Abstract
Recent studies indicated that mesenchymal stromal cells from bone marrow (bmMSC) differ in their osteogenic differentiation capacity compared to MSC from term placenta (pMSC). We extended these studies and investigated the expression of factors involved in regulation of bone metabolism in both cell types. To this end, MSC were expanded in vitro and characterized. The total transcriptome was investigated by microarrays, and for selected genes, the differences in gene expression were explored by quantitative reverse transcriptase-polymerase chain reaction, immunocytochemistry, and flow cytometry. We report that bmMSC and pMSC share expression of typical lineage surface markers, including CD73, CD90, CD105, and lack of CD14, CD34, and CD45. However, according to transcriptome analyses, they differ significantly in their expression of more than 590 genes. Factors involved in bone metabolism, including alkaline phosphatase (P<0.05), osteoglycin (P<0.05), osteomodulin (P<0.05), runt-related transcription factor 2 (Runx2) (P<0.04), and WISP2 (P<0.05), were expressed at significantly lower levels in pMSC, but twist-related protein 2 (Twist2) (P<0.0002) was expressed at significantly higher levels. The osteogenic differentiation capacity of pMSC was very low. The adipogenic differentiation was somewhat more prominent in bmMSC, while the chondrogenic differentiation seemed not to differ between bmMSC and pMSC, as determined by histochemical staining. However, expression and induction of peroxisome proliferator-activated receptor gamma-2 (PPARγ2) and Sox9, factors involved in early adipogenesis and chondrogenesis, respectively, were higher in bmMSC. We conclude that despite many similarities between bmMSC and pMSC, when expanded under identical conditions, they vary considerably with respect to their in vitro differentiation potential. For regenerative purposes, the choice of MSC may therefore influence the outcome of a treatment considerably.
Introduction
Human mesenchymal stromal cells (MSCs), sometimes referred to as mesenchymal stem cells, are multipotent cells found in different tissues of the adult body. They express a variety of cell surface antigens, including CD73, CD90, CD105, and CD146, but lack expression of antigens characteristic for hematopoietic or endothelial cells [1–3]. A unique MSC-defining epitope is not known yet [1,4,5]. The bright expression of nerve growth factor receptor (CD271), frizzled-9 (CD349), and tissue nonspecific alkaline phosphatase (TNAP) determined early differentiation stages of CD73+, CD90+, or CD105+ MSC [6]. In addition to these antigens, expression of the stage-specific embryonic antigen 4 (SSEA-4) or fibroblast activation protein-α (FAPA) were suggested as marker antigens for MSC [7,8]. However, other antigens such as CD146 (alias MCAM, MUC18), the sushi-containing domain 2 protein (susd2, alias W5C5 antigen) [9], or the molecule detectable by monoclonal antibody clone W12D1 (protein unknown) generated distinct histogram patterns, indicating that MSC prepared by conventional techniques present a complex mixture of cells [10,11].
Functionally different subsets of bone marrow-derived mesenchymal stromal cell (bmMSC) can be generated by fluorescence-activated cell sorting (FACS) ex vivo, yielding cells with a distinct differentiation or regeneration potential. In bmMSC, the CD271+CD56+ subset had a prominent chondrogenic potential, whereas the CD271+CD56− subset yielded more adipogenic cells. The osteogenic potential of bmMSC was high, but did not differ between the two subsets [12]. In contrast, the CD271+ fraction of periosteum-derived progenitor cells contained osteoblast precursors, which deposited a mineralized matrix, whereas the CD271− subset failed to do so [13]. Adipose tissue-derived mesenchymal stromal cells (atMSC) express CD34, an antigen found, for instance, on endothelial or hematopoietic precursor cells, but not on bmMSC or placenta-derived mesenchymal stromal cells (pMSC), and these CD34+ atMSC were osteogenic in vitro and in vivo [14]. MSC from another source, the synovial membrane, had a distinct chondrogenic potential [15], but failed to generate a stable cartilage ectopically [16]. Thus, in addition to the differences between the subsets of bmMSC enriched by monoclonal antibodies and FACS [12], MSC from other sources such as placenta or adipose tissue may differ in their gene expression patterns or in their regenerative potential as well [17,18].
We therefore explored some of the differences between bmMSC and pMSC in more detail and investigated the total transcriptome, expression of osteogenic factors, and the trilineage differentiation potential of bmMSC compared to pMSC in vitro. Here we report that these cells express runt-related transcription factor 2 (Runx2) and twist-related protein 2 (Twist2), key factors involved in osteogenic differentiation, at significantly different levels. We confirm a low osteogenic differentiation potential, but a rather normal adipogenic or chondrogenic potential of pMSC in vitro.
Materials and Methods
Preparation of MSCs
The bmMSC were prepared as described recently [19] and characterized as recommended by a consensus conference of the International Society for Cellular Therapy [1]. For this study, bmMSC were isolated from femoral aspirates of patients (n=16) undergoing a total hip replacement after written consent at the BG Centre of Trauma Surgery. The aspirates were washed with phosphate-buffered saline (PBS; PAA) and centrifuged (room temperature, 10 min at 150 g). The supernatant was discarded and the pellet was resuspended in PBS. The mononuclear cells in the suspension were enriched by density gradient centrifugation (Ficoll®; GE Healthcare; ρ=1.077, room temperature, 30 min at 400 g). The fraction of mononuclear cells was harvested from the interphase, washed once with PBS, and seeded in the MSC expansion medium in T75 flasks (BD Falcon).
The pMSC were isolated from human term placenta and characterized as described recently [2,10]. Healthy term placenta (n=14) was provided by the Department of Gynaecology and Obstetrics at UKT after consent from the mothers. In some experiments, the endometrial maternal part of the placenta was separated from the fetal part to enrich for maternal (pmMSC) and fetal (pfMSC) subsets of pMSC. The tissue was minced in small pieces and after triple washing with the Hank's balanced solution (PAA), the samples were proteolytically digested (1 h, 37°C, 12 U/mL collagenase type XI; Sigma-Aldrich; 2.4 U/mL dispase II; Roche). The digestion was stopped by addition of fetal calf serum (FCS) (0.1 vol; FCS Biochrom) and filtered through a sieve. Density gradient centrifugation was performed with the crude cell suspension obtained as described above. The mononuclear cells were collected and washed once with 1×PBS. The supernatant was discarded and the pellet was resuspended in MSC expansion media and seeded in T75 flasks (BD Falcon). The study was approved by the ethics committee.
Expansion and differentiation of MSC in vitro
MSC from individual donors were expanded in an expansion medium compliant with current good medical procedure (GMP) regulations as described [20]. Unless otherwise noted, MSC were harvested after two passages of in vitro culture and utilized for the different experiments. To explore their differentiation potential, cellular differentiation was induced in vitro.
For osteogenic differentiation, MSC were seeded at an inoculation density of 5×104 cells per six-well plate in the cell expansion medium for 7 days. Then, the expansion medium was replaced by the osteogenic induction medium containing high glucose DMEM (PAA), enriched 10% FCS (Biochrom), 100 μg/mL streptomycin, 100 U/mL penicillin (both Invitrogen), 10 mM β-glycerophosphate (Merck), 0.1 μM dexamethasone, and 0.17 mM ascorbic acid 2 phosphate (both Sigma-Aldrich). After 4 weeks of differentiation, cells were fixed with ice-cold methanol (VWR) and mineralization was determined by von Kossa staining [21].
For adipogenic differentiation, cells were also seeded in six-well plates. The adipogenic induction medium included high glucose DMEM (PAA), 10% FCS (Biochrom), 100 μg/mL streptomycin, 100 U/mL penicillin (both Invitrogen), 0.2 mM indomethacine (Calbiochem), 0.01 mg/mL insulin, 0.5 mM 3-isobutylxanthine, and 1 μM dexamethasone (both Sigma-Aldrich). After 28 days in the induction medium, the cells were washed and stained with Oil Red O [22].
For chondrogenic differentiation, the cells were seeded in round-bottom 96-well-plates at a density of 4×105 cells per well to allow microsphere formation. The chondrogenic induction medium was added, which consisted of high glucose DMEM (PAA), 0.17 mM ascorbic acid 2-phosphate, 100 μg/mL streptomycin, 100 U/mL penicillin, 0.1 μM dexamethasone, and 1×insulin-transferrin-selenite (ITS)+1 supplement (all from Sigma-Aldrich). After 4 weeks of induction, the microspheres were harvested, imbedded in Tissue Tek (Sakura), and stored at −70°C. For immunocytochemistry, samples were cyrosectioned (6 μM; Leica CM3050S) and stained with Alcian Blue [23].
Flow cytometry
All preparations of bmMSC and pMSC were characterized by microscopy and flow cytometry (FCM) as described recently [24]. The cells were detached with Accutase (PAA), washed with PFEA [PBS containing 2% FCS (Biochrom), 2 mM ethylenediaminetetraaceticacid (Merck), and 0.01% sodium azide (Merck)]. All antibodies were diluted in cold PFEA (4°C) and added to the cells (5×105 per sample) following the manufacturer's protocol. The cells were first incubated for 20 min at 4°C with Gamunex® (Talecris Biotherapeutics; 1:20 in PFEA), washed once with PFEA, and then stained with the antibodies. The cells were then washed with PBS and analyzed by FCM LSR II (BD Bioscience). To detect intracellular proteins by FCM, cells were permeabilized before antibody incubation (BD Bioscience; cytofix/cytoperm kit). Then, the anti-Runx2 (Cell Signaling) and anti-Twist-2 antibodies (Abcam) were added according to the manufacturer's protocol. The FCM data were analyzed with the DIVA® and FlowJo® software programs [25].
Determination of mitotic and respiratory activities by a modified MTT assay
Cells were seeded in 96-well plates (3,000 cells/well; Greiner Bio-One, Cellstar). The mitotic or respiratory activity was investigated using a cell proliferation assay (EZ4U; Biozol Diagnostic) according to the manufacturer's protocol. The extinction reading was accomplished with an ELISA reader (BioTek; EL800) using 450 nm wavelength. Data were evaluated with Excel®.
Investigation of the transcriptome
For investigation of gene expression, the total transcriptome of bmMSC and pMSC derived either from the maternal (pmMSC) or fetal (pfMSC) side of placenta was explored. The cells were expanded to the second passage, characterized as described above, and RNA was prepared according to the manufacturer's protocol (RNeasy Kit; Qiagen). For analysis of the total transcriptome, two sets of RNA samples were prepared from bmMSC (total n=11), pmMSC (total n=6), and pfMSC (total n=8) and the gene expression analysis was performed in two independent arrays by Affymetrix GeneChip® technology (using the Human Genome U133 Plus 2.0 arrays). The gene expression data was first normalized with the Robust Multichip Average method [25] followed by differential expression analyses using the microarray data analysis software Mayday [26]. Furthermore, we conducted a pathway analysis of the genes found to be differentially expressed using Ingenuity® Systems Ingenuity Pathway Analysis (IPA) (http://ingenuity.com) to identify signaling pathways involved in osteoblast differentiation or bone development. Ingenuity is a program that converts large data sets into networks containing direct and indirect relationships between genes based on known interactions in the literature. In addition, we used DAVID [27] to find enriched gene ontology terms related to the role of mesenchymal cells in osteogenesis in the set of differentially expressed genes.
For quantitative reverse transcriptase -polymerase chain reaction (RT-qPCR) of individual genes, RNA was extracted as described above and reverse transcription of RNA into cDNA was performed on 1 μg total RNA with oligo-(dT)n-priming (Advantage RT for PCR Kit; Clontech). RT-qPCR (LightCycler®; Roche) [28] utilizing commercially available primer pairs for the chondrogenic marker sex determining region Y-box 9 (Sox9; Qiagen), the osteogenic markers alkaline phosphatase (ALP), runt-related transcription factor 2 (Runx2), Twist homolog 2 (Twist2), and the adipogenic marker peroxisome proliferator-activated receptor gamma-2 (PPARγ2; all from Eurofins MWG Operon). Quantification of transcripts encoding GAPDH and serial dilutions of a recombinant DNA standard served as references in each PCR. The expression of the target genes was normalized to the expression of GAPDH, and amplifications were evaluated by the FitPoint (ΔΔCt−) method [28]. In addition, RT-qPCR of transcripts encoding β-actin, β-microglobulin, and ribosomal protein L13A (all primers form Qiagen) was employed to investigate if different media influenced the expression of GAPDH.
Immunocytochemistry
For immunofluorescent staining, the cells were seeded at a density of 1×104 cells per chamber on collagen type I-coated 8-well culture slides (BD Bioscience). After reaching 80% confluency, cells where fixed with ice-cold methanol and washed with PBS. Unspecific binding sites were blocked with 0.1% bovine serum albumin (BSA)/PBS, and incubated with the primary antibodies (10 μg/mL mouse-α-human anti-twist2 antibody; Abcam, rabbit-α-human runx2, clone D1H7; Cell Signaling, both at 1:50 in 0.1% BSA/PBS overnight at 4°C). The samples were washed twice with PBS and counterstained with secondary antibodies [fluorescein isothiocyanate -conjugated affiniPure F(ab′)2-fragement anti-mouse immunoglobulin G (IgG); Jackson ImmunoResearch; anti-rabbit IgG NL557 conjugated donkey IgG; R&D Systems, both at 1:100 in 0.1% BSA/PBS for 1 h in the dark at room temperature]. After washing twice with PBS, 4′,6-diamidino-2-phenylindol-dihydrochlorid (DAPI; Roche) staining was performed for 20 min in the dark at room temperature. Slides were mounted with coverslips using a fluorescence mounting medium (Dako), and explored by microspcopy (Zeiss Axiophot and Zeiss Axiovision software).
Statistics
Experimental data are presented as mean values±standard deviations. Statistical analyses were performed using a two-sided Student's t-test. Differences in gene expression levels yielding P-values less than 0.05 (after correction of the false discovery rate) and an absolute fold change of at least 4 were considered significant.
Results
Characterization of human term placenta-derived MSC
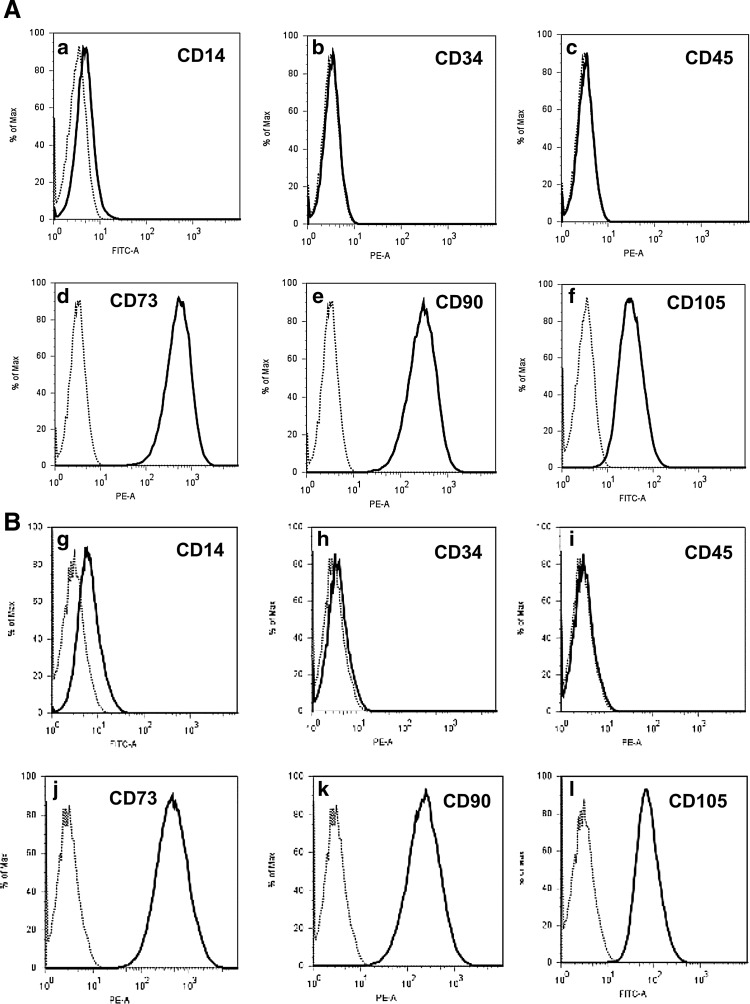
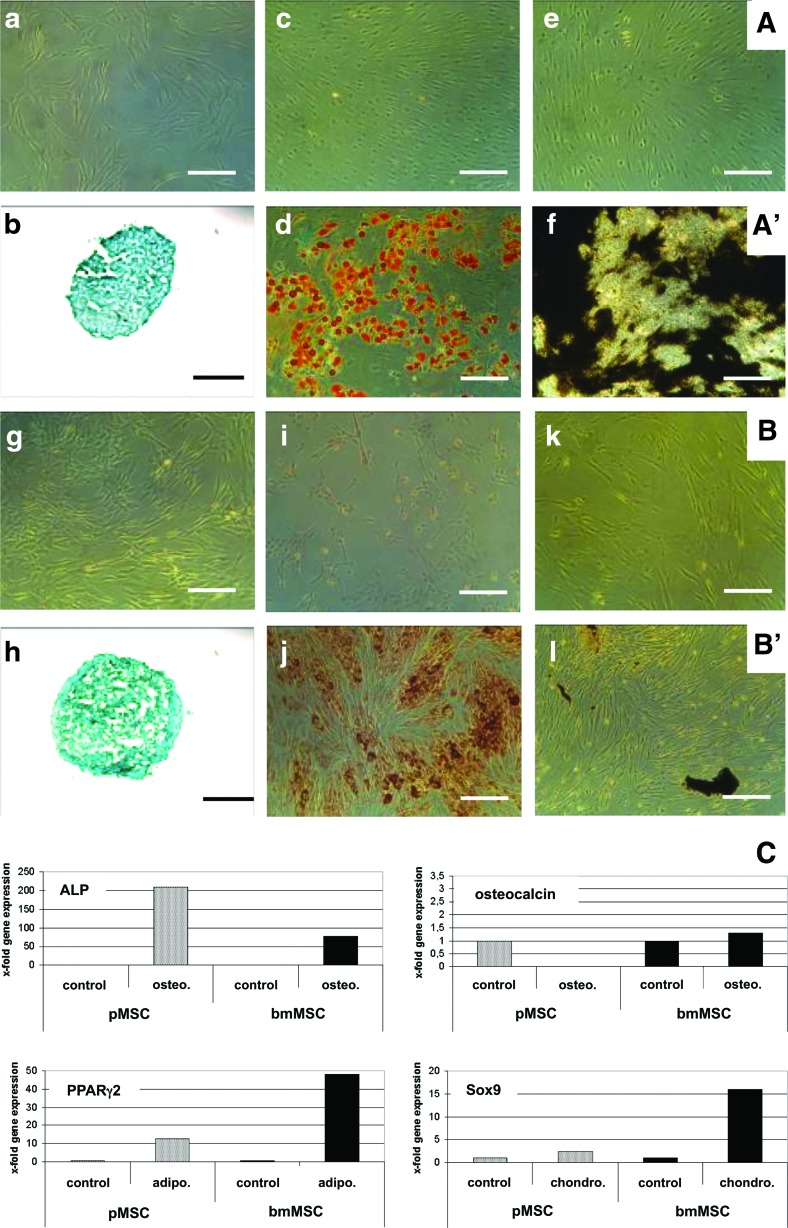
MSC from human bone marrow and from term placenta were isolated [10] and characterized [1,2] as described recently. The bmMSC expressed CD73, CD90, CD105, but lacked CD14, CD34, and CD45 (Fig. 1A[a–f]). There was no difference in the expression of these MSC markers between bmMSC and pMSC (Fig. 1A vs. B). The respiratory and/or proliferative activity of bmMSC and pMSC was investigated by a modified MTT assay. There was no significant difference in the proliferative or metabolic activity between bmMSC (mean 1.23 O.D.±0.3, n=3) and pMSC (mean 1.134 O.D.±0.17, n=3, P>0.06; data not shown) in early passages of MSC in in vitro culture. To investigate the differentiation potential of MSC, adipogenic, chondrogenic, and osteogenic differentiation was induced in vitro (Fig. 2). For immunocytochemical staining, MSCs were differentiated over 4 weeks and then reacted with different solutions according to the differentiation protocol employed. After this in vitro differentiation, there was no variance in chondrogenic microspheres generated from bmMSC or pMSC (Fig. 2b, h). The bright staining and larger size of the lipid vesicles indicated a somewhat higher adipogenic differentiation potential of bmMSC (Fig. 2d, j). However, the osteogenic differentiation potential of bmMSC (Fig. 2f) was more prominent compared to pMSC (Fig. 2l).
FIG. 1.
Expression of cell surface proteins on mesenchymal stromal cells (MSC). The bone marrow-derived mesenchymal stromal cells (bmMSC) (A) and placenta-derived mesenchymal stromal cells (pMSC) (B) were expanded in vitro and the expression of cell surface proteins was investigated by flow cytometry. According to consensus criteria, bmMSC and pMSC lack expression of CD14, CD34, CD45 (A[a–c], B[g–i]), but must express CD73, CD90, and CD105 (A[d–f], B[j–l]) [1,2].
FIG. 2.
Differentiation of bmMSC and pMSC in vitro. Before differentiation, the bmMSC (A[a]) and pMSC (B[g]) display a fibroblastic cell shape. The differentiation was induced in bmMSC (line A′) and pMSC (line B′) for 4 weeks in vitro and progress of chondrogenic, adipogenic, and osteogenic differentiation was explored by cytochemistry. Chondrogenic differentiation was detected by Alcian Blue (b, h). Adipogenic differentiation was detected by Oil Red O staining (d, j), and osteogenic differentiation by von Kossa staining (f, l). The corresponding controls were also stained with the suitable staining solutions, respectively (c, e, i, k). In contrast to bmMSC, efficient osteogenic differentiation could not be induced in pMSC [compare (f) vs. (l)]. The bars extend 250 μm. (C) To investigate differences in gene expression following differentiation, quantitative reverse transcriptase-polymerase chain reaction (RT-qPCR) was performed 7 days after induction of osteogenic differentiation [alkaline phosphatase (ALP) and osteocalcin], adipogenic (PPARγ2), or chondrogenic (Sox9) differentiation, respectively. MSC before differentiation served as controls. Black bars illustrate transcripts of bmMSC, and gray bars transcripts of pMSC. (C) Presents the x-fold transcript amounts of the corresponding differentiation marker gene relative to the transcripts in the MSC before differentiation as indicated (controls=1). Color images available online at www.liebertpub.com/scd
In addition, induction of differentiation was investigated by exploring the expression of transcripts encoding characteristic genes after 7 days of differentiation in vitro (Fig. 2C). On the transcription level, induction of PPARγ2 and Sox9 was considerably higher following adipogenic and chondrogenic differentiation, respectively, in bmMSC compared to that of pMSC (Fig. 2C). Although the relative increase of ALP encoding mRNA above controls was lower in differentiating bmMSC compared to pMSC (Fig. 2C), the absolute expression of ALP in bmMSC (mean 4.22×10−2) before differentiation was two logs above the steady state mRNA levels in pMSC (mean 2.78×10−4; and [10]), and it remained high during osteogenesis (bmMSC: mean 5.44×10−2 vs. pMSC: mean 4.88×10−4). Therefore, ALP is expressed at a much higher level in bmMSC and in differentiating bmMSC. In contrast to ALP, the late osteogenic factor osteocalcin was not significantly higher after 7 days of osteogenic differentiation of bmMSC suggesting that our bmMSC preparations did not contain a larger population of mature osteoblasts. In contrast, in pMSC, expression of osteocalcin was very low after 1 week of osteogenic differentiation, corroborating that pMSC fail to efficiently generate osteoblasts (Fig. 2C). A major effect of corticosteroids applied during adipogenic and osteogenic differentiation on expression of GAPDH in comparison to other transcript standards, β-actin, β-microglobulin, and ribosomal protein L13A, was not observed (data not shown).
Investigation of the transcriptome of MSC derived from bone marrow and from term placenta
We recently reported a significantly lower ALP expression in pMSC compared to bmMSC [10]. This is in line with our data reported here (Fig. 2C). We therefore hypothesized that additional factors may be expressed in bmMSC at different levels compared to pMSC. We investigated the whole transcriptome of bmMSC and pMSC before in vitro differentiation by gene array technology and confirmed the differences in gene expression by RT-qPCR. Overall, bmMSC generated a pattern of gene expression different from the patterns of placenta-derived fetal MSC (pfMSC) or placenta-derived maternal MSC (pmMSC), whereas the transcriptome of pfMSC and pmMSC was closely related (Fig. 3A). In particular, the mean expression correlation between the three groups was investigated. Identical cells show a mean correlation of 1.0. A lower number thus indicates less of a relationship. The mean expression correlation between pmMSC and pfMSC was 0.996 (data not shown), indicating a close relationship. In comparison, the mean correlation between pMSC (i.e., pfMSC and pmMSC combined) and bmMSC was 0.971 (data not shown) showing a decreased relationship.
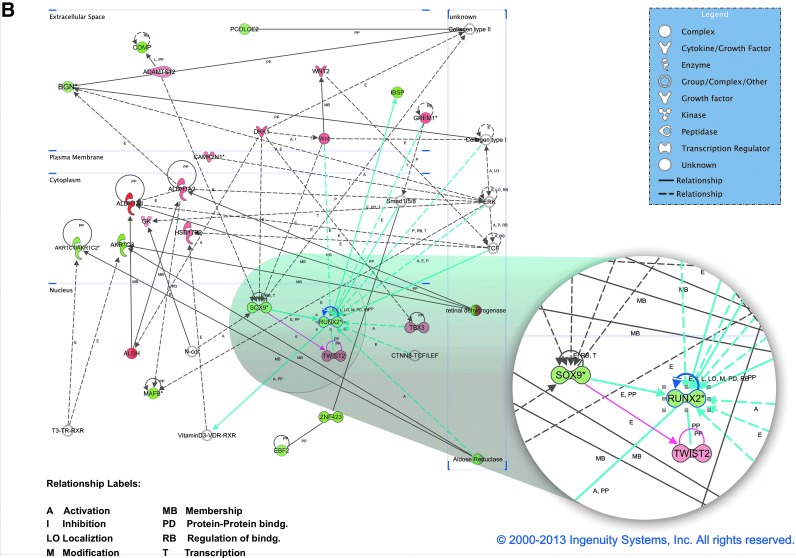
FIG. 3.

Investigation of the total transcriptome of bmMSC, placenta-derived maternal MSC (pmMSC), and placenta-derived fetal MSC (pfMSC). (A) Two independent sets of mRNA were prepared from bmMSC (four and seven donors), pmMSC (four and two donors), and pfMSC (four and four donors), reverse transcribed, labeled, and hybridized in two independent experiments to a GeneChip® (Human Genome U133 Plus 2.0 Array) representing all known human transcripts. The heatmap shows the expression of the 880 most variably expressed probe sets, representing about 600 different genes, between bmMSC, pmMSC, and pfMSC as indicated. Blue coloring indicates lower, yellow coloring indicates higher total expression. The values represent normalized absolute intensities on the log2 scale. (B) Functional annotation network from Ingenuity Pathway Analysis shows documented gene relationships among the genes in our data set. Biological findings are assigned to each gene and network based on the information in the Ingenuity Pathways Knowledge Base that was extracted from current scientific literature. Genes upregulated in bone marrow (green) or upregulated in placenta (red/pink) are colored accordingly. More intense colors represent higher gene expression. Blue lines represent relationships with Runx2, pink lines with Twist2, and black lines with all other genes. Color images available online at www.liebertpub.com/scd
More than 880 probe sets encompassing about 600 genes were differentially expressed between bmMSC compared to pMSC (i.e., pfMSC and pmMSC combined). Among them, several were genes associated with regulation of osteogenesis or bone metabolism (Tables 1 and 2). A functional analysis of enriched gene ontology categories revealed that adhesion, anatomical structures, and development are among the top significantly enriched biological processes. The top 10 molecular function categories involve calcium ion binding (56 genes) and signal transducer activity (76 genes).
Table 1.
Differences in Gene Expression in Naïve bmMSC Compared to Naïve pMSC
| Gene | Array difference | Main function |
|---|---|---|
| Alkaline phosphatase | 7.5-fold up | Mineralization of bone |
| Osteoglycin | 5.7-fold up | Induces ectopic bone formation |
| Osteomodulin | 4.3-fold up | Promotes attachment of osteoblasts |
| Runx2 | 5.3-fold up | Controls osteogenic differentiation |
| Twist-2 | 4.3-fold down | May inhibit osteoblast maturation |
| WISP2 | 48.5-fold up | Modulates bone turnover |
| WISP3 | 6.8-fold up | Essential for postnatal skeletal growth |
Representative genes involved in bone metabolism expressed at statistically significant different levels in bmMSC versus pMSC (P<0.05) are listed based on the evaluation of gene array data.
bmMSC, bone marrow-derived mesenchymal stromal cell; pMSC, placenta-derived mesenchymal stromal cell.
Table 2.
List of the Genes in Most Significantly Upregulated Canonical Pathways Associated with Runx2 and Twist2
| Category | Functions annotation | P-value | Molecules |
|---|---|---|---|
| Skeletal and muscular system development and function | Differentiation of osteoblasts | 2.03E-04 | BGN, DKK1, GREM1, RUNX2, TWIST2 |
| Cellular growth and proliferation | Proliferation of cells | 4.95E-04 | AKR1C1/AKR1C2, AKR1C3, ALDH1A1, ALDH1A2, BGN, CAMK2N1, COMP, DKK1, GREM1, IBSP, MAFB, RUNX2, SOX9, TBX3, TWIST2, WNT2, ZNF423 |
| Cellular development | Differentiation of cells | 1.36E-03 | AKR1C3, ALDH1A2, BGN, DKK1, EBF2, GREM1, IBSP, MAFB, RUNX2, SOX9, TBX3, TWIST2 |
| Cellular development | Differentiation of connective tissue cells | 2.16E-03 | BGN, DKK1, GREM1, RUNX2, SOX9, TWIST2 |
| Cell death and survival | Cell death | 5.15E-03 | ADAMTS12, ALDH1A1, ALDH1A2, BGN, COMP, DKK1, GREM1, IBSP, MAFB, RUNX2, SOX9, TBX3, TWIST2, WNT2, ZNF423 |
| Gene expression | Transcription of DNA | 6.21E-03 | DKK1, EBF2, GREM1, MAFB, RUNX2, SOX9, TBX3, TWIST2, ZNF423 |
| Cell death and survival | Apoptosis | 9.06E-03 | ALDH1A1, ALDH1A2, BGN, COMP, DKK1, GREM1, MAFB, RUNX2, SOX9, TBX3, TWIST2, WNT2, ZNF423 |
Pathways were expressed at statistically significant different levels in bmMSC versus pMSC data sets (P<0.05) and are listed based on gene array data analyzed by Ingenuity Pathway Analysis. Six pathways annotated to categories cancer and reproductive systems disease were omitted.
Among the factors expressed different between bmMSC and pMSC, transcription factor Runx2 was found elevated in bmMSC (Fig. 2C; Table 1). Runx2 is part of a major regulatory network, which includes other genes known to be involved in the development of the musculoskeletal anlagen (i.e., the initial clustering of embryonic cells from which a part or an organ develops) such as biglycan (BGN), cartilage oligomeric protein (COMP), integrin binding sialoprotein (IBSB), transcription factors Sox9 and Twist2 (Table 2). However, Twist2 was expressed at significantly lower levels in bmMSC versus pMSC (Table 1). To understand the underlying biology of Runx2 and Twist2 and their relationship with genes that were significantly up- or downregulated in our data sets (placenta MSC vs. bone marrow MSC), we used the IPA to compute a functional annotated network (i.e., documented gene relationships based on current literature among the genes from our data set) in which Runx2 and Twist2 are involved (Fig. 3B). This annotated network includes, for instance, the following factors relevant for bone and cartilage biology: BGN, type I and type II collagens, COMP, osteosarcoma marker EBF2, IBSB, transcription factor Sox9, and signaling factors Smad1/5/8 and elements of the wingless-related integration site (Wnt)-signaling pathway DKK1, Wnt2, and Wnt (Fig. 3B). Expression of the two transcription factors involved in the regulation of differentiation and maturation of osteoblasts, Runx2, and Twist2, was therefore investigated in more detail.
Expression of Runx2 and Twist2 in bmMSC and pMSC
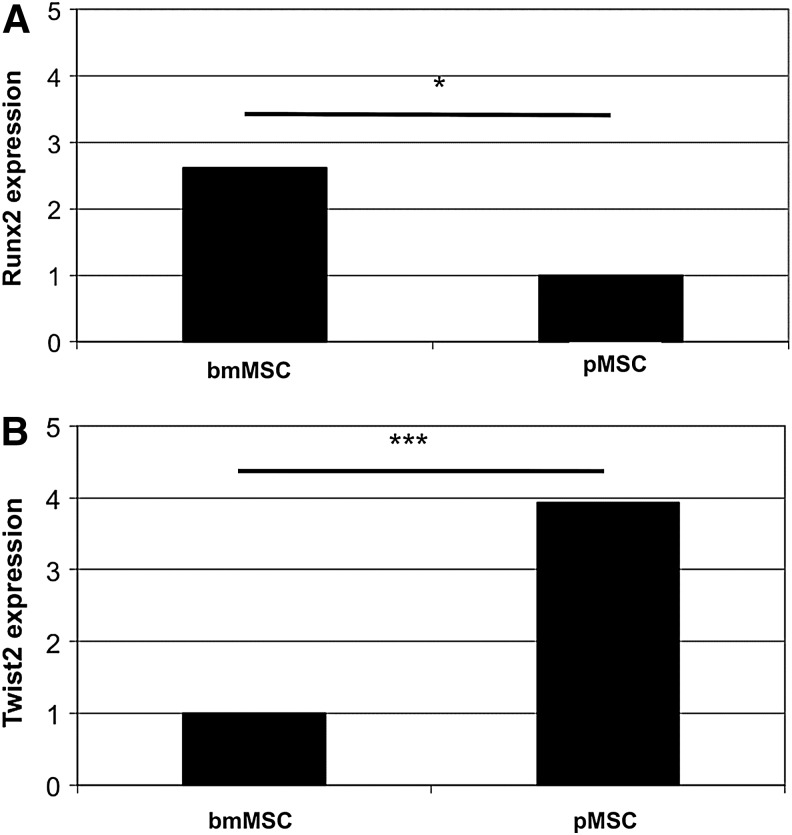
Additional MSC were expanded and expression of Runx2 and Twist2 was explored by RT-qPCR in the new samples. As expected from evaluation of the gene array analyses, in bmMSC, significantly more Runx2 encoding mRNA was found compared to pMSC (Fig. 4A, P<0.04), and bmMSC expressed significantly less Twist2 compared to pMSC (Fig. 4B, P<0.0002). To explore if these significant differences on the mRNA levels were translated into different amounts of these transcription factors on the protein level, immunocytochemistry was performed (Fig. 5).
FIG. 4.
Expression of runt-related transcription factor 2 (Runx2) and twist-related protein 2 (Twist2) encoding transcripts in bmMSC and pMSC. The cells were expanded and expression of Runx2 (A) and Twist2 (B) was investigated before differentiation by RT-qPCR. The bmMSC express significantly more Runx2 (2.6-fold, *P<0.04) compared to pMSC, and the pMSC express significantly more Twist2 compared to bmMSC (3.9-fold, ***P<0.0002).
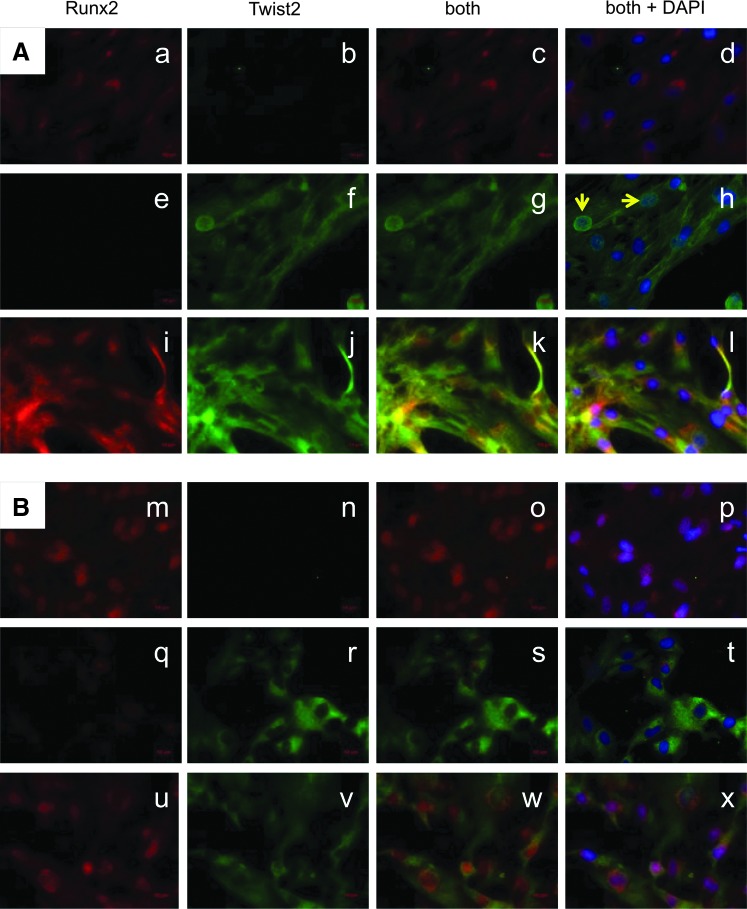
FIG. 5.
Detection of Runx2 and Twist2 in MSC by immunocytochemical staining. Expression and distribution of Runx2 and Twist2 proteins were investigated by immunocytochemistry/immunocytochemical in bmMSC (a–l) and pMSC (m–x) with antibodies as indicated. Nuclei were visualized by 4′,6-diamidino-2-phenylindol-dihydro-chlorid counterstaining and different areas of representative samples of MSC from three donors are shown. Expression of Runx2 (red, Cy3) is localized in and around the nuclei (d, l, p, x), whereas Twist2 [green, fluorescein isothiocyanate (FITC)] is spread all over the cytoplasm (h, t). In a few bmMSC, nuclear Twist2 was detected (h, arrows). There is no homogenous appearance of the distribution of Runx2- and/or Twist2-expressing cells across the samples. There are regions where only Runx2 (a, m) or Twist2 (f, r) is detected. In some areas, expression of both proteins is observed (k, w). Color images available online at www.liebertpub.com/scd
In both, bmMSC (Fig. 5A) and pMSC (Fig. 5B), Runx2 was enriched in and around the nuclei (Fig. 5a, i, m, u), whereas Twist2 appeared dispersed in the cytoplasm of bmMSC and pMSC (Fig. 5f, j, r, v). However, in some bmMSC, a nuclear concentration of Twist2 was observed (Fig. 5f) and nuclei appeared turquoise after counterstaining with DAPI (Fig. 5h). When overlaying both fluorescence channels, in some regions, a higher expression of Runx2 was observed (Fig. 5c, o), while in other regions, more Twist2 was detected (Fig. 5g, s). In other areas of bmMSC, the expression of Runx2 and Twist2 was similar and high (Fig. 5k). Overall, bmMSC samples appeared to contain more regions with prominent expression of both Runx2 and Twist2.
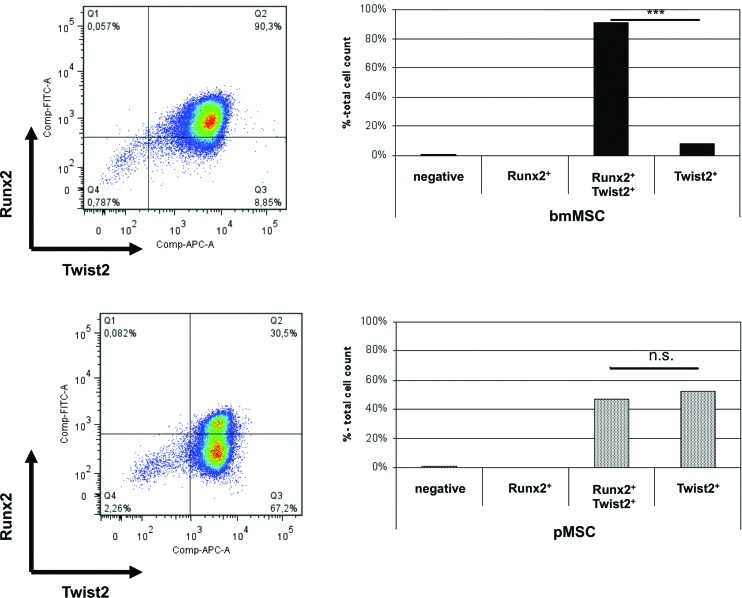
Immunocytochemistry is a qualitative method. To explore the differences in the expression of the Runx2 and Twist2 proteins in MSC by quantitative means, flow cytometry was performed (Fig. 6). On average, 91% of bmMSC showed bright intracellular staining of both transcription factors, Twist2 and Runx2, and 7.9% of bmMSC expressed only Twist2, while less than 1% expressed Runx2 alone. The difference in numbers of Runx2posTwist2pos and Runx2negTwist2pos was statistically significant (P<0.001, n=3 each). Double-negative (Runx2negTwist2neg) or Runx2posTwist2neg cells accounted for less that 2% of bmMSC. In contrast to bmMSC, in pMSC,. Runx2 (i.e., Runx2posTwist2pos plus Runx2posTwist2neg) was detected in fewer cells (average 46.8%, n=4). In addition, the mean fluorescence intensity (MFI) of the Runx2 staining was significantly lower in pMSC (MFI=450) compared to bmMSC (MFI=760, P<0.006, n≥3). For Twist2, this difference in MFI between bmMSC and pMSC was statistically significant as well (5877 vs. 3227, P<0.003, n≥3). Moreover, two subsets of pMSC were recorded: on average 52% of cells were Runx2negTwist2pos and 47% were Runx2posTwist2pos (difference in cell numbers not significant, n≥3 each). Runx2negTwist2neg or Runx2posTwist2neg cells accounted for less that 3% of pMSC. This strengthened our immunofluorescence results and corroborated that clearly more bmMSC express Runx2, and bmMSC express both transcription factors, Runx2 and Twist2, at higher levels.
FIG. 6.
Detection of intracellular Runx2 and Twist2 in MSC. The bmMSC (top panels) and pMSC (bottom panels) were expanded, characterized, and an aliquot of cells was permeabilized to allow staining of intracellular antigens. The cells were washed and analyzed by flow cytometry. Here two representative examples are shown. Expression of Twist2 is presented on X-axis (APC), expression of Runx2 on the Y-axis (FITC). Ninety percent of the bmMSC express Runx2 and Twist2, and only 9% Twist2 alone (quadrant Q3; upper left panel). The mean of the relative cell count of positive bmMSC was calculated from three individual flow cytometry experiments and the difference between numbers of Runx2posTwist2pos versus Runx2posTwist2neg was significant (***P<0.001; upper right panel). The majority of pMSC expressed Twist2 (quadrants Q2, Q3; lower left panel), although with a lower signal intensity compared with bmMSC (upper panel). While 30% of the pMSC were Runx2posTwist2pos, 67% were Runx2negTwist2pos (lower left panel). The mean of the relative cell counts of pMSC was calculated from three individual flow cytometry experiments, and the difference between numbers of Runx2posTwist2pos versus Runx2negTwist2pos was not significant (n.s., lower right panel). Color images available online at www.liebertpub.com/scd
We conclude that the expression of Twist2 and Runx2 differs significantly in bmMSC compared to pMSC, and the difference in expression of these transcription factors may contribute to the dissimilarities observed in the osteogenic differentiation between bmMSC and pMSC in vitro.
Discussion
In this study, we investigated the differences between bmMSC and pMSC and focused on the osteogenic differentiation potential of these cells. The rather low osteogenic potential of pMSC in comparison to bmMSC has been reported [18], and was associated at least, in part, with the differences in expression of ALP [10]. As ALP is the key enzyme utilized in the standard von Kossa staining method to detect mineralization of the matrix by osteoblasts, the prominent von Kossa staining generated from bmMSC could simply represent the differences in expression of ALP in bmMSC compared to pMSC. At the same time, expression of ALP on MSC, and on other stem cells as well, was suggested as an indicator for stemness [6,12,29,30]. Accordingly, within the bmMSC population, early stages of differentiation or maturation are detected [11,30,31]. However, mature osteoblasts express ALP as well, although less than bmMSC [32]. Monitoring osteogenesis by testing this enzyme on bmMSC may be therefore misleading, as contaminating osteoblasts may contribute to the ALP activity. Hence, we investigated the differentiation capacity of bmMSC and pMSC by investigating their total transcriptome in search of factors involved in regulation of osteogenesis rather than functional effector genes.
In the gene array data, a close relationship between the pfMSC and pmMSC was observed (Fig. 3). This could be caused by the techniques applied, since we separated the fetal from the maternal part of the placenta simply by slicing the tissue apart. However, the telomere lengths of pmMSC was significantly shorter compared to pfMSC (mean 20% shorter, P<0.029, Mann–Whitney test [33]). This confirmed that our methods for preparation of pfMSC and pmMSC enriched the expected types of cells. Moreover, the mean correlation of the transcriptome indicated a closer relationship between pmMSC and pfMSC (0.996), in comparison to pMSC and bmMSC (0.971). However, despite the differences in telomere lengths, genome, and transcriptome, the results also may suggest that pfMSC and pmMSC derived from the same tissue seem to be more closely related than MSC from different sources. Of course, differences in gender (bmMSC include male donors) or age (the age of bmMSC donors is 67±10 years, age of mothers in Tuebingen is on average 30±5 years, age of pfMSC 9 months) may account for differences as well. However, it is somewhat surprising that the pfMSC, although genetically clearly different from the pmMSC, share such a large portion of the transcriptome with pmMSC. This may indicate that the MSC niche has an influence on either the selection of MSC homing to these sites or may influence the cells with respect to gene expression and differentiation capacities. As the osteogenic differentiation capacity of pmMSC and pfMSC was rather low in both populations in comparison to bmMSC, and since tools such as antibodies for a defined separation of pmMSC and pfMSC by FACS, MACS, or alike are not at hand, we investigated the osteogenic differentiation potential of the total pMSC (pmMSC and pfMSC) in comparison to bmMSC.
The osteogenic differentiation of bmMSC is well known [1,17,34–36]. For atMSC and pMSC, osteogenic differentiation was described [2,14,37,38]. However, recent studies reported a rather weak osteogenic differentiation of pMSC [10,18]. These seemingly conflicting results regarding the differentiation capacities of pMSC could be explained at least, in part, by different protocols for isolation and expansion of the cells. For instance, changing the proteolytic enzymes (i.e., Dispase® and collagenases) influences the yield of pMSC isolated (unpublished observation), and may influence the relative amount of a given pMSC subset present in the bulk preparation. Contaminating osteoblasts in the bmMSC preparation could account for seemingly improved osteogenesis of bone marrow-derived cells. However, we can exclude this explanation as the expression and induction of the late osteogenic marker osteocalcin was rather low in bmMSC early after osteogenic stimulation (Fig. 2C). In addition, the composition of expansion media and the growth conditions have a major effect on MSC in vitro [19,20], and addition of platelet extract reportedly reduced the expression of the osteogenic markers, Ca-sensing receptor, and parathormone receptor on human MSC [39].
Moreover, it is well known that the osteogenic potential of MSC decreases over time of culture [40]. In our hands, bone marrow routinely yields fewer cells per donor and sample compared to placenta. Consequently, bmMSC require rather more population doublings to reach the number of cells needed for the experiments and still delivered a superior ostoegenic differentiation in vitro. However, significant differences in mitotic activities were not found between bmMSC and pMSC. Therefore, such technical differences between the preparation of bmMSC and pMSC seem not to account for the difference observed in osteogenesis.
One reason for the differences observed in osteogenesis could be the relationship between the two transcription factors Runx2 and Twist2. Runx2 is well known as an early osteogenic marker [41,42], and Runx2−/− mice lack normal bone formation [43]. Runx2 regulates expression of osteocalcin [44], a protein involved in matrix mineralization and calcium ion homeostasis [45,46], osteopontin (alias bone sialopotein-1), a polar linking protein in the extracellular matrix [47], and type I collagen, the major protein component of bone [48]. Growth factors such as bone morphogenic proteins (BMPs) and fibroblast growth factors (FGFs) activate the expression of Runx2, and Runx2 can negatively regulate its own expression [49]. We show that upon expansion in GMP-compliant media, bmMSC expressed Runx2 on transcript and protein levels significantly higher compared to pMSC and the MFI indicated a higher protein expression in bmMSCs as well (Figs. 4–6). Since bmMSC and pMSC were expanded in the same media, the concentration of BMPs or FGFs in the expansion media cannot account for differences in Runx2 expression in these cells. Another reason for the differences in Runx2 expression may be due to fluctuations of Runx2 during the cell cycle [50]. It has been shown that proliferating cells may contain more Runx2 [49]. Thus, proliferating Runx2-rich MSC should enter osteogenic differentiation more efficiently. Although others have shown that addition of steroids and corticosteroids to the differentiation media activated the proliferation of MSC in vitro and facilitated their osteogenic differentiation in a gender-dependent way [51,52], in the present study, we show that proliferation rates of bmMSC were not different in vitro compared to pMSC, excluding proliferation as a cause for elevated Runx2. However, experimental evidence from other laboratories provided evidence that a transient serum deprivation before induction of differentiation, which also causes a retardation of proliferation, could facilitate the differentiation of MSC in vitro [53]. In addition, supernatants from MSC transiently suppressed maturation of osteoblasts by downregulating expression of Runx2 [50]. Therefore, differences in the release of osteogenic factors from bmMSC in primary culture or during initiation of differentiation could account for their prominent osteogenic potential. However, a detailed investigation of osteogenic factors in bmMSC versus pMSC supernatants requires a whole set of additional experiments that are beyond the scope of this study.
Twist2, also called Dermo1, is involved in regulation of proliferation and cell lineage determination. In adipogenesis, Twist2 represses the activity of the adipocyte determination and differentiation factor 1 (ADD1) by binding to the ADD1 target sequence in gene promoter regions [54]. In osteoblasts, Twist1 and Twist2 are involved in the parathyroid hormone-dependent regulation of osteocalcin and the bone-related activating transcription factor 4 (ATF4) [54]. Twist proteins interact with the ATF4 protein and attenuate binding of ATF4 to the osteocalcin promoter. Moreover, the Twist proteins interact with Runx2 in gene regulation, and binding of Twist1 or Twist2 to Runx2 can block its function as a transcription factor, and thus block osteogenesis, but without affecting the expression of Runx2 itself [55]. At the same time, crossing Twist2−/− mice with Runx2+/− heterozygotes rescued their phenotype. Moreover, premature osteogenesis was observed in Twist2−/− knockout mice [55]. Despite a significantly higher expression of Twist2 encoding transcripts in pMSC (Fig. 4), the protein expression of Twist2 detected in pMSC by flow cytometry (MFI 3,227, Fig. 6) was lower compared to bmMSC (MFI 5,877, Fig. 6). Therefore, pMSC express the Twist2 protein at lower levels compared to bmMSC and about 50% of pMSC lack expression of the Runx2 protein, central factors needed for efficient osteogenesis. In agreement with our results, an elevated mRNA expression of Twist2 was reported in human decidua-derived pMSC [18]. A possible explanation for these seemingly conflicting results may be the modification of translation of Twist2 encoding mRNA by small RNA species [56]. Discordance between mRNA and protein expression data was reported in mouse embryonic development suggesting a post-transcriptional regulation of Twist [57]. In one study, interleukin-6 induced phosphorylation of the Twist protein and thereby increased its stability in human cancer cells [58]. Such post-transcriptional mechanisms may also work in human MSC with respect to Twist2.
In murine MSC, basic fibroblast growth factor (FGF2) inhibited the osteogenic and chondrogenic differentiation by inducing a significant expression of Twist2 and Sprouty4 mRNAs, while reducing Runx2 mRNA significantly. The Twist1 mRNA was not significantly changed [59]. At the same time, FGF2 caused nuclear aggregation of Twist1. However, the cellular redistribution of Twist2 by FGF2 was not investigated specifically [59]. Still, differences in FGF2 present in the MSC cultures could explain the differences in Twist2 and Runx2 expression, as well as the differences in the osteogensis of bmMSC compared to pMSC. However, a significant difference in FGF2 mRNA expression between bmMSC and pMSC was not detected by gene array (data not shown).
For human Twist, two functionally important nuclear localization signals were described, RKRR and KRGKK [60] and, together with expression of Bcl2, Twist2, and Snail, nuclear localization of Twist1 was considered a prognostic indicator for hepatocellular carcinoma [61]. Data on specific regulation of nuclear transport of Twist2 are sparse, but Twist1 and Twist2 share the KRGKK-motif (BLAST search: http://blast.ncbi.nlm.nih.gov). Therefore, Twist2 may utilize similar, if not the same pathways, to regulate its nuclear transport. Here we report that in pMSC, Twist2 was dispersed in the cytoplasm and not enriched around or in the nuclei, whereas in some bmMSC, Twist2 was recorded in or close to the nuclei (Fig. 5h). Thus, specific stimuli seem to be involved in nuclear translocation of Twist2 in MSC. However, the ostensible discrepancy between expression of Twist2 mRNA and protein in bmMSC versus pMSC in the context of their osteogenic differentiation potential seems to depend on a complex regulatory circuit, and may include phosphorylation, protein stability, and nuclear transport, but not only on the amounts of intracellular Twist protein. Clues resolving the issue must, however, await further investigations.
Summary and Conclusion
Despite many similarities, bmMSC and pMSC display distinct characteristics. Whereas chondrogenic differentiation and to a certain degree adipogenic differentiation were observed with pMSC, efficient osteogenic differentiation of pMSC was not observed. The differences in their osteogenic potential correlated with significant differences in expression of Runx2 and Twist2. In regenerative medicine, bmMSC seem to be ideal cells for bone repair, whereas MSC from other sources, including placenta could be better suited when mineralization results in adverse sequela for the patient treated.
Acknowledgments
The authors thank Torsten Kluba, MD, Falk Mittag, MD, and Nikolaus Wülker, MD (Department Orthop. Surg, UKT, Tuebingen, FRG) for providing samples of bone marrow, Tanja Abruzzese, Elisabeth Kienzle, and Stephanie Zug for excellent technical support, and André Hennig for help in evaluation of the Affymetrix® array data. This study was supported in part by a Fortune grant to M.L.H., by grants of the BMBF and DFG to W.K.A., and by funding through the departments involved.
Author Disclosure Statement
There is no conflict of interests to be disclosed for any of the authors of this manuscript.
References
- 1.Dominici M. Le Blanc K. Mueller I. Slaper-Cortenbach I. Marini F. Krause D. Deans R. Keating A. Prockop D. Horwitz E. Minimal criteria for defining multipotent mesenchymal stromal cells. International Society for Cellular Therapy position statement. Cytotherapy. 2006;8:315–317. doi: 10.1080/14653240600855905. [DOI] [PubMed] [Google Scholar]
- 2.Parolini O. Alviano F. Bagnara GP. Bilic G. Buhring HJ. Evangelista M. Hennerbichler S. Liu B. Magatti M, et al. Concise review: isolation and characterization of cells from human term placenta: outcome of the first International Workshop on Placenta Derived Stem Cells. Stem Cells. 2008;26:300–311. doi: 10.1634/stemcells.2007-0594. [DOI] [PubMed] [Google Scholar]
- 3.Tormin A. Li O. Brune JC. Walsh S. Schüz B. Ehinger M. Ditzel N. Kassem M. Scheding S. CD146 expression on primary nonhematopoietic bone marrow stem cells is correlated with in situ localization. Blood. 2011;117:5067–5077. doi: 10.1182/blood-2010-08-304287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chamberlain G. Fox J. Ashton B. Middleton J. Concise review: mesenchymal stem cells: their phenotype, differentiation capacity, immunological features, and potential for homing. Stem Cells. 2007;25:2739–2749. doi: 10.1634/stemcells.2007-0197. [DOI] [PubMed] [Google Scholar]
- 5.Kolf CM. Cho E. Tuan RS. Mesenchymal stromal cells. Biology of adult mesenchymal stem cells: regulation of niche, self-renewal and differentiation. Arthritis Res Ther. 2007;9:204. doi: 10.1186/ar2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buhring HJ. Battula VL. Treml S. Schewe B. Kanz L. Vogel W. Novel markers for the prospective isolation of human MSC. Ann N Y Acad Sci. 2007;1106:262–271. doi: 10.1196/annals.1392.000. [DOI] [PubMed] [Google Scholar]
- 7.Gang EJ. Bosnakovski D. Figueiredo CA. Visser JW. Perlingeiro RCR. SSEA-4 identifies mesenchymal stem cells from bone marrow. Blood. 2007;109:1743–1751. doi: 10.1182/blood-2005-11-010504. [DOI] [PubMed] [Google Scholar]
- 8.Bae S. Park CW. Son HK. Ju HK. Paik D. Jeon CJ. Koh GY. Kim J. Kim H. Fibroblast activation protein alpha identifies mesenchymal stromal cells from human bone marrow. Br J Haematol. 2008;142:827–830. doi: 10.1111/j.1365-2141.2008.07241.x. [DOI] [PubMed] [Google Scholar]
- 9.Sivasubramaniyan K. Harichandan A. Schumann S. Sobiesiak M. Lengerke C. Maurer A. Kalbacher H. Bühring HJ. Prospective isolation of mesenchymal stem cells from human bone marrow using novel antibodies directed against Sushi domain containing protein 2 (SUSD2) Stem Cells Dev. 2013;22:1944–1954. doi: 10.1089/scd.2012.0584. [DOI] [PubMed] [Google Scholar]
- 10.Pilz G. Ulrich C. Abruzzese T. Abele H. Schäfer R. Buhring HJ. Aicher WK. Human term placenta-derived mesenchymal stromal cells are less prone to osteogenic differentiation than bone marrow-derived mesenchymal stromal cells. Stem Cells Dev. 2011;20:635–646. doi: 10.1089/scd.2010.0308. [DOI] [PubMed] [Google Scholar]
- 11.Vogel W. Grünebach F. Messam CA. Kanz L. Brugger W. Bühring HJ. Heterogeneity among human bone marrow-derived mesenchymal stem cells and neural progenitor cells. Haematologica. 2003;88:126–133. [PubMed] [Google Scholar]
- 12.Battula VL. Treml S. Bareiss PM. Gieseke F. Roelofs H. de Zwart P. Müller I. Schewe B. Skutella T, et al. Isolation of functionally distinct mesenchymal stem cell subsets using antibodies against CD56, CD271 and mesenchymal stem cell antigen-1 (MSCA-1) Hematologica. 2009;94:19–30. doi: 10.3324/haematol.13740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alexander D. Schäfer F. Munz A. Friedrich B. Klein C. Hoffmann J. Bühring HJ. Reinert S. NGFR: a new osteogenic differentiation marker in mineralizing periosteal cells. Tissue Eng. 2009;15:715. [Google Scholar]
- 14.Zannettino ACW. Paton S. Arthur A. Khor F. Itescu S. Gimble JM. Gronthos S. Multipotential human adipose-derived stromal stem cells exhibit a perivascular phenotype in vitro and in vivo. J Cell Physiol. 2008;214:413–421. doi: 10.1002/jcp.21210. [DOI] [PubMed] [Google Scholar]
- 15.De Bari C. Dell'Accio F. Tylzanowski P. Luyten FP. Multipotent mesenchymal stem cells from adult synovial membrane. Arthritis Rheum. 2001;44:1928–1942. doi: 10.1002/1529-0131(200108)44:8<1928::AID-ART331>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 16.De Bari C. Dell'Accio F. Luyten FP. Failure of in vitro–differentiated mesenchymal stem cells from the synovial membrane to form ectopic stable cartilage in vivo. Arthritis Rheum. 2004;50:142–150. doi: 10.1002/art.11450. [DOI] [PubMed] [Google Scholar]
- 17.Aicher WK. Buhring HJ. Hart ML. Rolauffs B. Badke A. Klein G. Regeneration of cartilage and bone by defined subsets of mesenchymal stromal cells - potential and pitfalls. Adv Drug Deliv Rev. 2011;63:342–351. doi: 10.1016/j.addr.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 18.Kanematsu D. Shofuda T. Yamamoto A. Ban C. Ueda T. Yamasaki M. Kanemura Y. Isolation and cellular properties of mesenchymal cells derived from the decidua of human term placenta. Differentiation. 2011;82:77–88. doi: 10.1016/j.diff.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 19.Felka T. Schäfer R. Schewe B. Benz K. Aicher WK. Hypoxia reduces the inhibitpry effect of IL-1beta on chondrogenic differentiation of FCS-free expanded MSC. Osteoarthritis Cartilage. 2009;17:1368–1376. doi: 10.1016/j.joca.2009.04.023. [DOI] [PubMed] [Google Scholar]
- 20.Felka T. Schäfer R. deZwart P. Aicher WK. Animal serum-free differentiation of human mesenchymal stem cells. Cytotherapy. 2010;12:143–153. doi: 10.3109/14653240903470647. [DOI] [PubMed] [Google Scholar]
- 21.Hrapchak B. Theory and Practice of Histotechnology. Battelle Press; Columbus, OH: 1980. [Google Scholar]
- 22.Ellis R. Oil Red O Staining Protocol. www.ihcworld.com www.ihcworld.com
- 23.IHCworld. Alcian Blue Staining Protocol. www.ihcworld.com www.ihcworld.com
- 24.Pilz GA. Braun J. Ulrich C. Felka T. Warstat K. Ruh M. Schewe B. Abele H. Larbi A. Aicher WK. Human mesenchymal stromal cells express CD14 cross-reactive epitopes. Cytometry A. 2011;79:635–645. doi: 10.1002/cyto.a.21073. [DOI] [PubMed] [Google Scholar]
- 25.Bolstad BM. Irizarry RA. Astrand M. Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 26.Battke F. Symons S. Nieselt K. Mayday—integrative analytics for expression data. BMC Bioinformatics. 2010;11:121. doi: 10.1186/1471-2105-11-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang DW. Sherman BT. Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rasmussen R. Morrison T. Herrmann M. Wittwer C. Quantitative PCR by continuous fluorescence monitoring of a double strand DNA specific binding dye. Biochemica. 1998;2:8–11. [Google Scholar]
- 29.Tavakoli T. Xu X. Derby E. Serebryakova Y. Reid Y. Rao MS. Mattson MP. Ma W. Self-renewal and differentiation capabilities are variable between human embryonic stem cell lines I3, I6 and BG01V. BMC Cell Biol. 2009;10:44. doi: 10.1186/1471-2121-10-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sobiesiak M. Sivasubramaniyan K. Hermann C. Tan C. Orgel M. Treml S. Cerabona F. de Zwart P. Ochs U, et al. The mesenchymal stem cell antigen MSCA-1 is identical to tissue non-specific alkaline phosphatase. Stem Cells Dev. 2010;19:669–677. doi: 10.1089/scd.2009.0290. [DOI] [PubMed] [Google Scholar]
- 31.Sivasubramaniyan K. Lehnen D. Ghazanfari R. Sobiesiak M. Harichandan A. Mortha E. Petkova N. Grimm S. Cerabona F, et al. Phenotypic and functional heterogeneity of human bone marrow- and amnion-derived MSC subsets. Ann N Y Acad Sci. 2012;1266:94–106. doi: 10.1111/j.1749-6632.2012.06551.x. [DOI] [PubMed] [Google Scholar]
- 32.Manduca P. Sanguineti C. Pistone M. Boccignone E. Sanguineti F. Santolini F. Federici A. Differential expression of alkaline phosphatase in clones of human osteoblast-like cells. J Bone Miner Res. 1993;8:291–300. doi: 10.1002/jbmr.5650080306. [DOI] [PubMed] [Google Scholar]
- 33.Ulrich C. Diploma Thesis, Faculty for Biology. Eberhard Karls University; Tuebingen, Germany: 2009. Untersuchung der osteogenen Differenzierbarkeit von humanen adulten Stammzellen und von definierten Subpopulationen aus plazentalem Gewebe. [Google Scholar]
- 34.Friedenstein AJ. Chailakhyan CA. Gerasimov UV. Bone marrow osteogenic stem cells: in vitro cultivation and transplantation in diffusion chambers. Cell Tissue Kinet. 1987;20:263–272. doi: 10.1111/j.1365-2184.1987.tb01309.x. [DOI] [PubMed] [Google Scholar]
- 35.Pittenger MF. Mackay AM. Beck SC. Jaiswal RK. Douglas R. Mosca JD. Moorman MA. Simonetti DW. Craig S. Marshak DR. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 36.Pittenger MF. Mosca JD. McIntosh KR. Human mesenchymal stem cells: progenitor cells for cartilage, bone, fat and stroma. Curr Top Microbiol Immunol. 2000;251:3–11. doi: 10.1007/978-3-642-57276-0_1. [DOI] [PubMed] [Google Scholar]
- 37.Yumi F. Hideaki N. Daisuke S. Imiko H. Toshio K. Kohichiro T. Human placenta-derived cells have mesenchymal stem/progenitor cell potential. Stem Cells. 2004;22:649–658. doi: 10.1634/stemcells.22-5-649. [DOI] [PubMed] [Google Scholar]
- 38.Battula VL. Bareiss PM. Treml S. Conrad S. Albert I. Hojak S. Abele H. Schewe B. Just L. Skutella T. Buhring HJ. Human placenta and bone marrow derived MSC cultured in serum-free, b-FGF-containing medium express cell surface frizzled-9 and SSEA-4 and give rise to multilineage differentiation. Differentiation. 2007;75:279–291. doi: 10.1111/j.1432-0436.2006.00139.x. [DOI] [PubMed] [Google Scholar]
- 39.Ben Azouna N. Jenhani F. Regaya Z. Berraeis L. Ben Othman T. Ducrocq E. Domenech J. Phenotypical and functional characteristics of mesenchymal stem cells from bone marrow: comparison of culture using different media supplemented with human platelet lysate or fetal bovine serum. Stem Cell Res Ther. 2012;3:6. doi: 10.1186/scrt97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muraglia A. Cancedda R. Quarto R. Clonal mesenchymal progenitors from human bone marrow differentiate in vitro according to a hierarchical model. J Cell Sci. 2000;113:1161–1166. doi: 10.1242/jcs.113.7.1161. [DOI] [PubMed] [Google Scholar]
- 41.Lee M-H. Kim Y-J. Kim H-J. Park H-D. Kang A-R. Kyung H-M. Sung J-H. Wozney JM. Kim H-J. Ryoo H-M. BMP-2-induced Runx2 expression is mediated by Dlx5, and TGF-β1 opposes the BMP-2-induced osteoblast differentiation by suppression of Dlx5 expression. J Biol Chem. 2003;278:34387–34394. doi: 10.1074/jbc.M211386200. [DOI] [PubMed] [Google Scholar]
- 42.Yoda S. Suda N. Kitahara Y. Komori T. Ohyama K. Delayed tooth eruption and suppressed osteoclast number in the eruption pathway of heterozygous Runx2/Cbfa1 knockout mice. Arch Oral Biol. 2004;49:435–442. doi: 10.1016/j.archoralbio.2004.01.010. [DOI] [PubMed] [Google Scholar]
- 43.Komori T. Yagi H. Nomura S. Yamaguchi A. Sasaki K. Deguchi K. Shimizu Y. Bronson RT. Gao YH, et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 44.Sierra J. Villagra A. Paredes R. Cruzat F. Gutierrez S. Javed A. Arriagada G. Olate J. Imschenetzky M, et al. Regulation of the bone-specific osteocalcin gene by p300 requires Runx2/Cbfa1 and the vitamin D3 receptor but not p300 intrinsic histone acetyltransferase activity. Mol Cell Biol. 2003;23:3339–3351. doi: 10.1128/MCB.23.9.3339-3351.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marie PJ. Transcription factors controlling osteoblastogenesis. Arch Biochem Biophys. 2008;473:98–105. doi: 10.1016/j.abb.2008.02.030. [DOI] [PubMed] [Google Scholar]
- 46.Jang WG. Kim EJ. Kim DK. Ryoo HM. Lee KB. Kim SH. Choi HS. Koh JT. BMP2 protein regulates osteocalcin expression via Runx2-mediated Atf6 gene transcription. J Biol Chem. 2012;287:905–915. doi: 10.1074/jbc.M111.253187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang X. Meng X. Su X. Mauchley DC. Ao L. Cleveland JC., Jr. Fullerton DA. Bone morphogenic protein 2 induces Runx2 and osteopontin expression in human aortic valve interstitial cells: role of Smad1 and extracellular signal-regulated kinase 1/2. J Thorac Cardiovasc Surg. 2009;138:1008–1015. doi: 10.1016/j.jtcvs.2009.06.024. [DOI] [PubMed] [Google Scholar]
- 48.Ishida K. Yamaguchi M. Albumin regulates Runx2 and alpha1 (I) collagen mRNA expression in osteoblastic cells: comparison with insulin-like growth factor-I. Int J Mol Med. 2005;16:689–694. [PubMed] [Google Scholar]
- 49.Schroeder TM. Jensen ED. Westendorf JJ. Runx2: a master organizer of gene transcription in developing and maturing osteoblasts. Birth Defects Res C Embryo Today. 2005;75:213–225. doi: 10.1002/bdrc.20043. [DOI] [PubMed] [Google Scholar]
- 50.Sun J. Zhou H. Deng Y. Zhang Y. Gu P. Ge S. Fan X. Conditioned medium from bone marrow mesenchymal stem cells transiently retards osteoblast differentiation by downregulating Runx2. Cells Tissues Organs. 2012;196:510–522. doi: 10.1159/000339245. [DOI] [PubMed] [Google Scholar]
- 51.McCulloch CAG. Tenenbaum HC. Dexamethasone induces proliferation and terminal differentiation of osteogenic cells in tissue culture. Anat Rec. 1986;215:397–402. doi: 10.1002/ar.1092150410. [DOI] [PubMed] [Google Scholar]
- 52.Hong L. Sultana H. Paulius K. Zhang G. Steroid regulation of proliferation and osteogenic differentiation of bone marrow stromal cells: a gender difference. J Steroid Biochem Mol Biol. 2009;114:180–185. doi: 10.1016/j.jsbmb.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cordonnier T. Langonné A. Sohier J. Layrolle P. Rosset P. Sensébé L. Deschaseaux F. Consistent osteoblastic differentiation of human mesenchymal stem cells with bone morphogenetic protein 4 and low serum. Tissue Eng Part C Methods. 2011;17:249–259. doi: 10.1089/ten.TEC.2010.0387. [DOI] [PubMed] [Google Scholar]
- 54.Lee YS. Lee HH. Park J. Yoo EJ. Glackin CA. Choi YI. Jeon SH. Seong RH. Park SD. Kim JB. Twist2, a novel ADD1/SREBP1c interacting protein, represses the transcriptional activity of ADD1/SREBP1c. Nucleic Acids Res. 2003;31:7165–7174. doi: 10.1093/nar/gkg934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bialek P. Kern B. Yang X. Schrock M. Sosic D. Hong N. Wu H. Yu K. Ornitz DM, et al. A twist code determines the onset of osteoblast differentiation. Dev Cell. 2004;6:423–435. doi: 10.1016/s1534-5807(04)00058-9. [DOI] [PubMed] [Google Scholar]
- 56.Zeng Y. Wagner EJ. Cullen BR. Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol Cell. 2002;9:1327–1333. doi: 10.1016/s1097-2765(02)00541-5. [DOI] [PubMed] [Google Scholar]
- 57.Gitelman I. Twist protein in mouse embryogenesis. Dev Biol. 1997;189:205–214. doi: 10.1006/dbio.1997.8614. [DOI] [PubMed] [Google Scholar]
- 58.Su YW. Xie TX. Sano D. Myers JN. IL-6 stabilizes twist and enhances tumor cell motility in head and neck cancer cells through activation of casein kinase 2. PLoS One. 2011;6:e19412. doi: 10.1371/journal.pone.0019412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lai WT. Krishnappa V. Phinney DG. Fibroblast growth factor 2 (Fgf2) inhibits differentiation of mesenchymal stem cells by inducing Twist2 and Spry4, blocking extracellular regulated kinase activation, and altering Fgf receptor expression levels. Stem Cells. 2011;29:1102–1111. doi: 10.1002/stem.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Singh S. Gramolini AO. Characterization of sequences in human TWIST required for nuclear localization. BMC Cell Biol. 2009;10:47. doi: 10.1186/1471-2121-10-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhao N. Sun BC. Zhao XL. Liu ZY. Sun T. Qiu ZQ. Gu Q. Che N. Dong XY. Coexpression of Bcl-2 with epithelial-mesenchymal transition regulators is a prognostic indicator in hepatocellular carcinoma. Med Oncol. 2012;29:2780–2792. doi: 10.1007/s12032-012-0207-y. [DOI] [PubMed] [Google Scholar]