Abstract
Mammographic screening leads to frequent biopsies and concomitant overdiagnosis of breast cancer, particularly ductal carcinoma in situ (DCIS). Some DCIS lesions rapidly progress to invasive carcinoma whereas others remain indolent. Because we cannot yet predict which lesions will not progress, all DCIS is regarded as malignant, and many women are overtreated. Thus, there is a pressing need for a panel of molecular markers in addition to the current clinical and pathologic factors to provide prognostic information. Genomic technologies such as microarrays have made major contributions to defining sub-types of breast cancer. Next-generation sequencing (NGS) modalities offer unprecedented depth of expression analysis through revealing transcriptional boundaries, mutations, rare transcripts and alternative splice variants. NGS approaches are just beginning to be applied to DCIS. Here, we review the applications and challenges of NGS in discovering novel potential therapeutic targets and candidate biomarkers in the premalignant progression of breast cancer.
Keywords: Breast cancer, carcinoma in situ, deep sequencing, microarrays, network analysis, molecular markers
Breast ductal carcinoma in situ (DCIS)
The mortality from breast cancer has been declining for the past two decades [1]. This decline is thought to be due both to the introduction of widespread mammographic screening programs in the 1980s, resulting in earlier diagnosis and intervention [2–4], and the development and optimization of chemotherapy [5] and targeted therapies [6], although the relative contributions of each are hotly debated. Indeed, evidence now indicates that the contribution of screening to the decrease in death from breast cancer may be less than previously thought [7, 8]. For example, a recent review in The Lancet [9] concludes that screening in the U.K. only modestly reduces mortality (decrease of 43 deaths in 10,000 women screened for 20 years), and recommends that this should be balanced against the significant risk of overdiagnosis, in order to allow women to make informed decisions regarding screening. In the U.S., analysis of the Surveillance, Epidemiology, and End Results (SEER) database shows that as many as one-third of new breast cancer diagnoses (particularly that of DCIS) could represent overdiagnosis with “at best, only a small effect on the rate of death from breast cancer” [10]. This result is in line with a similar study using data from European countries, Canada, and Australia [11].
Overdiagnosis (Box 1) is a direct result of the use of mammography [7, 8] and is manifested by a dramatic apparent increase in DCIS. The National Institutes of Health Office of Medical Applications of Research commissioned a review on the incidence, treatment, and outcomes of DCIS to be used for the State of the Science Conference on the diagnosis and management of DCIS [12]. Their conclusions included that the incidence of DCIS has risen from 1.87 per 100,000 in 1973 to 32.5 per 100,000 in 2004, with the increase mostly accounted for by the introduction of screening mammography [12]. More recently it was reported that the overall incidence of early stage disease (DCIS and localized cancer combined) has doubled since 1976 [10]. At the same time, there is an insufficient decrease in the incidence of late stage disease to offset the increased detection of these early stage cancers [10]. This result strongly implies that mammographic screening has led to significant overdiagnosis of subclinical and indolent disease. Another interpretation from long-term follow-up is that mammographic screening is detecting a significant number of lesions that would not only have remained indolent, but would likely have spontaneously regressed [13]. According to analysis of a Swedish cohort, as many as one in five mammographically-detected breast cancers could spontaneously regress [13].
Box 1 Overdiagnosis vs. False Positives.
The term “overdiagnosis” refers to the detection of subclinical disease that, if left untreated, would never cause symptoms or death during the patient’s lifetime.
The term “false positive” describes the detection by a diagnostic or screening test of disease that is not really there.
Over the last decade, it has become widely accepted that there is indeed considerable overdiagnosis of both DCIS and invasive breast cancer. As many as one in three breast cancers (which includes DCIS) diagnosed in asymptomatic women by screening mammography may fit the description of being overdiagnosed.
Overdiagnosis leads to an apparent increase in incidence through the detection of subclinical indolent disease that would never have harmed the patient in her lifetime and was therefore rarely detected before the introduction of the screening test.
Overdiagnosis leads to overtreatment that does not benefit the patient and can only potentially harm her because overdiagnosed disease would, if left alone, never threaten the patient’s health within her lifetime and therefore does not require treatment.
The problems of overdiagnosis and consequent overtreatment have led some to question current mammography screening guidelines. In 2009, the United States Preventative Services Task Force recommended that the age at which mammographic screening begins be increased to 50 and the frequency be decreased to every two years in order to decrease overdiagnosis [156].
Any diagnosis of DCIS presents a challenge since we cannot yet distinguish cases that would remain indolent and not require aggressive treatment from cases that are likely to progress to life-threatening invasive ductal carcinoma (IDC) [14, 15]. A diagnosis of DCIS imposes a major burden, even in the absence of progression to IDC, as double mastectomy is an increasingly prevalent intervention [16]. A recent study presented at the 2012 American Society of Clinical Oncology Quality Care Symposium suggests that this major surgical procedure is almost certainly overused, particularly in the context of DCIS [17]. Molecular screening approaches are being considered, but so far only to inform decisions about post-surgery adjuvant treatments [18, 19]. At the 2011 San Antonio Breast Cancer meeting, Solin et al. reported that a subset of markers in the 21-gene Oncotype DX® test can predict likelihood of progression [20]. If the use of these markers spares those at low risk of progression to IDC from adjuvant radiation, then it will make a significant impact. Nonetheless, we still do not have molecular screens that can predict whether DCIS will progress, nor do we understand the pathways that control progression to IDC and thus might serve as therapeutic targets.
Models of DCIS progression to invasive breast cancer
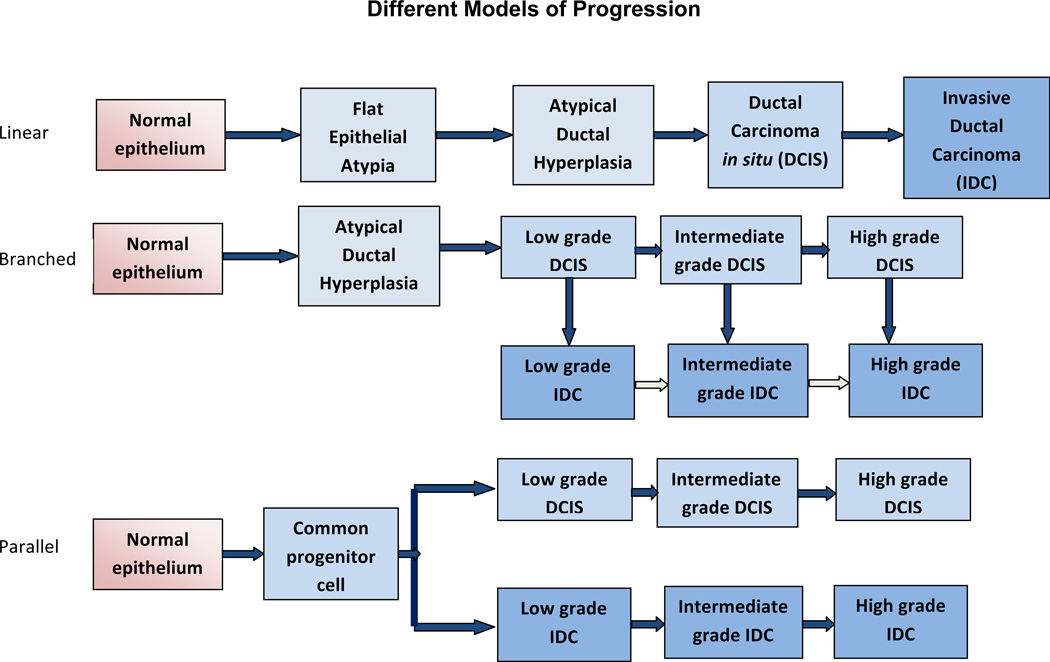
Several models have been proposed based on epidemiological, morphological and immunohistochemical studies to describe the progression from non-invasive DCIS to invasive breast carcinoma [Figure 1]. The traditional linear model of progression hypothesizes that the progression occurs sequentially in stages from normal epithelium to invasive carcinoma via hyperplasia and in situ carcinoma. Genomic and transcriptomic studies provide further support for this multistep progression model. In the classic “ductal” model proposed by Wellings and co-workers, flat epithelial atypia (FEA), atypical ductal hyperplasia (ADH) and DCIS are non-obligate precursors of IDC.
Figure 1. Models of Malignant Progression of Normal Breast Epithelium to Carcinoma.
Evidence from epidemiological, morphological and immunohistochemical studies has been used to develop several models to describe the progression from non-invasive DCIS to IDC. (Top) A linear model of progression that in sequential stages from normal epithelium to invasive carcinoma via hyperplasia and in situ carcinoma. The postulates of this hypothesis include that DCIS is a direct precursor of IDC and that atypical ductal hyperplasia is a direct precursor to low grade DCIS. In “non-linear” or “branched” models (Middle) DCIS is a progenitor of IDC, yet different grades of DCIS progress to corresponding grades of IDC. The low, intermediate and high grades can also be termed grades I, II, and III, respectively. There may also be further progression of IDC (pale arrows). The “parallel” model of progression (Bottom) hypothesizes that DCIS and IDC diverge from a common progenitor cell and progress independently through different grades in parallel.
The postulates of this theory are that DCIS is a direct precursor of IDC and that ADH is a direct precursor to low grade DCIS [21]. In “non-linear” or “branched” models DCIS is a progenitor of IDC, yet different grades of DCIS progress to corresponding grades of IDC. In contrast, the “parallel” model of progression of DCIS and IDC hypothesizes that DCIS and IDC diverge from a common progenitor cell and progress independently through different grades in parallel [22]. Strong evidence that DCIS and IDC develop from a common clonal origin and then can continue to evolve in parallel comes from a study of copy number variations in co-existing DCIS and IDC from patient samples [23] and from the long appreciated clinical observation that IDC is frequently accompanied by DCIS.
Experimental models of DCIS
Comprehensive molecular analyses of DCIS have been limited in part by the small size of DCIS lesions and thus tissue available for study as well as the few experimental model systems that recapitulate human disease [24]. Xenograft models in which human DCIS cells are directly injected into mammary fat pads or the flank of athymic nude mice are available [25]. These xenografts are amenable to hormonal manipulation and have been widely used to study the effects of estrogen and anti-estrogen therapies on epithelial cell proliferation [26, 27]. These models can be used for screening of chemopreventive or chemotherapeutic agents and for analysis of gene expression and signaling pathways. Despite their advantages, these xenograft models when used to study progression to invasive cancer do not fully recapitulate the complexity associated with the DCIS lesions. Even the humanized fat-pad transplantation models do not represent the true intraductal microenvironment. Nevertheless, sub-cutaneous implantation of MCF10.DCIS cells produces lesions that recapitulate human high-grade comedo DCIS by both histological and molecular markers [28]. Efforts are ongoing to develop better models of disease progression that represent the diversity of DCIS lesions within the context of innate surroundings. In this regard, a human in mouse intraductal (MIND) transplantation model has been developed using human DCIS cell lines such as MCF10.DCIS and SUM225 as well as primary human DCIS cells (FSK-H7) [29]. The MIND xenografts maintain histology and biomarker expression comparable to the original patient samples [30]. These models reflect some of the diversity of DCIS lesions because the MCF10.DCIS line represents basal-like DCIS whereas SUM225 and FSK-H7 derive from Her-2 overexpressing DCIS. The MIND system utilizes transplantation into the mammary ducts of immunocompromised NSG (NOD-SCID IL-2Rgamma null) mice for primary human cells. A major limitation of all xenograft models for DCIS to date is that the role of the interaction with immune cells in invasive progression cannot be assessed. The future application of humanized NSG mice to studies of DCIS may potentially address this drawback [31].
Transgenic mice are an alternative approach to study DCIS development and progression. Several models of transgenic mice are available that are pre-disposed to develop mammary carcinoma with various latencies due to induced expression of oncogenes, including Her-2, polyoma middle T antigen driven by mouse mammary tumor virus or SV40 large T antigen driven by the whey-acid protein promoter. As part of the progression of disease, these mice develop DCIS lesions with distinct morphological and cytological features that somewhat resemble those observed in DCIS patients. These transgenic models have improved our understanding of the genetic events and molecular mechanisms underlying DCIS progression [32–35] and offer opportunities to evaluate hypotheses for the role of DCIS in the progression of breast cancer [Figure 1].
Researchers have also focused efforts towards the development of cellular model systems that would recapitulate the architecture and complexity of DCIS in the human breast. Earlier studies for pre-clinical therapeutic identification and development were commonly based on conventional cell culture systems on plastic dishes. These monolayer cultures do not provide the environment as it exists in vivo, lacking natural cues for hallmarks of differentiated phenotypes such as polarity, and thus exhibiting altered cell signaling and gene expression [36]. Three dimensional (3D) matrices, such as reconstituted basement membrane (rBM), are more physiologically-relevant substrates and cancer cells grown in rBM exhibit responses and resistance to drugs that are closer to those observed in vivo [37–42]. Another advantage of 3D rBM cultures is that they provide a source for high quality RNA [43] without concomitant stromal cell RNA contamination. This is hard to achieve in the isolation of RNA from microscopic clinical DCIS specimens or from formalin-fixed paraffin-embedded (FFPE) tissues, although the use of laser capture and new sequencing technologies for fragmented samples are becoming available.
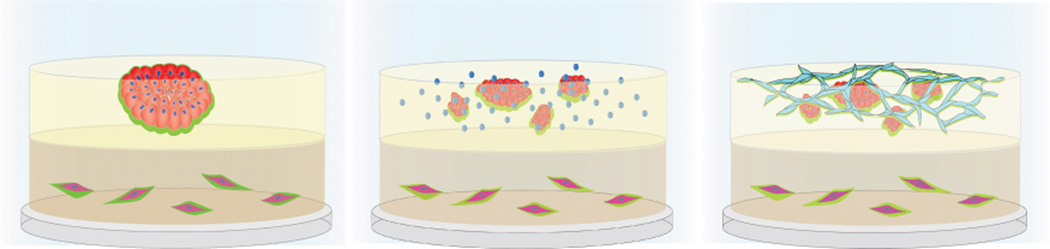
Heterotypic 3D co-culture models of DCIS, like MAME (mammary architecture and microenvironment engineering) [Figure 2], enable live-cell, real-time imaging of cell-cell and cell:matrix interactions. These models allow for analysis of interactions between the breast epithelial cells and various stromal cells like macrophages, fibroblasts and lymphatic and blood vessel microvascular endothelial cells. The individual cell populations can be manipulated and the contribution of each component to the progression to IDC can be characterized [44]. These models have: revealed roles for cathepsin B in pericellular proteolysis and invasiveness of premalignant epithelial and carcinoma cells [45]; identified a proteolytic pathway, involving caveolar localization of cathepsin B and urokinase plasminogen activator receptor (uPAR), in pericellular proteolysis and invasiveness of triple-negative inflammatory breast cancer cells [46]; demonstrated roles for cancer-associated fibroblasts (CAFs) and CAF secretion of HGF in increased proteolysis and invasiveness of DCIS cells [47]; and showed a role for acidic pH in pericellular proteolysis and invasiveness [48], which is linked to secretion of cathepsin B and induction of chronic autophagy [49]. Using similar heterotypic 3D co-culture models, Dang et al. [50] have shown differential interactions of basal and luminal subtypes with CAFs. They report that both basal and luminal type breast cancer cells when cultured alone formed noninvasive DCIS spheroids. In contrast, when co-cultured with CAFs, the basal-type showed invasive outgrowths and the luminal-type spheroids formed noninvasive duct-like structures [50].
Figure 2. Mammary Architecture and Microenvironment (MAME) Models for functional, live-cell imaging of DCIS-stromal interactions.
Left: MCF10.DCIS human DCIS cells in an upper layer of reconstituted basement membrane (rBM) + 2% rBM overlay form dysplastic structures. CAFs are in a lower layer of collagen I. Middle: Human macrophages are added to the rBM layer. Right: Human microvascular endothelial cells are added to the rBM layer.
Other recent developments include computer modeling of the biomechanical forces influencing growth and necrosis in comedo-type DCIS with the goal of improving the prognostic value of mammographic results [51], and a microfluidic 3D compartmentalized system that enables control of both spatial and temporal aspects within the microenvironment of cell cultures. This compartmentalized model system allows sampling of secreted molecules and facilitates inhibitor screening and studies of cell signaling pathways involved in the progression of DCIS [52]. Further optimization and validation of organotypic culture models will need to consider challenges such as adaptability to high-throughput screening.
Genomic analyses and expression profiling of DCIS
Identifying the genetic alterations and molecular mechanisms underlying the progression from premalignant DCIS lesions to invasive carcinoma may help to establish clinical biomarkers and also lead to identification of potential targets for therapeutic intervention. Although there have been fewer studies of DCIS [53–62] than IDC, it seems likely that the same chromosomal regions are amplified with comparable frequencies in DCIS as in IDC. Comparative genomic hybridization (CGH) analyses of DCIS and IDC lesions show that low and intermediate grade DCIS are characterized by chromosomal loss of 16q whereas 1q gain and 11q loss occurs at higher frequency in intermediate grade DCIS [63]. High grade DCIS, however, is more complex in terms of these alterations as shown by loss of 8p, 11q, 13q and 14q; gains in 1q, 5p, 8q and 17q; and amplifications of 17q12 (ErbB2/Her2) [64] and 11q13 (cyclin D1). Molecular cytogenetic analysis by CGH of several pre-invasive and invasive breast cancer cell lines revealed that the most common gains are found at 8q, 1q, 7q, 3q and 7p and the most common losses at Xp, 8p, 18q and Xq [65].
Advanced technology has facilitated interrogation of molecular events occurring at preinvasive stages of breast cancer. Several gene expression profiling studies of DCIS have been carried out using a combination of laser capture micro-dissection and microarrays [56, 61, 62, 66–68]. Serial analysis of gene expression found that the most dramatic transcriptome changes occur at the transition from normal epithelium to DCIS rather than from DCIS to invasive cancer [58]. This conclusion is supported by phenotypic and genomic analyses demonstrating that the molecular heterogeneity of breast ductal carcinomas is already established in in situ lesions [61], and by studies from co-existing DCIS and IDC [68]. Higher tumor grade and the presence of necrosis have been associated with greater gene expression variability and distinct transcriptional signatures [54, 66]. Hannemann et al. identified a gene expression classifier of 35 genes that differed between DCIS and IDC and a panel of 43 genes which further distinguished well and poorly differentiated DCIS [69].
Molecular markers of DCIS
Personalized approaches that combine molecular profiling and histological observations are being implemented to improve risk stratification and therapy for patients with DCIS. Using a combination of molecular and morphological features along with genomic and immunohistochemical data, most pre-invasive and invasive lesions can be stratified into a low-grade molecular pathway or a high-grade molecular pathway. The low-grade molecular subtype is characterized by loss of 16q, gain of 1q and expression of estrogen (ER) and progesterone (PR) receptors. The lesions of this subtype are referred to as ‘luminal’. Furthermore, depending upon the absence or presence of Her-2, they are classified as luminal A or luminal B, respectively. The high-grade variety has gain of 11q13, loss of 13q, amplification of 17q12 and infrequent expression of ER and PR. Lesions with these molecular and gene expression signatures are referred to as ‘basal’ or ‘Her-2’ depending upon the absence or presence of Her-2 expression, respectively [70–72]. Meta-analysis of microarray gene expression profiles of several triple negative breast cancer cases (TNBC) has revealed further intrinsic differences in gene signatures and ontologies. Based on gene expression profiles, Chen et al. [73] have developed a subtyping tool that can classify TNBC into six molecular subtypes: namely two basal-like (BL1 and BL2) subtypes, an immunomodulatory (IM) subtype, a mesenchymal (M) subtype, a mesenchymal stem-like (MSL) subtype and a luminal androgen receptor (LAR) subtype. The TNBC subtypes exhibit differential sensitivities to therapeutic agents and hence may explain differences in clinical behaviors of TNBC [73]. Analysis of publicly available gene expression datasets of DCIS cases using similar subtyping tools could provide an insight into the distinct genetic subtypes of DCIS that would require specific targeted agents to block progression to invasive cancer.
Some of the important molecular markers that have previously been identified in patients with DCIS are described in further detail below:
Her-2
Her-2 expression is primarily associated with high-grade comedo type DCIS, in the absence or presence of concomitant IDC, and hence is regarded as an independent prognostic factor [74]. Since Her-2 immunoreactivity has been reported to be significantly higher in DCIS cases than in IDC, Allred et al. proposed that Her-2/neu amplification plays a more important role in initiation than in progression of IDC [75, 76]. Her-2 status is regarded as an important prognostic and predictive marker with its overexpression predicting local recurrence [77].
Estrogen receptor (ER)
ER expression is inversely related to the grade of DCIS lesions [78] and treating DCIS that expresses ER with tamoxifen significantly reduces risk of subsequent breast cancer by 40%–50% [79].
Progesterone receptor (PR)
PR expression also has an inverse relationship to nuclear grade and its presence is associated with expression of ER and lack of comedo-necrosis in DCIS [80, 81].
Cyclin D1
Amplification of cyclin D1 has been observed in 10–18% cases of DCIS [82, 83]. There have, however, been conflicting reports on the correlation between expression of cyclin D1 and ER [84, 85]. There is not a correlation between cyclin D1 expression and risk of local recurrence [86].
Bcl-2
Bcl-2 has been shown to be an independent prognostic marker in early stages of breast carcinoma. It is present in the continuum of breast lesions from ADH to well-differentiated DCIS [87, 88] and its expression gradually decreases as lesions become more aggressive [89].
p53
Inactivating mutations of p53 have been observed in a large percentage (40%) of high-grade DCIS. Low-grade DCIS does not exhibit any alterations and the frequency of these mutations is very low (5%) in the intermediate grade lesions [90, 91].
Ki67
Well-differentiated DCIS lesions generally have lower expression of Ki67 whereas poorly differentiated lesions have higher levels. Quantitative assessment by automated image analysis of multiple DCIS tumor samples showed that invasion is associated with a significant increase in Ki67 expression and decreases of ER, PR, and Her-2 expression [92].
c-met and VEGF
Increased levels of c-met are associated with VEGFA and FGFR-1, all of which contribute to angiogenic processes [93]. High expression of VEGF correlates with biologic aggressiveness of DCIS lesions [94, 95]. Increased frequency of FGFR1 amplification is associated with progression of DCIS to IDC and poor prognosis [96].
Myc
Although increased myc expression is associated with poor prognosis in IDC, its role in preinvasive lesions is not clear. There have been conflicting reports regarding amplification of the c-myc gene in premalignant lesions of the breast. No c-myc amplification was observed in DCIS in two independent studies [83, 97], whereas another group reported c-myc amplification in DCIS lesions adjacent to invasive lesions [98]. Altintas et al. reported that high expression of c-myc in DCIS did not predict local recurrence [99].
Combinations
A direct positive relationship has been observed for the expression of ER, PR and Bcl-2 [100]. Ringberg et al. [101] suggest that a molecular signature with lack of ER and PR, Her2 over-expression, accumulation of p53, and high Ki67 expression is a strong predictor of local recurrence in DCIS. In a retrospective study of DCIS cases, DCIS lesions that were positive for p16, COX-2 and Ki67 expression are significantly associated with risk of subsequent invasive cancer. In contrast, DCIS lesions that either lacked ER, but were positive for ERBB2 and Ki67, or that lacked COX2, yet were positive for p16 and Ki67, are associated with recurrence of DCIS [102]. There are additional studies that associate p16 expression with progression [103, 104].
Other Markers
The results are conflicting on the status of other molecular markers such as TGFβ [105, 106], p27 and p21 and it is currently hard to interpret the roles of these molecules in DCIS progression [84]. Efforts are ongoing to decipher the molecular events associated with progression of DCIS. Lu et al. implicated 14-3-3-sigma in conjunction with Her2 in the progression to IDC [107]. Qi et al. analyzed miRNA expression patterns in pre-invasive and invasive lesions of the breast. They found a consistent increase at each successive stage in the expression of miR-21 along with its targets (PTEN, PCCD4 and TMI) [108]. Other examples of non-coding RNAs are given below.
The need for novel biomarkers in DCIS
Despite intensive research, robust biomarkers that would predict the risk of progression or recurrence of DCIS have not been discovered for screening patients. A comprehensive systematic review of studies on biological markers of DCIS over the past 10 years has been compiled by Lari and Kuerer [109]. They conclude that there is an unmet need to identify important markers in DCIS and that the study of biomarkers for DCIS is still in its infancy. Clinically useful biomarkers are difficult to define [110], and the only two well established biomarkers that have prognostic and predictive potential for breast cancer are ER status and Her-2 expression. Based on the expression of these markers, we can identify patients that will probably have worse or better outcome and select patients that will respond to therapy or treatment.
Transcriptome profiling by microarrays
Microarrays have been the technology of choice for most gene expression studies and have been extensively used for studying invasive breast cancer, with results being adapted for clinical use. This approach has also been used to advance the understanding of premalignant breast disease. Microarrays provide gene expression information in a high throughput manner and thus afford opportunities to identify the unique transcriptional fingerprint associated with each stage of disease progression.
Two microarray-derived tests for breast cancer are commercially available. Mammaprint™ is a 70-gene predictor for breast cancer outcome [111]. The Oncotype DX® assay [112] utilizes expression analysis of 21 genes for risk stratification and prognosis of breast cancer patients. These examples reflect the potential of microarrays to identify biomarkers of disease for screening patients. Two long-term, large-scale randomized trials (Trial Assigning Individualized Options for Treatment [TAILO]Rx and Microarray In Node- Negative and 1 to 3 Positive Lymph Node Disease May Avoid Chemotherapy [MINDACT]) are underway to determine the clinical utility of Oncotype DX® and Mammaprint™. Validation of novel biomarkers is essential before their application in routine clinical use and treatment related decisions. This is exemplified by the fact that two clinical trials in non-small cell lung cancer and one trial in breast cancer, which were based on genomic signatures thought to predict the outcome of chemotherapy, had to be terminated in 2011 due to insufficient prior validation [113, 114].
Next-generation sequencing (NGS)
Ultra-high-throughput massively parallel RNA sequencing (RNA-Seq) is a recently developed approach and rapidly emerging as a more powerful alternative platform to microarrays for whole genome expression profiling. Such NGS technologies offer many potential advantages as compared to microarrays [115]. First, they do not rely on prior sequence information as is required for the probes used for microarrays [116]. This allows the experimental design to be unrestricted. Second, the level of expression is assigned based on the entire transcript and not a few segments. The identification and quantification of gene expression at the whole genome level without a priori sequence knowledge is unbiased and provides higher confidence when novel targets and network pathways are discovered [117]. Third, sequencing instead of hybridization minimizes concern with regard to cross-hybridization. Microarray cross hybridization may happen if the probe sequences and the target transcript fragments are similar. Such hybridization noise may not be computationally solved by downstream data analysis. In comparison, if the optimal criteria are used during NGS alignment, misalignment or “in silico cross hybridization” can be effectively minimized. If two or more genome locations have very similar or identical sequences, the NGS short reads mapping to one location will also map to the other locations to produce in silico cross hybridization. The aligner can classify such reads as “multiple mapping reads” and they can still be further analyzed as this caveat is noted. Fourth, beyond gene expression analysis, NGS can also identify novel isoforms and exons, allele-specific expression, mutations, and fusion transcripts. Fifth, NGS data are obtained as digital signals that can be quantified, annotated and re-annotated to reflect the current genome consensus. These attributes make NGS ideal for the detection of differentially expressed transcripts. Using RNA-Seq, for example, Huber-Keener et al. defined gene expression alterations associated with anti-estrogen resistance by comparing the transcriptomes of breast cancer cells that are either sensitive or resistant to tamoxifen and identified differential expression of transcripts regulating ERα functions, cell cycle, transcription/translation and mitochondrial dysfunction [118].
NGS also enables the detection of rare transcripts, sequence mutations, transcriptional boundaries, alternative splice variants, differential polyadenylation, non-coding RNAs, and antisense transcripts [119–124]. Indeed, whole genome sequencing revealed the competing evolution of numerous sub-clones during early stages of breast cancer development [125], including identification of regions of localized hypermutation [126]. The application of NGS has similarly revealed that the breast cancer susceptibility genes BRCA1 and BRCA2 are associated with homologous patterns of somatic mutations that include both short deletions and base substitutions [126]. Microarray and PCR approaches have been used to identify splice variants in breast cancer samples [127–130]. To do this, estimates of the possible alternative splice variant sites are required before probes / primers can be designed and thus these methods can only test a definite number of candidate variant sites. In contrast, NGS can potentially detect all splice variants at the whole genome level. Further, with the advent of paired-end sequencing it is now possible to identify fusion genes that may encode for hybrid proteins with oncogenic potential like those identified in leukemias and lymphomas [131]. Paired-end sequencing of various breast cancer cell lines has identified multiple high frequency intra-chromosomal rearrangements and, to a lesser extent, inter-chromosomal rearrangements [132].
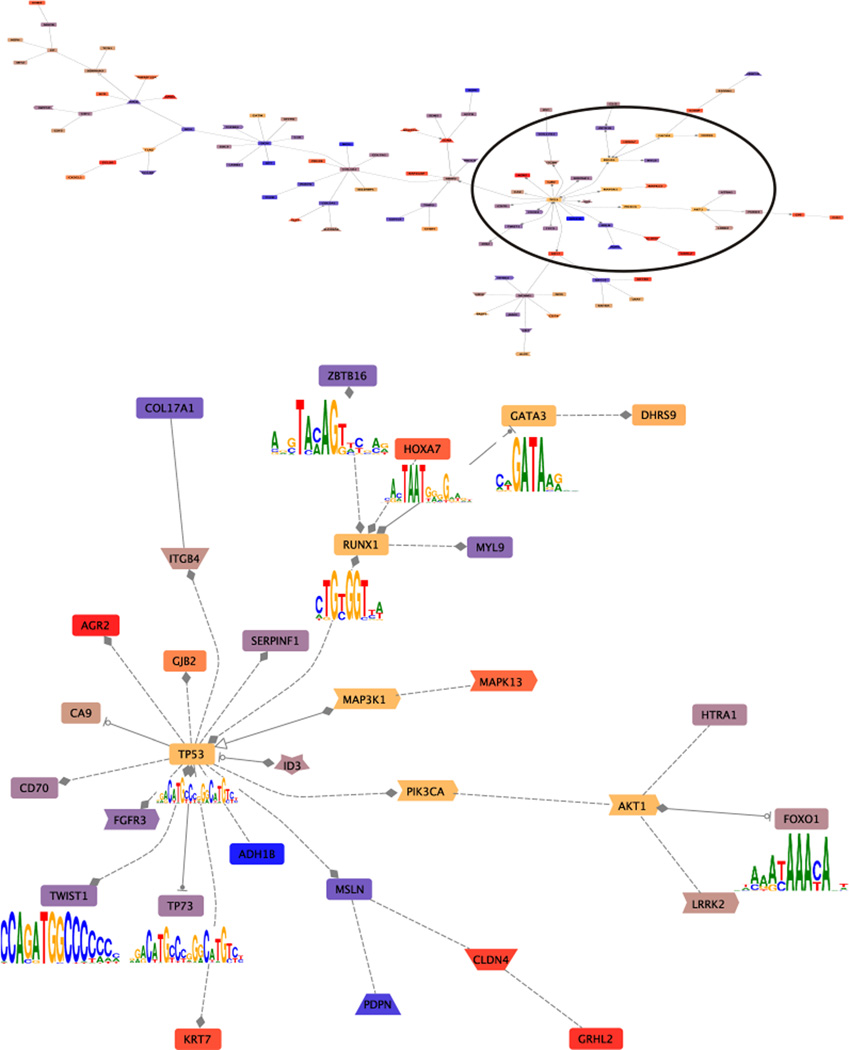
A recent issue of the journal Nature featured five studies using NGS approaches for whole genome analysis of breast cancer samples, producing many new insights on topics including copy number variations, new descriptions of driver and other mutations, and elevated mutation rates in treatment-resistant tumors [133–137]. Striking observations revealed by the use of NGS were that only approximately 36% of the gene mutations are detectably transcribed [137], and that many of the mutations would encode for truncated proteins [135]. The study by Banerji et al. is notable for its inclusion of whole-exome sequencing results from nine DCIS samples (basal, luminal A and B, and Her-2 sub-types [133], although the subset of results from the DCIS samples is not separately delineated from the entire set of 103 cancer and normal pairs. The overall data-set showed recurrent somatic mutations in five genes (PIK3CA, TP53, AKT1, GATA3 and MAP3K1), and recurrent mutations and deletions were discovered for CBFB and RUNX1. Three of these genes (TP53, GATA3, and RUNX1) encode transcription factors. These three genes were not subject to up or down-regulation in the available NGS data on DCIS models [43], but it is interesting to note that these factors can be resolved by co-citation networks [Figure 3] and are perhaps candidate drivers regulating some of the 295 differentially expressed genes observed in that analysis of DCIS. The immense amount of information provided by these studies, together with the additional data that will be further required to validate their clinical usefulness, point to a need for a sea change in our ability to organize and analyze clinical bioinformatic data [138].
Figure 3.
Co-citation interaction networks among the DCIS genes observed in NGS analysis of DCIS models [43]. The co-citation networks among 295 differentially expressed genes and PIK3CA TP53 AKT1 GATA3 MAP3K1 CBFB and RUNX1 7 were constructed using GePS with default parameters (Genomatix). The gene connections are based on previous published literature, i.e., the co-citations between two genes in the previous published literature that have been hand-curated. This network indicates the co-citations of the input genes. The up-regulated genes are shaded in warm colors (red, orange). The down-regulated genes are shaded cooler colors (blue, purple). CBFB does not appear in the network. PIK3CA TP53 AKT1 GATA3 MAP3K1, and RUNX17 are shaded yellow-brown. Of these TP53, GATA3, and RUNX1 encode transcription factors with the motif indicated. The upper panel shows the entire network formed. The circle indicates the region expanded in the lower panel. Note the number of genes which share transcription factor binding sites as indicated by the filled line terminators: diamonds indicate that gene A modulates gene B; arrowheads indicate that gene A activates gene B, and stopped circles that gene A inhibits gene B. This overview provides the opportunity to selectively evaluate the veracity of the resulting pathway.
The diversity of NGS techniques like DNase-Seq, ChiP-Seq (definition of factor binding sites) and RNA-Seq (transcript profiling) has enabled the success of ENCODE (The Encyclopedia of DNA Elements) project consortium. This now includes a compilation from 147 different cell types of 1,640 genome-wide data sets. These include transcription, transcription factor binding, DNase hypersensitive sites, chromatin structure, assembly and histone modification [139, 140]. This has provided new insights into the organization and regulation of the human genome. Sequencing primary and processed RNAs has revealed that three-quarters of the human genome can be transcribed and that genes are highly interlaced with overlapping transcripts that are transcribed from both DNA strands [141].
Cell-free circulating nucleic acids (CNA) have been recognized as potential biomarkers for the early detection and clinical monitoring of human breast cancers. Sequencing circulating nucleic acids in serum of patients with IDC showed that the quantities of specific cell-free transposable elements and endogenous retroviral DNA sequences in blood could distinguish early stage IDC from normal and nonmalignant controls. The sample size in this study was small (n=10) but the sensitivities and specificities suggest this could be a useful clinical tool if these results are borne out in larger trials [142].
Non-coding RNAs (ncRNA) are functional RNAs that play important roles in gene expression regulation at different transcriptional and translational levels. The relevance of small ncRNA to cancer, particularly to breast cancer, has mainly been studied using NGS. Micro RNA (miRNA) is the most widely studied class of ncRNAs. Genome-wide miRNA expression profiling of breast cancer patients by SOLiD sequencing observed that the expression of five miRNAs were altered at least five-fold. Two of them, miR-29a and miR-21, were identified as significantly increased in the serum of breast cancer patients [143]. Further analysis of serum genome-wide microRNAs revealed that miR-103, miR-23a, miR-29a, miR-222, miR-23b, miR-24 and miR-25 were coordinately up-regulated. Of this suite of miRNAs, miR-222 was significantly increased in the serum of breast cancer patients and has been proposed as a potential biomarker for breast cancer [144]. Also using NGS, a nine-microRNA signature has been identified that differentiates IDC from DCIS. Five miRNAs are associated with time to metastasis and overall survival of IDC patients [145].
It should be emphasized that different NGS techniques and applications require varied approaches and tools. The average length of a small ncRNA (18 – 30 nucleotides) is much shorter than an mRNA transcript. Extraction of small ncRNA and sequencing library preparation thus requires specific approaches. Sequence alignment tools that can effectively handle short length reads and trim the adapters are also necessary. For some ncRNAs, such as miRNA, potential target genes need to be identified during data analysis. Both RNA-Seq and ChIP-Seq demand deep sequencing and short read alignment. Optimized peak finding algorithms [146] such as MACS [147] or PeakSeq [148] are essential for analysis of ChIP-Seq data to identify the binding sites of the transcription factor of interest [149]. Comparing RNA-Seq with ChIP-Seq techniques, ChIP-Seq can help to determine how transcription factors and other chromatin-associated proteins interact with specific segment of the genome to regulate gene expression. The results can help to understand biological processes and diseases states, with the construction of coordinated, regulatory networks.
Expert Commentary
A combinatorial systems approach that includes empirical and computational techniques is needed to gain a better understanding of the complex biology of DCIS. Rather than studying the contribution of individual genes or a single pathway to DCIS evolution or progression, the focus needs to shift towards identifying the cross-talk among multi-component dysregulated pathways. The goal would be for NGS data to be leveraged to identify key components in regulatory pathways and important gene targets in DCIS through use of integrative approaches with tools like Oncomine [150] to overlay gene expression data, including that from the Cancer Genome Atlas [151], onto protein-protein interactions, signaling pathways and transcriptional-regulatory networks.
Construction of networks based on genotype-phenotype linkage, gene regulatory modules, functionally linked pathways and protein communication modules will help in elucidating the mechanisms responsible for DCIS progression. Computational systems biology approaches have revealed functional biological networks that could be targeted for therapy. This is shown by a recent network-based, integrative study that identified distinct driver-networks in ER+, Her2+ and TNBC breast cancer subtypes [152].
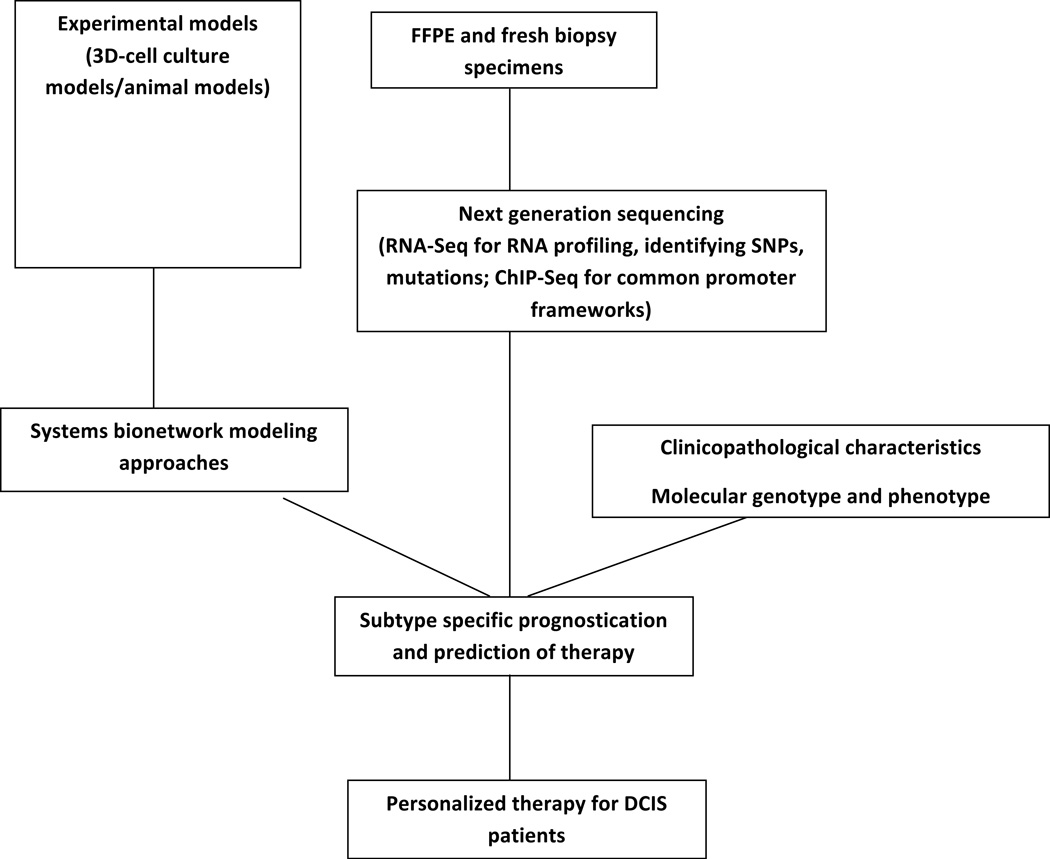
Further integration of genomic profiles with metabolomic, regulatory and signaling data will define functional networks and will aid in identifying candidate biomarkers in such networks. The NGS data can be explored to identify and analyze genes with similar expression patterns by exploring their promoter sequences to determine whether common promoter modules can be resolved. The common frameworks existing in the promoter regions, which can be verified by ChiP-Seq or resolved by DNase-1-Seq [153], can decipher regulatory connections among the different genes and may explain the functional regulation of co-expressed genes that have no detectable sequence similarities [154, 155]. With the continuing development of affordable NGS platforms, the construction of patient-specific signaling networks may become feasible. Analysis of those networks in individual patients could help in making risk predictions and treatment decisions that will transform current prognostic and treatment options [Figure 4].
Figure 4. Integrative flow chart for discovery of knowledge to personalize DCIS treatment therapy.
The DCIS samples for NGS may come from experimental models or clinical samples (FFPE or fresh biopsy). Genes with similar expression patterns could be identified by RNA-Seq and the common promoter frameworks verified by ChIP-Seq. Combining systems bionetwork modeling approaches with clinicopathological characteristics, molecular genotype and phenotype, NGS results will help to discover / predict personalized therapy for DCIS patients.
Future advances in prognostic, predictive and risk biomarkers of DCIS could be based on molecular events pertaining to the oxidative and pro-apoptotic stress or altered metabolic processes in the cancer cells or on analysis of the subpopulations of heterogenous DCIS lesions to distinguish indolent and aggressive forms. Given the complexity of the disease, a single or a small panel of molecular markers is not likely to be sufficient. Comprehensive approaches like whole genome based analyses that can identify gene expression and/or copy number changes, mutations, and epigenetic alterations could provide important information. The development of genetic and network-based biomarkers is needed to suitably predict the risk of progression in DCIS patients as well as their response to therapeutic regimens. All biomarkers, including those that will be derived from network analysis, require rigorous investigation to support their significance and applicability. Further validation of the predictive biomarkers that reflect specific mechanisms or aberrant signaling pathways may also lead to the development of targeted therapies like that of development of trastuzumab for Her-2.
Five-Year View
Many challenges still remain unresolved such as effective analytical tools for the enormous amounts of data generated by the sequencing technologies. Future directions should also explore the intratumoral heterogeneity of DCIS by sequencing of single cells and by subpopulation analysis. A complete picture will require analysis of stromal components and infiltrating cells as well. The integrated approaches that will take into account complete classification of diverse DCIS lesions are still being developed. Further advances in NGS and bioinformatic approaches, along with reduced costs and increased availability, should move us closer to personalized therapy for DCIS. Integration of the sequencing data from multiple collaborative teams with the available clinical data using computational tools and network modeling approaches may elucidate underlying driver mechanisms and result in identification of robust biomarkers that can stratify DCIS patients and eventually improve treatment decisions.
Key Issues.
DCIS was a rare diagnosis before the advent of mammographic screening. Its incidence has risen from 1.87 per 100 000 in the 1970s to 32.5 per 100,000 in 2004. Many of these lesions would likely have remained indolent or possibly even regressed spontaneously, but almost all DCIS is treated by surgical resection leading to a large burden of overtreatment.
Advances in expression profiling of breast cancer have led to increased understanding of subtypes of the disease and stratification of risk of recurrence and susceptibility to targeted therapeutics. This progress has yet to make a major impact on the treatment of DCIS.
Major unresolved questions include the profile of lesions that would remain indolent to allow discrimination from those that are likely to progress to invasive carcinoma. Identification of potential therapeutic targets in the former could lead to clinical trials of agents that would sustain dormancy or induce regression and so prevent malignancy. Identification of potential therapeutic targets in the latter could lead to clinical trials of agents for use as adjuncts to surgery.
Next-generation sequencing (NGS) offers unprecedented depth of analysis of gene expression that will reveal transcriptional boundaries, mutations, rare transcripts and alternative splice variants. NGS approaches are just beginning to be applied to DCIS.
Discoveries made through NGS will be leveraged through advances in bioinformatic analysis to reveal genetic and network-based biomarkers that predict the risk of progression in DCIS patients as well as their response to therapeutic regimens.
Validation of the predictive biomarkers that reflect specific mechanisms or aberrant signaling pathways may also lead to the development of targeted therapies
Acknowledgments
This work was supported in part by the National Institutes of Health through R01 CA131990 (BFS and RRM) and P30 CA22453 (support of Genomics and Microscopic Imaging and Cytometry Resources Core facilities). DHG is a recipient of an Advanced Clinical Research Award in Breast Cancer from the Conquer Cancer Foundation.
Footnotes
Financial Disclosure
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties. This review was written solely by the authors without any additional writing assistance.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA: a cancer journal for clinicians. 2012;62(1):10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Puliti D, Zappa M. Breast cancer screening: are we seeing the benefit? BMC Med. 2012;10:106. doi: 10.1186/1741-7015-10-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tabar L, Vitak B, Chen HH, Yen MF, Duffy SW, Smith RA. Beyond randomized controlled trials: organized mammographic screening substantially reduces breast carcinoma mortality. Cancer. 2001;91(9):1724–1731. doi: 10.1002/1097-0142(20010501)91:9<1724::aid-cncr1190>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 4.Kopans DB. Just the facts: mammography saves lives with little if any radiation risk to the mature breast. Health Phys. 2011;101(5):578–582. doi: 10.1097/HP.0b013e3182254e93. [DOI] [PubMed] [Google Scholar]
- 5.Peto R, Davies C, Godwin J, et al. Comparisons between different polychemotherapy regimens for early breast cancer: meta-analyses of long-term outcome among 100,000 women in 123 randomised trials. Lancet. 2012;379(9814):432–444. doi: 10.1016/S0140-6736(11)61625-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnston SR. The role of chemotherapy and targeted agents in patients with metastatic breast cancer. European journal of cancer. 2011;47(Suppl 3):S38–S47. doi: 10.1016/S0959-8049(11)70145-9. [DOI] [PubMed] [Google Scholar]
- 7.Gotzsche PC, Nielsen M. Screening for breast cancer with mammography. Cochrane Database Syst Rev. 2011;(1) doi: 10.1002/14651858.CD001877.pub4. CD001877. [DOI] [PubMed] [Google Scholar]
- 8.Gotzsche PC, Jorgensen KJ, Zahl PH, Maehlen J. Why mammography screening has not lived up to expectations from the randomised trials. Cancer Causes Control. 2012;23(1):15–21. doi: 10.1007/s10552-011-9867-8. [DOI] [PubMed] [Google Scholar]
- 9.Screening IUPoBC. The benefits and harms of breast cancer screening: an independent review. Lancet. 2012 doi: 10.1016/S0140-6736(12)61611-0. Epub ahead of printing. [DOI] [PubMed] [Google Scholar]
- 10. Bleyer A, Welch HG. Effect of three decades of screening mammography on breast-cancer incidence. The New England journal of medicine. 2012;367(21):1998–2005. doi: 10.1056/NEJMoa1206809. **Important recent study that emphasizes the prevalence of over-diagnosis of breast cancer. This study estimates that 1.3 million women in the U.S. have already been diagnosed with breast cancers that would not have become clinically significant.
- 11.Jorgensen KJ, Gotzsche PC. Overdiagnosis in publicly organised mammography screening programmes: systematic review of incidence trends. Bmj. 2009;339:b2587. doi: 10.1136/bmj.b2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Virnig BA, Tuttle TM, Shamliyan T, Kane RL. Ductal carcinoma in situ of the breast: a systematic review of incidence, treatment, and outcomes. J Natl Cancer Inst. 2010;102(3):170–178. doi: 10.1093/jnci/djp482. [DOI] [PubMed] [Google Scholar]
- 13.Zahl PH, Gotzsche PC, Maehlen J. Natural history of breast cancers detected in the Swedish mammography screening programme: a cohort study. The lancet oncology. 2011;12(12):1118–1124. doi: 10.1016/S1470-2045(11)70250-9. [DOI] [PubMed] [Google Scholar]
- 14. Sgroi DC. Preinvasive breast cancer. Annu Rev Pathol. 2010;5:193–221. doi: 10.1146/annurev.pathol.4.110807.092306. *Comprehensive review of premalignant breast cancer.
- 15.Espina V, Liotta LA. What is the malignant nature of human ductal carcinoma in situ? Nat Rev Cancer. 2011;11(s1):68–75. doi: 10.1038/nrc2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tuttle TM, Jarosek S, Habermann EB, et al. Increasing rates of contralateral prophylactic mastectomy among patients with ductal carcinoma in situ. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2009;27(9):1362–1367. doi: 10.1200/JCO.2008.20.1681. [DOI] [PubMed] [Google Scholar]
- 17.Hawley ST, Jagsi R, Katz SJ. Is contralateral prophylactic mastectomy (CPM) overused? Results from a population-based study. J Clin Oncol. 2012;30(Suppl 34) Abstr 26. [Google Scholar]
- 18.Rudloff U, Jacks LM, Goldberg JI, et al. Nomogram for predicting the risk of local recurrence after breast-conserving surgery for ductal carcinoma in situ. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2010;28(23):3762–3769. doi: 10.1200/JCO.2009.26.8847. [DOI] [PubMed] [Google Scholar]
- 19.Yi M, Meric-Bernstam F, Kuerer HM, et al. Evaluation of a breast cancer nomogram for predicting risk of ipsilateral breast tumor recurrences in patients with ductal carcinoma in situ after local excision. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2012;30(6):600–607. doi: 10.1200/JCO.2011.36.4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Solin LJ, Gray R, Baehner FL. A Quantitative Multigene RT-PCR Assay for Predicting Recurrence Risk after Surgical Excision Alone without Irradiation for Ductal Carcinoma In Situ (DCIS): A prospective Validation Study of the DCIS Score from ECOG E5194; CTRC-AACR San Antonio Breast Cancer Symposium; Dec, 2011. pp. 6–10. Ed.^(Eds) Abstract S4-6. [Google Scholar]
- 21.Wellings SR, Jensen HM. On the origin and progression of ductal carcinoma in the human breast. J Natl Cancer Inst. 1973;50(5):1111–1118. doi: 10.1093/jnci/50.5.1111. [DOI] [PubMed] [Google Scholar]
- 22.Sontag L, Axelrod DE. Evaluation of pathways for progression of heterogeneous breast tumors. J Theor Biol. 2005;232(2):179–189. doi: 10.1016/j.jtbi.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 23. Johnson CE, Gorringe KL, Thompson ER, et al. Identification of copy number alterations associated with the progression of DCIS to invasive ductal carcinoma. Breast Cancer Res Treat. 2012;133(3):889–898. doi: 10.1007/s10549-011-1835-1. *A detailed genomic study of synchronous DCIS and IDC that supports a model of parallel evolution from a common clonal origin.
- 24.Polyak K. Molecular markers for the diagnosis and management of ductal carcinoma in situ. J Natl Cancer Inst Monogr. 2010;2010(41):210–213. doi: 10.1093/jncimonographs/lgq019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller FR. Xenograft models of premalignant breast disease. J Mammary Gland Biol Neoplasia. 2000;5(4):379–391. doi: 10.1023/a:1009577811584. [DOI] [PubMed] [Google Scholar]
- 26.Brodie A, Sabnis G, Macedo L. Xenograft models for aromatase inhibitor studies. J Steroid Biochem Mol Biol. 2007;106(1-5):119–124. doi: 10.1016/j.jsbmb.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 27.Visscher DW, Nanjia-Makker P, Heppner G, Shekhar PV. Tamoxifen suppresses histologic progression to atypia and DCIS in MCFIOAT xenografts, a model of early human breast cancer. Breast Cancer Res Treat. 2001;65(1):41–47. doi: 10.1023/a:1006490000659. [DOI] [PubMed] [Google Scholar]
- 28. Hu M, Yao J, Carroll DK, et al. Regulation of in situ to invasive breast carcinoma transition. Cancer Cell. 2008;13(5):394–406. doi: 10.1016/j.ccr.2008.03.007. **A landmark study that combined models of DCIS and stromal cells with molecular profiling to determine interacting pathways that control malignant progression.
- 29. Behbod F, Kittrell FS, LaMarca H, et al. An intraductal human-in-mouse transplantation model mimics the subtypes of ductal carcinoma in situ. Breast Cancer Res. 2009;11(5):R66. doi: 10.1186/bcr2358. *An interesting approach to generate a more accurate model by growing cells from primary human DCIS tumors in the mammary ducts of the mouse.
- 30.Valdez KE, Fan F, Smith W, Allred DC, Medina D, Behbod F. Human primary ductal carcinoma in situ (DCIS) subtype-specific pathology is preserved in a mouse intraductal (MIND) xenograft model. J Pathol. 2011;225(4):565–573. doi: 10.1002/path.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pearson T, Greiner DL, Shultz LD. Humanized SCID mouse models for biomedical research. Curr Top Microbiol Immunol. 2008;324:25–51. doi: 10.1007/978-3-540-75647-7_2. [DOI] [PubMed] [Google Scholar]
- 32.Schulze-Garg C, Lohler J, Gocht A, Deppert W. A transgenic mouse model for the ductal carcinoma in situ (DCIS) of the mammary gland. Oncogene. 2000;19(8):1028–1037. doi: 10.1038/sj.onc.1203281. [DOI] [PubMed] [Google Scholar]
- 33.Tran-Thanh D, Buttars S, Wen Y, Wilson C, Done SJ. Cyclooxygenase-2 inhibition for the prophylaxis and treatment of preinvasive breast cancer in a her-2/neu mouse model. Cancer Prev Res (Phila) 2010;3(2):202–211. doi: 10.1158/1940-6207.CAPR-09-0181. [DOI] [PubMed] [Google Scholar]
- 34.Namba R, Young LJ, Abbey CK, et al. Rapamycin inhibits growth of premalignant and malignant mammary lesions in a mouse model of ductal carcinoma in situ. Clin Cancer Res. 2006;12(8):2613–2621. doi: 10.1158/1078-0432.CCR-05-2170. [DOI] [PubMed] [Google Scholar]
- 35.Ye Y, Qiu TH, Kavanaugh C, Green JE. Molecular mechanisms of breast cancer progression: lessons from mouse mammary cancer models and gene expression profiling. Breast Dis. 2004;19:69–82. doi: 10.3233/bd-2004-19109. [DOI] [PubMed] [Google Scholar]
- 36.Debnath J, Brugge JS. Modelling glandular epithelial cancers in three-dimensional cultures. Nat Rev Cancer. 2005;5(9):675–688. doi: 10.1038/nrc1695. [DOI] [PubMed] [Google Scholar]
- 37.Miller BE, Miller FR, Heppner GH. Factors affecting growth and drug sensitivity of mouse mammary tumor lines in collagen gel cultures. Cancer research. 1985;45(9):4200–4205. [PubMed] [Google Scholar]
- 38.Horning JL, Sahoo SK, Vijayaraghavalu S, et al. 3-D tumor model for in vitro evaluation of anticancer drugs. Molecular pharmaceutics. 2008;5(5):849–862. doi: 10.1021/mp800047v. [DOI] [PubMed] [Google Scholar]
- 39.Li Q, Chow AB, Mattingly RR. Three-dimensional overlay culture models of human breast cancer reveal a critical sensitivity to mitogen-activated protein kinase kinase inhibitors. J Pharmacol Exp Ther. 2010;332(3):821–828. doi: 10.1124/jpet.109.160390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hebner C, Weaver VM, Debnath J. Modeling morphogenesis and oncogenesis in three-dimensional breast epithelial cultures. Annu Rev Pathol. 2008;3:313–339. doi: 10.1146/annurev.pathmechdis.3.121806.151526. [DOI] [PubMed] [Google Scholar]
- 41.Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30(3):256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- 42.Kenny PA, Lee GY, Myers CA, et al. The morphologies of breast cancer cell lines in three-dimensional assays correlate with their profiles of gene expression. Mol Oncol. 2007;1(1):84–96. doi: 10.1016/j.molonc.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kaur H, Mao S, Li Q, et al. RNA-seq of human breast ductal carcinoma in situ models reveals aldehyde dehydrogenase isoform 5A1 as a novel potential target. PLoS One. 2012;7(12):e50249. doi: 10.1371/journal.pone.0050249. *The first application of NGS specifically to models of DCIS. The results emphasize the molecular changes that occur at the interface of DCIS with its microenvironment.
- 44. Sameni M, Anbalagan A, Olive MB, Moin K, Mattingly RR, Sloane BF. MAME models for 4D live-cell imaging of tumor: microenvironment interactions that impact malignant progression. J Vis Exp. 2012;(60) doi: 10.3791/3661. *Written and video protocols for establishing and live-cell imaging of 3D mammary architecture and microenvironment (MAME) co-culture models. This approach offers new opportunities for the study of DCIS.
- 45.Mullins SR, Sameni M, Blum G, Bogyo M, Sloane B, Moin K. Three-dimensional cultures model premalignant progression of human breast epithelial cells: role of cysteine cathepsins. Biol. Chem. doi: 10.1515/hsz-2012-0252. (in press; cover image) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Victor BC, Anbalagan A, Mohamed MM, Sloane BF, Cavallo-Medved D. Inhibition of cathepsin B activity attenuates extracellular matrix degradation and inflammatory breast cancer invasion. Breast Cancer Res. 2011;13(6):R115. doi: 10.1186/bcr3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jedeszko C, Victor BC, Podgorski I, Sloane BF. Fibroblast hepatocyte growth factor promotes invasion of human mammary ductal carcinoma in situ. Cancer Res. 2009;69(23):9148–9155. doi: 10.1158/0008-5472.CAN-09-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robey IF, Baggett BK, Kirkpatrick ND, et al. Bicarbonate increases tumor pH and inhibits spontaneous metastases. Cancer Res. 2009;69(6):2260–2268. doi: 10.1158/0008-5472.CAN-07-5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wojtkowiak JW, Rothberg JM, Kumar V, et al. Chronic autophagy is a cellular adaptation to tumor acidic pH microenvironments. Cancer Res. 2012;72(16):3938–3947. doi: 10.1158/0008-5472.CAN-11-3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dang TT, Prechtl AM, Pearson GW. Breast cancer subtype-specific interactions with the microenvironment dictate mechanisms of invasion. Cancer Res. 2011;71(21):6857–6866. doi: 10.1158/0008-5472.CAN-11-1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Macklin P, Edgerton ME, Thompson AM, Cristini V. Patient-calibrated agent-based modelling of ductal carcinoma in situ (DCIS): from microscopic measurements to macroscopic predictions of clinical progression. J Theor Biol. 2012;301:122–140. doi: 10.1016/j.jtbi.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sung KE, Yang N, Pehlke C, et al. Transition to invasion in breast cancer: a microfluidic in vitro model enables examination of spatial and temporal effects. Integr Biol (Camb) 2011;3(4):439–450. doi: 10.1039/c0ib00063a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abba MC, Drake JA, Hawkins KA, et al. Transcriptomic changes in human breast cancer progression as determined by serial analysis of gene expression. Breast Cancer Res. 2004;6(5):R499–R513. doi: 10.1186/bcr899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Adeyinka A, Emberley E, Niu Y, et al. Analysis of gene expression in ductal carcinoma in situ of the breast. Clinical cancer research : an official journal of the American Association for Cancer Research. 2002;8(12):3788–3795. [PubMed] [Google Scholar]
- 55.Castro NP, Osorio CA, Torres C, et al. Evidence that molecular changes in cells occur before morphological alterations during the progression of breast ductal carcinoma. Breast Cancer Res. 2008;10(5):R87. doi: 10.1186/bcr2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Emery LA, Tripathi A, King C, et al. Early dysregulation of cell adhesion and extracellular matrix pathways in breast cancer progression. The American journal of pathology. 2009;175(3):1292–1302. doi: 10.2353/ajpath.2009.090115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hannemann J, Velds A, Halfwerk JB, Kreike B, Peterse JL, van de Vijver MJ. Classification of ductal carcinoma in situ by gene expression profiling. Breast Cancer Res. 2006;8(5):R61. doi: 10.1186/bcr1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Porter D, Lahti-Domenici J, Keshaviah A, et al. Molecular markers in ductal carcinoma in situ of the breast. Molecular cancer research : MCR. 2003;1(5):362–375. [PubMed] [Google Scholar]
- 59.Schuetz CS, Bonin M, Clare SE, et al. Progression-specific genes identified by expression profiling of matched ductal carcinomas in situ and invasive breast tumors, combining laser capture microdissection and oligonucleotide microarray analysis. Cancer Res. 2006;66(10):5278–5286. doi: 10.1158/0008-5472.CAN-05-4610. [DOI] [PubMed] [Google Scholar]
- 60.Seth A, Kitching R, Landberg G, Xu J, Zubovits J, Burger AM. Gene expression profiling of ductal carcinomas in situ and invasive breast tumors. Anticancer Research. 2003;23(3A):2043–2051. [PubMed] [Google Scholar]
- 61.Vincent-Salomon A, Lucchesi C, Gruel N, et al. Integrated genomic and transcriptomic analysis of ductal carcinoma in situ of the breast. Clinical cancer research : an official journal of the American Association for Cancer Research. 2008;14(7):1956–1965. doi: 10.1158/1078-0432.CCR-07-1465. [DOI] [PubMed] [Google Scholar]
- 62.Muggerud AA, Hallett M, Johnsen H, et al. Molecular diversity in ductal carcinoma in situ (DCIS) and early invasive breast cancer. Mol Oncol. 2010;4(4):357–368. doi: 10.1016/j.molonc.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Buerger H, Otterbach F, Simon R, et al. Comparative genomic hybridization of ductal carcinoma in situ of the breast-evidence of multiple genetic pathways. The Journal of pathology. 1999;187(4):396–402. doi: 10.1002/(SICI)1096-9896(199903)187:4<396::AID-PATH286>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 64.Simpson PT, Gale T, Reis-Filho JS, et al. Columnar cell lesions of the breast: the missing link in breast cancer progression? A morphological and molecular analysis. Am J Surg Pathol. 2005;29(6):734–746. doi: 10.1097/01.pas.0000157295.93914.3b. [DOI] [PubMed] [Google Scholar]
- 65.Forozan F, Veldman R, Ammerman CA, et al. Molecular cytogenetic analysis of 11 new breast cancer cell lines. British journal of cancer. 1999;81(s8):1328–1334. doi: 10.1038/sj.bjc.6695007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ma XJ, Salunga R, Tuggle JT, et al. Gene expression profiles of human breast cancer progression. Proc Natl Acad Sci USA. 2003;100(10):5974–5979. doi: 10.1073/pnas.0931261100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schuetz CS, Bonin M, Clare SE, et al. Progression-specific genes identified by expression profiling of matched ductal carcinomas in situ and invasive breast tumors, combining laser capture microdissection and oligonucleotide microarray analysis. Cancer Res. 2006;66(10):5278–5286. doi: 10.1158/0008-5472.CAN-05-4610. [DOI] [PubMed] [Google Scholar]
- 68.Castro NP, Osorio CA, Torres C, et al. Evidence that molecular changes in cells occur before morphological alterations during the progression of breast ductal carcinoma. Breast Cancer Res. 2008;10(5):R87. doi: 10.1186/bcr2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hannemann J, Velds A, Halfwerk JB, Kreike B, Peterse JL, van de Vijver MJ. Classification of ductal carcinoma in situ by gene expression profiling. Breast Cancer Res. 2006;8(5):R61. doi: 10.1186/bcr1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Buerger H, Simon R, Schafer KL, et al. Genetic relation of lobular carcinoma in situ, ductal carcinoma in situ, and associated invasive carcinoma of the breast. Mol Pathol. 2000;53(3):118–121. doi: 10.1136/mp.53.3.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 72.Carey LA, Perou CM, Livasy CA, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. Jama. 2006;295(21):2492–2502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- 73. Chen X, Li J, Gray WH, et al. TNBCtype: A Subtyping Tool for Triple-Negative Breast Cancer. Cancer Inform. 2012;11:147–156. doi: 10.4137/CIN.S9983. * The use of expression profiling to sub-divide breast cancers should lead to improved prognostic and treatment decisions.
- 74.Makar AP, Desmedt EJ, De Potter CR, Vanderheyden JS, Schatteman EA. Neu(C-erbB-2) oncogene in breast cancer and its possible association with the risk of distant metastases. A retrospective study and review of literature. Acta Oncol. 1990;29(7):931–934. doi: 10.3109/02841869009096392. [DOI] [PubMed] [Google Scholar]
- 75.Allred DC, Clark GM, Molina R, et al. Overexpression of HER-2/neu and its relationship with other prognostic factors change during the progression of in situ to invasive breast cancer. Human pathology. 1992;23(9):974–979. doi: 10.1016/0046-8177(92)90257-4. [DOI] [PubMed] [Google Scholar]
- 76.Allred DC, Clark GM, Tandon AK, et al. HER-2/neu in node-negative breast cancer: prognostic significance of overexpression influenced by the presence of in situ carcinoma. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 1992;10(4):599–605. doi: 10.1200/JCO.1992.10.4.599. [DOI] [PubMed] [Google Scholar]
- 77.Han K, Nofech-Mozes S, Narod S, et al. Expression of HER2neu in ductal carcinoma in situ is associated with local recurrence. Clin Oncol (R Coll Radiol) 2012;24(3):183–189. doi: 10.1016/j.clon.2011.09.008. [DOI] [PubMed] [Google Scholar]
- 78.Kuerer HM, Albarracin CT, Yang WT, et al. Ductal carcinoma in situ: state of the science and roadmap to advance the field. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2009;27(2):279–288. doi: 10.1200/JCO.2008.18.3103. [DOI] [PubMed] [Google Scholar]
- 79.Allred DC, Anderson SJ, Paik S, et al. Adjuvant tamoxifen reduces subsequent breast cancer in women with estrogen receptor-positive ductal carcinoma in situ: a study based on NSABP protocol B-24. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2012;30(12):1268–1273. doi: 10.1200/JCO.2010.34.0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Claus EB, Chu P, Howe CL, et al. Pathobiologic findings in DCIS of the breast: morphologic features, angiogenesis, HER-2/neu and hormone receptors. Experimental and molecular pathology. 2001;70(3):303–316. doi: 10.1006/exmp.2001.2366. [DOI] [PubMed] [Google Scholar]
- 81.Barnes NL, Boland GP, Davenport A, Knox WF, Bundred NJ. Relationship between hormone receptor status and tumour size, grade and comedo necrosis in ductal carcinoma in situ. Br J Surg. 2005;92(4):429–434. doi: 10.1002/bjs.4878. [DOI] [PubMed] [Google Scholar]
- 82.Simpson JF, Quan DE, O'Malley F, Odom-Maryon T, Clarke PE. Amplification of CCND1 and expression of its protein product, cyclin D1, in ductal carcinoma in situ of the breast. The American journal of pathology. 1997;151(1):161–168. [PMC free article] [PubMed] [Google Scholar]
- 83.Vos CB, Ter Haar NT, Peterse JL, Cornelisse CJ, van de Vijver MJ. Cyclin D1 gene amplification and overexpression are present in ductal carcinoma in situ of the breast. The Journal of pathology. 1999;187(3):279–284. doi: 10.1002/(SICI)1096-9896(199902)187:3<279::AID-PATH240>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 84.Oh YL, Choi JS, Song SY, et al. Expression of p21Waf1, p27Kip1 and cyclin D1 proteins in breast ductal carcinoma in situ: Relation with clinicopathologic characteristics and with p53 expression and estrogen receptor status. Pathology international. 2001;51(2):94–99. doi: 10.1046/j.1440-1827.2001.01173.x. [DOI] [PubMed] [Google Scholar]
- 85.Lebeau A, Unholzer A, Amann G, et al. EGFR, HER-2/neu, cyclin D1, p21 and p53 in correlation to cell proliferation and steroid hormone receptor status in ductal carcinoma in situ of the breast. Breast cancer research and treatment. 2003;79(2):187–198. doi: 10.1023/a:1023958324448. [DOI] [PubMed] [Google Scholar]
- 86.Millar EK, Tran K, Marr P, Graham PH. p27KIP-1, cyclin A and cyclin D1 protein expression in ductal carcinoma in situ of the breast: p27KIP-1 correlates with hormone receptor status but not with local recurrence. Pathology international. 2007;57(4):183–189. doi: 10.1111/j.1440-1827.2007.02079.x. [DOI] [PubMed] [Google Scholar]
- 87.Meteoglu I, Dikicioglu E, Erkus M, et al. Breast carcinogenesis. Transition from hyperplasia to invasive lesions. Saudi Med J. 2005;26(12):1889–1896. [PubMed] [Google Scholar]
- 88.Siziopikou KP, Prioleau JE, Harris JR, Schnitt SJ. bcl-2 expression in the spectrum of preinvasive breast lesions. Cancer. 1996;77(3):499–506. doi: 10.1002/(SICI)1097-0142(19960201)77:3<499::AID-CNCR11>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 89.Mustonen M, Raunio H, Paakko P, Soini Y. The extent of apoptosis is inversely associated with bcl-2 expression in premalignant and malignant breast lesions. Histopathology. 1997;31(4):347–354. doi: 10.1046/j.1365-2559.1997.2710877.x. [DOI] [PubMed] [Google Scholar]
- 90.Walker RA, Jones JL, Chappell S, Walsh T, Shaw JA. Molecular pathology of breast cancer and its application to clinical management. Cancer metastasis reviews. 1997;16(1-2):5–27. doi: 10.1023/a:1005740222307. [DOI] [PubMed] [Google Scholar]
- 91.Done SJ, Eskandarian S, Bull S, Redston M, Andrulis IL. p53 missense mutations in microdissected high-grade ductal carcinoma in situ of the breast. Journal of the National Cancer Institute. 2001;93(9):700–704. doi: 10.1093/jnci/93.9.700. [DOI] [PubMed] [Google Scholar]
- 92.Sarode VR, Han JS, Morris DH, Peng Y, Rao R. A Comparative Analysis of Biomarker Expression and Molecular Subtypes of Pure Ductal Carcinoma In Situ and Invasive Breast Carcinoma by Image Analysis: Relationship of the Subtypes with Histologic Grade, Ki67, p53 Overexpression, and DNA Ploidy. Int J Breast Cancer. 2011;2011:217060. doi: 10.4061/2011/217060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gotte M, Kersting C, Radke I, Kiesel L, Wulfing P. An expression signature of syndecan-1 (CD138), E-cadherin and c-met is associated with factors of angiogenesis and lymphangiogenesis in ductal breast carcinoma in situ. Breast cancer research : BCR. 2007;9(1):R8. doi: 10.1186/bcr1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Viacava P, Naccarato AG, Bocci G, et al. Angiogenesis and VEGF expression in pre-invasive lesions of the human breast. The Journal of pathology. 2004;204(2):140–146. doi: 10.1002/path.1626. [DOI] [PubMed] [Google Scholar]
- 95.Hieken TJ, Farolan M, D'Alessandro S, Velasco JM. Predicting the biologic behavior of ductal carcinoma in situ: an analysis of molecular markers. Surgery. 2001;130(4):593–600. doi: 10.1067/msy.2001.116921. discussion 600-591. [DOI] [PubMed] [Google Scholar]
- 96.Jang MH, Kim EJ, Choi Y, et al. FGFR1 is amplified during the progression of in situ to invasive breast carcinoma. Breast Cancer Res. 2012;14(4):R115. doi: 10.1186/bcr3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Robanus-Maandag EC, Bosch CA, Kristel PM, et al. Association of C-MYC amplification with progression from the in situ to the invasive stage in C-MYC-amplified breast carcinomas. The Journal of pathology. 2003;201(1):75–82. doi: 10.1002/path.1385. [DOI] [PubMed] [Google Scholar]
- 98.Watson PH, Safneck JR, Le K, Dubik D, Shiu RP. Relationship of c-myc amplification to progression of breast cancer from in situ to invasive tumor and lymph node metastasis. Journal of the National Cancer Institute. 1993;85(11):902–907. doi: 10.1093/jnci/85.11.902. [DOI] [PubMed] [Google Scholar]
- 99.Altintas S, Lambein K, Huizing MT, et al. Prognostic significance of oncogenic markers in ductal carcinoma in situ of the breast: a clinicopathologic study. Breast J. 2009;15(2):120–132. doi: 10.1111/j.1524-4741.2009.00686.x. [DOI] [PubMed] [Google Scholar]
- 100.Provenzano E, Hopper JL, Giles GG, Marr G, Venter DJ, Armes JE. Biological markers that predict clinical recurrence in ductal carcinoma in situ of the breast. Eur J Cancer. 2003;39(5):622–630. doi: 10.1016/s0959-8049(02)00666-4. [DOI] [PubMed] [Google Scholar]
- 101.Ringberg A, Anagnostaki L, Anderson H, Idvall I, Ferno M. Cell biological factors in ductal carcinoma in situ (DCIS) of the breast-relationship to ipsilateral local recurrence and histopathological characteristics. Eur J Cancer. 2001;37(12):1514–1522. doi: 10.1016/s0959-8049(01)00165-4. [DOI] [PubMed] [Google Scholar]
- 102.Kerlikowske K, Molinaro AM, Gauthier ML, et al. Biomarker expression and risk of subsequent tumors after initial ductal carcinoma in situ diagnosis. Journal of the National Cancer Institute. 2010;102(9):627–637. doi: 10.1093/jnci/djq101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Van Zee KJ, Calvano JE, Bisogna M. Hypomethylation and increased gene expression of p16INK4a in primary and metastatic breast carcinoma as compared to normal breast tissue. Oncogene. 1998;16(21):2723–2727. doi: 10.1038/sj.onc.1201794. [DOI] [PubMed] [Google Scholar]
- 104.Liu T, Niu Y, Feng Y, et al. Methylation of CpG islands of p16(INK4a) and cyclinD1 overexpression associated with progression of intraductal proliferative lesions of the breast. Hum Pathol. 2008;39(11):1637–1646. doi: 10.1016/j.humpath.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 105.Gobbi H, Dupont WD, Simpson JF, et al. Transforming growth factor-beta and breast cancer risk in women with mammary epithelial hyperplasia. J Natl Cancer Inst. 1999;91(24):2096–2101. doi: 10.1093/jnci/91.24.2096. [DOI] [PubMed] [Google Scholar]
- 106.Walker RA, Dearing SJ. Transforming growth factor beta 1 in ductal carcinoma in situ and invasive carcinomas of the breast. Eur J Cancer. 1992;28(2-3):641–644. doi: 10.1016/s0959-8049(05)80116-9. [DOI] [PubMed] [Google Scholar]
- 107.Lu J, Guo H, Treekitkarnmongkol W, et al. 14-3-3zeta Cooperates with ErbB2 to promote ductal carcinoma in situ progression to invasive breast cancer by inducing epithelial-mesenchymal transition. Cancer Cell. 2009;16(3):195–207. doi: 10.1016/j.ccr.2009.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Qi L, Bart J, Tan LP, et al. Expression of miR-21 and its targets (PTEN, PDCD4, TM1) in flat epithelial atypia of the breast in relation to ductal carcinoma in situ and invasive carcinoma. BMC Cancer. 2009;9:163. doi: 10.1186/1471-2407-9-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lari SA, Kuerer HM. Biological Markers in DCIS and Risk of Breast Recurrence: A Systematic Review. J Cancer. 2011;2:232–261. doi: 10.7150/jca.2.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kern SE. Why your new cancer biomarker may never work: recurrent patterns and remarkable diversity in biomarker failures. Cancer research. 2012;72(23):6097–6101. doi: 10.1158/0008-5472.CAN-12-3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.van 't Veer LJ, Dai H, van de Vijver MJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415(6871):530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 112.Cobleigh MA, Tabesh B, Bitterman P, et al. Tumor gene expression and prognosis in breast cancer patients with 10 or more positive lymph nodes. Clinical cancer research : an official journal of the American Association for Cancer Research. 2005;11(24 Pt 1):8623–8631. doi: 10.1158/1078-0432.CCR-05-0735. [DOI] [PubMed] [Google Scholar]
- 113.Baggerly KA, Coombes KR. What information should be required to support clinical "omics" publications? Clin Chem. 2011;57(5):688–690. doi: 10.1373/clinchem.2010.158618. [DOI] [PubMed] [Google Scholar]
- 114.Ioannidis JP, Khoury MJ. Improving validation practices in "omics" research. Science. 2011;334(6060):1230–1232. doi: 10.1126/science.1211811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Desmedt C, Voet T, Sotiriou C, Campbell PJ. Next-generation sequencing in breast cancer: first take home messages. Current opinion in oncology. 2012;24(6):597–604. doi: 10.1097/CCO.0b013e328359554e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Vera JC, Wheat CW, Fescemyer HW, et al. Rapid transcriptome characterization for a nonmodel organism using 454 pyrosequencing. Mol Ecol. 2008;17(7):1636–1647. doi: 10.1111/j.1365-294X.2008.03666.x. [DOI] [PubMed] [Google Scholar]
- 117.Emrich SJ, Barbazuk WB, Li L, Schnable PS. Gene discovery and annotation using LCM-454 transcriptome sequencing. Genome Res. 2007;17(1):69–73. doi: 10.1101/gr.5145806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Huber-Keener KJ, Liu X, Wang Z, et al. Differential gene expression in tamoxifen-resistant breast cancer cells revealed by a new analytical model of RNA-Seq data. PLoS One. 2012;7(7):e41333. doi: 10.1371/journal.pone.0041333. * This report highlights the ability of NGS to identify transcriptome changes in tamoxifen-resistant breast cancer.
- 119.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5(7):621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 120.Barbazuk WB, Emrich SJ, Chen HD, Li L, Schnable PS. SNP discovery via 454 transcriptome sequencing. Plant J. 2007;51(5):910–918. doi: 10.1111/j.1365-313X.2007.03193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10(1):57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Costa V, Angelini C, De Feis I, Ciccodicola A. Uncovering the complexity of transcriptomes with RNA-Seq. J Biomed Biotechnol. 2010;2010:853916. doi: 10.1155/2010/853916. *An excellent review of RNA-Seq methodology and the challenges that arise from both biological and bioinformatic perspectives.
- 123.Sultan M, Schulz MH, Richard H, et al. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science. 2008;321(5891):956–960. doi: 10.1126/science.1160342. [DOI] [PubMed] [Google Scholar]
- 124.Passalacqua KD, Varadarajan A, Weist C, et al. Strand-Specific RNA-Seq Reveals Ordered Patterns of Sense and Antisense Transcription in Bacillus anthracis. PLoS One. 2012;7(8):e43350. doi: 10.1371/journal.pone.0043350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Nik-Zainal S, Van Loo P, Wedge DC, et al. The life history of 21 breast cancers. Cell. 2012;149(5):994–1007. doi: 10.1016/j.cell.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Nik-Zainal S, Alexandrov LB, Wedge DC, et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149(5):979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bemmo A, Dias C, Rose AA, Russo C, Siegel P, Majewski J. Exon-level transcriptome profiling in murine breast cancer reveals splicing changes specific to tumors with different metastatic abilities. PLoS One. 2010;5(s8):e11981. doi: 10.1371/journal.pone.0011981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Tsunoda T, Inada H, Kalembeyi I, et al. Involvement of large tenascin-C splice variants in breast cancer progression. The American journal of pathology. 2003;162(6):1857–1867. doi: 10.1016/S0002-9440(10)64320-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ferreira EN, Rangel MC, Galante PF, et al. Alternative splicing enriched cDNA libraries identify breast cancer-associated transcripts. BMC Genomics. 2010;11(Suppl 5):S4. doi: 10.1186/1471-2164-11-S5-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Omenn GS, Yocum AK, Menon R. Alternative splice variants, a new class of protein cancer biomarker candidates: findings in pancreatic cancer and breast cancer with systems biology implications. Dis Markers. 2010;28(4):241–251. doi: 10.3233/DMA-2010-0702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer. 2007;7(4):233–245. doi: 10.1038/nrc2091. [DOI] [PubMed] [Google Scholar]
- 132.Stephens PJ, McBride DJ, Lin ML, et al. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature. 2009;462(7276):1005–1010. doi: 10.1038/nature08645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Banerji S, Cibulskis K, Rangel-Escareno C, et al. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature. 2012;486(7403):405–409. doi: 10.1038/nature11154. **This report uses NGS to define somatic mutations and rearrangements in a large number of breast cancer genomes, and includes a subset of nine DCIS samples.
- 134.Ellis MJ, Ding L, Shen D, et al. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature. 2012;486(7403):353–360. doi: 10.1038/nature11143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Stephens PJ, Tarpey PS, Davies H, et al. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486(7403):400–404. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Curtis C, Shah SP, Chin SF, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486(7403):346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Shah SP, Roth A, Goya R, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486(7403):395–399. doi: 10.1038/nature10933. *This study emphasizes the complexity of TNBC disease. Comparison of genomic data on somatic single nucleotide variants (i.e., mutations) with RNA-Seq data indicated that many mutated genes are not transcribed.
- 138.Gray J, Druker B. Genomics: the breast cancer landscape. Nature. 2012;486(7403):328–329. doi: 10.1038/486328a. [DOI] [PubMed] [Google Scholar]
- 139.Thurman RE, Rynes E, Humbert R, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489(7414):75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Neph S, Vierstra J, Stergachis AB, et al. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature. 2012;489(7414):83–90. doi: 10.1038/nature11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Djebali S, Davis CA, Merkel A, et al. Landscape of transcription in human cells. Nature. 2012;489(7414):101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Beck J, Urnovitz HB, Mitchell WM, Schutz E. Next generation sequencing of serum circulating nucleic acids from patients with invasive ductal breast cancer reveals differences to healthy and nonmalignant controls. Molecular cancer research : MCR. 2010;8(3):335–342. doi: 10.1158/1541-7786.MCR-09-0314. *This study uses NGS to detect and quantify nucleic acids in the circulation of patients with IDC.
- 143.Wu Q, Lu Z, Li H, Lu J, Guo L, Ge Q. Next-generation sequencing of microRNAs for breast cancer detection. J Biomed Biotechnol. 2011;2011:597145. doi: 10.1155/2011/597145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wu Q, Wang C, Lu Z, Guo L, Ge Q. Analysis of serum genome-wide microRNAs for breast cancer detection. Clinica chimica acta; international journal of clinical chemistry. 2012;413(13-14):1058–1065. doi: 10.1016/j.cca.2012.02.016. [DOI] [PubMed] [Google Scholar]
- 145.Volinia S, Galasso M, Sana ME, et al. Breast cancer signatures for invasiveness and prognosis defined by deep sequencing of microRNA. P Natl Acad Sci USA. 2012;109(8):3024–3029. doi: 10.1073/pnas.1200010109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kharchenko PV, Tolstorukov MY, Park PJ. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nature biotechnology. 2008;26(12):1351–1359. doi: 10.1038/nbt.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Feng J, Liu T, Qin B, Zhang Y, Liu XS. Identifying ChIP-seq enrichment using MACS. Nature protocols. 2012;7(9):1728–1740. doi: 10.1038/nprot.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Rozowsky J, Euskirchen G, Auerbach RK, et al. PeakSeq enables systematic scoring of ChIP-seq experiments relative to controls. Nature biotechnology. 2009;27(1):66–75. doi: 10.1038/nbt.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Landt SG, Marinov GK, Kundaje A, et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012;22(9):1813–1831. doi: 10.1101/gr.136184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Rhodes DR, Yu J, Shanker K, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6(1):1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zhang J, Baran J, Cros A, et al. International Cancer Genome Consortium Data Portal--a one-stop shop for cancer genomics data. Database (Oxford) 2011;2011 doi: 10.1093/database/bar026. bar026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Dutta B, Pusztai L, Qi Y, et al. A network-based, integrative study to identify core biological pathways that drive breast cancer clinical subtypes. Br J Cancer. 2012;106(6):1107–1116. doi: 10.1038/bjc.2011.584. *This report describes the development of bioinformatic methodology to identify driving networks in three clinical subtypes of breast cancer.
- 153.Pique-Regi R, Degner JF, Pai AA, Gaffney DJ, Gilad Y, Pritchard JK. Accurate inference of transcription factor binding from DNA sequence and chromatin accessibility data. Genome Res. 2011;21(3):447–455. doi: 10.1101/gr.112623.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Werner T. Cluster analysis and promoter modelling as bioinformatics tools for the identification of target genes from expression array data. Pharmacogenomics. 2001;2(1):25–36. doi: 10.1517/14622416.2.1.25. [DOI] [PubMed] [Google Scholar]
- 155.Edelmann SL, Nelson PJ, Brocker T. Comparative promoter analysis in vivo: identification of a dendritic cell-specific promoter module. Blood. 2011;118(11):e40–e49. doi: 10.1182/blood-2011-03-342261. [DOI] [PubMed] [Google Scholar]
- 156.Screening for breast cancer: U.S. Preventive Services Task Force recommendation statement. Ann Intern Med. 2009;151(10):716–726. doi: 10.7326/0003-4819-151-10-200911170-00008. W-236. [DOI] [PubMed] [Google Scholar]