Abstract
Aeromonas species are well distributed in freshwater environments, and their natural susceptibility to antimicrobials renders them interesting candidates for the survey of antimicrobial resistance in freshwater milieu. Water samples were collected from Kat and Tyume rivers in the Eastern Cape province of South Africa, and a total of 45 isolates identified as Aeromonas species were recovered from the two rivers. All Aeromonas isolates were resistant to oxacillin, penicillin, clindamycin, cephalothin, vancomycin, and rifamycin, while appreciable susceptibilities (89.3 : 94.1%, 82.1 : 94.1%, 85.7 : 88.2%, and 92.9 : 88.2%) were observed against ciprofloxacin, chloramphenicol, nitrofurantoin, and gentamicin from Kat and Tyume rivers, respectively. Multiple antibiotic resistance (MAR) indices ranged from 0.016 to 0.044 for the two rivers. Class 1 integron was detected in about 20% of the isolates, and all the isolates except one showed ability to produce biofilm in vitro as weak producers (53.33%), moderate producers (15.56%), and strong producers (28.9%). This investigation provides a baseline data on antibiotic resistance as well as the adhesive characteristics of Aeromonas isolates from Tyume and Kat rivers in the Eastern Cape province of South Africa.
1. Introduction
Aeromonas species are Gram-negative, rod-shaped, non-spore-forming, facultatively anaerobic bacteria that occur ubiquitously and autochthonously in aquatic environments. The Aeromonas genus has undergone a number of taxonomic and nomenclature revisions in the past two decades [1]. Initially, Aeromonas was placed in the family Vibrionaceae, but successive phylogenetic analyses revealed that Aeromonas is not closely related to Vibrios and resulted in moving Aeromonas to a new family, the Aeromonadaceae [2, 3]. Aeromonads share in common many biochemical characteristics with members of the Enterobacteriaceae; however, they are easily differentiated by being oxidase positive.
Aeromonas species are agents of infection in fish [4] and are associated with human diarrheal diseases and wound infections which may result due to contact with contaminated water [5, 6]. Wound infections may become severe and systemic. Aeromonas is also associated with sepsis, respiratory tract, eye and other systemic infections [7]. In fish, Aeromonas causes bacterial infections which may pose relatively high resistance to antibiotics including clinically relevant cases and diseases [8]. Treatment of Aeromonas infection is usually with the use of antibiotics; however, antimicrobial resistance can make these infections difficult to treat.
The ubiquity of Aeromonas species in aquatic ecosystems and their natural susceptibility to antimicrobials render them interesting candidates for the survey of antimicrobial resistance in freshwater environments [9, 10]. Freshwater streams are usually receptors of many industrial, domestic and agricultural wastes, which could contain antimicrobial agents and antimicrobial-resistant bacteria [10, 11]. Due to diverse microbial population in such ecosystems, freshwater environment provides favourable conditions for the spread of antimicrobial resistance.
Aquatic bacteria such as Aeromonas may become reservoirs of antibiotic resistance determinants as a result of influents from diverse sources entering into the river and as such may transfer these antibiotic resistance determinants to other aquatic organisms including pathogenic bacteria [10]. In the United States, a study has shown that several rivers are becoming major reservoirs of antibiotic resistance microorganisms [12]. With widespread commerce and global travel, antibiotic resistant organisms can spread across the globe. The occurrence and distribution of Aeromonas in aquatic ecosystems, its emerging significance as a contaminant of water, and the pathogenic potential mediated by mesophilic Aeromonas species are all of public health concern [13, 14]. The ability of bacteria to develop multiple-drug resistance result in part from their ability to acquire new antibiotic resistance genes. Mobile elements called integrons determine a site-specific recombination system that is responsible for the acquisition of many antibiotic resistance determinants [15, 16].
Bacterial adherence to surfaces is one of the initial steps leading to biofilm formation and is therefore a significant microbiological event in medicine and the environment [17, 18]. Aeromonas hydrophila can attach to and form biofilms on polystyrene, glass surface, stainless steel, and polyvinyl chloride[19]. Aeromonas can also attach to solid surfaces and form biofilms in aquatic environments. The presence of Aeromonas in biofilm samples from water distribution systems in South Africa has been documented [20]. Biofilm is an irreversible growth of a combination of bacterial micro-colonies on surfaces entrenched in extracellular polysaccharide matrix [21, 22]. The formation of biofilm results in resistance of bacteria to antimicrobial drugs and persistent infections [21] which can lead to the severity of several bacterial diseases affecting both human [23] and animal health [24].
Although Aeromonas species are well distributed in freshwater habitat and have been assigned as an emerging threat to human health [3, 25], no data exists about the antibiotic susceptibility profiles and adhesive properties of aeromonads from Kat and Tyume rivers in the Eastern Cape province of South Africa. The Food and Agriculture Organization/World Health Organization (FAO/WHO) commission recommends that to prevent waterborne diseases in developing countries, aquatic environments having direct impact on human populations should be characterized physically, chemically, and microbiologically. In view of this recommendation, and as part of our surveillance of reservoir of antibiotic resistant commensal bacteria, this present study aimed to (i) evaluate the levels of antimicrobial resistance in aeromonads isolates from Kat and Tyume rivers, (ii) determine the presence of class 1 and 2 integron associated gene cassette, and (iii) evaluate their biofilm forming capabilities.
2. Materials and Methods
2.1. Study Area, Sampling and Processing of Samples
Kat river is located in a semiurban location at geographical coordinates: S32°46.547′ E026°38.456′ while Tyume river is located in a rural community at geographical coordinates: S32°47.279′ E026° 50.520′ in the Eastern Cape province of South Africa. Water samples were collected four times at random between April 2011 and March 2012. Water samples were collected in duplicates using 2 L bottles placed on ice and transported to the laboratory for analysis. Hundred microliter (100 μL) was spread on several Glutamate phenol(GSP) agar (biolab, merck SA); on the other hand, 500 μL was inoculated into 145 mL sterile nutrient broth and incubated in a rotary incubator at 150 rpm overnight at 36°C. At the end of the incubation period, a loopful of the culture was spread and/or streaked on GSP agar, and all agar plates were incubated at 36°C for 24 h. Typical yellow colonies on GSP agar were purified using the same media. Pure colonies were transferred unto nutrient agar plates and subjected to oxidase and catalase tests. Oxidase and catalase positive isolates were further screened for biochemical characteristics using API 20NE kit. The strips were then read, and final identification was made using API lab plus software (bioMerieux, Marcy l'Etoile, France).
2.2. Antimicrobial Susceptibility Testing
Isolates were subcultured on nutrient agar plates incubated for 24 h at 36°C. Colonies were picked from the agar plates, and suspended in normal saline (0.85% w/v), and adjusted to an A560 value of 0.12 ± 0.02 (0.5 McFarland standard). The bacterial suspension was spread on the Mueller Hinton agar plates using a sterile swab stick, allowed to dry, and impregnated with antibiotic disk. The antibiotics used were as follows: ciprofloxacin (5 μg), trimethoprim (5 μg), chloramphenicol (3 μg), penicillins (10 μg), clindamycins (2 μg), ofloxacin (5 μg), ampicillin-sulbactam (20 μg), oxacillin (1 μg). ampicillin (25 μg), gentamicin (10 μg), nalidixic acid (30 μg), cefotaxime (30 μg), nitrofurantoin (300 μg), sulfamethoxazole (25 μg), cephalothin (30 μg), erythromycin (15 μg), tetracycline (10 μg), minocycline (30 μg), vancomycin (30 μg), and rifampicin (5 μg). Disks were purchased from Mast Diagnostics (Mast Group Merseyside UK). Plates were incubated at 36°C for 24 h. Diameters of the zones of inhibition were measured and interpreted, as susceptible, intermediate or resistant according to the Clinical Laboratory Standard Guidelines [26]. The frequency of antibiotic-resistant Aeromonas isolates was calculated by the following equation: A/B × 100%, where A is the number of isolates resistant to an antibiotic and B is the total number of isolates from the sample. The multiple antibiotic resistance (MAR) index of each samples was estimated by the following equation: a/(b × c), where a represents the aggregate antibiotic resistance score of all isolates from the sample, b represents the number of antibiotics, and c represents the number of isolates from the sample as outlined in [27–29].
2.3. PCR Detection of Integrons
DNA was extracted following the method described elsewhere [30, 31]. Template DNA was stored at −20°C until it was ready for use. The primer used for the detection of class 1 and class 2 integron is shown in Table 1. The PCR conditions were as follows: initial denaturation at 94°C for 2 min followed by 30 cycles of denaturation at (95°C for 45 s), annealing (56°C for 1 min), extension (72°C for 90 s), and a final extension at 72°C for 10 min.
Table 1.
Primer set for the detection of class 1 and 2 integron.
2.4. Biofilm Formation Assay
Quantitatively, biofilm formation among Aeromonas isolates was assessed using microtitre plate method described by Stepanovic et al. [32] and Odeyemi et al. [22] with modification. Wells of 96 flat bottomed microtiter plates were filled with 200 μL of Tryptone Soy Broth (TSB) and inoculated with 20 μL of Aeromonas isolates grown overnight and standardized to 0.5 McFarland standard. Plates were incubated at 37°C for 24 h. Positive control wells contained Aeromonas hydrophila ATCC 7699, and the negative control wells contained uninoculated Tryptone Soy Broth. Contents of each well were aspirated and washed three times with sterile phosphate-buffered saline (PBS). After air-drying, wells were stained with 200 μL of 1% crystal violet for 30 min. The wells were carefully washed with distilled water to remove the excess stain. Plates were allowed to dry at room temperature. Dye bound to adherent cells was resolubilized with 150 μL of absolute ethanol. Microplate reader (Synergy mx Biotek, USA) was used to read the plates at 570 nm wavelength. Average optical density (OD) of each duplicate result was taken including positive and negative controls. Isolates were categorized as nonbiofilm producer (ODi < ODc), weak (ODc < ODi < 0.1), moderate (ODi = 0.1 < 0.12) and strong (ODi > 0.12) producers according to the modified methods of [33, 34].
3. Statistical Analysis
Susceptibility data were compared by using a Chi-square test with SPSS software for Windows, version 17.0. Both susceptibility and resistance were calculated as percentages with 95% confidence intervals. A P value < 0.05 was considered to be statistically significant.
4. Results
A total of 45 isolates (17 from Kat river and 28 from Tyume river) were identified as Aeromonas hydrophila/caviae. The isolates were evaluated for their antibiotic susceptibilities. Generally, all isolates were resistant to oxacillin, penicillin, clindamycin, cephalothin, vancomycin, and rifamycin, while over 80% of the isolates were susceptible to ciprofloxacin, chloramphenicol, gentamicin, and nitrofurantoin (Table 2). Isolates from both rivers showed a diversified resistance trend to the same antibiotics. For example, all the Aeromonas isolates from Tyume river were resistant to ofloxacin, while 89.3% of isolates from Kat river were susceptible to ofloxacin (Table 2). Also, 70.6% of the isolates from Tyume river were resistant against tetracycline, while 82.1% of the isolates from Kat river were susceptible to the antibiotic. Similarly, Aeromonas isolates showed 94.1% resistance and 60.7% susceptibility against trimethoprim and 82.3% resistance and 78.6% susceptibility against cefotaxime from Tyume and Kat rivers, respectively, as shown in Table 2.
Table 2.
Antibiogram of Aeromonas isolates recovered from Kat river and Tyume river.
| Antibiotics | Kat river (%) | Tyume river (%) | ||||
|---|---|---|---|---|---|---|
| S | I | R | S | I | R | |
| Ciprofloxacin | 89.3 | 0 | 10.7 | 94.1 | 5.9 | 0 |
| Trimethoprim | 60.7 | 32.1 | 7.14 | 5.9 | 0 | 94.1 |
| Chloramphenicol | 82.1 | 17.9 | 0 | 94.1 | 5.9 | 0 |
| Penicillin | 0 | 0 | 100 | 0 | 0 | 100 |
| Clindamycin | 0 | 0 | 100 | 0 | 0 | 100 |
| Ofloxacin | 89.3 | 7.14 | 3.6 | 0 | 0 | 100 |
| Ampicillin-sulbactam | 3.6 | 0 | 96.4 | 0 | 0 | 100 |
| Oxacillin | 0 | 0 | 100 | 0 | 0 | 100 |
| Ampicillin | 0 | 0 | 100 | 5.9 | 0 | 94.1 |
| Gentamicin | 92.9 | 0 | 7.14 | 88.2 | 0 | 11.8 |
| Nalidixic acid | 14.3 | 3.6 | 82.1 | 70.6 | 0 | 29.4 |
| Cefotaxime | 78.6 | 14.3 | 7.14 | 11.8 | 5.9 | 82.3 |
| Nitrofurantoin | 85.7 | 3.6 | 10.7 | 88.2 | 11.8 | 0 |
| Sulfamethoxazole | 0 | 3.6 | 96.4 | 0 | 0 | 100 |
| Cephalothin | 0 | 0 | 100 | 0 | 0 | 100 |
| Erythromycin | 7.14 | 57.1 | 35.7 | 58.8 | 5.9 | 35.2 |
| Tetracycline | 82.1 | 21.4 | 7.14 | 17.7 | 11.8 | 70.6 |
| Minocycline | 42.9 | 57.1 | 0 | 23.5 | 11.8 | 64.7 |
| Vancomycin | 0 | 0 | 100 | 0 | 0 | 100 |
| Rifampicin | 0 | 0 | 100 | 0 | 0 | 100 |
The frequency of antibiotic resistance among the Aeromonas isolates from Kat and Tyume rivers is shown in Figure 1. The MAR index of Kat river ranged from 0.026 to 0.044 with a mean value of 0.034. The highest MAR index was observed against only one isolate (MAR 0.044), while six isolates exhibited MAR index of 0.029. Aeromonas isolates from Tyume river demonstrated MAR index which ranged from 0.016 to 0.029 with a mean value of 0.0214. Twelve isolates exhibited the lowest MAR index of 0.016. PCR amplification of class 1 and class 2 integron detected class 1 integron in 20% of Aeromonas isolates, while class 2 integron was not detected.
Figure 1.
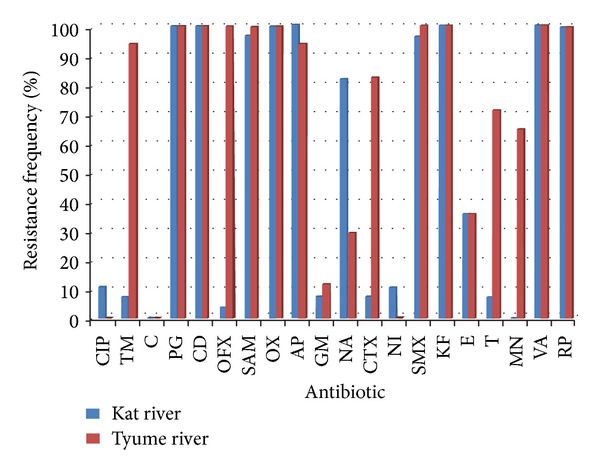
The Frequency of antibiotic-resistant Aeromonas isolates from Kat and Tyume rivers. CIP: ciprofloxacin, TM: trimethoprim, C: chloramphenicol, PG: penicillin, CD: clindamycin, OFX: ofloxacin, SAM: ampicillin-sulbactam, OX: oxacillin, AP: ampicillin, GM: gentamicin, NA: nalidixic acid, CTX: cefotaxime, NI: nitrofurantoin, SMX: sulfamethoxazole, KF: cephalothin, E: erythromycin, T: tetracycline, MN: minocycline, VA: vancomycin, RP: rifamycin.
Aeromonas in this study was able to form biofilms in vitro on a polystyrene microtitre plate. Optical densities and biofilm production status of Aeromonas isolates from Kat and Tyume rivers are as shown in Table 3. Isolates were categorized into four groups according to the criteria as described above. Weak producers of biofilm which make up 53.33% of the isolates were highest among the groups classified. Thirteen isolates (28.9%) were strong producers while, 7 (15.56%) demonstrated moderate capabilities for biofilm production (Figure 2). Table 4 shows the adherent characteristics of Aeromonas isolates from Tyume and Kat rivers.
Table 3.
Optical densities and biofilm production status of Aeromonas species isolated from both Kat and Tyume rivers.
| Isolate code | Isolate source (river) | Mean ODi ± SD | Biofilm producing status |
|---|---|---|---|
| Control (TSB) | 0.069 ± 0.003 | ||
| ISKAE001 | Kat | 0.084 ± 0.006 | Weak |
| ISKAE002 | Kat | 0.0725 ± 0.015 | Weak |
| ISKAE003 | Kat | 0.092 ± 0.021 | Weak |
| ISKAE004 | Kat | 0.095 ± 0.013 | Weak |
| ISKAE005 | Kat | 0.102 ± 0.024 | Moderate |
| ISKAE006 | Kat | 0.073 ± 0.004 | Weak |
| ISKAE007 | Kat | 0.076 ± 0.037 | Weak |
| ISKAE008 | Kat | 0.128 ± 0.056 | Strong |
| ISKAE009 | Kat | 0.123 ± 0.035 | Strong |
| ISKAE010 | Kat | 0.098 ± 0.009 | Weak |
| ISKAE011 | Kat | 0.085 ± 0.004 | Weak |
| ISKAE012 | Kat | 0.119 ± 0.028 | Strong |
| ISKAE020 | Kat | 0.083 ± 0.016 | Weak |
| ISKWA003 | Kat | 0.097 ± 0.006 | Weak |
| ISKWA007 | Kat | 0.073 ± 0.027 | Weak |
| ISKWA013 | Kat | 0.092 ± 0.045 | Weak |
| ISKWA034 | Kat | 0.096 ± 0.03 | Weak |
| ISKJA001 | Tyume | 0.105 ± 0.019 | Moderate |
| ISKJA006 | Tyume | 0.137 ± 0.018 | Strong |
| ISKJA008 | Tyume | 0.080 ± 0.009 | Weak |
| ISKJA011 | Tyume | 0.132 ± 0.04 | Strong |
| ISKJA014 | Tyume | 0.074 ± 0.015 | Weak |
| ISKJA015 | Tyume | 0.14 ± 0.002 | Strong |
| ISKJA016 | Tyume | 0.092 ± 0.021 | Weak |
| ISKJA017 | Tyume | 0.093 ± 0.005 | Weak |
| ISKJA018 | Tyume | 0.152 ± 0.007 | Strong |
| ISKJA019 | Tyume | 0.134 ± 0.012 | Strong |
| ISKJA 021 | Tyume | 0.142 ± 0.009 | Strong |
| ISKJA 030 | Tyume | 0.065 ± 0.003 | Non-producer |
| ISKJA 031 | Tyume | 0.093 ± 0.015 | Weak |
| ISKJA 032 | Tyume | 0.188 ± 0.002 | Strong |
| ISKJA 033 | Tyume | 0.163 ± 0.007 | Strong |
| ISKJA 038 | Tyume | 0.078 ± 0.006 | Weak |
| ISKJA 053 | Tyume | 0.082 ± 0.007 | Weak |
| ISKJA054 | Tyume | 0.092 ± 0.004 | Weak |
| ISKJA 055 | Tyume | 0.119 ± 0.013 | Moderate |
| ISKJA 056 | Tyume | 0.089 ± 0.002 | Weak |
| ISKJA 062 | Tyume | 0.098 ± 0.008 | Weak |
| ISKJA 064 | Tyume | 0.104 ± 0.009 | Moderate |
| ISKWA 033 | Tyume | 0.12 ± 0.002 | Moderate |
| ISKWA 035 | Tyume | 0.117 ± 0.011 | Moderate |
| ISKWA 060 | Tyume | 0.13 ± 0.007 | Strong |
| ISKXA 063 | Tyume | 0.095 ± 0.004 | Weak |
| ISKXA 074 | Tyume | 0.149 ± 0.003 | Strong |
| ISKXA 076 | Tyume | 0.114 ± 0.017 | Moderate |
Figure 2.
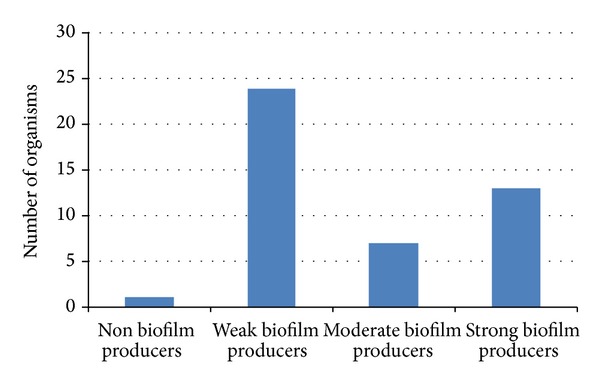
Occurrence of biofilm production of Aeromonas isolates in microtitre plates.
Table 4.
Adherence capability of Aeromonas isolates from Kat and Tyume rivers.
| River | No. of isolates | Adherence properties | |||
|---|---|---|---|---|---|
| NA | WA | MA | SA | ||
| Kat | 17 | 0 | 13 | 1 | 3 |
| Tyume | 28 | 1 | 11 | 6 | 10 |
NA: nonadherent, WA: weakly adherent, MA: moderately adherent, and SA: strongly adherent.
5. Discussion
The presence of multidrug resistant bacteria in surface water is a major public health burden as drug resistant bacteria could be transferred to humans by means of drinking contaminated water which subsequently contributes to the spread and persistence of antibiotic resistance bacteria in general population and environment [29]. Aeromonas species are well distributed in freshwater ecosystems, and their potential to enter distribution systems increases as a result of ineffective water treatment [35]. The result of this investigation demonstrates the presence of multidrug resistant aeromonads in the two rivers assayed. Absolute resistance was observed against several antibiotics with no complete susceptibility to any; however, appreciable susceptibility was observed against ciprofloxacin, chloramphenicol, and nitrofurantoin. Considerable susceptibility was observed for the aminoglycosides tested (gentamicin) with higher susceptibility occurring with isolates from Kat river. Our findings corroborate with the study carried out by Abulhamd [1], which reports susceptibility of Aeromonas species isolated from environmental water source to gentamicin. Similar findings of aminoglycosides susceptibility from water sources have been observed [36].
Resistance to the penicillin group of antibiotics was observed which can be attributed to Aeromonas instinctive resistance to penicillins especially to ampicillin due to the production of β-lactamases. Among the cephalosporins tested, 100% resistance was observed against cephalothin, relative susceptibility was observed against cefotaxime with isolates from Kat river, and insignificant susceptibility with Aeromonas isolates from Tyume river. Bizani and Brandelli [36] reported 100% resistance of Aeromonas species isolated from water distribution system used in a bovine abattoir; however in contrast to our observation, other studies have documented a rare incidence of cephalosporin resistance from Aeromonas isolated from water sources [37, 38].
Aeromonas isolates from both sources showed similar behaviours to some antibiotics; for instance, isolates from both sources showed high level of susceptibility against chloramphenicol, and none was resistant. Similarly, none of the isolates were susceptible to sulfamethoxazole or cephalothin, while the isolates demonstrated a diversified variation in their susceptibility and resistance against trimethoprim and nalidixic acids. These differences in resistance patterns to antibiotics of the isolates from Kat and Tyume rivers could be as a result of sampling locations (rural and semiurban), anthropogenic activities, and other environmental factors.
Variable susceptibility against the quinolones was observed for Aeromonas isolates from both sources, but ofloxacin showed a distinct pattern as it showed 89% susceptibility with isolates from Kat river and absolute resistance against isolates from Tyume river. The reason for these differences may be attributed to enzymatic conduction and selective environmental pressure of Aeromonas isolates from these two settings. The presence of multidrug resistant Aeromonas hydrophila from aquatic animals including fish [39], eel, and catfish [40, 41] could be as a result of widespread distribution of multidrug resistance among A. hydrophila in aquatic (freshwater) habitat; hence, the need for continuous surveillance of emerging and resistance pool of antibiotic resistance determinant.
Integrons are elements that encode a site-specific recombination system that identifies and captures mobile gene cassettes and are closely related to multiple resistances of environmental microorganisms [42]. The role of class 1 integrons in conferring antibiotic resistance to clinical isolates of different bacterial strains is well documented [43–45]. Studies have also shown link between incidence of class 1 integron and antibiotic resistance in bacterial pathogens such as Escherichia coli and Aeromonas salmonicida [46, 47]. The incidences of integron-bearing Aeromonas isolates in our study suggest that these isolates are potential contaminants and that have the possibility for horizontal gene transfer exits. Lukkana et al. [41] has documented the presence of class 1 integron in Aeromonas hydrophilia isolated from Nile Tilapia, in Thailand. Rosser and Young [48] documented the incidence of class 1 integrons in 3.6% of bacteria isolated from the Tay estuary, and Lin and Biyela [49] reported the presence of class 1 integron in 58% of Enterobacteriaceae isolated from Mhlathuze river in KwaZulu-Natal, Republic of South Africa, and this may imply a wide distribution and persistence of class 1 integron in South African aquatic milieu. The presence of integron in a wide variety of bacteria and in different habitat substantiates the horizontal mobility and stability of this gene capture system [50].
Biofilm-producing bacteria have been shown to be associated with numerous human diseases and capable of colonizing a wide range of environments. In aquatic environment, microbial adhesion initiates biofilm formation, exacerbates contamination, reduces the aesthetic quality of the water body, and reduces microbiological safety through augmented survival of pathogens [18, 51]. The result obtained in this study is consistent with the observation of Saidi et al. [52] who found high percentage (95%) of biofilm forming Aeromonas species isolated from a river near the seacoast of Monastir, Tunisia. Also, our findings are also similar to the result documented by Odeyemi et al. [22] who found Aeromonas isolates from estuary in Malaysia to be biofilm formers with high percentage of weak biofilm producers and strong producers trailing as the second in occurrence. This is the first carried out study on the adhesive properties of environmental A. hydrophila strains isolated from aquatic source in the Eastern Cape province of South Africa. Ability of Aeromonas to form biofilm in aquatic environment enhances the recycling of nutrients and minerals in aquatic environment, thereby promoting the growth of potential pathogen in the aquatic milieu. Motility and flagella play a vital role in adhesion, biofilm formation, and colonization of several pathogenic bacteria, such as Aeromonas hydrophila [52, 53]. Our previous study has shown that Aeromonas isolates from the studied microhabitat possess some virulent potential [31], and the ability of these virulent strains to form biofilm in aquatic environment may enhance an elevated waterborne dispersal capacity, an attribute that has been related to an elevated risk of bacterial transmission and infectivity [54]. Biofilm formation and development by microorganisms play a vital role in the pathogenesis of a disease; hence, the biofilm forming capability demonstrated in vitro by Aeromonas isolates from Tyume and Kat rivers further suggests enhanced pathogenic status of these isolates.
6. Conclusion
This study provides a baseline data on the antibiotic resistance profile of Aeromonas species isolated from Kat and Tyume rivers in the Eastern Cape province of South Africa. The result of this study reveals that the antibiotic resistance patterns of Aeromonas species isolated from the two rivers and the incidence of class 1 integron suggest the possibility of horizontal gene transfer of antibiotic resistance determinants in these isolates. The result of this study shows that Aeromonas strains from Kat and Tyume rivers have ability to bind to surfaces and form biofilms which is of public health significance as biofilm formation results in resistance of bacteria to conventional antimicrobial agents and persistent infections.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
Acknowledgment
The authors are grateful to the University of Fort Hare for the financial support.
References
- 1.Abulhamd AT. Characterization of Aeromonas hydrophila isolated from aquatic environments using phenotypic and genotyping methods. Research Journal of Agriculture and Biological Sciences. 2009;5(6):923–931. [Google Scholar]
- 2.Colwell RR, MacDonell MT, De Ley J. Proposal to recognize the family Aeromonadaceae fam. nov. International Journal of Systematic Bacteriology. 1986;36(3):473–477. [Google Scholar]
- 3.Igbinosa IH, Igumbor EU, Aghdasi F, Tom M, Okoh AI. Emerging Aeromonas species infections and their significance in public health. The Scientific World Journal. 2012;2012:13 pages. doi: 10.1100/2012/625023.625023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saavedra MJ, Guedes-Novais S, Alves A, et al. Resistance to β-lactam antibiotics in Aeromonas hydrophila isolated from rainbow trout (Oncorhynchus mykiss) International Microbiology. 2004;7(3):207–211. [PubMed] [Google Scholar]
- 5.Kühn I, Allestam G, Huys G, et al. Diversity, persistence, and virulence of Aeromonas strains isolated from drinking water distribution systems in Sweden. Applied and Environmental Microbiology. 1997;63(7):2708–2715. doi: 10.1128/aem.63.7.2708-2715.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Joseph SW. Aeromonas gastrointestinal disease: a case study in causation? In: Austin B, Altwegg M, Gosling PJ, Joseph SW, editors. The Genus Aeromonas. Chichester, UK: John Wiley & Sons; 1996. pp. 311–326. [Google Scholar]
- 7.Janda JM, Duffey PS. Mesophilic aeromonads in human disease: current taxonomy, laboratory identification, and infectious disease spectrum. Reviews of Infectious Diseases. 1988;10(5):980–997. doi: 10.1093/clinids/10.5.980. [DOI] [PubMed] [Google Scholar]
- 8.Figueras MJ, Alperi A, Saavedra MJ, et al. Clinical relevance of the recently described species Aeromonas aquariorum . Journal of Clinical Microbiology. 2009;47(11):3742–3746. doi: 10.1128/JCM.02216-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huddleston JR, Zak JC, Jeter RM. Antimicrobial susceptibilities of Aeromonas spp. isolated from environmental sources. Applied and Environmental Microbiology. 2006;72(11):7036–7042. doi: 10.1128/AEM.00774-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gordon L, Cloeckaert A, Doublet B, et al. Complete sequence of the floR-carrying multiresistance plasmid pAB5S9 from freshwater Aeromonas bestiarum . Journal of Antimicrobial Chemotherapy. 2008;62(1):65–71. doi: 10.1093/jac/dkn166. [DOI] [PubMed] [Google Scholar]
- 11.Halling-Sørensen B, Nors Nielsen S, Lanzky PF, Ingerslev F, Holten Lützhøft HC, Jørgensen SE. Occurrence, fate and effects of pharmaceutical substances in the environment: a review. Chemosphere. 1998;36(2):357–393. doi: 10.1016/s0045-6535(97)00354-8. [DOI] [PubMed] [Google Scholar]
- 12.Ash RJ, Mauck B, Morgan M. Antibiotic resistance of gram-negative bacteria in rivers, United States. Emerging Infectious Diseases. 2002;8(7):713–716. doi: 10.3201/eid0807.010264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borrell N, Figueras MJ, Guarro J. Phenotypic identification of Aeromonas genomospecies from clinical and environmental sources. Canadian Journal of Microbiology. 1998;44(2):103–108. doi: 10.1139/w97-135. [DOI] [PubMed] [Google Scholar]
- 14.Joseph SW, Carnahan AM. Update on the genus Aeromonas . ASM News. 2000;66:218–223. [Google Scholar]
- 15.Hall RM, Stokes HW. Integrons: novel DNA elements which capture genes by site-specific recombination. Genetica. 1993;90(2-3):115–132. doi: 10.1007/BF01435034. [DOI] [PubMed] [Google Scholar]
- 16.Hall RM, Collis CM. Mobile gene cassettes and integrons: capture and spread of genes by site-specific recombination. Molecular Microbiology. 1995;15(4):593–600. doi: 10.1111/j.1365-2958.1995.tb02368.x. [DOI] [PubMed] [Google Scholar]
- 17.Bayoudh S, Ponsonnet L, Ouada HB, Bakhrouf A, Othmane A. Bacterial detachment from hydrophilic and hydrophobic surfaces using a microjet impingement. Colloids and Surfaces A. 2005;266(1–3):160–167. [Google Scholar]
- 18.Simões LC, Azevedo N, Pacheco A, Keevil CW, Vieira MJ. Drinking water biofilm assessment of total and culturable bacteria under different operating conditions. Biofouling. 2006;22(2):91–99. doi: 10.1080/08927010600598603. [DOI] [PubMed] [Google Scholar]
- 19.Kirov SM, Castrisios M, Shaw JG. Aeromonas flagella (polar and lateral) are enterocyte adhesins that contribute to biofilm formation on surfaces. Infection and Immunity. 2004;72(4):1939–1945. doi: 10.1128/IAI.72.4.1939-1945.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.September SM, Els FA, Venter SN, Brözel VS. Prevalence of bacterial pathogens in biofilms of drinking water distribution systems. Journal of Water and Health. 2007;5(2):219–227. [PubMed] [Google Scholar]
- 21.Donlan RM. Biofilm formation: a clinically relevant microbiological process. Clinical Infectious Diseases. 2001;33(8):1387–1392. doi: 10.1086/322972. [DOI] [PubMed] [Google Scholar]
- 22.Odeyemi OA, Asmat A, Usup G. Antibiotics resistance and putative virulence factors of Aeromonas hydrophila isolated from estuary. Journal of Microbiology, Biotechnology and Food Sciences. 2012;1(6):1339–1357. [Google Scholar]
- 23.Probert HM, Gibson GR. Bacterial biofilms in the human gastrointestinal tract. Current Issues in Intestinal Microbiology. 2002;3(2):23–27. [PubMed] [Google Scholar]
- 24.Melchior MB, Vaarkamp H, Fink-Gremmels J. Biofilms: a role in recurrent mastitis infections? Veterinary Journal. 2006;171(3):398–407. doi: 10.1016/j.tvjl.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 25.Figueras MJ, Suarez-Franquet A, Chacón MR, et al. First record of the rare species Aeromonas culicicola from a drinking water supply. Applied and Environmental Microbiology. 2005;71(1):538–541. doi: 10.1128/AEM.71.1.538-541.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clinical and Laboratory Standards Institute (CLSI) Methods for Dilution of Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically: Approved Standard M7-A7. 7th edition. Wayne, Pa, USA: Clinical and Laboratory Standards Institute; 2006. [Google Scholar]
- 27.Krumperman PH. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Applied and Environmental Microbiology. 1983;46(1):165–170. doi: 10.1128/aem.46.1.165-170.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miranda CD, Zemelman R. Antimicrobial multiresistance in bacteria isolated from freshwater Chilean salmon farms. Science of the Total Environment. 2002;293(1–3):207–218. doi: 10.1016/s0048-9697(02)00022-0. [DOI] [PubMed] [Google Scholar]
- 29.Tao R, Ying GG, Su HC, Zhou HW, Sidhu JPS. Detection of antibiotic resistance and tetracycline resistance genes in Enterobacteriaceae isolated from the Pearl rivers in South China. Environmental Pollution. 2010;158(6):2101–2109. doi: 10.1016/j.envpol.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3rd edition. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 31.Igbinosa IH, Okoh AI. Detection and distribution of putative virulence associated genes in Aeromonas species from freshwater and wastewater treatment plant. Journal of Basic Microbiology. 2013 doi: 10.1002/jobm.201200351. [DOI] [PubMed] [Google Scholar]
- 32.Stepanovic S, Vukovic D, Davic I, Savić B, Švabić-Vlahović M. A modified microtiter-plate test for quantification of staphylococcal biofilm formation. Journal of Microbiological Methods. 2000;40(2):175–179. doi: 10.1016/s0167-7012(00)00122-6. [DOI] [PubMed] [Google Scholar]
- 33.Cevahir N, Demir M, Kaleli I, Gurbuz M, Tikvesli S. Evaluation of biofilm production, gelatinase activity, and mannose-resistant hemagglutination in Acinetobacter baumannii strains. Journal of Microbiology, Immunology and Infection. 2008;41(6):513–518. [PubMed] [Google Scholar]
- 34.Hassan A, Usman J, Kaleem F, Omair M, Khalid A, Iqbal M. Evaluation of different detection methods of biofilm formation in the clinical isolates. Brazilian Journal of Infectious Diseases. 2011;15(4):305–311. [PubMed] [Google Scholar]
- 35.Holmes P, Niccolls LM, Sartory DP. The ecology of mesophilic Aeromonas in the aquatic environment. In: Austin B, Altwegg M, Gosling PJ, Joseph S, editors. The Genus Aeromonas. New York, NY, USA: Wiley; 1996. pp. 127–150. [Google Scholar]
- 36.Bizani D, Brandelli A. Antimicrobial susceptibility, hemolysis, and hemagglutination among Aeromonas spp. isolated from water of a bovine abattoir. Brazilian Journal of Microbiology. 2001;32(4):334–339. [Google Scholar]
- 37.Miranda CD, Castillo G. Resistance to antibiotic and heavy metals of motile aeromonads from Chilean freshwater. Science of the Total Environment. 1998;224(1–3):167–176. doi: 10.1016/s0048-9697(98)00354-4. [DOI] [PubMed] [Google Scholar]
- 38.Goñi-Urriza M, Capdepuy M, Arpin C, Raymond N, Pierre Caumette CQ. Impact of an urban effluent on antibiotic resistance of riverine Enterobacteriaceae and Aeromonas spp. Applied and Environmental Microbiology. 2000;66(1):125–132. doi: 10.1128/aem.66.1.125-132.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hatha M, Vivekanandhan AA, Julie Joice G, Christol C. Antibiotic resistance pattern of motile aeromonads from farm raised fresh water fish. International Journal of Food Microbiology. 2005;98(2):131–134. doi: 10.1016/j.ijfoodmicro.2004.05.017. [DOI] [PubMed] [Google Scholar]
- 40.Penders J, Stobberingh EE. Antibiotic resistance of motile aeromonads in indoor catfish and eel farms in the southern part of The Netherlands. International Journal of Antimicrobial Agents. 2008;31(3):261–265. doi: 10.1016/j.ijantimicag.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 41.Lukkana M, Wongtavatchai J, Chuanchuen R. Class 1 integrons in Aeromonas hydrophila isolates from farmed Nile Tilapia (Oreochromis nilotica) Journal of Veterinary Medical Sciences. 2012;74(4):435–440. doi: 10.1292/jvms.11-0441. [DOI] [PubMed] [Google Scholar]
- 42.Ma L, Zhang XX, Cheng S, et al. Occurrence, abundance and elimination of class 1 integrons in one municipal sewage treatment plant. Ecotoxicology. 2011;20(5):968–973. doi: 10.1007/s10646-011-0652-y. [DOI] [PubMed] [Google Scholar]
- 43.Kücken D, Feucht HH, Kaulfers PM. Association of qacE and qacEΔ1 with multiple resistance to antibiotics and antiseptics in clinical isolates of Gram-negative bacteria. FEMS Microbiology Letters. 2000;183(1):95–98. doi: 10.1111/j.1574-6968.2000.tb08939.x. [DOI] [PubMed] [Google Scholar]
- 44.White PA, Rawlinson WD. Current status of the aadA dfr gene cassette families. Journal of Antimicrobial Chemotherapy. 2001;47(4):495–496. doi: 10.1093/jac/47.4.495. [DOI] [PubMed] [Google Scholar]
- 45.Leverstein-Van Hall MA, Blok HEM, Donders ART, Paauw A, Fluit AC, Verhoe J. Multidrug resistance among enterobacteriaceae is strongly associated with the presence of integrons and is independent of species or isolate origin. Journal of Infectious Diseases. 2003;187(2):251–259. doi: 10.1086/345880. [DOI] [PubMed] [Google Scholar]
- 46.Sørum H, L’Abée-Lund TM, Solberg A, Wold A. Integron-containing IncU R plasmids pRAS1 and pAr-32 from the fish pathogen Aeromonas salmonicida . Antimicrobial Agents and Chemotherapy. 2003;47(4):1285–1290. doi: 10.1128/AAC.47.4.1285-1290.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gaze WH, Abdouslam N, Hawkey PM, Wellington EMH. Incidence of class 1 integrons in a quaternary ammonium compound-polluted environment. Antimicrobial Agents and Chemotherapy. 2005;49(5):1802–1807. doi: 10.1128/AAC.49.5.1802-1807.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rosser SJ, Young HK. Identification and characterization of class 1 integrons in bacteria from an aquatic environment. Journal of Antimicrobial Chemotherapy. 1999;44(1):11–18. doi: 10.1093/jac/44.1.11. [DOI] [PubMed] [Google Scholar]
- 49.Lin J, Biyela PT. Convergent acquisition of antibiotic resistance determinants amongst the Enterobacteriaceae isolates of the Mhlathuze River, KwaZulu-Natal (RSA) Water SA. 2005;31(2):257–260. [Google Scholar]
- 50.Dubois V, Parizano MP, Arpin C, Coulange L, Bezian MC, Quentin C. High genetic stability of integrons in clinical isolates of Shigella spp. of worldwide origin. Antimicrobial Agents and Chemotherapy. 2007;51(4):1333–1340. doi: 10.1128/AAC.01109-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tsai YP. Simulation of biofilm formation at different assimilable organic carbon concentrations under lower flow velocity condition. Journal of Basic Microbiology. 2005;45(6):475–485. doi: 10.1002/jobm.200510583. [DOI] [PubMed] [Google Scholar]
- 52.Saidi N, Snoussi M, Usai D, Zanetti S, Bakhrouf A. Adhesive properties of Aeromonas hydrophila strains isolated from Tunisian aquatic biotopes. African Journal of Microbiology Research. 2011;5(31):5644–5655. [Google Scholar]
- 53.Rabaan AA, Gryllos I, Tomás JM, Shaw JG. Motility and the polar flagellum are required for Aeromonas caviae adherence to HEp-2 cells. Infection and Immunity. 2001;69(7):4257–4267. doi: 10.1128/IAI.69.7.4257-4267.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fux CA, Costerton JW, Stewart PS, Stoodley P. Survival strategies of infectious biofilms. Trends in Microbiology. 2005;13(1):34–40. doi: 10.1016/j.tim.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 55.Nawaz M, Khan SA, Khan AA, et al. Detection and characterization of virulence genes and integrons in Aeromonas veronii isolated from catfish. Food Microbiology. 2010;27(3):327–331. doi: 10.1016/j.fm.2009.11.007. [DOI] [PubMed] [Google Scholar]


