Abstract
The gut mucosa hosts large numbers of activated lymphocytes, exposed to stimuli from diet, microbiota and pathogens. Although CD4+ T cells are crucial for defense, intestinal homeostasis precludes exaggerated response towards luminal contents, harmful or not. We investigated mechanisms used by CD4+ T cells to avoid excessive activation within the intestine. Using genetic tools to label and interfere with T cell development transcription factors we show that CD4+ T cells acquired CD8-lineage transcription factor Runx3 while losing CD4-lineage transcription factor ThPOK along with their TH17 differentiation and colitogenic potential, in a transforming growth factor-β (TGF-β) and retinoic-acid-dependent manner. These results show a remarkable plasticity in the CD4+ T cell lineage that allows chronic exposure to luminal antigens without pathological inflammation.
INTRODUCTION
Environmental cues are part of differentiation processes of all cell types, including T cells. Environmental cues such as cytokine milieu, influence mature CD4+ T cells to differentiate into various subsets that have multiple functional roles in the periphery, including proper control of infections (helper T cells) and prevention of progressive immune activation (regulatory T cells or Treg). On the other hand, mature CD8+ T cells are primarily cytotoxic (CTL), being essential in the protection against intracellular pathogens.
The transcription factor ThPOK (also known as Zbtb7b and cKrox) drives CD4+ T cell development from double-positive precursors while CD8+ T cell development primarily requires the expression of Runx3 and the zinc-finger transcription factor MAZR (also called PATZ1 or Zfp278)1–3. These transcription factors bind to each other and their dynamic interaction ultimately determines thymic T cell fate. In this regard, ablation of the Runx complex in developing thymocytes results in derepression of the Zbtb7b (here called Thpok) gene at the double-positive stage and a severe reduction of mature CD8 single-positive T cells in the periphery4. Conversely, inactivation of Thpok by conditional deletion, hypomorphic expression or loss-of-function HD mutation results in a near absence of peripheral CD4+ T cells5–7.
In the intestine, where a large amount of diverse antigens can be constantly perceived as stimuli, the immune system developed particular pathways to deal with this rich luminal content without generating progressive inflammation8. While Treg and other regulatory cells can be found in the intestinal tissue, not much is known about cell-intrinsic mechanisms that regulate CD4+ T helper function at this environmental intersection.
Peripheral mature CD4+ and CD8+ T cells express ThPOK and Runx3, respectively, in a mutually exclusive fashion3,5. However, ThPOK expression by CD4+ T cells may not be as stable as previously thought, since intestinal CD4+ T cells show consistent post-thymic downregulation of ThPOK9. To address whether such pattern was associated with changes in Runx3 expression by intestinal CD4+ T cells, we analyzed ThPOK and Runx3 expression using green fluorescent protein (GFP) or yellow fluorescent protein (YFP)-knockin reporter strains, respectively3,5. We observed that both reduced expression of ThPOK and high expression of Runx3 were associated with changes toward the CD8 lineage and reduced TH17 differentiation. ThPOK loss-of-function experiments resulted in dampening of CD4+ T cell inflammatory potential, although it did not directly regulate TH17 differentiation. On the other hand, Runx3 loss-of-function resulted in higher expression of ThPOK by intestinal CD4+ T cells and enhanced TH17 differentiation. These experiments provide mechanistic evidence of how transcription factors involved in T cell lineage choice continue to play a decisive role in cell function in the periphery.
RESULTS
Reciprocal expression of ThPOK and Runx3 by CD4+ T cells
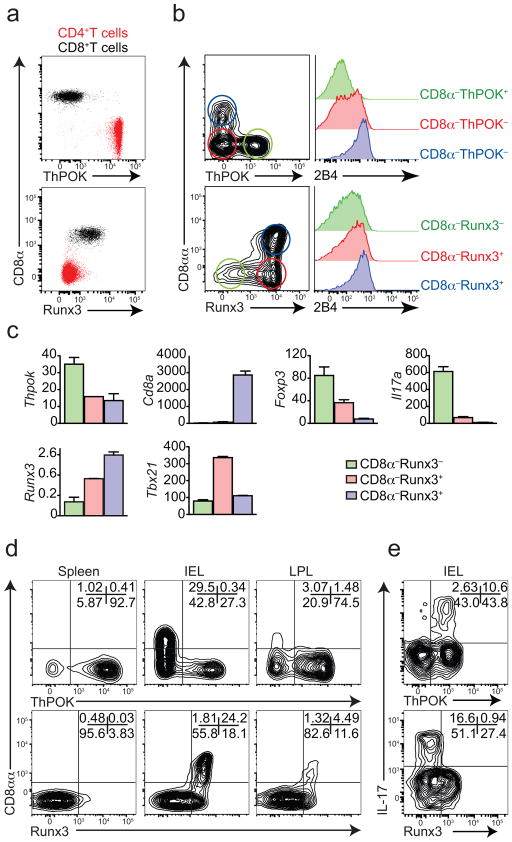
We used knockin reporters for both Thpok and Runx3 and found that while these transcription factors are expressed by CD4+ and CD8+ T cells, respectively, in peripheral tissues (Fig. 1a), intestinal CD4+ T cells do not follow the same pattern (Fig. 1b). The majority of CD4+ T intraepithelial lymphocytes (IELs) expressed modest ThPOK but high amounts of the distal promoter-derived long isoform of Runx3 (ref. 5) (Fig. 1b, c). Upregulation of Runx3 by CD4+ T cells was directly associated to CD8αα expression (CD8α+CD8β−) (Fig. 1b, c). Furthermore, acquisition of Runx3 paralleled upregulation of the natural killer (NK)- and CTL-related molecule 2B4 (CD244) (Fig. 1b, c) and also Tbx21 (encoding T-bet). In contrast, Runx3hi CD4 IEL showed low expression of Thpok, Foxp3 and interleukin 17A (Il17a) (Fig. 1c), resembling gene signature of CD4+ IEL populations that lose ThPOK expression9, which suggests that this pathway diverts CD4+ T cells towards IELs or innate-like cytotoxic T cells10–12. To extend this analysis to a pro-inflammatory setting, we used the T cell-transfer model of colitis13. Pure naïve (CD25−CD62LhiCD44lo) CD4+ T cells were sorted from Thpok-GFP (as ThPOK+ naïve CD4+ T cells) or Runx3-YFP (as Runx3− naïve CD4+ T cells) reporter animals and adoptively transferred into Rag1−/− animals. The expansion of CD4+ T cell populations in the lymphopenic host induces severe colitis and inflammatory cytokine expression by CD4+ T cells, particularly interferon-γ (IFN-γ) and IL-1713,14. Consistent with a post-thymic event, CD4+ T cells upregulated Runx3 upon migration to the intestinal environment, particularly in the intraepithelial compartment (Fig. 1d). As observed in the steady state, CD8αα expression by CD4+ T cells was restricted to ThPOKlo or Runx3hi populations (Fig. 1d). Additionally, only cells that sustained high amounts of ThPOK expression9, or expressed little or no Runx3 were able to produce IL-17 (Fig. 1e). This data indicates that gut environmental cues lead to a distinct program of CD4+ T cells, in which loss of ThPOK and acquisition of Runx3 are both associated with a suppression of features of CD4+ T helper cells and differentiation towards a CTL or IEL-like phenotype.
Figure 1. ThPOK and Runx3 are reciprocally regulated at the intestinal tissue.
(a, b) ThPOK or Runx3 expression by CD45+TCRβ+ cells from spleen (a) or CD45+TCRβ+CD4+ cells from small intestine epithelium (intraepithelial lymphocytes, IEL) (b) of naive Thpok-GFP (top) or Runx3-YFP (bottom) reporter mice. On the respective right sides, histograms for 2B4 (CD244) expression by ThPOK+CD8α−CD4+, ThPOK−CD8α CD4+ and ThPOK−CD8α+CD4+ (top) or Runx3+CD8α−CD4+, Runx3−CD8α CD4+ and Runx3−CD8α+CD4+ (bottom). Plots are representative of at least three independent experiments (n=5 per group). (c) Relative mRNA expression of genes in sIEL sorted from naïve Runx3-YFP reporter as in b. Pooled data from n=3 representative of two independent experiments with similar results (error bars, s.e.m. of triplicates). (d, e) Sorted naïve CD4+ T cells isolated from spleen of Thpok-GFP (top) or Runx3-YFP (bottom) were adoptively transferred to Rag1−/− recipients and mice were analyzed 40 days after transfer. (d) CD8αα and ThPOK (top) or Runx3 (bottom) expression by CD45+TCRβ+CD4+ from spleen, small intestine IEL and lamina propria (LPL). (e) ThPOK (top) or Runx3 (bottom) and intracellular IL-17A expression by CD45+TCRβ+CD4+ from small intestine IELs. Plots are representative of three independent experiments (n=5 per group).
TGF-β and RA signaling in intestinal CD4+ T cells
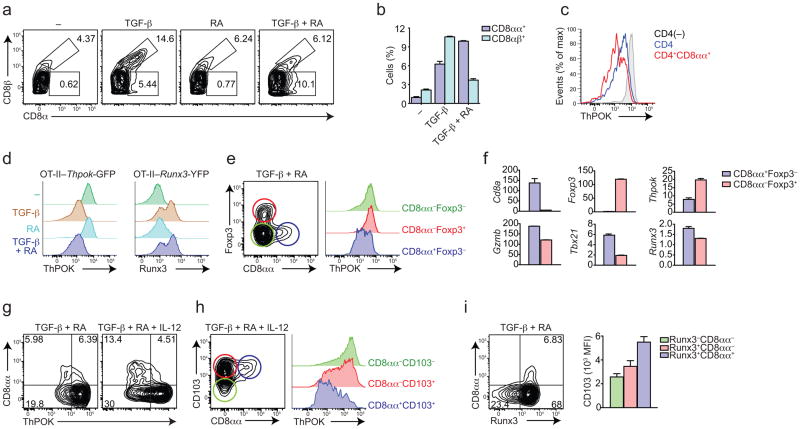
The integrin CD103 (αE) is highly expressed by IELs and the heterodimer αEβ7 binds to E-cadherin expressed by intestinal epithelial cells. CD103 expression can be modulated by TGF-β signaling, via Runx3 (ref. 15,16). Retinoic acid (RA) is another environmental factor that modulates CD4+ T cell differentiation and migration to the intestine, and it is known to synergize with TGF-β for example by inducing Foxp3-expressing Treg (iTreg) cells, and by enhancing T-bet expression in TGF-β plus IL-6 conditions, suppressing TH17 differentiation17–19. We used both in vitro and in vivo models to evaluate the environmental cues involved in the modulation of ThPOK and Runx3 expression by CD4+ T cells. Initially, ovalbumbin (OVA)-specific TCR transgenic CD4+ T cells (OT-II) were cultured with splenic dendritic cells (DCs) and OVA peptide in the presence of soluble cytokines. As previously described20, exogenous TGF-β induced some expression of CD8α in CD4+ T cells (Fig. 2a). However, while TGF-β preferentially induced CD8αβ, the combination of TGF-β and RA induced mostly CD8αα (CD8β−) OT-II cells (Fig. 2a, b). To address whether these factors were involved in the peripheral modulation of T cell-lineage transcription factors, we interbred OT-II mice with Thpok-GFP or Runx3-YFP mice. Indeed, in vitro induced CD4+CD8αα (CD8β−) cells showed reduced ThPOK expression (Fig. 2c). Addition of either TGF-β alone or the combination of TGF-β and RA efficiently suppressed ThPOK expression while enhancing that of Runx3 (Fig. 2d). The induction of CD4+CD8αα paralleled induction of Foxp3, mostly in a reciprocal manner (Fig. 2e). However, only CD4+CD8αα cells showed less ThPOK, while iTreg cells maintained high amounts of Thpok-GFP expression (Fig. 2e, f). Consistent with their naturally occurring IEL counterparts, in vitro TGF-β plus RA-induced CD4+CD8αα cells expressed more Tbx21, Gzmb (encoding granzyme B) and Runx3 (Fig. 2f). Since intestinal T cells are chronically stimulated, we restimulated OT-II cells initially primed with TGF-β plus RA. To address whether increased Tbx21 expression by ThPOKlo or Runx3hi CD4+ T cells correlated with deviation towards CD4+CD8αα phenotype, we performed secondary stimulation in the presence of IL-12, a potent inducer of T-bet21. We found that this condition mirrored intestinal CD4+ T cells found in naïve animals, in which a substantial part of CD4+ T cells downmodulated ThPOK and upregulated CD8αα, CD103 and Runx3 (Fig. 2h, i). Additionally, overexpression of long-form Runx3 in TH17 cells resulted in increased Tbx21 expression while enforced Tbx21 expression reciprocally resulted in upregulation of long-form Runx3 by TH17 cells (Supplementary Fig. 1a, b). In either case, IL-17 production was suppressed (Supplementary Fig. 1c, d).
Figure 2. TGF-β and RA concomitantly induce Treg and ThPOKloRunx3hiCD4+ T cells.
Sorted naïve Vα2+CD4+ T cells isolated from reporter OT-II mice (Thpok-GFP, a-d, g and h; Runx3-YFP, d and i; Thpok-GFP–Foxp3-RFP, e and f) were co-cultured with DCs, OVA peptide and indicated cytokines. (a, b) CD8αα and CD8αβ expression by T cells. Graph depicts the average frequency (error bars, s.e.m. of duplicates). (c) ThPOK expression by T cells stimulated without additional cytokines (grey) or by gated CD4 (blue) and CD4+CD8αα (red) stimulated in the presence of TGF-β. (d) ThPOK (left) or Runx3 (right) expression by T cells. (e) CD8αα and Foxp3 expression by T cells. Histograms show ThPOK expression by gated CD8α+Foxp3− (blue), CD8α−Foxp3+ (red), and CD8α−Foxp3− (green) populations. (f) Relative mRNA expression of genes in cells gated from e (error bars, s.e.m. of duplicates). (g, h) Cells were cultured in the presence of TGF-β+RA and re-stimulated in the presence of TGF-β+RA or IL-12. CD8αα and ThPOK (g) and CD103 (h) expression by T cells. Histograms show ThPOK expression by gated CD8α+CD103+ (blue), CD8α−CD103+ (red) and CD8α−CD103− (green) populations. (i) Runx3 and CD8αα expression by T cells. Graph depicts the average (error bars, s.e.m. of duplicates) of CD103 mean-fluorescence-intensity (MFI) from gated cells. Data are representative of at least three independent experiments. How many mice per independent experiment? P values for the graphs – if only three mice, might be better to show the individual mouse values rather than as bars.
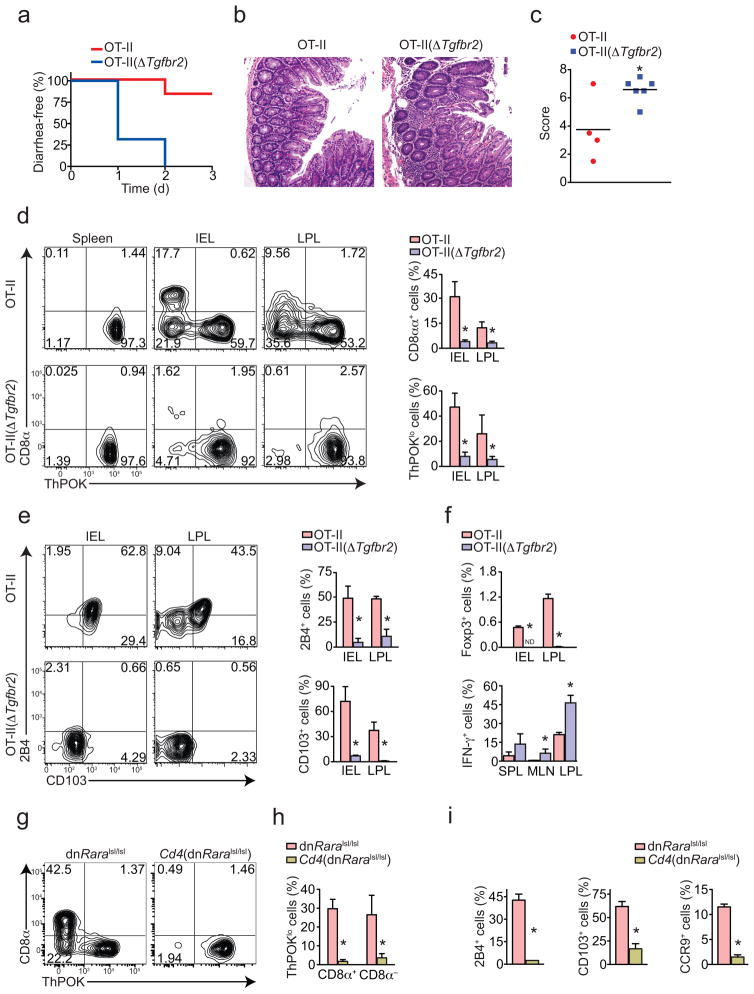
Next we addressed the in vivo role of TGF-β and RA signaling in the modulation of the peripheral CD4+ T cell lineage. To this end, we used conditional knockout Cd4-Cre–Tgfbr2fl/fl mice22 in a Thpok-GFP reporter background. To avoid systemic inflammation and premature death induced by the lack of TGF-β signaling in CD4+ T cells22, we backcrossed these mice into OT-II–Rag1−/− (crossing hereafter called OT-II(ΔTgfbr2)). As expected, OT-II(ΔTgfbr2) showed a normal lifespan and no signs of inflammation (data not shown). To induce naïve T cell differentiation and migration to the intestinal tissue, we kept OT-II and OT-II(ΔTgfbr2) mice on an OVA-containing diet for 7 days23. OT-II(ΔTgfbr2) but not wild-type OT-II mice showed signs of both small and large intestine inflammation with polymorphonuclear infiltrate and epithelial cell damage, and developed severe diarrhea, indicating that oral tolerance was impaired (Fig. 3a–c). We found that while in wild-type OT-II mice about 50% of CD4+ T cells in the IEL compartment downregulated ThPOK, lack of TGF-β signaling resulted in only about 10% of ThPOK loss and greatly reduced CD8αα expression within the CD4+ T cell compartment (Fig. 3d). Conversely, OVA feeding induced high frequency of CD103+ and 2B4+ CD4+ T cells in wild-type OT-II, but not in OT-II(ΔTgfbr2) mice (Fig. 3e). The lack of CD103 upregulation is consistent with a reduced Runx3 induction in OVA-fed OT-II(ΔTgfbr2) mice. This regimen induced modest numbers of Foxp3-expressing cells in the gut, but absence of TGF-βRII abolished the Treg development (Fig. 3f). Although we did not observe induction of IL-17 after OVA exposure in either group (data not shown), OT-II(ΔTgfbr2) mice produced substantial IFN-γ (Fig. 3f). These results establish a role for TGF-β signaling in the tolerance to food-derived antigens that goes beyond iTreg induction, suggesting that modulation of ThPOK and Runx3 may also be required to avoid excessive immune response.
Figure 3. TGF-β and RA signaling in intestinal CD4+ T cells are required for ThPOK downmodulation and CD8α expression in vivo.
(a–f) OT-II or OT-II(ΔTgfbr2) mice were fed with OVA-containing chow for 7 days and cells analyzed by flow cytometry 3 days later. (a) Frequency of diarrhea-free mice after oral OVA challenge. (b) Hematoxylin and eosin staining of the proximal colon. Original magnification, 20x. (c) Histological scores of the colon (each symbol represents one mouse; bar, mean). CD8α and ThPOK (d), 2B4 and CD103 (e), and intracellular IFN-γ and Foxp3 (f) expression by CD45+Vα2+CD4+CD8β− cells from spleen (SPL), mesenteric lymph nodes (MLN), small intestine IEL and LPL. Graphs depict frequency of gated cells in each tissue (error bars, s.e.m.). Data are representative of two independent experiments (n=4 to 6 per group). (g–i) CD8α, 2B4, CD103, CCR9, and ThPOK expression by CD45+TCRβ+CD4+CD8β − cells isolated from the small intestine IEL of naïve control Cd4-Cre− or Cd4-Cre+ dominant negative Raralsl/lsl –Thpok-GFP mice. Graphs depict frequency among gated cells (error bars, s.e.m.). Plots are representative of three independent experiments (n=3 to 4 per group). * p<0.05.
To assess the role of RA signaling in the plasticity of the CD4 lineage in vivo, we used a strain carrying a dominant-negative form of the retinoic acid receptor RAR-α, RAR403, downstream of a loxP-flanked STOP cassette (dnRaralsl/lsl)24. These mice were crossed with Cd4-Cre (Cd4(dnRaralsl/lsl)) and with Thpok-GFP reporter mice to allow visualization of ThPOK expression after disruption RAR-α signaling in T cells. We compared CD4 IEL populations between Cd4-Cre− and Cd4-Cre+ littermates and found that while about 60–70% of CD4 IELs were ThPOKlo in the Cd4-Cre− mice, less than 5% of CD4+ T cells downregulated ThPOK in Cd4-Cre+ mice (Fig. 3g, h). Consistently, Cd4(dnRaralsl/lsl) mice lacked CD4+CD8αα cells in the IEL compartment (Fig. 3g, h). The expression of other molecules associated with intestinal modulation of ThPOK and Runx3, such as 2B4 and CD103 was also significantly dampened in the presence of the dnRAR-α (Fig. 3i). In accordance with the role of RA signaling in gut homing of T cells25, expression of CCR9 was also suppressed (Fig. 3i). In addition to their previously described role in regulating Treg and effector CD4+ T cell differentiation17,19,20,26–29, the above data establishes a role for TGF-β and RA in the modulation of Runx3 and ThPOK expression by CD4+ T cells in vivo.
Thpok is required for CD4+ T cell-induced inflammation
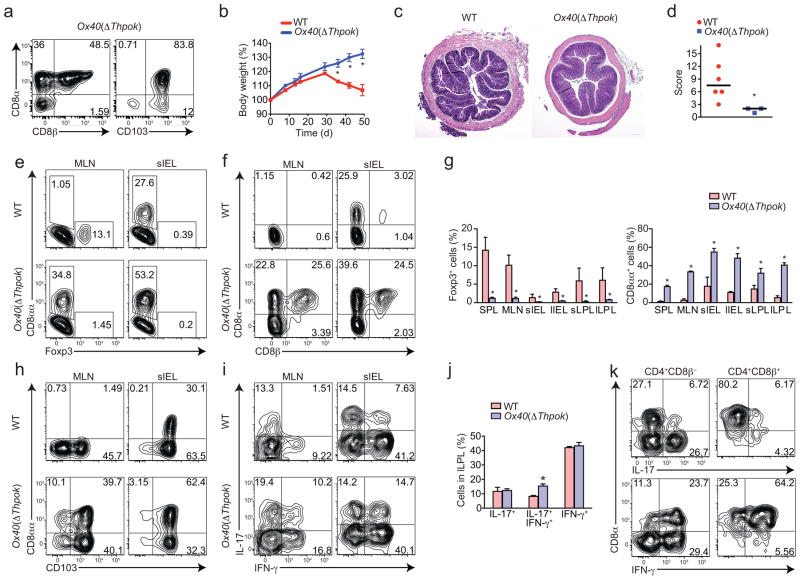
Physiological loss of ThPOK in the intestine observed in intact mice as well as after T cell transfer was inversely correlated with TH17 differentiation and directly correlated with acquisition of a CTL-like gene signature9. To address whether ThPOK loss per se was required for these effects, we forced ThPOK downmodulation in CD4+ T cells in vivo. Given that ThPOK is required for thymic CD4+ T cell differentiation, usual Cre drivers (CD4, LCK) crossed with Thpok-floxed allele (Thpokfl/fl) mice impair CD4+ T cell development6. To circumvent this issue, we developed different strategies. First, we crossed Thpokfl/fl with Tnfrsf4 (here called Ox40)-Cre mice. OX40 expression does not occur during developmental stages of CD4+ T cells, but is restricted to Treg and activated CD4+ T cell30. Therefore, the expression of Cre recombinase in the Ox40-Cre– Thpokfl/fl (Ox40(ΔThpok)) mice allows CD4+ T cells development, as confirmed by similar CD4/CD8 T cell ratio in the periphery when compared to Ox40-Cre–Thpokfl/+ or wild-type mice (Supplementary Fig. 2a). In contrast, Ox40(ΔThpok) mice showed a high frequency of CD4+ IELs expressing CD103, CD8αβ and CD8αα, reinforcing the idea that ThPOK loss is an important driving event in this program (Fig. 4a). Since in naïve wild-type animals CD8α is generally expressed in CD8αα homodimers, it is likely that CD4+CD8αβ cells developed as consequence of incomplete differentiation towards CD4+CD8αα phenotype, which requires intestinal environment. To study the consequences of the loss of ThPOK in activated CD4+ T cells in vivo, we transferred naïve CD4+ T cells from Ox40(ΔThpok) mice into Rag1−/− hosts. We found that wild-type, but not Ox40(ΔThpok) CD4+ T cells induced severe colitis and wasting disease (Fig. 4b–d). The lack of disease in Ox40(ΔThpok) recipient was not caused by reduced proliferation or expansion of donor cells, since similar cell numbers were recovered from mice recipient of naïve wild-type CD4+ T cells donors (Supplementary Fig. 2b). We analyzed Foxp3 expression by donor cells to address the possibility that Ox40(ΔThpok) cells preferentially converted into Treg cells, which also would result in protection from disease13. Instead, we found that Ox40(ΔThpok) donor T cells did not efficiently upregulate Foxp3, particularly in the lamina propria and in peripheral tissues (Fig. 4e, g). On the other hand, Ox40(ΔThpok) transfer resulted in increased differentiation of CD4+CD8α T cells (CD8αβ and CD8αα) (Fig. 4f, g and Supplementary Fig. 2c). Confirming that Ox40(ΔThpok) cells underwent CD4+CD8αα programming, possibly mediated by Runx3 upregulation, we found increased CD103 expression by these cells (Fig. 4h). However, we did not find significant reduction in the frequency of IL-17+ or IFN-γ+ intestinal CD4+ T cells isolated from mice recipients of Ox40(ΔThpok) cells (Fig. 4i). In fact, IL-17 and IFN-γ double-producing cells were increased in mice that received Ox40(ΔThpok) CD4+ T cells (Fig. 4j). Nonetheless, IL-17 production was mostly restricted to CD8α − cells and IFN-γ producing cells were mostly CD8α+ (Fig. 4k). To independently confirm that activated CD4+ T cells lacking ThPOK expression are less prone to induce intestinal inflammation, we adoptively transferred in vitro activated Thpokfl/fl CD4+ T cells transduced with a Cre-GFP-containing retroviral construct into Rag1−/− recipients (Supplementary Fig. 2d, e). Contrary to mock-transduced, CD4+ T cells with ThPOK deletion were not able to induce wasting disease, expressed abundant CD8αα and exhibited no significant reduction in IL-17 expression (Supplementary Fig. 2f–h).
Figure 4. ThPOK loss by activated CD4+ T cells hinders colitis development.
(a) CD8α, CD8β and CD103 expression by CD45+TCRβ+CD4+ cells isolated from the small intestine IEL of a naive Ox40(ΔThpok) mouse. Plots are representative of n=5 mice per group. (b–k) Sorted naïve CD4+ T cells isolated from spleen of WT or Ox40(ΔThpok) mice were adoptively transferred to Rag1 −/− recipients and mice were analyzed 40 to 50 days later. (b) Body weight of recipient mice. (c) Hematoxylin and eosin staining of the proximal colon of recipient mice. Original magnification, 4x. (d) Histological scores of the colon from recipient mice (each symbol represents one mouse; bars, mean). CD8α, CD8β and intracellular Foxp3 (e–g), CD103 (h) and intracellular IL-17 and IFN-γ expression (i, j) by CD45+TCRβ+CD4+ cells isolated from the indicated tissues of recipient mice. Graphs depict frequency of gated cells in the different tissues (error bars, s.e.m.). (k) CD8α and intracellular IL-17 and IFN-γ expression by CD45+TCRβ+CD4+ CD8β − or CD8β+ cells isolated from the large intestine LPL of mice recipients of Ox40(ΔThpok)naïve CD4+ T cells. Data are representative of two independent experiments (n=3 to 6 per group). * p<0.05.
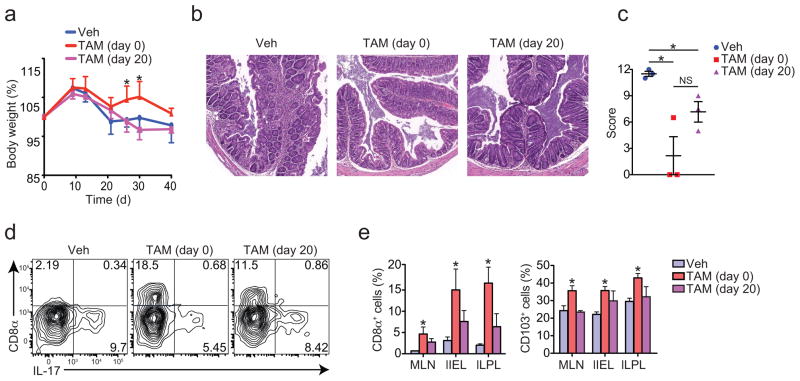
We next asked if deletion of Thpok during the course of inflammatory responses would also suppress colitis. We interbred Thpokfl/fl with ROSA26-CreERT2 mice to generate an inducible Cre strain (ThpokERT2) that allows temporal regulation of gene expression yet avoiding embryonic deletion. Administration of increasing doses of tamoxifen (TAM) to CD4+ T cells from ThpokERT2 mice resulted in gradual loss of ThPOK in vitro (Supplementary Fig. 2i). We transferred naïve CD4+ T cells from ThpokERT2 mice into Rag1−/− hosts. Administration of TAM immediately after transfer, but not 20 days later significantly prevented wasting disease in Rag1−/− recipients (Fig. 5a). However, both groups treated with TAM showed marked reduction in the intestinal inflammation when compared to control animals (Fig. 5b, c). PCR analysis of isolated intestinal cells confirmed Thpok deletion in vivo (data not shown). In addition, consistent with a role for ThPOK in preventing CD4+CD8αα differentiation, TAM administration starting at day 0 or day 20 led to enhanced differentiation of these cells (Fig. 5d, e). However, again we failed to detect changes in the IL-17 or IFN-γ production by CD4+ T cells with enforced ThPOK deletion (Supplementary Fig. 2j). We concluded that ThPOK is involved in the maintenance of CD4+ T cell inflammatory capacity while preventing the differentiation of the CTL-program, although additional factors must act concomitantly to suppress TH17 differentiation.
Figure 5. Continuous ThPOK expression is required for inflammatory activity of CD4+ T cells.
(a-f) Sorted naïve CD4+ T cells isolated from spleen of ThpokERT2 mice were adoptively transferred to Rag1−/− recipients and mice were analyzed 40 days later. Recipient mice were administered tamoxifen (TAM) intraperitoneally three times, every three days, starting on day 0 or day 20 after transfer. (a) Body weight of recipient mice (error bars, s.e.m.). (b) Hematoxylin and eosin staining of the proximal colon of recipient mice. Original magnification, 10x. (c) Histological scores of recipient mice (each symbol represents one mouse; bars, mean). (d, e) CD8α, CD103 and intracellular IL-17 expression by CD45+TCRβ+CD4+ cells. Graphs depict frequency of gated cells in the different tissues (error bars, s.e.m.). Data are representative of two independent experiments (n=3 per group). * p<0.05.
Runx3 modulates ThPOK in peripheral CD4+ T cells
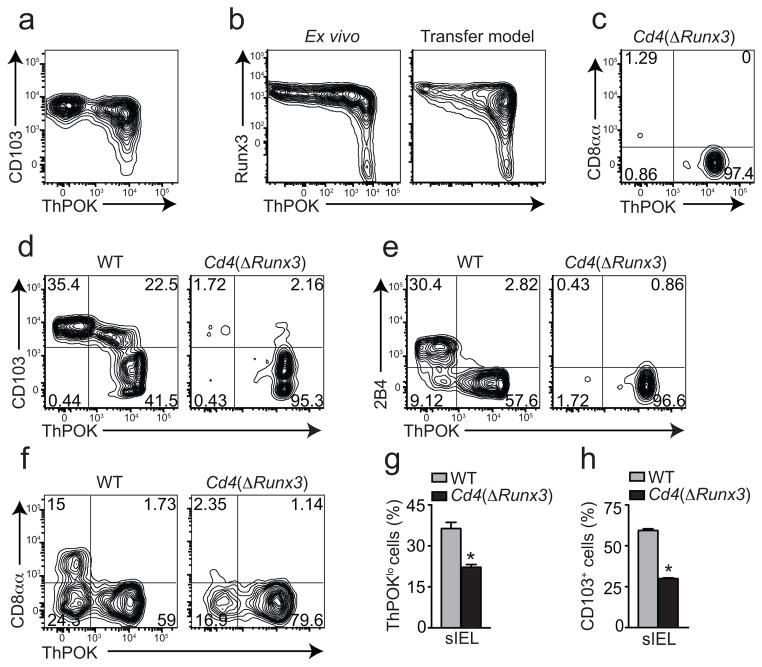
The above functional implications of ThPOK downregulation prompted us to examine its underlying mechanisms. Runx3 binding to the transcriptional silencer elements of the Thpok gene leads to suppression of ThPOK expression by developing DP thymocytes, directing them to the CD8 lineage4. Moreover, concurring with the role for Runx3 also in peripheral regulation of CD4+ T cells, we found that ThPOKlo CD4+ T cells were CD103+, while some ThPOKhi cells contained both CD103+ and CD103− cells, suggesting that Runx3-induced CD103 upregulation precedes ThPOK loss by intestinal CD4+ T cells (Fig. 6a). To address whether Runx3 upregulation indeed occurred in ThPOKhi cells, we interbred Runx3-YFP–with Thpok-GFP reporter mice and analyzed their intestinal CD4+ T cells. While part of the CD4 cells was Runx3hiThPOKhi, we did not observe Runx3loThPOKlo CD4+ T cells (Fig. 6b), suggesting that upregulation of Runx3 occurred before CD4+ T cells downregulated ThPOK expression. Similar results were obtained after transferring naïve CD4+ T cells from Runx3-YFP–Thpok-GFP mice into Rag1−/− recipients (Fig. 6b). The development of Runx3hiThPOKhi could possibly represent an initial event in CD4+CD8αα development.
Figure 6. Runx3 upregulation precedes ThPOK downmodulation and CD8αα expression by intestinal CD4+ T cells.
(a) ThPOK and CD103 expression by CD45+TCRβ+CD4+ cells isolated from the small intestine epithelium of a naïve Thpok-GFP mouse. Plot is representative of three independent experiments (n=5). (b) Runx3 and ThPOK expression by CD45+TCRβ+CD4+ cells isolated from the small intestine epithelium of naïve Runx3-YFP–Thpok-GFP mice or 40 days following adoptive transfer of sorted naïve CD4+ T cells isolated from spleen of Runx3-YFP–Thpok-GFP mice to Rag1−/− recipients. Data are representative of three independent experiments (n=5). (c) ThPOK and CD8α expression by CD45+TCRβ+CD4+CD8β − cells isolated from small intestine epithelium of naïve Cd4(ΔRunx3)–Thpok-GFP mice. (d, e) ThPOK, CD103 and 2B4 expression by CD45+TCRβ+CD4+ cells isolated from the small intestine epithelium of naïve Thpok-GFP (WT) or Cd4(ΔRunx3)–Thpok-GFP mice. (c–e) Data are representative of two independent experiments (n=3 per group). (f–h) Sorted naïve CD4+ T cells isolated from spleen of Thpok-GFP (WT) or Cd4(ΔRunx3)–Thpok-GFP mice were adoptively transferred to Rag1−/− recipients and mice were analyzed 40 days later. (f) CD8αα and ThPOK expression by CD45+TCRβ+CD4+CD8β − cells isolated from small intestine epithelium of recipient mice. Graphs depict frequency of ThPOKlo (g) and CD103+ (h) cells gated as in f (error bars, s.e.m.). Data are representative of two independent experiments (n=4 per group). * p<0.05.
To directly investigate Runx3 requirement for ThPOK loss by intestinal CD4+ T cells, we crossed Runx3 T cell conditional knockout mice Cd4-Cre–Runx3fl/fl (Cd4(ΔRunx3)) with Thpok-GFP reporter mice. Cd4(ΔRunx3) animals have normal CD4+ T cell development although CD8+ T cell development is significantly impaired31. Consistent with a role for Runx3 in directly suppressing ThPOK expression in intestinal CD4+ T cells, we found that Cd4(ΔRunx3)–Thpok-GFP animals show a severe reduction in the ThPOKlo CD4+ IEL population, as well as in the frequency of CD8αα, CD103 and 2B4 expressing CD4+ T cells (Fig. 6c–e). These data provides evidence that Runx3 is required for ThPOK downmodulation in intestinal CD4+ T cells.
To independently confirm the requirement for Runx3 in the peripheral downmodulation of ThPOK during inflammation, we adoptively transferred naïve Cd4(ΔRunx3) Thpok-GFP T cells into Rag1−/− hosts and analyzed recipient mice 4–6 weeks after transfer. We observed that Cd4(ΔRunx3)–Thpok-GFP donor cells did not undergo CD4+CD8αα pathway, and a reduced frequency of ThPOKlo CD4+ T cells was found in the intestine of recipient mice when compared to wild-type Thpok-GFP donors (Fig. 6f–h). However, the fact that roughly 15% of Cd4(ΔRunx3)–Thpok-GFP T cells lost ThPOK expression during colitis suggests that during chronic inflammation additional factors, such as MAZR9, also contribute to the modulation of ThPOK. Altogether, the above data establishes a role for Runx3 in the modulation of ThPOK expression by peripheral CD4+ T cells.
Runx3 modulates intestinal inflammatory responses
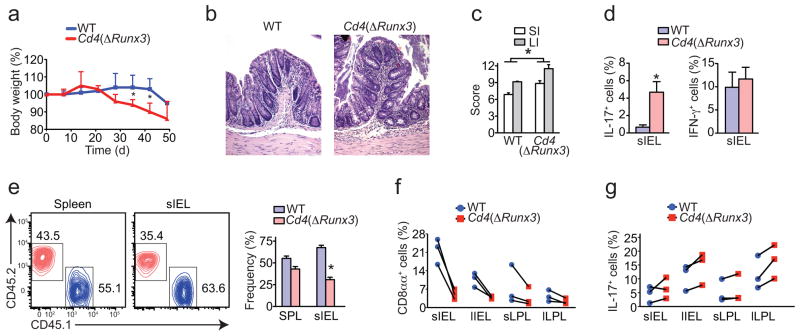
Next, we investigated whether Runx3 plays a role in CD4+ T helper function in vivo, particularly in the development of colitis. Similarly to the experiments described above, we transferred naïve CD4+ T cells from Cd4(ΔRunx3)–Thpok-GFP mice into Rag1−/− recipients. Mice receiving Cd4(ΔRunx3)–Thpok-GFP cells showed accelerated wasting disease (Fig. 7a). Mice recipient of both wild-type and Cd4(ΔRunx3)–Thpok-GFP cells eventually developed intestinal inflammation, although histopathological analysis of mice recipient of Cd4(ΔRunx3)–Thpok-GFP showed increased crypt abscesses and neutrophil infiltrate, the later directly associated with TH17 responses (Fig. 7b, c). Consistent with previous experiments, transferred Cd4(ΔRunx3)–Thpok-GFP cells showed reduced ThPOK loss and CD103 expression (data not shown). Additionally, in the absence of Runx3, IL-17 expression was significantly increased while IFN-γ production was not altered (Fig. 7d). To specifically investigate cell-autonomous effects of Runx3-deficiency in CD4+ T cells, we co-transferred naïve CD4+ T cells from wild-type CD45.1 and Cd4(ΔRunx3)–Thpok-GFP CD45.2 into Rag1−/− hosts at 1:1 ratio and analyzed mice 6 weeks after transfer. We found a similar survival and expansion of Cd4(ΔRunx3)–Thpok-GFP and wild-type donor CD4+ T cells in peripheral tissue but at a moderately reduced ratio in the intestine, indicating that the accelerated disease in Cd4(ΔRunx3)–Thpok-GFP was not due to increased gut homing or proliferation of donor cells (Fig. 7e). Analysis of CD8αα and IL-17 expression confirmed the single-transfer data and indicated a cell-autonomous role for Runx3 in modulating CD4+ T cell differentiation (Fig. 7f, g).
Figure 7. Runx3 expression by CD4+ T cells inversely correlates with their inflammatory potential.
(a–g) Sorted naïve CD4+ T cells isolated from spleen of Thpok-GFP (WT) or Cd4(ΔRunx3)–Thpok-GFP mice were adoptively transferred to Rag1−/− recipients and mice were analyzed 40 to 50 days later. (a) Body weight of recipient mice (error bars, s.e.m.). (b) Hematoxylin and eosin staining of the proximal colon of recipient mice. Original magnification, 20x. (c) Histological scores of small intestine and colon of recipient mice (error bars, s.e.m.). (d) Frequency of IL-17 and IFN-γ expressing cells among CD45+TCRβ+CD4+ cells isolated from the small intestine epithelium of recipient mice (error bars, s.e.m.). Data are representative of three independent experiments (n= 5 per group). (e–g) Sorted naïve CD4+ T cells isolated from spleen of WT CD45.1 or Cd4(ΔRunx3) CD45.2 mice were adoptively co-transferred at a ratio 1:1 to Rag1−/− recipients and mice were analyzed 40 days later (n=3 per group). (e) CD45.1 and CD45.2 expression by TCRβ+CD4+ cells isolated from spleen or small intestine epithelium of recipient mice. (f, g) Frequency of CD8α (CD8β−) and IL-17 expressing cells among TCRβ+CD4+ CD45.1 or CD45.2 cells isolated from the small and large intestine IEL and LPL of recipient mice. Connecting lines represent each individual recipient mouse. * p<0.05.
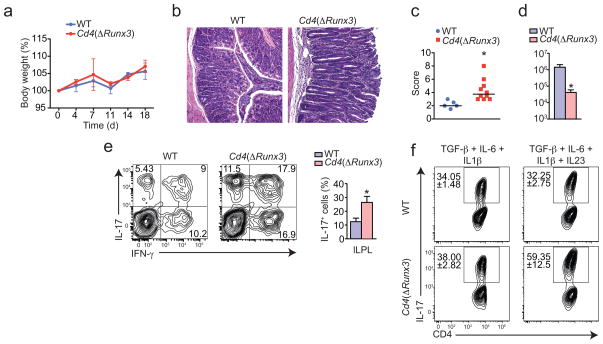
Although TH17 responses can be detrimental in some models of colitis, they are required for the control of several extracellular bacteria infections, particularly at the mucosal surfaces32. Therefore, we investigated whether Cd4(ΔRunx3) animals would show altered responses to the extracellular bacteria Citrobacter rodentium, a murine correspondent of enteropathogenic Escherichia coli32. We found that although both wild-type and Cd4(ΔRunx3)strains showed similar weight loss after infection (Fig. 8a), histopathological analysis of the colon from infected Cd4(ΔRunx3) mice showed increased inflammation, with transmural infiltrate and epithelial cell damage, likely result of exaggerated inflammatory TH17 responses (Fig. 8b, c). Indeed, Cd4(ΔRunx3) cleared Citrobacter more efficiently, with reduced pathogen burden, and showed increased frequency of IL-17-producing CD4+ T cells (Fig. 8d, e). Consistently, naïve Cd4(ΔRunx3) T cells were more efficient in differentiating towards pathogenic TH17 (induced by low TGF-β, IL-1β, IL-6 and IL-23), but not towards non-pathogenic TH17 cells (induced by TGF-β and IL-6)33,34 than wild-type CD4+ T cells (Fig. 8f). The above data indicate that in addition to its effects on ThPOK expression, Runx3 expression by CD4+ T cells may be involved in repression of the TH17 pathway. To confirm the inverse association between Runx3 expression and severity of colitis and also TH17 differentiation, we overexpressed Runx3 in primary CD4+ T cells using retrovirus encoding long isoform of Runx3-IRES-GFP (CD4(Runx3)) and transferred to Rag1−/− hosts. Contrary to Cd4(ΔRunx3)cells, CD4(Runx3)induced wasting disease with slower kinetics in recipient mice (Supplementary Fig. 3a). Additionally, CD4(Runx3)donor cells expressed more CD103 and CD8αα but produced reduced amounts of IL-17 when compared to mock-transduced cells (Supplementary Fig. 3b, c). We conclude that upregulation of Runx3, rather than ThPOK loss per se, drives suppression of pro-inflammatory TH17 responses. Altogether, the combined effects of Runx3 and ThPOK loss result in substantial changes in CD4+ T cell function, deviating from classical helper cells toward CTL-like cells with reduced capacity to induce tissue damage and inflammation.
Figure 8. Cd4(ΔRunx3) mice show increased resistance to Citrobacter rodentium infection.
(a–e) WT or Cd4(ΔRunx3) mice were orally infected with C. rodentium and analyzed 18 days post infection. (a) Body weight of mice (error bars, s.e.m.). (b) Hematoxylin and eosin staining of the proximal colon of infected mice. Original magnification, 20x. (c) Histological scores of proximal colon of infected mice (each symbol represents one mouse from two combined experiments; bars, mean) (d) Colony-forming-unit (CFU) of C. rodentium from fecal pellets of infected mice (error bars, s.e.m.). (e) Intracellular IL-17 and IFN-γ expression by CD45+TCRβ+CD4+ cells isolated from the large intestine LPL. Graph depicts frequency of IL-17 producing cells (error bars, s.e.m.). Data are representative of three independent experiments (n=3 to 5 per group). (f) Sorted naïve CD4+ T cells isolated from WT or Cd4(ΔRunx3) mice were co-cultured for 4.5 days with splenic DCs and soluble anti-CD3ε in the presence of the indicated cytokines. Plots show intracellular IL-17 expression by gated CD4+ T cells (values, mean and s.e.m. of duplicate wells). Data are representative from three independent experiments. * p<0.05.
DISCUSSION
The intestinal tissue offers a unique environment for the immune system. It represents the main site for nutrient absorption while forming the largest surface exposed to environmental allergens. It interacts with beneficial microbiota in the midst of invading pathogens. Here we describe an alternative fate for CD4+ T cells that migrate to the intestinal tissue, particularly to the intraepithelial compartment, where cues such as TGF-β and RA are required for a post-thymic suppression of the transcription factor ThPOK and upregulation of Runx3. Cells that undergo this pathway show gene signature resembling CTL or innate-like lymphocytes10–12 and conversely, these intestinal CD4+ T cells showed reduced expression of molecules associated with CD4+ helper T cells.
We addressed whether this drastic change in the CD4 program was driven by ThPOK downregulation, Runx3 upregulation, or both. Previous studies have demonstrated that forced suppression of ThPOK expression leads to derepression of CTL program in CD4+ T cells5–7. The extensive in vivo data presented here showed that modulation of ThPOK in CD4+ T cells is a physiological process and has consequences for T cell differentiation and function. Using several novel approaches to delete Thpok post-thymically, we demonstrated the causal relationship between expression of ThPOK and suppression of CTL program in CD4+ T cells in vivo. More importantly, we showed that loss of ThPOK in activated CD4+ T cells dampened their inflammatory potential. Nevertheless, unlike ThPOKlo CD4 IELs, activated CD4+ T cells with “floxed” Thpok showed similar capacity to produce IL-17, even though they became inefficient to cause colitis. Indeed, the role of TH17 cells in the transfer model of colitis is controversial. Although generally associated with inflammation35, some populations of IL-17-producing T cells are also associated with regulatory immune responses, including those in colitis models33,34,36,37. It is possible that mutual expression of ThPOK and Runx3 differentially regulates “non-pathogenic” versus “pathogenic” TH17 cells33,34, possibility supported by our in vitro data. It should be noted that Ox40(ΔThpok) CD4+ T cells did not cause colitis even though Treg conversion was impaired, which points to the relevance of this pathway to CD4+ T cell function. It is possible that induced CD4+CD8αα cells could suppress inflammation in a cell-extrinsic manner, i.e., they are regulatory, although we have not found evidence for that. Additionally, upregulation of CTL and NK-related genes in Thpok-depleted CD4+ T cells and their increased tendency to divert to the CD4+CD8αα pathway could per se explain their decreased potential to cause intestinal inflammation.
In contrast to the reduced capacity of Thpok-deficient CD4+ T cells to induce colitis, we also showed that Runx3-deficient CD4+ T cells induce aggravated wasting disease with increased TH17 differentiation. However, Cd4(ΔRunx3)mice were more efficient at C. rodentium clearance while sustaining increased inflammatory infiltrate, known to be a TH17 or TH22-dependent process32. This is a remarkable example of how intestinal immune responses must constantly deal with two faces of inflammation: protection and tissue damage. Since Thpok depletion experiments did not result in marked reduction in IL-17 production by CD4+ T cells, we concluded that Runx3 upregulation, rather than ThPOK downregulation, drives the suppression of TH17 differentiation. Therefore, Runx3 could play a role both in the initiation of this program, suppressing ThPOK expression, and downstream by suppressing TH17 differentiation, contrasting with Runx1, positively associated with TH17 development38,39. Cooperation between Runx3 and T-bet was proposed to mediate activation of Ifng in TH1 cells40 and our data indicate that this cooperation could also play a role in the suppression of TH17 differentiation.
What are the mechanisms for physiological ThPOK downregulation in CD4+ T cells? Identification of regulatory elements of the Thpok gene and demonstration of Runx3 binding sites at its silencers, which suppresses Thpok transcription, served as a basis for the current understanding of T cell lineage decision 4,41. Here, we described a physiological process that results in a post-thymic suppression of ThPOK via Runx3 upregulation in intestinal CD4+ T cells. We provided mechanistic insights into this process, which at least in part are mediated by TGF-β and RA signaling. We also provided ample evidence for the physiological role for this TGF-β-dependent pathway, from modulating susceptibility to intestinal infections, development of wasting disease and colitis, to oral tolerance. The later however, is also influenced by TGF-β-induced peripheral Treg cells23,42. Although we currently do not understand the mechanisms that direct naïve T cells towards CD4+CD8αα versus iTreg cells, our data indicate that differential expression of ThPOK, Runx3 and T-bet are involved and we are currently investigating this possibility.
This study provides functional relevance and mechanistic evidence for a continuous role of T-cell development transcription factors in the periphery, composing a tightly regulated process that drastically changes CD4+ T cell activity in a site-specific manner. The functional studies with induced changes in ThPOK and Runx3 expression raise the possibility that disruption of this pathway may result in uncontrolled CD4+ T cell responses. Reinforcement of this pathway, in contrast, leads to attenuated intestinal inflammation and increased susceptibility to extracellular bacteria infection. It is also possible however, that excessive triggering of the CTL-function on CD4+ T cells results in breakdown of the epithelial layer as observed in celiac disease, but paradoxically could also increase resistance to viral infections that shut down the MHC-I presentation pathway. Factors that influence this pathway in one way or another are a potential target for therapeutic interventions in inflammatory disorders.
ONLINE METHODS
Mice
C57BL/6 CD45.1 and CD45.2, OT-II TCR-transgenic, Rag1−/−, Thpokfl/fl, Cd4-Cre, Rosa26-CreERT2, Ox40-Cre and Foxp3-RFP mice were purchased from the Jackson Laboratories and maintained in our facilities. Tgfbr2fl/fl mice were obtained from NCI. Dominant negative Raralsl/lsl and Runx3-YFP reporter mice were generously provided by R. Noelle (Dartmouth)24 and D. Littman (NYU), respectively. Several of these lines were interbred in our facilities to obtain the final strains described in the text. Mice were maintained at the Rockefeller University animal facilties under specific pathogen-free conditions and sentinel mice from the Rag1−/− mice colony were tested to be negative for Helicobacter spp. and C. rodentium. Mice were used at 7–12 weeks of age for most experiments. Animal care and experimentation were consistent with the NIH guidelines and were approved by the Institutional Animal Care and Use Committee at the Rockefeller University.
Antibodies and flow cytometry analysis
Fluorescent-dye-conjugated antibodies were purchased from BD-Pharmingen (anti-CD4, 550954; anti-CD25, 553866; anti-Thy1.1, 561401; anti-CD103, 557495; anti-CD244.2, 553306 anti-IL-17a, 559502; anti-T-bet, 561312) or eBioscience (anti-CD8α, 56-0081; anti-CD44, 56-0441; anti-CD45.1, 25-0453; anti-CD45.2, 47-0454; anti-CD62L, 48-0621; anti-TCR-β, 47-5961; anti-IFN-γ, 25-7311; anti-Foxp3, 17-5773; anti-Vα2, 48-5812). Flow cytometry data was acquired on a LSR-II flow cytometer (Becton Dickinson) and analyzed using FlowJo software package (Tri-Star). Intracellular staining of Foxp3 and Granzyme B was conducted using Foxp3 Mouse Regulatory T cell Staining Kit (eBioscience). For flow cytometric analysis of cytokine-secreting cells, cells were incubated in the presence of 100ng/ml PMA (Sigma), 500ng/ml Ionomycin (Sigma) for 4.5h and 10μg/ml brefeldin A (BFA) (Sigma) for the last 2.5h prior staning. Cell populations were first stained with antibodies against the indicated cell surface markers, followed by permeabilization in Fix/Perm buffer, and intracellular staining in Perm/Wash buffer (BD Pharmingen).
In vitro T-cell culture
Naive (defined as CD4+CD25−CD62hiCD44lo) T cells were sorted using FACS Aria cell sorter flow cytometer (Becton Dickinson) and cultured for 4.5 days in 96 well plates pre-coated with 2μg/ml of anti-CD3ε (17A2) and 1μg/ml of soluble anti-CD28 (37.51). Cells were then stimulated with indicated cytokines (10ng of IL-1β, 20ng of IL-6, 10ng of IL-12, 10ng of IL-23, 10nM of RA, 2ng of TGF-β (Treg), 0.2ng of TGF-βTH17). OT-II cells were co-cultured with CD11c + splenic DCs in the presence of 500nM of OVA peptide and indicated cytokines. For re-stimulation experiments, cells were cultured for 4.5 days as above and resuspended in new media containing the indicated cytokines for another 72h.
Retroviral transduction of CD4+ T cells
Retroviral vectors for Runx3, or Cre were previously described31,41. Retroviral vectors for T-bet were generously provided by L. Glimcher (Cornell) and were previously described39. Transduction of CD4+ T cells with retroviral vectors was performed as previously described31,41. Briefly, sorted naïve T cells were in vitro activated with 2μg/ml of plate-bound anti-CD3ε and 1μg/ml of soluble anti-CD28 for 36–48h. Cells were then spin transduced with retrovirus for 2h at 1250g.
Quantitative PCR
(q)PCR was performed as previously described17. RPL32 housekeeping gene was used to normalize samples. Primers used: Thpok-forward 5′-ATGGGATTCCAATCAGGTCA-3′, Thpok-reverse 5′-TTCTTCCTACACCCTGTGCC-3′; Tbx21-forward 5′-ATCCTGTAATGGCTTGTGGG-3′, Tbx21-reverse 5′-TCAACCAGCACCAGACAGAG-3′; Rpl32-forward 5′-GAAACTGGCGGAAACCCA-3′, Rpl32-reverse 5′-GGATCTGGCCCTTGAACCTT-3′; Cd8a-forward 5′-ACTGCAAGGAAGCAAGTGGT-3′, Cd8a-reverse 5′-CACCGCTAAAGGCAGTTCTC-3′; RUNX3L-forward 5′-ACAGCATCTTTGACTCCTTCC-3′, RUNX3L-reverse 5′-TGTTCTCGCCCATCTTGC-3′; FOXP3-forward 5′-CCCATCCCCAGGAGTCTTG-3′, FOXP3-reverse 5′-ACCATGACTAGGGGCACTGTA-3′; Il17a-forward 5′-TGAGAGCTGCCCCTTCACTT-3′, Il17a-reverse 5′-ACGCAGGTGCAGCCCA-3′.
Experimental colitis model
Colitis was induced after transfer of 5 × 105 sorted naïve T cells into Rag1−/− mice, as previously described17. For co-transfer experiments, 2.5 × 105 sorted naïve T cells from Cd4(ΔRunx3) CD45.2 mice were injected together with 2.5 × 105 sorted naïve T cells from C57BL/6 CD45.1 mice. Transferred mice were monitored regularly for signs of disease including weight loss, hunched over appearance, pilo-erection of the coat and diarrhea, and analyzed at various times after the initial transfer or when they reached 80% of their initial weight. Colitis was blindly scored (max. 17) according to the following combination of parameters: extension of inflammation (0 -none, 1 - lamina propria, 2 - submucosal, 3 - transmural), degree of inflammation (0 -none, 1 - mild, 2 - moderate, 3 - severe), abnormal crypt morphology (0 – 3), neutrophil infiltrate (0 – 4), goblet cell loss (0 - none, 1 - moderate, 2 - severe), mucosal erosion or ulcer (0 or 1), crypt abscess (0 or 1).
Tamoxifen treatment
For in vitro treatment, sorted naïve T cells were cultured with 10nM of 4-hydroxytamoxifen (Sigma) for 4 days. DNA and RNA were extracted to confirm treatment efficiency. For in vivo treatment, mice were intraperitoneally (IP) injected with 2 mg of tamoxifen-citrate (Acros) dissolved in vegetable oil (Sigma) at 10mg/ml. Mice were injected IP every 3 days for a total of 6mg of tamoxifen per animal.
Citrobacter rodentium infection
Mice were infected with 2 × 108 of C. rodentium per animal, as previously described32. Bacteria were inoculated by gavage in recipient mice in a total volume of 200μl. After infection, mice were followed daily for weight loss and colony forming units in feces and liver. Mice were sacrificed and analyzed 18 days after infection. Inflammation induced by C. rodentium infection was blindly scored (max. 14) according to the following combination of parameters: extension of inflammation (0 -none, 1 - lamina propria, 2 - submucosal, 3 - transmural), severity of epithelial injury (0 –3), goblet cell loss (0 or 1), mucosal hypertrophy (0 or 1), neutrophil infiltrate (0 –3), mononuclear infiltrate (0 – 3).
Oral OVA treatment
Mice were fed with regular chow containing 1% chicken-ovalbumin (OVA) for 7 days and analyzed 3 days later. To measure diarrhea, animals were daily challenged by gavage with 50mg of OVA solution (250mg/ml) until diarrhea was detected. Mice were classified as positive when signs of diarrhea were detected two consecutive times after challenge. Tissue inflammation was scored as described for colitis experiments.
Preparation of intraepithelial and lamina propria lymphocytes
Intraepithelial and lamina propria lymphocytes were isolated as previously described 17.
Statistics
Statistical analysis was performed in GraphPad Prism software. Data was analyzed by applying one-way ANOVA or unpaired Student’s t-test whenever necessary. For analysis of histological scores non-parametric Mann Whitney test was used. A P value of less than 0.05 was considered significant.
Supplementary Material
Acknowledgments
We are indebted to Klara Velinzon and Yelena Shatalina for sorting cells, members of the Nussenzweig, Steinman, Marraffini, Tavazoie labs and The Rockefeller University employees for continuous assistance. We thank R. Noelle (Dartmouth) and D. Littman (NYU) for generously providing dnRaralsl/lsl and Runx3-YFP strains, respectively, and L. Glimcher (Cornell) for Tbx21 vectors. We thank L. Marraffini, S. Tavazoie, G. Kim (LIAI), M. Kronenberg (LIAI), H. Cheroutre (LIAI), and members of our laboratory, particularly L. Feighery for discussions and critical reading and editing of the manuscript. D.M. is supported by an Ellison Medical Foundation New Scholar Award in Aging, an Irma T. Hirschl Award, a Crohn’s & Colitis Foundation of America Senior Research Award, a National Institutes of Health NIH R01 DK093674-01 grant and by the Leona M. and Harry B. Helmsley Charitable Trust. F.A.C-P. is supported by FAPESP and CNPq (Brazil).
Footnotes
The authors have no conflicting financial interests.
AUTHOR CONTRIBUTIONS
D.M. conceived and supervised this study. B.S.R. and D.M. designed experiments. B.S.R., A.R. and D.M. performed experiments. B.S.R. prepared figures and helped with manuscript preparation. F.A.C-P. analyzed and scored intestinal tissue for inflammation and helped with the manuscript preparation. I.T. provided mouse strains, constructs for over-expression of genes and helped with manuscript preparation. D.M. wrote the paper.
References
- 1.He X, Park K, Kappes DJ. The role of ThPOK in control of CD4/CD8 lineage commitment. Annu Rev Immunol. 2010;28:295–320. doi: 10.1146/annurev.immunol.25.022106.141715. [DOI] [PubMed] [Google Scholar]
- 2.Xiong Y, Bosselut R. CD4-CD8 differentiation in the thymus: connecting circuits and building memories. Curr Opin Immunol. 2012;24:139–145. doi: 10.1016/j.coi.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sakaguchi S, et al. The zinc-finger protein MAZR is part of the transcription factor network that controls the CD4 versus CD8 lineage fate of double-positive thymocytes. Nat Immunol. 2010;11:442–448. doi: 10.1038/ni.1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Setoguchi R, et al. Repression of the transcription factor Th-POK by Runx complexes in cytotoxic T cell development. Science. 2008;319:822–825. doi: 10.1126/science.1151844. [DOI] [PubMed] [Google Scholar]
- 5.Egawa T, Littman DR. ThPOK acts late in specification of the helper T cell lineage and suppresses Runx-mediated commitment to the cytotoxic T cell lineage. Nat Immunol. 2008;9:1131–1139. doi: 10.1038/ni.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He X, et al. The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature. 2005;433:826–833. doi: 10.1038/nature03338. [DOI] [PubMed] [Google Scholar]
- 7.Wang L, et al. The zinc finger transcription factor Zbtb7b represses CD8-lineage gene expression in peripheral CD4+ T cells. Immunity. 2008;29:876–887. doi: 10.1016/j.immuni.2008.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheroutre H, Lambolez F, Mucida D. The light and dark sides of intestinal intraepithelial lymphocytes. Nat Rev Immunol. 2011;11:445–456. doi: 10.1038/nri3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mucida D, et al. Transcriptional reprogramming of mature CD4 T helper cells generates distinct MHC class II restricted cytotoxic T lymphocytes. Nat Immunol. 2013 doi: 10.1038/ni.2523. Doi:10.1038.niXXXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denning TL, et al. Mouse TCRalphabeta+CD8alphaalpha intraepithelial lymphocytes express genes that down-regulate their antigen reactivity and suppress immune responses. J Immunol. 2007;178:4230–4239. doi: 10.4049/jimmunol.178.7.4230. [DOI] [PubMed] [Google Scholar]
- 11.Yeh JH, Sidhu SS, Chan AC. Regulation of a late phase of T cell polarity and effector functions by Crtam. Cell. 2008;132:846–859. doi: 10.1016/j.cell.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 12.Yamagata T, Mathis D, Benoist C. Self-reactivity in thymic double-positive cells commits cells to a CD8 alpha alpha lineage with characteristics of innate immune cells. Nat Immunol. 2004;5:597–605. doi: 10.1038/ni1070. [DOI] [PubMed] [Google Scholar]
- 13.Ahern PP, et al. Interleukin-23 drives intestinal inflammation through direct activity on T cells. Immunity. 2010;33:279–288. doi: 10.1016/j.immuni.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sujino T, et al. Regulatory T cells suppress development of colitis, blocking differentiation of T-helper 17 into alternative T-helper 1 cells. Gastroenterology. 2011;141:1014–1023. doi: 10.1053/j.gastro.2011.05.052. [DOI] [PubMed] [Google Scholar]
- 15.Shi MJ, Stavnezer J. CBF alpha3 (AML2) is induced by TGF-beta1 to bind and activate the mouse germline Ig alpha promoter. J Immunol. 1998;161:6751–6760. [PubMed] [Google Scholar]
- 16.Grueter B, et al. Runx3 regulates integrin alpha E/CD103 and CD4 expression during development of CD4−/CD8+ T cells. J Immunol. 2005;175:1694–1705. doi: 10.4049/jimmunol.175.3.1694. [DOI] [PubMed] [Google Scholar]
- 17.Mucida D, et al. Reciprocal TH17 and regulatory T cell differentiation mediated by retinoic acid. Science. 2007;317:256–260. doi: 10.1126/science.1145697. [DOI] [PubMed] [Google Scholar]
- 18.Mucida D, et al. Retinoic acid can directly promote TGF-beta-mediated Foxp3(+) Treg cell conversion of naive T cells. Immunity. 2009;30:471–472. doi: 10.1016/j.immuni.2009.03.008. author reply 472–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takahashi H, et al. TGF-beta and retinoic acid induce the microRNA miR-10a, which targets Bcl-6 and constrains the plasticity of helper T cells. Nat Immunol. 2012;13:587–595. doi: 10.1038/ni.2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Konkel JE, et al. Control of the development of CD8alphaalpha+ intestinal intraepithelial lymphocytes by TGF-beta. Nat Immunol. 2011;12:312–319. doi: 10.1038/ni.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lazarevic V, Glimcher LH. T-bet in disease. Nat Immunol. 2011;12:597–606. doi: 10.1038/ni.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li MO, Sanjabi S, Flavell RA. Transforming growth factor-beta controls development, homeostasis, and tolerance of T cells by regulatory T cell-dependent and -independent mechanisms. Immunity. 2006;25:455–471. doi: 10.1016/j.immuni.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 23.Mucida D, et al. Oral tolerance in the absence of naturally occurring Tregs. J Clin Invest. 2005;115:1923–1933. doi: 10.1172/JCI24487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rajaii F, Bitzer ZT, Xu Q, Sockanathan S. Expression of the dominant negative retinoid receptor, RAR403, alters telencephalic progenitor proliferation, survival, and cell fate specification. Dev Biol. 2008;316:371–382. doi: 10.1016/j.ydbio.2008.01.041. [DOI] [PubMed] [Google Scholar]
- 25.Iwata M, et al. Retinoic acid imprints gut-homing specificity on T cells. Immunity. 2004;21:527–538. doi: 10.1016/j.immuni.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 26.Mucida D, Park Y, Cheroutre H. From the diet to the nucleus: vitamin A and TGF-beta join efforts at the mucosal interface of the intestine. Semin Immunol. 2009;21:14–21. doi: 10.1016/j.smim.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DePaolo RW, et al. Co-adjuvant effects of retinoic acid and IL-15 induce inflammatory immunity to dietary antigens. Nature. 2011;471:220–224. doi: 10.1038/nature09849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hall JA, et al. Essential role for retinoic acid in the promotion of CD4(+) T cell effector responses via retinoic acid receptor alpha. Immunity. 2011;34:435–447. doi: 10.1016/j.immuni.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pino-Lagos K, et al. A retinoic acid-dependent checkpoint in the development of CD4+ T cell-mediated immunity. J Exp Med. 2011;208:1767–1775. doi: 10.1084/jem.20102358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klinger M, et al. Thymic OX40 expression discriminates cells undergoing strong responses to selection ligands. J Immunol. 2009;182:4581–4589. doi: 10.4049/jimmunol.0900010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Naoe Y, et al. Repression of interleukin-4 in T helper type 1 cells by Runx/Cbf beta binding to the Il4 silencer. J Exp Med. 2007;204:1749–1755. doi: 10.1084/jem.20062456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mangan PR, et al. Transforming growth factor-beta induces development of the T(H)17 lineage. Nature. 2006;441:231–234. doi: 10.1038/nature04754. [DOI] [PubMed] [Google Scholar]
- 33.Ghoreschi K, et al. Generation of pathogenic T(H)17 cells in the absence of TGF-beta signalling. Nature. 2010;467:967–971. doi: 10.1038/nature09447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee Y, et al. Induction and molecular signature of pathogenic TH17 cells. Nat Immunol. 2012;13:991–999. doi: 10.1038/ni.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leppkes M, et al. RORgamma-expressing Th17 cells induce murine chronic intestinal inflammation via redundant effects of IL-17A and IL-17F. Gastroenterology. 2009;136:257–267. doi: 10.1053/j.gastro.2008.10.018. [DOI] [PubMed] [Google Scholar]
- 36.O’Connor W, Jr, et al. Nat Immunol. 2009;10:603–609. doi: 10.1038/ni.1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ono Y, et al. T-helper 17 and Interleukin-17-Producing Lymphoid Tissue Inducer-Like Cells Make Different Contributions to Colitis in Mice. Gastroenterology. 2012;143:1288–1297. doi: 10.1053/j.gastro.2012.07.108. [DOI] [PubMed] [Google Scholar]
- 38.Zhang F, Meng G, Strober W. Interactions among the transcription factors Runx1, RORgammat and Foxp3 regulate the differentiation of interleukin 17-producing T cells. Nat Immunol. 2008;9:1297–1306. doi: 10.1038/ni.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lazarevic V, et al. T-bet represses T(H)17 differentiation by preventing Runx1-mediated activation of the gene encoding RORgammat. Nat Immunol. 2011;12:96–104. doi: 10.1038/ni.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Djuretic IM, et al. Transcription factors T-bet and Runx3 cooperate to activate Ifng and silence Il4 in T helper type 1 cells. Nat Immunol. 2007;8:145–153. doi: 10.1038/ni1424. [DOI] [PubMed] [Google Scholar]
- 41.Muroi S, et al. Cascading suppression of transcriptional silencers by ThPOK seals helper T cell fate. Nat Immunol. 2008;9:1113–1121. doi: 10.1038/ni.1650. [DOI] [PubMed] [Google Scholar]
- 42.Curotto de Lafaille MA, et al. Adaptive Foxp3+ regulatory T cell-dependent and -independent control of allergic inflammation. Immunity. 2008;29:114–126. doi: 10.1016/j.immuni.2008.05.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.