Abstract
Porphyria cutanea tarda (PCT) arises from decreased hepatic activity of uroporphyrinogen decarboxylase (UROD). Both genetic and environmental factors interplay in the precipitation of clinically overt PCT, but these factors may vary between different geographic areas. Decreased activity of UROD in erythrocytes was used to identify patients with UROD mutations among a group of 130 Spanish PCT patients. Nineteen patients (14.6%) were found to harbor a mutation in the UROD gene. Eight mutations were novel: M1I, 5del10, A22V, D79N, F84I, Q116X, T141I and Y182C. Five others were previously described: F46L, V134Q, R142Q, P150L and E218G. The new missense mutations and P150L were expressed in Escherichia coli. D79N and P150L resulted in proteins that were localized to inclusion bodies. The other mutations produced recombinant proteins that were purified and showed reduced activity (range: 2.3–73.2% of wild type). These single amino acid changes were predicted to produce complex structural alterations and/or reduced stability of the enzyme. Screening of relatives of the probands showed that 37.5% of mutation carriers demonstrated increased urinary porphyrins. This study emphasizes the role of UROD mutations as a strong risk factor for PCT even in areas where environmental factors (hepatitis C virus) have been shown to be highly associated with the disease.
Keywords: porphyria, UROD gene, UROD protein
The porphyrias are a group of metabolic diseases that arise from disturbances in heme biosynthesis (1). Porphyria cutanea tarda (PCT; OMIM 176100), the most common form, arises from a deficiency of uroporphyrinogen decarboxylase (UROD), the fifth enzyme of the heme synthesis pathway (2). In overt PCT, there is accumulation of uroporphyrinogen and other polycarboxylated intermediates in the liver that are oxidized to the corresponding porphyrins. These are then released from the liver and circulate in plasma where they become phototoxic on sun-exposed areas, leading to formation of the bullous lesions and hypertrichosis associated with the clinical phenotype of PCT (3).
At least two variants of the disease exist, designated types I and II. The most common form is type I (sporadic PCT, S-PCT) in which UROD deficiency is restricted to the liver. In type II (familial PCT, F-PCT), transmitted as an autosomal dominant trait, mutations in the UROD gene are present and enzyme activity is reduced in all tissues. Variable clinical expression is seen in F-PCT and in the absence of aggravating factors is rarely sufficient to trigger an active phenotype.
Several environmental factors have been shown to precipitate overt clinical PCT in both F-PCT and S-PCT, including alcohol abuse, chronic hepatitis C virus (HCV) infection and use of oral estrogens (3). Mutations in the hemochromatosis gene (HFE) facilitate hepatic siderosis that is recognized as a significant risk factor for developing PCT (4).
In overt PCT, the catalytic activity of UROD in the liver decreases to ~20% of normal without an apparent change in protein level. Similar observations have been reported in chemically induced porphyria in rodents. The conclusion from these studies was that a heat-stable competitive inhibitor of UROD is formed in vivo by a cytochrome P450-dependent mechanism (5, 6).
The human UROD protein (accession number NP_000365) is a cytosolic homodimer that catalyzes the sequential decarboxylation of four acetate chains of uroporphyrinogen to form the tetracarboxylic coproporphyrinogen. The amino acid sequence is highly conserved among species, and the crystal structures of both human and tobacco UROD have been solved (7, 8). The two structures are very similar with respect to the overall fold.
The human UROD gene has been cloned, sequenced and mapped to the short arm of chromosome 1 (1p34). The gene is ~3.4 kb and has 10 exons (9). Approximately 70 different mutations have been identified in UROD that associate with F-PCT or the homozygous variant of PCT named hepatoerythropoietic porphyria [HEP; (10)].
In this study, we examined a series of Spanish PCT patients, mainly from the northeastern Mediterranean area (Barcelona). In this region, the high prevalence of chronic HCV infection has been shown to significantly contribute to the appearance of S-PCT (11). Among this cohort of PCT patients, we characterized new mutations in the UROD gene, studied the pathogenicity of the new mutants and explored the complex structural and functional consequences of single amino acid substitutions in the UROD protein. We also screened family members in search of silent mutation carriers that may present alterations in the porphyrin excretion profile.
Materials and methods
Subjects studied
We studied 130 patients with suspected PCT who were referred to the Dermatology Unit, Hospital Clinic of Barcelona (Spain) during the period 2004–2007 for diagnosis or follow-up. There were 100 men and 30 women, Spanish Caucasians, living mainly in the area of Barcelona. A diagnosis of PCT was established on clinical grounds and confirmed by biochemical analysis of urine and feces. The mean age was 61 (range: 25–86 years). The average age at the clinical onset of disease was 44.8 (range: 22–74 years), with no significant differences between men and women.
All patients showed a positive response to standard treatment of PCT. A clinical history and a lifestyle questionnaire were given to all patients with data including age at onset of the cutaneous symptoms, associated diseases, family history, estrogen use and drug and alcohol consumption. Additional biochemical and genetic data for most of these patients were available including serum markers of HCV infection, markers of hepatic iron status and presence or absence of mutations in the HFE gene (C282Y and H63D). All patients were asked to enroll in a study that would include further biochemical analysis of urine porphyrins, erythrocyte UROD activity and sequencing of the UROD gene.
Additionally, 39 relatives of 12 probands were also genotyped and their urinary porphyrins were examined.
The study was conducted in accordance with the Declaration of Helsinki principles and was approved by the Ethics Committee of the Hospital Clinic of Barcelona. Written consent was signed by all patients and relatives studied.
Biochemical characterization of PCT in patients and relatives
Urinary porphyrin excretion pattern was analyzed by reverse-phase high-pressure liquid chromatography (HPLC) according to Lim and Peters (12).
Separation of porphyrins and isomers was achieved using an analytical column of 250 × 4.6 mm, 5-µm particle size (BDS-Hypersil; Shandon HPLC, Cheshire, UK). Each porphyrin and isomer fraction was quantified independently in urine and referred to nmol/mmol creatinine. Urinary porphyrin excretion profiles were classified as being either normal (total porphyrins in urine <35 nmol/mmol creatinine and normal distribution: coproporphyrin III > I, coproporphyrin I + III > uroporphyrins, and heptacarboxyl porphyrin III<0.2 nmol/mmol creatinine) or abnormal [total porphyrins >35 nmol/mmol creatinine and significant deviations from the normal distribution (13)].
Erythrocytes UROD activity
UROD activity in erythrocytes was measured essentially according to McManus et al. (14). Briefly, pentacarboxyl porphyrin I (Porphyrin Products, Logan, Utah) was reduced to pentacarboxylic porphyrinogen I (PPI) with a sodium mercury amalgam (Sigma-Aldrich), and red blood cells lysates were incubated adding PPI as a substrate of UROD, for 30 min at pH 6.0, 37°C, under nitrogen. The substrate concentration used was ~10-fold greater than the Km of UROD for PPI. The reaction was stopped by adding 0.5 ml of a 1:1 mixture of trichloroacetic acid (10%) and dimethyl sulfoxide containing 0.7 µmol of mesoporphyrin per liter as internal standard. The amount of coproporphyrinogen formed from PPI was assessed by HPLC according to the method described above. UROD activity is reported as pmoles of coproporphyrinogen formed per hour per mg of hemoglobin (U/mgHb).
Sequencing of the UROD loci
Genomic DNA was extracted from whole blood samples following the salting-out procedure (15). We analyzed exons 1–10 of the UROD gene and the associated splice donor and acceptor sites. The primers used for exon amplification are available from the authors.
Polymerase chain reaction products were purified using the GFX™ PCR DNA and gel band purification kit (Amersham Biosciences, Uppsala, Sweden) and automatically sequenced using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA) and an ABI3100 automatic sequencer (Applied Biosystems). Direct sequencing was performed using primers available from the authors.
Expression of UROD proteins
The expression plasmid pHT77 contains the human UROD complementary DNA under the control of the T7 inducible promoter (16). Missense mutations in the UROD gene identified in this study were introduced into pHT77 using the QuikChange II Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA). The mutations were confirmed by sequencing.
UROD proteins were expressed in Rosetta2 (DE3) pLysS (Novagen, Madison, WI) in 2-l cultures by autoinduction. Cells were harvested and resuspended in 50 mM Tris at pH 8.0 and 100 mM NaCl and allowed to lyse for 30 min on ice. The lysate was sonicated for three times for 30 s for 1.5 min total. The cell debris was removed by centrifugation and the supernatant was filtered through a 0.8-µm filter. The UROD was purified on a 1 ml Ni-NTA (Qiagen, Valencia, CA) column using a step gradient with buffers containing 50 mM NaPO4 at pH 7.0, 500 mM NaCl, 5% glycerol and 2.5 mM β-mercaptoethanol with or without 250 mM imidazole. Peak fractions containing UROD were pooled, concentrated to 1–6 mg/ml and dialyzed against 4 l 50 mM Tris at pH 6.8, 150 mM NaCl, 5% glycerol and 1 mM β-mercaptoethanol. Proteins were assayed for enzymatic activity as previously described (17).
Results
Identification of F-PCT patients
Erythrocyte UROD activity was measured in all PCT patients. Selection of candidates for sequencing the UROD loci and the identification of individuals harboring UROD mutations were performed as follows.
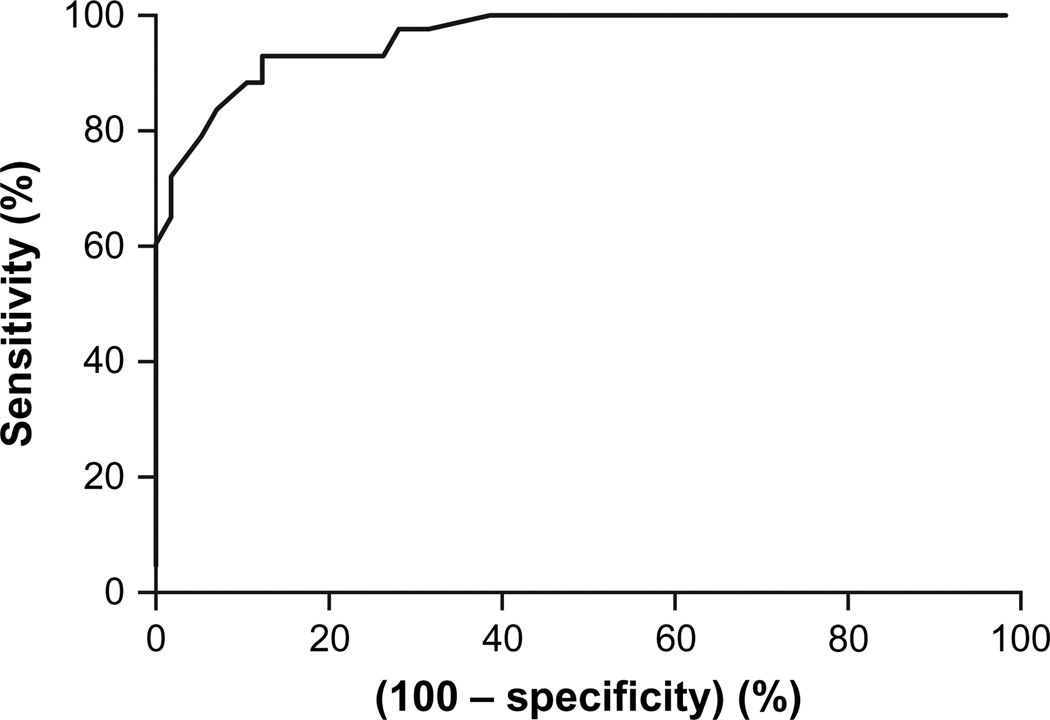
A preliminary receiver operating characteristic (ROC) curve was constructed for UROD activity in erythrocytes based on the UROD genotyping (presence or absence of mutation) as a reference (18). UROD activity of the first 20 individuals (including 10 patients and 10 family members) who were found to harbor a mutation within the UROD gene was plotted against enzyme activity of 21 control individuals who were negative for a mutant UROD allele (including 10 family members and 11 unrelated controls).
The area under the curve was 0.983 (SE: 0.016; 95% CI: 0.914–0.998). A provisional cutoff value for UROD activity was set at 38 U/mgHb (sensitivity: 96.7; 95% CI: 82.7–99.4; specificity: 88.6; 95% CI: 73.2–96.7). From there on, all new consecutive PCT patients, with erythrocyte UROD activity below the cutoff point, were sequenced (n = 6; all were found to be carriers). All the patients who showed UROD activity between 38 U/mgHb (cutoff) and 43 U/mgHb (cutoff + 15; sensitivity: 100; 95% CI: 88.3–100) were also sequenced (n = 25; 3 were found to be carriers). Fifteen additional patients with high UROD activity (all UROD >43 U/mgHb) were randomly selected and genotyped. None of these patients were found to be carriers of mutant UROD alleles. This allowed classification of the patients into F-PCT (n = 19; 12 women and 7 men; all genotyped and with a confirmed mutation) and S-PCT (n = 111; 18 women and 93 men; 37 genotyped and being wild type and the rest with high UROD activity and not genotyped). All the relatives (n = 39) were directly genotyped. At the conclusion of the study, a new ROC curve was constructed based on all individuals sequenced and with UROD measured (n = 95; Fig. 1). The cutoff point was adjusted and set at 39 U/mgHb (sensitivity: 93.00; 95% CI: 80.94–98.5; specificity: 87.7; 95% CI: 75.3–94.9).
Fig. 1.
Receiver operating characteristic curve for UROD activity in erythrocytes. Area under the curve (95% CI): 0.9625 (0.9314–0.9935).
Consequently, the proportion of F-PCT among this series of Spanish PCT patients was 14.6%. No patients with a family history of PCT and normal UROD activity (PCT type III) were identified in this study.
Identification of mutations within the UROD gene
Sequencing of the UROD loci of the F-PCT patients allowed identification of 13 different mutations within the UROD gene including 8 novel mutations (Table 1).
Table 1.
Mutations found in the UROD gene among porphyria cutanea tarda patients
| Mutation | Exon | Nucleotide position | Number of cases | Conserved | Reference |
|---|---|---|---|---|---|
| M1I | 1 | 3G>A | 1 | T | Present study |
| 5del10 | 1 | 5 | 1 | Present study | |
| A22V | 2 | 65C>T | 1 | L | Present study |
| F46L | 4 | 138T>I | 2 | H | Ged et al. (2002) |
| D79N | 4 | 235G>A | 1 | T | Present study |
| F84I | 4 | 250T>A | 1 | H | Present study |
| Q116X | 5 | 346C>T | 1 | Present study | |
| V134Q | 5 | 399-401TGC>CCA | 2 | L | Meguro et al. (1994) |
| T141I | 5 | 422C>T | 1 | L | Present study |
| R142Q | 5 | 425G>A | 2 | H | Cappellini et al. (2001) |
| P150L | 5 | 449C>T | 1 | H | Martinez et al. (2002) |
| Y182C | 6 | 645A>G | 1 | H | Present study |
| E218G | 7 | 653A>G | 4 | H | Méndez et al. (2007) |
H, highly conserved; L, low conservation among divergent species; T, totally conserved.
Expression of new missense mutations in prokaryotes
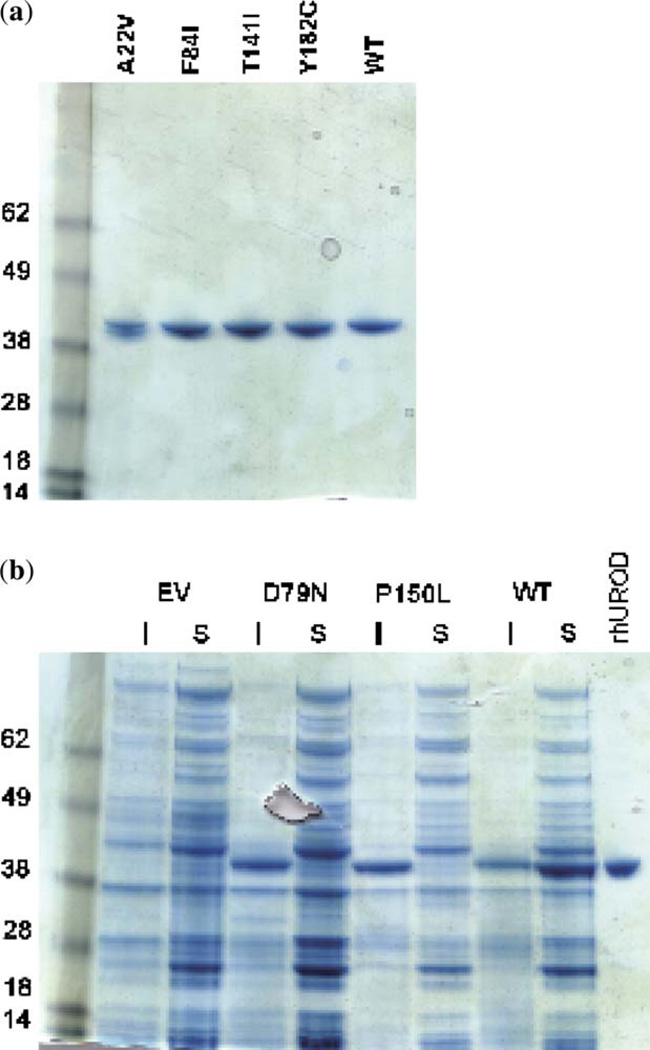
Previously uncharacterized missense mutations were cloned into an expression system and purified for additional analysis. The P150L mutation that had been initially found in Italy but not expressed in prokaryotes was incorporated into this analysis (19). Recombinant UROD proteins were recovered from four of the mutants: A22V, F84I, T141I and Y182C (Fig. 2a). Two of the mutants, D79N and P150L, were expressed in the Escherichia coli to significant levels; however, the proteins were not present in the soluble fraction and were not studied further (Fig. 2b). The activities of the mutant proteins were measured using uroporphyrinogen I and uroporphyrinogen III as substrates. The activities for each are shown in Table 2.
Fig. 2.
Expression and purification of UROD proteins in Escherichia coli. (a) Recombinant proteins were purified using an Ni-NTA resin and separated on a 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) gel. Recombinant human UROD runs at an apparent molecular weight of 42 kDa. The UROD without a mutation (wild type, WT) is run for comparison. (b) The insoluble pellet (I) and the soluble supernatant (S) were separated on a 10% SDS-PAGE gel; D79N, P150L and WT were compared with the purified recombinant human UROD (rhUROD). The UROD protein from D79N and P150L is present in the insoluble fraction, while WT is primarily present in the soluble fraction. EV, empty vector control.
Table 2.
Activity of UROD proteins
| Mutation | Uroporphyrinogen I activitya |
SD | Uroporphyrinogen III activityb |
SD |
|---|---|---|---|---|
| A22V | 62.6 | 12.3 | 73.2 | 14.5 |
| F84I | 1.7 | 1.3 | 2.3 | 1.4 |
| T141I | 21.3 | 2.9 | 26.7 | 4.0 |
| Y182C | 40.0 | 10.8 | 65.2 | 10.0 |
Activity of UROD mutant proteins toward uroporphyrinogen I.
Activity of UROD mutant proteins toward uroporphyrinogen III. Activity shown as percentage of wild-type activity.
Additional risk factors
Additional risk factors for PCT were investigated among this group of F-PCT patients. Twelve out of 19 patients (63%) presented at least one identifiable risk factor in addition to the UROD gene mutation. Three patients were heterozygous for the HFE C282Y allele and other three were homozygous for the H63D allele; three other patients presented anti-HCV antibodies in blood and other three confirmed the use of estrogens either for oral contraception or menopausal symptoms. Daily alcohol consumption >40 g was found in 6 out of 7 men and in 1 out of 12 women.
Comparison between F-PCT and S-PCT patients showed that the only environmental risk factor which was significantly different between both groups was chronic HCV infection (15% and 75%, respectively).
Family study
We genotyped 39 relatives belonging to 12 different F-PCT families. Mutations in UROD were detected in 16 of the relatives genotyped (41%). On examination, 6 of 16 (37.5%) revealed increased urinary porphyrins and a typical PCT pattern (Table 3). Only three of them presented dermatological manifestations, thus documenting the existence of a number of PCT family carriers with only biochemical abnormalities.
Table 3.
Screening of family members
| Family | Mutation | Number screeneda |
Number of carriersb |
Urinary porphyrinsc |
|---|---|---|---|---|
| 1 | V134Q | 5 | 3 | 0 |
| 2 | V134Q | 6 | 2 | 1 |
| 3 | R142Q | 4 | 1 | 0 |
| 4 | E218G | 2 | 1 | 0 |
| 5 | E218G | 1 | 0 | 0 |
| 6 | M1I | 2 | 2 | 2 |
| 7 | D79N | 3 | 1 | 0 |
| 8 | F46L | 1 | 0 | 0 |
| 9 | P150L | 2 | 2 | 1 |
| 10 | R142Q | 2 | 2 | 0 |
| 11 | A22V | 1 | 0 | 0 |
| 12 | F46L | 7 | 2 | 2 |
Number of family members genotyped.
Family members carriers of UROD mutation.
Family members with increased urinary porphyrins and porphyria cutanea tarda pattern.
Discussion
The proportion of PCT patients classified as F-PCT, based on the presence of a mutant UROD allele, has differed based on geographic and ethnic factors. According to published reports that include UROD genotyping, this may range from 24% found among Danish PCT patients to 50% found among a series of patients from Chile (20, 21). Our study found a somewhat lower percentage of F-PCT (14.6%), which may be in accordance with previous studies that found a high prevalence of HCV infection among the general Spanish population and a probable major role of HCV infection in the triggering of PCT in this geographic zone (11, 22).
The prevalence of UROD mutations among the general population is unknown but is most likely very low. Therefore, the finding of UROD mutations in 14.6% of our patients, even if lower to that found in other studies, clearly shows that harboring a UROD mutation is a major risk factor for developing PCT. Moreover, the existence of a significant number of family carriers with increased urinary porphyrins (Table 3) reinforces this notion. However, given the frequent coexistence of other risk factors among the F-PCT patients (at least 63% in our series), the inherited UROD mutation may be viewed as one factor among a complex series of events that ultimately lead to the clinical precipitation of the disease.
One of the previously reported mutations, the E218G mutation, has been observed in Spanish PCT patients unrelated to the four probands reported in this study, thus suggesting a relatively high frequency of this mutation among Spanish F-PCT carriers (23).
The F46L mutation, found in two independent probands of our series, had been previously reported in patients with HEP and also in a PCT patient from Chile (21, 24, 25). One study reported an HEP patient with a unique urinary excretion pattern with a predominance of pentacarboxylated porphyrins (25). This observation was not confirmed in the F-PCT patients reported in this study. Both our patients with the F46L mutation presented with typical urine porphyrin profiles with a predominance of uroporphyrin and heptacarboxylated porphyrins.
The V134Q mutation has also been reported both in HEP and in PCT patients (20, 26, 27). Its appearance in a different series of patients is also striking as the amino acid change involved is distant from the enzyme active site and the purified recombinant UROD yielded a nearly normal activity when engineered in prokaryotes (27). This suggests that this mutation creates a UROD protein that may be rapidly degraded in vivo.
Mutations initially identified in HEP patients were considered to rarely precipitate overt disease when present as simple heterozygotes (3). One study reported the existence of an HEP pedigree member who was heterozygous for the F46L allele and presented a normal erythrocyte UROD activity (24).
Nonetheless, our results as a whole tend to confirm an increasing number of HEP mutations being found also among heterozygous F-PCT patients.
The M1I substitution (c.3G>A) is a novel mutation found in this study, although a different change in the same codon, M1T, has been previously reported (20). The absence of an initial methionine codon within the messenger RNA (mRNA) Kozac consensus sequence is predicted to lead to a loss of translation from the mutant allele.
Nucleotide sequencing of a proband identified a C>T substitution at nucleotide position 346 in exon 5 (Table 1). The predicted effect is a change from glutamine at amino acid position 116 to a stop codon (CAG > TAG), designated Q116X. This may yield a truncated protein of 115 amino acids or nonsense-mediated mRNA decay. A similar effect can be predicted from a deletion of 10 bp initiated at the fifth nucleotide of exon 1 (5del10; Table 1).
Our study identified several new missense mutations. Characterization of these (plus the already reported P450L) (28) included heterologous expression and biochemical characterization.
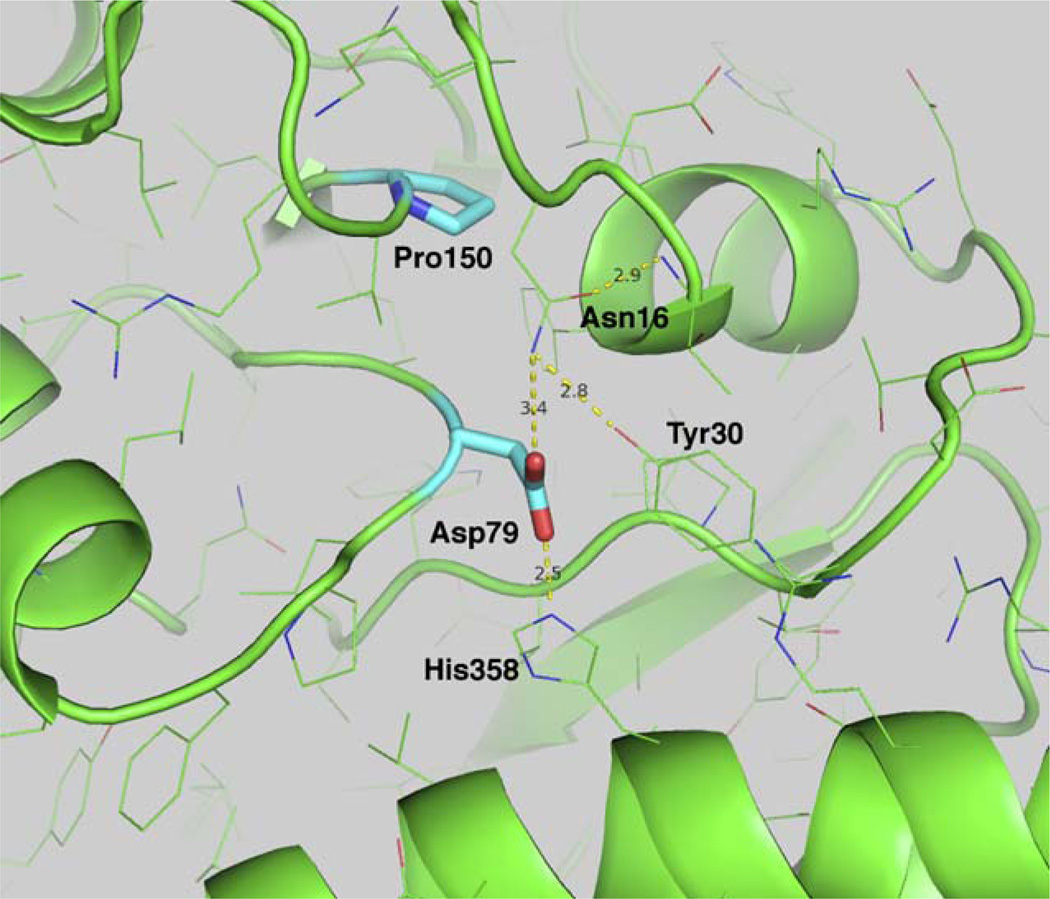
The two mutants that failed to produce a soluble protein are located at the base of the (β/α)8-barrel. The aspartic acid at position 79 (D79N mutant) is totally conserved across 13 species of UROD (7). The extensive hydrogen bonding network of D79, to H358, N16 and Y30 is important to correctly position the terminus of the helix as it transitions into the β-sheet. Both carboxylate oxygen atoms participate in the formation of the hydrogen bonding network (Fig. 3). The proline in position 150 is also situated at the base of an α-helix that turns sharply up into one of the β-sheets that forms the core of the barrel. Proline at position 150 (P150L) is highly conserved, and disruption of either of these two leads to a misfolded protein that is trafficked to inclusion bodies.
Fig. 3.
Location of UROD mutants at the base of the TIM barrel. Mutants D79N and P150L destabilize the core of the barrel and lead to misfolding of the proteins. The hydrogen bonding distances of D79 are shown as dashed yellow lines.
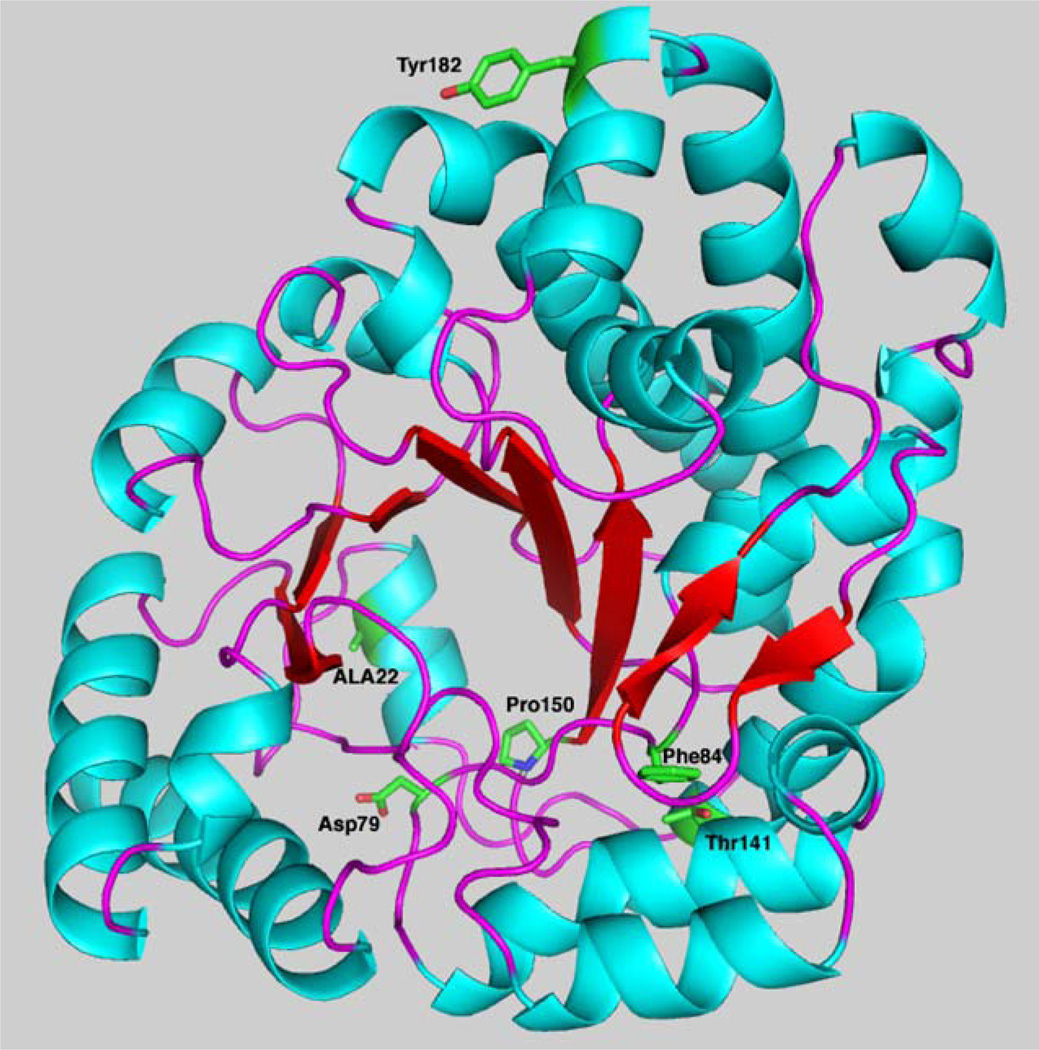
The four remaining mutants produce a protein that is soluble but has reduced catalytic activity (Table 2). The alteration of alanine 22 to a valine is a minor structural shift (Fig. 4). The residue is buried in at the base of the barrel, and the additional bulk of the valine residue may be sufficient to alter the active site geometry. The minimal loss in catalytic activity supports this; however, this alteration may decrease the stability of the protein thereby reducing the total amount of UROD available for catalysis. Residue 84 is one of several phenylalanines that forms the base of the active site (Fig. 4). This network of hydrophobic aromatic residues forms a platform that allows the substrate to adopt the domed configuration that positions the four pyrrole nitrogens to hydrogen bond to aspartic acid 86.
Fig. 4.
Location of UROD missense mutants identified in this study. A monomer of UROD with β-sheets in red, α-helices in blue and stands in purple is shown in the cartoon. The side chains of amino acids mutated are labeled and shown in green.
Threonine in position 141 is positioned with the side chain exposed to the interior of the structure. The larger sidespin of isoleucine is unable to be accommodated in this environment. Compensatory changes in the structure would likely be required to adjust to the bulk of the isoleucine. The tyrosine in position 182 is located on the dimer interface and has few inter- or intramolecular interactions. The loss in catalytic activity may be due to subtle alterations in the structure that affect interaction between monomers.
The family study showed that a significant proportion of mutation carrier relatives present abnormal and increased urinary excretion of porphyrins. Only a few of them presented with clinical symptoms, suggesting that the mutations may induce only subtle biochemical manifestations in some individuals. The study also confirms that a high proportion of UROD mutation carriers never develop overt PCT symptoms and that the factors that may lead to clinical expression in the patients remain in many cases obscure.
Acknowledgements
We are grateful to Maria Sala for their skilled technical assistance. This study was supported by Spanish ‘Fondo de Investigación Sanitaria’ (grant PI06/0150) and the NIH RO-1 DK20503 to J. P.
Footnotes
The authors state no conflict of interest.
References
- 1.Kappas A, Sassa S, Galbraith RA, Nordmann Y. The porphyrias. In: Scriver CR, Beaudet AL, Sly WS, Valle D, editors. The metabolic and molecular bases of inherited disease. Vol. 2. New York, NY: McGraw-Hill; 1995. pp. 2103–2160. [Google Scholar]
- 2.Kushner JP. The enzymatic defect in porphyria cutanea tarda. N Engl J Med. 1982;306(13):799–800. doi: 10.1056/NEJM198204013061309. [DOI] [PubMed] [Google Scholar]
- 3.Elder GH. Porphyria cutanea tarda. Semin Liver Dis. 1998;18(1):67–75. doi: 10.1055/s-2007-1007142. [DOI] [PubMed] [Google Scholar]
- 4.Bulaj ZJ, Phillips JD, Ajioka RS, et al. Hemochromatosis genes and other factors contributing to the pathogenesis of porphyria cutanea tarda. Blood. 2000;95(5):1565–1571. [PubMed] [Google Scholar]
- 5.Smith AG. Porphyria caused by chlorinated AH receptor ligands and associated mechanisms of liver injury and cancer. In: Kadish KM, Smith KM, Guilard R, editors. The porphyrin handbook: medical aspects of porphyria. Vol 14. San Diego, CA: Academic Press; 2003. pp. 169–210. [Google Scholar]
- 6.Phillips JD, Bergonia HA, Reilly CA, Franklin MR, Kushner JP. A porphomethene inhibitor of uroporphyrinogen decarboxylase causes porphyria cutanea tarda. Proc Natl Acad Sci U S A. 2007;104(12):5079–5084. doi: 10.1073/pnas.0700547104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whitby FG, Phillips JD, Kushner JP, Hill CP. Crystal structure of human uroporphyrinogen decarboxylase. EMBO J. 1998;17(9):2463–2471. doi: 10.1093/emboj/17.9.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martins BM, Grimm B, Mock HP, Huber R, Messerschmidt A. Crystal structure and substrate binding modeling of the uroporphyrinogen-III decarboxylase from Nicotiana tabacum. Implications for the catalytic mechanism. J Biol Chem. 2001;276(47):44108–44116. doi: 10.1074/jbc.M104759200. [DOI] [PubMed] [Google Scholar]
- 9.Romana M, Dubart A, Beaupain D, Chabret C, Goossens M, Romeo PH. Structure of the gene for human uroporphyrinogen decarboxylase. Nucleic Acids Res. 1987;15(18):7343–7356. doi: 10.1093/nar/15.18.7343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.The Human Gene Mutation Database (HGMD), Institute of Medical Genetics in Cardiff. [Retrieved July 2008]; from http://www.hgmd.cf.ac.uk/ac/index.php. [Google Scholar]
- 11.Herrero C, Vicente A, Bruguera M, et al. Is hepatitis C virus infection a trigger of porphyria cutanea tarda? Lancet. 1993;341(8848):788–789. doi: 10.1016/0140-6736(93)90562-u. [DOI] [PubMed] [Google Scholar]
- 12.Lim CK, Peters TJ. Urine and faecal porphyrin profiles by reversed-phase high-performance liquid chromatography in the porphyrias. Clin Chim Acta. 1984;139(1):55–63. doi: 10.1016/0009-8981(84)90192-x. [DOI] [PubMed] [Google Scholar]
- 13.Smith SG, Rao KR, Jackson AH. The prophyrins of normal human urine, with a comparison of the excretion pattern in porphyria cutanea tarda. Int J Biochem. 1980;12(5–6):1081–1084. doi: 10.1016/0020-711x(80)90216-5. [DOI] [PubMed] [Google Scholar]
- 14.McManus J, Blake D, Ratnaike S. An assay of uroporphyrinogen decarboxylase in erythrocytes. Clin Chem. 1988;34(11):2355–2357. [PubMed] [Google Scholar]
- 15.Miller SA, Dykes DD, Polesky H. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16(3):12–15. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Phillips JD, Whitby FG, Kushner JP, Hill CP. Characterization and crystallization of human uroporphyrinogen decarboxylase. Protein Sci. 1997;6(6):1343–1346. doi: 10.1002/pro.5560060624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41(1):207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 18.Zweig MH, Campbell G. Receiver-operating characteristic (ROC) plots: a fundamental evaluation tool in clinical medicine. Clin Chem. 1993;39(4):561–577. [PubMed] [Google Scholar]
- 19.Cappellini MD, Martinez di Montemuros F, Tavazzi D, et al. Seven novel point mutations in the uroporphyrinogen decarboxylase (UROD) gene in patients with familial porphyria cutanea tarda. Hum Mutat. 2001;17(4):350. doi: 10.1002/humu.35. [DOI] [PubMed] [Google Scholar]
- 20.Christiansen L, Brøns-Poulsen J, Hørder M, Brock A, Petersen NE. Expression and characterization of six clinically relevant uroporphyrinogen decarboxylase gene mutations. Scand J Clin Lab Invest. 2005;65(3):227–235. doi: 10.1080/00365510510013631. [DOI] [PubMed] [Google Scholar]
- 21.Poblete-Gutiérrez P, Mendez M, Wiederholt T, et al. The molecular basis of porphyria cutanea tarda in Chile: identification and functional characterization of mutations in the uroporphyrinogen decarboxylase gene. Exp Dermatol. 2004;13(6):372–379. doi: 10.1111/j.0906-6705.2004.00163.x. [DOI] [PubMed] [Google Scholar]
- 22.Bruguera M, Forns X. Hepatitis C in Spain. Med Clin (Barc) 2006;127(3):113–117. doi: 10.1157/13090276. [DOI] [PubMed] [Google Scholar]
- 23.Méndez M, Poblete-Gutiérrez P, García-Bravo M, et al. Molecular heterogeneity of familial porphyria cutanea tarda in Spain: characterization of 10 novel mutations in the UROD gene. Br J Dermatol. 2007;157(3):501–507. doi: 10.1111/j.1365-2133.2007.08064.x. [DOI] [PubMed] [Google Scholar]
- 24.Ged C, Ozalla D, Herrero C, et al. Description of a new mutation in hepatoerythropoietic porphyria and prenatal exclusion of a homozygous fetus. Arch Dermatol. 2002;138(7):957–960. doi: 10.1001/archderm.138.7.957. [DOI] [PubMed] [Google Scholar]
- 25.Armstrong DK, Sharpe PC, Chambers CR, Whatley SD, Roberts AG, Elder GH. Hepatoerythropoietic porphyria: a missense mutation in the UROD gene is associated with mild disease and an unusual porphyrin excretion pattern. Br J Dermatol. 2004;151(4):920–923. doi: 10.1111/j.1365-2133.2004.06101.x. [DOI] [PubMed] [Google Scholar]
- 26.Meguro K, Fujita H, Ishida N, et al. Molecular defects of uroporphyrinogen decarboxylase in a patient with mild hepatoerythropoietic porphyria. J Invest Dermatol. 1994;102(5):681–685. doi: 10.1111/1523-1747.ep12374134. [DOI] [PubMed] [Google Scholar]
- 27.Phillips JD, Parker TL, Schubert HL, Whitby FG, Hill CP, Kushner JP. Functional consequences of naturally occurring mutations in human uroporphyrinogen decarboxylase. Blood. 2001;98(12):3179–3185. doi: 10.1182/blood.v98.12.3179. [DOI] [PubMed] [Google Scholar]
- 28.Martinez di Montemuros F, Di Pierro E, Patti E, et al. Molecular characterization of porphyrias in Italy: a diagnostic flow-chart. Cell Mol Biol (Noisy-le-grand) 2002;48(8):867–876. [PubMed] [Google Scholar]