Abstract
The higher potential efficacy of alpha-particle radiopharmaceutical therapy lies in the 3 to 8-fold greater biological effectiveness (RBE) of alpha particles relative to photon or beta-particle radiation. This greater RBE, however, also applies to normal tissue, thereby reducing the potential advantage of high RBE. Since alpha particles typically cause DNA double strand breaks (DSBs), targeting tumors that are defective in DSB repair effectively increases the RBE, yielding a secondary, RBE-based differentiation between tumor and normal tissue that is complementary to conventional, receptor-mediated tumor targeting. In some triple negative breast cancers (TNBC, ER−/PR−/HER-2−), germline mutation in BRCA-1, a key gene in homologous recombination (HR) DSB repair, predisposes patients to early onset of breast cancer. These patients have few treatment options once the cancer has metastasized. In this study, we investigated the efficacy of alpha particle emitter, 213Bi labeled anti-EGFR antibody, Cetuximab, in BRCA-1 defective TNBC. 213Bi-Cetuximab was found to be significantly more effective in the BRCA-1 mutated TNBC cell line HCC1937 than BRCA-1 competent TNBC cell MDA-MB-231. siRNA knockdown of BRCA-1 or DNA-PKcs, a key gene in non-homologous end joining (NHEJ) DSB repair pathway, also sensitized TNBC cells to 213Bi-Cetuximab. Furthermore, the small molecule inhibitor of DNA-PKcs, NU7441, sensitized BRCA-1 competent TNBC cells to alpha particle radiation. Immunofluorescent staining of γH2AX foci and comet assay confirmed that enhanced RBE is caused by impaired DSB repair. These data offer a novel strategy for enhancing conventional receptor-mediated targeting with an additional, potentially synergistic radiobiological targeting that could be applied to TNBC.
Keywords: Alpha-particle, radiopharmaceutical therapy, triple negative breast cancer
Introduction
Radioimmunotherapy of established large solid tumors has not achieved clinical success (1, 2), partly because radiolabeled antibodies are not able to penetrate and deliver sufficient doses (typically less than 30 Gy) to elicit responses (3, 4). Accordingly, radioimmunotherapy of solid tumors is best implemented when the tumor size is small or, ideally, at a very early stage when the tumors are still microscopic clusters of malignant cells (5, 6). Contrarily, in non-Hodgkin’s lymphoma patients, equivalent or even lower tumor doses are able to elicit objective responses (7). Several factors contribute to this differential response, these include the unique biological efficacy of anti-CD20 antibody, uniform and high expression of the CD20 antigen, and the ready accessibility of lymphoma cells to radiolabeled antibodies (8). More importantly, compared to solid tumors, non-Hodgkin’s lymphoma cells are exquisitely sensitive to radiation with typical D0 values in the range of 1.3 to 1.8 Gy without an appreciable shoulder on the survival curves (9). Genetic analysis has revealed that the increased radiosensitivity of non-Hodgkin’s lymphoma cells can be attributed to impaired DNA repair due to inactivation of ATM, p53 and DNA-PKcs genes (10).
Solid tumors are often associated with defects in the DNA Damage Response (DDR) pathways and loss of function in DDR. Germline mutations in these genes cause genomic instability and predispose patients to the development of cancer (11). For example, 5–10% of hereditary breast cancer (12), 10–15% of ovarian cancer (13) and 5–10% of pancreatic cancers (14) are caused by mutations in BRCA-1/2, key genes involved in DSB repair responses. Familial form of colorectal cancer (about 3 to 4%), hereditary non-polyposis colorectal cancer (HNPCC), is associated with defective mutations in DNA mismatch repair (MMR) genes, such as MSH2 and MLH1 (15). In glioblastoma, promoter methylation on O6-methylguanine-DNA methyltransferase (MGMT) gene in base alkylation reversion repair was found on 40% of patients and is a reliable predictor for clinical outcome (16).
The differential DNA damage response between normal tissue cells with intact DNA repair and DNA repair defective tumors cells can be utilized to further enhance the efficacy of highly potent alpha particle radiopharmaceutical therapy. Alpha particles travel a short distance (<100 µm) and deposit highly focused energy along their tracks (80 keV/µm) enabling a single track to generate DSB (17). Eukaryotic cells can repair DSBs through two main pathways, homologous recombination (HR) and non-homologous end joining (NHEJ) (18). Here, we hypothesized that targeted alpha particle radiation is more effective against solid tumors that contain somatic loss of function mutations in genes involved in HR and NHEJ pathways and examined the feasibility of radiobiological targeting that may complement or synergize with conventional receptor-mediated targeting. The BRCA-1 defective TNBC model was used to evaluate this approach.
Breast cancer is a heterogeneous disease where tumors display highly varied histopathological features, gene expression profiles, response to therapy and prognosis even though they arise from the same organ. Studies in genome wide gene expression profiles have established four breast cancer types: luminal (type A and B), HER-2 positive, normal breast like and basal like (19). Among the four types of breast cancer, basal like breast cancers constitute about 15% of all breast cancers and are typically ER−, PR− and HER-2−. Triple negative breast cancer is poorly differentiated and highly aggressive; patients with TNBC are almost twice as likely as other breast cancer patients to develop distant metastasis and, therefore, suffer shorter survival (20). BRCA-1 defective tumors often belong to TNBC and share many clinical and pathological features (21). Importantly, most TNBC has high expression of EGFR (22) making it an ideal target for alpha radioimmunotherapy.
Our previous studies have shown that alpha particle emitter, 213Bi labeled, anti-rat HER-2/neu monoclonal antibody 7.16.4 prolongs the survival of HER-2/neu transgenic mice (neuN) bearing syngeneic breast cancer bone metastasis and lung metastasis but did not lead to cure (23, 24). In this study, we first evaluated the efficacy of alpha emitter 213Bi labeled anti-EGFR monoclonal antibody (Cetuximab) in TNBC cells with mutated BRCA-1. Then, we tested inhibition of DNA-PKcs, a key enzyme in the NHEJ pathway, to sensitize 213Bi-Cetuximab. This treatment strategy was then evaluated in a mouse model of TNBC metastases.
Materials and methods
Breast cancer cell lines and reagents
Four triple negative human breast cancer cell lines, MDA-MB-231, MDA-MB-436, MDA-MB-468, HCC1937 (BRCA-1 defective) and a control cell line MCF-7 (ER+, PR+, HER-2−) were obtained from the American Type Culture Collection (ATCC, Manassas, VA). The cell lines were authenticated and tested by ATCC using short tandem repeat profiling and karyotyping tests. The cell lines were grown in RPMI 1640 media containing 10% fetal bovine serum, 0.5% penicillin/streptomycin (Invitrogen, Carlsbad, CA), 1% L-glutamine, 1% nonessential amino acids, 1% sodium pyruvate, 0.02% gentamicin, and 0.2% insulin (Sigma, St. Louis, MO) and maintained at 37 °C in 5% CO2. Anti-human EGFR monoclonal antibody, Cetuximab, was obtained from Eli Lilly & Co. (Indianapolis, IN) under a Material Transfer Agreement (MTA). Anti-human phospho-Histone H2AX (Ser139) monoclonal antibody was purchased from Millipore (Billerica, MA). Alexa fluor 488 conjugated goat anti-mouse antibody was purchased from Invitrogen. DNA-PKcs inhibitor NU7441 was purchased from Tocris Bioscience (Ellisville, MO).
Antibody radiolabeling with 213Bi and 111In
Cetuximab was conjugated to SCN-CHX-A”-DTPA as described before (23). 225Ac was provided by the Institute for Transuranium Elements (Karlsruhe, Germany) (25, 26) and 213Bi was eluted from an 225Ac/213Bi generator built in-house (27). Cetuximab conjugated to the chelate was incubated with 213Bi (10 mCi/mg) for 8 min in a reaction buffer (pH 4.5) containing 3 M ammonium acetate (Fisher Scientific) and 150 mg/ml L-ascorbic acid (Sigma) preheated to 37 °C. The radiolabeling reaction was quenched with 1 µl of 100 mmol/L EDTA and radiolabeled Cetuximab was purified by size exclusion Microspin G-25 column (GE Healthcare, Buckinghamshire, UK). Cetuximab was also radiolabeled with 111In (PerkinElmer, Waltham, MA) according to a published procedure (28). The reaction efficiency and purity of the radioimmunoconjugates was determined with instant thin layer chromatography (ITLC) using silica gel impregnated paper (Agilent Technologies, Lake Forest, CA). The immunoreactivity of 13Bi-Cetuximab was evaluated by incubating 5 ng of 213Bi-Cetuximab with excess antigen binding sites (1× 107 MDA-MB-231 cells) twice on ice for 30 min each time. Antibody immunoreactivity was calculated as the percentage of 213Bi-Cetuximab bound to the cells.
EGFR expression measured by flow cytometry and Scatchard analysis
Expression of EGFR on the four TNBC cells was determined by FITC labeled Cetuximab using FACS Calibur (Becton Dickinson Biosciences, San Jose, CA). The EGFR expression level was also quantified by Scatchard analysis. Briefly, 1×106 cells were incubated with serial dilutions of 111In-Cetuximab (0.01 to 4.0 µg/ml) for 45 mins at 4 °C. Cells were washed with PBS three times before both cell pellets and supernants were collected and counted with a gamma counter (CompuGamma CS, Pharmacia). The equilibrium binding curve of bound/free antibody versus bound antibody concentration was fitted and the number of binding sites, Bmax, and antibody dissociation constant, KD, were calculated.
siRNA knockdown of BRCA-1, DNA-PKcs and RT-PCR
The siRNA knockdown studies are performed mainly to account for impact of the receptor number and cell sizes on cell survival on DSBs induction by radiolabeled antibody. siRNAs targeting BRCA-1 and DNA-PKcs as well as non-targeting scrambled siRNA were obtained from Ambion (Austin, TX). Twenty-four hours before transfection, TNBC cells were plated into 24-well plates at a density of 5×104 cells per well in cell culture media without antibiotics. Cells were transfected with Lipofectamine (Invitrogen) in OPTI-MEM media (Invitrogen) at a siRNA concentration of 20 pmol/well (add 100 µl of siRNA:Lipofectamine™2000 complexes to 0.5 ml medium). Forty eight hours after transfection, breast cancer cells were examined for knockdown of gene expression by RT-PCR. Total cellular RNA was isolated with PerfectPure RNA Cultured Cell Kit (5 Prime, Gaithersburg, MD) according to the manufacturer’s instruction. For cDNA synthesis, 1µg of RNA was reverse transcribed using qScript cDNA Synthesis Kit (Quanta Bioscience, Gaithersburg, MD). BRCA-1 and DNA-PKcs mRNA level were measured by quantitative real-time PCR using SYBR Green PCR Master Mix on an ABI Prism 7900HT Sequence Detection System (Applied Biosystem, Carlsbad, CA). The primers for BRCA-1 are: forward GGCTATCCTCTCAGAGTGACATTTTA, reverse GCTTTATCAGGTTATGTTGCA TGGT; for DNA-PKcs are: forward CCACTTACAGGATCATAGCGACGAATGC, reverse CGTGCGGGTAGTTATCAGTGATTT. Beta-actin mRNA level was used as control and the primers are: forward ACCAACTGGGACGACATGGAG, reverse GTGAGGATCTTCATGAG GTAGTC.
Specific kill of triple negative breast cancer cells by 213Bi-Cetuximab
Survival curves of TNBC cells after treatment with 213Bi-Cetuximab were measured by colony formation assay. The four TNBC cells and MCF-7 cells were plated in 24 well plates. Two days after cell plating, they were incubated with 213Bi-Cetuximab (concentration from 1.0 to 8.0 µCi/ml) overnight in duplicates and transferred to petri dishes for colony growth. All survival curves studies were repeated again to confirm the findings. To investigate the effects of BRCA-1 and DNA-PKcs on cell kill by alpha radiation, TNBC cells were transfected with siRNAs targeting BRCA-1 and DNA-PKcs. 48 hrs after transfection, cells were treated with 213Bi-Cetuximab overnight and transferred to petri dishes for colony growth. To compare the effect of small molecule inhibitor of DNA-PKcs on cell kill by alpha radiation, TNBC cells were treated with NU7441 (1.0 µM) one hour before 213Bi-Cetuximab treatment and cells were continuously incubated with NU7441 for 16 hrs before they were transferred for colony formation (29).
Immunofluorescent staining of γH2AX foci as a biomarker for DSBs
DSBs induced by 213Bi-Cetuximab were measured by immunofluorescent staining of phosphorylated histone H2AX (γH2AX). TNBC and MCF-7 cells were treated with 8 µCi 213Bi-Cetuximab. At various time points (20mins, 1, 2, 4, 6, 24hr) after initiation of 213Bi-Cetuximab treatment, cells were washed with PBS and fixed with 2% paraformaldehyde. After incubation with 0.5% NP-40 to permeabilize cell membrane, γH2AX was detected by mouse anti-human phosphorylated histone H2AX (Ser139) and Alexa Fluor 488 conjugated goat anti-mouse IgG. Cell nuclei were stained with Hoechst 33342 (Invitrogen). The fluorescent images were examined and captured by a Nikon E800 fluorescent microscope equipped with NIS-Element software (Nikon, Tokyo, Japan). The number of gamma H2AX foci per cell was counted (>50 cells per data point).
For TNBC cells transfected with siRNA targeting either DNA-PKcs or BRCA-1, the number of γH2AX foci was counted at 1 and 24 hr after treatment with 213Bi-Cetuximab. Additionally, to compare the effect of small molecule DNA-PKcs inhibitor NU7441 on DSB repair after alpha radiation, TNBC cells were treated with NU7441 as described before with 213Bi-Cetuximab and the number of γH2AX foci was quantified at 1 and 24 hr after treatment.
Comet Assay and cell cycle analysis
MDA-MB-231 and HCC1937 cells were incubated with 213Bi-Cetuximab (8µCi/mL) in cell culture media containing 2% FBS at 37°C. Cells were harvested for comet assays under neutral condition at 1 or 24 hr after initiation of incubation using the Trevigen CometAssay kit (Trevigen, Gaithersburg, MD). Briefly, cells were mixed with low melting point agarose and plated on microscope slides and allowed to gel. Cells were then lysed for 1 hr at 4 °C followed by rinse in TBE buffer (10.8 % [w/v] tris base, 5.5% [w/v] boric acid, 0.93% [w/v] EDTA). After electrophoresis in TBE buffer for 30 min at 1 V/cm, slides were washed in water and dehydrated with ethanol, air dried overnight and then treated with SYBR Green for DNA staining. Comets were imaged using a Zeiss Imager.Z1 fluorescent microscopy (Carl Zeiss AG, Oberkochen, Germany) and analyzed using the software CometScore (Autocomet.com). The Olive Tail Moment (OTM) was calculated as the average of at least 70 comets.
For cell cycle assay, at 1 or 24 hr after 213Bi-Cetuximab treatment, cells were harvested and fixed in 70% ethanol overnight. After PBS wash, cells were stained with 0.02 mg/ml propidium iodide (Sigma) in 0.1% (v/v) Triton X-100/PBS containing 0.2 mg RNase A (Sigma). Flow cytometry was performed on a FACSCalibur and data were analyzed with Cell Quest Pro (BD Bioscience).
Biodistribution of radiolabeled Cetuximab in EGFR positive TNBC
Nude mice (5 mice per group) were injected with 1×106 MDA-MB-231 cells subcutaneously in the mammary fat pad region. Ten weeks after tumor inoculation (tumor size 0.5–1.0 cm in diameter), mice were injected i.v. with approximately 20 µCi 111In-Cetuximab and were then sacrificed at 1, 3, 6, 24, 48, and 72 h post-injection. Major organs, including blood, heart, lungs, liver, spleen, kidney, stomach, intestine, muscle, femur and tumor were collected and counted in the gamma counter. The uptake of 111In-Cetuximab in the tumor and organs was decay corrected to the time of injection and calculated as %ID/g ± standard deviation.
Dosimetry
Biodistribution data from 111In-Cetuximab were converted to 213Bi-Cetuximab biodistribution based on our previous findings that radioisotope properties of 111In is a good surrogate for 213Bi antibody kinetics (23). An exponential expression was fitted to the time activity data, and the cumulative activity in each normal organ was calculated by analytic integration of the fitted expression. Absorbed doses were calculated as described in Sgouros et al. (19). Absorbed dose is thus calculated as D = (Ã× Δα + Ã×Δe)/M, wherein D is absorbed dose, à is the total number of disintegrations in an organ or tumor, M is the weight of the organ, and the mean energy emitted per nuclear transition for alpha particles, Δα, and electrons, Δe, is 1.33 ×10−12 and 1.05× 10−13 Gy kg/Bq-s, respectively. For cell level dosimetry, the absorbed dose to the TNBC cells treated with non-binding antibody was calculated as the absorbed dose to the irradiated media volume assuming all alpha particle energies are absorbed locally: where ΔEBi213 is the mean alpha-particle and electron energy per decay (Gy-kg/Bq-s), t1 is the time of treatment, A0 is the initial activity, ρ is the density of the cell (assuming water equivalent at 1.0 g/cm3), V0 is the volume of the treatment and λ is the decay constant for 213Bi. The absorbed dose was divided by two since the cells are attached to the bottom of the tissue culture plates and are assumed to receive half of the radiation from above them. The absorbed dose to EGFR positive TNBC cells targeted by 213Bi-Cetuximab was calculated using a cellular S factor (30, 31) for 213Bi, the measured number of EGF receptors per cell and assuming receptor saturation at 1 hr after generator elution. , where SBi-213, N←CS is the cellular S factor, SA0 is the specific activity and N is the number of EGF receptors per cell. The sizes of cell and cell nuclei were measured by fluorescent microscopy (Nikon 80i) and analyzed with NIS-Element imaging analysis software (Nikon, Tokyo, Japan) after cells were stained with Hoechst 33342 (Invitrogen). Cell and nucleus radius of MDA-MB-231 cell were measured as 9.2± 0.8 and 6.4 ±0.8 µm, respectively.
Statistical analysis
The statistical significance of differences between two groups was analyzed with two-way ANOVA and Kaplan-Meier survival analysis using MedCalc (MedCalc. Software). Differences with P values <0.05 were considered statistically significant.
Results
EGFR expression, radiolabeling and antibody immunoreactivity
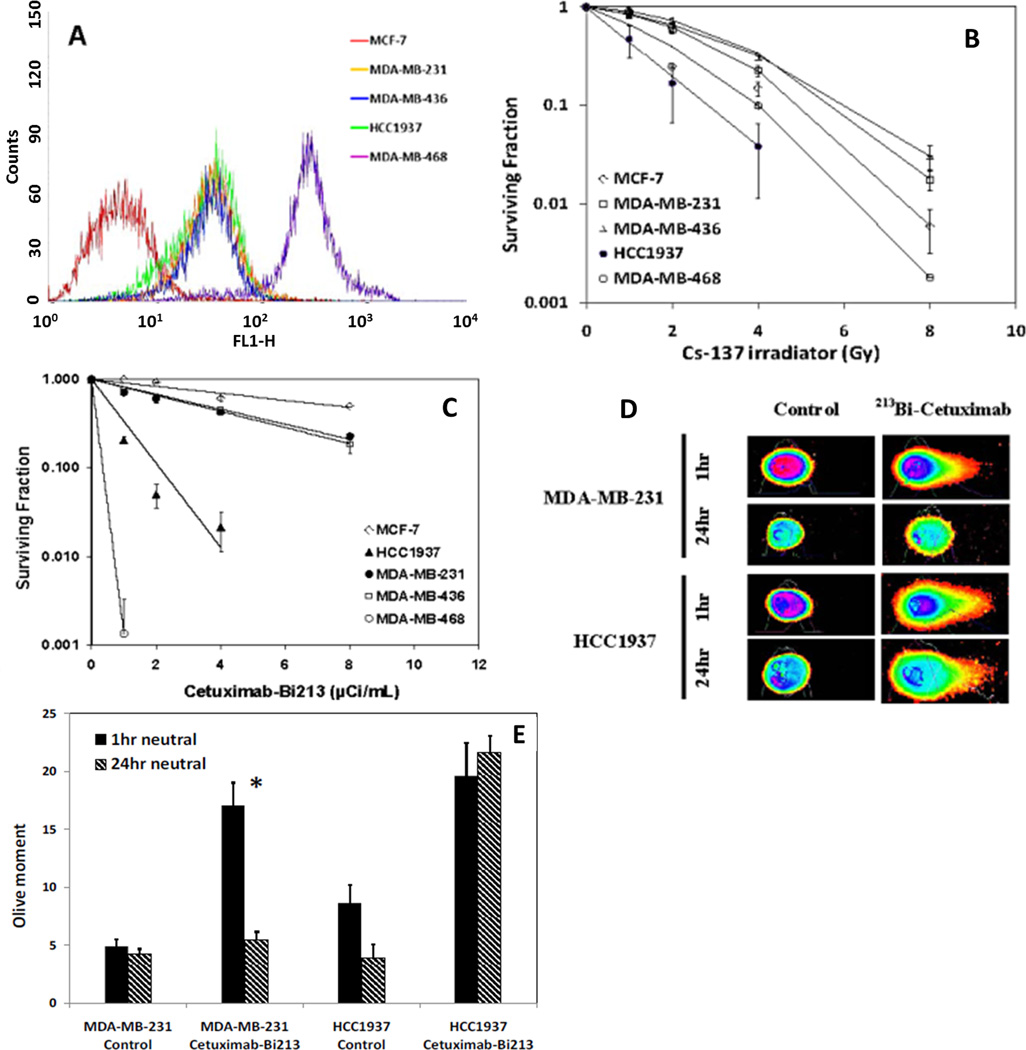
Flow cytometry with Cetuximab-FITC found EGFR expression on all four TNBC cell lines, but not on MCF-7 cells (Figure 1A). MDA-MB-468 had the highest expression level. The results of Scatchard analysis using 111In-Cetuximab are shown in tabular form in Table 1, with increased EGFR expression on MDA-MB-436, MDA-MB-231, HCC1937 and MDA-MB-468 cells. Also shown on Table 1 are the KD values of radiolabeled Cetuximab for these cell lines, which are similar to values obtained with unlabeled antibody. Reaction efficiency and purity after size exclusion purification of 213Bi labeled Cetuximab was 93.5% ± 1.7% (n=7) and 97.2% ± 0.4% (n=4) as determined by ITLC. Both reaction efficiency and purity of 111In labeled Cetuximab were routinely over 98%. The fraction of 111In-Cetuximab that is able to bind MDA-MB-231 cells in the immunoreactivity assay was 89.7%.
Figure 1.
Radiosensitivity of TNBC cell lines. A) Flow cytometry found high expression of EGFR by all four TNBC cells (MDA-MB-231, MDA-MB-436, HCC1937, MDA-MB-468). MCF-7 cell has very low level expression of EGFR and was used as negative control in the studies. B) Cell survival curves of TNBC cells and MCF-7 cells after treatment by 137Cs (gamma) radiation. BRCA-1 defective HCC1937 was found to be the most radiosensitive cell line. MDA-MB-468 cells are also relatively sensitive to 137Cs radiation when delivered at 0.5 Gy per minute. C) Cell survival curves of TNBC cells and MCF-7 cells after treatment by alpha particle emitter labeled 213Bi-Cetuximab. BRCA-1 defective HCC1937 cells are more sensitive to 213Bi-Cetuximab compared to MDA-MB-231 and MDA-MB-436 cells. MDA-MB-468 cells are the most sensitive among the TNBC cells treated. D) DNA damage caused by 213Bi-Cetuximab at 1hr after treatment and DNA damage repair at 24 hr after treatment in MDA-MB-231 and HCC937 cells as assessed by neutral Comet assay. Representative images showing head and tail fluorescent intensities of MDA-MB-231 and HCC1937 cells. E) Quantification of comet tail Olive moments at 1 hr and 24 hr following treatments.
Table 1.
Dissociation constant (KD)and receptor density for the cell lines used in this study.
| Cell line | KD (nM) | EGFR/cell |
|---|---|---|
| MDA-MB-231 | 0.8 | 2.8 × 105 |
| MDA-MB-436 | 0.2 | 0.8 × 105 |
| HCC1937 | 0.8 | 4.5 × 105 |
| MDA-MB-468 | 2.0 | 2.7 × 106 |
In vitro cytotoxicity of 213Bi-Cetuximab to TNBC cells and immunofluorescent staining of γH2AX
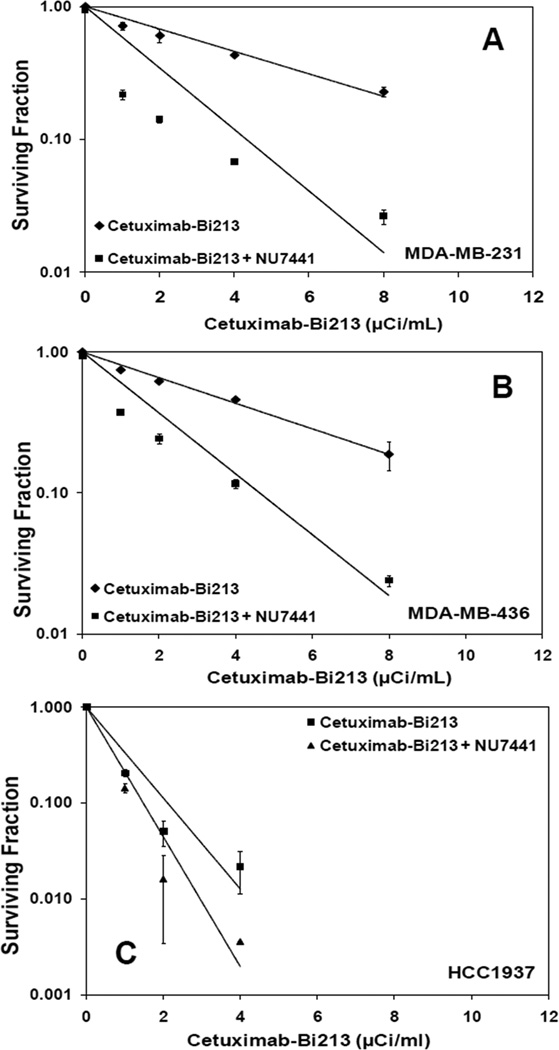
213Bi-Cetuximab kills EGFR expressing TNBC cells effectively (Figure 1C). The activity concentrations that can kill 50% (ED50) of MDA-MB-231 and MDA-MB-436 cells are 3.2 and 3.5 µCi/ml, compared to 7.8 µCi/ml in EGFR negative MCF-7 cells. Noticeably, the radiosensitivity of BRCA-1 defective HCC1937 cells to 213Bi-Cetuximab is significantly enhanced with an ED50 of 0.63 µCi/ml. MDA-MB-468 cells are the most sensitive to 213Bi-Cetuximab treatment with an ED50 of 0.50 µCi/ml. Neutral Comet assay showed that DSB are repaired in MDA-MB-231 cells but not in BRCA-1 defective HCC1937 cells at 24 hr after treatment (Figure 1D). Two way ANOVA test showed significant decrease in Olive moment in MDA-MB-231 cells from 1 hr to 24 hr (*P<0.0001) but not in HCC1937 cells (P=0.88). The difference in Olive moment between MDA-MB-231 and HCC1937 cells is also statistically significant at 1 hr (P=0.02) and 24 hr (P<0.001) (Figure 1E). Pre-treating TNBC cells with the DNA-PKcs inhibitor NU7441 (1.0 µM) further improved the efficacy of 213Bi-Cetuximab for TNBC cells (Figure 2A, B, C), with ED50 of 0.61, 0.77, 0.58 µCi/ml for MDA-MB-231, MDA-MB-436, HCC1937 cells, respectively.
Figure 2.
DNA-PKcs inhibitor NU7441 enhances cytotoxicity of 213Bi-Cetuximab in TNBC cells. Cell survival curves of A) MDA-MB-231, B) MDA-MB-436 and C) HCC1937 after treatment by 213Bi-Cetuximab or 213Bi-Cetuximab plus NU7441.
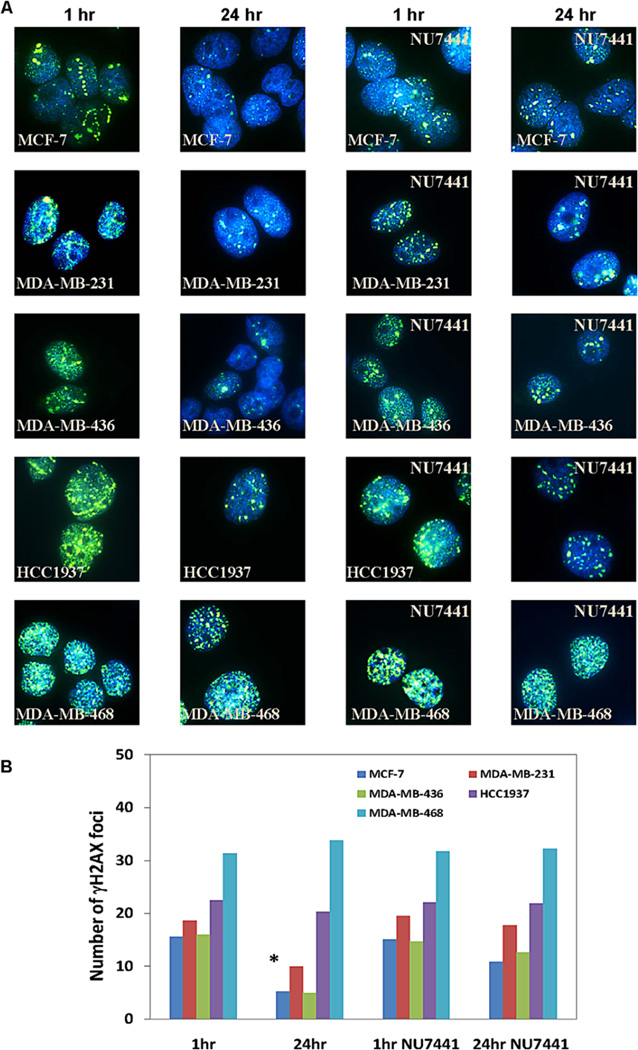
Immunofluorescent staining of γH2AX at 1 hr and 24 hrs after treatment with 213Bi-Cetuximab is shown in Figure 3. Unlike random γH2AX foci that are usually formed after the cells are irradiated with gamma-rays (data not shown), the γH2AX foci typically formed noticeable straight line tracks within cell nuclei after treatment by 213Bi-Cetuximab, indicating DSBs caused by alpha particle tracks (Fig.3a, for example). The time course of γH2AX foci formation after alpha radiation treatment was evaluated at 20 mins, 1, 2, 4, 6 and 24 hrs after initiation of treatment. The number of γH2AX foci peaked at 1hr and decreased gradually thereafter (data not shown). The reduction in γH2AX foci from 1 to 24 h was therefore used as a measure of DSB repair capacity after 213Bi-Cetuximab treatment. In MCF-7, MDA-MB-231, MDA-MB-436 cells, the number of γH2AX foci per cell all decreased significantly from 15.5±4.8 (1hr) to 5.2±2.4 (24hr), 18.7±4.7 (1hr) to 10.0±5.1 (24hr) and 16.0±3.8 (1hr) to 4.9±2.3 (24 hr), respectively. In contrast in BRCA-1 defective HCC1937 cells and MDA-MB-468 cells, there was no significant change in the number of γH2AX foci at 1hr (22.5±6.6 and 31.3±8.0) and 24 hr (20.3±6.5 and 33.8±10.3) (Figure 3B), strongly suggesting compromised, if not complete lack of, DSB repair in these two cell lines.
Figure 3.
A) Representative images showing immunefluorescently stained γH2AX foci at 1hr and 24hr after treatment with 213Bi-Cetuximab or 213Bi-Cetuximab plus NU7441 in MCF-7 and the four TNBC cell lines studied. B) Quantification of number of γ-H2AX foci at 1hr and 24hr after treatment by 213Bi-Cetuximab or 213Bi-Cetuximab plus NU7441. Two way ANOVA test showed that there was a statistical significant decrease (*) of DSBs foci in MDA-MB-231 (P<0.001), MDA-MB-436 (P<0.001) and MCF-7 (P<0.001) cells from 1hr to 24 hr after treatment with 213Bi-Cetuximab alone, while such decrease was not observed in HCC1937 (P=0.13) and MDA-MB-468 (P=0.63) cells. NU7441 treatment significantly inhibited the decrease of γH2AX foci from 1hr to 24 hr in MDA-MB-231 (P=0.43) and MDA-MB-436 (P=0.68) cells, but did not affect HCC1937 and MDA-MB-468 cells.
Treatment of MCF-7, MDA-MB-231 and MDA-MB-436 cells by the DNA-PKcs inhibitor NU7441 and 213Bi-Cetuximab abolished the significant decrease of γH2AX foci from 1hr to the 24-hour period, likely a result of DSB repair inhibition (Figure 3). For the two cell lines already sensitive to 213Bi-Cetuximab, HCC1937 and MDA-MB-468, adding NU7441 to 213Bi-Cetuximab did not affect the number of γH2AX foci from 1hr to 24hr. (Figure 3B).
siRNA knockdown of BRCA-1 or DNA-PKcs enhanced radiosensitivity of TNBC cells
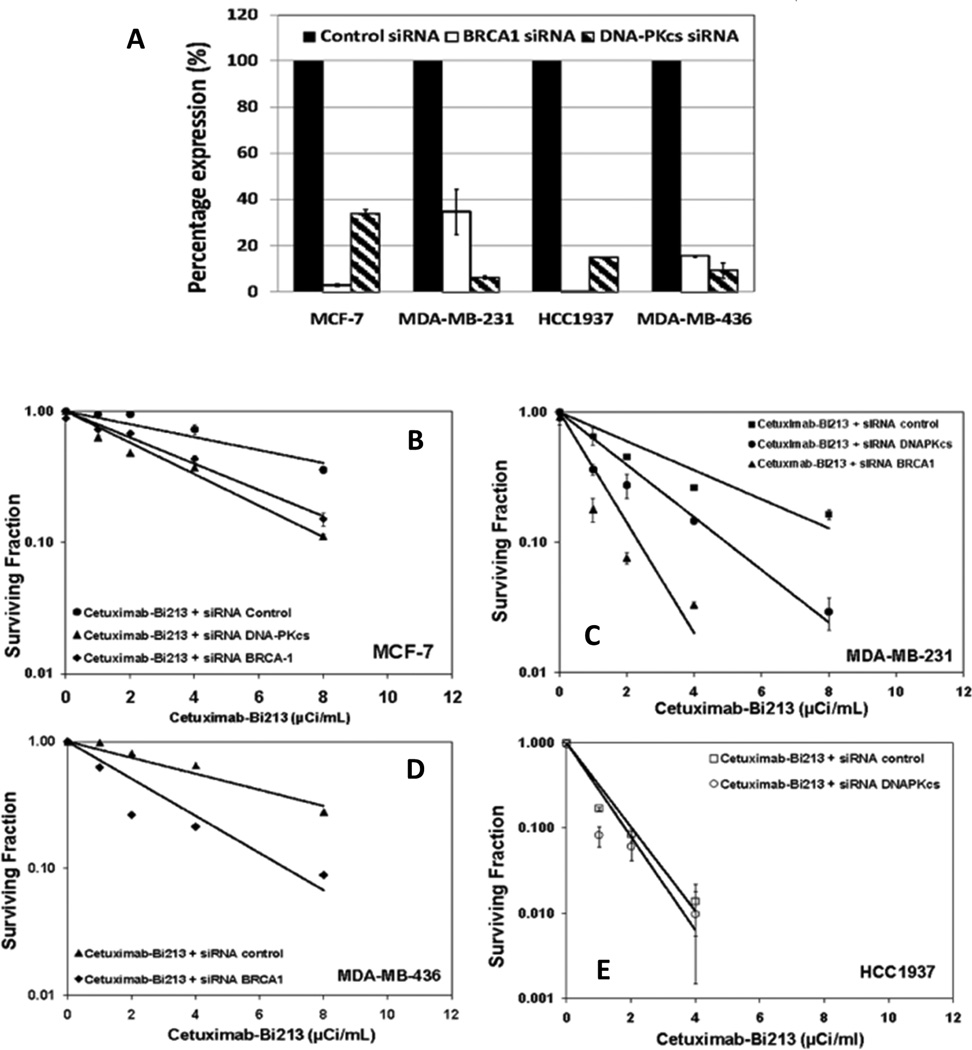
Gene expression of either BRCA-1 or DNA-PKcs was down-regulated in the four TNBC cells, MCF-7, MDA-MB-231, MDA-MB-436 and HCC1937 after transfection of siRNA targeting either BRCA-1 or DNA-PKcs as confirmed by real time RT-PCR (Figure 4A). Knockdown of either DNA-PKcs or BRCA-1 improved the radiosensitivity of TNBC cells to 213Bi-Cetuximab. In MCF-7 cells, the ED50 was reduced from 6.5 µCi/ml from control siRNA to 3.4 and 1.9 µCi/ml in cells knocked down with BRCA-1 or DNA-PKcs siRNA, respectively (Figure 4B). In MDA-MB-231 cells treated with 213Bi-Cetuximab, the ED50 improved significantly from 1.8 µCi/ml in cells transfected by negative control scrambled siRNA to 0.56 µCi/ml in cells transfected by siRNA targeting BRCA-1 and to 0.78 µCi/ml in cells transfected by siRNA targeting DNA-PKcs (Figure 4C). Likewise, siRNA knockdown of BRCA-1 in MDA-MB-436 cells enhanced its radiosensitivity to 213Bi-Cetxumiab with an ED50 change from 5.6 to 1.4 µCi/ml (Figure 4D). In the BRCA-1 mutated HCC1937 cells, knockdown of DNA-PKcs slightly reduced the ED50 from 0.60 to 0.53 µCi/ml, consistent with that observed in the parental cells treated with DNA-PKcs inhibitor NU7441 and 213Bi-Cetuximab.
Figure 4.
siRNA knockdown of BRCA-1 (HR) and DNA-PKcs (NHEJ) enhances the radiosensitivity of breast cancer cells: A) Real time PCR confirmed siRNA knockdown of BRCA-1 and DNA-PKcs in MCF-7, MDA-MB231, MDA-MB4236 and HCC1937 cells. Cell survival curves are shown for B). MCF-7 cells; C) MDA-MB-231 cells; D) MDA-MB436 cells; E) HCC1937 cells; to alpha particle radiation delivered by 213Bi-Cetuximab.
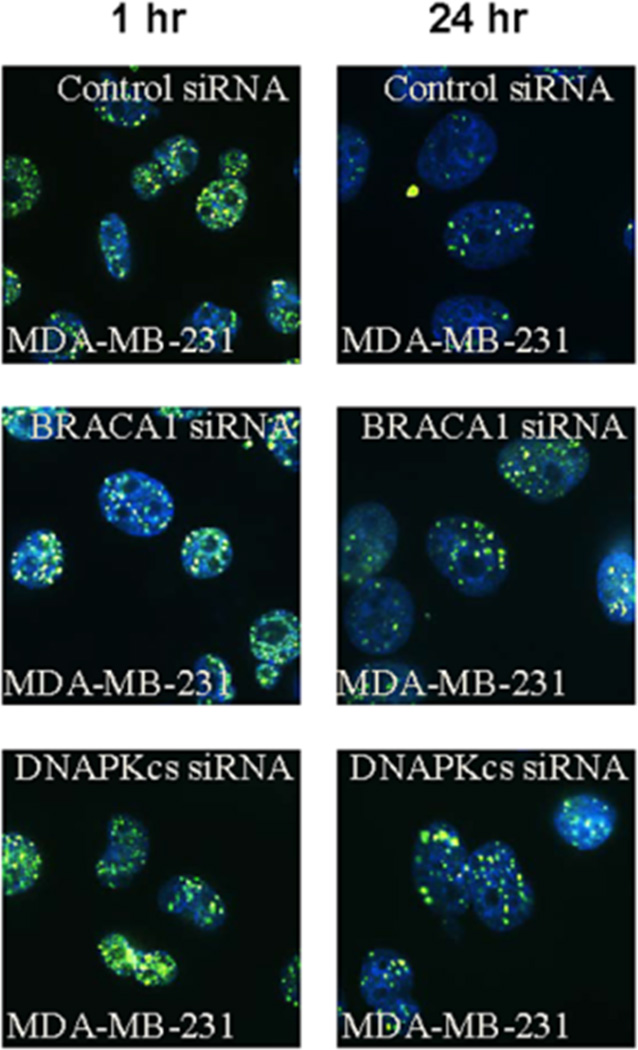
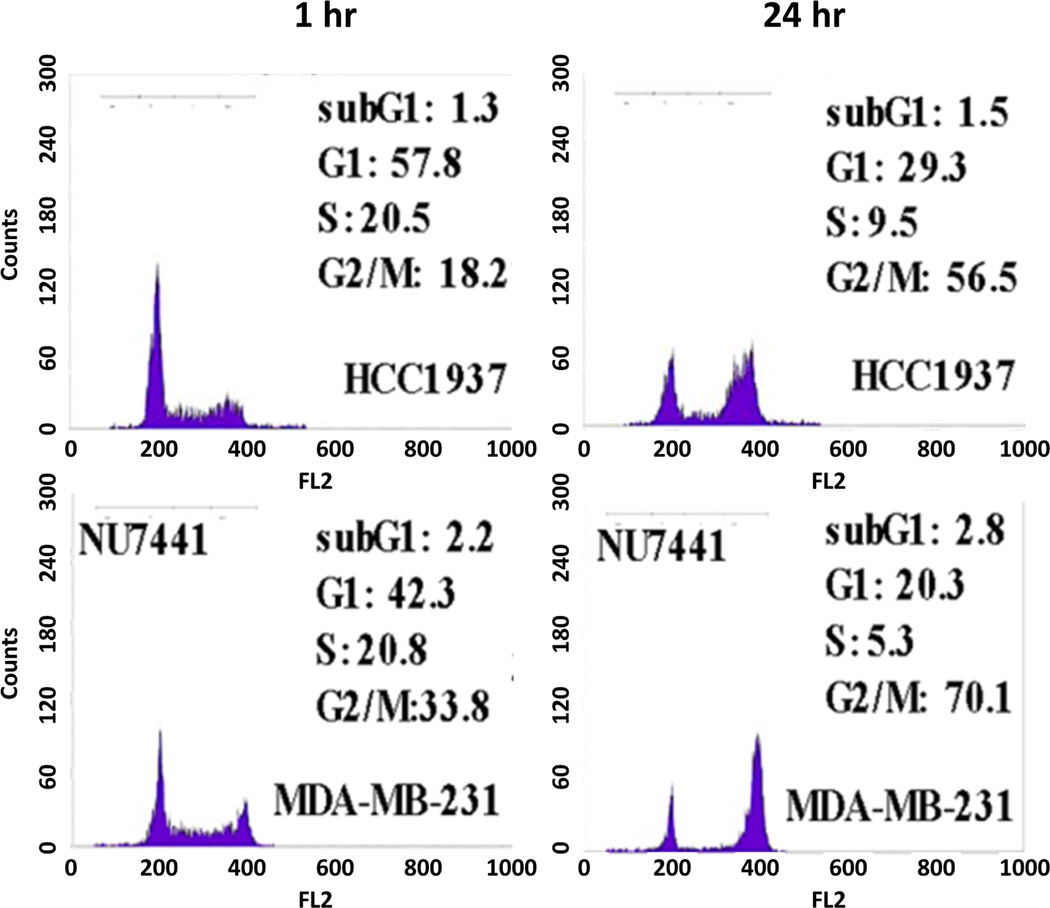
Immunofluorescent staining of γH2AX foci confirmed the inhibition of DSB repair in MDA-MB-231 cells transfected with siRNA targeting DNA-PKcs or BRCA-1 (Figure 5). Cell cycle analysis showed that both TNBC cells, MDA-MB-231 and HCC1937, were arrested in G2/M phase after treatment with 213Bi-Cetuximab. Inhibiting DNA-PKcs in MDA-MB-231 cells further enhanced the G2/M arrest after 213Bi-Cetuximab treatment (Figure 6).
Figure 5.
γ-H2AX immunofluorescent staining shows that knockdown of BRCA-1 and DNA-PKcs genes inhibited the resolution of γ-H2AX at 24 hour in MDA-MB-231 cells.
Figure 6.
Cell cycle analysis showed alpha particle induced cell cycle arrest in G2/M phase in BRCA-1 defective HCC1937 and NU7441 treated MDAMB231 cells.
Biodistribution of 111In-Cetuximab in EGFR positive TNBC tumors
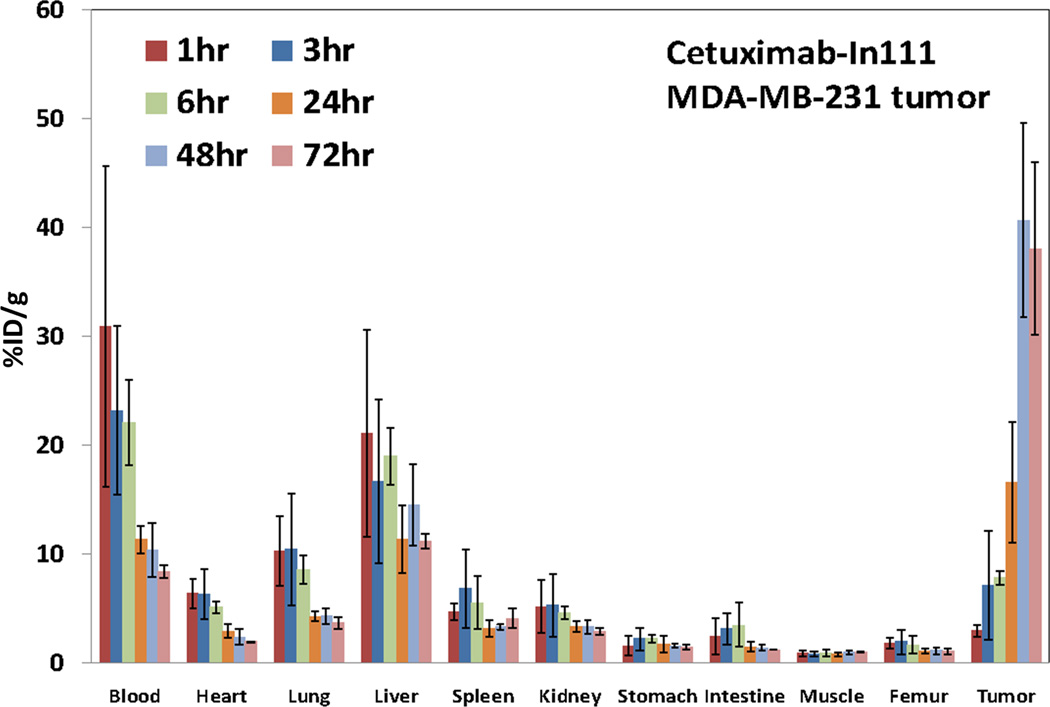
The biodistribution of 111In-Cetuximab in nude mice bearing a subcutaneous MDA-MB-231 tumor is shown in Figure 7. 111In-Cetuximab was cleared slowly from blood with an effective half-life of 40.8 hours and accumulated in the MDA-MB-231 tumors; %ID/g in the tumors increased from 2.9 ± 0.5% at 1hr to its peak of 40.6 ± 8.9% at 48 hr after injection. Tumor uptake persisted at 72 hr at 38.0 ± 7.9 %ID/g. Like most antibodies, liver is the normal organ after blood with the highest uptake of 21.1 ± 9.5%ID/g at 1 hr. Consistent with the finding that Cetuximab does not cross react with mouse EGFR, no significant uptake in other normal organs was observed. Finally, 111In-Cetuximab cleared slowly from major organs with half-life ranging from 40.8 to 173.3 hr.
Figure 7.
Biodistribution of 111In-Cetuximab in nude mice (5 mice per group) bearing subcutaneous MDA-MB-231 tumors. Mice were sacrificed at 1, 3, 6, 24, 48, 72 hr after injection of radiolabeled Cetuximab. Tumor uptake of Cetuximab peaked at 48 hrs post injection.
Dosimetry
Absorbed doses from 120 µCi 213Bi-Cetuximab to normal organs are shown in Table 2. The blood absorbed dose from alpha particles is 5.23 Gy and liver has the highest normal organ absorbed dose with 4.28 and 0.22 Gy from alpha and beta particle emissions, respectively. These results are consistent with our observations in other dosimetric studies of alpha particle emitter labeled intact antibodies (23). The RBE for MDA-MB-231 cells based on calculations using of non-specific and specific antibody showed that non-specific 213Bi-Rituximab gives an RBE of 3.8 using 37% cell survival as biological endpoint and 137Cs γ-rays as reference, while a similar RBE of 3.7 was found for 213Bi-Cetuximab. When DSB repair genes DNA-PKcs and BRCA-1 are knocked down by siRNA, the RBEs increased significantly to 8.6 and 15.6 respectively (Table 3).
Table 2.
Tissue absorbed doses
| Tissue | Absorbed dose (Gy) | |
|---|---|---|
| α-particle | electron | |
| Blood | 5.23 | 0.27 |
| Heart | 1.34 | 0.07 |
| Lung | 2.14 | 0.11 |
| Liver | 4.28 | 0.22 |
| Spleen | 1.21 | 0.06 |
| Kidney | 1.15 | 0.06 |
| Stomach | 0.37 | 0.02 |
| Intestine | 0.66 | 0.03 |
| Muscle | 0.19 | 0.01 |
| Femur | 0.40 | 0.02 |
Table 3.
Radiosensitivity (D0) and relative biological efficacy (RBE) of the MDA-MB-231cell line under different exposure and DNA repair pathway inhibition conditions.
| Agent, manipulation | D0 (Gy) | RBE* |
|---|---|---|
| 213Bi-Rituximab (irrelevant Ab) | 0.84 | 3.8 |
| 213Bi-Cetuximab | 0.87 | 3.7 |
| 213Bi-Cetuximab ,siRNA scrambled control | 0.69 | 4.7 |
| 213Bi-Cetuximab, siRNA DNA-PKcs-/DNA-PKcs- | 0.37 | 8.6 |
| 213Bi-Cetuximab, siRNA BRCA1-/BRCA1- | 0.21 | 15.6 |
RBE is reported using 37% cell survival as the biological endpoint and Cs-137 gamma rays as the reference radiation.
Discussion
In this study we showed that targeting alpha particle radiation (213Bi) with anti-EGFR antibody Cetuximab is highly effective in killing BRCA-1 defective TNBC cells where DSB repair is compromised. We further showed that inhibiting DSB repair pathways, either the HR pathway via BRCA-1 or NHEJ pathway via DNA-PKcs, preferentially sensitizes breast cancer cells with intact DSB repair function to alpha particles. Thus, similar to the concept of prescreening HER-2 positive breast cancer before treatment with Trastuzumab, one could envision, in the era of cancer genomics, pre-screening breast tumors with loss of function mutations in DSB repair genes would increase the efficacy of targeted alpha radioimmunotherapy.
Targeted alpha particle emitters are currently under clinical investigations for leukemia (32) ovarian cancer (33), malignant melanoma (34) and glioblastoma (35) because of their high potency. The recent clinical success of Radium-223 (Alpharadin) in castration-resistant prostate cancer with bone metastases (36) supports further development of alpha particle emitters and highlights the importance of a unique biological mechanism in the highly specific accumulation of targeted radioisotopes. EGFR is highly expressed on a variety of epithelial cancers, 99mTc, 64Cu, 86Y, 89Zr anti-EGFR antibodies (Cetuximab, Panitumumab) have been extensively investigated as imaging agents in colorectal cancer, squamous cell carcinoma of head and neck (SCCHN) and lung cancer (37–39). As radiotherapeutics, anti-EGFR antibodies have been labeled with both beta emitters 90Y, 177Lu and alpha emitter, 213Bi, and were found effective in several preclinical models of SCCHN, bladder and colorectal cancer (40, 41). Few clinical trials have been launched partially due to the prevalent expression of EGFR on normal epithelial cells that lead to skin and gastrointestinal toxicity observed in Cetuximab trials (42). A clinical study of intravenous 125I labeled anti-EGFR antibody 425 with radiotherapy in patients with glioblastoma found no survival benefit compared to radiotherapy alone (43). Targeting TNBC cells with high EGFR expression level and defective DSB repair pathways can potentially open a therapeutic window that leads to clinical responses with tolerated toxicity to normal tissues.
Tumors that have homologous loss-of-function defects at DSB repair pathway genes often arise from the normal tissues that are heterozygous and preserve DNA repair function. Nieuwenhuis et al. and others measured the rejoining of DNA breaks in normal fibroblast and lymphocytes cells with heterozygous BRCA-1/BRCA-2 mutations after X-ray radiation and found no defect in their ability to repair DNA breaks (44). In external beam radiotherapy, multiple studies were not able to link pathogenic heterozygous mutations in DNA damage response genes such as ATM, BRCA-1, BRCA2 and RAD50 to normal tissue radiosensitivity (45). These findings suggest that high LET alpha-particle radiation could be targeted to treat tumors with homozygous loss-of-function in DNA repair while sparing normal tissues with heterozygous DNA repair genes.
In cells deficient in NHEJ and HR, we found that compromised repair of DSB lesions, rather than initially produced DSBs by alpha particle radiation determines cell survival. This is consistent with heavy ion beam studies. Irradiation of glioblastoma cells MO59J deficient in DNA-PKcs with heavy ion beam found no reduction of γ-H2AX foci after 21 hrs (46). In Ku80 deficient (NHEJ deficient) CHO cells treated with alpha particles, significantly more γ-H2AX foci were present (58.4% to 69.5%) 2 hrs after radiation compared to normal CHO cells (36.5%-42.8%) (47).
Cell survival curves showed that 213Bi-Cetuximab is most efficacious in BRCA-1 defective HCC1937 cells and BRCA-1 knockdown MDA-MB-231 cells. In TNBC patients that are not BRCA-1 defective, agents that lead to BRCA-1 inhibition combined with targeted alpha radiotherapy would be useful. Until recently, only a handful of BRCA-1 inhibitors, however, are under early stage evaluation primarily due to low specificity since many of these protein-protein interactions, unlike enzyme active sites, also have large, multiple contact points and poorly defined topology (48). Many other DSB repair inhibitors are currently available for combination with targeted alpha therapy for TNBC including the DNA-PKcs inhibitor used in this study (29), Ataxia-Telangiectasia Mutated Kinase (ATM) inhibitor and Rad51 inhibitors (49).
Poly (ADP-ribose) polymerase (PARP-1) inhibitors are also under clinical investigation for breast and ovarian cancer patients with BRCA-1 mutations under the synthetic lethality hypothesis (21). A phase II trial showed that PARP-1 inhibitor iniparib in combination with gemcitabine and carboplatin significantly improved progression free survival and overall survival in TNBC patients (50). The subsequent phase III trial, however, failed to confirm its survival benefit (51). Since alpha particle emitter radiolabeled antibody is targeting the same DNA repair deficiency as PARP-1 inhibitors, these clinical results suggested the potential of alpha radioimmunotherapy in TNBC patients but at the same time warrant more careful preclinical studies of, for example, the correlation of EGFR expression level, mutation rate with their deficiency in BRCA-1 and response to alpha radiation.
An interesting observation in this study is that MDA-MB-468 cells were exquisitely sensitive to the treatment of 213Bi-Cetuximab, even more so than the BRCA-1 defective HCC1937 cells. One of the main reasons probably is its high expression of EGFR receptors (almost ten times higher) compared to other TNBC cells lines, potentially delivering significantly higher alpha doses to the tumor cells. In addition, the cell survival curve with gamma-rays showed that the MDA-MB-468 cells are sensitive to radiation and exhibit a diminished shoulder region, consistent with its radiosensitivity to UV radiation attributed to the deficient RB gene and deficiency in recovery of mRNA synthesis (52). The possible dis-regulation of DSB repair in MDA-MB-468 cells remains to be studied.
We have performed initial in vivo evaluation of 213Bi-Cetuximab in a mouse model of TNBC. Due to poor tumorigenesis of the HCC1937 cells, however, we cannot interpret the data reliably. We are currently developing a more robust mouse model to evaluate the efficacy of alpha radioimmunotherapy in tumors with homozygous loss-of-function and toxicity in normal tissues with heterozygous at DSB repair genes.
In conclusion, we have shown that TNBC can be targeted by alpha particle emitter labeled anti-EGFR antibody. The defective DSB repair machinery in a subset of TNBC can be uniquely suitable for alpha radiation treatment. New approaches combining inhibitors of DSB repair pathways and internal alpha particle emitters targeting remain to be explored. Finally, the safety of such an approach in patients with heterozygous expression of DSB repair genes needs to be evaluated.
Acknowledgments
Grant Support: This work was supported by NIH Grant R01 CA 113797 (G. Sgouros) and Maryland Stem Cell Research Fund (MSCRF) grant 2009-MSCRFE-0109-00 (H. Song).
Abbreviations List
- RBE
Relative biological effectiveness
- TNBC
Triple negative breast cancer
- HR
homologous recombination
- NHEJ
non-homologous end joining
- DDR
DNA damage response
- PARP
Poly (ADP-ribose) polymerase
Footnotes
Disclosure of Potentail Conflicts of Interest: None
References
- 1.Goldenberg DM. Targeted therapy of cancer with radiolabeled antibodies. J Nucl Med. 2002;43:693–713. [PubMed] [Google Scholar]
- 2.Song H, Sgouros G. Radioimmunotherapy of solid tumors: searching for the right target. Curr Drug Deliv. 8:26–44. doi: 10.2174/156720111793663651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pai-Scherf LH, Carrasquillo JA, Paik C, Gansow O, Whatley M, Pearson D, et al. Imaging and phase I study of 111In- and 90Y-labeled anti-LewisY monoclonal antibody B3. Clin Cancer Res. 2000;6:1720–1730. [PubMed] [Google Scholar]
- 4.Sharkey RM, Goldenberg DM, Murthy S, Pinsky H, Vagg R, Pawlyk D, et al. Clinical evaluation of tumor targeting with a high-affinity, anticarcinoembryonic-antigen-specific, murine monoclonal antibody, MN-14. Cancer. 1993;71:2082–2096. doi: 10.1002/1097-0142(19930315)71:6<2082::aid-cncr2820710625>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 5.Ballangrud AM, Yang WH, Palm S, Enmon R, Borchardt PE, Pellegrini VA, et al. Alpha-Particle Emitting Atomic Generator (Actinium-225)-Labeled Trastuzumab (Herceptin) Targeting of Breast Cancer Spheroids: Efficacy versus HER2/neu Expression. Clin Cancer Res. 2004;10:4489–4497. doi: 10.1158/1078-0432.CCR-03-0800. [DOI] [PubMed] [Google Scholar]
- 6.Ballangrud AM, Yang WH, Charlton DE, McDevitt MR, Hamacher KA, Panageas KS, et al. Response of LNCaP spheroids after treatment with an alpha-particle emitter (213Bi)-labeled anti-prostate-specific membrane antigen antibody (J591) Cancer Res. 2001;61:2008–2014. [PubMed] [Google Scholar]
- 7.Sgouros G, Squeri S, Ballangrud AM, Kolbert KS, Teitcher JB, Panageas KS, et al. Patient-specific, 3-dimensional dosimetry in non-Hodgkin's lymphoma patients treated with 131I-anti-B1 antibody: assessment of tumor dose-response. J Nucl Med. 2003;44:260–268. [PubMed] [Google Scholar]
- 8.Hernandez MC, Knox SJ. Radiobiology of radioimmunotherapy with 90Y ibritumomab tiuxetan (Zevalin) Semin Oncol. 2003;30:6–10. doi: 10.1053/j.seminoncol.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 9.Johansson L, Carlsson J, Nilsson K. Radiosensitivity of human B-lymphocytic lymphomas in vitro. Int J Radiat Biol Relat Stud Phys Chem Med. 1982;41:411–420. doi: 10.1080/09553008214550451. [DOI] [PubMed] [Google Scholar]
- 10.M'Kacher R, Bennaceur A, Farace F, Lauge A, Plassa LF, Wittmer E, et al. Multiple molecular mechanisms contribute to radiation sensitivity in mantle cell lymphoma. Oncogene. 2003;22:7905–7912. doi: 10.1038/sj.onc.1206826. [DOI] [PubMed] [Google Scholar]
- 11.Lord CJ, Ashworth A. The DNA damage response and cancer therapy. Nature. 2012;481:287–294. doi: 10.1038/nature10760. [DOI] [PubMed] [Google Scholar]
- 12.Fackenthal JD, Olopade OI. Breast cancer risk associated with BRCA1 and BRCA2 in diverse populations. Nat Rev Cancer. 2007;7:937–948. doi: 10.1038/nrc2054. [DOI] [PubMed] [Google Scholar]
- 13.Risch HA, McLaughlin JR, Cole DE, Rosen B, Bradley L, Kwan E, et al. Prevalence and penetrance of germline BRCA1 and BRCA2 mutations in a population series of 649 women with ovarian cancer. Am J Hum Genet. 2001;68:700–710. doi: 10.1086/318787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goggins M, Schutte M, Lu J, Moskaluk CA, Weinstein CL, Petersen GM, et al. Germline BRCA2 gene mutations in patients with apparently sporadic pancreatic carcinomas. Cancer Res. 1996;56:5360–5364. [PubMed] [Google Scholar]
- 15.Lynch HT, de la Chapelle A. Hereditary colorectal cancer. N Engl J Med. 2003;348:919–932. doi: 10.1056/NEJMra012242. [DOI] [PubMed] [Google Scholar]
- 16.Esteller M, Garcia-Foncillas J, Andion E, Goodman SN, Hidalgo OF, Vanaclocha V, et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000;343:1350–1354. doi: 10.1056/NEJM200011093431901. [DOI] [PubMed] [Google Scholar]
- 17.McDevitt MR, Sgouros G, Finn RD, Humm JL, Jurcic JG, Larson SM, et al. Radioimmunotherapy with alpha-emitting nuclides. Eur J Nucl Med. 1998;25:1341–1351. doi: 10.1007/s002590050306. [DOI] [PubMed] [Google Scholar]
- 18.Kasparek TR, Humphrey TC. DNA double-strand break repair pathways, chromosomal rearrangements and cancer. Semin Cell Dev Biol. 2011;22:886–897. doi: 10.1016/j.semcdb.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 19.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 20.Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, Sawka CA, et al. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res. 2007;13:4429–4434. doi: 10.1158/1078-0432.CCR-06-3045. [DOI] [PubMed] [Google Scholar]
- 21.Anders CK, Winer EP, Ford JM, Dent R, Silver DP, Sledge GW, et al. Poly(ADP-Ribose) polymerase inhibition: "targeted" therapy for triple-negative breast cancer. Clin Cancer Res. 2010;16:4702–4710. doi: 10.1158/1078-0432.CCR-10-0939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song H, Shahverdi K, Huso DL, Esaias C, Fox J, Liedy A, et al. 213Bi (alpha-emitter)-antibody targeting of breast cancer metastases in the neu-N transgenic mouse model. Cancer Res. 2008;68:3873–3880. doi: 10.1158/0008-5472.CAN-07-6308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song H, Hobbs RF, Vajravelu R, Huso DL, Esaias C, Apostolidis C, et al. Radioimmunotherapy of breast cancer metastases with alpha-particle emitter 225Ac: comparing efficacy with 213Bi and 90Y. Cancer Res. 2009;69:8941–8948. doi: 10.1158/0008-5472.CAN-09-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Apostolidis C, Molinet R, Rasmussen G, Morgenstern A. Production of Ac-225 from Th-229 for targeted alpha therapy. Anal Chem. 2005;77:6288–6291. doi: 10.1021/ac0580114. [DOI] [PubMed] [Google Scholar]
- 26.Zielinska B, Apostolidis C, Bruchertseifer F, Morgenstern A. An improved method for the production of Ac-225/Bi-213 from Th-229 for targeted alpha therapy. Solvent Extraction and Ion Exchange. 2007;25:339–349. [Google Scholar]
- 27.McDevitt MR, Finn RD, Sgouros G, Ma D, Scheinberg DA. An 225Ac/213Bi generator system for therapeutic clinical applications: construction and operation. Appl Radiat Isot. 1999;50:895–904. doi: 10.1016/s0969-8043(98)00151-1. [DOI] [PubMed] [Google Scholar]
- 28.Song H, Shahverdi K, Huso DL, Wang Y, Fox JJ, Hobbs RF, et al. An immunotolerant HER-2/neu transgenic mouse model of metastatic breast cancer. Clin Cancer Res. 2008;14:6116–6124. doi: 10.1158/1078-0432.CCR-07-4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao Y, Thomas HD, Batey MA, Cowell IG, Richardson CJ, Griffin RJ, et al. Preclinical evaluation of a potent novel DNA-dependent protein kinase inhibitor NU7441. Cancer Res. 2006;66:5354–5362. doi: 10.1158/0008-5472.CAN-05-4275. [DOI] [PubMed] [Google Scholar]
- 30.Howell RW, Rao DV, Bouchet LG, Bolch WE, Goddu SM. MIRD Cellular S Values: Reston (VA) Society of Nuclear Medicine. 1997 [Google Scholar]
- 31.Hamacher KA, Den RB, Den EI, Sgouros G. Cellular dose conversion factors for alpha-particle--emitting radionuclides of interest in radionuclide therapy. J Nucl Med. 2001;42:1216–1221. [PubMed] [Google Scholar]
- 32.Rosenblat TL, McDevitt MR, Mulford DA, Pandit-Taskar N, Divgi CR, Panageas KS, et al. Sequential cytarabine and alpha-particle immunotherapy with bismuth-213-lintuzumab (HuM195) for acute myeloid leukemia. Clin Cancer Res. 2010;16:5303–5311. doi: 10.1158/1078-0432.CCR-10-0382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andersson H, Cederkrantz E, Back T, Divgi C, Elgqvist J, Himmelman J, et al. Intraperitoneal alpha-particle radioimmunotherapy of ovarian cancer patients: pharmacokinetics and dosimetry of (211)At-MX35 F(ab')2--a phase I study. J Nucl Med. 2009;50:1153–1160. doi: 10.2967/jnumed.109.062604. [DOI] [PubMed] [Google Scholar]
- 34.Allen BJ, Singla AA, Rizvi SM, Graham P, Bruchertseifer F, Apostolidis C, et al. Analysis of patient survival in a Phase I trial of systemic targeted alpha-therapy for metastatic melanoma. Immunotherapy. 2011;3:1041–1050. doi: 10.2217/imt.11.97. [DOI] [PubMed] [Google Scholar]
- 35.Kneifel S, Cordier D, Good S, Ionescu MC, Ghaffari A, Hofer S, et al. Local targeting of malignant gliomas by the diffusible peptidic vector 1,4,7,10-tetraazacyclododecane-1-glutaric acid-4,7,10-triacetic acid-substance p. Clin Cancer Res. 2006;12:3843–3850. doi: 10.1158/1078-0432.CCR-05-2820. [DOI] [PubMed] [Google Scholar]
- 36.Nilsson S, Franzen L, Parker C, Tyrrell C, Blom R, Tennvall J, et al. Bone-targeted radium-223 in symptomatic, hormone-refractory prostate cancer: a randomised, multicentre, placebo-controlled phase II study. Lancet Oncol. 2007;8:587–594. doi: 10.1016/S1470-2045(07)70147-X. [DOI] [PubMed] [Google Scholar]
- 37.Nayak TK, Regino CA, Wong KJ, Milenic DE, Garmestani K, Baidoo KE, et al. PET imaging of HER1-expressing xenografts in mice with 86Y-CHX-A"-DTPA-cetuximab. Eur J Nucl Med Mol Imaging. 37:1368–1376. doi: 10.1007/s00259-009-1370-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nayak TK, Garmestani K, Milenic DE, Brechbiel MW. PET and MRI of metastatic peritoneal and pulmonary colorectal cancer in mice with human epidermal growth factor receptor 1-targeted 89Zr-labeled panitumumab. J Nucl Med. 53:113–120. doi: 10.2967/jnumed.111.094169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schechter NR, Wendt RE, 3rd., Yang DJ, Azhdarinia A, Erwin WD, Stachowiak AM, et al. Radiation dosimetry of 99mTc-labeled C225 in patients with squamous cell carcinoma of the head and neck. J Nucl Med. 2004;45:1683–1687. [PubMed] [Google Scholar]
- 40.Pfost B, Seidl C, Autenrieth M, Saur D, Bruchertseifer F, Morgenstern A, et al. Intravesical alpha-radioimmunotherapy with 213Bi-anti-EGFR-mAb defeats human bladder carcinoma in xenografted nude mice. J Nucl Med. 2009;50:1700–1708. doi: 10.2967/jnumed.109.065961. [DOI] [PubMed] [Google Scholar]
- 41.Milenic DE, Wong KJ, Baidoo KE, Ray GL, Garmestani K, Williams M, et al. Cetuximab: preclinical evaluation of a monoclonal antibody targeting EGFR for radioimmunodiagnostic and radioimmunotherapeutic applications. Cancer Biother Radiopharm. 2008;23:619–631. doi: 10.1089/cbr.2008.0493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van Cutsem E, Kohne CH, Hitre E, Zaluski J, Chang Chien CR, Makhson A, et al. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med. 2009;360:1408–1417. doi: 10.1056/NEJMoa0805019. [DOI] [PubMed] [Google Scholar]
- 43.Wygoda Z, Kula D, Bierzynska-Macyszyn G, Larysz D, Jarzab M, Wlaszczuk P, et al. Use of monoclonal anti-EGFR antibody in the radioimmunotherapy of malignant gliomas in the context of EGFR expression in grade III and IV tumors. Hybridoma (Larchmt) 2006;25:125–132. doi: 10.1089/hyb.2006.25.125. [DOI] [PubMed] [Google Scholar]
- 44.Nieuwenhuis B, Van Assen-Bolt AJ, Van Waarde-Verhagen MA, Sijmons RH, Van der Hout AH, Bauch T, et al. BRCA1 and BRCA2 heterozygosity and repair of X-ray-induced DNA damage. Int J Radiat Biol. 2002;78:285–295. doi: 10.1080/09553000110097974. [DOI] [PubMed] [Google Scholar]
- 45.Andreassen CN, Alsner J. Genetic variants and normal tissue toxicity after radiotherapy: a systematic review. Radiother Oncol. 2009;92:299–309. doi: 10.1016/j.radonc.2009.06.015. [DOI] [PubMed] [Google Scholar]
- 46.Karlsson KH, Stenerlow B. Focus formation of DNA repair proteins in normal and repair-deficient cells irradiated with high-LET ions. Radiation Research. 2004;161:517–527. doi: 10.1667/rr3171. [DOI] [PubMed] [Google Scholar]
- 47.Kinashi Y, Takahashi S, Kashino G, Okayasu R, Masunaga S, Suzuki M, et al. DNA double-strand break induction in Ku80-deficient CHO cells following Boron Neutron Capture Reaction. Radiation Oncology. 2011;6 doi: 10.1186/1748-717X-6-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pessetto ZY, Yan Y, Bessho T, Natarajan A. Inhibition of BRCT(BRCA1)-phosphoprotein interaction enhances the cytotoxic effect of olaparib in breast cancer cells: a proof of concept study for synthetic lethal therapeutic option. Breast Cancer Res Treat. doi: 10.1007/s10549-012-2079-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang F, Motlekar NA, Burgwin CM, Napper AD, Diamond SL, Mazin AV. Identification of Specific Inhibitors of Human RAD51 Recombinase Using High-Throughput Screening. Acs Chemical Biology. 2011;6:628–635. doi: 10.1021/cb100428c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.O'Shaughnessy J, Osborne C, Pippen JE, Yoffe M, Patt D, Rocha C, et al. Iniparib plus chemotherapy in metastatic triple-negative breast cancer. N Engl J Med. 364:205–214. doi: 10.1056/NEJMoa1011418. [DOI] [PubMed] [Google Scholar]
- 51.O'Shaughnessy J, Schwartzberg LS, Danso MA, Rugo HS, Miller K, Yardley R, et al. A randomized phase III study of iniparib (BSI-201) in combination with gemcitabine/carboplatin (G/C) in metastatic triple-negative breast cancer (TNBC) J Clin Oncol. 2011;29:S1007. [Google Scholar]
- 52.Billecke CA, Ljungman ME, McKay BC, Rehemtulla A, Taneja N, Ethier SP. Lack of functional pRb results in attenuated recovery of mRNA synthesis and increased apoptosis following UV radiation in human breast cancer cells. Oncogene. 2002;21:4481–4489. doi: 10.1038/sj.onc.1205546. [DOI] [PubMed] [Google Scholar]