Abstract
Human Protein Reference Database (HPRD) is a rich resource of diverse features of human proteins, which are experimentally proven. Protein information in HPRD includes protein–protein interactions, post-translational modifications, enzyme/substrate relationships, disease associations, tissue expression, and subcellular localization of human proteins. Although, protein-protein interaction data from HPRD has been widely used by scientific community, its phosphoproteome data has not been exploited up to the potential. HPRD is one of the largest documentation of human phosphoproteins in the public domain. Currently, phosphorylation data in HPRD comprises of 95,016 phosphosites mapped on to 13,041 proteins. Additionally, enzyme-substrate reactions responsible for 5,930 phosphorylation events were also curated. Significant improvements in technologies and high-throughput platforms in biomedical investigations, led to exponential increase of biological data and phosphoproteomic data in the recent years. Human Proteinpedia, a community annotation portal developed by us, has also led to significant increase in phosphoproteomic data in HPRD. A large number of phosphorylation events have been mapped on to reference sequences available in HPRD and Human Proteinpedia along with associated protein features. This will provide a platform for systems biology approaches to determine role of protein phosphorylation in protein function, cell signaling, biological processes and their implications in human diseases. This review aims to provide a composite view of phosphoproteomic data pertaining to human proteins in HPRD and Human Proteinpedia.
Keywords: Protein database, biocuration, kinases, phosphatases
Introduction
Post-translational modifications (PTMs) are crucial for protein stability, sorting and function. Commonly found PTMs include phosphorylation, acetylation, glycosylation, methylation, proteolytic cleavage, GPI anchor, palmitoylation, sumoylation and ubiquitination. Phosphorylation of proteins is mediated by a class of enzymes called protein kinases, which enable transfer of phosphate groups from ATP to the hydroxyl group of either serine, threonine, or tyrosine residues of their protein substrates 1. Protein phosphorylation is a reversible and dynamic process which plays a critical role in growth, function and development of living cells 2. Protein phosphorylation and dephosphorylation events are known to regulate several receptor mediated signal transduction pathways and assume switch-on and –off role in enzymatic activity, protein stability, protein-protein interactions (PPIs) and subcellular localization 3-5. Protein kinases and phosphatases recognize their substrates by means of a set of conserved residues among substrates sequences called phosphomotifs 6. There are more than 518 protein kinases in humans including approximately 400 serine/threonine kinases and ∼90 protein tyrosine kinases 7-9. Similarly, there are an estimated number of 30-40 serine/threonine phosphatases and 109 tyrosine phosphatases 9-11. Dual specificity kinases or phosphatases are capable of acting on both serine/threonine and tyrosine residues within the same substrate 12, 13. As phosphorylation and dephosphorylation events are known to regulate activity of several enzymes and mediate signaling pathways, a manually curated phosphorylation data resource would allow ‘systems level’ discoveries by facilitating analyses of protein networks and signaling pathways in different biological contexts including human diseases 14. This is especially relevant because, dysregulation of protein phosphorylation events and signaling pathways has already been implicated in several diseases such as cancer, neurodegenerative disorders including Alzheimer's disease, dementias, disorders of learning and memory 15-21.
Traditional methods used for identification of phosphorylated proteins involved radioactive labeling of proteins with 32P-labeled ATP followed by SDS-PAGE or high-performance liquid chromatography followed by Edman sequencing to determine the site of phosphorylation 22. However, this technique is cumbersome and requires large amounts of pure protein samples. Western blotting using phosphosite-specific antibodies is a method that is gaining popularity but remains limited because of the restricted availability of such antibodies 23. High-resolution mass spectrometry (MS) has evolved as the method of choice for analysis of phosphoproteome in complex protein samples owing to its sensitivity and high-throughput nature 24. Such experiments often involve protein isolation followed by phosphoproteome enrichment 25-27. Subsequently, the enriched fractions are resolved by liquid chromatography and analyzed by MS 28-30.
Several high-throughput studies have substantially increased the data pertaining to the human phosphoproteome in this decade. Olsen et al. reported 6,600 phosphorylation sites from 10,000 phosphopeptides on 2,244 proteins in HeLa cells stimulated with epidermal growth factor (EGF). Quantitative proteomics approach followed in this study combined SILAC for quantitation and titanium dioxide (TiO2) chromatography for phosphopeptide enrichment 31. Nagaraj et al. identified 5,359 and 6,845 phosphosites using TiO2 affinity-based enrichment of HeLa S3 cell lysates using two different modes of fragmentation - collision induced dissociation and high-energy collision-induced dissociation, respectively 32. In another study, 2,225 non-redundant phosphosites from 1,023 proteins were obtained from human liver by enrichment of phosphopeptides using immobilized metal ion affinity chromatography microspheres – IMAC33. Oyama and coworkers performed tyrosine phosphoproteome analysis of EGF-stimulated A431 cells and identified 3,730 phosphopeptides 34. Li and coworkers used pTyr-100 and 4G10 phosphotyrosine antibodies to enrich phosphotyrosine proteome in human Hep3B and MHCC97H cell lines 35. As the data obtained in these large-scale studies are voluminous, the phosphopeptides information usually ends up in supplementary files of the research articles. It is difficult for an individual investigator to benefit from these data, because of difficulty in accessing them, non-uniformity in data presentation and file formats. For a systems level analysis, it will be helpful if phosphoproteome data derived from diverse biological systems and experimental platforms are curated uniformly in a centralized resource. The essential information for phosphoproteome may include the residues and sites of phosphorylation, name and reference sequence of the protein that these phosphosites were mapped onto, experimental methods for enrichment and analysis with links to the research articles. HPRD and Human Proteinpedia, as community resources, serve this function. In HPRD, a large number of phosphorylation and dephosphorylation reactions were included from targeted low-throughput studies in addition to the phosphoproteome data derived from high-throughput analyses. These resources also allow researchers to perform complex queries in the context of other protein features of proteins. Phosphorylation data information from these databases can be freely downloaded. The information in these resources can be used to undertake systems approaches to study phosphorylation especially in the light of newer initiatives to understand human proteins and biological systems.
Protein features annotated in Human Protein Reference Database
HPRD (http://www.hprd.org) is a comprehensive resource of several types of protein information. It allows for the integration of data from diverse experimental platforms pertaining to different protein features in the context of an individual protein. HPRD links each curated information to the original research article in which it is described. It documents various protein features such as PPIs, PTMs, enzyme-substrate relationships, disease associations, subcellular localization, tissue expression, biological motifs/domains derived from a variety of experimental platforms. HPRD contains curated information for a non-redundant set of 30,047 human proteins including 19,653 proteins and 10,394 additional isoforms as reported in the RefSeq database 36. Curated information in HPRD is currently comprised of 39,194 unique PPIs, 109,513 PTMs, 112,158 tissue expression annotations, and 22,490 subcellular localization entries which are linked to 59,194 published research articles. It also contains annotations pertaining to isoform-specific subcellular localization (1,117) and tissue expression (5,499). The graphic user interface of HPRD displays an image for each protein sequence, which depicts biological motifs and protein domains along with the PTMs in the context of each protein sequence. HPRD has implemented Gene Ontology in describing subcellular localization, molecular function and biological process. PPI data in HPRD has been used in the standardization of Proteomics Standard Initiative-Molecular Interaction (PSI-MI) vocabularies 37. Data annotated in HPRD is freely accessible for academic community in PSI-MI, XML and tab-delimited formats.
Annotation of phosphorylation data in HPRD
HPRD follows a manual curation strategy to annotate phosphorylation and dephosphorylation data, which are described in several low- and high-throughput research articles. Each phosphosite is mapped to the reference sequence provided in the HPRD, using phosphopeptide sequence data provided in the article. Only unique phosphosites which map to a single human protein are considered for inclusion. Data is entered using software called BioBuilder, developed previously by our group 38. Biobuilder has built-in consistency checks to prevent redundant entries and annotation of wrong residue and site information. The annotated information will be reflected in HPRD protein page for visualization.
Figure 1 shows the molecule page of insulin receptor in HPRD, which provides links to research articles describing phosphorylation sites and residues. For example, phosphorylated residues marked in peptides DGGSSpLGFK and APESpEELMEFEDMNVPLDR as Ser-1275 and Ser-1309 in the respective literature39, 40. However, when we map these peptides to corresponding RefSeq protein sequence (accession # NP_000199.2), these position were matched to Ser-1314 and Ser-1348. These sequence matched sites have been annotated in HPRD. In addition, the enzymes directly responsible for phosphorylation or dephosphorylation reactions are also documented. Three proteins, protein kinase C alpha (PRKCA), epsilon (PRKCE) and delta (PRKCD) have been shown to phosphorylate the opioid receptor (OPRD1) at Ser-344 41. Elongation factor 2 kinase (EEF2K) and Phosphatase 2A, catalytic subunit, alpha (PPP2CA), respectively, have been reported to mediate phosphorylation and dephosphorylation reactions on Thr-57 and Thr-59 of Elongation factor 2 (EEF2)42.
Figure 1. Annotation and display of phosphorylation data in HPRD.
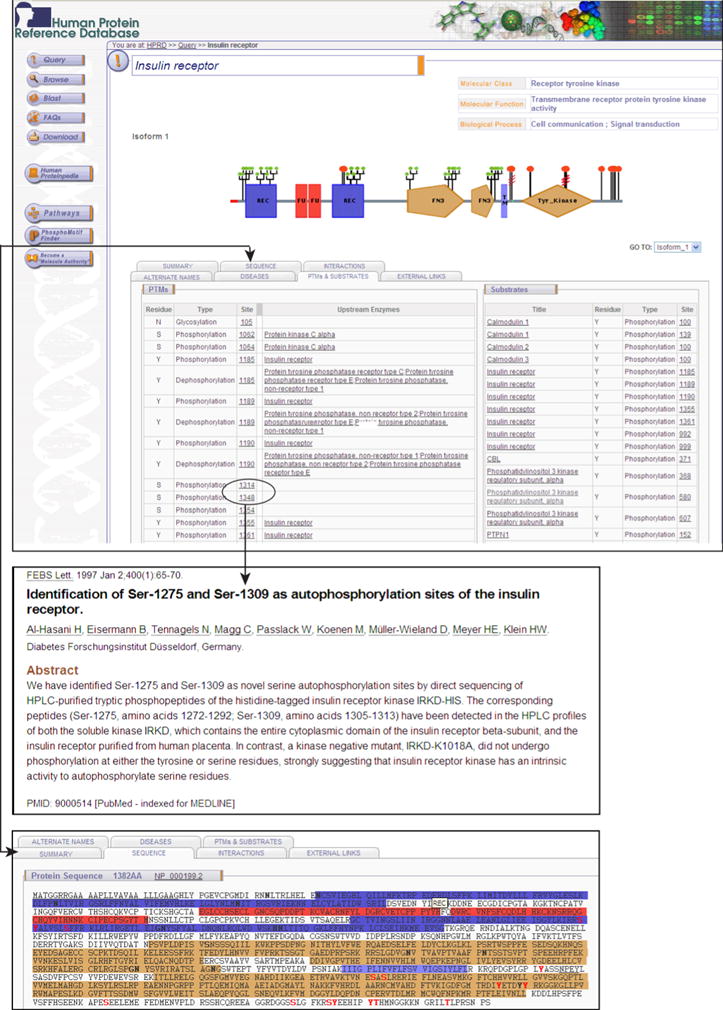
The molecule page for insulin receptor in Human Protein Reference Database is shown. Phosphorylation data is annotated through BioBuilder tool which is used to store and manage complex protein data. Each phosphorylation site is hyperlinked to the corresponding research articles that describes it. Wherever known, the upstream enzyme that phosphorylates a given site is also annotated.
Analysis of the phosphoproteome data in HPRD
Improvements in experimental platforms in biology have resulted in a great increase in biological information including phosphorylation. Table 1 provides a list of large-scale studies that cataloged a large number of phosphopeptides in humans along with a summary of information regarding the cell line/tissue used for the experiments, type of mass spectrometer employed, methods used for enrichment and the number of identified phosphosites.
Table 1.
Phosphorylation data from previous high throughput studies
| Citation | Number of phosphorylation sites identified | Cell line/tissue used | Method used | Mass spectrometer used | |
|---|---|---|---|---|---|
| 1 | Nagaraj N et al. 75, 2010 | 16,000 | HeLa cells | TiO2 affinity enrichment | LTQ Orbitrap |
| 2 | Han G et al. 33, 2010 | 2,225 | Liver | Ti4+-IMAC microspheres | Finnigan LTQ linear ion trap |
| 3 | Nie S et al 76, 2010 | 1,356 | HeLa cells | TiO2 affinity enrichment | LTQ Orbitrap |
| 4 | Malik R et al. 77, 2009 | 1,940 | HeLa S3 cells | TiO2 affinity enrichment | nanoLC-Orbitrap |
| 5 | Olsen JV et al. 78, 2010 | 20,443 | HeLa S3 cells | TiO2 affinity enrichment or IMAC | LTQ Orbitrap |
| 6 | Pan C et al. 79, 2009 | 4,009 | HeLa cells | TiO2 affinity enrichment | LTQ Orbitrap |
| 7 | Daub H et al. 80, 2008 | 1,007 | HeLa S3 cells | TiO2 affinity enrichment | LTQ Orbitrap |
| 8 | Wang B et al. 81, 2008 | 1,068 | Human mitotic spindles | TiO2 affinity enrichment | Hybrid linear ion trap/Orbitrap |
| 9 | Olsen JV et al. 31, 2006 | 6,600 | HeLa cells | TiO2 affinity enrichment | LTQ Orbitrap |
| 10 | Dephoure N et al. 82, 2008 | 14,000 | HeLa cells | TiO2 affinity enrichment or IMAC | LTQ Orbitrap |
| 11 | Beausoleil SA et al. 83, 2004 | 2,002 | HeLa cells | Differential Net Solution Charge State | LCQ-DECA XP |
| 12 | Beausoleil SA et al. 84, 2006 | 1,079 | HeLa cells | Differential Net Solution Charge State | LTQ-FT hybrid linear Ion trap–7 Tesla FTICR |
| 13 | Kyono Y et al. 85, 2008 | 1,803 | HeLa cells | Immobilized metal affinity chromatography | QSTAR XL |
| 14 | Sugiyama N et al. 86, 2007 | 1,646 | HeLa cells | Aliphatic hydroxy acid-modified metal oxide chromatography | QSTAR XL, LTQ Orbitrap |
| 15 | Molina H et al 28, 2007 | 1,435 | 293T cells | TiO2 affinity enrichment | 3-D ion trap with ETD |
| 16 | Brill LM et al. 87, 2009 | 2,546 | Human embryonic stem cells | Immobilized metal affinity chromatography | LTQ linear ion trap |
| 17 | Ruse CI et al. 88, 2008 | 1,037 | HEK-293 cells, HeLa cells | Ba2+/pH/acetone precipitation | LTQ Orbitrap |
| 18 | Gauci S et al. 89, 2009 | 5,036 | HEK-293 | Low-pH SCX | LTQ Orbitrap |
| 19 | Van Hoof D et al. 90, 2009 | 3,067 | Human embryonic stem cells | TiO2 affinity enrichment | LTQ Orbitrap |
| 20 | Han G et al. 91, 2010 20166139 |
2,225 | Liver | Ti4+-IMAC microspheres | LTQ linear ion trap |
| 21 | Raijmakers R et al. 92, 2010 | 1,020 | Human leukocyte | TiO2 affinity enrichment | Q-TOF |
| 22 | McNulty DE et al. 93, 2008 | 1,000 | HeLa cells | IMAC enrichment | LC/MSD Trap XCT Ultra |
| 23 | Yu LR et al. 94, 2007 | 1,034 | HeLa cells | TiO2 affinity enrichment | LCQ-Deca XP |
| 24 | Sui S et al. 95, 2008 | 1,035 | Chang liver cell line | TiO2 affinity enrichment | Quadrupole-TOF |
| 25 | Tsai CF et al. 96, 2008 | 2,360 | Human non-small lung carcinoma cell line (H1299) | pH/acid controlled IMAC enrichment | LC-Q-TOF, QSTAR Pulsar i |
| 26 | Li QR et al. 97, 2009 | 3,159 | HeLa cells | TiO2 affinity enrichment | LTQ Orbitrap |
| 27 | Schreiber TB et al. 98, 2010 | 5,300 | A498 cell line | TiO2 affinity enrichment | LTQ Orbitrap |
| 28 | Yan GR et al. 99 2009 | 1,117 | Gastric cancer cells (SGC-7901) | TiO2 affinity enrichment | LTQ Orbitrap |
| 29 | Xiao K et. al. 100 2010 | 1,555 | HEK293 cells | IMAC enrichment | LTQ Orbitrap XL |
| 30 | Santamaria A et.al. 101 2010 | 3,894 | HeLa S3 | TiO2 affinity enrichment | LTQ Orbitrap |
Phosphorylation data were annotated based on the sites of modification and the modified residues. Figure 2A depicts distribution of phosphorylation data in HPRD derived from in vivo and in vitro experiments. HPRD has 95,016 annotated phosphorylation sites, of which 88,250 phosphosites are identified based on in vivo analysis alone, 2,678 from in vitro experiments and 4,088 phosphorylation sites by both methods. Among the annotated phosphorylated sites, 66,050 (69%) are on serine, 20,652 (22%) on threonine, 8,314 (9%) on tyrosine.
Figure 2. Overview of phosphoserine, phosphothreonine and phosphotyrosine data in HPRD.
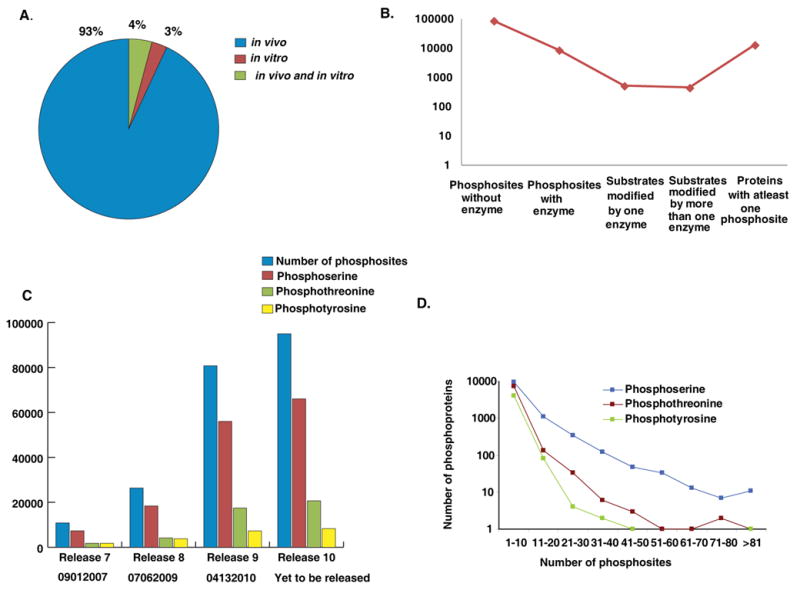
A) Distribution of phosphorylation data in HPRD derived from in vivo and in vitro experiments B) Fraction of phosphosites in HPRD with an annotated upstream enzyme; C) Exponential increase of HPRD phosphorylation data over the last four releases; D) Distribution of protein groups based on number of phosphoserine, phosphothreonine and phosphotyrosine sites they harbor.
While annotating phosphorylation data in HPRD, we have also documented enzymes responsible for 5,930 phosphorylation events. We realized that enzymes responsible for only a minor fraction of all phosphorylation events have been determined so far (Figure 2B). The annotation of enzymes for these PTMs has also resulted in the documentation of 2,118 unique enzyme-substrate relationships. We found 447 substrates, which are known to be phosphorylated by more than one enzyme and 517 substrates modified by only one enzyme. Overall, in HPRD, 13,014 proteins are annotated with at least one phosphorylation site. Figure 2C shows an exponential increase of HPRD phosphorylation data over the last four releases, which is mainly due to an increase in high-throughput phosphoproteomic analysis of proteins by mass spectrometry. Since HPRD release 7 in the year 2007, we have added approximately 84,165 phosphosites which contains 58,738 phosphoserine, 18,888 phosphothreonine and 6,539 phosphotyrosine on human proteins. The distribution of phosphorylation events on phosphoproteins with respect to serine, threonine and tyrosine is illustrated in Figure 2 (D). Of these, only 1,368 phosphorylation reactions have been defined in the context of human signaling pathways, as curated in NetPath (http://www.netpath.org) indicating that the large majority of phosphosites have still not been explored for their role in cell signaling. Thus, phosphorylation data contained in HPRD should provide a platform for biomedical investigators to further evaluate the role of these phosphosites and proteins in cell signaling and in diseases.
Proteomic data
Curation of heterogeneous proteomic data from multiple sources into a uniform format is quite a challenging task. Large-scale phosphorylation data is usually provided as supplementary files with no uniformity in data presentation. If most of investigators deposited their data into a centralized resource in a predefined format, it could be used more easily by the broader biology community. Therefore, we developed distributed annotation system protocols to annotate human protein data so as to allow proteomic investigators a platform to directly submit their data in a single resource; we named it Human Proteinpedia (http://www.humanproteinpedia.org/). Users can submit their data by uploading batch files, FTP or email. It utilizes protein information in HPRD as a scaffold and maps all proteomic data entered by the users in Human Proteinpedia to HPRD. The data submitted to Human Proteinpedia can be viewed through HPRD in the context of an individual protein molecule. It also allows laboratories to share human proteomic data obtained from multiple experimental platforms and increases the speed of data dissemination. Users can also share both unpublished and published human proteomic data. Data thus submitted will remain linked to the investigators and the laboratories. This resource has entries pertaining to 34,500 PPIs, 24,152 PTMs, 2,900 subcellular localization and 192,557 cell lines/tissues expression. Figure 3 displays the HPRD molecule page of microtubule associated protein 4 (MAP4) which promotes microtubule assembly.
Figure 3. Integration of phosphoprotein data from Human Proteinpedia into HPRD.
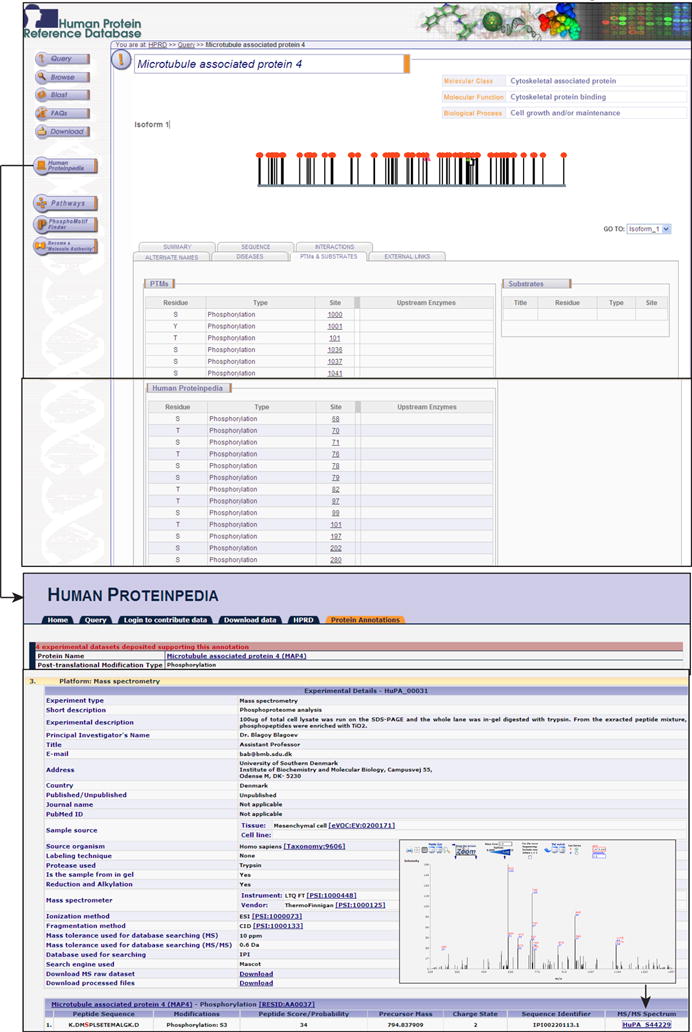
Microtubule associated protein 4 molecule page is shown in HPRD with links to Human Proteinpedia. PTMs page displays phosphorylation sites from HPRD and Human Proteinpedia. The upper right panel shows description of the peptide score, experimental description, peptide identification data, precursor mass, charge state, sequence identifier, algorithm, MS/MS spectrum, ionization method, fragmentation method and mass tolerance used for database searching. The hyperlink to MS/MS spectrum is given in lower right panel that helps users to inspect the spectrum manually.
Users of Human Proteinpedia have submitted 60 novel phosphorylation sites of MAP4 in HPRD and the data on PTMs can be viewed in a separate table in PTMs page in HPRD. It provides more insights into phosphorylation data from mass spectrometry by giving option of adding peptide score, experimental description, peptide identification data, precursor mass, charge state, sequence identifier, algorithm, MS/MS spectrum, ionization methods, fragmentation methods and mass tolerance used for database searching. All peptides are linked to their respective MS/MS spectrum which can be visualized using Spectrum Viewer43. Phosphopeptides derived from multiple experimental platforms and enrichment strategies will help researchers to determine a combination of strategies to be used to maximize the discovery in phosphoproteomics. There are over 1,960,352 phosphopeptides deposited in Human Proteinpedia. It becomes important source of phosphoproteome information and can drive meta-analysis of human phosphoproteome. Several phosphopeptides in Human Proteinpedia have multiple line of evidence as they have been submitted by more than one group. In addition to enriching phosphopeptide data in the public domain, multiple sources of evidence gathered from various labs on different mass spectrometry platforms validates these phosphopeptides.
Human protein phosphorylation resources
Biological databases pertaining to various post-translational modifications are important for investigating biological and pathway information in the cells. Several public databases collect published phosphorylation data disseminated in the scientific literature and provide researchers access to their curated datasets. These usually reference the original publication and the experimental method that determined every individual phosphorylation. HPRD is richer in human phosphosites as compared to other publicly available repositories for human proteins. For instance, Phospho.ELM version 9.0 (http://phospho.elm.eu.org/) contains data detailing 8,718 substrate proteins from different species covering phosphorylation of 3,370 tyrosine, 31,754 serine and 7,449 threonine instances44 . Phospho3D version 2 (http://www.phospho3d.org/) provides three-dimensional structures of phosphorylation sites 45, 46 PHOSIDA (http://www.phosida.com) is an archive of 24,262 phosphosites on 8,283 proteins from Homo sapiens47. LymPHOS (http://www.lymphos.org) describes 342 phosphorylation sites in human T-Lymphocytes 48 and RESID (http://www.ebi.ac.uk/RESID/) has collection of structures for modified proteins 50. PhosphoSitePlus (http://www.phosphosite.org) has reported 97,589 non-redundant sites from all organisms with phospho-antibody information. dbPTM 2.0 (http://dbptm.mbc.nctu.edu.tw/) has 36,466 PTM sites from Swiss-Prot, Phospho.ELM, O-GLYCBASE and UbiProt, which were categorized by the PTM types, among which 22,363 are phosphorylation sites 50. SysPTM (http://www.biosino.org.cn/SysPTM/) contains 117,349 experimentally determined PTMs sites on 33,421 proteins involving nearly 50 PTMs types from many species, curated from public resources 51. One of the potential uses (use) of human protein phosphorylation resources will be in analyzing top/middle-down data from mass spectrometry. ProSight PTM 2.0 is a web-based tool that allows characterization of intact proteins using top-down tandem mass spectrometry approach. This tool utilizes proteomic databases annotated with known or predicted PTMs information including phosphorylation to identify and characterize intact proteins. The algorithm makes use of post-translational modifications annotation to calculate intact protein mass by considering each PTMs in isolation as well as in combination with others to characterize different protein forms using top-down approach. Each of these databases has their own unique features with large variations in the type and depth of annotation.
PhosphoMotif Finder
Several software or algorithms are available, which can predict phosphorylation motifs in the protein sequences. These include NetPhos (http://www.cbs.dtu.dk/ws/ws.php?entry=NetPhos) 52, GPS: group-based phosphorylation site 53, Scansite (http://scansite.mit.edu) 54, PredPhospho (http://www.ngri.re.kr/proteo/PredPhospho.htm) 55, KinasePhos (http://kinasephos.mbc.nctu.edu.tw/) 56, NetworKIN (http://www.networkin.info/search.php) 57, 58, DISPHOS 1.3 (http://www.ist.temple.edu/disphos/ ), ELM (http://www.elm.eu.org/) 59, PhoScan (http://bioinfo.au.tsinghua.edu.cn/phoscan/) 60, pkaPS (http://mendel.imp.ac.at/sat/pkaPS/) 61, PPSP (Prediction of PK-specific Phosphorylation site, http://ppsp.biocuckoo.org/) 62, NetPhorest (http://netphorest.info/) 63 and Motif-X (http://motif-x.med.harvard.edu/) algorithms also identify novel and known phosphorylation motifs 64. However, these software do not simply map literature based experimentally proven phosphomotifs onto a specific protein query. We developed a software called PhosphoMotif Finder in order to address this vacuum and integrated it in HPRD. It currently contains 324 phosphorylation based motifs described in the literature65. We categorized phosphomotifs into phosphorylation-based substrate motifs and phosphorylation based binding motifs. The former category of motifs will be recognized kinases and phosphatases and the latter will provide scaffold for other proteins to bind to phosphorylated motifs. PhosphoMotif Finder has 170 serine/threonine kinase substrate motifs, 5 serine/threonine phosphatase substrate motifs, 17 serine/threonine binding motifs, 50 tyrosine kinase substrate motifs, 19 tyrosine phosphatase substrate motifs and 63 binding motifs.
Figure 4 shows 52 known tyrosine phosphorylation motifs in vascular endothelial growth factor receptor 3 (VEGF receptor 3). It displays position of the motif in the query sequence and its features mentioned in the existing literature. This compendium is useful for the biomedical research community to identify potential phosphosites in a hypothetical protein, novel phosphorylation motifs, enzyme association and potential protein interactors of a phosphorylated protein. PhosphoMotif Finder can be useful tool for designing experiments to characterize phosphorylation reactions of a set of proteins. Phosphomotif Finder will be updated regularly as new phosphomotifs are described.
Figure 4. PhosphoMotif Finder in HPRD.
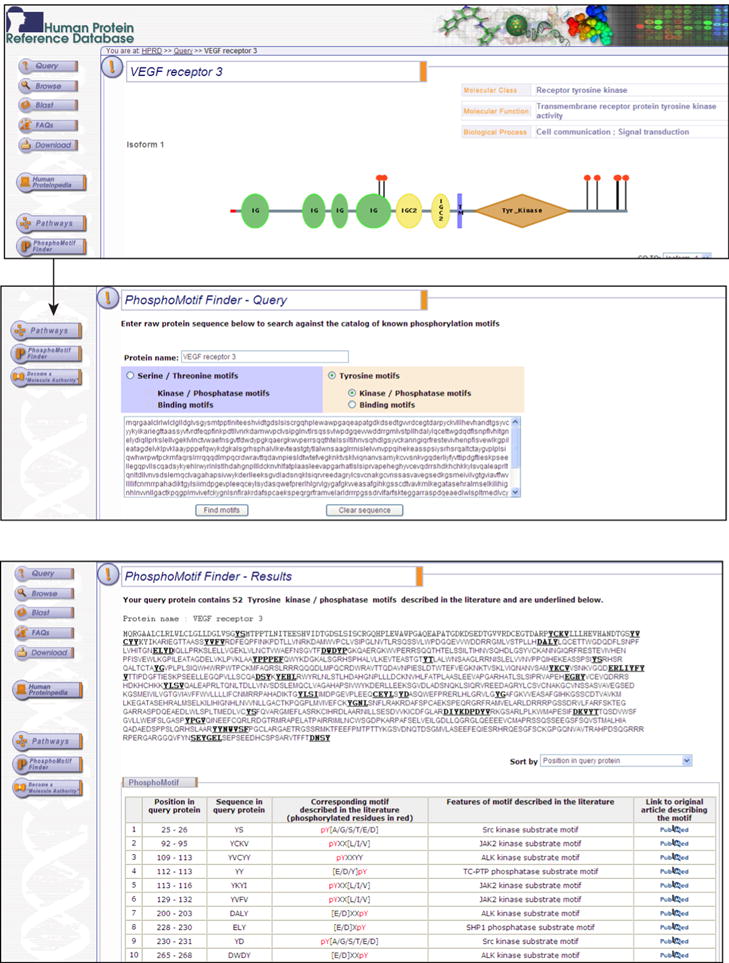
The screenshots show PhosphoMotif Finder software that can be accessed through HPRD. PhosphoMotif Finder output for VEGF receptor 3 sequence query is shown.
Utilization of Phosphoproteome Data from HPRD
The phosphorylation data from HPRD has been used by scientific community to develop new prediction strategies, to compare experimental datasets, to enrich other databases and to perform targeted or specific bioinformatics analyses. SysPTM (http://www.sysbio.ac.cn/SysPTM) has used PTMs data from HPRD along with other databases for PTMBlast, PTMPhylog, PTMCluster and PTMPathway 51. Yang et al. introduced PhosphoPOINT, which integrates data from HPRD as well as from other resources 67. RegPhos (http://regphos.mbc.nctu.edu.tw/) has used HPRD phosphorylation sites to predict phosphorylation network with respect to subcellular localization 67. HPRD data has been used to create a resource of protein tyrosine phosphatases, which includes tyrosine-specific and dual-specificity phosphatases 68. The Pathway Palette (http://blaispathways.dfci.harvard.edu/Palette.html) generates a PPIs network of mass spectrometry derived data based on information curated in HPRD and other databases 69. Short linear motifs (SLiMs) (http://bioware.ucd.ie/∼slimdisc/slimfinder/conmasking/), a software designed to find functional microdomains in proteins which are important in many biological processes, used data from HPRD 70. Yachie et al. obtained data of serine kinase and phosphatase motifs data from HPRD for the evaluation of relationship of amino acid sequences of phosphoproteins and positions of phosphosites 71. Bioinformatics analyses of phosphoproteome data from HPRD have led to many useful biological interpretations in protein signaling. Cao et al. found 80 tyrosine, 83 serine and 19 threonine phosphorylation sites in stimulated Jurkat cells to understand the signaling pathways involved in T cell. They used HPRD data to identify novel phosphorylation sites 72. Amanchy et al. identified 23 novel c-Src kinase substrates involved in PDGF signaling using SILAC 73. Tang et al. used HPRD data while investigating phosphoproteins in Wnt signaling 74. However, HPRD helps researchers to use data to maximize the discovery in proteomics.
Conclusions
HPRD and Human Proteinpedia serve as repositories of diverse features of human proteins. The major fraction of published human phosphoproteome has already been incorporated into these resources because of our ongoing curation efforts. Human Proteinpedia provides a list of phosphopeptides identified by mass spectrometry. PhosphoMotif Finder allows mapping of phosphomotifs in a given protein. These datasets together can help in designing phospho-specific antibodies and peptide arrays. Datasets obtained from different experimental methods and platforms can be used to analyze the efficiency of these technologies and to decide on a methodology that suits most for any investigation in phosphoproteomics. A minority of phosphorylation reactions and phosphosites described in HPRD have been studied with their relevance to cell signaling and protein activity. Targeted studies can be designed to explore the biological role of these phosphorylation events. Phosphoproteomic data available in these resources will provide baseline data for further biomedical investigations.
Acknowledgments
We thank the Department of Biotechnology (DBT), Government of India for research support to the Institute of Bioinformatics. Harsha Gowda is a Wellcome Trust/DBT India Alliance Early Career Fellow. A.P. was supported by NIH Roadmap grant “Technology Center for Networks and Pathways” (U54 RR 020839) and W81XWH-06-1-0428 from the Department of Defense for Proteomic and Functional Analysis of Fibroblasts in Breast Cancer. Dr. T. S. Keshava Prasad is supported by research grant on “Development of Infrastructure and a Computational Framework for Analysis of Proteomic Data” from DBT, Government of India, New Delhi, India.
Abbreviations
- SCX
strong cation exchange chromatography
- LC-MS/MS
liquid chromatography based tandem mass spectrometry
- PPIs
Protein-protein interactions
- PTMs
Post-translational modifications
References
- 1.Campbell D, Morrice N. J Biomol Tech. 2002;13:119–130. [PMC free article] [PubMed] [Google Scholar]
- 2.Hunter T. Cell. 2000;100:113–127. doi: 10.1016/s0092-8674(00)81688-8. [DOI] [PubMed] [Google Scholar]
- 3.Manning BD, Cantley LC. Sci STKE. 2002;2002:pe49. doi: 10.1126/stke.2002.162.pe49. [DOI] [PubMed] [Google Scholar]
- 4.Pandey A, Mann M. Nature. 2000;405:837–846. doi: 10.1038/35015709. [DOI] [PubMed] [Google Scholar]
- 5.Pawson T. Cell. 2004;116:191–203. doi: 10.1016/s0092-8674(03)01077-8. [DOI] [PubMed] [Google Scholar]
- 6.Braconi Quintaje S, Orchard S. Mol Cell Proteomics. 2008;7:1409–1419. doi: 10.1074/mcp.R700001-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang H, Zha X, Tan Y, Hornbeck PV, Mastrangelo AJ, Alessi DR, Polakiewicz RD, Comb MJ. J Biol Chem. 2002;277:39379–39387. doi: 10.1074/jbc.M206399200. [DOI] [PubMed] [Google Scholar]
- 8.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 9.Hunter T. Curr Opin Cell Biol. 2009;21:140–146. doi: 10.1016/j.ceb.2009.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi Y. Sci China C Life Sci. 2009;52:135–146. doi: 10.1007/s11427-009-0018-3. [DOI] [PubMed] [Google Scholar]
- 11.Alonso A, Sasin J, Bottini N, Friedberg I, Friedberg I, Osterman A, Godzik A, Hunter T, Dixon J, Mustelin T. Cell. 2004;117:699–711. doi: 10.1016/j.cell.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 12.Adayev T, Chen-Hwang MC, Murakami N, Lee E, Bolton DC, Hwang YW. Biochemistry. 2007;46:7614–7624. doi: 10.1021/bi700251n. [DOI] [PubMed] [Google Scholar]
- 13.Kentrup H, Becker W, Heukelbach J, Wilmes A, Schurmann A, Huppertz C, Kainulainen H, Joost HG. J Biol Chem. 1996;271:3488–3495. doi: 10.1074/jbc.271.7.3488. [DOI] [PubMed] [Google Scholar]
- 14.Williamson BL, Marchese J, Morrice NA. Mol Cell Proteomics. 2006;5:337–346. doi: 10.1074/mcp.M500210-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.Idriss HT. Cancer Chemother Pharmacol. 2004;54:101–104. doi: 10.1007/s00280-004-0778-1. [DOI] [PubMed] [Google Scholar]
- 16.Mi K, Johnson GV. Curr Alzheimer Res. 2006;3:449–463. doi: 10.2174/156720506779025279. [DOI] [PubMed] [Google Scholar]
- 17.Harsha HC, Jimeno A, Molina H, Mihalas AB, Goggins MG, Hruban RH, Schulick RD, Kamath U, Maitra A, Hidalgo M, Pandey A. J Proteome Res. 2008;7:4651–4658. doi: 10.1021/pr800139r. [DOI] [PubMed] [Google Scholar]
- 18.Byers LA, Heymach JV. Clin Lung Cancer. 2007;(8 Suppl 2):S79–85. doi: 10.3816/clc.2007.s.006. [DOI] [PubMed] [Google Scholar]
- 19.Ebi H, Tomida S, Takeuchi T, Arima C, Sato T, Mitsudomi T, Yatabe Y, Osada H, Takahashi T. Cancer Res. 2009;69:4027–4035. doi: 10.1158/0008-5472.CAN-08-3403. [DOI] [PubMed] [Google Scholar]
- 20.Mansuy IM, Shenolikar S. Trends Neurosci. 2006;29:679–686. doi: 10.1016/j.tins.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 21.Ryder J, Su Y, Ni B. Cell Signal. 2004;16:187–200. doi: 10.1016/j.cellsig.2003.07.004. [DOI] [PubMed] [Google Scholar]
- 22.McLachlin DT, Chait BT. Curr Opin Chem Biol. 2001;5:591–602. doi: 10.1016/s1367-5931(00)00250-7. [DOI] [PubMed] [Google Scholar]
- 23.Macek B, Mann M, Olsen JV. Annu Rev Pharmacol Toxicol. 2009;49:199–221. doi: 10.1146/annurev.pharmtox.011008.145606. [DOI] [PubMed] [Google Scholar]
- 24.Aebersold R, Mann M. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 25.Maguire PB, Wynne KJ, Harney DF, O'Donoghue NM, Stephens G, Fitzgerald DJ. Proteomics. 2002;2:642–648. doi: 10.1002/1615-9861(200206)2:6<642::AID-PROT642>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 26.Rush J, Moritz A, Lee KA, Guo A, Goss VL, Spek EJ, Zhang H, Zha XM, Polakiewicz RD, Comb MJ. Nat Biotechnol. 2005;23:94–101. doi: 10.1038/nbt1046. [DOI] [PubMed] [Google Scholar]
- 27.Zheng H, Hu P, Quinn DF, Wang YK. Mol Cell Proteomics. 2005;4:721–730. doi: 10.1074/mcp.M400077-MCP200. [DOI] [PubMed] [Google Scholar]
- 28.Molina H, Horn DM, Tang N, Mathivanan S, Pandey A. Proc Natl Acad Sci U S A. 2007;104:2199–2204. doi: 10.1073/pnas.0611217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bose R, Molina H, Patterson AS, Bitok JK, Periaswamy B, Bader JS, Pandey A, Cole PA. Proc Natl Acad Sci U S A. 2006;103:9773–9778. doi: 10.1073/pnas.0603948103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harsha HC, Molina H, Pandey A. Nat Protoc. 2008;3:505–516. doi: 10.1038/nprot.2008.2. [DOI] [PubMed] [Google Scholar]
- 31.Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 32.Nagaraj N, D'Souza RC, Cox J, Olsen JV, Mann M. J Proteome Res. 9:6786–6794. doi: 10.1021/pr100637q. [DOI] [PubMed] [Google Scholar]
- 33.Han G, Ye M, Liu H, Song C, Sun D, Wu Y, Jiang X, Chen R, Wang C, Wang L, Zou H. Electrophoresis. 31:1080–1089. doi: 10.1002/elps.200900493. [DOI] [PubMed] [Google Scholar]
- 34.Oyama M, Kozuka-Hata H, Tasaki S, Semba K, Hattori S, Sugano S, Inoue J, Yamamoto T. Mol Cell Proteomics. 2009;8:226–231. doi: 10.1074/mcp.M800186-MCP200. [DOI] [PubMed] [Google Scholar]
- 35.Li H, Ren Z, Kang X, Zhang L, Li X, Wang Y, Xue T, Shen Y, Liu Y. BMC Cancer. 2009;9:366. doi: 10.1186/1471-2407-9-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Edgar R, Federhen S, Feolo M, Geer LY, Helmberg W, Kapustin Y, Khovayko O, Landsman D, Lipman DJ, Madden TL, Maglott DR, Miller V, Ostell J, Pruitt KD, Schuler GD, Shumway M, Sequeira E, Sherry ST, Sirotkin K, Souvorov A, Starchenko G, Tatusov RL, Tatusova TA, Wagner L, Yaschenko E. Nucleic Acids Res. 2008;36:D13–21. doi: 10.1093/nar/gkm1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Orchard S, Hermjakob H. Brief Bioinform. 2008;9:166–173. doi: 10.1093/bib/bbm061. [DOI] [PubMed] [Google Scholar]
- 38.Navarro JD, Talreja N, Peri S, Vrushabendra BM, Rashmi BP, Padma N, Surendranath V, Jonnalagadda CK, Kousthub PS, Deshpande N, Shanker K, Pandey A. BMC Bioinformatics. 2004;5:43. doi: 10.1186/1471-2105-5-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Al-Hasani H, Eisermann B, Tennagels N, Magg C, Passlack W, Koenen M, Muller-Wieland D, Meyer HE, Klein HW. FEBS Lett. 400:1997. 65–70. doi: 10.1016/s0014-5793(96)01342-7. [DOI] [PubMed] [Google Scholar]
- 40.Tennagels N, Telting D, Parvaresch S, Maassen JA, Klein HW. Biochem Biophys Res Commun. 2001;282:387–393. doi: 10.1006/bbrc.2001.4589. [DOI] [PubMed] [Google Scholar]
- 41.Xiang B, Yu GH, Guo J, Chen L, Hu W, Pei G, Ma L. J Biol Chem. 2001;276:4709–4716. doi: 10.1074/jbc.M006187200. [DOI] [PubMed] [Google Scholar]
- 42.Redpath NT, Price NT, Severinov KV, Proud CG. Eur J Biochem. 1993;213:689–699. doi: 10.1111/j.1432-1033.1993.tb17809.x. [DOI] [PubMed] [Google Scholar]
- 43.Vizcaino JA, Cote R, Reisinger F, Barsnes H, Foster JM, Rameseder J, Hermjakob H, Martens L. Nucleic Acids Res. 2010;38:D736–742. doi: 10.1093/nar/gkp964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dinkel H, Chica C, Via A, Gould CM, Jensen LJ, Gibson TJ, Diella F. Nucleic Acids Res. 2011;39:D261–267. doi: 10.1093/nar/gkq1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zanzoni A, Ausiello G, Via A, Gherardini PF, Helmer-Citterich M. Nucleic Acids Res. 2007;35:D229–231. doi: 10.1093/nar/gkl922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zanzoni A, Carbajo D, Diella F, Gherardini PF, Tramontano A, Helmer-Citterich M, Via A. Nucleic Acids Res. doi: 10.1093/nar/gkq936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gnad F, Ren S, Cox J, Olsen JV, Macek B, Oroshi M, Mann M. Genome Biol. 2007;8:R250. doi: 10.1186/gb-2007-8-11-r250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ovelleiro D, Carrascal M, Casas V, Abian J. Proteomics. 2009;9:3741–3751. doi: 10.1002/pmic.200800701. [DOI] [PubMed] [Google Scholar]
- 49.Garavelli JS. Proteomics. 2004;4:1527–1533. doi: 10.1002/pmic.200300777. [DOI] [PubMed] [Google Scholar]
- 50.Lee TY, Hsu JB, Chang WC, Wang TY, Hsu PC, Huang HD. BMC Res Notes. 2009;2:111. doi: 10.1186/1756-0500-2-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li H, Xing X, Ding G, Li Q, Wang C, Xie L, Zeng R, Li Y. Mol Cell Proteomics. 2009;8:1839–1849. doi: 10.1074/mcp.M900030-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miller ML, Blom N. Methods Mol Biol. 2009;527:299–310. doi: 10.1007/978-1-60327-834-8_22. x. [DOI] [PubMed] [Google Scholar]
- 53.Zhou FF, Xue Y, Chen GL, Yao X. Biochem Biophys Res Commun. 2004;325:1443–1448. doi: 10.1016/j.bbrc.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 54.Obenauer JC, Cantley LC, Yaffe MB. Nucleic Acids Res. 2003;31:3635–3641. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim JH, Lee J, Oh B, Kimm K, Koh I. Bioinformatics. 2004;20:3179–3184. doi: 10.1093/bioinformatics/bth382. [DOI] [PubMed] [Google Scholar]
- 56.Wong YH, Lee TY, Liang HK, Huang CM, Wang TY, Yang YH, Chu CH, Huang HD, Ko MT, Hwang JK. Nucleic Acids Res. 2007;35:W588–594. doi: 10.1093/nar/gkm322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Linding R, Jensen LJ, Ostheimer GJ, van Vugt MA, Jorgensen C, Miron IM, Diella F, Colwill K, Taylor L, Elder K, Metalnikov P, Nguyen V, Pasculescu A, Jin J, Park JG, Samson LD, Woodgett JR, Russell RB, Bork P, Yaffe MB, Pawson T. Cell. 2007;129:1415–1426. doi: 10.1016/j.cell.2007.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Linding R, Jensen LJ, Pasculescu A, Olhovsky M, Colwill K, Bork P, Yaffe MB, Pawson T. Nucleic Acids Res. 2008;36:D695–699. doi: 10.1093/nar/gkm902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gould CM, Diella F, Via A, Puntervoll P, Gemund C, Chabanis-Davidson S, Michael S, Sayadi A, Bryne JC, Chica C, Seiler M, Davey NE, Haslam N, Weatheritt RJ, Budd A, Hughes T, Pas J, Rychlewski L, Trave G, Aasland R, Helmer-Citterich M, Linding R, Gibson TJ. Nucleic Acids Res. 38:D167–180. doi: 10.1093/nar/gkp1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li T, Li F, Zhang X. Proteins. 2008;70:404–414. doi: 10.1002/prot.21563. [DOI] [PubMed] [Google Scholar]
- 61.Neuberger G, Schneider G, Eisenhaber F. Biol Direct. 2007;2:1. doi: 10.1186/1745-6150-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xue Y, Li A, Wang L, Feng H, Yao X. BMC Bioinformatics. 2006;7:163. doi: 10.1186/1471-2105-7-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Miller ML, Jensen LJ, Diella F, Jorgensen C, Tinti M, Li L, Hsiung M, Parker SA, Bordeaux J, Sicheritz-Ponten T, Olhovsky M, Pasculescu A, Alexander J, Knapp S, Blom N, Bork P, Li S, Cesareni G, Pawson T, Turk BE, Yaffe MB, Brunak S, Linding R. Sci Signal. 2008;1:ra2. doi: 10.1126/scisignal.1159433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schwartz D, Gygi SP. Nat Biotechnol. 2005;23:1391–1398. doi: 10.1038/nbt1146. [DOI] [PubMed] [Google Scholar]
- 65.Amanchy R, Periaswamy B, Mathivanan S, Reddy R, Tattikota SG, Pandey A. Nat Biotechnol. 2007;25:285–286. doi: 10.1038/nbt0307-285. [DOI] [PubMed] [Google Scholar]
- 66.Yang CY, Chang CH, Yu YL, Lin TC, Lee SA, Yen CC, Yang JM, Lai JM, Hong YR, Tseng TL, Chao KM, Huang CY. Bioinformatics. 2008;24:i14–20. doi: 10.1093/bioinformatics/btn297. [DOI] [PubMed] [Google Scholar]
- 67.Lee TY, Bo-Kai Hsu J, Chang WC, Huang HD. Nucleic Acids Res. doi: 10.1093/nar/gkq970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gandhi TK, Chandran S, Peri S, Saravana R, Amanchy R, Prasad TS, Pandey A. DNA Res. 2005;12:79–89. doi: 10.1093/dnares/12.2.79. [DOI] [PubMed] [Google Scholar]
- 69.Askenazi M, Li S, Singh S, Marto JA. Proteomics. 10:1880–1885. doi: 10.1002/pmic.200900723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Davey NE, Shields DC, Edwards RJ. Bioinformatics. 2009;25:443–450. doi: 10.1093/bioinformatics/btn664. [DOI] [PubMed] [Google Scholar]
- 71.Yachie N, Saito R, Sugahara J, Tomita M, Ishihama Y. Mol Cell Proteomics. 2009;8:1061–1071. doi: 10.1074/mcp.M800466-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cao L, Yu K, Salomon AR. Adv Exp Med Biol. 2006;584:277–288. doi: 10.1007/0-387-34132-3_19. [DOI] [PubMed] [Google Scholar]
- 73.Amanchy R, Zhong J, Hong R, Kim JH, Gucek M, Cole RN, Molina H, Pandey A. Mol Oncol. 2009;3:439–450. doi: 10.1016/j.molonc.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tang LY, Deng N, Wang LS, Dai J, Wang ZL, Jiang XS, Li SJ, Li L, Sheng QH, Wu DQ, Li L, Zeng R. Mol Cell Proteomics. 2007;6:1952–1967. doi: 10.1074/mcp.M700120-MCP200. [DOI] [PubMed] [Google Scholar]
- 75.Nagaraj N, D'Souza RC, Cox J, Olsen JV, Mann M. J Proteome Res. doi: 10.1021/pr100637q. [DOI] [PubMed] [Google Scholar]
- 76.Nie S, Dai J, Ning ZB, Cao XJ, Sheng QH, Zeng R. J Proteome Res. 9:4585–4594. doi: 10.1021/pr100632h. [DOI] [PubMed] [Google Scholar]
- 77.Malik R, Lenobel R, Santamaria A, Ries A, Nigg EA, Korner R. J Proteome Res. 2009;8:4553–4563. doi: 10.1021/pr9003773. [DOI] [PubMed] [Google Scholar]
- 78.Olsen JV, Vermeulen M, Santamaria A, Kumar C, Miller ML, Jensen LJ, Gnad F, Cox J, Jensen TS, Nigg EA, Brunak S, Mann M. Sci Signal. 3:ra3. doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- 79.Pan C, Olsen JV, Daub H, Mann M. Mol Cell Proteomics. 2009;8:2796–2808. doi: 10.1074/mcp.M900285-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Daub H, Olsen JV, Bairlein M, Gnad F, Oppermann FS, Korner R, Greff Z, Keri G, Stemmann O, Mann M. Mol Cell. 2008;31:438–448. doi: 10.1016/j.molcel.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 81.Wang B, Malik R, Nigg EA, Korner R. Anal Chem. 2008;80:9526–9533. doi: 10.1021/ac801708p. [DOI] [PubMed] [Google Scholar]
- 82.Dephoure N, Zhou C, Villen J, Beausoleil SA, Bakalarski CE, Elledge SJ, Gygi SP. Proc Natl Acad Sci U S A. 2008;105:10762–10767. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Beausoleil SA, Jedrychowski M, Schwartz D, Elias JE, Villen J, Li J, Cohn MA, Cantley LC, Gygi SP. Proc Natl Acad Sci U S A. 2004;101:12130–12135. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP. Nat Biotechnol. 2006;24:1285–1292. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- 85.Kyono Y, Sugiyama N, Imami K, Tomita M, Ishihama Y. J Proteome Res. 2008;7:4585–4593. doi: 10.1021/pr800305y. [DOI] [PubMed] [Google Scholar]
- 86.Sugiyama N, Masuda T, Shinoda K, Nakamura A, Tomita M, Ishihama Y. Mol Cell Proteomics. 2007;6:1103–1109. doi: 10.1074/mcp.T600060-MCP200. [DOI] [PubMed] [Google Scholar]
- 87.Brill LM, Xiong W, Lee KB, Ficarro SB, Crain A, Xu Y, Terskikh A, Snyder EY, Ding S. Cell Stem Cell. 2009;5:204–213. doi: 10.1016/j.stem.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ruse CI, McClatchy DB, Lu B, Cociorva D, Motoyama A, Park SK, Yates JR., 3rd J Proteome Res. 2008;7:2140–2150. doi: 10.1021/pr800147u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gauci S, Helbig AO, Slijper M, Krijgsveld J, Heck AJ, Mohammed S. Anal Chem. 2009;81:4493–4501. doi: 10.1021/ac9004309. [DOI] [PubMed] [Google Scholar]
- 90.Van Hoof D, Munoz J, Braam SR, Pinkse MW, Linding R, Heck AJ, Mummery CL, Krijgsveld J. Cell Stem Cell. 2009;5:214–226. doi: 10.1016/j.stem.2009.05.021. [DOI] [PubMed] [Google Scholar]
- 91.Ho TC, Chen SL, Shih SC, Wu JY, Han WH, Cheng HC, Yang SL, Tsao YP. Am J Pathol. 2010;177:1798–1811. doi: 10.2353/ajpath.2010.091085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Raijmakers R, Kraiczek K, de Jong AP, Mohammed S, Heck AJ. Anal Chem. 82:824–832. doi: 10.1021/ac901764g. [DOI] [PubMed] [Google Scholar]
- 93.McNulty DE, Annan RS. Mol Cell Proteomics. 2008;7:971–980. doi: 10.1074/mcp.M700543-MCP200. [DOI] [PubMed] [Google Scholar]
- 94.Yu LR, Zhu Z, Chan KC, Issaq HJ, Dimitrov DS, Veenstra TD. J Proteome Res. 2007;6:4150–4162. doi: 10.1021/pr070152u. [DOI] [PubMed] [Google Scholar]
- 95.Sui S, Wang J, Yang B, Song L, Zhang J, Chen M, Liu J, Lu Z, Cai Y, Chen S, Bi W, Zhu Y, He F, Qian X. Proteomics. 2008;8:2024–2034. doi: 10.1002/pmic.200700896. [DOI] [PubMed] [Google Scholar]
- 96.Tsai CF, Wang YT, Chen YR, Lai CY, Lin PY, Pan KT, Chen JY, Khoo KH, Chen YJ. J Proteome Res. 2008;7:4058–4069. doi: 10.1021/pr800364d. [DOI] [PubMed] [Google Scholar]
- 97.Li QR, Ning ZB, Tang JS, Nie S, Zeng R. J Proteome Res. 2009;8:5375–5381. doi: 10.1021/pr900659n. [DOI] [PubMed] [Google Scholar]
- 98.Schreiber TB, Mausbacher N, Keri G, Cox J, Daub H. Mol Cell Proteomics. 9:1047–1062. doi: 10.1074/mcp.M900486-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yan GR, Xiao CL, Yin XF, Zhou HH, Tan ZL, He QY. J Proteome Res. 2009 doi: 10.1021/pr900385t. [DOI] [Google Scholar]
- 100.Xiao K, Sun J, Kim J, Rajagopal S, Zhai B, Villén J, Haas W, Kovacs JJ, Shukla AK, Hara MR, Hernandez M, Lachmann A, Zhao S, Lin Y, Cheng Y, Mizuno K, Ma'ayan A, Gygi SP, Lefkowitz RJ. Proc Natl Acad Sci U S A. 2010;107:15299–15304. doi: 10.1073/pnas.1008461107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Santamaria A, Wang B, Elowe S, Malik R, Zhang F, Bauer M, Schmidt A, Silljé HH, Körner R, Nigg EA. Mol Cell Proteomics. 2011;10(1) doi: 10.1074/mcp.M110.004457. M110.004457. [DOI] [PMC free article] [PubMed] [Google Scholar]


