Abstract
The aim of this study was to determine the effect of subinhibitory concentrations (sub-MICs) of ciprofloxacin, amikacin and colistin on biofilm formation, motility, curli fimbriae formation by planktonic and biofilm cells of E. coli strains isolated from the urine of patients with various urinary system infections. Quantification of biofilm formation was carried out using a microtiter plate assay and a spectrophotometric method. Bacterial enumeration was used to assess the viability of bacteria in the biofilm. Curli expression was determined by using YESCA agar supplemented with congo red. Using motility agar the ability to move was examined. All the antibiotics used at sub-MICs reduced biofilm formation in vitro, decreased the survival of bacteria, but had no effect on the motility of planktonic as well as biofilm cells. The inhibitory effect of sub-MICs of antimicrobial agents on curli fimbriae formation was dependent on the form in which the bacteria occurred, incubation time and antibiotic used. Our results clearly show that all the three antibiotics tested reduce biofilm production, interfere with curli expression but do not influence motility. This study suggests that ciprofloxacin, amikacin and colistin may be useful in the treatment of biofilm-associated infections caused by E. coli strains.
Keywords: biofilm, sub-MIC, ciprofloxacin, amikacin, colistin
Introduction
Escherichia coli is the predominant pathogen causing nosocomial urinary tract infections associated with indwelling bladder catheter (Warren 2001). The major virulence factors associated with such infections are the ability of microorganisms to adhere to medical devices and to subsequently form biofilm. Biofilm formation is initiated by the attachment of a single planktonic cell to a surface. Multiplication and the development of microcolonies separated by water-filled channels follow. Curli fibers and motility may play an important role during the initial interaction of bacterial cell with the surface, neutralizing repulsive forces which prevent the surface colonization (Pratt and Kolter, 1998; Prigent-Combaret et al., 2001).
Sub-minimum inhibitory concentrations (sub-MICs) of various antibiotics have been shown to both stimulate and/or impede E. coli biofilm formation in human infections (Boehm et al., 2009; Bret and Di Martino, 2004). Thus, broadening our current knowledge of the effect that sub-lethal concentrations of antimicrobial agents elicit on biofilms formed by E. coli strains is of clinical interest.
The purpose of this study was to evaluate the ability of E. coli strains isolated from urine of patients with urinary tract infection to attach on polystyrene and grow as a biofilm. We also investigated the effect of sub-MICs of ciprofloxacin, amikacin, and colistin on biofilm formation. Moreover, we examined motility and curli formation in both planktonic and biofilm phenotypes of E. coli strains, both in presence and absence of sub-inhibitory levels of antibiotics.
Materials and Methods
Bacterial strains
Escherichia coli strains (n = 100) were isolated from urine samples collected in the period 2008–2009 from patients suffered from the lower urinary tract infections (urethra and bladder), hospitalized in Korczak Pediatric Center of Lower Silesia in Wroclaw. They were not catheterized. The strains were maintained on Mueller-Hinton agar slopes (Oxoid) at 4 °C. The API-20E test (bioMerieux, Poland) for the identification of bacteria was used.
Selection of motile and fimbriae curli-producing strains
Motility test
All strains (n = 100) were grown overnight at 37 °C in TSB, than were stab-inoculated into Motility Agar and incubated at 37 °C for 24 h. Non-motile bacteria grew only where they were inoculated. Motile bacteria grew along the stab and also swam away from the stabbed area.
Detection of curli fibers
Each motile strain was grown overnight in TSB at 37 °C and then inoculated onto plate containing YESCA agar (10 g casamino acids, 1 g yeast extract, and 20 g agar per 1 liter) supplemented with 20 mg/L of congo red (CRI). Curli-producing E. coli bound congo red dye and formed red colonies, whereas curli-negative bacteria formed white colonies (Hammar et al., 1995).
Antibiotics
Three antimicrobial agents with different bacterial cell targets were used in this study: ciprofloxacin lactate (Proxacin©, Warsaw, Poland), amikacin disulfate salt (Biodacyna©, Warsaw, Poland), and colistin sodium methanesulfonate (Colistin©, Warsaw, Poland).
Planktonic antimicrobial susceptibility testing
MIC assay against three antimicrobial agents was performed in Mueller-Hinton broth (MHB; Oxoid) according to CLSI guidelines for broth microdilution susceptibility testing (CLSI 2006).
Biofilm formation assay and quantification
The biofilm formation assay was performed according to O’Toole and Kolter (1998) with slight modifications. Twenty microliters of each diluted culture (1–2×108 cfu/mL) was inoculated into each of six wells of a 96-well polystyrene plate containing 200 μL of MHB. After incubation for 24, 48 and 72 h at 37 °C the wells were rinsed thoroughly with phosphate buffered saline (PBS) in order to remove nonadherent bacteria. Bacterial cells bound to the walls of the wells were stained with 1% (w/v) crystal violet (Sigma-Aldrich, Poznan, Poland) for 15 min, and then rinsed thoroughly with PBS. Afterwards the dye bound to the adherent bacterial cells was resolubilized with 95% (v/v) ethanol. The optical density (OD) of each well was measured at 495 nm using a plate reader (ANALCO-GBG STAT-FAX 2100). In each plate four wells were used as blanks containing MHB medium only. On the basis of ODs of bacterial biofilms, E. coli strains were classified into four categories (Stepanovic et al., 2004). The cut-off OD (ODc) was defined as three standard deviations (SD) above the mean OD of the negative control. Strains were classified as follows: OD ≤ ODc no biofilm producer; ODc < OD ≤ 2 x ODc weak biofilm producer; 2 x ODc < OD ≤ 4 x ODc moderate biofilm producer; 4 x ODc < OD strong biofilm producer. In our study the ODc value was 0.004.
Count of live bacteria in biofilm
Count of live bacteria in biofilm was performed according to Okajima et al. (2006). Briefly, after incubation (24, 48 and 72 h at 37 °C) as described above, each bacterial culture was washed three times with PBS to remove nonadherent bacteria. Bacterial biofilm was transferred into a microtube containing 10 μL of PBS. The tubes were centrifuged for 2 min at 2500 rpm to separate cells from the biofilm matrix. The number of colony-forming units (cfu) was assessed by plating serial dilutions on Mueller-Hinton agar. Each experiment was conducted in triplicate.
Effect of sub-MICs of antimicrobial agents on biofilm formation and count of live bacteria in biofilm
Effect of sub-MICs of antimicrobial agents on biofilm formation was performed according to Di Bonaventura et al. (2004). Briefly, each drug was tested at 0.5x MIC to study its effect on biofilm formation. Various concentrations of antimicrobial agents prepared in 200 μL of MHB were added to microtiter wells containing 20 μL of diluted culture of bacteria. After 24, 48 and 72 h of incubation, quantitation of biofilms was measured as described in biofilm formation assay. Drug-free medium was used in control wells. The viability of biofilm cells was determined by plate counts (Okajima et al., 2006).
Effect of sub-MICs of antimicrobial agents on curli expression of planktonic cells
Effect of sub-MICs of antimicrobial agents on curli expression of planktonic cells was performed according to Hammar et al. (1995). Briefly, each strain was grown overnight with the sub-MICs of antibiotics and then inoculated onto plate containing YESCA agar supplemented with congo red (CRI) and the same sub-MICs. Curli-producing E. coli bound congo red dye and formed red colonies on CRI, whereas curli-negative bacteria formed white colonies. Control cultures contained no antibiotics.
Effect of sub-MICs of antimicrobial agents on curli expression of biofilm cells
Biofilm was grown in MHB in the presence and absence of 0.5x MIC of the antibiotic for 24, 48 and 72 h at 37 °C as described above. Each bacterial culture was washed three times with PBS to remove nonadherent bacteria. Bacterial biofilm was transferred into a microtube containing 10 μL of PBS. The tubes were centrifuged for 2 min at 2500 rpm to separate the cells from biofilm matrix. Bacterial suspension was plated on CRI agar. The plates were incubated for 24 h at 37 °C. Control cultures contained no antibiotics.
Effect of sub-MICs of antimicrobial agents on motility of planktonic cells
Assays were performed on each strain grown overnight with the sub-MICs of antibiotics. Using a sterile toothpick, bacteria cells were stab-inoculated into Motility Agar containing the same sub-MICs and incubated at 37 °C for 24 h. Non-motile bacteria grew only where they were inoculated. Motile bacteria grew along the stab and also swam away from the stabbed area. Control cultures contained no antibiotics (Vranes et al., 2001).
Effect of sub-MICs of antimicrobial agents on motility of biofilm cells
Following 24-, 48- and 72-h incubation at 37 °C in the presence and absence of 0.5x MIC of antibiotic, each bacterial culture was washed three times with PBS to remove nonadherent cells. Bacterial biofilm was placed into a microtube containing 10 μL of PBS. The tubes were centrifuged for 2 min at 2500 rpm to separate the cells from biofilm matrix. Bacterial culture was inoculated into the tube with Motility Agar. Control cultures contained no antibiotics.
Statistical analysis
Data were the mean value ± SD from seven samples from three independent experiments. Differences between the mean were analyzed by one-way analysis of variance (ANOVA, STATISTICA 9 PL). Statistical significance was considered when p was < 0.05.
Results
Bacterial motility and curli fimbriae production
Seventy-three of one hundred examined strains were motile. Ten of them produced curli fibers. For further experiments only motile and fimbriae curli-producing strains (n = 10) were selected: E. coli No 8, 9, 10, 11, 31, 864, 906, 1148, 1583, 5579.
Biofilm formation and viable cell counts
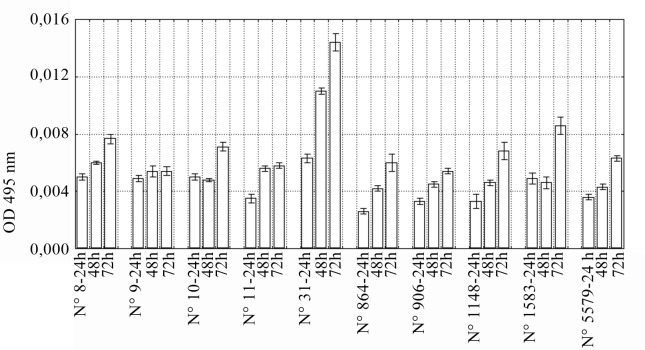
The mean kinetics of 10 selected E. coli strains to form biofilm on the surface of polystyrene wells after 24-, 48- and 72-h incubation is illustrated in Figure 1. The ODs of the 24-h biofilm ranged from 0.0026 to 0.0063. Five strains (E. coli No 8, 9, 10, 31, and 1583) produced weak biofilm (0.004 < OD ≤ 0.008). After 48-h incubation all isolates were able to form biofilm. Nine of E. coli isolates produced weak biofilm and only one strain (E. coli No 31) produced moderate biofilm (0.008 < OD ≤ 0.016). After 72-h incubation only E. coli No 31 and 1583 were moderate biofilm producers. Eight remaining isolates still created weak biofilm.
Figure 1.
Biofilm production by E. coli strains. Values represent means from three independent experiments. Bars indicate standard deviations.
For the next stage of our research, five E. coli strains (No 8, 9, 10, 31, and 1583), which produced biofilm after 24-h incubation were selected. In the 24-h biofilm unexposed to antibiotics the average number of living bacterial cells was 9.4×109 cfu/mL. This value decreased to 5.4×109 cfu/mL and 4.7×109 cfu/mL in 48-h and 72-h biofilms, respectively.
Antimicrobial susceptibility testing
In the next stage of study we determined the MIC of antibiotics suppressing growth of E. coli strains No 8, 9, 10, 31, and 1583. The MICs of ciprofloxacin, amikacin and colistin were in the range 0.007–32.0 μg/mL, 1.0–4.0 μg/mL and 0.062–0.5 μg/mL, respectively (Table 1).
Table 1.
MIC values (μg/mL) of antimicrobial agents against E. coli strains.
| Strain | CIP | AN | CL |
|---|---|---|---|
| E. coli No 8 | 0.25 | 1.0 | 0.5 |
| E. coli No 9 | 32.0 | 1.0 | 0.5 |
| E. coli No 10 | 0.007 | 1.0 | 0.125 |
| E. coli No 31 | 0.007 | 1.0 | 0.125 |
| E. coli No 1583 | 0.031 | 4.0 | 0.062 |
CIP: ciprofloxacin; AN: amikacin; CL: colistin.
Effect of sub-MICs of antimicrobial agents on biofilm formation and count of live bacteria in biofilm
The activities of antimicrobial agents tested at concentration 0.5x MIC against total biomass of E. coli biofilm are shown in Table 2. Ciprofloxacin, amikacin and colistin significantly (p < 0.05) reduced the optical density of the biofilms of E. coli (ODs ≤ 0.004). Amikacin was the most effective antibiotic in eradication of biofilm. In the presence of amikacin the optical density of the biofilms was decreased to 25, 26, and 27% in comparison to the controls. The exposure of bacteria to ciprofloxacin resulted in reduction of the optical density to 75, 38 and 25%, whereas treatment with colistin reduced the optical density to 73, 24, and 10%.
Table 2.
Effect of 0.5x MIC of antibiotics on biofilm formation. Results are the mean ODs for 5 selected E. coli strains.
| Time of incubation [h] | Biofilm formation | |||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Control | CIP | AN | CL | |||||
|
|
|
|
|
|||||
| OD(± SD) | [%] | OD(± SD) | [%] | OD(± SD) | [%] | OD(± SD) | [%] | |
| 24 | 0.0048 (± 0.0001) | 100 | 0.0036 (± 0.0001) | 75 | 0.0012 (± 0.0001) | 25 | 0.0035 (± 0.0001) | 73 |
| 48 | 0.0061 (± 0.0001) | 100 | 0.0023 (± 0.0001) | 38 | 0.0016 (± 0.0002) | 26 | 0.0015 (± 0.0001) | 24 |
| 72 | 0.0086 (± 0.0002) | 100 | 0.0022 (± 0.0003) | 25 | 0.0023 (± 0.0001) | 27 | 0.0009 (± 0.0001) | 10 |
CIP: ciprofloxacin; AN: amikacin; CL: colistin.
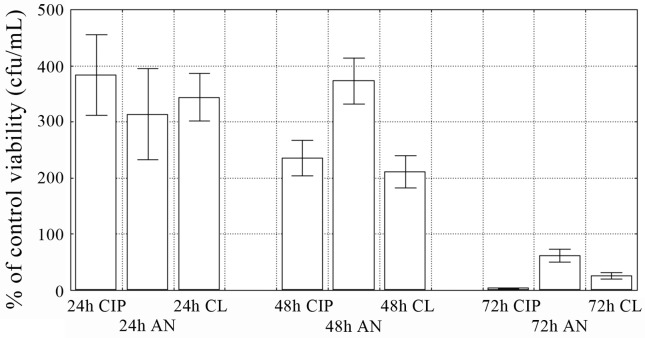
During the same experiments the number of living bacterial cells in the biofilm exposed to 0.5x MIC of antibiotics was examined (Figure 2). In 24-h and 48-h biofilms the number of bacteria significantly (p < 0.05) increased, regardless of the antibiotic used. The values were in the range from 314.0 to 384.0%, and 211.0 to 373.0% in 24-h and 48-h biofilm, respectively. The antibacterial effect of all antimicrobials was observed only after 72-h incubation (p < 0.05). The percentage of living bacterial cells were 61.9 for amikacin, 26.1 for colistin and 3.8 in case of ciprofloxacin.
Figure 2.
The number of live bacterial cells in biofilm in the presence of 0.5x MIC of antibiotics. Values represent the mean percentage of control viability (cfu/mL) for 5 selected E. coli strains. Bars indicate standard deviations. CIP: ciprofloxacin; AN: amikacin; CL: colistin.
Effect of sub-MICs of antimicrobial agents on curli expression of planktonic and biofilm cells
We also examined the effect of sub-MICs of amikacin, ciprofloxacin and colistin on curli fibers formation of planktonic and biofilm forms. As seen in Table 3 the 0.5x MIC of ciprofloxacin suppressed fimbriation of planktonic cells, except E. coli No 9. All strains grown in 0.5x MIC of amikacin produced curli fibers. Three out of five investigated strains demonstrated absence of curli fimbriae after contact with 0.5x MIC of colistin.
Table 3.
Effect of 0.5x MIC of antibiotics on curli fibers production of planktonic and biofilm forms of E. coli strains.
| Strain | Planktonic forms | Biofilm forms | |||||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||
| Control | CIP | AN | CL | Time of incubation [h] | Control | CIP | AN | CL | |
| E. coli No 8 | + | − | + | − | 24 | + | + | + | + |
| 48 | + | − | − | − | |||||
| 72 | + | − | − | − | |||||
|
| |||||||||
| E. coli No 9 | + | + | + | − | 24 | + | + | − | + |
| 48 | + | − | − | − | |||||
| 72 | + | − | − | − | |||||
|
| |||||||||
| E. coli No 10 | + | − | + | + | 24 | + | + | + | + |
| 48 | + | + | + | + | |||||
| 72 | + | + | + | − | |||||
|
| |||||||||
| E. coli No 31 | + | − | + | + | 24 | + | + | + | + |
| 48 | + | + | + | + | |||||
| 72 | + | + | + | + | |||||
|
| |||||||||
| E. coli No 1583 | + | − | + | − | 24 | + | + | + | + |
| 48 | + | + | − | + | |||||
| 72 | + | − | − | − | |||||
+: existing; −: not existing; CIP: ciprofloxacin; AN: amikacin; CL: colistin.
In contrast to planktonic forms, E. coli strains growing in 24-h biofilm in the presence of 0.5x MIC of antibiotics retained their ability to curli fimbriae formation. The exception was E. coli No 9 strain which lost this ability during 24-h incubation at 0.5x MIC of amikacin. The inhibition of fimbriae formation was also found in E. coli No 8 and 9 isolates after 48-h incubation at all antibiotic suspensions and in E. coli No 1583 exposed to 0.5x MIC of amikacin. After 72-h incubation in the presence of all the antibiotics E. coli No 1583 as well as E. coli No 8 and 9 strains produced no curli fibers. The exposure of E. coli No 10 and 31 rods to 0.5x MIC of antibiotics did not affect the ability to curli synthesis at all.
Effect of sub-MICs of antimicrobial agents on motility of planktonic and biofilm cells
In the next part of experiments we have determined motility of planktonic cells treated with sub-MICs of ciprofloxacin, amikacin, and colistin. The exposure of E. coli strains to sub-MICs of all antibiotics did not inhibit the motility of planktonic cultures. Bacteria isolated from the biofilm cultures treated with sub-MICs of ciprofloxacin, amikacin, and colistin did not lose the ability to move. All bacterial strains from 24- and 48-h biofilm remained motile, even after exposure to antimicrobial agents. Examined rods growing in 72-h control samples of biofilm became nonmotile.
Discussion
More than 50% of all microbial infections have now been associated with the biofilm formation (Costerton et al., 1999). Several bacterial surface structures are known to be involved in biofilm creation (Pratt and Kolter, 1998; Van Houdt and Michiels, 2005; Ulett et al., 2007) Our research study demonstrates that motile strains possessing curli fimbriae form biofilm on the surface of the wells of microtiter plates. Similar result was obtained by Pratt and Kolter (1998). They showed that motility promotes biofilm development of the E. coli 2K1056 strain. As opposed to our and Pratt and Kolter’s (1998) results, Prigent-Combaret et al. (2000) indicated that flagella and motility are not involved in either bacterial attachment or subsequent biofilm development of curli-producing E. coli strains. However, the same authors emphasised that these results may be caused by the differences in growth conditions. Cai et al. (2010) concluded that twitching motility is not a prerequisite for the formation of mature biofilm and the ability to biofilm production depends among others on environmental conditions in which bacteria are grown.
Curli fibers produced by E. coli, Salmonella, Citrobacter, and Enterobacter species act as cell-to-surface and cell-to-cell adhesins (Prigent-Combaret et al., 2000). The results published by Jackson et al. (2002) and Jonas et al. (2007) showed that curli fimbriae play significant role in biofilm formation. However, the expression of curli is temperature- and other growth conditions-dependent (Kikuchi et al., 2005). At 37 °C bacteria expressed curli when grown on solid abiotic surfaces, but not on agar plates or when they were shaken. Curli were expressed at 25 °C independently from growth conditions. It has been reported that enterohemorrhagic and enterotoxigenic E. coli strains as well as sepsis isolates express curli at 26 °C and curli creation is weaker when bacteria are grown at 37 °C (Olsen et al., 1993). Our results indicate that E. coli strains growing at 37 °C on YESCA agar plates produce curli fibers.
In current research the expression of curli and motility in E. coli biofilms have been examined. Our results show that bacterial cells growing in the biofilm retain the ability to curli fimbriae formation. It confirms their putative role in bacteria autoaggregation within biofilm. In 24- and 48-h biofilm, the examined rods were motile, while in 72-h biofilm this trait was lost. Prigent-Combaret et al. (1999) also observed that the motility was suppressed during production of biofilm. In contrast, Klausen et al. (2003) showed that in many bacterial species flagella play a role not only in the initial phase of biofilm formation but also in the later phases of biofilm architecture development. In later stages of biofilm maturation, motility is a significant feature which allows bacteria to leave their current place of location and swim toward another sites of colonization. It probably means that bacteria before leaving of mature biofilm, must induce specific signals to re-stimulate the production of flagella (Klausen et al., 2003).
In our study we also examined the number of living cells at different stages of biofilm development. We established that the fraction of living cells was maximal in the 24-h biofilm whereas it decreased in later phases of biofilm maturation. Tresse et al. (2003) and Hola et al. (2006) obtained similar results. This phenomenon may be associated with the limited diffusion of the nutrient and oxygen through the layer of the biofilm (Hola et al., 2006). The almost total consumption of oxygen and glucose in the surface layers creates anaerobic niches deeper in the biofilm where the cells downregulate into an extremely slow-growing or non-growing state in order to survive (Leid et al., 2002).
Bacteria forming biofilm have slower metabolism and are subjected to phenotype changes, which in consequence increases their resistance and virulence. It may be the main cause of the recurrent urinary tract infections, despite antimicrobial treatment. Patients may be exposed to subinhibitory concentrations of antibiotics for many reasons, including underdosing, poor compliance, the fluctuation of drug concentrations between doses. The sub-MICs can inhibit the bacterial growth and modify their virulence factors (Wojnicz and Jankowski, 2007; Wojnicz and Cisowska, 2009). Antimicrobial concentrations below MIC may be encountered clinically during the treatment and prevention of biofilm-related medical device infections.
Therefore, it seems advisable to check the effect of sub-MICs of ciprofloxacin, amikacin and colistin on biofilm production, curli synthesis and motility of planktonic and biofilm cultures. The results of our study presented in Table 2 show that when E. coli strains were grown in the presence of sub-MIC of antibiotics the inhibition of biofilm formation was observed. Similar results were also obtained by other researchers (Cerca et al., 2005; Majtan et al., 2008). Interesting was the fact that the number of live bacterial cells in 24- and 48-hours biofilms formed in the presence of sub-MIC of antibiotics were higher than that of unexposed controls. It is probably associated with the so-called persister cells residing in the biofilm. Persisters are known to be largely responsible for high levels of biofilm tolerance to antimicrobials. Keren et al. (2004) estimated that tolerance of E. coli to ampicillin and ofloxacin was dependent on growth stages of persister cells.
As shown in currant studies carried out on planktonic forms of E. coli strains, the sub-therapeutic concentrations of ciprofloxacin and colistin, but not amikacin, inhibited curli production in some isolates. As demonstrated in Table 3, the ability to curli expression by rods growing in biofilm in the presence sub-MICs of antibiotics varied and in some cases was dependent on the incubation time. However, in the two strains tested, none of the antibiotics inhibited curli synthesis. Moreover no antibiotic affected the movement ability of either planktonic or biofilm cells.
These results may mean that antimicrobial agents do not always change the phenotype of bacterial cells growing in biofilm structure.
In conclusion, our study shows that sub-MICs of ciprofloxacin, amikacin and colistin reduce the ability to biofilm production on microtiter plates. The effect of sub-lethal doses on curli fimbriae synthesis is dependent on the form in which bacteria occur (planktonic or biofilm). However, obtained results indicate that the presence of antibiotics does not influence motility in any bacterial vegetation form. The exception were bacterial cells isolated from 72-h biofilm treated with sub-MICs of all antibiotics. They lost the ability to move. Clearly, more studies are required to investigate further the action mechanisms of sub-MICs of antibiotics against planktonic and biofilm bacteria populations.
Acknowledgments
Authors are grateful to Professor Andrzej Hendrich for reading the manuscript and helpful discussions.
References
- Boehm A, Steiner S, Zaehringer F, Casanova A, Hamburger F, Ritz D, Keck W, Ackermann M, Schirmer T, Jenal U. Second messenger signaling governs Escherichia coli biofilm induction upon ribosomal stress. Mol Microbiol. 2009;72:1500–1516. doi: 10.1111/j.1365-2958.2009.06739.x. [DOI] [PubMed] [Google Scholar]
- Bret L, Di Martino P. Effect of ceftazidime, amikacin and ciprofloxacin on biofilm formation by some enterobacterial clinical isolates. Chemotherapy. 2004;50:255–259. doi: 10.1159/000081947. [DOI] [PubMed] [Google Scholar]
- Cai Y, Wang R, An M, Liang B. Iron-depletion prevents biofilm formation in Pseudomonas aeruginosa through twitching motility and quorum sensing. Braz J Microbiol. 2010;41:37–41. doi: 10.1590/S1517-83822010000100008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerca N, Martins S, Sillankorva S, Jefferson KK, Pier GB, Oliveira R, Azeredo J. Effects of growth in the presence of subinhibitory concentrations of dicloxacillin on Staphylococcus epidermidis and Staphylococcus haemolyticus biofilms. Appl Environ Microbiol. 2005;71:8677–8682. doi: 10.1128/AEM.71.12.8677-8682.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLSI. Proc. 16th Informational Supplement. M100. Clinical and Laboratory Standards Institute; Wayne: 2006. Performance standards for antimicrobial susceptibility Testing; S16 pp. [Google Scholar]
- Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: A common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- Di Bonaventura G, Spedicato I, D’Antonio D, Robuffo I, Piccolomini R. Biofilm formation by Stenotrophomonas maltophilia: modulation by quinolones, trimethoprim-sulfamethoxazole, and ceftazidime. Antimicrob Agents Chemother. 2004;48:151–160. doi: 10.1128/AAC.48.1.151-160.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammar M, Arnqvist A, Bian Z, Olsen A, Normark S. Expression of two csg operons is required for production of fibronectin- and congo red-binding curli polymers in Escherichia coli K-12. Mol Microbiol. 1995;18:661–670. doi: 10.1111/j.1365-2958.1995.mmi_18040661.x.. [DOI] [PubMed] [Google Scholar]
- Hola V, Ruzicka F, Votava M. The dynamics of Staphylococcus epidermidis biofilm formation in relation to nutrition, temperature, and time. Scripta Medica (Brno) 2006;79:169–174. [Google Scholar]
- Jackson DW, Suzuki K, Oakford L, Simecka JW, Hart ME, Romeo T. Biofilm formation and dispersal under the influence of the global regulator CsrA of Escherichia coli. J Bacteriol. 2002;184:290–301. doi: 10.1128/JB.184.1.290-301.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas K, Tomenius H, Kader A, Normark S, Römling U, Belova LM, Melefors Ö. Roles of curli, cellulose and BapA in Salmonella biofilm morphology studied by atomic force microscopy. BMC Microbiology. 2007;7:70. doi: 10.1186/1471-2180-7-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren I, Kaldalu N, Spoering A, Wang Y, Lewis K. Persister cells and tolerance to antimicrobials. FEMS Microbiol Lett. 2004;230:13–18. doi: 10.1016/S0378-1097(03)00856-5. [DOI] [PubMed] [Google Scholar]
- Kikuchi T, Mizunoe Y, Takade A, Naito S, Yoshida S. Curli fibers are required for development of biofilm architecture in Escherichia coli K-12 and enhance bacterial adherence to human uroepithelial cells. Microbiol Immunol. 2005;49:875–884. doi: 10.1111/j.1348-0421.2005.tb03678.x. [DOI] [PubMed] [Google Scholar]
- Klausen M, Heydorn A, Ragas P, Lambertsen L, Aaes-Jørgensen A, Molin S, Tolker-Nielsen T. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol Microbiol. 2003;48:1511–1524. doi: 10.1046/j.1365-2958.2003.03525.x. [DOI] [PubMed] [Google Scholar]
- Leid JG, Shirtliff ME, Costerton JW, Stoodley AP. Human leukocytes adhere to, penetrate, and respond to Staphylococcus aureus biofilms. Infect Immun. 2002;70:6339–6345. doi: 10.1128/IAI.70.11.6339-6345.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majtan J, Majtanova L, Xu M, Majtan V. In vitro effect of subinhibitory concentrations of antibiotics on biofilm formation by clinical strains of Salmonella enterica serovar Typhimurium isolated in Slovakia. J Appl Microbiol. 2008;104:1294–1301. doi: 10.1111/j.1365-2672.2007.03653.x. [DOI] [PubMed] [Google Scholar]
- O’Toole GA, Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol. 1998;30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- Okajima Y, Kobayakawa S, Tsuji A, Tochikubo T. Biofilm formation by Staphylococcus epidermidis on intraocular lens material. Invest Ophthalmol Vis Sci. 2006;47:2971–2975. doi: 10.1167/iovs.05-1172. [DOI] [PubMed] [Google Scholar]
- Olsen A, Arnqvist A, Hammar M, Sukupolvi S, Normark S. The RpoS sigma factor relieves H-NS-mediated transcriptional repression of csgA, the subunit gene of fibronectin-binding curli in Escherichia coli. Mol Microbiol. 1993;7:523–536. doi: 10.1111/j.1365-2958.1993.tb01143.x. [DOI] [PubMed] [Google Scholar]
- Pratt LA, Kolter R. Genetic analysis of Escherichia coli biofilm formation: Roles of flagella, motility, chemotaxis and type I pili. Mol Microbiol. 1998;30:285–293. doi: 10.1046/j.1365-2958.1998.01061.x. [DOI] [PubMed] [Google Scholar]
- Prigent-Combaret C, Brombacher E, Vidal O, Ambert A, Lejeune P, Landini P, Dorel C. Complex regulatory network controls initial adhesion and biofilm formation in Escherichia coli via regulation of the csgD gene. J Bacteriol. 2001;183:7213–7223. doi: 10.1128/JB.183.24.7213-7223.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigent-Combaret C, Prensier G, Le Thi TT, Vidal O, Lejeune P, Dorel C. Developmental pathway for biofilm formation in curli-producing Escherichia coli strains: Role of flagella, curli and colanic acid. Environ Microbiol. 2000;2:450–464. doi: 10.1046/j.1462-2920.2000.00128.x. [DOI] [PubMed] [Google Scholar]
- Prigent-Combaret C, Vidal O, Dorel C, Lejeune P. Abiotic surface sensing and biofilm-dependent regulation of gene expression in Escherichia coli. J Bacteriol. 1999;181:5993–6002. doi: 10.1128/jb.181.19.5993-6002.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanovic S, Cirkovic I, Ranin L, Svabic-Vlahovic M. Biofilm formation by Salmonella spp. and Listeria monocytogenes on plastic surface. Lett Appl Microbiol. 2004;38:428–432. doi: 10.1111/j.1472-765X.2004.01513.x. [DOI] [PubMed] [Google Scholar]
- Tresse O, Lescob S, Rho D. Dynamics of living and dead bacterial cells within a mixed-species biofilm during toluene degradation in a biotrickling filter. J Appl Microbiol. 2003;94:849–854. doi: 10.1046/j.1365-2672.2003.01914.x. [DOI] [PubMed] [Google Scholar]
- Ulett GC, Valle J, Beloin C, Sherlock O, Ghigo JM, Schembri MA. Functional analysis of antigen 43 in uropathogenic Escherichia coli reveals a role in long-term persistence in the urinary tract. Infect Immun. 2007;75:3233–3244. doi: 10.1128/IAI.01952-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Houdt R, Michiels CW. Role of bacterial cell surface structures in Escherichia coli biofilm formation. Res Microbiol. 2005;156:626–633. doi: 10.1016/j.resmic.2005.02.005. [DOI] [PubMed] [Google Scholar]
- Vranes J, Schönwald S, Sterk-Kuzmanovic N, Ivanic B. Low virulence of Escherichia coli strains causing exacerbation of chronic pyelonephritis. Acta Clin Croat. 2001;40:165–170. [Google Scholar]
- Warren JW. Catheter-associated urinary tract infections. Int J Antimicrob Agents. 2001;17:299–303. doi: 10.1016/s0924-8579(00)00359-9. [DOI] [PubMed] [Google Scholar]
- Wojnicz D, Jankowski S. Effects of subinhibitory concentrations of amikacin and ciprofloxacin on the hydrophobicity and adherence to epithelial cells of uropathogenic Escherichia coli strains. Int J Antimicrob Agents. 2007;29:700–704. doi: 10.1016/j.ijantimicag.2007.01.007. [DOI] [PubMed] [Google Scholar]
- Wojnicz D, Cisowska A. Composition of the outer membrane proteins of Escherichia coli strains in relation to serum susceptibility after exposure to subinhibitory concentrations of amikacin and ciprofloxacin. Int J Antimicrob Agents. 2009;33:579–582. doi: 10.1016/j.ijantimicag.2008.12.006. [DOI] [PubMed] [Google Scholar]