This communication describes a simple method for patterning paper to create well-defined, millimeter-sized channels, comprising hydrophilic paper bounded by hydrophobic polymer. We believe that this type of patterned paper will become the basis for low-cost, portable, and technically simple multiplexed bioassays. We demonstrate this capability by the simultaneous detection of glucose and protein in 5 μL of urine. The assay system is small, disposable, easy to use (and carry), and requires no external equipment, reagents, or power sources. We believe this kind of system is attractive for uses in less-industrialized countries, in the field, or as an inexpensive alternative to more advanced technologies already used in clinical settings.[1-4]
The analysis of biological fluids is necessary for monitoring the health of populations,[2] but these measurements are difficult to implement in remote regions such as those found in less-industrialized countries, in emergency situations, or in home health-care settings.[3] Conventional laboratory instruments provide quantitative measurements of biological samples, but they are unsuitable for these situations since they are large, expensive, and require trained personnel and considerable volumes of biological samples.[2] Other bioassay platforms provide alternatives to more expensive instruments,[5-7] but the need remains for a platform that uses small volumes of sample and that is sufficiently inexpensive to be used widely for measuring samples from large populations.
We believe that paper may serve as a particularly convenient platform for running bioassays in the remote situations locations. As a prototype for a mthod we believe to be particularly promosing, we patterned photoresist onto chromatography paper to form defined areas of hydrophilic paper separated by hydrophobic lines or “walls”; these patterns provide spatial control of biological fluids and enable fluid transport, without pumping, due to capillary action in the millimeter-sized channels produced. This method for patterning paper makes it possible to run multiple diagnostic assays on one strip of paper, while still using only small volumes of a single sample. In a fully developed technology, patterned photoresist would be replaced by an appropriate printing technology, but patterning paper with photoresist is: i) convenient for prototyping these devices, and ii) a useful new micropatterning technology in its own right.
We patterned chromatography paper with SU-8 2010 photoresist as shown in Figure 1a and as described below: we soaked a 7.5-cm diameter piece of chromatography paper in 2 mL of SU-8 2010 for 30 s, spun it at 2000 rpm for 30 s, and then baked it at 95 °C for 5 min to remove the cyclopentanone in the SU-8 formula. We then exposed the photoresist and paper to 405 nm UV light (50 mW/cm2) for 10 s through a photo-mask (CAD/Art Services, Inc.) that was aligned using a mask aligner (OL-2 Mask Aligner, AB-M, Inc). After exposure, we baked the paper a second time at 95 °C for 5 min to cross-link the exposed portions of the resist. The unpolymerized photoresist was removed by soaking the paper in propylene glycol monomethyl ether acetate (PGMEA) (5 min), and by washing the pattern with propan-2-ol (3 × 10 mL). The paper was more hydrophobic after it was patterned, presumably due to residual resist bound to the paper, so we exposed the entire surface to an oxygen plasma for 10 s at 600 millitorr (SPI Plasma-Prep II, Structure Probe, Inc) to increase the hydrophilicity of the paper (Figures 2a and 2b).
Figure 1.
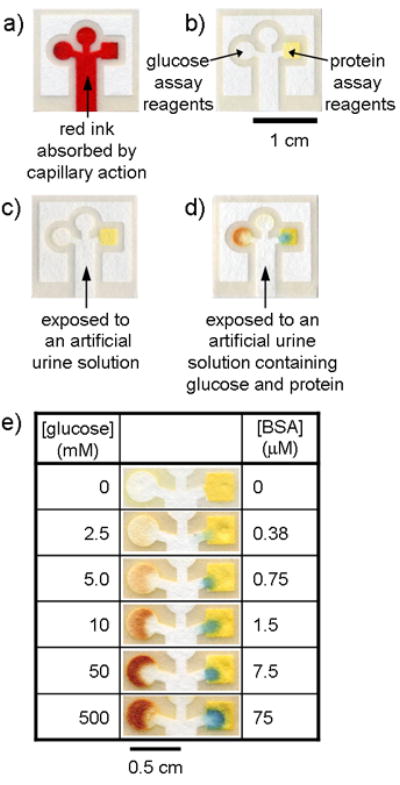
Chromatography paper patterned with photoresist. The darker lines are cured photoresist; the lighter areas are unexposed paper. (a) Patterned paper after absorbing 5 μL of Waterman red ink by capillary action. The central channel absorbs the sample by capillary action and the pattern directs the sample into three separate test areas. (b) Complete assay after spotting the reagents. The square region on the right is the protein test and the circular region on the left is the glucose test. The circular region on the top was used as a control well and was spotted with either iodide but no enzyme solution, or with enzyme solution but no iodide. (c) Negative control for glucose (left) and protein (right) using 5 μL of an artificial urine solution.[18] (d) Positive assay for glucose (left) and protein (right) using 5 μL of a solution that contained 550 mM glucose and 75 μM BSA in an artificial urine solution. The control well was spotted with the potassium iodide solution, but not with the enzyme solution. A similar control containing the enzyme solution, but not the iodide, gave identical results (data not shown). (e) Glucose and protein detection assays using varying concentrations of glucose and BSA.
Figure 2.
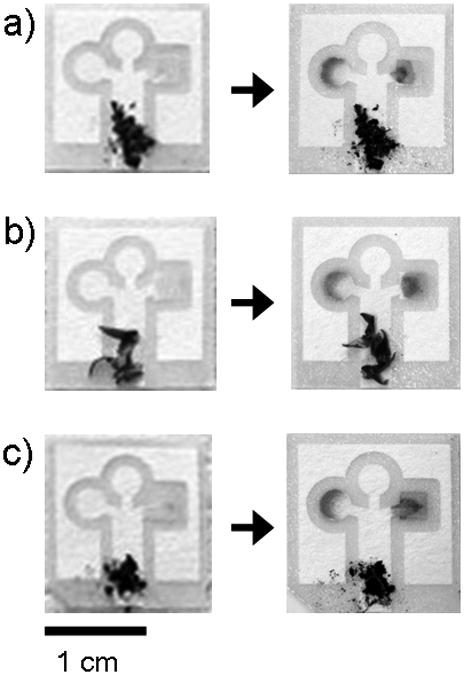
Assays contaminated with (a) dirt, (b) plant pollen, and (c) graphite powder. The pictures were taken before and after running an artificial urine solution that contained 550 mM glucose and 75 μM BSA. The particulates do not move up the channels during the assay.
The patterned paper can be derivatized for biological assays by adding appropriate reagents to the test areas (Figures 1b and 2b). In this communication, we demonstrate the method by detecting glucose and protein,[8] but the surface should be suitable for measuring many other analytes as well.[7] The glucose assay is based on the enzymatic oxidation of iodide to iodine,[9] where a color change from clear to brown is associated with the presence of glucose.[10] The protein assay is based on the color change of tetrabromophenol blue (TBPB) when it ionizes and binds to proteins;[11] a positive result in this case is indicated by a color change from yellow to blue.
For the glucose assay, we spotted 0.3 μL of a 0.6 M solution of potassium iodide, followed by 0.3 μL of a 1:5 horseradish peroxidase/glucose oxidase solution (15 units of protein per mL of solution). For the protein assay, we spotted 0.3 μL of a 250-mM citrate buffer (pH 1.8) in a well separate from the glucose assay, and then layered 0.3 μL of a 3.3 mM solution of tetrabromophenol blue (TBPB) in 95% ethanol over the citrate buffer. The spotted reagents were allowed to air dry at room temperature. This pre-loaded paper gave consistent results for the protein assay regardless of storage temperature and time (when stored for 15 d both at 0 °C and at 23 °C, wrapped in aluminum foil). The glucose assay was sensitive to storage conditions, and showed decreased signal for assays run 24 h after spotting the reagents (when stored at 23 °C); when stored at 0 °C, however, the glucose assay was as sensitive after day 15 as it was on day 1.
We measured artificial samples of glucose and protein in clinically relevant ranges (2.5-50 mM for glucose and 0.38-7.5 μM for bovine serum albumin (BSA))[12, 13] by dipping the bottom of each test strip in 5 μL of a pre-made test solution (Figure 2d). The fluid filled the entire pattern within ca. one minute, but the assays required 10-11 min for the paper to dry and for the color to fully develop.[14] In all cases, we observed color changes corresponding roughly in intensity to the amount of glucose and protein in the test samples, where the lowest concentrations define the lower limits to which these assays can be used (Figure 2e). For comparison, commercially-available dipsticks detect glucose at concentrations as low as 5 mM[7, 9] and protein as low as 0.75 μM;[6, 15] these limits indicate that these paper-based assays are comparable in sensitivity to commercial dipstick assays. Our assay format also allows for the measurement of multiple analytes.
This paper-based assay is suitable for measuring multiple samples in parallel and in a relatively short period of time. For example, in one trial, one researcher was able to run 20 different samples (all with 550 mM glucose and 75 μM BSA) within 7.5 min (followed by another 10.5 min for the color to fully develop). An 18-min assay of this type—one capable of measuring two analytes in 20 different sample—may be efficient enough to use in high-throughput screens of larger sample pools.
In the field, samples will not be measured under sterile conditions, and dust and dirt may contaminate the assays. The combination of paper and capillary action provides a mechanism for separating particulates from a biological fluid. As a demonstration, we purposely contaminated the artificial urine samples with quantities of dirt, plant pollen, and graphite powder at levels higher than we might expect to see in the samples in the field. These particulates do not move up the channels under the action of capillary wicking, and do not interfere with the assay (Figure 3).
Paper strips have been used in biomedical assays for decades because they offer an inexpensive platform for colorimetric chemical testing.[1] Patterned paper has characteristics that lead to miniaturized assays that run by capillary action (e.g., without external pumping), with small volumes of fluids. These methods suggest a path for the development of simple, inexpensive, and portable diagnostic assays that may be useful in remote settings, and in particular, in less-industrialized countries where simple assays are becoming increasingly important for detecting disease and monitoring health,[16, 17], for environmental monitoring, in veterinary and agricultural practice and for other applications.
Supplementary Material
Figure S1. (a) Device run with an artificial urine solution containing 75 mM glucose and 50 μM BSA, (b) device run with an artificial urine solution containing 50 μM BSA, (c) device run with an artificial urine solution containing 75 mM glucose.
Scheme 1.
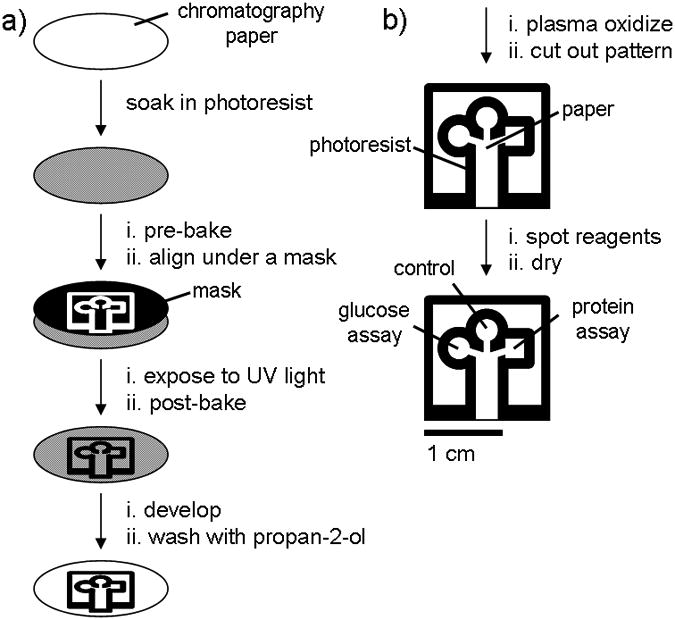
Schematic diagram depicting the method for patterning paper into millimeter-sized channels: (a) Photolithography was used to pattern SU-8 photoresist embedded into paper; (b) The patterned paper was modified for bioassays.
Footnotes
This research was supported by the National Institutes of Health (NIH) (GM065364). We used the Materials Research Science and Engineering Centers (MRSEC) shared facilities supported by the National Science Foundation (NSF) under award no. DMR-0213805. This work also was supported by a predoctoral fellowship from NSF (A.W.M), a Damon Runyon Cancer Research Foundation Fellowship (DRG-1805-04) (S.T.P), and an Alexander and Margaret Stewart Trust Award and Massachusetts Technology Transfer Council Grant (M.J.B.).
References
- 1.von Lode P. Clin Biochem. 2005;38:591–606. doi: 10.1016/j.clinbiochem.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 2.Mabey D, Peeling RW, Ustianowski A, Perkins MD. Nat Rev Microbiol. 2004;2:231–240. doi: 10.1038/nrmicro841. [DOI] [PubMed] [Google Scholar]
- 3.Daar AS, Thorsteinsdottir H, Martin DK, Smith AC, Nast S, Singer PA. Nat Genet. 2002;32:229–232. doi: 10.1038/ng1002-229. [DOI] [PubMed] [Google Scholar]
- 4.Willis RC. Anal Chem. 2006;78:5261–5265. doi: 10.1021/ac069435g. [DOI] [PubMed] [Google Scholar]
- 5.Sia SK, Linder V, Parviz BA, Siegel A, Whitesides GM. Angew Chem Int Ed Engl. 2004;43:498–502. doi: 10.1002/anie.200353016. [DOI] [PubMed] [Google Scholar]
- 6.Cypress Diagnostics, Urine-9 product insert. http://www.diagnostics.be/products/clinical/quicktest/Urine/urine-9.htm.
- 7.Craig Medical, URS-10 professional urinalysis test strip product insert
- 8.The proposed platform was designed for the qualitative detection of protein and glucose in the clinical ranges of these analytes in urine. The lowest concentrations that we detected were 0.38 μM for protein and 1 mM for glucose. Detection lower concentrations of these analytes will require more expensive equipment and reagents than we used, and we think would make the urinalysis assay less appealing for use in less industrialized countries.
- 9.Peele JD, Jr, Gadsden RH, Crews R. Clin Chem. 1977;23:2242–2246. [PubMed] [Google Scholar]
- 10.In the glucose assay, glucose is oxidized by glucose oxidase in the presence of water and oxygen to give gluconic acid and hydrogen peroxide. The hydrogen peroxide is then reduced to water by horseradish peroxidase with concomitant oxidation of iodide to iodine.
- 11.Pugia MJ, Lott JA, Profitt JA, Cast TK. J Clin Lab Anal. 1999;13:180–187. doi: 10.1002/(SICI)1098-2825(1999)13:4<180::AID-JCLA7>3.0.CO;2-R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuen VG, McNeill JH. J Pharmacol Toxicol Methods. 2000;44:543–546. doi: 10.1016/s1056-8719(01)00117-4. [DOI] [PubMed] [Google Scholar]
- 13.Mogensen CE. N Engl J Med. 1984;310:356–360. doi: 10.1056/NEJM198402093100605. [DOI] [PubMed] [Google Scholar]
- 14.It takes 10 minutes for the color to develop because it takes this long for the paper to dry, and for the reactions to produce an observable amount of product. The glucose assay is based on the enzymatic oxidation of iodide to iodine, and it takes several minutes for a detectable amount of iodine to accumulate. Iodine continues to accumulate until the paper dries, at which point the enzymatic reaction stops. The protein assay gives a color change as BSA contacts the reagents, but the intensity of this color increases until the paper is dry.
- 15.Sasaki M, Pugia MJ, Parker DR, Kuromoto K, Furukawa I, Konishi I. J Clin Lab Anal. 1999;13:246–250. doi: 10.1002/(SICI)1098-2825(1999)13:5<246::AID-JCLA10>3.0.CO;2-W. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mabey D, Peeling RW, Perkins MD. Sex Transm Infect. 2001;77:397–398. doi: 10.1136/sti.77.6.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yager P, Edwards T, Fu E, Helton K, Nelson K, Tam MR, Weigl BH. Nature. 2006;442:412–418. doi: 10.1038/nature05064. [DOI] [PubMed] [Google Scholar]
- 18.An artificial urine solution was prepared according to the recipe provided by Brooks and Keevil (T. Brooks, C. W. Keevil, Lett. Appl. Microbiol. 1997, 24, 203-206). The artificial urine solution was 1.1 mM lactic acid, 2.0 mM citric acid, 25 mM sodium bicarbonate, 170 mM urea, 2.5 mM calcium chloride, 90 mM sodium chloride, 2.0 mM magnesium sulfate, 10 mM sodium sulfate, 7.0 mM potassium dihydrogen phosphate, 7.0 mM di-potassium hydrogen phosphate, and 25 mM ammonium chloride all mixed in Millipore water. The pH of the solution was adjusted to 6.0 by addition of 1.0 M hydrochloric acid.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. (a) Device run with an artificial urine solution containing 75 mM glucose and 50 μM BSA, (b) device run with an artificial urine solution containing 50 μM BSA, (c) device run with an artificial urine solution containing 75 mM glucose.


