Abstract
The field of regenerative medicine has increasingly recognized the importance to be inspired by developmental processes to identify signaling pathways crucial for 3D organogenesis and tissue regeneration. Here, we aimed at recapitulating the first events occurring during limb development (ie, cell condensation and expansion of an undifferentiated mesenchymal cell population) to prime 3D cultures of human bone marrow-derived mesenchymal stromal/stem cells (hBM-MSC) toward the chondrogenic route. Based on embryonic development studies, we hypothesized that Wnt3a and fibroblast growth factor 2 (FGF2) induce hBM-MSC to proliferate in 3D culture as an undifferentiated pool of progenitors (defined by clonogenic capacity and expression of typical markers), retaining chondrogenic potential upon induction by suitable morphogens. hBM-MSC were responsive to Wnt signaling in 3D pellet culture, as assessed by significant upregulation of main target genes and increase of unphosphorylated β-catenin levels. Wnt3a was able to induce a five-fold increase in the number of proliferating hBM-MSC (6.4% vs. 1.3% in the vehicle condition), although total DNA content of the 3D construct was decreasing over time. Preconditioning with Wnt3a improved transforming growth factor-β1 mediated chondrogenesis (30% more glycosaminoglycans/cell in average). In contrast to developmental and 2D MSC culture models, FGF2 antagonized the Wnt-mediated effects. Interestingly, the CD146+ subpopulation was found to be more responsive to Wnt3a. The presented data indicate a possible strategy to prime 3D cultures of hBM-MSC by invoking a “developmental engineering” approach. The study also identifies some opportunities and challenges to cross-fertilize skeletal development models and 3D hBM-MSC culture systems.
Introduction
In the last years, regenerative medicine has found inspiration from the field of developmental biology, leading to the so called “developmental engineering” paradigm, namely the engineering of developmental processes for inducing tissue regeneration [1,2]. The understanding and recapitulation of key signaling pathways are expected to lead to enhanced tissue repair by a molecular control over expansion and differentiation of stem/progenitor cells [2,3].
In the context of the skeletal system, we and others have recently defined a strategy to generate bone tissue by instructing human expanded bone marrow-derived mesenchymal stromal/stem cells (hBM-MSC) toward the endochondral route [4,5]. The process exemplifies a developmental engineering paradigm in that it recapitulates the temporal sequence of events occurring during limb development, namely (i) cellular condensation, chondrogenesis, and hypertrophic differentiation, (ii) formation of a bony collar, (iii) matrix remodeling, (iv) vascularization, (v) bone matrix deposition over the resorbed cartilaginous template, and (vi) formation of a complete bone organ, including hematopoietic elements [6,7]. Upon chondrogenic and hypertrophic activation, the endochondral process proceeds in a semi-autonomous and self-regulated manner, comparable to normal embryonic development and leading to efficient hBM-MSC induction toward the osteoblastic lineage [4].
However, one important process not recapitulated yet is the initial 3D expansion of the generated mesenchyme prior to the endochondral commitment of hBM-MSC aggregates. Molecular regulation of this process could lead to enhanced formation of hypertrophic cartilage, and to the generation of structures with higher and more controlled degree of spatial order. In this study, we thus focused on the first events occurring during limb development, namely cell condensation and expansion of an undifferentiated mesenchymal cell pool, while maintaining as a target read-out of the 3D system the capacity to generate a cartilage anlage.
Developmental genetic models suggest Wnt and fibroblast growth factor (FGF) pathways as key regulators during mesenchymal cell growth [8,9]. Wnt canonical pathway activation regulates the segregation of skeletal stem cells from the undifferentiated mesenchyme while supporting their expansion. Wnt exposure maintains mesenchymal cells in a proliferative state, while the exit from the signal, along the elongation of the tissue (ie, increase in distance from Wnt source), allows for the chondrogenic differentiation to occur [2]. Also FGFs have key instructive roles in mesenchymal cell proliferation and lineage specification [10], and in skeletal patterning [11] during the first phases of limb development.
Previous studies using 2D culture models indicate that the combination of Wnt3a and FGFs signals strongly promotes the growth of chicken and mouse limb progenitor cells, while maintaining their undifferentiated state and multilineage differentiation capacity [9]. Inoue et al. [12] also reported the same synergistic effect of the two factors on proliferation of murine adult retinal stem/progenitor cells. The role of Wnt-canonical [13–15] and FGF [16,17] pathway activation has been largely investigated for hBM-MSC proliferation and maintenance of multipotency, but to the best of our knowledge only in plastic dishes, where the feature of 3D mesenchyme expansion cannot be recapitulated.
In analogy with limb bud development, we here hypothesized that Wnt3a and FGF2 induce hBM-MSC to expand in a 3D culture system as an undifferentiated pool of progenitors capable to produce clonogenic strains, maintaining the expression of typical mesenchymal stromal markers and competent to undergo chondrogenesis. We thus investigated whether supplementation of Wnt3a and/or FGF2 during the early phase of 3D culture of hBM-MSC modulates the extent of cell proliferation/division, phenotype, clonogenicity, and cartilage formation capacity.
Materials and Methods
All reagents were purchased from Gibco (Life Technologies) if not otherwise indicated.
Cell isolation and expansion
Human mesenchymal stem/stromal cells cultures derived from bone marrow aspirates (hBM-MSC, n=10, age: 34.9±3.3. male:female=7:3) were established as already described [18]. hBM-MSC were expanded for about 7 doublings (average from n=10 donors) in alpha-MEM containing 10% fetal bovine serum (FBS), 4.5 mg/mL D-glucose, 0.1 mM nonessential amino acids, 1 mM sodium pyruvate, 100 mM HEPES buffer, 100 UI/mL penicillin, 100 μg/mL streptomycin, and 0.29 mg/mL L-glutamate (complete medium), further supplemented with 5 ng/mL FGF2 (R&D) to maintain an undifferentiated state [19].
3D pellet cultures
To establish 3D cultures, aliquots of 2.5×105 cells/0.25 mL hBM-MSC were centrifuged at 1,100 rpm for 5 min in 1.5 mL polypropylene conical tubes (Sarstedt) to form pellets. Pellets were cultured for 1 week in DMEM with 2% of FBS [20], 1 mM sodium pyruvate, 100 mM HEPES buffer, 100 UI/mL penicillin, 100 μg/mL streptomycin, 0.29 mg/mL L-glutamate, 0.1 mM ascorbic acid 2-phosphate (Sigma), with or without 20 ng/mL rhWnt3a (wingless-type MMTV integration site family member3a; R&D), 100 ng/mL rhDkk1 (Dickkopf 1; R&D), and 5 ng/mL rhFGF2 during the first 3 or 7 days. The use of 2% FBS, resembling the extracellular matrix components present in the environment of limb development, was selected based on preliminary experiments, indicating that this minimal amount of serum (i) supports cell survival in 3D without impairing later chondrogenesis, (ii) does not reduce the DNA content as compared to 10% FBS and (iii) does not mask the effect of the tested morphogens. For chondrogenic differentiation, pellets were cultured for two additional weeks in serum-free medium supplemented with 10−7 M dexamethasone and 10 ng/mL transforming growth factor-β1 (TGF-β1; R&D). Media were changed twice a week.
Clonogenicity tests
To determine the colony forming efficiency following 3D culture, pellets were digested in 0.05% Trypsin-EDTA 1× at 37 °C for 15 min to obtain a single cell suspension. hBM-MSC were then plated in Petri dishes at a density of 200 cells/cm2 and cultured in complete medium without any growth factor for 15 days, with medium changes twice a week. Afterward, cells were washed with phosphate-buffered saline (PBS), fixed in PFA 4%, and stained with Crystal Violet (Sigma) for 10 min.
CD146 sorting
Either passaged hBM-MSC or cells upon 3D pellet culture were washed and resuspended in a solution containing PBS pH 7.2, 0.5% bovine serum albumin (BSA), and 2 mM EDTA. At this stage, cells were magnetically labeled and then sorted using a CD146 MicroBead Kit (Milteny Biotech) following the manufacturer's protocol (Large Columns; Milteny Biotech). Unsorted or positively selected (CD146+) hBM-MSCs were then used for following experiments.
Real-time polymerase chain reaction
Total RNA was extracted from pellets using TRIzol® (Life Technologies) and cDNA was generated as previously described [21]. The PCR master mix was based on AmpliTaq Gold DNA polymerase (Perkin Elmer/Applied Biosystems). TaqMan® Gene Expression or on-Demand assays (Life Technologies) were used on a ABI 7900 Fast Real-time PCR System (Life Technologies) for 40 cycles to measure gene expression of Axin2 (Hs00610344_m1), CyclinD1 (Hs00765553_m1), p21 (Hs00355782_m1), Dkk1 (Hs00183740_m1), Ki67 (Hs01032443_m1), and Sox9 (Hs00165814_m1), BMP4 (Hs00191626_m1), IHH (Hs01081800_m1), Col2a1 (Hs00264051_m1), Col10a1 (Hs00166657_m1) using GAPDH (Hs99999905_m1) as the housekeeping gene.
Cell proliferation assays
To measure the number of hBM-MSC in the S-phase of the cell cycle a 5-ethynyl-2′-deoxyuridine (Edu)-based assay was used according to the manufacturer's protocol (Molecular Probes, Life Technologies). Briefly, 10 μM Edu was added to the pellet cultures for 24 h at 48 h and 6 days; the samples were then fixed and incorporation of Edu and quantification of Edu+ cells were determined via flow cytometry or immunofluorescence staining, as described below.
To assess mitotic divisions, a Cell Proliferation Kit based on carboxyfluorescein diacetate succinimidyl ester (CFSE) (Molecular Probes) was used according to the manufacturer's recommendations. Briefly, before 3D aggregate preparation, cell suspension was incubated with 1 μM CFSE in PBS at 37°C for 10 min in the dark. Unspecific bindings were blocked by incubating the cells with human serum albumin for 5 min at 37°C. Cells were then washed thrice with PBS before starting the 3D pellet culture. Some samples were treated with colchicine (100 ng/mL; Invitrogen, Life Technologies) to inhibit cell division and thus providing a negative control for the further quantification analysis. At specific time points, the 3D aggregates were analyzed by flow cytometry.
High-throughput microscopy
hBM-MSC (n=2 donor) were cultured in monolayer (2D) in a 96-well plate at 3,000 cells/cm2 for 72 h and exposed to different concentrations of Wnt3a (5–40 ng/mL), FGF2 (1–20 ng/mL), or their combination. Two hours before stopping the experiment, 10 μM Edu was added to the wells. The cells were then stained according to the manufacturer's protocol, using DAPI as counterstaining. Afterward, the 96-well plate was read by using the Operetta High Throughput Imaging System (Perkin Elmer). A total of 45 fields/well were acquired with a 20× magnification objective. The total number of cells and the% of EDU+ cells (n=135, 45 fields per well, and three wells per condition) were calculated by means of the Columbus Image Data Storage and Analysis System (Perkin Elmer).
Glycosaminoglycans and DNA quantification
Pellets were digested with 1 mg/mL protease K in 50 mM Tris with 1 mM EDTA, 1 mM iodoacetamide, and 10 mg/mL pepstatin-A for 15 h at 56°C. Glycosaminoglycans (GAG) amount was spectrophotometrically measured using 1,9-dimethylmethylene blue chloride dye [22]. Results were normalized to DNA levels, which were assessed by CyQuant® Cell Proliferation Assay Kit (Molecular Probes).
Histology, immunohistochemistry, and immunofluorescence
Pellets after in vitro cultures were fixed in 4% paraformaldehyde, dehydrated, and embedded in paraffin. Five micrometer sections were cut by means of a Microm HM400 microtome. GAG accumulation, cell morphology, and matrix organization was evaluated with safranin-O staining (Fluka, Sigma) with hematoxylin counterstaining. Immunohistochemical analysis was performed to detect the CD146 and collagen type II expression. Upon rehydration in ethanol series, sections were digested using respectively heat-mediated antigen retrieval step (95°C for 30 min) and enzymatic digestion (with hyaluronidasae and pronase) [23]. The immunobinding was detected with biotinylated secondary antibodies and by Vectastatin ABC kit (Dako). The red signal was developed with Fast red kit (Dako) according to the manufacturer's instructions, with hematoxylin as counterstaining. Negative controls were included for each analysis, omitting the primary antibodies.
Immunofluorescence for TUNEL and Edu staining was performed on sections of 3D samples following the manufacturer's indications (Molecular Probe, Life Technologies), using Hoechst as counterstaining. Histological, immunohistochemical and immunofluorescence sections were analyzed by means of an Olympus BX-61 microscope. The percentage of Edu+ cells over the total number of Hoechst+ cells was calculated by means of ImageJ software (NIH).
Cytofluorimetric analysis
Pellets were enzymatically treated by 0.05% Trypsin-EDTA 1× for 15 min at 37°C to obtain a single cell suspension (modified from Ref. [4]). The harvested cells were then washed, maintained in 1% BSA for 10 min at room temperature, and then incubated in the dark at 4°C for 15 min with a fluorochrome-conjugated antibody for CD90, CD105, CD271, and CD146 (Becton Dickinson) or the correspondent IgG control. For the intracellular staining, hBM-MSC were fixed and permeabilized by incubation in Flow Cytometry Fixation/Permeabilization Buffer I (R&D) for 30 min at 4°C. Unphoshorylated β-catenin was detected by incubating the cells with 10 μg/mL of Mouse IgG1 anti-active β-catenin (Millipore) for 30 min at 4°C. An isotype control was added by incubating cells with 10 μg/mL of Mouse IgG1 anti-CD1c (R&D). Finally, cells were resuspended in a Rat Anti-mouse IgG1 FITC-conjugated secondary antibody solution for 15 min at 4°C. Edu+ cells were detected by using Alexa Fluor 647-conjugated Click-iT Edu Flow Cytometry Assay Kit® (Molecular Probes), according to the manufacturer's instruction. Cells were finally analyzed using an FACSCalibur flow cytometer (Becton Dickinson). FlowJo Proliferation Platform (Treestar) was used to analyze the results of the cell division assay with CFSE stained cells.
Statistical analysis
Data are presented as mean±standard deviation. The number of donors used is specified for each experiment. For each donor and experimental condition, at least triplicate samples were used for each assessment. Statistical analysis was performed using ANOVA, followed by unpaired or nonparametric t-tests (after assessing the normality of distribution of the collected data), by means of Prism® software (GraphPad Software).
Results
Selection of Wnt3a and FGF2 doses
Monolayer expanded hBM-MSC (n=2 donors) were exposed to different doses and combinations of Wnt3a (5–40 ng/mL) and FGF2 (1–20 ng/mL). Both factors (alone or in combination) positively influenced cell proliferation in a dose-dependent manner (data not shown) and were most effective at concentrations of 20 and 5 ng/mL respectively, which were thus selected for subsequent 3D experiments.
Activation of Wnt signaling in 3D hBM-MSC cultures
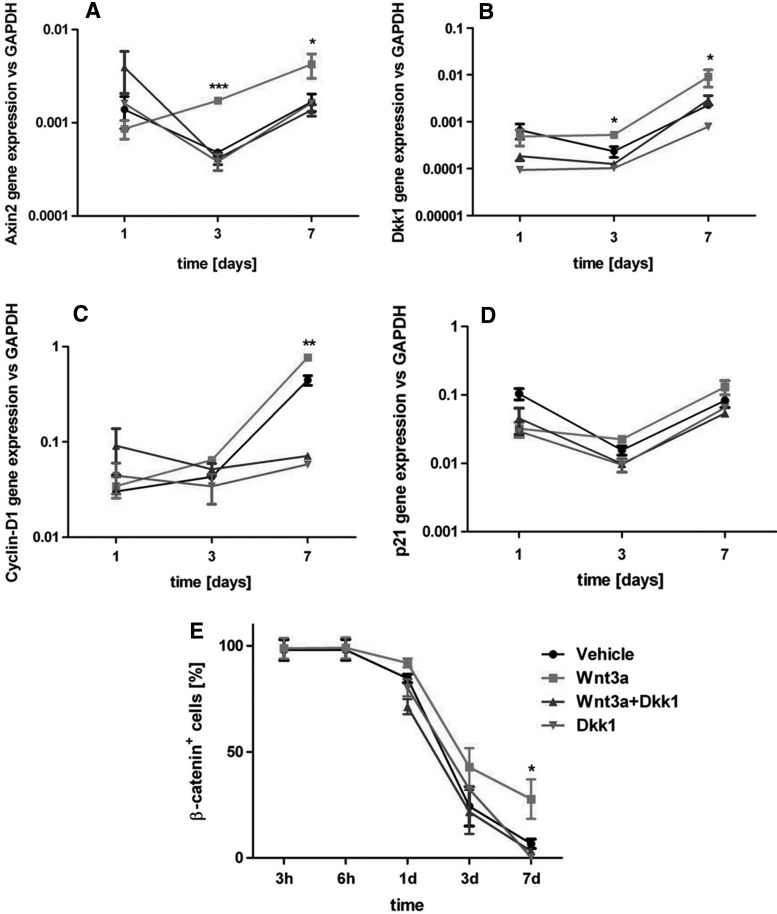
hBM-MSC (n=6 donors) were cultured as 3D pellets and exposed to 20 ng/mL of Wnt3a for up to 7 days. Figure 1 shows the temporal expression profiles of genes involved in the Wnt pathway, such as Axin2 (Fig. 1A) and Dkk1 (Fig. 1B), and in the cell cycle progression, namely Cyclin-D1 (Fig. 1C) and p21 (Fig. 1D). Wnt-stimulated hBM-MSC increased overtime relative expression of all the genes but p21, with statistically significant differences from vehicle starting from day 3, thus indicating the Wnt pathway activation and entering into the cell cycle.
FIG. 1.
hBM-MSC responsiveness to Wnt signaling in 3D. (A–D) Gene expression profile for (A) Axin2, (B) Dkk1, (C) Cyclin-D1, and (D) p21. (E) Expression overtime of the unphosphorylated form of β-catenin assessed via FACS and quantified as the percentage of β-catenin+ cells respect to the total number of cells. Statistical significance between the Wnt3a group and all other conditions at the corresponding time point: *P<0.05, **P<0.01, ***P<0.001. hBM-MSC, human bone marrow-derived mesenchymal stromal/stem cells.
We then assessed the presence of the unphosphorylated form of β-catenin, which, after Wnt canonical pathway activation, is known to accumulate into the nucleus [24]. Upon 3D condensation in pellets, β-catenin was spontaneously expressed by almost the totality of hBM-MSC (Fig. 1E), whereas it started to decrease 24 h after the pellet formation. Starting from day 3, a higher amount of active β-catenin could be detected in the Wnt3a group, with a statistically significant difference after 7 days of stimulation (Fig. 1E). All the effects of Wnt3a discussed above were totally blocked by the addition of Dkk1, a known Wnt pathway inhibitor, ultimately demonstrating the specificity of the effect of Wnt3a (Fig. 1A–E).
Effect of Wnt3a and FGF2 on hBM-MSC proliferation in 3D culture
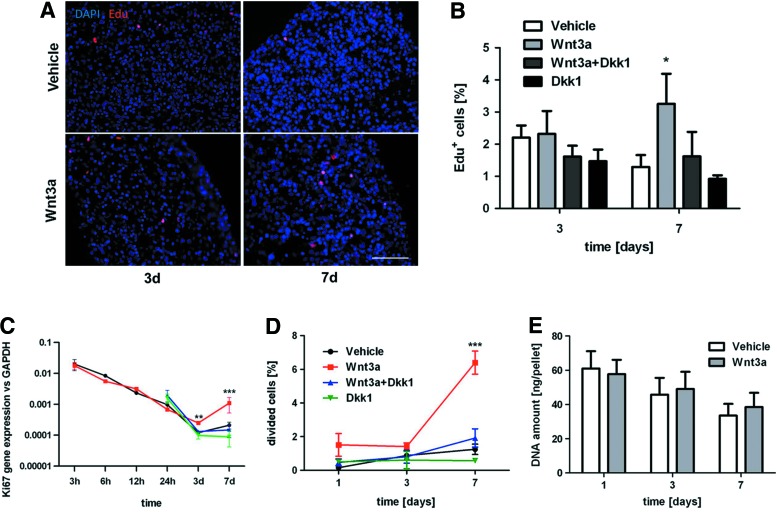
The effect of Wnt3a on the proliferation of hBM-MSC in a 3D pellet culture (n=3 donors) was tested with different readouts. Figure 2A shows a representative picture of an Edu incorporation assay, indicating the scattered presence of Edu+, proliferating cells in the micromass. A significantly higher percentage of Edu+ cells was found at day 7 in the Wnt3a-treated group (Fig. 2B), in line with the established Wnt pathway activation timing (Fig. 1). These results were consistent with the Ki67 gene expression trend (Fig. 2C) and with the percentage of cells that underwent division (6.40%±0.69% divided cells in Wnt3a-treated group compared to 1.25%±0.31% in vehicle condition at day 7) calculated by CFSE incorporation (Fig. 2D). As described, relevant differences in terms of proliferation-related gene expression and cell number were seen starting from day 3, after the lag phase frequently described for 3D cellular culture and caused by cell recovery or cell loss in suspension [25]. The specificity of hBM-MSC response to the Wnt canonical pathway was further confirmed by the blocking effect obtained with the addition of Dkk1 in the 3D cultures.
FIG. 2.
Wnt3a-stimulated hBM-MSC 3D proliferation. (A) Immunofluorescence for Edu at 3 and 7 days of hBM-MSC stimulated or not with Wnt3a. Scale bar: 100 μm. (B) Edu+ hBM-MSC quantification, calculated as the percentage of Edu+ cells with respect to the total number of Hoechst+ cells. (C) Ki67 gene expression profile overtime up to 7 days. (D) Percentage of cells that underwent division, calculated by carboxyfluorescein diacetate succinimidyl ester assay. (E) DNA quantification at 1, 3, and 7 days of pellets stimulated or not with Wnt3a. Statistical significance between the Wnt3a group and all other conditions at the corresponding time point: *P<0.05, **P<0.01, ***P<0.001. Color images available online at www.liebertpub.com/scd
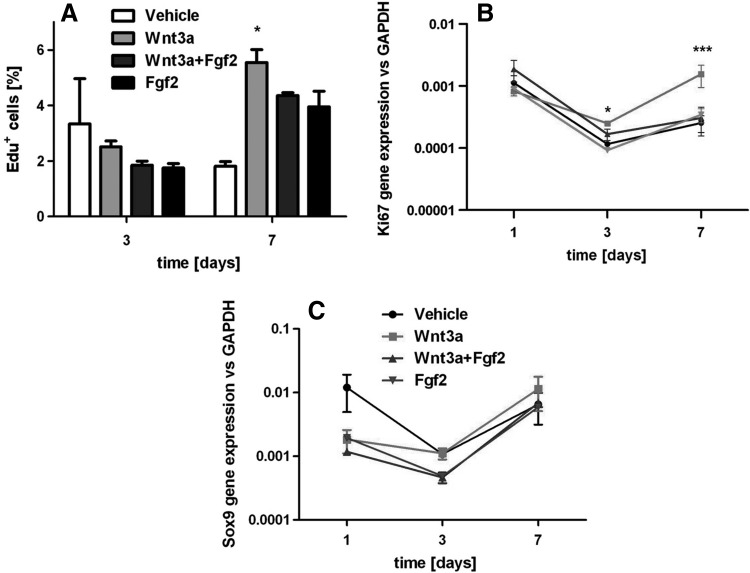
Importantly, the 3D pellets displayed an overtime decrease in the DNA amount both in the vehicle and Wnt3a experimental conditions (Fig. 2E), in line with a reduction (respectively 61.8%±1.6% and 52.9%±2.9%) of the pellet diameter from day 1 to 7. Thus, Wnt3a was able to enhance hBM-MSC proliferation in 3D cultures, though to a relatively limited extent and not resulting in a macroscopic increase in pellet size. Massive cell death caused by apoptosis could be excluded as not detected within the cellular aggregates by immunofluorescence upon TUNEL Assay (data not shown). The supplementation of FGF2 in the 3D culture system (n=3 donors) not only had no effect on the percentage of Edu+ proliferating cells (Fig. 3A) but also antagonized the effect of Wnt3a, as displayed by the Ki67 gene expression profile (Fig. 3B).
FIG. 3.
(A, B) FGF2 does not have a synergistic effect on hBM-MSC 3D proliferation. (A) Edu+ hBM-MSC quantification, calculated as the percentage of Edu+ cells with respect to the total number of Hoechst+ cells starting. (B) Ki67 gene expression profile overtime up to 7 days. (C) Sox9 gene expression after 7 days of treatment with Wnt3a and/or FGF2. Statistical significance between the Wnt3a group and all other conditions at the corresponding time point: *P<0.05, ***P<0.001. FGF2, fibroblast growth factor 2.
Chondrogenesis of hBM-MSC following Wnt3a removal
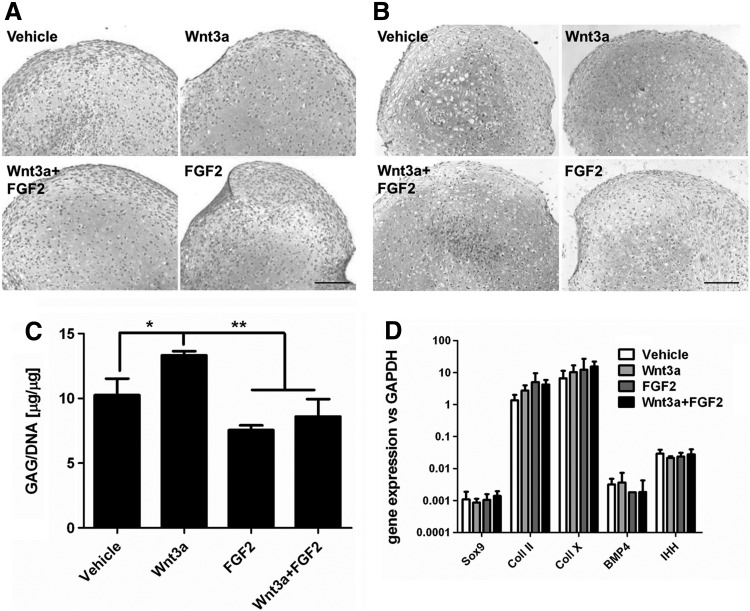
The chondrogenic potential of hBM-MSC in the 3D culture system after Wnt3a and/or FGF2 exposure was evaluated to verify their capacity to progress toward the formation of the cartilage anlage (n=4 donors). Wnt3a and/or FGF2 preconditioning did not modulate the expression of Sox9 (Fig. 3C). When TGF-β-mediated chondrogenesis was induced, Wnt3a preconditioned samples deposited a higher quality of cartilaginous matrix than the other groups, as indicated by increased Safranin-O and collagen type II staining intensity and confirmed by an increase in GAG deposition ranging from 12.4% to 51.8% (average 29.9%±3.8%) (Fig. 4A–C). However, no significant differences were found in the expression profile of typical chondrogenic genes (Fig. 4D). In the absence of TGF-β1 supplementation, chondrogenesis did not occur in any of the conditions. Noteworthy, the addition of FGF2 antagonized the beneficial effect of Wnt3a (Fig. 4A–C).
FIG. 4.
Chondrogenesis upon Wnt3a and/or FGF2 removal. (A) SafraninO staining. Scale bar: 100 μm. (B) Immunohistochemistry for collagen type II. Scale bar: 100 μm. (C) Quantification of GAG/DNA ratio of cartilaginous pellets. (D) Gene expression profile for Sox9, Coll II, and Coll X, BMP4, and Indian Hedgehog (IHH). *P<0.05, **P<0.01. GAG, glycosaminoglycans; Coll II, collagen type II; Coll X, collagen type X; BMP, bone morphogenetic protein.
Response to Wnt3a by CD146+ hBM-MSC
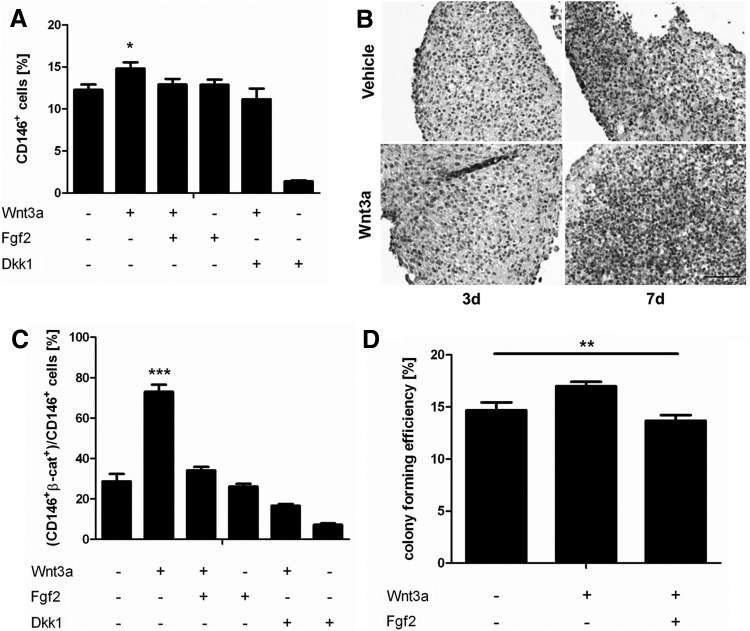
Several typical MSC markers were cytofluorimetrically tracked along Wnt and FGF stimulation in the 3D culture system (n=4 donors). No differences among the experimental groups were detected in the expression of typical mesenchymal markers, including CD90, CD105, and CD271 [26] (data not shown). Instead, the percentage of hBM-MSC expressing CD146, a putative marker for early, self-renewing mesenchymal progenitors [27–29] was significantly increased after 7 days of stimulation by Wnt3a with respect to both vehicle and FGF2 conditions (n=5) (Fig. 5A). The increase was blocked by the addition of FGF2 or by the inhibition of the Wnt canonical pathway with Dkk1. CD146+ cells were uniformly distributed over the 3D constructs (Fig. 5B). To further correlate CD146+ cells with Wnt-responding cells, cells dissociated after 7 days of pellet culture were cytofluorimetrically assessed by double staining for CD146/β-catenin. Figure 5C displays that 60.9% of the CD146+ cells were also positive for active β-catenin in the Wnt3a-treated group, with a 2.3-fold increase with respect to the vehicle condition. This difference resulted in a statistically significant higher clonogenicity (n=4) of the dissociated cells stimulated with Wnt3a (Fig. 5D).
FIG. 5.
Wnt3a-mediated CD146 modulation in hBM-MSC. CD146 expression for 3D pellets stimulated with Wnt3a and/or FGF2 assessed via (A) FACS and (B) immunohistochemistry. Scale bar: 100 μm. (C) Quantification via FACS of CD146/β-catenin double-positive hBM-MSC normalized with respect to the total number of CD146+ cells. (D) Percentage of clonogenic cells upon Wnt3a and/or FGF2 3D stimulation. Statistical significance between the Wnt3a group and all other conditions: *P<0.05, **P<0.01, ***P<0.001.
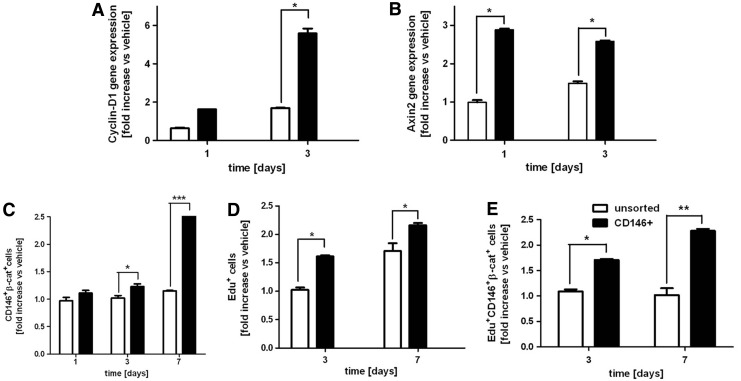
At this stage, we hypothesized that the CD146-enriched population could be more responsive to Wnt-canonical pathway activation. To test this hypothesis, unsorted or CD146 positively-sorted hBM-MSC (three donors, at least triplicate for each experimental condition) were cultured in 3D pellets up to 1 week with or without the addition of Wnt3a. Figure 6 shows that Wnt3a-stimulated CD146+ hBM-MSC activated both CyclinD1 and Axin2 gene expression faster and to a higher extent as compared to the unsorted population (Fig. 6A, B). Moreover, a 1.3-fold higher number of CD146/β-catenin double-positive cells were generated starting from day 3 with respect to the unsorted population (Fig. 6C). Moreover, Wnt3a stimulation also increased the number of Edu+ proliferating cells in the CD146-positively sorted population (Fig. 6D). This effect was even more evident when referred to the responding subpopulation of CD146+/β-catenin+ hBM-MSC, as shown in Fig. 6E. Taken together, these data suggest that the expression of CD146 correlates with responsiveness to Wnt canonical pathway activation.
FIG. 6.
(A–C) CD146-enriched hBM-MSC responsiveness to Wnt3a. (A, B) Gene expression profile of Wnt3a-stimulated CD146+ and unsorted hBM-MSC populations for (A) CyclinD1 and (B) Axin2 gene expression. (C) Quantification via FACS of CD146/β-catenin double-positive cells for CD146+ and unsorted hBM-MSC populations. (D, E) Wnt3a-stimulated CD146+ hBM-MSC 3D proliferation. (D) Percentage of Edu+ proliferating cells with respect to the total number of cells in the CD146-positively sorted and unsorted populations. (E) Quantification via FACS of CD146+/β-catenin+/Edu+ hBM-MSC. *P<0.05, **P<0.01, ***P<0.001.
Discussion
In this study, we demonstrated for the first time (i) the responsiveness of expanded hBM-MSC to Wnt3a exposure in a 3D culture model, (ii) the increase in the fraction of proliferating cells by Wnt canonical pathway activation in the 3D cellular constructs, and (iii) the enhanced in vitro chondrogenesis of Wnt3a stimulated cells upon switch to a chondrogenic medium including TGF-β1. Interestingly, the CD146+ clonogenic subpopulation was more responsive to Wnt pathway activation and more prompt to exert a Wnt3a-mediated response in terms of 3D proliferation.
Wnt3a was previously reported to enhance bone morphogenetic protein 2 (BMP2)-mediated chondrogenesis in a multipotent mesenchymal cell line [30], and to increase the proliferation rate [14,15] of hBM-MSC, expanding a clonogenic subpopulation. However, these studies have been performed using typical 2D culture models, which (i) expose hBM-MSC to an infinitely stiff, nonphysiological substrate [31] and (ii) are not permissive to chondrogenesis, unless cells are then transferred to a pellet culture system [32]. Our findings confirmed that also in a 3D culture setting, relevant to mimic the developmental steps of mesenchyme condensation and growth, Wnt3a represents a pivotal cue to regulate the proliferation and differentiation capacity of hBM-MSC. On the other hand, the model allowed to identify so far unaddressed limits and challenges.
In fact, despite the responsiveness of hBM-MSC to Wnt3a and the associated increase in the fraction of proliferating cells, the resulting 3D construct did not increase in size or cell number, in contrast to what is observed for the expansion of condensating mesenchyme during limb development. This seems to be different from other stem cell-based models of organogenesis (ie, optic cup and gut crypt), where self-organization of 3D morphogenesis has been successfully recapitulated in vitro [33,34]. The finding could be related to some features of the 3D culture model, to the lack of other key signaling ligands, and/or to the intrinsic biological difference between expanded hBM-MSC and mesenchymal limb progenitors, as discussed in the following paragraphs.
The use of a suitable 3D culture system is fundamental to allow cells to respond to environmental/signaling cues. In our experimental model, the condensation induced in 3D pellets might have negatively influenced cell expansion by imposing too tight cell-to-cell contacts, possibly interfering with cell proliferation. For example, cell density may have resulted into a construct elasticity not matching the corresponding native tissue, thereby disregulating cell proliferation and fate determination programs [35]. Moreover, the uniform delivery of Wnt3a and FGF2 in the medium, as opposed to the time-dependent establishment of spatial gradients during limb development, might have determined an uncontrolled morphogen distribution within the developing tissue, resulting into a heterogeneous and suboptimal cell response. Further studies might thus need to target the combined use of “spacer” scaffolds (eg, hydrogels) with morphogen delivery from a spatially confined source (eg, microbead).
Alternatively, 3D tissue expansion of hBM-MSC might require additional signaling factors. Contrary to murine and chick models [8,9] and to the observed results obtained with hBM-MSC in monolayer (data not shown) where FGFs synergistically act with Wnt pathway, in our 3D culture conditions FGF2 seemed either to antagonize or to have no additive effect with the Wnt3a. On one hand, the 3D environment may play a role in differential responsiveness toward supplemented factors [36]. On the other hand, as suggested by Oldershaw and colleagues [37], the expansion of mesenchymal progenitors densely packed in 3D nodules, which precedes the complete chondrogenic differentiation of human embryonic stem cells, requires the stimulation by Wnt3a and FGF2, along with Activin-A and/or BMP4. Thus, next investigations might include selected members of the TGF-β superfamily in the cocktail of growth factors to guarantee higher 3D cell survival and expansion and to prime hBM-MSC toward a chondrogenic fate.
Our findings may also imply that hBM-MSC markedly differ from limb mesenchymal cells, so that their expansion and chondrogenesis cannot be instructed by directly recapitulating developmental cues. Wnt ligands are known to activate cell entry into the cell cycle in a way that depends on the state of commitment of the responder population [38] and that is based on the balance between Sox9 and Cbfa1 [2,39]. The constitutively high levels of expression of these key transcriptional factors in expanded hBM-MSC [40] suggests a more restricted cell commitment than in mesenchymal limb progenitors and consequently the limited 3D growth in response to Wnt signaling. Accordingly, it is conceivable that only early progenitors within the heterogeneous hBM-MSC population would similarly respond to embryonic mesenchyme upon Wnt3a stimulation. In support of this hypothesis, we found that a CD146-positive subpopulation was more consistently and robustly activated by Wnt3a. Interestingly, despite the plethora of features associated with hBM-MSC, the general consensus associates CD146 with bona fide stem cells, able to self-renew and undergo multilineage differentiation [28,29,41]. Our results are in line with a key principle proposed by 3D organogenesis pioneers [33,34], namely that working with a defined homogenous cell population is pivotal to achieve control over signaling pathways and finally tissue morphogenesis. Despite the preliminary nature of our descriptive observations, our findings would prompt for further investigations, possibly at clonal level [42], aimed at correlating the potential to manipulate the fate of hBM-MSC by Wnt signaling with the stage of cell differentiation and associated marker expression.
Conclusions
We have demonstrated that hBM-MSC could respond to key morphoregulatory factors (ie, Wnt3a) mimicking the first events of endochondral route, but to a limited extent. This work represents an additional proof of principle that certain limb development paradigms can be recapitulated with adult cells, though with significant differences. The identification of crucial nodes to control and manipulate hBM-MSC fate and, ultimately, engineer self-regulated cellular processes will be an important milestone for advances in skeletal tissue regeneration and in turn will offer invaluable human cellular models to developmental biologists.
Acknowledgment
This work was partially funded by the Swiss National Science Foundation program “Sinergia” (SNF grant no. CRSII3_136179/1).
Author Disclosure Statement
The authors declare no conflict of interest.
References
- 1.Lenas P. Moos M. Luyten FP. Developmental engineering: a new paradigm for the design and manufacturing of cell-based products. Part I: from three-dimensional cell growth to biomimetics of in vivo development. Tissue Eng Part B Rev. 2009;15:381–394. doi: 10.1089/ten.TEB.2008.0575. [DOI] [PubMed] [Google Scholar]
- 2.Leucht P. Minear S. Ten Berge D. Nusse R. Helms JA. Translating insights from development into regenerative medicine: the function of Wnts in bone biology. Semin Cell Dev Biol. 2008;19:434–443. doi: 10.1016/j.semcdb.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 3.Abo A. Clevers H. Modulating WNT receptor turnover for tissue repair. Nat Biotechnol. 2012;30:835–836. doi: 10.1038/nbt.2361. [DOI] [PubMed] [Google Scholar]
- 4.Scotti C. Tonnarelli B. Papadimitropoulos A. Scherberich A. Schaeren S. Schauerte A. Lopez-Rios J. Zeller R. Barbero A. Martin I. Recapitulation of endochondral bone formation using human adult mesenchymal stem cells as a paradigm for developmental engineering. Proc Natl Acad Sci U S A. 2010;107:7251–7256. doi: 10.1073/pnas.1000302107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jukes JM. Both SK. Leusink A. Sterk LM. van Blitterswijk CA. de Boer J. Endochondral bone tissue engineering using embryonic stem cells. Proc Natl Acad Sci U S A. 2008;105:6840–6845. doi: 10.1073/pnas.0711662105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kronenberg HM. Developmental regulation of the growth plate. Nature. 2003;423:332–336. doi: 10.1038/nature01657. [DOI] [PubMed] [Google Scholar]
- 7.Scotti C. Piccinini E. Takizawa H. Todorov A. Bourgine P. Papadimitropoulos A. Barbero A. Manz MG. Martin I. Engineering of a functional bone organ through endochondral ossification. Proc Natl Acad Sci U S A. 2013;110:3997–4002. doi: 10.1073/pnas.1220108110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maruyama T. Deng AJ CX. Hsu W. The balance of WNT and FGF signaling influences mesenchymal stem cell fate during skeletal development. Sci Signal. 2010;3:ra40. doi: 10.1126/scisignal.2000727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.ten Berge D. Brugmann SA. Helms JA. Nusse R. Wnt and FGF signals interact to coordinate growth with cell fate specification during limb development. Development. 2008;135:3247–3257. doi: 10.1242/dev.023176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benazet JD. Bischofberger M. Tiecke E. Gonçalves A. Martin JF. Zuniga A. Naef F. Zeller R. A self-regulatory system of interlinked signaling feedback loops controls mouse limb patterning. Science. 2009;323:1050–1053. doi: 10.1126/science.1168755. [DOI] [PubMed] [Google Scholar]
- 11.Yu K. Ornitz DM. FGF signaling regulates mesenchymal differentiation and skeletal patterning along the limb bud proximodistal axis. Development. 2008;135:483–491. doi: 10.1242/dev.013268. [DOI] [PubMed] [Google Scholar]
- 12.Inoue T. Kagawa T. Fukushima M. Shimizu T. Yoshinaga Y. Takada S. Tanihara H. Taga T. Activation of canonical Wnt pathway promotes proliferation of retinal stem cells derived from adult mouse ciliary margin. Stem Cells. 2006;24:95–104. doi: 10.1634/stemcells.2005-0124. [DOI] [PubMed] [Google Scholar]
- 13.Etheridge SL. Spencer GJ. Heath DJ. Genever PG. Expression profiling and functional analysis of Wnt signaling mechanisms in mesenchymal stem cells. Stem Cells. 2004;22:849–860. doi: 10.1634/stemcells.22-5-849. [DOI] [PubMed] [Google Scholar]
- 14.Boland GM. Perkins G. Hall DJ. Tuan RS. Wnt3a promotes proliferation and suppresses osteogenic differentiation of adult human mesenchymal stem cells. J Cell Biochem. 2004;93:1210–1230. doi: 10.1002/jcb.20284. [DOI] [PubMed] [Google Scholar]
- 15.Baksh D. Tuan RS. Canonical and non-canonical Wnts differentially affect the development potential of primary isolate of human bone marrow mesenchymal stem cells. J Cell Physiol. 2010;212:817–826. doi: 10.1002/jcp.21080. [DOI] [PubMed] [Google Scholar]
- 16.Di Maggio N. Mehrkens A. Papadimitropoulos A. Schaeren S. Heberer M. Banfi A. Martin I. Fibroblast growth factor-2 maintains a niche-dependent population of self-renewing highly potent non-adherent mesenchymal progenitors through FGFR2c. Stem Cells. 2012;30:1455–1464. doi: 10.1002/stem.1106. [DOI] [PubMed] [Google Scholar]
- 17.Coutu DL. François M. Galipeau J. Inhibition of cellular senescence by developmentally regulated FGF receptors in mesenchymal stem cells. Blood. 2011;117:6801–6812. doi: 10.1182/blood-2010-12-321539. [DOI] [PubMed] [Google Scholar]
- 18.Martin I. Muraglia A. Campanile G. Cancedda R. Quarto R. Fibroblast growth factor-2 supports ex vivo expansion and maintenance of osteogenic precursors from human bone marrow. Endocrinology. 1997;138:4456–4462. doi: 10.1210/endo.138.10.5425. [DOI] [PubMed] [Google Scholar]
- 19.Bianchi G. Banfi A. Mastrogiacomo M. Nozzato R. Luzzato L. Cancedda R. Quarto R. Ex vivo enrichment of mesenchymal cell progenitors by fibroblast growth factor 2. Exp Cell Res. 2003;287:98–105. doi: 10.1016/s0014-4827(03)00138-1. [DOI] [PubMed] [Google Scholar]
- 20.Sabatino MA. Santoro R. Gueven S. Jaquiery C. Wendt DJ. Martin I. Moretti M. Barbero A. Cartilage graft engineering by co-culturing primary human articular chondrocytes with human bone marrow stromal cells. J Tissue Eng Regen Med. 2012 doi: 10.1002/term.1661. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 21.Strobel S. Loparic M. Wendt D. Schenk AD. Candrian C. Lindberg RLP. Moldovan F. Barbero A. Martin I. Anabolic and catabolic responses of human articular chondrocytes to varying oxygen percentages. Arthritis Res Ther. 2010;12:R34. doi: 10.1186/ar2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farndale RW. Buttle DJ. Barrett AJ. Improved quantitation and discrimination of sulphated glycosaminoglycans by use of dimethylmethylene blue. Biochim Biophys Acta. 1986;883:173–177. doi: 10.1016/0304-4165(86)90306-5. [DOI] [PubMed] [Google Scholar]
- 23.Dickhut A. Pelttari K. Janicki P. Wagner W. Eckstein V. Egermann M. Richter W. Calcification or dedifferentiation: requirement to lock mesenchymal stem cells in a desired differentiation stage. J Cell Physiol. 2009;219:219–226. doi: 10.1002/jcp.21673. [DOI] [PubMed] [Google Scholar]
- 24.Ling L. Nurcombe V. Cool SM. Wnt signaling controls the fate of mesenchymal stem cells. Gene. 2009;433:1–7. doi: 10.1016/j.gene.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 25.Grayson WL. Zhao F. Izadpanah R. Bunnell B. Ma T. Effects of hypoxia on human mesenchymal stem cell expansion and plasticity in 3D constructs. J Cell Physiol. 2006;207:331–339. doi: 10.1002/jcp.20571. [DOI] [PubMed] [Google Scholar]
- 26.Churchman SM. Ponchel F. Boxall SA. Cuthbert R. Kouroupis D. Roshdy T. Giannoudis PV. Emery P. McGonagle D. Jones EA. Transcriptional profile of native CD271+ multipotential stromal cells: evidence for multiple fates, with prominent osteogenic and Wnt pathway signaling activity. Arthritis Rheum. 2012;64:2632–2643. doi: 10.1002/art.34434. [DOI] [PubMed] [Google Scholar]
- 27.Russell KC. Phinney DG. Lacey MR. Barrilleaux BL. Meyertholen KE. O'Connor KC. In vitro high-capacity assay to quantify the clonal heterogeneity in trilineage potential of mesenchymal stem cells reveals a complex hierarchy of lineage commitment. Stem Cells. 2010;28:788–798. doi: 10.1002/stem.312. [DOI] [PubMed] [Google Scholar]
- 28.Sacchetti B. Funari A. Michienzi S. Di Cesare S. Piersanti S. Saggio I. Tagliafico E. Ferrari S. Robey PG. Riminucci M. Bianco P. Self-renewing osteoprogenitors in bone marrow sinusoids can organize a hematopoietic microenvironment. Cell. 2007;131:324–336. doi: 10.1016/j.cell.2007.08.025. [DOI] [PubMed] [Google Scholar]
- 29.Bianco P. Gehron-Robey P. Simmons PJ. Mesenchymal stem cells: revisiting history, concepts, and assays. Cell Stem Cell. 2008;2:313–318. doi: 10.1016/j.stem.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fischer L. Boland G. Tuan RS. Wnt-3A enhances bone morphogenetic protein-2-mediated chondrogenesis of murine C3H10T1/2 mesenchymal cells. J Biol Chem. 2002;277:30870–30878. doi: 10.1074/jbc.M109330200. [DOI] [PubMed] [Google Scholar]
- 31.Engler AJ. Sen S. Sweeney HL. Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–89. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 32.Johnstone B. Hering TM. Caplan AI. Goldberg VM. Yoo JU. In vitro chondrogenesis of bone marrow-derived mesenchymal progenitor cells. Exp Cell Res. 1998;238:265–72. doi: 10.1006/excr.1997.3858. [DOI] [PubMed] [Google Scholar]
- 33.Sasai Y. Eiraku M. Suga H. In vitro organogenesis in three dimensions: self-organising stem cells. Development. 2012;139:4111–4121. doi: 10.1242/dev.079590. [DOI] [PubMed] [Google Scholar]
- 34.Sasai Y. Cytosystems dynamics in self-organization of tissue architecture. Nature. 2013;493:318–326. doi: 10.1038/nature11859. [DOI] [PubMed] [Google Scholar]
- 35.Gilbert PM. Havenstrite KL. Magnusson KE. Sacco A. Leonardi NA. Kraft P. Nguyen NK. Thrun S. Lutolf MP. Blau HM. Substrate elasticity regulates skeletal muscle stem cell self-renewal in culture. Science. 2010;329:1078–1081. doi: 10.1126/science.1191035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen X. Thibeault SL. Response of fibroblasts to transforming growth factor-β1 on two-dimensional and in three-dimensional hyaluronan hydrogels. Tissue Eng Part A. 2012;18:2528–2538. doi: 10.1089/ten.tea.2012.0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oldershaw RA. Baxter MA. Lowe ET. Bates N. Grady LM. Soncin F. Brison DR. Hardingham TE. Kimber SJ. Directed differentiation of human embryonic stem cells toward chondrocytes. Nat Biotechnol. 2011;28:1187–1194. doi: 10.1038/nbt.1683. [DOI] [PubMed] [Google Scholar]
- 38.Logan CY. Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 39.Day TF. Guo X. Garrett-Beal L. Yang Y. Wnt/beta-catenin signaling in mesenchymal progenitors controls osteoblast and chondrocyte differentiation during vertebrate skeletogenesis. Dev Cell. 2005;2005;8:739–750. doi: 10.1016/j.devcel.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 40.Delorme B. Ringe J. Pontikoglu C. Gaillard J. Langonnè A. Sensebè L. Noel D. Jorgensen C. Häupl T. Charbord P. Specific lineage-priming of bone marrow mesenchymal stem cells provides the molecular framework for their plasticity. Stem Cells. 2009;27:1142–1151. doi: 10.1002/stem.34. [DOI] [PubMed] [Google Scholar]
- 41.Bianco P. Back to the future: moving beyond “mesenchymal stem cells”. J Cell Biochem. 2011;112:1713–1721. doi: 10.1002/jcb.23103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bianco P. Cao X. Frenette PS. Mao JJ. Robey PG. Simmons PJ. Wang CY. The meaning, the sense and the significance: translating the science of mesenchymal stem cells into medicine. Nat Med. 2013;19:35–42. doi: 10.1038/nm.3028. [DOI] [PMC free article] [PubMed] [Google Scholar]