Abstract
Human immunodeficiency virus type-1 (HIV-1) reverse transcriptase (RT) has a unique tight binding to dNTP substrates. Structural modeling of Ala-114 of HIV-1 RT suggests that longer side chains at this residue can reduce the space normally occupied by the sugar moiety of an incoming dNTP. Indeed, mutations at Ala-114 decrease the ability of RT to synthesize DNA at low dNTP concentrations and reduce the dNTP-binding affinity (Kd) of RT. However, the Kd values of WT and A114C RT remained equivalent with an acylic dNTP substrate. Finally, mutant A114 RT HIV-1 vectors displayed a greatly reduced transduction in nondividing human lung fibroblasts (HLFs), while WT HIV-1 vector efficiently transduced both dividing and nondividing HLFs. Together these data support that the A114 residue of HIV-1 RT plays a key mechanistic role in the dNTP binding of HIV-1 RT and the unique viral infectivity of target cell types with low dNTP pools.
Keywords: HIV-1 RT, dNTP binding pocket, cell type specific replication
Introduction
Human immunodeficiency virus type-1 (HIV-1) belongs to the genus lentivirus in the family Retroviridae. Most retroviruses in this family, such as alpha, beta, gamma and deltaretroviruses are oncoretroviruses, which exclusively infect dividing cells. Lentiviruses however, productively infect both dividing CD4+ T cells and nondividing cells such as macrophages, which have been shown to act as long-term HIV-1 viral reservoirs (Igarashi et al., 2001; Lewis, Hensel, and Emerman, 1992; Lewis and Emerman, 1994; Weinberg et al., 1991). Due to this dual cellular tropism, the HIV-1 encoded DNA polymerase, reverse transcriptase (RT), must function in two distinct dNTP substrate environments. The first, activated CD4+T cells, have a cellular dNTP concentration around 1.4-2.5μM; the second, macrophages, have a much lower concentration of about ∼40nM (Diamond et al., 2004; Kennedy et al., 2010). Our earlier pre-steady-state kinetic analysis reported that HIV-1 RT has a high binding affinity to dNTPs with a Kd near 1μM, while the Kd value of murine leukemia virus (MuLV) RT was found to be approximately 40μM (Skasko et al., 2005; Weiss et al., 2004). We also observed that HIV-1 variants harboring RT mutants with a decreased dNTP binding affinity, failed to replicate in macrophages and cell types harboring low dNTP concentrations. However, these mutant RTs do not significantly alter the viral replication in cells containing high dNTP concentrations (Jamburuthugoda et al., 2008; Weiss, Bambara, and Kim, 2002). Therefore, these studies suggested that the tighter dNTP binding affinity of RT contributes to the unique replication capability of HIV-1 in nondividing cells containing limited dNTP pools.
The DNA polymerase active site of HIV-1 RT contains a series of residues that are responsible for the binding and/or incorporation of dNTPs onto the 3′ end of a primer (Harris et al., 1998). The residues A113, A114, Y115, Q151, and M184 are found near the 3′ OH of an incoming dNTP (Figure 1) and are all highly conserved in HIV-1 RT as well as other lentiviral RTs (Cases-González and Menéndez-Arias, 2005; Huang et al., 1998; Lowe et al., 1991; Pandey et al., 1996). Y115 (green) appears to play a key role as a gate, discriminating dNTPs from rNTPs (Boyer et al., 2000; Gao et al., 1997; Joyce, 1997; Martín-Hernández, Domingo, and Menéndez-Arias, 1996). Mutations at residue M184 (purple), which naturally contributes to mismatch selectivity, are frequently found in HIV-1 drug resistant patients (Menéndez-Arias, 2008; Pandey et al., 1996; Ray, Basavapathruni, and Anderson, 2002; Svedhem et al., 2007; Yang et al., 2008; Yoshimura et al., 1999). M184I, for example, is a transient 3TC resistant mutation, ultimately resulting in M184V (Frost et al., 2000; Mulder, Harari, and Simon, 2008; Svedhem et al., 2007). It was found that the beta branched side chain of isoleucine sterically blocks the entry of 3TC into the active site of RT (Sarafianos et al., 1999). We have shown that the M184I mutation also alters the binding of RT to dNTPs, raising the Kd to 56μM, which is similar to the Kd value of MuLV RT (Skasko et al., 2005). The M184I mutation was also found to change the tropism of an HIV-1 vector, preventing it from transducing human macrophages (Jamburuthugoda et al., 2008; Skasko et al., 2005). We observed a similar outcome with Q151N, a non-clinical RT mutation (light blue) (Diamond et al., 2004). The glutamine at 151 interacts with the 3′OH of an incoming dNTP through a hydrogen bond; however, when mutated to asparagine, the Kd of RT for substrate is raised to 257μM (Weiss, Bambara, and Kim, 2002). HIV-1 vectors carrying this mutation also fail to transduce primary human macrophages but maintain a significant level of transduction in cells with high dNTP pools. Principally, this work suggests that the manipulation of certain residues within the active site of RT can impact the dNTP binding affinity through different mechanisms, which result in altered HIV-1 cellular tropism (Diamond et al., 2004).
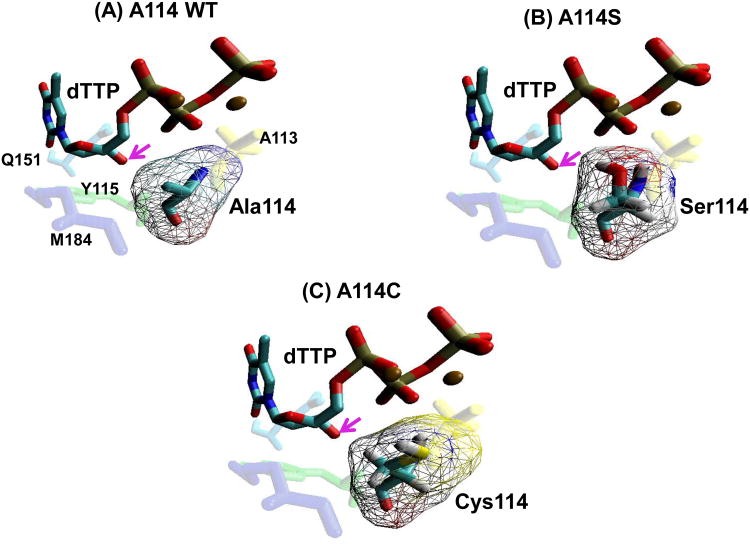
Figure 1. Structural modeling of the HIV-1 RT residue 114 mutants at the dNTP-binding pocket.
(A) The position of the HIV-1 RT A114 residue (teal) in relation to an incoming dTTP substrate (blue/red), and Y115 (green), A113 (yellow), M184 (purple) and Q151 (light blue), is shown using coordinate set 1rtd.pdb from the Protein Data Bank (Huang et al., 1998). (B) and (C) The positions of the S114 and C114 mutant residues were constructed by the program Pymol (Schrödinger) that manually selected the optimal rotamer for serine or cysteine at residue 114 of HIV-1 reverse transcriptase from Huang et-al 1999 (Huang et al., 1998). These were then minimized with AMBER 9.0 with 500 cycles (250 steepest descent) with implicit solvent. Arrows indicate the 3′ OH of the incoming dTTP. Volume of amino acids were calculated as described previously (Zamyatnin, 1984).
The A114 residue contributes a nonspecific Van der Waals interaction with the incoming dNTP, unlike Q151, which interacts specifically with the sugar moiety of the dNTP (3′ OH) (Huang et al., 1998; Menéndez-Arias, 2008). The Stanford HIV Database, includes 19 isolates carrying mutations at A114 however, most isolates have been previously shown to be detrimental to RTs activity (Halvas, Svarovskaia, and Pathak, 2000; Rhee et al., 2003). A steady-state study demonstrated that a decrease in the Km values of RT could be correlated with the size of the mutated side chain present at this residue (Cases-González and Menéndez-Arias, 2005). However, the effect of mutations at A114 on the dNTP binding of HIV-1 RT has not been characterized. Structurally, A114 is located in a pocket that binds the 3′ OH of an incoming dNTP, which is in close proximity to the dNTPs sugar moiety (Huang et al., 1998). In this study we hypothesize that mutations at residue 114 of RT to amino acids with larger side chains may reduce the volume of the HIV-1 dNTP-binding pocket, resulting in a decreased space for the sugar moiety of normal dNTPs to reside, but conceivably not dNTP analogs lacking the intact sugar moiety. The requirement of a certain volume in the active site of RT for an incoming dNTP would represent unique mechanism of substrate binding; unlike the mutation Q151N, that decreases the dNTP binding through a loss of interaction or M184I, which results in a steric clash with the incoming dNTP. We tested this hypothesis to understand the molecular interaction of HIV-1 RT with its dNTP substrate, which plays a key role in HIV-1 proviral DNA synthesis and viral cellular tropism.
Materials and Methods
Chemicals and cell lines
Oligonucleotides and primers were purchased from Integrated DNA Technology. E. coli, XL1 Blues (Invitrogen), were used for the construction of plasmids and BL21 (Novagen, WI) for the overexpression of HIV-1 RT. MRC5 human lung fibroblasts (HLFs) were a kind gift from Dr. Toru Tokimoto (University of Rochester). The 293FTs used for production of viral vectors were purchased from invitrogen. dTTP was purchased from USB Affymatrix and acyclic dTTP from New England Biolabs. Primers were labeled with [γ-32 P] ATP (Amersham Biosciences).
Molecular modeling
Mutations were made with Pymol (Schrödinger) manually selecting the optimal rotamer for serine or cysteine at residue 114 of HIV-1 reverse transcriptase from (Huang et al., 1998). These were then energy minimized with AMBER 9.0 with 500 cycles (250 steepest descent) with implicit solvent. Mutations were visualized with VMD with the residues 113, 115, 151, and 184 (Humphrey, Dalke, and Schulten, 1996). The distances from the Cβ or hydroxyl group of the amino acid at 114 of RT from the 3′OH of the dTTP were calculated with the minimized PDB models using (Sirus 1.2).
Construction and purification of HIV-1 RT residue 114 mutant proteins
Mutations inresidue 114 of the HIV-1 RT gene were introduced through the amplification of pET28containing HXB2 HIV-1 RT by PCR-based site directed mutagenesis (Stragene). RT homodimers derived from HIV-1 (NL4-3) were hexahistidine-tagged (Kim, 1997; Weiss et al., 2004) and expressed using an overexpression system in BL21 E. coli. RT was purified using Ni2+ chelation chromatography as described previously (Diamond et al., 2004; Weiss et al., 2004). 1L cultures produced 1.5μg of purified p66/p66 homodimers. The purified RT was analyzed by 10% SDS-polyacrylamide gel using 1.5μg of bovine serum albumin (Sigma), as a control.
Primer extension reaction of HIV-1 RT
The primer extension assay was conducted as previously described (Diamond et al., 2004). Briefly, a 38-mer RNA template (5′AAGCUUGGCUGCAGAAUAUUGCUAGCGGGAAUUCGGCGCG-3′) was annealed to a 17-mer extend T primer (5′ CCGAATTCCCGCTAGCAATATTC 3′), 32P-labled at the 5′ end, with a T4 polynucleotide kinase. Reactions with a final volume of 20μl contained 20nM of the template/primer (T/P) and all 4 dNTPs in varying concentrations (250, 100, 25, 5, 1, 0.5, 0.25, and 0.1μM). Reactions were incubated at 37°C for 5 min then quenched with 40mM EDTA. These reaction conditions allow multiple rounds of primer extension. The amount of RT used in the titration reaction was determined by the dilution of RT that produced 50% T/P extension at 250μM dNTP concentration. We also preformed a single round primer extension reaction. In this reaction only one of the following nucleotides were added, dTTP, or acyTTP, in concentrations described in the figure legends.
Steady-State kinetic analysis of HIV-1 RT proteins
The reaction conditions of the steady-state kinetic analysis were identical to the primer extension reactions. The reactions were repeated with eight dNTP concentrations of dTTP (1000, 400, 200, 80, 16, 6.4, 3.2, 0.28 μM and acyTTP (300, 200, 150, 75, 40, 20, 5, 1 μM). Single nucleotide incorporation was measured on a polyacylamide-urea gel and quantitated by phosphorimaging analysis using OptiQuant software (PerkinElmer Life Sciences). The kcat and Km values were determined using the Micheaelis-Menten equation.
Pre-steady-State kinetic analysis of HIV-1 RT proteins
To determine the concentration of active WT and A114C RT, single-turnover burst assays were performed as described before (Jamburuthugoda et al., 2008). Briefly 100nM of a 5′ end 32P-labeled 23-mer extend-T primer and 200nM of cold primer were annealed to a 38-mer RNA template and extended with 100nM WT or A114C RT in presence of 800μM dTTP for single round incorporation and rapidly quenched with EDTA at different time points using the Kintek RFQ3 machine. The samples were then analyzed on a 14% sequencing gel under denaturing conditions and the percent of extended product was quantified (Quantity One 1-D analysis software, Biorad). The product formation at each time point was plotted and fitted to Eq 1, which determined the concentration of active RT (amplitude of the burst). The pre-steady-state reactions with 200nM active proteins and 50 nM T/P complexes were performed to assess the observed rates of product formation (kobs) at five or six time points in duplicate per dNTPs concentration. This was performed for each dNTPs (dTTP, dATP, dCTP, dGTP) and acyTTP, a dNTP analog. The kobs values were then fitted to Eq. 2 to yield kd and kpol. The data was curve-fitted with nonlinear regression to Eq. 1 and 2 with KaleidaGraph as described previously (Weiss et al., 2004).
| (Eq. 1) |
| (Eq. 2) |
HIV-1 vector construction and cell transduction
The D3HIV-GFP virus expresses eGFP and all HIV-1 NL4-3 proteins except Nef and Env. It was pseudotyped with VSV-G as previously described (Jamburuthugoda et al., 2008). The construction of the A114S, A114C, and A114V mutants were carried out using a site directed mutagenesis kit (Stragene). 293FTs (Invitrogen) were transfected with A114S, A114C, A114V or WT pD3HIV-GFP and pVSV-G. Cells were washed after 24 hours. Supernatant was collected at 48 and 72 hours after transfection. The collected supernatant was spun in an ultra centrifuge (Beckman) with a SW-28 rotor at 22k for 2 hours. The viral pellets were collected from each spin and treated with DNase and stored at -80°C. The vectors were normalized by p24 levels, which were determined by an enzyme-linked immunosorbent assay system (PerkinElmer).
HLFs were grown in either media containing 0.2% FBS (serum starved) or 10% FBS for 48 hours before transduction. Two hours post transduction HLFs were treated with 10μg/ml of Polybrene (Sigma) and 1mM dNs. Vectors were normalized to equal p24 amounts (5 × 105 pg/mL). 24 hours after transduction HLFs were washed with DPBS and re-treated with 1mM of dNs. Cells were washed again at 48 hours and harvested. The cells were monitored for GFP expression by fluorescent activated cell sorting using the FL-1 channel of a C6 Flow Cytometer (Accuri). Percent transduction was determined using FlowJo software (version 8.8.5, Tree Star, Inc). Statistical analysis and graphing was preformed with Prism 4 (GraphPad Software Inc). The transduction experiments were repeated in triplicate with the same batch of viral vector.
Results
Structural view of the wild type and mutant residues at HIV-1 RT position 114
First, in order to define the position of the methyl side chain of the HIV-1 RT A114 residue with respect to the incoming dNTP, we constructed a model of the dNTP-binding pocket of RT in a ternary complex bound to template/primer and dNTP substrate using a previously determined crystal structure (Huang et al., 1998). As illustrated in Figure 1A, the methyl side chain of A114 (teal blue) lies near the sugar moiety of the incoming dTTP (light blue/red). More specifically, the methyl side chain of A114 protrudes from the surface of the dNTP-binding pocket toward the sugar moiety of the incoming dNTP, forming a distance of 3.37Å between the Cβ of A114 and the 3′OH (pink arrows) of the incoming dNTP (Cases-González and Menéndez-Arias, 2005). This unique geometric position of A114 led us to hypothesize that an increased side chain volume at this residue would conflict with the sugar moiety of an incoming dNTP. Indeed, the reduced space for the sugar moiety of a dNTP appears to be inevitable after calculating the distance of A114S to the 3′OH, 1.96Å. However, the distance between the mutated residue and the 3′OH does not account for alterations in volume of the binding pocket. After modeling the positions of A114S (Fig. 1B), A114C (Fig. 1C), and volume (TableS1) of other additional amino acids, it is apparent that by incrementally extending the side chain of residue 114, the volume of the 3′OH binding pocket decreases. Therefore, we hypothesized that mutations at residue 114 with longer side chains may decrease the high dNTP-binding affinity of HIV-1 RT, ultimately leading to a reduction in the capability of HIV-1 to replicate in cells harboring low dNTP concentrations.
dNTP concentration dependent DNA polymerization by WT and A114 mutant HIV-1 RT proteins
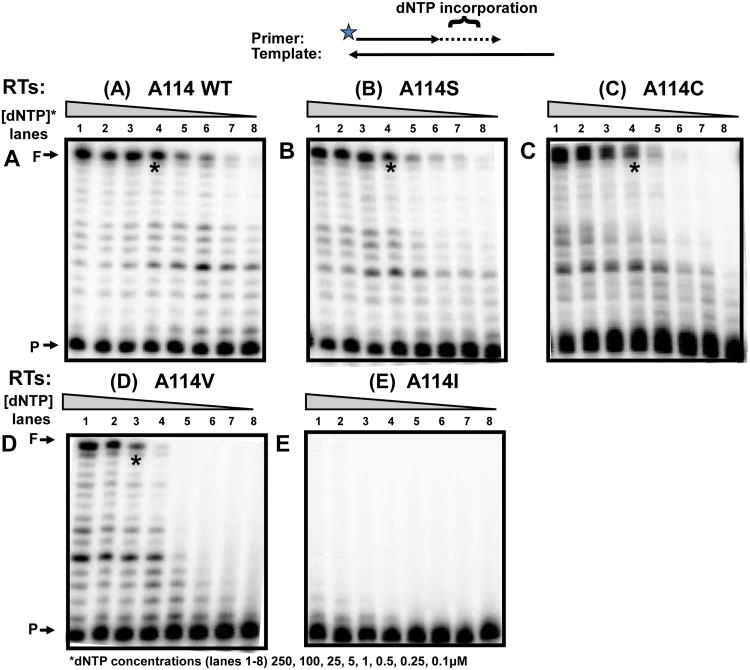
To test our hypothesis, we constructed and purified a series of HIV-1 NL4-3 RT A114 mutant proteins with increasingly longer side chains: A114S, A114C, A114V and A114I. First, we investigated the dNTP concentration dependent DNA polymerase activity of wild type (WT) and the A114 HIV-1 RT mutants using a primer extension assay for steady-state multiple dNTP incorporations. We expected that mutations at this residue may restrict the entry of a dNTP and thus would display reduced polymerase activities at lower dNTP concentrations, when compared to WT HIV-1 RT. This assay uses a 5′ end 32P-labled DNA 23-mer primer (P) annealed to a 38-mer RNA template (T) and varying concentrations of dNTPs. These reactions were repeated in triplicate (data not shown) with representative gels shown in Figure 2. The RT protein concentrations for this assay were normalized by an activity that gave ∼50% extension of the primer at a saturating dNTP concentration (250μM), under standard reaction conditions (5 min and 37oC). This reaction was then repeated with gradually decreasing dNTP concentrations. As shown in Figure 2A, WT A114 HIV-1 RT generated substantial amounts of the fully extended products (F) even at 5μM (lane 4), and continued showing detectable dNTP incorporation as low as 0.1 μM. However, the A114S (Fig. 2B) and A114C (Fig. 2C) RT mutants, with slightly longer side chains, demonstrated reduced activity compared to WT RT at 0.25μM (see lane 4 and “*”). The activity of A114V was greatly reduced even at a 2.5μM dNTP concentration (Fig. 2D). These results were expected due to the large addition at 114 of the beta branched side chain of valine, compared to the relatively minimal alterations of an OH or SH group addition with the mutations A114S or A114C. Lastly, when the residue 114 was mutated to an even larger side chain, A114I, RT failed to display any detectable primer extension even at 250μM (Fig. 2E). This can be compared to A114V, which still exhibited a detectable activity at high dNTP concentrations. This experiment demonstrates that WT RT is efficient at reverse transcription in both high and low dNTP concentrations; however, mutations at the residue 114 of HIV-1 RT with increasing side chain sizes gradually decrease the capability of RT to polymerize DNA, specifically at low dNTP concentrations. This supports the prediction made in the structural model in Figure 1 and is consistent with a previously published steady-state analysis of A114 RT mutants (Cases-González and Menéndez-Arias, 2005).
Figure 2. dNTP concentration dependent reverse transcription activity of HIV-1 RT variants.
Steady-state incorporation rates of dNTPs by WT RT or A114 mutants were measured by using a 5′ end 32P-labled 23-mer A primer (P) annealed to a 38-mer RNA template was extended by HIV-1 RT proteins, (A) WT A114, (B) A114S, (C) A114C, (D) A114V, and (E) A114I showing ∼50% of primer extension with 250μM dNTPs (lane 1) at 37°C for 5 min. The reactions were repeated with decreasing dNTP concentrations (lanes 1-8: 100, 25, 5, 1, 0.5, 0.25, and 0.1 μM). F: 38 nt fully extended products. “*”: Lane 4 with 5μM dNTP.
Steady-state activity of HIV-1 RT and A114 mutants with dTTP and acyTTP
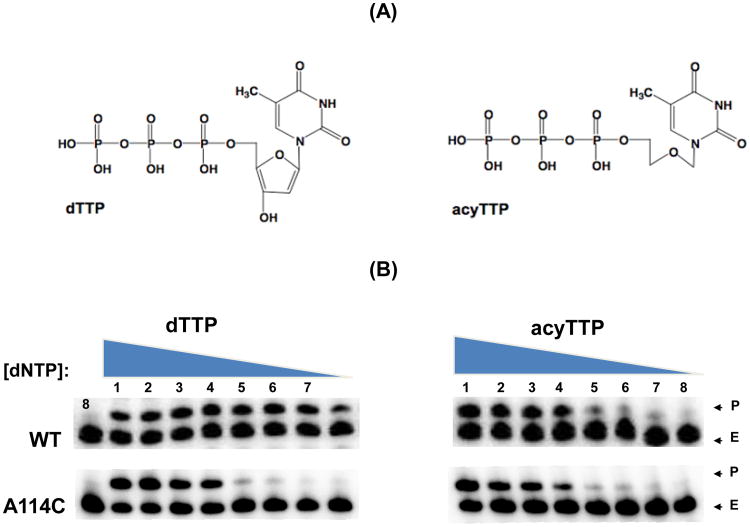
The model in Figure 1 predicts that bulky mutant side chains at the residue 114 could reduce the volume of the dNTP substrate-binding pocket, specifically the space where the sugar moiety resides. This reduction of available volume maybe responsible for the decreased rate of dNTP incorporation observed at low dNTP concentrations (Fig. 2) and the increased Km values (Cases-González and Menéndez-Arias, 2005; Huang et al., 1998). Next, we tested our hypothesis by decreasing the sugar moiety size of the incoming nucleotide substrate. We predicted that the HIV-1 RT mutants with bulkier side chains might have an enhanced capability to incorporate a dNTP analog with a much smaller sugar moiety lacking the 2′ and 3′ carbons of the sugar ring, acyTTP (Figure 3A), which would require less space (Jamburuthugoda et al., 2005), compared to normal dNTPs. We found that both WT and A114C RT have equal primer template extension at low concentrations of acyTTP; however, A114C RT displayed reduced extension at low dTTP concentrations when compared to WT (Figure 3B). This further suggests an increased side chain volume at residue 114 will conflict directly with the 3′OH of an incoming dNTP.
Figure 3. Incorporation comparison of WT and A114C HIV-1 RT with dNTP and acyTTP analog containing a modified sugar moiety.
The incorporation of either acyTTP or dTTP by A114C or WT RT was measured. (A) Chemical structure of dTTP and acyTTP. (B) Incorporation comparison of WT and A114C with dTTP and acyTTP. A 5′ end 32P labeled 23-mer T primer annealed to the 38-mer RNA template was extended by WT and A114C RTs. Concentrations of the nucleotides were 250,100, 25, 5, 1, 0.5, 0.25, and 0.1 μM (lanes 1-8). C: No substrate control, P: Primer, E: Extended product.
We next determined steady-state Km and Kcat values for WT and residue 114 mutants, for quantitative comparison of single nucleotide incorporation kinetics for dTTP and acyTTP with a 5′ end 32P labeled 23-mer T primer annealed to the 38-mer RNA template. First, as shown in Table 1, the Km values of A114S, A114C and A114V RTs with normal dTTP incorporation continue to incrementally increase by 18, 25 and 278-folds, compared to the Km value of wild type RT, which is consistent with the previous observation (Menéndez-Arias, 2008). However, when the same analysis was conducted with acyTTP incorporation, A114S, A114C and A114V RT displayed only 10, 3 and 10-fold increase in Km, when compared to wild type. This lower increase of the A114 mutant RT Km values with acyTTP compared to dTTP is consistent with our hypothesis; it further demonstrates that the entry of acyTTP into the dNTP binding pocket is less affected by the larger side chain volumes at residue 114, compare to the entry of dTTP. This is most likely due to the large sugar moiety volume of the natural dTTP compared to the acyTTP, which requires a much smaller space in the dNTP-binding pocket than the normal dTTP. In addition, this further suggests that the absence of the 2′ and 3′ carbons space in acyTTP, complements the large side chain volumes of the A114C and A114V mutant RTs, supporting the idea that the side chain of HIV-1 RT residue 114 lies proximal to the 2′ and 3′ carbons of the incoming dNTP substrate. However, A114 was unable to incorporate acyTTP even at a 200μM dNTP concentration (data not shown), suggesting that the volume of this side chain may conflict with more than just the ribose ring of an incoming dNTP.
Table 1.
Steady-state kinetics of WT and residue 114 mutant HIV-1 RT proteins with dTTP and acyTTP.
| Enzyme | dNTPs | Ŧ kcat (min-1) | Km (μM) (Fold Change Compared to WT) | Ŧ kcat/Km (μM-1·min-1) |
|---|---|---|---|---|
| WT | dTTP | 1.91 ± 0.09 | 0.29 ± 0.07 (1×) | 6.61 |
| acyTTP | 1.91 ± 0.05 | 0.97 ± 0.28 (1×) | 1.97 | |
| A114S | dTTP | 7.92 ± 0.26 | 5.36 ± 1.94 (18×) | 1.48 |
| acyTTP | 7.74 ± 0.93 | 9.6 ± 2.73 (10×) | 0.81 | |
| A114C | dTTP | 12.95 ± 1.19 | 7.14 ± 2.73 (25×) | 1.81 |
| acyTTP | 11.95 ± 0.56 | 2.54 ± 0.76 (3×) | 4.70 | |
| A114V | dTTP | 1.38 ± 0.02 | 80.60 ± 44.97 (278×) | 0.02 |
| acyTTP | 1.13 ± 0.15 | 9.37 ± 3.05 | 0.12 |
kcat = (Vmax/enzyme concentration) ×10-3
Pre-steady-state kinetic analysis for the dNTP binding affinity of WT and A114C mutant HIV-1 RT proteins
Next, we tested if mutations at the A114 residue affected the dNTP-binding step of DNA polymerization. To test this we performed a pre-steady-state kinetic analysis and measured the dNTP binding affinity (Kd) and conformational change/catalysis (kpol) of WT RT and the mutant A114C that displayed a milder defect in steady-state kinetics. First, the active site concentrations of WT and A114C RT protein were determined with an excess of T/P using a rapid quench technology. This experiment is done to ascertain the concentration of active protein present in the WT and A114C RT preparations (Fig. 4A). The dNTP concentration dependent pre-steady state rates (kobs) were then determined with excess of RT protein for all four dNTPs and acyTTP. The experimentally determined kobs values were plotted to calculate the Kd and kpol values of WT and A114C proteins with all four dNTPs, as described in the materials and methods section. As shown In Table 2, the A114C mutant demonstrated a 2.5 - 4.8 fold higher Kd than WT RT, with little difference in the kpol values. This further supports the idea that mutations at A114 reduce the dNTP binding affinity of HIV-1 RT. Moreover, as shown in Table 2 and Figure 4C, both the kpol and Kd of A114C compared to WT were not affected when acyTTP, a dNTP analog that lacks 2′ and 3′ carbons of sugar ring, was used as a substrate. This indicates that unlike dTTP, bulky side chains at position 114 do not affect the entrance of acyTTP. Taken together, observations from the pre-steady-state data strongly suggest that introducing bulkier side chains at position 114 reduce the dNTP binding affinity of HIV-1 RT, due to a volume requirement of the incoming dNTP sugar moiety.
Figure 4. Active site determination and dNTP titration curves of WT and A114C HIV-1 RT proteins.
(A) Pre-steady and steady-state kinetics of WT and A114C HIV-1 RT incorporating TTP. Reactions were carried out at the indicated times by mixing together pre-bound RT (100nM)•T/P (100nM) with 800μM TTP under rapid quench conditions (see Materials and Methods). The data was fit into the burst equation (Eq. 1 in Materials and Methods) as indicated by the solid line, which provides a measure of the active concentration of RT. (B) and (C) dNTP and acyTTP (respectively) titration curves of WT and A114C RTs under the pre-steady-state conditions. kobs was measured at varying concentrations with four different primers for each of the four nucleotides and acyTTP. Results were fit into Eq. 2 (indicated by the solid line) to calculate the corresponding values of kpol and Kd, which are summarized in Table 2 for all five substrates.
Table 2.
Pre-steadv-state kinetic values of WT and A114C RT proteins with four dNTPs.
| Enzyme | dNTPs | kpol(sec-1) | Kd (μM) (Fold difference) | kpol/Kd (μM-1sec1) |
| WT | dTTP | 85.7 ±7.3 | 3.4 ±0.6 | 25.2 |
| dGTP | 39.8 ±1.4 | 1.7 ± 0.3 | 23.4.8 | |
| dCTP | 138.8 ±12.3 | 0.6 ±0.3 | 231.3 | |
| dATP | 69.6 ±5.4 | 1.2 ±0.4 | 58.0 | |
| acyTTP | 14.12 ± 1.2 | 6.9 ±2.7 | 2.1 | |
| A114C | dTTP | 242.3 ±41.1 | 8.4 ±4.0 (2.5×) | 28.8 |
| dGTP | 33.3 ±2.9 | 8.1 ±2.1 (4.8×) | 4.1 | |
| dCTP | 261.2 ±18.9 | 2.1 ±0.6 (3.5×) | 124.3 | |
| dATP | 50.5 ±8.5 | 4.3 ±2.1 (3.6×) | 11.7 | |
| acyTTP | 18.2 ±1.6 (1.3×) | 5.7 ±1.4 (1.2×) | 3.2 |
HIV-1 transduction in human lung fibroblasts with different dNTP concentrations
Both pre-steady-state and steady-state kinetic analysis confirmed that mutations at 114 reduce the efficiency of RTs DNA synthesis at low dNTP concentrations when compared to WT (Fig. 2), which is a result of RTs reduced dNTP binding when a mutant side chain with a larger volume exists at residue114 (Table 2). Thus, these biochemical observations further predict a dNTP concentration dependence of RT during proviral DNA synthesis that would impact vector transduction in cells containing low dNTP concentrations, but not in cells containing high dNTP concentrations.
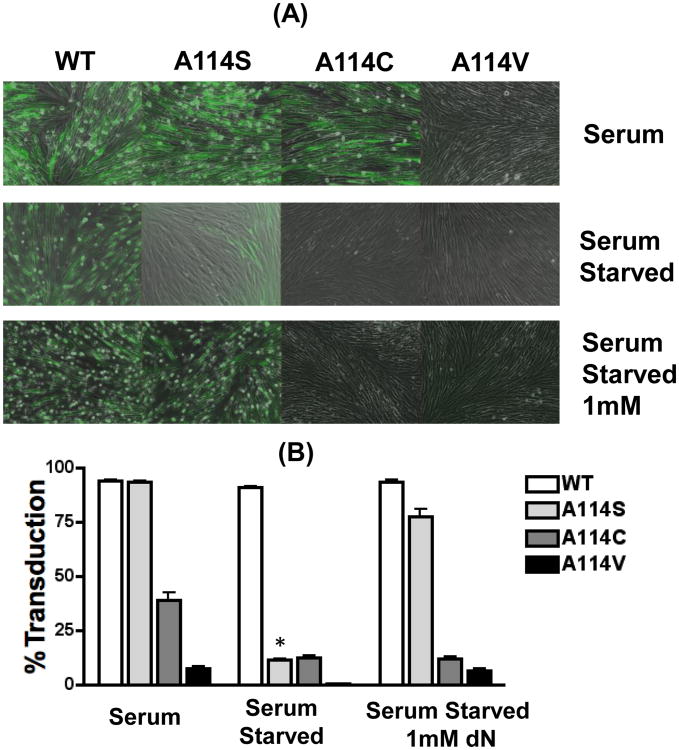
To test this prediction, we carried out a HIV-1 transduction study using human lung fibroblasts (HLFs). HLFs have been used as a model system for both dividing and nondividing cells as their dNTP concentrations can be controlled through cell cycle manipulation (Jamburuthugoda et al., 2008; Skasko and Kim, 2008) or deoxynucleoside (dN) treatment (Jamburuthugoda, Chugh, and Kim, 2006). Dividing HLFs cultured in 10% serum have an intracellular dNTP concentration of approximately 150-300nM (Jamburuthugoda, Chugh, and Kim, 2006) which is higher than the dNTP concentration found in human macrophages (∼50nM), but much lower than activated CD4+ T cells (1.4-2.5μM) (Charneau, Alizon, and Clavel, 1992; Diamond et al., 2004). Treating dividing and nondividing HLFs with deoxynucleosides (dNs) elevates their cellular dNTP concentrations to 50μM and ∼1μM, respectively. By altering the dNTP concentrations in HLFs, we were able to control the conditions for viral DNA synthesis of HIV-1 transduction (Jamburuthugoda et al., 2008). We introduced the A114S, A114C or A114V mutation into the pol gene of a HIV-1 vector. Equal p24 amounts of all vectors were used to transduce to HLFs grown under three culture conditions: 1) 10% serum, 2) serum starved, or 3) serum starved with 1mM dN treatment. The cells were visualized in both bright and dark fields (Fig. 5A), and the transduction efficiency was determined by FACS analysis for GFP expression (Fig. 5B). As shown in Figure 5A, WT RT vector was able to efficiently transduce HLFs under al three conditions, indicating that WT HIV-1 RT can efficiently synthesize DNA even at low dNTP concentrations found in the nondividing HLFs. The A114S vector displayed WT levels of transduction in dividing HLFs (grown in 10% serum: Fig. 5A and B). However, the A114S vector showed significantly reduced transduction in serum starved HLFs (Fig. 5A and B “*”). This suggests that the transduction of a HIV-1 vector carrying the A114S mutation is specifically affected in cells containing low dNTP concentration, but not in dividing HLFs with high cellular dNTP concentrations. Indeed, the reduced transduction of the A114S vector could be rescued by 1mM dN treatment in serum starved HLFs (Fig. 5A and 5B). Further indicating the impaired transduction capability of the A114S vector was due to the limited cellular dNTP pools in serum starved cells (Aquaro et al., 2005). Unlike A114S and WT, the A114C vector displayed reduced transduction in HLFs grown in serum. This transduction decreased further in serum-starved cells and was unable to be rescued by dN treatment. This again indicates a larger dNTP-binding defect generated with this mutation when compared to WT or A114S. The A114V failed to transduce HFLs grown in serum-starved media and demonstrated almost undetectable transduction levels in HLFs grown in serum or serum-starved media supplemented with dNs. Taken all together, this indicates that the transduction of an HIV-1 vector can be reduced by increasing the side chain volume present at A114 and that this transduction can be further decreased by limiting the cellular dNTP concentration.
Figure 5. Transduction of human lung fibroblasts with HIV-1 vectors containing WT and A114S RTs.
HLFs were transduced with WT or A114 mutant vectors and monitored for GFP expression. (A) HLFs were grown in media supplemented with 10% serum, serum starved, or serum starved with 1 mM dNs. Cells were transduced using an equal p24 amount (5.5 × 105 pg/mL) of WT, A114S, A114C or A114V D3HIV-GFP vectors. The transduction of the vectors was monitored after 48 for GFP expression by microscope in bright and dark/GFP fields. (B) The cells were harvested for FACS analyzed for the GFP positive cells. The experiment was conducted in triplicate. “*” Indicates significance as preformed by a t test with a P value of <0.0001, between the conditions serum and serum starved.
Discussion
Nucleoside RT inhibitors (NRTIs) are one of the key anti-retroviral agents for the treatment of HIV-1 infected patients. NRTIs enter the dNTP-binding pocket of HIV-1 RT during reverse transcription through competition with cellular dNTP substrates. Thus, it is important to understand the molecular basis of HIV-1 RT natural substrate selection during viral replication. Importantly, HIV-1 uniquely replicates both in activated dividing CD4+ T cells and terminally differentiated nondividing macrophages, which contributes to its pathogenesis. It has been shown that these two natural target cell types harbor vastly different cellular dNTP pools; macrophages have 20-50nM dNTPs, which is a much lower concentration than the Kd of most known cellular DNA polymerases, whereas T cells contain around 1.4-2.5 μM dNTPs (Diamond et al., 2004). It has also been shown that other DNA viruses replicating in macrophages (i.e. herpes simplex virus (Ho, Hui, and Lam, 2004; Kaplitt and Pfaff, 1996; Okada et al., 2002; Sulpizi et al., 2001) require their own dNTP synthesis machinery (i.e. thymidine kinase) to provide the virus with an adequate supply of dNTP substrates for DNA replication. HIV-1 and other lentiviruses lack this viral dNTP biosynthesis machinery. This raises the question of how lentiviruses such as HIV-1 synthesize their proviral DNA in macrophages, which contain limited dNTP pools. Our previous steady and pre-steady-state kinetic analysis revealed that lentiviral RTs are capable of synthesizing DNA in the low dNTP concentrations found in macrophages, while in the same condition oncoretroviral RTs fail. This unique capability of HIV-1 has been attributed to RTs tight binding to dNTPs (Diamond et al., 2004). Thus, it is intriguing to elucidate the mechanistic and structural features of the HIV-1 RT dNTP-binding pocket.
Molecular manipulation of both the HIV-1 RT residue 114 and the incoming dNTP substrate support that there is a mechanistic volume requirement between the side chain at residue 114 and the sugar moiety of the incoming dNTP substrate. In addition the mutations that were generated in this study had minimal impact on the processivity of RT with the exception of A114V (Figure 1S). This suggests that the A114V mutations may result in larger structural changes that affect RTs interaction with the template primers as well as dNTPs. We have previously demonstrated that the Q151 residue, which directly contacts the 3′ OH of the incoming dNTP via a hydrogen bond, plays an important role in the tight dNTP binding affinity of HIV-1 RT (Weiss, Bambara, and Kim, 2002). The Q151N mutation was found to significantly reduced not only the dNTP binding affinity of RT but also the transduction of HIV-1 vectors bearing the Q151N mutation in cells containing low dNTP concentrations (Jamburuthugoda, Chugh, and Kim, 2006). As shown in Figure 5 we further demonstrated that mutations at residue 114 impact vector transduction specifically in cells containing low dNTP concentrations.
Interestingly, as shown in Table 2, the A114C mutation prompted a larger increase in the Kd values of purine nucleotide substrates when compared to pyrimidine nucleotides. One simple explanation for this differential effect on these two groups of nucleotide substrates could be that the larger base moieties of purines require an increased space in the active site of RT, compared to the pyrimidine nucleotides. The mutation A114C might reduces this space, causing a decrease in the binding of purine substrates to RT and an increase in their Kd. A second explanation can be generated through past studies done with other DNA polymerases. Its been previously shown that the nucleotide selectivity can be affected by residues at or around the steric gate in the polymerase active site, which allows polymerases to discriminate between natural/correct dNTPs and nonnatural/incorrect dNTPs (DeLucia et al., 2006). Therefore, it is possible that the A114C mutation structurally influences the steric gate residue of HIV-1 RT, Tyr115 (Klarmann et al., 2007).
Our biochemical and virological results support that like the Q151N HIV-1 RT mutant, the A114 RT mutants enzymatically mimic oncoretroviral RTs such as MuLV RT and Avian Sarcoma Virus (ASV) RT which both efficiently synthesize DNA only at the high dNTP concentrations found in dividing cells (Lewis and Emerman, 1994). However, while the Q151N HIV-1 RT mutation reduces the dNTP binding affinity through a lost interaction with the sugar moiety of a dNTP substrate, the bulky mutations at 114 appears to reduce the dNTP binding affinity by occupying the space in the pocket where the sugar moiety of dNTP normally binds RT. This is mechanistically supported by the observation with acyTTP incorporation.
This study demonstrates a unique way to modulate the entry of a dNTP substrate within the dNTP-binding pocket of HIV-1 RT. Furthermore, our HIV-1 vector experiment confirmed that the reduction of the RT dNTP binding affinity is one way to make HIV-1 mimic oncoretroviruses in terms of their distinct cell type tropism.
Supplementary Material
Acknowledgments
This work was supported by NIH AI049781 (B.K.) and NIH T32AI049815(SVCH).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aquaro S, Svicher V, Ceccherini-Silberstein F, Cenci A, Marcuccilli F, Giannella S, Marcon L, Caliò R, Balzarini J, Perno CF. Limited development and progression of resistance of HIV-1 to the nucleoside analogue reverse transcriptase inhibitor lamivudine in human primary macrophages. J Antimicrob Chemother. 2005;55(6):872–8. doi: 10.1093/jac/dki104. [DOI] [PubMed] [Google Scholar]
- Boyer PL, Sarafianos SG, Arnold E, Hughes SH. Analysis of mutations at positions 115 and 116 in the dNTP binding site of HIV-1 reverse transcriptase. P Natl Acad Sci Usa. 2000;97(7):3056–61. doi: 10.1073/pnas.97.7.3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cases-González CE, Menéndez-Arias L. Nucleotide specificity of HIV-1 reverse transcriptases with amino acid substitutions affecting Ala-114. Biochem J. 2005;387(Pt 1):221–9. doi: 10.1042/BJ20041056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charneau P, Alizon M, Clavel F. A second origin of DNA plus-strand synthesis is required for optimal human immunodeficiency virus replication. Journal of Virology. 1992;66(5):2814–20. doi: 10.1128/jvi.66.5.2814-2820.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLucia AM, Chaudhuri S, Potapova O, Grindley ND, Joyce CM. The properties of steric gate mutants reveal different constraints within the active sites of Y-family and A-family DNA polymerases. J Biol Chem. 2006;281(37):27286–91. doi: 10.1074/jbc.M604393200. [DOI] [PubMed] [Google Scholar]
- Diamond TL, Roshal M, Jamburuthugoda VK, Reynolds HM, Merriam AR, Lee KY, Balakrishnan M, Bambara RA, Planelles V, Dewhurst S, Kim B. Macrophage tropism of HIV-1 depends on efficient cellular dNTP utilization by reverse transcriptase. J Biol Chem. 2004;279(49):51545–53. doi: 10.1074/jbc.M408573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost SD, Nijhuis M, Schuurman R, Boucher CA, Brown AJ. Evolution of lamivudine resistance in human immunodeficiency virus type 1-infected individuals: the relative roles of drift and selection. Journal of Virology. 2000;74(14):6262–8. doi: 10.1128/jvi.74.14.6262-6268.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G, Orlova M, Georgiadis MM, Hendrickson WA, Goff SP. Conferring RNA polymerase activity to a DNA polymerase: a single residue in reverse transcriptase controls substrate selection. P Natl Acad Sci Usa. 1997;94(2):407–11. doi: 10.1073/pnas.94.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halvas EK, Svarovskaia ES, Pathak VK. Role of murine leukemia virus reverse transcriptase deoxyribonucleoside triphosphate-binding site in retroviral replication and in vivo fidelity. Journal of Virology. 2000;74(22):10349–58. doi: 10.1128/jvi.74.22.10349-10358.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris D, Kaushik N, Pandey PK, Yadav PN, Pandey VN. Functional analysis of amino acid residues constituting the dNTP binding pocket of HIV-1 reverse transcriptase. J Biol Chem. 1998;273(50):33624–34. doi: 10.1074/jbc.273.50.33624. [DOI] [PubMed] [Google Scholar]
- Ho IA, Hui KM, Lam PY. Glioma-specific and cell cycle-regulated herpes simplex virus type 1 amplicon viral vector. Hum Gene Ther. 2004;15(5):495–508. doi: 10.1089/10430340460745810. [DOI] [PubMed] [Google Scholar]
- Huang H, Chopra R, Verdine GL, Harrison SC. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science. 1998;282(5394):1669–75. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K. VMD - Visual Molecular Dynamics. Journal of Molecular Dynamics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Igarashi T, Brown CR, Endo Y, Buckler-White A, Plishka R, Bischofberger N, Hirsch V, Martin MA. Macrophage are the principal reservoir and sustain high virus loads in rhesus macaques after the depletion of CD4+ T cells by a highly pathogenic simian immunodeficiency virus/HIV type 1 chimera (SHIV): Implications for HIV-1 infections of humans. P Natl Acad Sci Usa. 2001;98(2):658–63. doi: 10.1073/pnas.021551798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamburuthugoda VK, Chugh P, Kim B. Modification of human immunodeficiency virus type 1 reverse transcriptase to target cells with elevated cellular dNTP concentrations. J Biol Chem. 2006;281(19):13388–95. doi: 10.1074/jbc.M600291200. [DOI] [PubMed] [Google Scholar]
- Jamburuthugoda VK, Guo D, Wedekind JE, Kim B. Kinetic evidence for interaction of human immunodeficiency virus type 1 reverse transcriptase with the 3′-OH of the incoming dTTP substrate. Biochemistry. 2005;44(31):10635–43. doi: 10.1021/bi050611+. [DOI] [PubMed] [Google Scholar]
- Jamburuthugoda VK, Santos-Velazquez JM, Skasko M, Operario DJ, Purohit V, Chugh P, Szymanski EA, Wedekind JE, Bambara RA, Kim B. Reduced dNTP Binding Affinity of 3TC-resistant M184I HIV-1 Reverse Transcriptase Variants Responsible for Viral Infection Failure in Macrophage. Journal of Biological Chemistry. 2008;283(14):9206. doi: 10.1074/jbc.M710149200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce CM. Choosing the right sugar: how polymerases select a nucleotide substrate. P Natl Acad Sci Usa. 1997;94(5):1619–22. doi: 10.1073/pnas.94.5.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplitt MG, Pfaff DW. Viral Vectors for Gene Delivery and Expression in the CNS. Methods. 1996;10(3):343–50. doi: 10.1006/meth.1996.0112. [DOI] [PubMed] [Google Scholar]
- Kennedy EM, Gavegnano C, Nguyen L, Slater R, Lucas A, Fromentin E, Schinazi RF, Kim B. Ribonucleoside triphosphates as substrate of human immunodeficiency virus type 1 reverse transcriptase in human macrophages. J Biol Chem. 2010;285(50):39380–91. doi: 10.1074/jbc.M110.178582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B. Genetic selection in Escherichia coli for active human immunodeficiency virus reverse transcriptase mutants. Methods. 1997;12(4):318–24. doi: 10.1006/meth.1997.0485. [DOI] [PubMed] [Google Scholar]
- Klarmann GJ, Eisenhauer BM, Zhang Y, Gotte M, Pata JD, Chatterjee DK, Hecht SM, Le Grice SF. Investigating the “steric gate” of human immunodeficiency virus type 1 (HIV-1) reverse transcriptase by targeted insertion of unnatural amino acids. Biochemistry. 2007;46(8):2118–26. doi: 10.1021/bi061772w. [DOI] [PubMed] [Google Scholar]
- Lewis P, Hensel M, Emerman M. Human immunodeficiency virus infection of cells arrested in the cell cycle. EMBO J. 1992;11(8):3053–8. doi: 10.1002/j.1460-2075.1992.tb05376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis PF, Emerman M. Passage through mitosis is required for oncoretroviruses but not for the human immunodeficiency virus. Journal of Virology. 1994;68(1):510–6. doi: 10.1128/jvi.68.1.510-516.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe DM, Parmar V, Kemp SD, Larder BA. Mutational analysis of two conserved sequence motifs in HIV-1 reverse transcriptase. FEBS Lett. 1991;282(2):231–4. doi: 10.1016/0014-5793(91)80484-k. [DOI] [PubMed] [Google Scholar]
- Martín-Hernández AM, Domingo E, Menéndez-Arias L. Human immunodeficiency virus type 1 reverse transcriptase: role of Tyr115 in deoxynucleotide binding and misinsertion fidelity of DNA synthesis. EMBO J. 1996;15(16):4434–42. [PMC free article] [PubMed] [Google Scholar]
- Menéndez-Arias L. Mechanisms of resistance to nucleoside analogue inhibitors of HIV-1 reverse transcriptase. Virus Res. 2008;134(1-2):124–46. doi: 10.1016/j.virusres.2007.12.015. [DOI] [PubMed] [Google Scholar]
- Mulder LC, Harari A, Simon V. Cytidine deamination induced HIV-1 drug resistance. P Natl Acad Sci Usa. 2008;105(14):5501–6. doi: 10.1073/pnas.0710190105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada T, Nomoto T, Shimazaki K, Lijun W, Lu Y, Matsushita T, Mizukami H, Urabe M, Hanazono Y, Kume A, Muramatsu S, Nakano I, Ozawa K. Adeno-associated virus vectors for gene transfer to the brain. Methods. 2002;28(2):237–47. doi: 10.1016/s1046-2023(02)00228-1. [DOI] [PubMed] [Google Scholar]
- Pandey VN, Kaushik N, Rege N, Sarafianos SG, Yadav PN, Modak MJ. Role of methionine 184 of human immunodeficiency virus type-1 reverse transcriptase in the polymerase function and fidelity of DNA synthesis. Biochemistry. 1996;35(7):2168–79. doi: 10.1021/bi9516642. [DOI] [PubMed] [Google Scholar]
- Ray AS, Basavapathruni A, Anderson KS. Mechanistic studies to understand the progressive development of resistance in human immunodeficiency virus type 1 reverse transcriptase to abacavir. J Biol Chem. 2002;277(43):40479–90. doi: 10.1074/jbc.M205303200. [DOI] [PubMed] [Google Scholar]
- Rhee SY, Gonzales MJ, Kantor R, Betts BJ, Ravela J, Shafer RW. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res. 2003;31(1):298–303. doi: 10.1093/nar/gkg100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarafianos SG, Das K, Clark AD, Ding J, Boyer PL, Hughes SH, Arnold E. Lamivudine (3TC) resistance in HIV-1 reverse transcriptase involves steric hindrance with beta-branched amino acids. P Natl Acad Sci Usa. 1999;96(18):10027–32. doi: 10.1073/pnas.96.18.10027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The PyMOL Molecular Graphics System 1.2r3pre: LLC
- Skasko M, Kim B. Compensatory role of human immunodeficiency virus central polypurine tract sequence in kinetically disrupted reverse transcription. Journal of Virology. 2008;82(15):7716–20. doi: 10.1128/JVI.00120-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skasko M, Weiss KK, Reynolds HM, Jamburuthugoda VK, Lee K, Kim B. Mechanistic differences in RNA-dependent DNA polymerization and fidelity between murine leukemia virus and HIV-1 reverse transcriptases. J Biol Chem. 2005;280(13):12190–200. doi: 10.1074/jbc.M412859200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulpizi M, Schelling P, Folkers G, Carloni P, Scapozza L. The rational of catalytic activity of herpes simplex virus thymidine kinase. a combined biochemical and quantum chemical study. J Biol Chem. 2001;276(24):21692–7. doi: 10.1074/jbc.M010223200. [DOI] [PubMed] [Google Scholar]
- Svedhem V, Bergroth T, Lidman K, Sönnerborg A. Presence of M184I/V in minor HIV-1 populations of patients with lamivudine and/or didanosine treatment failure. HIV Med. 2007;8(8):504–10. doi: 10.1111/j.1468-1293.2007.00502.x. [DOI] [PubMed] [Google Scholar]
- Weinberg JB, Matthews TJ, Cullen BR, Malim MH. Productive human immunodeficiency virus type 1 (HIV-1) infection of nonproliferating human monocytes. J Exp Med. 1991;174(6):1477–82. doi: 10.1084/jem.174.6.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss KK, Bambara RA, Kim B. Mechanistic role of residue Gln151 in error prone DNA synthesis by human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT). Pre-steady state kinetic study of the Q151N HIV-1 RT mutant with increased fidelity. J Biol Chem. 2002;277(25):22662–9. doi: 10.1074/jbc.M200202200. [DOI] [PubMed] [Google Scholar]
- Weiss KK, Chen R, Skasko M, Reynolds HM, Lee K, Bambara RA, Mansky LM, Kim B. A role for dNTP binding of human immunodeficiency virus type 1 reverse transcriptase in viral mutagenesis. Biochemistry. 2004;43(15):4490–500. doi: 10.1021/bi035258r. [DOI] [PubMed] [Google Scholar]
- Yang G, Wang J, Cheng Y, Dutschman GE, Tanaka H, Baba M, Cheng YC. Mechanism of inhibition of human immunodeficiency virus type 1 reverse transcriptase by a stavudine analogue, 4′-ethynyl stavudine triphosphate. Antimicrob Agents Chemother. 2008;52(6):2035–42. doi: 10.1128/AAC.00083-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura K, Feldman R, Kodama E, Kavlick MF, Qiu YL, Zemlicka J, Mitsuya H. In vitro induction of human immunodeficiency virus type 1 variants resistant to phosphoralaninate prodrugs of Z-methylenecyclopropane nucleoside analogues. Antimicrob Agents Chemother. 1999;43(10):2479–83. doi: 10.1128/aac.43.10.2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamyatnin AA. Amino acid, peptide, and protein volume in solution. Annu Rev Biophys Bioeng. 1984;13:145–65. doi: 10.1146/annurev.bb.13.060184.001045. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.