Abstract
Taste receptor cells encounter chemicals in foods and transmit this information to the gustatory neurons, which convey it further to the gustatory relay nuclei in the lower brainstem. Characterizing neurons involved in the transmission of gustatory information in the peripheral and central nervous systems helps us better understand how we perceive and discriminate tastes. However, it is difficult to anatomically identify them. Using cell-type-specific promoters/enhancers and a transneuronal tracer, we generated transgenic mice to visualize neurons in the gustatory neural pathways. We observed the tracer in the neurons of cranial sensory ganglia and the nucleus of the solitary tract in the medulla where gustatory neurons project. The tracer was also distributed in the reticular formation and several motor nuclei in the medulla that have not been recognized as gustatory ascending pathways. These transgenic mice revealed gustatory relay neurons in the known gustatory ascending pathway and an unexpected, thus presumably novel, neural circuit of gustatory system.
Keywords: taste cell, gustatory neural pathway, genetic tracing
Sensing external chemicals is important for animals to survive in nature. Thus, chemosensory systems, such as gustatory and olfactory systems, are common in animals. Taste plays a central role in detecting chemical information in foods and leads to one of two feeding behaviors: acceptance or avoidance. To make this decision, animals have neural mechanisms to discriminate tastes.
Chemical information received by taste receptor cells in the oropharyngeal epithelia is perceived in the primary gustatory cortex.1–3) However, the mechanism to discriminate taste qualities (e.g., sweet, bitter) is still controversial. One set of experiments showed that gustatory cortical neurons respond to multiple taste qualities in a rapid timescale,4) and another showed that gustatory cortical neurons are tuned to single taste qualities and are distributed in separate areas topographically in a taste-quality-dependent manner.5) Controversial results were also reported on the responses to taste stimuli in other gustatory relay nuclei in the lower brainstem.6,7)
Recent molecular genetic studies have revealed taste receptors and taste receptor cells responsible for specific tastes. A heterooligomer of T1R2 and T1R3 (also known as Tas1r2 and Tas1r3, respectively) functions as a sweet receptor,8–10) a heterooligomer of T1R1 (also known as Tas1r1) and T1R3 functions as an umami (amino acid) receptor,10) and T2Rs (also known as Tas2rs) function as bitter receptors.11–13) Also, the cells expressing PKD2L1 and Car4 function as sour- and carbonation-sensing cells.14,15) One conclusion from these studies is that taste receptor cells are tuned to evoke single taste modalities/qualities, such as sweet, umami, bitter, and sour taste.16) This indicates that taste substances are discriminated at the receptor cell level; thus, why many gustatory relay neurons respond to multiple taste stimuli from periphery to cortex is unknown.
To help interpret the data of physiological experiments, we aimed to anatomically identify gustatory relay neurons in the peripheral and central nervous systems. As a first step, we generated transgenic mice to visualize gustatory neural pathways using cell type-specific promoters/enhancers and the transneuronal tracer wheat germ agglutinin (WGA).17–19) This review summarizes the knowledge obtained from genetic tracing studies of gustatory neural pathways originating from taste receptor cells.
I. Transneuronal Genetic Tracer
Transneuronal tracers, such as the plant lectin WGA, are powerful tools to study the neural wiring in the peripheral and central nervous systems. To analyze specific neural circuits of interest, it is imperative to know the origin of the transneuronal tracer. However, physically administering the tracer to the cells of interest is difficult.
An advantage of proteinaceous transneuronal tracers like WGA and the WGA ortholog barley lectin is that they can be encoded in the genes which enables them to be expressed strictly in cells of interest, using a promoter/enhancer of the cell-specific gene.20,21) To efficiently induce mature WGA and barley lectin in the secretary pathway in mammalian cells, genetic mutants with truncated C-terminal propeptides have been used.20) WGA can be immunohistochemically detected using anti-WGA antibodies that are commercially available,20,21) which makes WGA more efficient as a genetic tracer than is barley lectin for immunohistochemical detection.
The transneuronal transport of WGA is passive. Thus, how far along the circuit WGA is traceable depends on the strength of the promoter and the convergence or divergence of the neural circuits. Also, although transcellular transport of WGA depends on intercellular “contact,” it is not unidirectional—WGA can be transported both anterogradely and retrogradely. It is also not limited to synaptic transport—WGA can also be a transcellular or transneuronal tracer, depending on the context. These characteristics are advantageous in labeling ascending neural pathways for taste information from taste cells that lack synaptic structures, such as sweet, umami, and bitter taste cells, but have adjoining nerve processes.22)
II. Ascending Gustatory Pathway in Nervous Systems
Taste cells activated by taste substances transmit chemical information to peripheral sensory neurons called gustatory neurons, whose cell bodies are located in the VIIth, IXth, and Xth cranial sensory ganglia (also called geniculate, petrosal, and nodose ganglia, respectively).1) The gustatory neurons innervate gustatory relay neurons in the nucleus of the solitary tract (NST) in the medulla oblongata. Gustatory information is then transmitted to neurons in the parabrachial nucleus (except in humans, where this step is bypassed), then to the thalamic gustatory area, and gustatory cortex (Fig. 1).
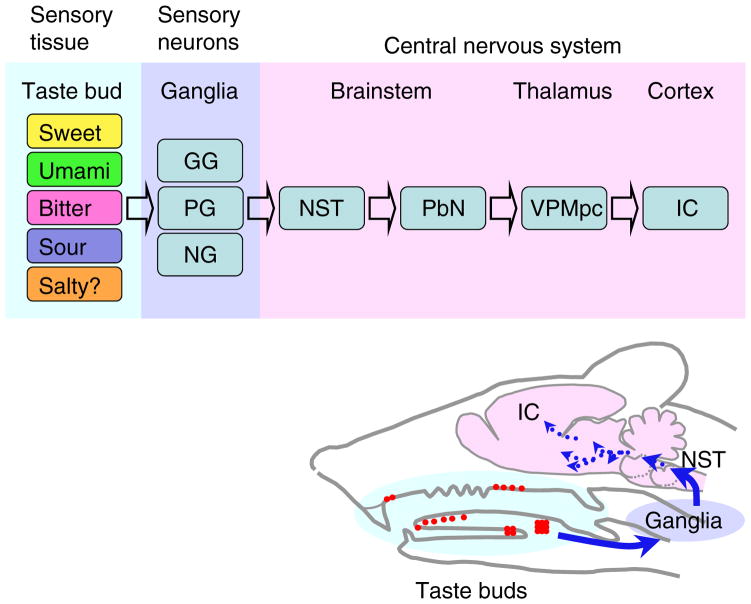
Fig. 1. Ascending Gustatory Neural Pathway.
Chemicals (taste substances) in foods detected by sensory cells in taste buds distributed in the oropharyngeal epithelia are recognized as tastes in the insular cortex (IC), which is the primary gustatory cortex. Between the peripheral sensory tissue and the IC, many neurons mediate gustatory information, beginning with the geniculate (GG), petrosal (PG), and nodose (NG) ganglia. Gustatory information from the nucleus of the solitary tract (NST) in the lower brainstem reaches the parabrachial nucleus (PbN; except in primates, including humans, which bypass this step) and then the parvocellular division of ventral posteromedial nucleus of the thalamus (VPMpc), and from there arrive in the IC.
Taste buds distributed in the anterior oral cavity are innervated by chorda tympani and greater superficial petrosal nerves from geniculate ganglia, and those in the posterior oral cavity are innervated by glossopharyngeal nerves from petrosal ganglia.3) A small number of taste buds in the epiglottis are innervated by superior laryngeal nerves from nodose ganglia.3) The topographic map of taste buds along the anterior-posterior axis is observed in the NST as a rostral-caudal axis.23) However, details of topographic maps across taste qualities in the ganglia and the nuclei of ascending gustatory pathway in the central nervous system remain elusive.
III. Transgenic Mice Expressing WGA under the Control of Taste-Related Gene Promoters/Enhancers
1) Using t1r3-WGA and t2r5-WGA transgenic mice to visualize sweet/umami and bitter neural pathways, respectively
Transgenic mice lacking taste cells with synapses can nonetheless perceive tastes received by these taste cells.14) Therefore, the taste cells lacking synapses, such as sweet, umami, and bitter taste cells, should still be able to transmit gustatory information, probably directly to the gustatory neurons. Consistent with this hypothesis, nerve processes adjoin each of these cell types.22)
T1R3 is a component of both sweet and umami receptors, and T2R5 (also known as Tas2r105) functions as a bitter receptor. Using promoters/enhancers of t1r3 and t2r5 genes, we generated transgenic mice that express WGA specifically in sweet/umami taste cells (t1r3-WGA mice) and in bitter taste cells (t2r5-WGA mice).17,18) In the t1r3-WGA mice, WGA was detected along the known ascending gustatory pathway in the neurons of geniculate and nodose/petrosal ganglia and NST, but not in the parabrachial nucleus, where NST neurons project17) (Fig. 2). In t2r5-WGA mice, WGA was detected in the peripheral sensory ganglia but not in the central nuclei, not even in the NST, probably due to weakness of the T2R5 gene promoter.18) These data strongly suggest that sensory neurons innervate sweet, umami, and bitter taste cells from an anatomical perspective.
Fig. 2. Gustatory Neural Circuits Revealed by Genetic Tracing: Distribution of WGA in t1r3-WGA and pkd1l3-WGA Transgenic Mice.
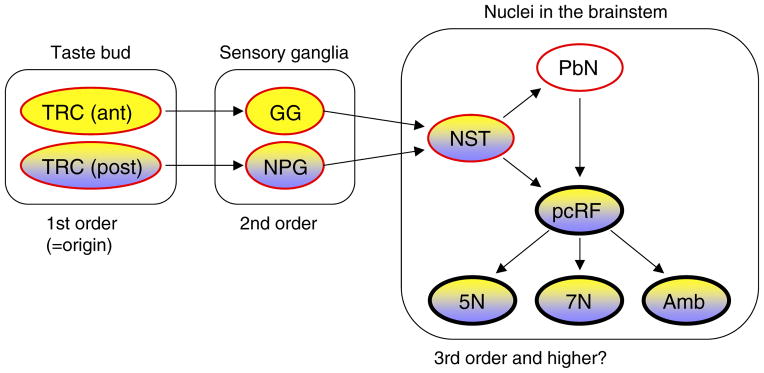
Yellow and purple indicate the tissues/nuclei where WGA immunoreactivity was detected in the t1r3-WGA and pkd1l3-WGA mice, respectively. The red outlines indicate tissues/nuclei of known ascending gustatory pathways, and arrows indicate known neural connectivity. The promoters/enhancers of t1r3 and pkd1l3 genes induced the expression of WGA mRNA faithfully in the cells where t1r3 and pkd1l3 mRNAs are endogenously expressed, and thus the WGA was observed only in taste buds of the posterior oral cavity in the pkd1l3-WGA mice. FuP, fungiform papilla; FoP, foliate papilla; CvP, circumvallate papilla; GG, geniculate ganglion; NPG, nodose/petrosal ganglia; NST, nucleus of the solitary tract; PbN, parabrachial nucleus; pcRF, parvocellular the reticular formation; 5N, trigeminal motor nucleus; 7N, facial motor nucleus; Amb, ambiguous nucleus.
2) Using pkd1l3-WGA transgenic mice to visualize the sour neural pathway from the posterior tongue
Pkd1l3 is expressed in a subset of sour cells, and its distribution is confined to the taste buds of the circumvallate and foliate papillary structures in the posterior oral cavity.14,24) Using promoter/enhancer of the pkd1l3 gene, we generated transgenic mice that express WGA specifically in these sour taste cells (pkd1l3-WGA mice).19) Of the cranial sensory ganglia in the pkd1l3-WGA transgenic mice, WGA was predominantly detected in the nodose/petrosal ganglia. However, WGA was also clearly but much less frequently detected in the geniculate ganglia. This is consistent with previous studies showing that some taste buds in the foliate papillae, presumably the anterior portion, are innervated by the chorda tympani nerve.25) WGA was detected in the NST but not in the parabrachial nucleus, as was the case with t1r3-WGA transgenic mice.
IV. Distribution of WGA in the Brainstem
WGA immunoreactivity was observed in the transgenic mice in the brainstem, in addition to the NST. Intriguingly, the distribution of WGA was not restricted to the nuclei known to be involved in the transmission of gustatory information. The presence of WGA in the spinal trigeminal nucleus and the principal nucleus of the trigeminal nerve in the t1r3-WGA mice led us to identify intrinsic expression of Tas1r3 taste protein in solitary chemosensory cells distributed in respiratory epithelia and their ascending pathway via trigeminal neurons.17) In addition to the sensory nuclei mediating gustatory and trigeminal information to the higher central neurons, the neurons in the reticular formation and several motor nuclei exhibited WGA distribution. It is important to note that these unexpected WGA distributions in the reticular formation and motor nuclei were observed in both the t1r3-WGA and pkd1l3-WGA mice, although the frequency of WGA-positive neurons in the brainstem nuclei differed.17,19) Although the neurons in these regions are associated with routes of reflexes such as vomiting and swallowing, why WGA labeled the descending (reflex) pathway from gustatory relay neurons in the NST but not the parabrachial nucleus neurons, the ascending target of gustatory relay neurons in the NST, is unknown. The labeling of neurons in the reticular formation and motor nuclei in the t1r3-WGA and pkd1l3-WGA mice might indicate as yet unidentified gustatory neural circuits or connectivity.
V. Perspective
Genetic tracing of ascending gustatory pathways originating from taste receptor cells enabled us to identify specific neurons mediating ascending gustatory information in peripheral ganglia and in the brainstem. These studies also revealed potentially newly identified pathways: gustatory relay nuclei higher than NST, neural pathways from whole bitter, sour, and salty taste cells, and, most important, the relationship of multiple taste modalities among gustatory relay neurons in the ganglia and central nuclei. Identifying marker genes would help elucidate these new findings. For example, a gene specifically and strongly expressed in whole bitter cells could enable us to trace whole relay neurons of the taste of interest, at least in the peripheral ganglia, and a gene expressed in gustatory neurons in the ganglia or gustatory relay neurons in the NST would provide clues to identify higher-order gustatory relay neurons in the central nervous system. Engineering genetic tracers will enable us to visualize and compare multiple gustatory neural pathways in the same mouse. To answer the question of why WGA was detected in the reticular formation and motor nuclei, further analyses, such as viral tracing, need to be carried out. Anatomical identification of gustatory relay neurons through these findings will provide new insights that, combined with physiological characterization, will contribute to understanding of how animals can perceive and distinguish multiple tastes.
Acknowledgments
This research was predominantly carried out at the Department of Applied Biological Chemistry of Graduate School of Agricultural and Life Sciences at the University of Tokyo. I thank all my collaborators, especially Professor Emeritus Keiko Abe (University of Tokyo), Senior Team Leader Dr. Yoshihiro Yoshihara (RIKEN Brain Science Institute), Dr. Makoto Ohmoto (Monell Chemical Senses Center), and Dr. Yoshiro Ishimaru (University of Tokyo), for their pertinent advice and expert experimental support. I would also like to thank members of Prof. Abe’s, Dr. Yoshihara’s, and my laboratories for encouraging this research and miscellaneous support. This research was supported in part by Grant-in-Aid for Young Scientists (A) from the Japan Society for the Promotion of Science and by research grant DC011143 from the National Institute on Deafness and Other Communication Disorders, National Institutes of Health.
Abbreviations
- WGA
wheat germ agglutinin
- NST
nucleus of the solitary tract
Footnotes
This review was written in response to the author’s receipt of the Japan Society for Bioscience, Biotechnology, and Agrochemistry Award for the Encouragement of Young Scientists in 2009.
References
- 1.Buck LB. In: Smell and taste: the chemical senses. 4. Kandel ER, Schwartz JH, Jessell TM, editors. New York: McGraw-Hill; 2000. pp. 625–647. [Google Scholar]
- 2.Saper CB. In: Brain stem, reflexive behavior, and the cranial nerves. 4. Kandel ER, Schwartz JH, Jessell TM, editors. New York: McGraw-Hill; 2000. pp. 873–888. [Google Scholar]
- 3.Lundy RF, Jr, Norgren R. In: Gustatory system. 3. Paxinos G, editor. San Diego: Elsevier Academis Press; 2004. pp. 891–921. [Google Scholar]
- 4.Stapleton JR, Lavine ML, Wolpert RL, Nicolelis MAL, Simon SA. J Neurosci. 2006;26:4126–4138. doi: 10.1523/JNEUROSCI.0092-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen X, Gabitto M, Peng Y, Ryba NJP, Zuker CS. Science. 2011;333:1262–1266. doi: 10.1126/science.1204076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lemon CH, Smith DV. J Neurophysiol. 2005;94:3719–3729. doi: 10.1152/jn.00700.2005. [DOI] [PubMed] [Google Scholar]
- 7.Geran LC, Travers SP. J Neurophysiol. 2006;96:2513–2527. doi: 10.1152/jn.00607.2006. [DOI] [PubMed] [Google Scholar]
- 8.Bachmanov AA, Li X, Reed DR, Ohmen JD, Li S, Chen Z, Tordoff MG, Jong PJd, Wu C, West DB, Chatterjee A, Ross DA, Beauchamp GK. Chem Senses. 2001;26:925–933. doi: 10.1093/chemse/26.7.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nelson G, Hoon MA, Chandrashekar J, Zhang Y, Ryba NJP, Zuker CS. Cell. 2001;106:381–390. doi: 10.1016/s0092-8674(01)00451-2. [DOI] [PubMed] [Google Scholar]
- 10.Zhao GQ, Zhang Y, Hoon MA, Chandrashekar J, Erlenbach I, Ryba NJP, Zuker CS. Cell. 2003;115:255–266. doi: 10.1016/s0092-8674(03)00844-4. [DOI] [PubMed] [Google Scholar]
- 11.Adler E, Hoon MA, Mueller KL, Chandrashekar J, Ryba NJP, Zuker CS. Cell. 2000;100:693–702. doi: 10.1016/s0092-8674(00)80705-9. [DOI] [PubMed] [Google Scholar]
- 12.Matsunami H, Montmayeur J-P, Buck LB. Nature. 2000;404:601–604. doi: 10.1038/35007072. [DOI] [PubMed] [Google Scholar]
- 13.Mueller KL, Hoon MA, Erlenbach I, Chandrashekar J, Zuker CS, Ryba NJP. Nature. 2005;434:225–229. doi: 10.1038/nature03352. [DOI] [PubMed] [Google Scholar]
- 14.Huang AL, Chen X, Hoon MA, Chandrashekar J, Guo W, Tränkner D, Ryba NJP, Zuker CS. Nature. 2006;442:934–938. doi: 10.1038/nature05084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chandrashekar J, Yarmolinsky D, Buchholtz Lv, Oka Y, Sly W, Ryba NJP, Zuker CS. Science. 2009;326:443–445. doi: 10.1126/science.1174601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chandrashekar J, Hoon MA, Ryba NJP, Zuker CS. Nature. 2006;444:288–294. doi: 10.1038/nature05401. [DOI] [PubMed] [Google Scholar]
- 17.Ohmoto M, Matsumoto I, Yasuoka A, Yoshihara Y, Abe K. Mol Cell Neurosci. 2008;38:505–517. doi: 10.1016/j.mcn.2008.04.011. [DOI] [PubMed] [Google Scholar]
- 18.Ohmoto M, Maeda N, Abe K, Yoshihara Y, Matsumoto I. Biochem Biophys Res Commun. 2010;400:734–738. doi: 10.1016/j.bbrc.2010.08.139. [DOI] [PubMed] [Google Scholar]
- 19.Yamamoto K, Ishimaru Y, Ohmoto M, Matsumoto I, Asakura T, Abe K. J Neurochem. 2011;119:497–506. doi: 10.1111/j.1471-4159.2011.07443.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoshihara Y, Mizuno T, Nakahira M, Kawasaki M, Watanabe Y, Kagamiyama H, Jishage K-i, Ueda O, Suzuki H, Tabuchi K, Sawamoto K, Okano H, Noda T, Mori K. Neuron. 1999;22:33–41. doi: 10.1016/s0896-6273(00)80676-5. [DOI] [PubMed] [Google Scholar]
- 21.Horowitz LF, Montmayeur J-P, Echelard Y, Buck LB. Proc Natl Acad Sci USA. 1999;96:3194–3199. doi: 10.1073/pnas.96.6.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clapp TR, Yang R, Stoick CL, Kinnamon SC, Kinnamon JC. J Comp Neurol. 2004;468:311–321. doi: 10.1002/cne.10963. [DOI] [PubMed] [Google Scholar]
- 23.Hamilton RB, Norgren R. J Comp Neurol. 1984;222:560–577. doi: 10.1002/cne.902220408. [DOI] [PubMed] [Google Scholar]
- 24.Ishimaru Y, Inada H, Kubota M, Zhuang H, Tominaga M, Matsunami H. Proc Natl Acad Sci USA. 2006;103:12569–12574. doi: 10.1073/pnas.0602702103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamamoto T, Kawamura Y. Chem Senses. 1975;1:241–244. [Google Scholar]