Abstract
Deadenylation is the major step in triggering mRNA decay and results in mRNA translation inhibition in eukaryotic cells. Therefore, it is plausible that deadenylation also induces the mRNP remodeling required for formation of GW bodies or RNA processing bodies (P-bodies), which harbor translationally silenced mRNPs. In this chapter, we discuss several examples to illustrate the roles of deadenylation in regulating gene expression. We highlight several lines of evidence indicating that even though non-translatable mRNPs may be prepared and/or assembled into P-bodies in different ways, deadenylation is always a necessary, and perhaps the earliest, step in mRNA decay pathways that enable mRNP remodeling required for P-body formation. Thus, deadenylation and the participating deadenylases are not simply required for preparing mRNA substrates; they play an indispensable role both structurally and functionally in P-body formation and regulation.
Keywords: GW body, mRNA turnover, poly(A), microRNA, deadenylation, P-body, translational control
11.1. Introduction
Regulation of the abundance and translation of messenger RNAs (mRNAs) is important for controlling gene expression because mRNAs carry genetic information from the nucleus to the cytoplasm, where they can be translated into proteins. mRNAs associate with a number of proteins, and the resulting mRNA-protein complexes (mRNPs) undergo a series of remodeling events that impact and/or are influenced by the translation and mRNA decay machineries (Moore 2005; Shyu and Chen 2009). The components of most mRNPs are in dynamic equilibrium with the translational pool, allowing rapid shifts between translation, storage, and degradation (Balagopal and Parker 2009; Kedersha et al. 2005; Wilkinson and Shyu 2001). Thus, the metabolism and functions of mRNAs can be regulated through mRNP remodeling to accommodate various cellular processes.
Some protein components of particular mRNPs may promote assembly of microscopically visible, cytoplasmic granules, such as the GW or RNA processing bodies (P-bodies), which harbor translationally silenced mRNPs (Eystathioy et al. 2002; Eystathioy et al. 2003; Sheth and Parker 2003; van Dijk et al. 2002). While many mRNA decay factors are linked to P-bodies, the recent finding that deadenylation (i.e., shortening of the 3′ poly(A) tail of mRNAs) is prerequisite for P-body formation has opened up new aspects of P-body dynamics and regulation (Zheng et al. 2008). Deadenylation is the major step triggering mRNA decay in eukaryotic cells (Chen and Shyu 2011). The poly(A) tail and associated poly(A)-binding protein (PABP) interacts with the 5′ m7G-cap/cap-binding complex to form a closed loop that enhances translation initiation and protects the mRNA ends from nuclease attack (Jacobson 1996; Sachs 2000). Thus, deadenylation can impact an mRNA not only by inducing its degradation but also by reducing its translatability. Given that deadenylation serves as an important control point for both mRNA degradation and translational silencing, it is not surprising that regulation of deadenylation provides a key means of controlling eukaryotic gene expression. In this chapter, we discuss the importance of deadenylation in regulating gene expression and how deadenylation may impact P-bodies.
11. 2. Deadenylation
The 3′ termini of all mRNAs except histone mRNAs contain a poly(A) tail. The length of the poly(A) tail plays a critical role in determining mRNA stability and translation efficiency (Jacobson and Peltz 1996; Wickens et al. 1997; Wilusz et al. 2001). The poly(A) tails of newly synthesized mRNAs entering the cytoplasm are 200–250 nucleotides (nt) long; the poly(A) tails subsequently undergo deadenylation at different rates until their length reaches 10–60 nt (Baker 1993; Brawerman 1981). Recent identification and characterization of at least eight distinct deadenylases in metazoa highlights the diverse biological functions of deadenylases and thus of deadenylation in gene regulation (Dupressoir et al. 2001; Goldstrohm and Wickens 2008; Zuo and Deutscher 2001).
11. 2. 1. Deadenylation is the major step triggering mRNA decay
RNA destabilizing elements exert their effects mainly by inducing accelerated deadenylation, thereby leading to rapid mRNA decay in mammalian cells (Chen and Shyu 2011). Computational modeling of eukaryotic mRNA turnover also suggests that changes in levels of mRNA are tightly linked to the rate of deadenylation (Cao and Parker 2001). Thus, deadenylation provides a critical point for regulation of gene expression. We propose that the deadenylation rate of any given mRNA in a specific cellular environment reflects interactions among several key components, including deadenylases, the poly(A)-PABP complex, mRNA element-binding complexes, and factors that modulate these interactions.
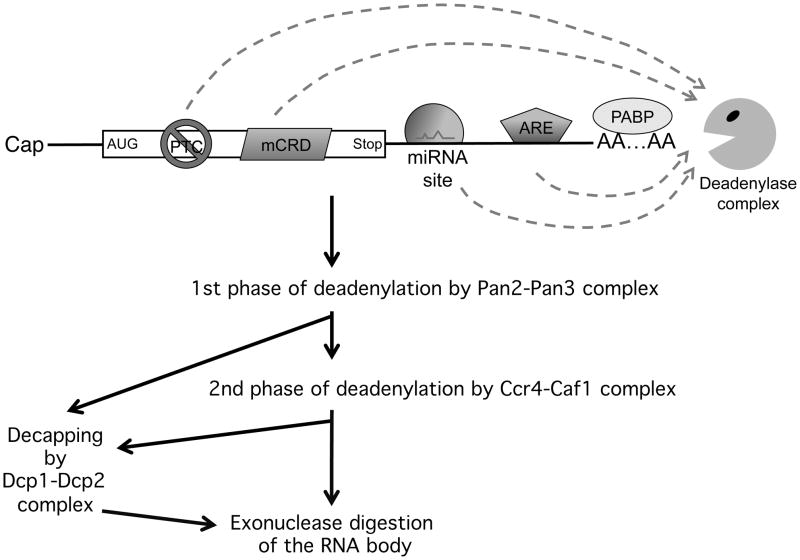
All major modes of mammalian mRNA decay observed thus far are triggered by deadenylation (Chen and Shyu 2011). These include decay directed by AU-rich elements (AREs) in the 3′ untranslated region (UTR) (e.g., (Chen et al. 1994; Shyu et al. 1991)), decay mediated by destabilizing elements in protein-coding regions (e.g., (Chang et al. 2004; Grosset et al. 2000)), decay of nonsense-containing mRNA (i.e., NMD) (Chen and Shyu 2003), the microRNA (miRNA)-mediated decay (miRMD) (Chen et al. 2009; Wu et al. 2006), and the default decay of stable messages lacking destabilizing elements (Fig. 1). Mammalian mRNA deadenylation involves two consecutive phases mediated by the Pan2-Pan3 and the Ccr4-Caf1 complexes, respectively (Fig. 1). Decapping takes place after deadenylation and may serve as a backup trigger for mRNA decay if initial deadenylation is compromised (Yamashita et al. 2005; Zheng et al. 2008). Compromising both Ccr4-Caf1 and Dcp2 activities essentially halts various modes of mRNA decay with concomitant accumulation of stable intermediates containing ~110 nt of poly(A) tail (Chen et al. 2009; Yamashita et al. 2005; Zheng et al. 2008). While it remains unknown as to whether deadenylation-independent decapping operates in mammalian cells, it is clear that the major route for mRNA decay in mammalian cells is triggered by deadenylation followed by decapping and 5′ to 3′ exonuleolytic digestion of the RNA body. Thus, even though the mechanisms for mRNA degradation differ in yeast and in mammals, the major mRNA decay pathway is highly conserved (Chen and Shyu 2003; Muhlrad and Parker 1994). As a process that is reversible deadenylation may serve as an important checkpoint before an mRNA is committed to elimination, which makes deadenylation an important step for regulation of gene expression during a variety of biological processes, such as embryogenesis, cell growth, and cell differentiation.
Figure 1. Deadenylation and major mRNA decay pathways in mammalian cells.
Sequence elements are depicted for nonsense-mediated decay (NMD) triggered by a premature termination codon (PTC), decay mediated by the c-fos coding-region determinant (mCRD), miRNA-mediated decay, and decay mediated by AU-rich elements (ARE). The poly(A) tail, the poly(A)-binding protein (PABP) and the deadenylase complexes are shown at the 3′ end. The four RNA destabilizing elements or mutations and their cognate binding complexes have distinct paths (shown as dashed lines) to recruit deadenylation machinery and thus accelerate deadenylation, which occurs in two phases. In mammalian cells, decapping does not occur until the end of the first or second phase of deadenylation. Note that the slow, default decay of stable mRNAs lacking recognized destabilizing elements is also triggered by deadenylation.
11. 2. 2. The role of deadenylation in miRNA-mediated gene silencing
Gene silencing is one mechanism to ensure that proteins are expressed at proper levels and miRNAs contribute to gene silencing mainly by accelerating deadenylation to promote rapid decay of their mRNA targets (Chen and Shyu 2011). Here, we discuss some new findings on the mechanism of miRNA-mediated mRNA decay to illustrate the importance of deadenylation in controlling eukaryotic gene expression.
miRNAs induce degradation of mRNA targets in many eukaryotic cells, including those from humans, C. elegans, Drosophila, and zebrafish (Bagga et al. 2005; Behm-Ansmant et al. 2006; Chen et al. 2009; Giraldez et al. 2006; Wu et al. 2006). Accumulating evidence from kinetic studies supports the idea that miRNAs destabilize target mRNAs through deadenylation and subsequent decapping and 5′ to 3′ exonucleolytic digestion (Chen et al. 2009; Piao et al. 2010). For example, poly(A) length assays indicated that miRNAs mediate deadenylation of a wide array of targets in a variety of systems. In zebrafish, miR-430 mediates the deadenylation of hundreds of maternal transcripts early in embryonic development (Giraldez et al. 2006). Results from a recent study using mouse P19 embryonic carcinoma cells (Wu and Belasco 2005) demonstrated that lin-28 mRNA, whose levels decrease during retinoic acid-induced neuronal differentiation, is deadenylated through the action of miR-125, a miRNA whose levels increase during differentiation (Wu et al. 2006). Deadenylation mediated by miRNAs has also been demonstrated in mammalian and Drosophila cell-free extracts (Fabian et al. 2009; Iwasaki et al. 2009; Wakiyama et al. 2007).
The precursor-product relationships of mRNAs targeted by let-7 miRNA for degradation were directly demonstrated by using a transcriptional pulsing approach to analyze mRNA decay kinetics combined with a strategy to block specific endogenous decay enzymes (Chen et al. 2009; Wu et al. 2006). These studies were able to trap mRNA intermediates during decay directed by let-7 via either miRNA-mediated decay or siRNA-mediated decay in mouse fibroblasts. The results showed that let-7 routes target mRNAs to the major cytoplasmic mRNA decay pathway, in which degradation of mRNA is triggered by deadenylation involving Pan2-Pan3 and Ccr4-Caf1 complexes followed by decapping via the Dcp1-Dcp2 complex (Chen et al. 2009). Moreover, tethering AGO proteins or GW182 protein, components of the miRNA-induced silencing complex (miRISC) required for gene silencing (Fabian et al. 2010), promoted decay of target mRNA by triggering rapid deadenylation (Chen et al. 2009). These observations indicate that promoting rapid decay by triggering deadenylation is an intrinsic property of miRISC in mammalian cells. Consistent with this notion is the observation that deadenylation is a widespread feature of miRNA regulation (Eulalio et al. 2009).
11. 3. Deadenylation and P-bodies
P-bodies are dynamic cytoplasmic foci in eukaryotic cells that contain non- translatable mRNAs as well as proteins involved in translational inhibition and mRNA decay (Eulalio et al. 2007a; Franks and Lykke-Andersen 2008; Kedersha and Anderson 2007; Parker and Sheth 2007). As deadenylation triggers mRNA degradation and also leads to inhibition of mRNA translation in eukaryotic cells, it is plausible that deadenylation induces the mRNP remodeling required for P-body formation. Although non-translatable mRNPs can follow different paths leading to assembly into P-bodies, several lines of evidence indicate that deadenylation is always a necessary and perhaps the earliest step that begins the mRNP remodeling required for P-body formation.
11. 3. 1. Deadenylation is required for P-body formation
The roles of deadenylation and its participating enzymes and factors in the formation of P-bodies were unclear until recently and thus were largely ignored for a long time after the cytoplasmic foci were discovered. Also contributing to earlier neglect of deadenylation were observations that P-bodies are enriched in factors involved in decapping and 5′ to 3′ decay and that deadenylases are not readily found in yeast P-bodies (Sheth and Parker 2003). Furthermore, an earlier study in yeast reported that deletion of the major deadenylase complex had much less effect on P-bodies than did deletion of factors related to decapping and 5′ to 3′ decay (Teixeira and Parker 2007). This led to the idea that deadenylation is not critical for P-body formation. However, observations that Ccr4 co-localizes with P-bodies and that siRNA-mediated knockdown of Ccr4 blocked P-body formation in HeLa human cells (Andrei et al. 2005; Cougot et al. 2004) hinted at an important role for deadenylation in P-body assembly. Yet, it was unresolved whether Ccr4 deadenylase activity or just the Ccr4 protein was required for P-body formation. It thus remained unclear, especially in mammalian cells, whether deadenylation is important for P-body formation.
With the recent findings that deadenylation is the major step triggering mRNA decay in yeast and mammalian cells (Cao and Parker 2001; Chen and Shyu 2011), the impact of deadenylation on P-bodies can no longer be overlooked, especially as P-bodies have been found to contain many mRNA decay factors and the corresponding mRNA substrates (Eulalio et al. 2007a; Franks and Lykke-Andersen 2008; Kedersha and Anderson 2007; Parker and Sheth 2007). In 2008, Zheng et al. (Zheng et al. 2008) reported that P-bodies contain all the major mammalian deadenylases, including the Pan2-Pan3 and Ccr4-Caf1 complexes, thereby linking P-bodies with all major mRNA decay factors except the exosome components. Other results from the same study (Zheng et al. 2008) indicated that deadenylation is required for P-body formation in mammalian cells. First, impairment of deadenylation by knocking down Caf1 led to loss of P-bodies. Second, a dominant negative mutant of Caf1 inhibited both deadenylation and P-body formation, demonstrating that the effect on P-bodies involved loss of deadenylation activity rather than loss of the Caf1 protein per se. Further, co-expression of wild-type Ccr4 with the Caf1 mutant to rescue deadenylation activity restored P-body formation. In contrast, puromycin, which increases the pool of non-translatable mRNAs and thus promotes P-body formation (Cougot et al. 2004; Eulalio et al. 2007b; Yang and Bloch 2007) in control cells, did not induce P-body formation when deadenylation was blocked by expressing the Caf1 mutant or by knocking down Caf1.
Blocking deadenylation impairs P-body formation but the converse is not true. Evidence on this point came from the observation that knockdown of Pan3, a key component of P-bodies, impairs P-body formation but has little effect on deadenylation and decay of ARE-containing transcripts and miRNA targeted mRNAs (Zheng et al. 2008). Thus, deadenylation does not require P-body formation.
With a linkage established between deadenylation and P-body formation, it seems possible that poly(A)-shortened mRNAs are major components of mRNPs in P-bodies. Thus, although non-translatable mRNAs may arrive at P-bodies by different pathways, deadenylation is always required for P-body formation.
11. 3. 2. Deadenylation factors play different roles in P-body formation
siRNA-mediated gene knockdown experiments show that knocking down Pan3 impairs P-body formation (Zheng et al. 2008), indicating an essential role for Pan3 in P-body formation. Moreover, Pan3 helps enrich Pan2, Ccr4, and Caf1 in P-bodies (Zheng et al. 2008). In contrast, knocking down Pan2 had little effect on P-body formation (Zheng et al. 2008), indicating that Pan2 is dispensable for P-body formation. This is consistent with the observation that expression of a catalytically inactive mutant of Pan2 had no effect on P-bodies in mouse fibroblasts (Zheng et al. 2008). siRNA-mediated knockdown of Caf1 inhibited P-body formation in NIH 3T3 cells (Zheng et al. 2008) and Ccr4 knockdown blocked P-body formation in HeLa cells (Andrei et al. 2005). Also, as mentioned in section 14. 3. 1, over-expression of Caf1 catalytic mutant inhibited deadenylation and also blocked P-body formation; restoring deadenylation by co-expression of wild-type Ccr4 with the Caf1 mutant led to reappearance of P-bodies (Zheng et al. 2008). Collectively, these observations indicate that P-body formation in mammalian cells requires the deadenylase activity of the Ccr4-Caf1 complex.
It is noteworthy that even though the Ccr4p-Pop2p complex is the major deadenylase in yeast, deletion of either Ccr4p or Pop2p (yeast Caf1) had only a small effect on P-bodies (Teixeira and Parker 2007), suggesting that yeast and mammalian cells have different mechanisms for P-body formation. Consistent with this notion, it was reported that yeast, but not human, P-body components are more likely to contain Q/N-rich aggregation-prone regions (Reijns et al. 2008), which may help accumulate the associated mRNPs into P-bodies.
The observation that Pan3 is physically required for P-body formation in mammalian cells and its knockdown has little effect on deadenylation (Zheng et al. 2008) argues for an additional role of Pan3 in mammalian P-body formation. This is further substantiated by the finding that Pan3 greatly enhances the localization of other P-body components to P-bodies (Zheng et al. 2008). Thus, deadenylation and the participating deadenylases are not simply required for preparing mRNA substrates; they play an indispensable role both structurally and functionally in P-body formation and regulation.
11. 3. 3. Deadenylation triggers mRNP remodeling for P-body formation
Although P-bodies harbor translationally silenced mRNPs, formation of P-bodies is not simply a consequence of increasing the pool of untranslatable mRNAs. Instead, P-body formation requires an active deadenylation process. This requirement was demonstrated by experiments using puromycin, a translation inhibitor that releases ribosomes from mRNAs and enhances P-body formation (Cougot et al. 2004; Eulalio et al. 2007b; Yang and Bloch 2007). When deadenylation was inhibited by either overexpressing a Caf1 dominant-negative mutant or by knocking down Caf1, P-body formation was impaired with or without puromycin treatment (Zheng et al. 2008). This indicates that deadenylation does something more than simply enhancing P-body formation by rendering the mRNAs ribosome-free because the poly(A) tail is known to promote translation initiation (Franks and Lykke-Andersen 2008). Thus, even though an mRNA needs to be ribosome-free to enter or form P-bodies (Cougot et al. 2004; Sheth and Parker 2003), the importance of deadenylation in P-body formation cannot be simply explained as a block on recruitment of ribosomes to the mRNAs. Instead, deadenylation is required for a distinct process, which may involve dissociation of factors that could prevent mRNPs from joining P-bodies. One candidate for such a factor is PABP.
Through its interactions with various decay and translation factors in the eukaryotic cytoplasm, PABP plays a central role in mRNA turnover and in translation (Mangus et al. 2003). Several PABP-interacting proteins are reported to co-localize with P-bodies. For example, PABP’s interaction with Pan3 enhances Pan2 nuclease activity (Boeck et al. 1996; Mangus et al. 2004; Uchida et al. 2004). This suggests that the first phase of deadenylation mediated by the Pan2-Pan3 deadenylase complex is PABP-dependent. However, PABP itself is not a component of P-bodies (Kedersha et al. 2005; Zheng et al. 2008). Thus, it is possible that mRNPs go through the first phase of deadenylation and subsequent dissociation of PABPs before participating in P-body formation. An important implication is that association with PABPs may inhibit mRNPs from joining existing P-bodies or nucleating P-body formation. In this interpretation, deadenylation helps mRNPs become PABP-free for P-body assembly. This is consistent with the observation that mRNAs competent for translation normally carry a poly(A) tail that is long enough to associate with PABPs (Mangus et al. 2003).
Other factors that are important for efficient translation of mRNAs may also dissociate from mRNPs during the remodeling process triggered by deadenylation. For example, eIF4G, a translation activator, can simultaneously interact with both the cap-binding protein eIF4E and PABP to circularize poly(A)+ mRNAs (Jacobson 1996; Sachs 2000). The resulting closed-loop conformation enhances translation initiation and provides an effective way to prevent mRNAs from degradation and P-body localization. Deadenylation would disrupt the binding sites for factors that stabilize the closed-loop structure, which increases the untranslatable pool of mRNPs and promotes P-body formation.
The mRNP remodeling triggered by deadenylation may also promote recruitment to the mRNP of translation repressors or other P-body components, such as decapping complexes (Coller and Parker 2004; Franks and Lykke-Andersen 2008). This notion is supported by the observation that deadenylation promotes the interaction between the poly(A)-shortened mRNAs and the Lsm1–7 complex, a decapping activator that contains some P-body components and prefers to bind the 3′ end of deadenylated mRNAs (Chowdhury and Tharun 2008). Based on these current observations, it is plausible that deadenylation triggers mRNP remodeling that is critical for P-body assembly.
11. 3. 4. Deadenylation may occur in P-bodies
Some observations raise a question as to whether deadenylation occurs in P-bodies. For example, both the Pan2-Pan3 and the Ccr4-Caf1 deadenylase complexes, the two main deadenylase complexes responsible for cytoplasmic poly(A)-shortening in eukaryotes, can co-localize with P-bodies of mammalian cells (Andrei et al. 2005; Cougot et al. 2004; Zheng et al. 2008). In yeast, both Ccr4p and Pop2p (yeast Caf1) co-localized with P-bodies when decapping or 5′ to 3′ mRNA decay was inhibited (Teixeira and Parker 2007). One interpretation of these observations is that the Ccr4-Caf1 complex transits through P-bodies quickly and localizes to P-bodies under restricted conditions. Interestingly, the detection of poly(A)+ mRNAs in yeast P-bodies during glucose deprivation and in stationary phase (Brengues and Parker 2007) suggests that some mRNAs in P-bodies have poly(A) tails that are longer than oligo(A). This is consistent with the observation that yeast mRNAs can be released from P-bodies and recruited to polysomes for translation in response to stress (Sheth and Parker 2006). One possibility is that some translationally inhibited poly(A)+ mRNAs are in P-bodies for temporary storage; these mRNAs can subsequently be deadenylated, decapped, and degraded in P-bodies, or be released for translation before complete deadenylation.
11. 4. A model linking deadenylation and P-body formation
Based on the current information, we have devised a model for the linkage between deadenylation and P-body formation in mammalian cells (Fig. 2). The 3′ poly(A) tail-PABP complex stimulates poly(A) shortening by the Pan2-Pan3 complex but inhibits the activity of the Ccr4-Caf1 complex (Chen et al. 2002; Tucker et al. 2002). Thus, the first phase of deadenylation is initiated when PABPs on the mRNA poly(A) tail interact with Pan3 to recruit the Pan2 deadenylase. After the poly(A) tail is significantly shortened by Pan2, the remaining bound PABPs are less effective in inhibiting the deadenylase activity of the Ccr4-Caf1 complex, allowing the second phase of deadenylation to occur. During the first phase and/or early second phase of deadenylation, factors for efficient translation (such as PABP, eIF4G, ribosomes, etc.) may dissociate from the mRNPs, which would allow mRNPs to reversibly associate with P-bodies. At this stage, the mRNPs could either be released from P-bodies for translation or remain in the P-bodies to be further deadenylated by Ccr4-Caf1, resulting in recruitment of the Lsm1–7 complex and other decapping factors. The resultant mRNPs would constitute the core of P-bodies and their mRNA components would be decapped and degraded. Alternatively, the second phase of deadenylation mediated by Ccr4-Caf1 and the subsequent recruitment of decapping complex and related factors could occur outside P-bodies. In this case, the remodeled mRNPs would aggregate into P-bodies. Thus, it is possible that the mRNPs could not form the cores of P-bodies or aggregate into P-bodies when the deadenylase activity of Ccr4-Caf1 is inhibited. The proposed model helps explain why the deadenylation complexes co-localize with P-bodies, why deadenylation is required for P-body formation, why PABP, eIF4G, and ribosomes are not enriched in P-bodies, and why some mRNAs released from P-bodies can still be translated.
Figure 2.
Working model linking deadenylation and P-body formation.
Future research addressing the key changes in mRNP composition at each stage of the life of an mRNA after it arrives in the cytoplasm will be crucial for understanding what targets an mRNP to P-bodies. Along this line, determining when and how PABPs dissociate from an mRNP, a particularly critical unresolved issue, may further elucidate the importance of mRNP remodeling and the link between deadenylation and P-bodies.
Acknowledgments
We thank many colleagues who have contributed in various ways over the years to the content described in this chapter, and to Drs. Richard Kulmacz and Julia Lever for critical reading of the manuscript. Work in our laboratory was supported by the U.S. National Institutes of Health, the Houston Endowment, Inc., and the Sandler Program for Asthma Research.
References
- Andrei MA, Ingelfinger D, Heintzmann R, Achsel T, Rivera-Pomar R, Luhrmann R. A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA. 2005;11:717–727. doi: 10.1261/rna.2340405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, Pasquinelli AE. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553– 563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- Baker EJ. Control of poly(A) length. In: Belasco JG, Brawerman G, editors. Control of messenger RNA stability. Academic Press; San Diego: 1993. [Google Scholar]
- Balagopal V, Parker R. Polysomes, P bodies and stress granules: states and fates of eukaryotic mRNAs. Current Opinion in Cell Biology. 2009;21:403–408. doi: 10.1016/j.ceb.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 2006;20:1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeck R, Tarun S, Jr, Rieger M, Deardorff JA, Muller-Auer S, Sachs AB. The yeast Pan2 protein is required for poly(A)-binding protein-stimulated poly(A)-nuclease activity. J Biol Chem. 1996;271:432–438. doi: 10.1074/jbc.271.1.432. [DOI] [PubMed] [Google Scholar]
- Brawerman G. The role of the poly(A) sequence in mammalian messenger RNA. Crit Rev Biochem. 1981;10:1–38. doi: 10.3109/10409238109114634. [DOI] [PubMed] [Google Scholar]
- Brengues M, Parker R. Accumulation of polyadenylated mRNA, Pab1p, eIF4E, and eIF4G with P-bodies in Saccharomyces cerevisiae. Mol Biol Cell. 2007;18:2592–2602. doi: 10.1091/mbc.E06-12-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao D, Parker R. Computational modeling of eukaryotic mRNA turnover. RNA. 2001;7:1192–1212. doi: 10.1017/s1355838201010330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang T-C, Yamashita A, Chen C-YA, Yamashita Y, Zhu W, Durdan S, Kahvejian A, Sonenberg N, Shyu A-B. UNR, a new partner of poly(A)-binding protein, plays a key role in translationally coupled mRNA turnover mediated by the c-fos major coding-region determinant. Genes Dev. 2004;18:2010–2023. doi: 10.1101/gad.1219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C-YA, Shyu A-B. Rapid deadenylation triggered by a nonsense codon precedes decay of the RNA body in a mammalian cytoplasmic nonsense-mediated decay pathway. Mol Cell Biol. 2003;23:4805–4813. doi: 10.1128/MCB.23.14.4805-4813.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C-YA, Zheng D, Xia Z, Shyu A-B. Ago-TNRC6 triggers microRNA-mediated decay by promoting two deadenylation steps. Nat Struct Mol Biol. 2009;16:1160–1166. doi: 10.1038/nsmb.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Chen TM, Shyu AB. Interplay of two functionally and structurally distinct domains of the c-fos AU-rich element specifies its mRNA-destabilizing function. Mol Cell Biol. 1994;14:416–426. doi: 10.1128/mcb.14.1.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CYA, Shyu AB. Mechanisms of deadenylation-dependent decay. Wiley Interdisciplinary Reviews - RNA. 2011;2:167–183. doi: 10.1002/wrna.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Chiang Y-C, Denis CL. CCR4, a 3′-5′ poly(A) RNA and ssDNA exonuclease, is the catalytic component of the cytoplasmic deadenylase. EMBO J. 2002;21:1414–1426. doi: 10.1093/emboj/21.6.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury A, Tharun S. lsm1 mutations impairing the ability of the Lsm1p-7p-Pat1p complex to preferentially bind to oligoadenylated RNA affect mRNA decay in vivo. RNA. 2008;14:2149–2158. doi: 10.1261/rna.1094208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller J, Parker R. Eukaryotic mRNA decapping. Annu Rev Biochem. 2004;74:861–890. doi: 10.1146/annurev.biochem.73.011303.074032. [DOI] [PubMed] [Google Scholar]
- Cougot N, Babajko S, Seraphin B. Cytoplasmic foci are sites of mRNA decay in human cells. J Cell Biol. 2004;165:31–40. doi: 10.1083/jcb.200309008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupressoir A, Morel A-P, Barbot W, Loireau M-P, Corbo L, Heidmann T. Identification of four families of yCCR4- and Mg2+-dependent endonuclease-related proteins in higher eukaryotes, and characterization of orthologs of yCCR4 with a conserved leucine-rich repeat essential for hCAF1/hPOP2 binding. BMC Genomics. 2001;2:9. doi: 10.1186/1471-2164-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Behm-Ansmant I, Izaurralde E. P bodies: at the crossroads of post-transcriptional pathways. Nat Rev Mol Cell Biol. 2007a;8:9–22. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- Eulalio A, Behm-Ansmant I, Schweizer D, Izaurralde E. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol Cell Biol. 2007b;27:3970–3981. doi: 10.1128/MCB.00128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Huntzinger E, Nishihara T, Rehwinkel J, Fauser M, Izaurralde E. Deadenylation is a widespread effect of miRNA regulation. RNA. 2009;15:21–32. doi: 10.1261/rna.1399509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eystathioy T, Chan EKL, Tenenbaum SA, Keene JD, Griffith K, Fritzler MJ. A phosphorylated cytoplasmic autoantigen, GW182, associates with a unique population of human mRNAs within novel cytoplasmic speckles. Mol Biol Cell. 2002;13:1338–1351. doi: 10.1091/mbc.01-11-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eystathioy T, Jakymiw A, Chan EKL, Seraphin B, Cougot N, Fritzler MJ. The GW182 protein colocalizes with mRNA degradation associated proteins hDcp1 and hLSm4 in cytoplasmic GW bodies. RNA. 2003;9:1171–1173. doi: 10.1261/rna.5810203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian MR, Mathonnet G, Sundermeier T, Mathys H, Zipprich JT, Svitkin YV, Rivas F, Jinek M, Wohlschlegel J, Doudna JA, Chen C-YA, Shyu A-B, Yates JR, III, Hannon GJ, Filipowicz W, Duchaine TF, Sonenberg N. Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Molecular Cell. 2009;35:868–880. doi: 10.1016/j.molcel.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- Franks TM, Lykke-Andersen J. The control of mRNA decapping and P-body formation. Molecular Cell. 2008;32:605–615. doi: 10.1016/j.molcel.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- Goldstrohm AC, Wickens M. Multifunctional deadenylase complexes diversify mRNA control. Nat Rev Mol Cell Biol. 2008;9:337–344. doi: 10.1038/nrm2370. [DOI] [PubMed] [Google Scholar]
- Grosset C, Chen C-YA, Xu N, Sonenberg N, Jacquemin-Sablon H, Shyu A-B. A mechanism for translationally coupled mRNA turnover: Interaction between the poly(A) tail and a c-fos RNA coding determinant via a protein complex. Cell. 2000;103:29–40. doi: 10.1016/s0092-8674(00)00102-1. [DOI] [PubMed] [Google Scholar]
- Iwasaki S, Kawamata T, Tomari Y. Drosophila Argonaute 1 and Argonaute 2 employ distinct mechanisms for translational repression. Mol Cell. 2009;34:58–67. doi: 10.1016/j.molcel.2009.02.010. [DOI] [PubMed] [Google Scholar]
- Jacobson A. Poly(A) metabolism and translation: The closed-loop Model. In: Hershey JWB, Mathews MB, Sonenberg N, editors. Tanslational Control. Cold Spring Harbor Laboratory Press; Plainview: 1996. pp. 451–480. [Google Scholar]
- Jacobson A, Peltz SW. Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annual Review Biochem. 1996;65:693–739. doi: 10.1146/annurev.bi.65.070196.003401. [DOI] [PubMed] [Google Scholar]
- Kedersha N, Anderson P. Mammalian stress granules and processing bodies. Methods Enzymol. 2007;431:61–81. doi: 10.1016/S0076-6879(07)31005-7. [DOI] [PubMed] [Google Scholar]
- Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, Scheuner D, Kaufman RJ, Golan DE, Anderson P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangus DA, Evans MC, Agrin NS, Smith M, Gongidi P, Jacobson A. Positive and negative regulation of poly(A) nuclease. Mol Cell Biol. 2004;24:5521–5533. doi: 10.1128/MCB.24.12.5521-5533.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangus DA, Evans MC, Jacobson A. Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 2003;4:233. doi: 10.1186/gb-2003-4-7-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005;309:1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- Muhlrad D, Parker R. Premature translational termination triggers mRNA decapping. Nature. 1994;370:578–581. doi: 10.1038/370578a0. [DOI] [PubMed] [Google Scholar]
- Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Piao X, Zhang X, Wu L, Belasco JG. CCR4-NOT Deadenylates mRNA associated with RNA-induced silencing complexes in human cells. Mol Cell Biol. 2010;30:1486–1494. doi: 10.1128/MCB.01481-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijns MAM, Alexander RD, Spiller MP, Beggs JD. A role for Q/N-rich aggregation-prone regions in P-body localization. J Cell Sci. 2008;121:2463–2472. doi: 10.1242/jcs.024976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs AB. Physical and functional interactions between the mRNA cap structure and the poly(A) tail. In: Sonenberg N, Hershey JWB, Mathews MB, editors. Translational control of gene expression. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 2000. [Google Scholar]
- Sheth U, Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U, Parker R. Targeting of aberrant mRNAs to cytoplasmic processing bodies. Cell. 2006;125:1095–1109. doi: 10.1016/j.cell.2006.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyu A-B, Belasco JG, Greenberg MG. Two distinct destabilizing elements in the c-fos message trigger deadenylation as a first step in rapid mRNA decay. Genes Dev. 1991;5:221–232. doi: 10.1101/gad.5.2.221. [DOI] [PubMed] [Google Scholar]
- Shyu AB, Chen AC-Y. Regulation of mRNA turnover. In: Bradshaw RA, Dennis EA, editors. Handbook of Cell Signaling. Post-transcriptional control. 2. Elsevier; San Diego: 2009. pp. 2311–2315. [Google Scholar]
- Teixeira D, Parker R. Analysis of P-body assembly in Saccharomyces cerevisiae. Mol Biol Cell. 2007;18:2274–2287. doi: 10.1091/mbc.E07-03-0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker M, Staples RR, Valencia-Sanchez MA, Muhlrad D, Parker R. Ccr4p is the catalytic subunit of a Ccr4p/Pop2p/Notp mRNA deadenylase complex in Saccharomyces cerevisiae. EMBO J. 2002;21:1427–1436. doi: 10.1093/emboj/21.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida N, Hoshino S-i, Katada T. Identification of a human cytoplasmic poly(A) nuclease complex stimulated by poly(A)-binding protein. J Biol Chem. 2004;279:1383–1391. doi: 10.1074/jbc.M309125200. [DOI] [PubMed] [Google Scholar]
- van Dijk E, Cougot N, Meyer S, Babajko S, Wahle E, Seraphin B. Human Dcp2: a catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J. 2002;21:6915–6924. doi: 10.1093/emboj/cdf678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakiyama M, Takimoto K, Ohara O, Yokoyama S. Let-7 microRNA-mediated mRNA deadenylation and translational repression in a mammalian cell-free system. Genes Dev. 2007;21:1857–1862. doi: 10.1101/gad.1566707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickens M, Anderson P, Jackson RJ. Life and death in the cytoplasm: messages from the 3′ end. Curr Opin Genet Dev. 1997;7:220–232. doi: 10.1016/s0959-437x(97)80132-3. [DOI] [PubMed] [Google Scholar]
- Wilkinson MF, Shyu AB. Multifunctional regulatory proteins that control gene expression in both the nucleus and the cytoplasm. Bioessays. 2001;23:775–787. doi: 10.1002/bies.1113. [DOI] [PubMed] [Google Scholar]
- Wilusz CW, Wormington M, Peltz SW. The cap-to-tail guide to mRNA turnover. Nature Reviews Mol Cell Biol. 2001;2:237–246. doi: 10.1038/35067025. [DOI] [PubMed] [Google Scholar]
- Wu L, Belasco JG. Micro-RNA regulation of the mammalian lin-28 gene during neuronal differentiation of embryonal carcinoma cells. Mol Cell Biol. 2005;25:9198–9208. doi: 10.1128/MCB.25.21.9198-9208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Fan J, Belasco JG. MicroRNAs direct rapid deadenylation of mRNA. PNAS. 2006;103:4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A, Chang TC, Yamashita Y, Zhu W, Zhong Z, Chen CY, Shyu AB. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat Struct Mol Biol. 2005;12:1054–1063. doi: 10.1038/nsmb1016. [DOI] [PubMed] [Google Scholar]
- Yang W-H, Bloch DB. Probing the mRNA processing body using protein macroarrays and “autoantigenomics”. RNA. 2007;13:704–712. doi: 10.1261/rna.411907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D, Ezzeddine N, Chen C-YA, Zhu W, He X, Shyu A-B. Deadenylation is prerequisite for P-body formation and mRNA decay in mammalian cells. J Cell Biol. 2008;182:89–101. doi: 10.1083/jcb.200801196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Y, Deutscher MP. Exoribonuclease superfamilies: structural analysis and phylogenetic distribution. Nucl Acids Res. 2001;29:1017–1026. doi: 10.1093/nar/29.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]