Abstract
The Wnt1-Cre transgenic mouse line is extensively used in the study of the development of the neural crest and its derivatives and the midbrain. The Wnt1 gene has important developmental roles in formation of the midbrain–hindbrain boundary, regulation of midbrain size, and neurogenesis of ventral midbrain dopaminergic (mDA) neurons. Here, we report that Wnt1-Cre transgenic mice exhibit phenotypes in multiple aspects of midbrain development. Significant expansion of the midbrain and increased proliferation in the developing inferior colliculus is associated with ectopic expression of Wnt1. Marked elevation of Wnt1 expression in the ventral midbrain is correlated with disruption of the differentiation program of ventral mDA neurons. We find that these phenotypes can be attributed to ectopic expression of Wnt1 from the Wnt1-Cre transgene leading to the ectopic activation of canonical Wnt/β-catenin signaling. Since these caveats could complicate the utility of Wnt1-Cre in some developmental circumstances, we report a new Wnt1-Cre2 transgenic mouse line that can serve the same purposes as the original without the associated phenotypic complications. These studies reveal an important caveat to a widely-used reagent, provide an improved version of this reagent, and indicate that the original Wnt1-Cre transgenic mouse line may be useful as a gain of function model for interrogating Wnt signaling mechanisms in multiple aspects of midbrain development.
Keywords: Neural crest, Wnt1-Cre, Midbrain, Wnt signaling, Dopaminergic neurons, Craniofacial
Introduction
The Wnt1-Cre mouse transgenic line is widely used for interrogating gene function and cell lineage relationships in a variety of developmental contexts including early neural crest migration, secondary palate, calvaria, cardiac outflow tract, and midbrain development (Brewer et al., 2004; Chai et al., 2000; Ito et al., 2003; Jiang et al., 2000, 2002; Katayama et al., 2011; Merrill et al., 2006; Tallquist and Soriano, 2003; Yoshida et al., 2008). Its utility is based on the fact that the Wnt1 gene is highly expressed in the dorsal neural tube prior to the emigration of the neural crest and in the midbrain–hindbrain boundary (MHB) at the initiation of midbrain development. The Wnt1-Cre line, in combination with various Cre reporters, has thus been heavily employed for lineage tracing studies of neural crest cells and the midbrain.
Wnt1 has multiple roles in midbrain development. Inactivation of the Wnt1 gene in mice resulted in loss of the entire mid/hindbrain region (McMahon and Bradley, 1990; Thomas and Capecchi, 1990). Ectopic expression of Wnt1 throughout the CNS under the control of the HoxB4 enhancer dramatically increased mitogenesis without altering patterning (Dickinson et al., 1994). Further, when Wnt1 was ectopically expressed under the control of the endogenous engrailed1 promoter in En1Wnt1 mice, pattern formation in the MHB was maintained, but increased proliferation and drastic overgrowth of the caudal-dorsal midbrain were observed (Panhuysen et al., 2004).
Ventral midbrain dopaminergic (mDA) neurons develop in close proximity to the MHB, a source of Wnt1 expression, and ultimately modulate a broad range of neural processes, including movement, cognition and reward; their dysfunction leads to severe neurological disorders such as Parkinson's disease (PD). Therefore, understanding the molecular control of mDA differentiation is critical in the ultimate goal of therapeutic mDA neuron production, engraftment and function. The mDA differentiation program can be divided into three distinct steps (Prakash and Wurst, 2006b). First, induction of a progenitor field within the neuroepithelium that is competent to generate mDA precursors occurs between E8.5–E10.5 in the mouse and is characterized by the expression of general proneural genes such as the Ngn2 transcription factor. Second, the specification of postmitotic mDA precursors marked by expression of markers such as the nuclear receptor family member Nurr1 occurs at E10.5–E12.5. Finally, terminal differentiation begins by E12.5 and is characterized by the expression of tyrosine hydroxylase (TH), the rate-limiting enzyme in DA synthesis (Prakash and Wurst, 2006a, 2006b). Wnt signaling has been shown to have critical roles in the second and third steps of mDA neurogenesis, but has not been implicated in the first (Prakash et al., 2006; Tang et al., 2009). It is clear that Wnt1 is involved in establishment of the committed mDA progenitor domain because ectopic Wnt1 expression in the ventral hindbrain induced ectopic mDA neurons and because its expression was necessary for ectopic induction of mDA neurons by Fgf8 and Shh in ventral forebrain explants (Prakash et al., 2006). Loss of the obligate downstream Wnt signaling mediator β-catenin within the ventral midbrain in Shh-Cre; β-cateninfl/fl embryos led to a dramatic reduction in mDA neurogenesis without changes in ventral midbrain patterning, whereas the later loss of β-catenin in differentiating mDA neurons revealed a role in the final step of mDA differentiation (Tang et al., 2009). Interestingly, hyperactivation of the pathway within the ventral midbrain in Shh-Cre; β-cateninEx3 embryos resulted in a loss of mDA neurons, possibly by preventing cell cycle exit and differentiation of Ngn2-expressing committed progenitors to postmitotic progenitors (Tang et al., 2010). In these embryos, the induction of an mDA-competent progenitor field occurred and expression of Ngn2 and Nurr1 were slightly expanded. Taken together, these studies established the importance of Wnt signaling during midbrain development. During our studies of gene function in neural crest, we discovered that midbrain development is perturbed by the Wnt1-Cre transgene; since this transgene is so widely used, we investigated these phenotypes in greater depth.
Materials and methods
Genotyping
DNA was isolated from either tail or embryonic yolk sac samples, depending upon embryonic stage. qPCR was performed for the Wnt1-Cre transgene using primers in the second and third introns of Wnt1 to determine quantity of transgene relative to the wild-type locus: 5′-CCA-CCT-CTT-CGG-CAA-GAT-CG-3′ forward and 5′-GCT-AGA-AAG-AAT-CTG-GTG-CTG-ACC-3′ reverse. This genotyping was conducted using Bambi as a reference gene for copy number.
Histology, immunofluorescence and in situ hybridization
Embryos for histology were fixed in Bouin's fixative, dehydrated through graded ethanol concentrations and embedded in paraffin. Sections were subsequently cut at 7 µm thickness and stained with hematoxylin and eosin. Embryos for immunofluorescence and in situ hybridization were fixed in 4% PFA, dehydrated to 25% sucrose and embedded in OCT; sections were cut at a thickness of 12 µm. Immunostaining was performed according to standard protocols using antibodies recognizing tyrosine hydroxylase (1:250; Millipore cat# AB152), BrdU (1:200; Abcam AB6326), Lef1 (1:75; Cell Signaling Technology product# 2230), and CyclinD1 (1:200; ThermoScientific cat# RB-010-P*). For BrdU experiments, pregnant dams were injected with 50 µg BrdU per gram body weight at 20 or 30 min prior to euthanasia for E12.5 or E15.5 embryos, respectively. In situ hybridization was performed according to standard protocols. In situ hybridized slides were counterstained with Orange G (Fig. 3; Supplementary Fig. 3; Supplementary Fig. 4I–K) or Nuclear Fast Red (Supplementary Fig. 1). The Wnt1 in situ probe hybridizes to base pairs 437–852 on the Wnt1 mRNA (RefSeq NM_021279), corresponding to the entire second exon and roughly 60% of the third exon.
Fig. 3.
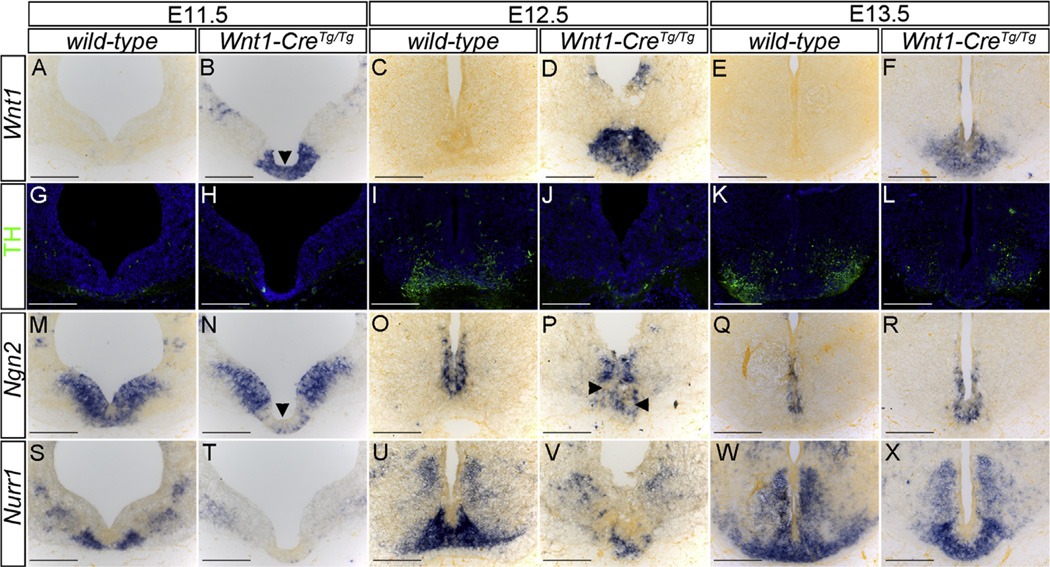
Elevated Wnt1 expression in the ventral midbrain disrupts the mDA differentiation program. (A–F) In situ hybridization of coronal sections of the ventral midbrain reveals a dramatic elevation in Wnt1 expression in the ventral midbrain region of Wnt1-CreTg/Tg embryos (B,D,F) compared with wild-type (A,C,E). (G–L) Immunostaining of sections adjacent to A–F with an antibody to tyrosine hydroxylase (TH) reveals a nearly complete absence of mDA neurons in E12.5 Wnt1-CreTg/Tg embryos (J) compared to wild-type (I). Dramatically reduced TH staining is observed in E13.5 Wnt1-CreTg/Tg embryos (L) compared to wild-type (K). (M–R) In situ hybridization of sections adjacent to A–F reveals the absence of expression of Ngn2 within the Wnt1 misexpression domain in E11.5 Wnt1-CreTg/Tg embryos (arrowheads in B,N) compared to wild-type (M). At E12.5, reduced Ngn2 expression is observed within this domain in Wnt1-CreTg/Tg embryos (arrowheads in P) compared to wild-type (O), whereas Ngn2 expression appears to have recovered at E13.5 (Q,R). In situ hybridization of sections adjacent to A–F reveals a complete loss of Nurr1 expression in Wnt1-CreTg/Tg embryos compared to wild-type at E11.5 (S,T) and E12.5 (U,V). Nearly complete recovery of Nurr1 expression is observed at E13.5 (W,X). Scale bars: 200 µm.
RT-PCR, qRT-PCR and western blots
Total RNA was extracted from snap frozen E12.5 and E13.5 embryos, according to Invitrogen's TRIzol Reagent protocol. cDNA was synthesized using Invitrogen's First-Strand Synthesis kit using random hexamers to prime the reverse transcriptase. Wnt1-Cre transcript sequencing was performed on RT-PCR products obtained using the following primer sets: Wnt1 5′utr 5′-CTC-ATT-GTC-TGT-GGC-CCT-GAC-C-3′ forward with Cre 5′-ACG-CCT-GGC-GAT-CCC-TGA-AC-3′ reverse, and Cre 5′-CGC-TGG-AGT-TTC-AAT-ACC-GG-3′ forward with Wnt1 exon 3 5′-CGG-AGG-TGA-TTG-CGA-AGA-TG-3′ reverse. Wnt1-Gal4 transcript sequencing was performed on RT-PCR products obtained according to the above scheme, replacing Cre primers with the following Gal4 primers: 5′-GTC-GGC-AAA-TAT-CGC-ATG-CTT-G-3′ reverse and 5′-TTC-AAA-ACC-ACT-GTC-ACC-TGG-3′ forward. Expression analysis of total Wnt1 was carried out by qRT-PCR with Wnt1 primers in the third and fourth exons (e3e4): 5′-CAT-CTT-CGC-AAT-CAC-CTC-CG-3′ forward and 5′-GTG-GCA-TTT-GCA-CTC-TTG-G-3′ reverse. Endogenous Wnt1 expression was quantified using a Wnt1 primer pair in the first and second exons (e1e2): 5′-ACA-GTC-GTC-AGA-ACC-GCA-GC-3′ forward and 5′-TCA-ACA-GGT-TCG-TGG-AGG-AGG-C-3′ reverse. These experiments were conducted using GAPDH as a reference gene. For protein quantification, E14.5 heads were homogenized in NP40 buffer and subjected to 10% SDS-PAGE and western blotting according to standard techniques. Blots were probed with Wnt1 (Abcam cat# ab15251) and non-muscle myosin heavy chain IIA (Covance cat# PRB-440) antibodies and analyzed using an ImageQuant LAS 4000 scanner and software (GE).
Generation of Wnt1-Cre2 transgenic mice
Wnt1 promoter and enhancer sequences were isolated from the pWEXP3C plasmid (Danielian and McMahon, 1996). The 1.3 kb Wnt1 5′ promoter sequence was isolated by digestion with NotI and EcoRV from the pWEXP3C plasmid and placed upstream of a cDNA encoding Cre recombinase, followed by a 5.5 kb BglII fragment that includes the Wnt1 3′ enhancer. Transgenic lines were backcrossed to 129S4 mice and examined for Cre activity by crossing to ROSA26R reporter mice (Soriano, 1999). Allele specific genotyping of Wnt1-Cre2 transgenic mice was performed by PCR with primers (5′-ACA-GCG-AAC-CAT-GCT-GCC-TG-3′) and (5′-CAT-GTC-CAT-CAG-GTT-CTT-GC-3′) giving a 465 bp product. The Wnt1-Cre2 mice (129S4-Tg(Wnt1-cre)1Sor/J) are being deposited to the Jackson Laboratory Mouse Repository and will be available under Stock# 22137.
Results and discussion
Abnormal Wnt1 expression and its phenotypic consequences in Wnt1-Cre mice
Over the course of our studies, we came to appreciate that Wnt1-CreTg/+ embryos display a marked enlargement of the midbrain beginning at E11.5, concomitant with dilation of the midbrain aqueduct (Fig. 1A–F). At E15.5, the inferior colliculi were thin and expanded, with increased CyclinD1 expression and overt cell proliferation as measured by BrdU incorporation, phenotypes that were exacerbated in Wnt1-CreTg/Tg brains (Supplementary Fig. 1A–C,G–I). Moreover, at E12.5, Wnt1-CreTg/+ embryos also displayed a reduction in TH-expressing mDA neurons, which were nearly completely absent from Wnt1-CreTg/Tg embryos (Supplementary Fig. 2A–C). These phenotypes were completely penetrant and consistent in Wnt1-Cre mice obtained directly from Dr. A.P. McMahon or from the Jackson Laboratories (Tg(Wnt1-cre)11Rth Tg(Wnt1-GAL4)11Rth/J), on 129S4 and C57Bl/6J backgrounds.
Fig. 1.
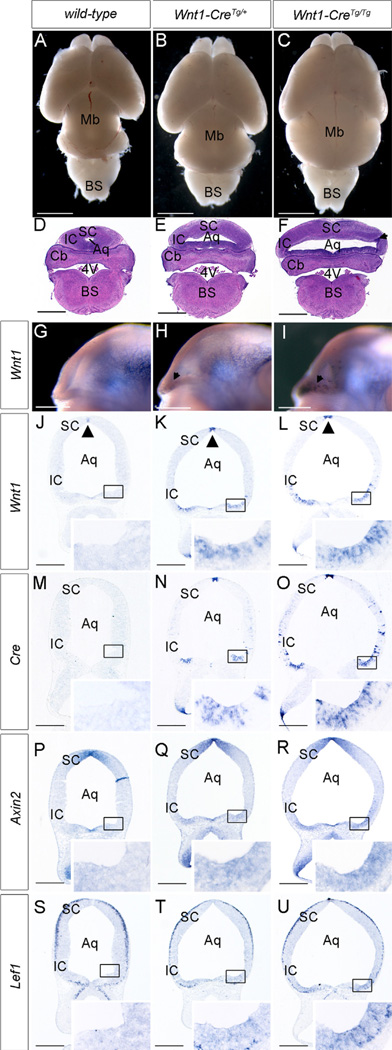
Wnt1 misexpression activates ectopic canonical Wnt/β-catenin signaling and disrupts midbrain development in Wnt1-Cre transgenic mice. (A–C) Whole mount view of P0 brains reveals slightly expanded midbrain in Wnt1-CreTg/+ mice (B) and dramatically expanded midbrain in Wnt1-CreTg/Tg mice (C) compared with wild-type (A). (D–F) Histological coronal sections of E18.5 mouse brains show dramatic expansion of the midbrain in Wnt1-CreTg/+ mice (E) and more severe expansion in Wnt1-CreTg/Tg mice (F). This expansion mostly affects the inferior colliculus and is accompanied by its thinning and increased nuclear density (arrowhead in F). (G–I) Whole mount in-situ hybridization of E12.5 embryos reveals punctate misexpression of Wnt1 around the caudolateral edge of the midbrain–hindbrain boundary in Wnt1-CreTg/+ (arrows in H) and Wnt1-CreTg/Tg embryos (arrows in I) compared with wild-type (G) (J–L) In situ hybridization of E12.5 coronal sections reveals ectopic Wnt1 expression in the caudolateral aspect of the mesencephalon in Wnt1-CreTg/+ embryos (K), with increased intensity and extent in Wnt1-CreTg/Tg embryos (L) (inset boxes in J–L). The expression of Wnt1 in the dorsal aspect of the midbrain is also increased in intensity at these stages (arrowheads, J–L). (M–U) In situ hybridization of sections adjacent to J–L reveals Cre expression (M–O), Axin2 upregulation (P–R), and Lef1 upregulation (S–U) in the caudolateral aspect of the mesencephalon of Wnt1-CreTg/+ (N,Q,T) and Wnt1-CreTg/Tg (O,R,U) embryos in regions corresponding to those with ectopic expression of Wnt1. Scale bars: (A–C) 2 mm, (D–F) 1 mm, (G–U) 500 µm. (SC) superior colliculus, (IC) inferior colliculus, (Aq) aqueduct, (Cb) cerebellum, (4 V) fourth ventricle, (BS) brain stem.
To understand the basis for these phenotypes, we examined Wnt1 expression by in-situ hybridization and found that at E12.5, Wnt1 consistently exhibited elevated expression in the dorsal neural tube and ectopic expression in the caudolateral neuroectoderm of the mesencephalon (Fig. 1G–L). In addition, Wnt1 exhibited expanded expression in more rostral sections of the ventral midbrain, adjacent to regions with altered TH staining (Fig. 3A–F; Supplementary Fig. 2D–F). Furthermore, the pattern of misexpression of Wnt1 was identical to that of the Cre transcript in Wnt1-Cre embryos and correlated with the positions of midbrain expansion and mDA neuron loss (Fig. 1M–O; Supplementary Fig. 2A–C). These results indicate that Wnt1-Cre transgenic mice exhibit ectopic upregulation of Wnt1 that correlates with the midbrain phenotypes that we observe.
Ectopic expression of Wnt1 from the Wnt1-Cre transgene
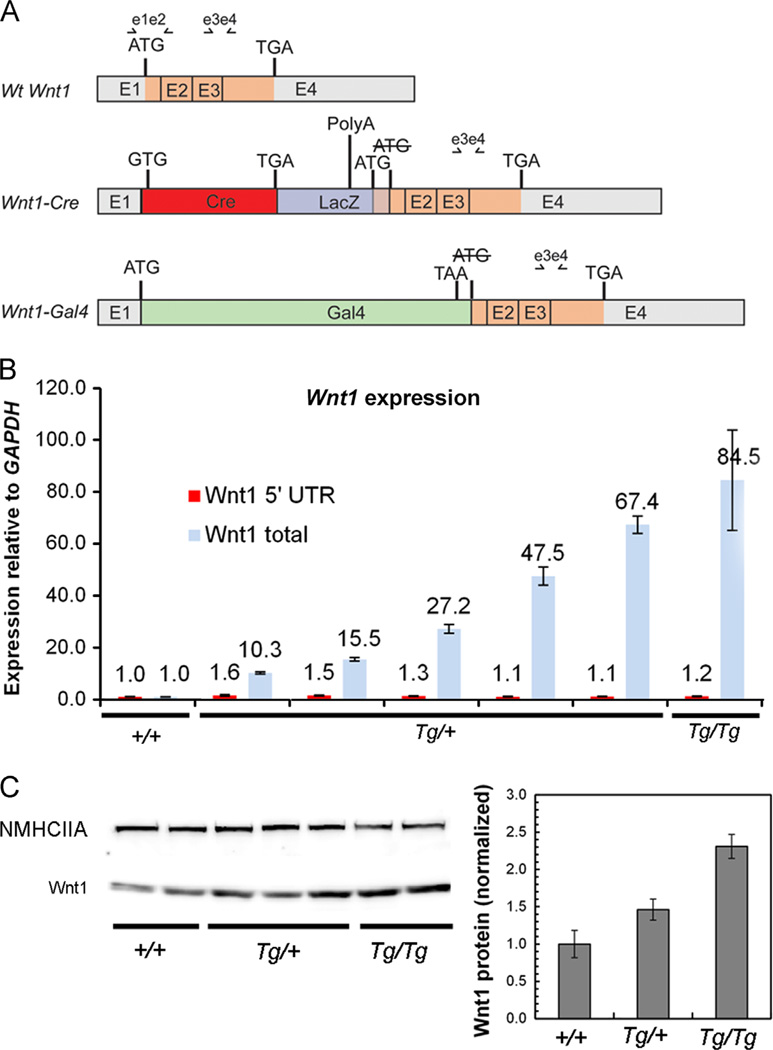
The Wnt1-Cre transgene was generated by inserting a Cre recombinase cDNA between a 1.3 kb upstream element including the promoter and the Wnt1 coding sequence, destroying the Wnt1 initiation ATG in the process. This was followed by a 5.5 kb downstream genomic sequence that carries Wnt1 enhancer elements (Danielian et al., 1998). This transgene was co-injected with a Wnt1-Gal4 transgene constructed using a similar strategy (http://jaxmice.jax.org/strain/003829.html). To determine the basis for the misexpression of full length Wnt1, we performed a series of RT-PCR sequence experiments, identifying two transgene-specific species of Wnt1 transcript in Wnt1-CreTg embryos (Fig. 2A). The first of these included the 5′ UTR of Wnt1 followed by Cre and the 3′ part of exon 1 spliced normally to the remaining 3′ exons. Although the Wnt1 ATG initiator codon was mutated to ATC, we found an in-frame ATG located in the vector sequence 165 bp upstream of the usual Wnt1 start codon. This reading frame is predicted to encode a protein with 55 additional N-terminal amino acids, which would be cleaved as part of the signal sequence. A second transgene-specific transcript instead incorporated the Gal4 gene sequence into the first exon of Wnt1 but did not possess an in-frame ATG that could explain Wnt1 expression.
Fig. 2.
Molecular basis for ectopic Wnt1 expression in Wnt1-CreTg embryos. (A) Schematic representation of wild-type Wnt1 transcript and two Wnt1-CreTg-specific Wnt1 transcripts. A Wnt1-Cre chimeric transcript carrying the complete Cre open reading frame, followed by part of the LacZ gene, also has an open reading frame predicted to encode the entire Wnt1 transcript plus 165 5′ base-pairs derived from the LacZ sequence. The Wnt1-Gal4 chimeric transcript is not predicted to encode a complete Wnt1 open reading frame. (B) Quantitative RT-PCR using primers recognizing all three Wnt1 transcripts (e3e4 in A) indicates variable overexpression of Wnt1 transcript in E13.5 embryonic Wnt1-CreTg heads. Quantitative RT-PCR using primers recognizing only the wild-type Wnt1 transcript indicate that endogenous Wnt1 expression remains similar between genotypes (e1e2 in A). Error bars represent standard deviation from qRT-PCR performed on individual embryos in triplicate. (C) Western blot analysis of Wnt1 from E14.5 embryonic head extracts reveals a 1.5-fold upregulation of Wnt1 protein in Wnt1-CreTg/+ embryos, and an approximately 2.3-fold upregulation of Wnt1 protein in Wnt1-CreTg/Tg embryos. Non-muscle myosin heavy chain IIA was used as a loading control, and error bars represent standard error.
Using quantitative RT-PCR, we found that total Wnt1 transcript was between 10.3 and 67.4 fold more abundant in E14.5 Wnt1-CreTg/+ embryonic heads and 84.5 fold more abundant in Wnt1-CreTg/Tg than in controls (Fig. 2B). To test whether this transgene might also be affecting expression of the endogenous Wnt1 locus, we performed quantitative RT-PCR to differentiate between endogenous Wnt1 transcript and total transcript and found that endogenous Wnt1 transcript remained similar to wild-type levels in Wnt1-CreTg embryos. Western blot analysis of E14.5 embryonic head lysates confirmed an increase in Wnt1 protein correlating with genotype (Fig. 2C). Since Wnt1 is typically an activator of canonical Wnt/β-catenin signaling, we examined the expression of the downstream transcriptional targets Axin2 and Lef1; these two genes were also upregulated in the regions of ectopic Wnt1 upregulation (Fig. 1P–U; Supplementary Fig. 1D–I; Supplementary Fig. 2G–I). Taken together, these results indicate that ectopic Wnt1 expressed from the Wnt1-Cre transgene activates canonical Wnt/β-catenin signaling, possibly leading to the midbrain phenotypes that we observe.
Ectopic Wnt signaling perturbs the first step of the mDA differentiation program
To begin to determine the developmental mechanisms by which elevated Wnt1 expression in the ventral midbrain (vMb) disrupts mDA neurogenesis, we examined expression of key markers in a developmental sequence from E11.5 to E13.5. At each of these stages, dramatically elevated Wnt1 expression was observed in the vMb of Wnt1-CreTg/Tg embryos, correlating with loss of TH-expressing mDA neurons (Fig. 3A–L) and increased cell proliferation in the vMb of E12.5 Wnt1-CreTg/Tg embryos (Supplementary Fig. 3D–F). Examination of the generic proneural marker Ngn2 revealed a striking loss of expression in Wnt1-CreTg/Tg at E11.5, specifically within the domain of Wnt1 overexpression (Fig. 3B,N). Whereas the domain of Ngn2 expression was still perturbed within the domain of Wnt1 overexpression at E12.5, nearly complete recovery of expression was observed at E13.5, at which point a few TH-positive mDA neurons had also formed (Fig. 3O–R). The expression of Nurr1, a marker of postmitotic mDA progenitors and a gene necessary for the expression of TH in mature mDA neurons (Prakash andWurst, 2006b), was completely absent from the vMb at E11.5 and dramatically reduced in E12.5 Wnt1-CreTg/Tg embryos compared with control (Fig. 3S–V). Again, recovery of Nurr1 expression was observed by E13.5, when some TH-expressing neurons were apparent (Fig. 3W–X). Taken together, these results provide evidence that elevated Wnt/β-catenin signaling in the vMb of Wnt1-CreTg/Tg embryos causes extended maintenance of a proliferative, mDA-incompetent domain. This idea is consistent with findings that treatment of rat ventral midbrain precursor cultures with Wnt1-conditioned media increased the total number of neurons by affecting proliferation and not the proportion of TH-positive cells (Castelo-Branco et al., 2003). Our results suggest that Wnt/β-catenin signaling levels must be tightly controlled to allow the correct initiation of an mDA-competent precursor domain in the vMb and indicate that the Wnt1-CreTg/Tg mouse line may be a useful tool in the detailed study of this process.
Generation and characterization of a new Wnt1-Cre transgenic mouse
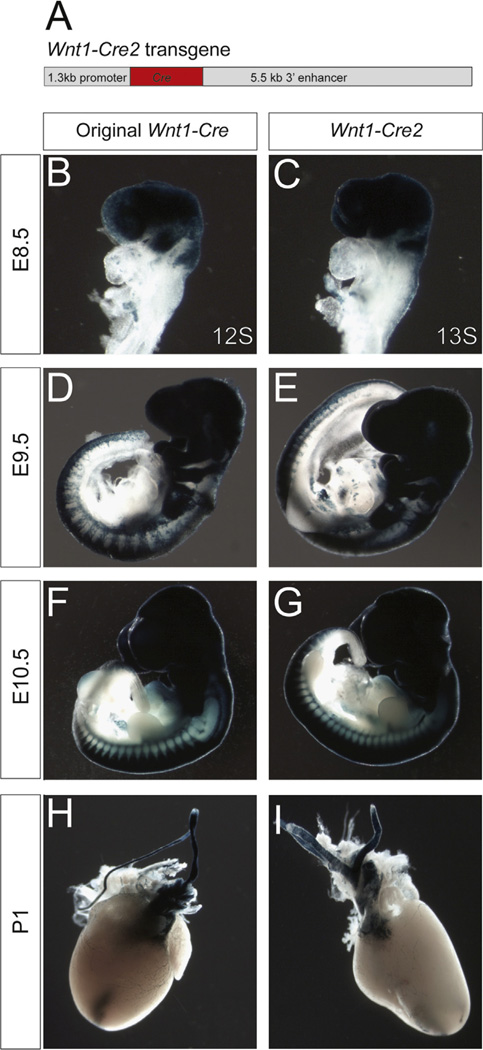
Because the Wnt1-Cre transgene has been so transformative for studies of neural crest and midbrain development, we adapted the original strategy for the generation of this transgene to generate a new Wnt1-Cre2 transgenic mouse line (129S4-Tg(Wnt1-cre)1Sor/J) that would avoid the complications of ectopic Wnt1 expression. This line includes the same 1.3 kb 5′ promoter sequence, Cre, and a 5.5 kb 3′ enhancer sequence without the Wnt1 gene sequence itself. This transgene is similar to a transgenic line that has been previously shown to exhibit Wnt1-restricted LacZ reporter activity (Echelard et al., 1994). We screened founders by PCR and verified insertion of the Wnt1-Cre2 transgene in four of these lines by Southern blot (data not shown). Crossing these new Wnt1-Cre2 founders with ROSA26R Cre reporter mice indicated that these transgenic lines recapitulate Cre recombinase activity in both the cardiac and cranial neural crest (Fig. 4; Supplementary Fig. 4C–F). More detailed characterization of one of these lines revealed similar recombination patterns to the original Wnt1-Cre in the branchial arches and cardiac outflow tract (Fig. 4B–I; Supplementary Fig. 4C–F), two tissues with well-characterized neural crest contributions. Although some recombination was also detected in branchial arch and frontonasal epithelium of Wnt1-Cre2; ROSA26R+/− embryos, closer examination revealed similar epithelial activity of the original Wnt1-Cre transgenic line (Supplementary Fig. 4A–B). This pattern of staining has been found to be reproducible over four backcross generations to 129S4, indicating that expression of Cre is stable in this transgenic line. Importantly, the Wnt1-Cre2 line does not exhibit the ectopic Wnt1 expression or midbrain enlargement (Supplementary Fig. 4I–K) present in the original Wnt1-Cre transgenic mouse. Taken together, these observations indicate that the Wnt1-Cre2 line will be a suitable replacement to achieve deletion in the neural crest cell lineages.
Fig. 4.
The new Wnt1-Cre2 transgenic mouse line mediates recombination in the neural crest. (A) Schematic representation of the new Wnt1-Cre2 transgenic construct. (B–I) β-galactosidase staining of Wnt1-CreTg/+; ROSA26R+/− embryos from the original line (B,D,F,H) compared with newly generated Wnt1-Cre2Tg/+; ROSA26R+/− transgenic mice (C,E,G,I), in which the Wnt1 coding sequence is not included in the transgene, at progressive stages of development. Comparable staining is observed in the neuroepithelium and migrating neural crest of E8.5 embryos in Wnt1-Cre2Tg/+; ROSA26R+/− (C) compared to the original Wnt1-CreTg/+; ROSA26R+/− (B). No overt difference was seen at E9.5 (D,E) or E10.5 (F,G), or in whole mount β-galactosidase staining of hearts at P1 (H,I).
Conclusion
The generation of the Wnt1-Cre transgene was pioneering, and its broad application in midbrain and neural crest studies has transformed multiple fields of developmental biology. As such, it is critical to note the caveat of ectopic activation of Wnt/β-catenin signaling that we describe here. These phenotypes are consistent with known roles for Wnt/β-catenin signaling in these contexts and indicate that ectopic activation of Wnt/β-catenin signaling may pose a confounding factor for some studies involving this mouse line (Dickinson et al., 1994; Panhuysen et al., 2004; Prakash et al., 2006; Tang et al., 2009, 2010). The Wnt1-Cre line may continue to prove useful in contexts that have been verified to lack ectopic activation of Wnt/β-catenin signaling. This promoter construct has also been used to drive the expression of other genes during midbrain development (Danielian and McMahon, 1996; Lee et al., 1997); we have no information on whether those studies would be influenced by the findings presented here. Finally, the effect of misexpression of Wnt1 on mDA neurons may make Wnt1-Cre transgenic mice a useful model for studying the differentiation of mDA neurons with the purpose of designing restorative therapies for Parkinson's disease.
Supplementary Material
Acknowledgments
We thank Tuba Nemati for her assistance with sample preparation, Kevin Kelley and the Mt. Sinai Mouse Genetics Facility for transgenic mice, and Nancy Ann Oberheim and our laboratory colleagues for helpful advice, discussions, and comments on the manuscript. This work was supported by NIH/NIDCR grants to J.O.B. (R00 DE020855) and P.S. (RO1 DE22363).
Footnotes
Appendix A. Supporting information
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.ydbio.2013.04.026.
References
- Brewer S, Feng W, Huang J, Sullivan S, Williams T. Wnt1-Cre-mediated deletion of AP-2alpha causes multiple neural crest-related defects. Dev. Biol. 2004;267:135–152. doi: 10.1016/j.ydbio.2003.10.039. [DOI] [PubMed] [Google Scholar]
- Castelo-Branco G, Wagner J, Rodriguez FJ, Kele J, Sousa K, Rawal N, Pasolli HA, Fuchs E, Kitajewski J, Arenas E. Differential regulation of midbrain dopaminergic neuron development by Wnt-1, Wnt-3a, and Wnt-5a. Proc. Natl. Acad. Sci. USA. 2003;100:12747–12752. doi: 10.1073/pnas.1534900100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Y, Jiang X, Ito Y, Bringas P, Jr, Han J, Rowitch DH, Soriano P, McMahon AP, Sucov HM. Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development. 2000;127:1671–1679. doi: 10.1242/dev.127.8.1671. [DOI] [PubMed] [Google Scholar]
- Danielian PS, McMahon AP. Engrailed-1 as a target of the Wnt-1 signalling pathway in vertebrate midbrain development. Nature. 1996;383:332–334. doi: 10.1038/383332a0. [DOI] [PubMed] [Google Scholar]
- Danielian PS, Muccino D, Rowitch DH, Michael SK, McMahon AP. Modification of gene activity in mouse embryos in utero by a tamoxifeninducible form of Cre recombinase. Curr. Biol. 1998;8:1323–1326. doi: 10.1016/s0960-9822(07)00562-3. [DOI] [PubMed] [Google Scholar]
- Dickinson ME, Krumlauf R, McMahon AP. Evidence for a mitogenic effect of Wnt-1 in the developing mammalian central nervous system. Development. 1994;120:1453–1471. doi: 10.1242/dev.120.6.1453. [DOI] [PubMed] [Google Scholar]
- Echelard Y, Vassileva G, McMahon AP. Cis-acting regulatory sequences governing Wnt-1 expression in the developing mouse CNS. Development. 1994;120:2213–2224. doi: 10.1242/dev.120.8.2213. [DOI] [PubMed] [Google Scholar]
- Ito Y, Yeo JY, Chytil A, Han J, Bringas P, Jr, Nakajima A, Shuler CF, Moses HL, Chai Y. Conditional inactivation of Tgfbr2 in cranial neural crest causes cleft palate and calvaria defects. Development. 2003;130:5269–5280. doi: 10.1242/dev.00708. [DOI] [PubMed] [Google Scholar]
- Jiang X, Iseki S, Maxson RE, Sucov HM, Morriss-Kay GM. Tissue origins and interactions in the mammalian skull vault. Dev Biol. 2002;241:106–116. doi: 10.1006/dbio.2001.0487. [DOI] [PubMed] [Google Scholar]
- Jiang X, Rowitch DH, Soriano P, McMahon AP, Sucov HM. Fate of the mammalian cardiac neural crest. Development. 2000;127:1607–1616. doi: 10.1242/dev.127.8.1607. [DOI] [PubMed] [Google Scholar]
- Katayama K, Melendez J, Baumann JM, Leslie JR, Chauhan BK, Nemkul N, Lang RA, Kuan CY, Zheng Y, Yoshida Y. Loss of RhoA in neural progenitor cells causes the disruption of adherens junctions and hyperproliferation. Proc. Natl. Acad. Sci. USA. 2011;108:7607–7612. doi: 10.1073/pnas.1101347108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SM, Danielian PS, Fritzsch B, McMahon AP. Evidence that FGF8 signalling from the midbrain–hindbrain junction regulates growth and polarity in the developing midbrain. Development. 1997;124:959–969. doi: 10.1242/dev.124.5.959. [DOI] [PubMed] [Google Scholar]
- McMahon AP, Bradley A. The Wnt-1 (int-1) proto-oncogene is required for development of a large region of the mouse brain. Cell. 1990;62:1073–1085. doi: 10.1016/0092-8674(90)90385-r. [DOI] [PubMed] [Google Scholar]
- Merrill AE, Bochukova EG, Brugger SM, Ishii M, Pilz DT, Wall SA, Lyons KM, Wilkie AO, Maxson RE., Jr Cell mixing at a neural crest-mesoderm boundary and deficient ephrin-Eph signaling in the pathogenesis of craniosynostosis. Hum. Mol. Genet. 2006;15:1319–1328. doi: 10.1093/hmg/ddl052. [DOI] [PubMed] [Google Scholar]
- Panhuysen M, Vogt Weisenhorn DM, Blanquet V, Brodski C, Heinzmann U, Beisker W, Wurst W. Effects of Wnt1 signaling on proliferation in the developing mid-/hindbrain region. Mol. Cell. Neurosci. 2004;26:101–111. doi: 10.1016/j.mcn.2004.01.011. [DOI] [PubMed] [Google Scholar]
- Prakash N, Brodski C, Naserke T, Puelles E, Gogoi R, Hall A, Panhuysen M, Echevarria D, Sussel L, Weisenhorn DM, Martinez S, Arenas E, Simeone A, Wurst W. A Wnt1-regulated genetic network controls the identity and fate of midbrain-dopaminergic progenitors in vivo. Development. 2006;133:89–98. doi: 10.1242/dev.02181. [DOI] [PubMed] [Google Scholar]
- Prakash N, Wurst W. Development of dopaminergic neurons in the mammalian brain. Cell. Mol. Life Sci. 2006a;63:187–206. doi: 10.1007/s00018-005-5387-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash N, Wurst W. Genetic networks controlling the development of midbrain dopaminergic neurons. J. Physiol. 2006b;575:403–410. doi: 10.1113/jphysiol.2006.113464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Tallquist MD, Soriano P. Cell autonomous requirement for PDGFRalpha in populations of cranial and cardiac neural crest cells. Development. 2003;130:507–518. doi: 10.1242/dev.00241. [DOI] [PubMed] [Google Scholar]
- Tang M, Miyamoto Y, Huang EJ. Multiple roles of beta-catenin in controlling the neurogenic niche for midbrain dopamine neurons. Development. 2009;136:2027–2038. doi: 10.1242/dev.034330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang M, Villaescusa JC, Luo SX, Guitarte C, Lei S, Miyamoto Y, Taketo MM, Arenas E, Huang EJ. Interactions of Wnt/beta-catenin signaling and sonic hedgehog regulate the neurogenesis of ventral midbrain dopamine neurons. J. Neurosci. 2010;30:9280–9291. doi: 10.1523/JNEUROSCI.0860-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas KR, Capecchi MR. Targeted disruption of the murine int-1 protooncogene resulting in severe abnormalities in midbrain and cerebellar development. Nature. 1990;346:847–850. doi: 10.1038/346847a0. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Vivatbutsiri P, Morriss-Kay G, Saga Y, Iseki S. Cell lineage in mammalian craniofacial mesenchyme. Mech. Dev. 2008;125:797–808. doi: 10.1016/j.mod.2008.06.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.