MR. Galanin and addiction. Cell Mol Life Sci. 2008;65:1872–1879. doi: 10.1007/s00018-008-8151-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hawes JJ. Brunzell DH. Narasimhaiah R. Langel U. Wynick D. Picciotto MR. Galanin protects against behavioral and neurochemical correlates of opiate reward. Neuropsychopharmacology. 2008;33:1864–1873. doi: 10.1038/sj.npp.1301579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holmes A. Picciotto MR. Galanin: a novel therapeutic target for depression, anxiety disorders and drug addiction? CNS Neurol Disord Drug Targets. 2006;5:225–232. doi: 10.2174/187152706776359600. [DOI] [PubMed] [Google Scholar]
- 22.Skofitsch G. Sills MA. Jacobowitz DM. Autoradiographic distribution of 125I-galanin binding sites in the rat central nervous system. Peptides. 1986;7:1029–1042. doi: 10.1016/0196-9781(86)90133-6. [DOI] [PubMed] [Google Scholar]
- 23.O'Donnell D. Ahmad S. Wahlestedt C. Walker P. Expression of the novel galanin receptor subtype GALR2 in the adult rat CNS: distinct distribution from GALR1. J Comp Neurol. 1999;409:469–481. [PubMed] [Google Scholar]
- 24.Burazin TC. Larm JA. Ryan MC. Gundlach AL. Galanin-R1 and -R2 receptor mRNA expression during the development of rat brain suggests differential subtype involvement in synaptic transmission and plasticity. Eur J Neurosci. 2000;12:2901–2917. doi: 10.1046/j.1460-9568.2000.00184.x. [DOI] [PubMed] [Google Scholar]
- 25.Mennicken F. Hoffert C. Pelletier M. Ahmad S. O'Donnell D. Restricted distribution of galanin receptor 3 (GalR3) mRNA in the adult rat central nervous system. J Chem Neuroanat. 2002;24:257–268. doi: 10.1016/s0891-0618(02)00068-6. [DOI] [PubMed] [Google Scholar]
- 26.Belfer I. Hipp H. Bollettino A, et al. Alcoholism is associated with GALR3 but not two other galanin receptor genes. Genes Brain Behav. 2007;6:473–481. doi: 10.1111/j.1601-183X.2006.00275.x. [DOI] [PubMed] [Google Scholar]
- 27.Swanson CJ. Blackburn TP. Zhang X, et al. Anxiolytic- and antidepressant-like profiles of the galanin-3 receptor (Gal3) antagonists SNAP 37889 and SNAP 398299. Proc Natl Acad Sci USA. 2005;102:17489–17494. doi: 10.1073/pnas.0508970102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Belfer I. Hipp H. McKnight C, et al. Association of galanin haplotypes with alcoholism and anxiety in two ethnically distinct populations. Mol Psychiatry. 2006;11:301–311. doi: 10.1038/sj.mp.4001768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mitsukawa K. Lu X. Bartfai T. Galanin, galanin receptors and drug targets. Cell Mol Life Sci. 2008;65:1796–1805. doi: 10.1007/s00018-008-8153-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith KE. Walker MW. Artymyshyn R, et al. Cloned human and rat galanin GALR3 receptors. Pharmacology and activation of G-protein inwardly rectifying K+ channels. J Biol Chem. 1998;273:23321–23326. doi: 10.1074/jbc.273.36.23321. [DOI] [PubMed] [Google Scholar]
- 31.Lu X. Lundstrom L. Langel U. Bartfai T. Galanin receptor ligands. Neuropeptides. 2005;39:143–146. doi: 10.1016/j.npep.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 32.Lang R. Berger A. Santic R, et al. Pharmacological and functional characterization of galanin-like peptide fragments as potent galanin receptor agonists. Neuropeptides. 2005;39:179–184. doi: 10.1016/j.npep.2004.12.015. [DOI] [PubMed] [Google Scholar]
- 33.Packiarajan M. 2,4,6-Triaminopyrimidines for the treatment of depression and/or anxiety. U.S. Patent. 2005 6,936,607. [Google Scholar]
- 34.Packiarajan M. Use of GAL3 antagonist for treatment of depression and/or anxiety, compounds useful in such methods. U.S. Patent. 2008 7,465,750. [Google Scholar]
- 35.Ciambrone GJ. Liu VF. Lin DC. McGuinness RP. Leung GK. Pitchford S. Cellular dielectric spectroscopy: a powerful new approach to label-free cellular analysis. J Biomol Screen. 2004;9:467–480. doi: 10.1177/1087057104267788. [DOI] [PubMed] [Google Scholar]
- 36.Benke D. Mechanisms of GABAB receptor exocytosis, endocytosis, and degradation. Adv Pharmacol. 2010;58:93–111. doi: 10.1016/S1054-3589(10)58004-9. [DOI] [PubMed] [Google Scholar]
- 37.Dong C. Filipeanu CM. Duvernay MT. Wu G. Regulation of G protein-coupled receptor export trafficking. Biochim Biophys Acta. 2007;1768:853–870. doi: 10.1016/j.bbamem.2006.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hamdan FF. Audet M. Garneau P. Pelletier J. Bouvier M. High-throughput screening of G protein-coupled receptor antagonists using a bioluminescence resonance energy transfer 1-based beta-arrestin2 recruitment assay. J Biomol Screen. 2005;10:463–475. doi: 10.1177/1087057105275344. [DOI] [PubMed] [Google Scholar]
- 39.Brothers SP. Saldanha SA. Spicer TP, et al. Selective and brain penetrant neuropeptide y y2 receptor antagonists discovered by whole-cell high-throughput screening. Mol Pharmacol. 2010;77:46–57. doi: 10.1124/mol.109.058677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Titus S. Neumann S. Zheng W, et al. Quantitative high-throughput screening using a live-cell cAMP assay identifies small-molecule agonists of the TSH receptor. J Biomol Screen. 2008;13:120–127. doi: 10.1177/1087057107313786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saldanha SA. Brothers SP. Spicer T, et al. National Institutes of Health; Bethesda, MD: 2010. Probe Report for NPY-Y2 Receptor Antagonists. Probe Reports from the NIH Molecular Libraries Program. [PubMed] [Google Scholar]