Abstract
Proper cell division is essential for growth and development of uni- and multicellular organisms. The fungal septation initiation network (SIN) functions as kinase cascade that connects cell cycle progression with the initiation of cytokinesis. Miss-regulation of the homologous Hippo pathway in animals results in excessive cell proliferation and formation of tumors, underscoring the conservation of both pathways. How SIN proteins interact and transmit signals through the cascade is only beginning to be understood. Moreover, our understanding of septum formation and its regulation in filamentous fungi, which represent the vast majority of the fungal kingdom, is highly fragmentary. We determined that a tripartite kinase cascade, consisting of CDC-7, SID-1 and DBF-2, together with their regulatory subunits CDC-14 and MOB-1, is important for septum formation in the model mold Neurospora crassa. DBF-2 activity and septum formation requires auto-phosphorylation at Ser499 within the activation segment and phosphorylation of Thr671 in the hydrophobic motif by SID-1. Moreover, SID-1-stimulated DBF-2 activity is further enhanced by CDC-7, supporting a stepwise activation mechanism of the tripartite SIN kinase cascade in fungi. However, in contrast to the situation described for unicellular yeasts, the localization of the entire SIN cascade to spindle pole bodies is constitutive and cell cycle independent. Moreover, all SIN proteins except CDC-7 form cortical rings prior to septum initiation and localize to constricting septa. Thus, SIN localization and activity regulation significantly differs in unicellular versus syncytial ascomycete fungi.
Introduction
Proper cell division is essential for growth and development of uni- and multicellular organisms. The fission yeast septation initiation network (SIN) has been identified as a tripartite kinase cascade that connects cell cycle progression with the initiation of cytokinesis [1]. Miss-regulation of the homologous Hippo pathway in animals results in excessive proliferation and formation of tumors [2]. Activation of the SIN by the spindle pole body (SPB)-associated ras superfamily GTPase Spg1 results in recruitment of the STE kinase Cdc7 to SPBs. Two additional SPB-associated kinases Sid1 and Sid2, with their respective regulatory subunits Cdc14 and Mob1 are part of the SIN. Localization studies indicate a hierarchical order of Cdc7 – Sid1 – Sid2, although biochemical evidence for a linear cascade is lacking [3]. Phosphorylation of the phosphatase Clp1 by the nuclear DBF2-related (NDR) effector kinase Sid2 promotes mitotic exit by counteracting the function of the cyclin kinase Cdc2 [4]. In addition, the active Sid2-Mob1 kinase complex, yet not the other SIN components, relocates to the cell cortex, where it is involved in the assembly and constriction of the contractile actomyosin ring that triggers septum formation [5,6].
The related mitotic exit network (MEN) of budding yeast has a similar composition and functions in an analogous manner [7]. However, two significant differences exist. First, the MEN lacks a homolog of the fission yeast kinase Sid1, and the effector kinase Dbf2p is directly phosphorylated by the Cdc7 homolog Cdc15p [8]. The Dbf2p phospho-sites identified in this study correspond to sites highly conserved in other fungal nuclear NDR kinases [9,10], suggesting that Dbf2p activation involves activation segment auto-phosphorylation and phosphorylation of its C-terminal hydrophobic motif through Cdc15p. Second, budding yeast MEN mutants arrest late in the mitotic cell cycle, while the fission yeast SIN is not essential for mitotic exit, and SIN mutants generate aseptate and multinucleated cells. This is primarily based on the different morphologies of both yeasts and the fact that the MEN, but not the SIN, is an essential part of a checkpoint that monitors spindle position in anaphase [7].
Filamentous fungi represent the vast majority of the fungal kingdom. However, despite the importance of septum formation for growth and differentiation of molds, our understanding of septum formation and its regulation in molds is highly fragmentary [11,12]. In contrast to the total separation of mother and daughter cells in unicellular yeasts, the hyphal network formed by filamentous fungi is compartmentalized by incomplete septa. The septal pores enable communication and transport of metabolic components, RNA and organelles between adjacent cells. This controlled segmentation of hyphal units is the basis for the morphological complexity achieved by fungi and for the success of the fungal kingdom [13,14]. Genetic and biochemical analysis in the model molds Neurospora crassa and Aspergillus nidulans revealed that cell cycle-dependent signals of a subset of competent mitotic nuclei within the multinucleate hyphal compartments activate the SIN [15–18]. This, in turn, is critical for the cortical localization of a RHO-4 – BUD-3 GTPase module to sites of future septum formation. The RHO-4 – BUD-3 complex recruits a second RHO-4 – RGF-3 GTPase module, which is required for formin-dependent generation of the contractile actomyosin ring and its subsequent constriction [19–21].
Although most components of the SIN are present in the genomes of both model molds [22,23], a mechanistic understanding of the SIN in the multinuclear hyphal context of filamentous fungi is currently lacking. Mutants in the currently characterized A. nidulans SIN kinase SEPH (Cdc7 homolog) and the MOBA kinase adaptor (Mob1 homolog) are aseptate [24,25]. Similarly, the DBF-2 – MOB-1 complex is essential for septum formation in N. crassa [26]. The A. nidulans SIN proteins MOBA and SIDB (Dbf2 homolog) localize to SPBs and constricting septa [25,27]. However, in contrast to both yeasts, where SPB association of the SIN kinases is essential for activation of the NDR effector kinase and subsequent recruitment of the NDR-Mob1 complex to the cell cortex, SIN activation in A. nidulans does not require SPB association [27]. Already these limited data available in molds suggest major differences in the regulation of the SIN in unicellular versus syncytial fungi. Moreover, a mechanistic picture how SIN proteins interact and transmit signals through the cascade to trigger CAR assembly and constriction is only beginning to emerge in fungi. In this study, we characterized the central SIN network in N. crassa, and provide biochemical evidence for a stepwise activation of a tripartite kinase cascade, consisting of CDC-7, SID-1 and DBF-2.
Results
A tripartite SIN cascade is important for septum formation and localizes constitutively to SPBs and septa
BLAST searches of the N crassa genome with S. pombe and A. nidulans SIN proteins identified homologs for all components of the SIN network except one of the scaffold proteins, which is poorly conserved among different species (Table 1). Mutants defective in predicted components of the tripartite kinase cascade were available as heterokaryotic strains as part of the Neurospora Genome project. Crosses of ∆NCU01335 and ∆NCU04096 with wild type resulted in the expected segregation of the hygromycin cassette used for the gene deletions [28], and the hygromycin-resistent progeny produced thin and aseptate hyphae, which frequently lysed (Figure 1 A, B). Thus, we concluded that NCU01335 and NCU04096 function as part of the SIN, and the proteins were designated CDC-7 and SID-1, respectively.
Table 1. (Predicted) SIN components in yeasts and filamentous fungi.
| Protein feature | S. pombe | S. cerevisiae | N. crassa * | A. nidulans * |
|---|---|---|---|---|
| Polo kinase | Plo1 | Cdc5p | NCU09258 | PLKA |
| GTPase | Spg1 | Tem1p | NCU08878 | AN7206 |
| two component GAP | Cdc16 | Bub2p | NCU03237 | BUBA |
| Byr4 | Bfa1p | NCU11967 | BYRA | |
| STE kinase | Cdc7 | Cdc15 | NCU01335 | SEPH |
| GC kinase | Sid1 | / | NCU04096 | AN8033 |
| GC kinase adaptor | Cdc14 | / | NCU06636 | AN0655 |
| NDR kinase | Sid2 | DBF-2p | DBF-2 | SIDB |
| NDR kinase adaptor | Mob1 | Mob1p | MOB-1 | MOBA |
| Leucin-rich scaffold | Cdc11 | Nud1p | NCU03545 | SEPK |
| Coiled coil scaffold | Sid4 | ? | ? | SNAD |
Generic NCUxxxxx and ANxxxx nomenclature indicates uncharacterized proteins
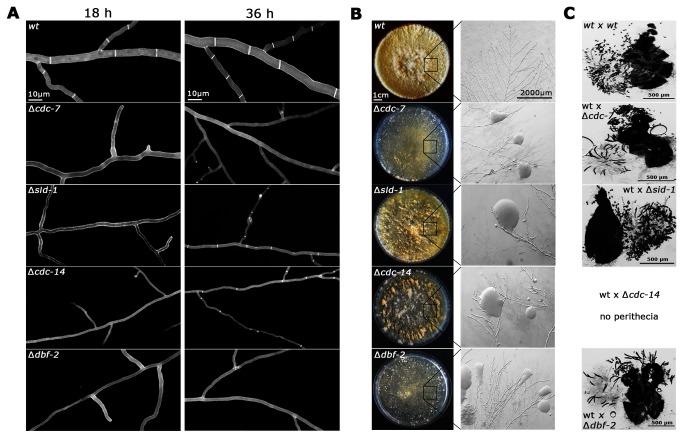
Figure 1. N. crassa SIN components are required for septum formation but display distinct mutant characteristics.
(A) Deletion strains defective in the indicated SIN components generate thin and aseptate hypha in young colonies (18 h time point). In older colonies, the septation defects were suppressed in ∆sid-1 and ∆cdc-14 strains (36 h time point). Cell wall and septa were labeled with Calcofluor White. (B) SIN mutants showed cytoplasmic leakage (magnified inserts), but, due to the fast ability to septate, ∆sid-1 and ∆cdc-14 generated abundant aerial mycelium and asexual spores (conidia; plate morphology). (C) SIN mutants displayed distinct abnormalities during sexual development. wt x ∆ crosses with ∆cdc-7(het) and ∆dbf-2(het) resulted in the frequent formation of large, banana-shaped ascospores. In contrast, wt x ∆sid-1(het) progeny morphology was normal, while crosses of wt x ∆cdc-14(het) produced no mature perithecia .
As previously described for ∆dbf-2 and ∆mob-1 [26], ∆cdc-7 colonies started to show sectors within 1-2 days, in which septa occurred. Also the formation of aerial hyphae and the initiation of the asexual developmental program and production of conidia were strictly dependent on the ability to form septa. Back-crosses of septum-forming ∆cdc-7 colonies (and of ∆sid-1 or ∆cdc-14 colonies; see below) with wild type resulted in two types of hygromycin-resistent progeny: aseptate germlings that produced septa only at later stages of colony development and germlings that formed septa with frequencies that were similar to those of wild type germlings. When we compared the frequency of suppressor occurrence between the different strains, we noted that ∆sid-1 behaved differently than ∆cdc-7, ∆dbf-2 and ∆mob-1 in that septa appeared much faster in this mutant, resulting in the formation of abundant aerial mycelium and sporulation (Figure 1 A, B). Thus, we also analyzed a deletion strain defective for its predicted regulatory subunit NCU06636/CDC-14, which is essential for Sid1 function in fission yeast [1]. ∆cdc-14 germlings were initially aseptate, but produced septa with frequencies comparable to ∆sid-1 and faster than the other SIN deletion strains (Figure 1 A, B). In support of different defects produced by ∆sid-1 and ∆cdc-14 versus ∆cdc-7, ∆dbf-2 and ∆mob-1, we also noted that the morphology of sexual progeny generated in wt x ∆sid-1 crosses was normal, while wt x ∆cdc-14 crosses did not result in mature perithecia (Figure 1 C). This contrasted with the generation of large, banana-shaped ascospores produced in wt x ∆ crosses with ∆cdc-7 as we have previously shown for ∆dbf-2 and ∆mob-1 [26].
Next, we generated strains expressing GFP-fusion proteins to investigate the cellular distribution of these SIN proteins. All constructs were controlled by the ccg-1 promoter and targeted to the his-3 locus in the respective deletion strain in order to confirm functionality of the fusion proteins. In order to test for potential effects of ectopic over-expression, we also modified the endogenous locus of dbf-2 to allow expression of DBF-2-GFP under the control of its endogenous regulatory elements. Although ccg-1 driven GFP-DBF-2 was expressed at ca. 3-fold higher levels, resulting in enhanced cytoplasmic background fluorescence (Figure S1), we observed no differences in the localization pattern of DBF-2 in the two strains (data not shown). We determined that the entire SIN cascade associated with septa (Figure 2 A). DBF-2, SID-1 and CDC-14 accumulated first as cortical ring at the cell cortex prior to the initiation of septum constriction and remained associated with the septal pore of the mature septum after completion of the septation process (Movies S1, S2, S3). We were unable to detect CDC-7-GFP at the cell cortex and a clear CDC-7 signal was only visible at the septal pore of the mature septum. The failure to observe CDC-7 at early stages of septum formation is consistent with the low expression level of ccg-1 driven CDC-7-GFP (Figure S1). Alternatively, this may indicate that CDC-7 only associates with the septum after constriction. Moreover, all SIN components associated with spindle pole bodies (SPBs) in a constitutive manner and independently of the cell cycle state (Figure 2 B).
Figure 2. N. crassa SIN components localize to SPBs and septa (A).
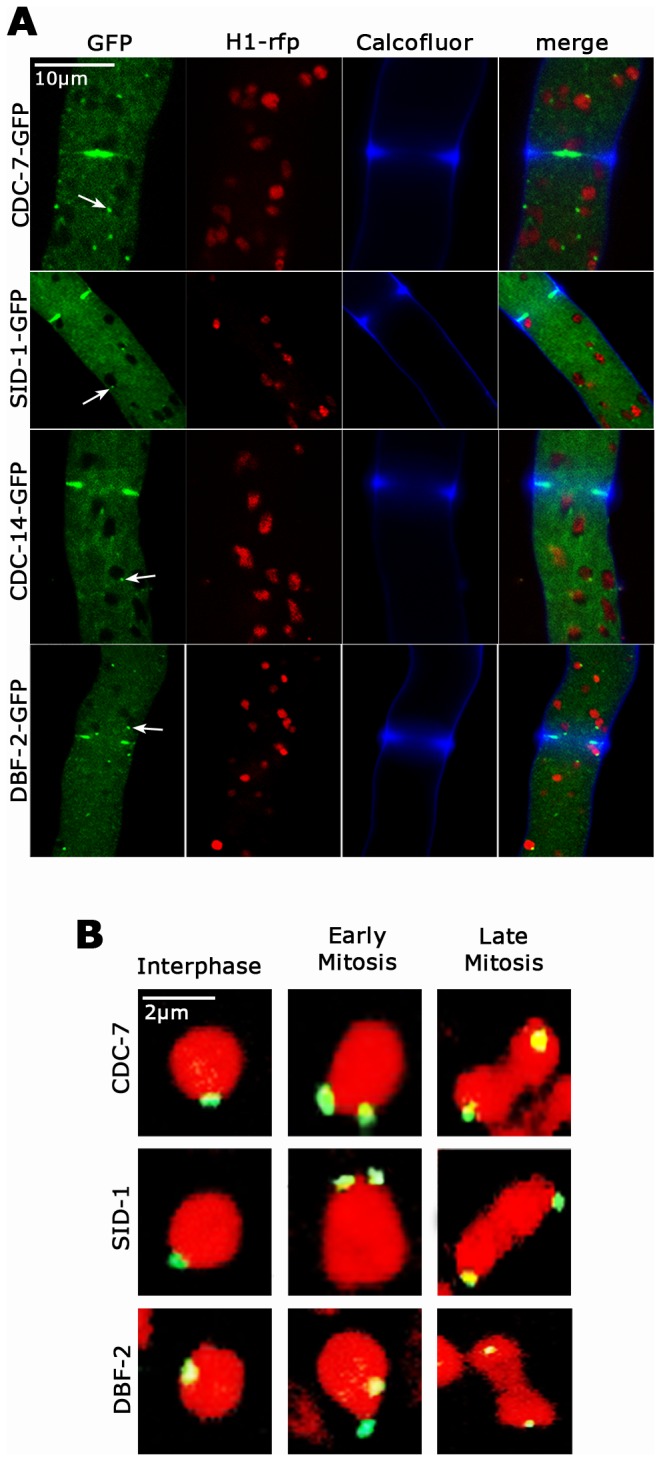
Functional GFP fusion proteins of CDC-7, SID-1, CDC-14 and DBF-2 localized to spindle pole bodies (arrows) and as constricting rings at forming septa. Nuclei were labeled with histone H1-RFP, the cell wall was stained with Calcofluor White. (B) The localization of the three SIN kinases CDC-7, SID-1 and DBF-2 to SPBs is constitutive and cell cycle independent. The three SIN kinases associate with SPBs of interphase nuclei as well as during early and late mitotic stages (as indicated by nuclear morphology). Nuclei were labeled with histone H1-RFP.
CDC-7-dependent activation of DBF-2 occurs through SID-1
Activation of Dbf2p in S. cerevisiae involves direct phosphorylation by the Cdc7 homolog Cdc15p [8]. In contrast, the S. pombe SIN cascade likely acts in the order Cdc7-Sid1-Sid2, but biochemical evidence for a step-wise activation of these kinases is lacking [3,29]. In order to determine the functional relationship between the three N. crassa SIN kinases and to provide a mechanism for DBF-2 activation, we tested if CDC-7 interacted with SID-1 and/or DBF-2. Reciprocal co-immunoprecipitation (co-IP) experiments of CDC-7-GFP and HA-SID-1 from cell extracts co-expressing both functionally tagged proteins proved a stable interaction of the two kinases (Figure 3 A). In contrast, we were unable to show interactions between CDC-7 and DBF-2 or SID-1 and DBF-2 in co-IP experiments performed under identical conditions (data not shown), suggesting that these interactions may be more dynamic.
Figure 3. CDC-7-dependent activation of DBF-2 occurs through SID-1 (A).
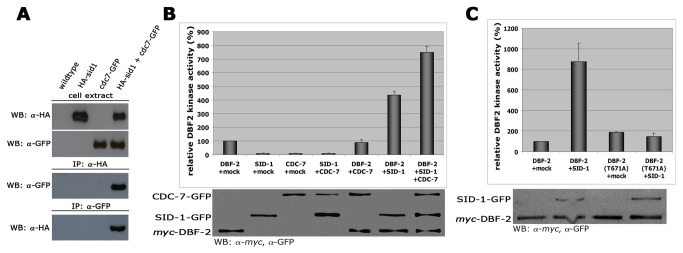
Reciprocal co-immunoprecipitation experiments of CDC-7-GFP and HA-SID-1 from cell extracts co-expressing both functionally tagged proteins indicate interaction of the two kinases. (B) In vitro DBF-2 activity assays revealed that addition of separately purified SID-1 stimulate DBF-2. SID-1-dependent stimulation of DBF-2 was further increased by addition of CDC-7 to the reaction. As control, CDC-7 alone did not stimulate DBF-2 (n = 5). To confirm equal precipitation of protein used for the kinase reactions, the kinase-sepharose pellet after the kinase reaction was boiled in Laemmli buffer and the supernatant used to determine equal protein abundance by SDS-PAGE and Western blot. (C) SID-1 was able to stimulate DBF-2, but not DBF-2(T671A). Western blot analysis of the precipitated proteins was used to determine comparable kinase levels (n = 5).
Wild type DBF-2 precipitated from N. crassa extracts displayed activity towards a synthetic peptide encompassing the consensus NDR kinase target motif, which we had previously used for in vitro activity assays with the related NDR kinase COT-1 [30]. Using this assay, we determined that precipitated SID-1 enhanced the activity of separately purified DBF-2 (Figure 3 B). If the N. crassa SIN functions as tripartite, stepwise kinase cascade, we hypothesized that SID-1-dependent stimulation of DBF-2 should be further increased by addition of CDC-7 – a prediction that we were able to confirm in in vitro assays of individually precipitated components (Figure 3 B). Control experiments with SID-1 or CDC-7 precipitates proved the specificity of this assay for the NDR kinase and support the interpretation that that SID-1 is required for transmitting CDC-7-dependent signals towards DBF-2.
Activation of NDR kinases requires phosphorylation of a C-terminal hydrophobic motif [9,31,32]. We determined that SID-1-dependent stimulation of DBF-2 was only possible with purified wild type DBF-2, but not with DBF-2(T761A), which contained a threonine to alanine substitution of the predicted hydrophobic motif phosphorylation site (Figure 3 C). Phosphorylation experiments coupled with mass-spectrometric analysis further supported Thr671 phosphorylation of DBF-2 by SID-1: we were only able to identify phosphopeptides of the hydrophobic motif of DBF-2 precipitated under high-stringency conditions when we co-incubated DBF-2 with separately purified SID-1 in in vitro kinase reactions (Figure S2). Based on these results, we conclude that SID-1 phosphorylates DBF-2 at Thr671.
Dual phosphorylation of DBF-2 is required for kinase activity and septum formation
In order to further dissect the phospho-regulation of DBF-2, we characterized strains expressing DBF-2 variants harboring point mutations in the predicted auto-phosphorylation and hydrophobic motif sites Ser499 and Thr671, respectively. Both, the Ser499 to alanine (inactive mimic) and to glutamate (active mimic) substitutions were nonfunctional, and these kinase variants were unable to complement the septation defects of ∆dbf-2 (Figure 4 A). Of the Thr671 to alanine/glutamate substitutions, DBF-2(T671E), but not DBF-2(T671A) was functional and complemented the deletion strain. DBF-2 variants harboring either of the two Ser499 substitutions displayed in vitro kinase activities reduced to ca. 1/3 of the wild type DBF-2 control (33±6% and 30±12% for DBF-2(S499A) and DBF-2(S499E), respectively; n = 5; Figure 4 B). In contrast, the kinase activity of DBF-2(T671A) was slightly increased (200±7%; n = 5), while DBF-2(T671E) displayed >30-fold increased activity (3300±240%; n = 5). Modification of homologous residues affected the interaction of the S. pombe NDR kinase Sid2 with Mob1 [29]. However, MOB-1 binding was not affected by these modifications in N. crassa, and wild type MOB-1 levels co-precipitated with each DBF-2 variant (Figure 4 B). We also performed in vitro phosphorylation experiments coupled with mass-spectrometric analysis of kinase variants precipitated under high-stringency conditions to confirm Ser499 as auto-phosphorylation site. Strong phosphophorylation of Ser499 in tryptic peptides generated from wild type DBF-2 and hyperactive DBF-2(T671E), but not kinase-dead DBF-2(D422A) indicated that Ser499 is the primary site of DBF-2 auto-phosphorylation (Figure S2). Moreover, we noticed phosphorylation of multiple S/T residues in the N-terminal, non-catalytic region of the kinase in DBF-2(T671E).
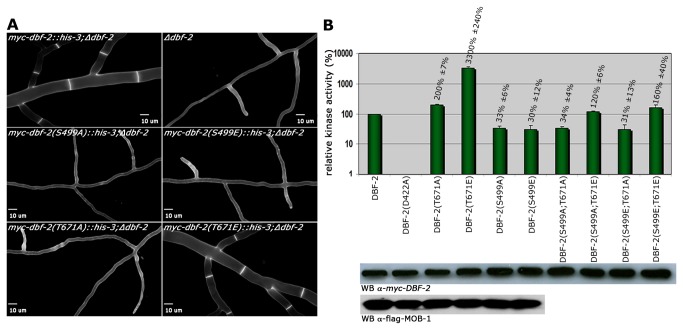
Figure 4. Dual phosphorylation of DBF-2 is required for kinase activity and septum formation.
(A) Functional characterization of two conserved phosphorylation sites of DBF-2. The phosphomimetic DBF-2(T671E) variant complemented Δdbf-2, while substitution of Ser499 to alanine and glutamate and Thr671 to alanine did not. Cell wall and septa were labeled with Calcofluor White. (B) Kinase activity and MOB-1 interaction pattern of the indicated DBF-2 variants. Hydrophobic motif phosphorylation of Thr671 was required for maximal kinase activity, while either modification of Ser499 within the activation segment reduced DBF-2 activity to ca. 30% of the wild type DBF-2 control. Phospho-site double mutant analysis indicated that substitution of Thr671 to glutamate in a S499A and S499E background could only partly restore kinase activity. Precipitated DBF-2 variants were assayed in vitro using the synthetic NDR kinase peptide (KKRNRRLSVA) as substrate (n = 5). Western Blot analysis indicated equal precipitation of the co-activator protein MOB-1 with DBF-2 activation segment and hydrophobic motif variants.
The fact that both Ser499 modifications reduced DBF-2 activity in vitro and were nonfunctional in vivo may suggest that dynamic modification of the activation segment (i.e. the regulated phosphorylation/dephosporylation) could modulate DBF-2 function. We generated various combinations of double mutants in order to further explore this possibility (Figure 4 B). None of these double modifications resulted in functional protein, and complementation of the ∆dbf-2 defects failed with all constructs (data not shown). Kinase assays revealed that the two Thr671 to alanine variants DBF-2(S499A;T671A) and DBF-2(S499E;T671A) displayed reduced activities, which were similar to the individual Ser499 mutations (34±4% and 31±13% of wild type DBF-2, respectively; n = 5). Substitution of the hydrophobic motif threonine with glutamate in a S499A and S499E background increased kinase activity 3.6- and 5.3-fold compared to the respective Ser499-modified protein (n = 5). Thus, phosphorylation of Thr671 can partly overcome the lack of activation segment modification, but the stable modification of Ser499 prevented full activation of DBF-2.
Discussion
Regulation of septum formation and the composition of the SIN in filamentous fungi is only beginning to be unraveled. The phenotypic and biochemical analysis of predicted SIN components allowed the identification of the SIN kinase cascade CDC-7, SID-1 and DBF-2 together with their respective regulatory subunits CDC-14 and MOB-1, and the characterization of their hierarchical relationship in N. crassa. Based on in vitro kinase assays, we provide the first biochemical evidence that SID-1 activates DBF-2 through hydrophobic motif phosphorylation, analogous to POD-6-dependent phosphorylation of COT-1 [32,33] or activation of related fungal and animal NDR kinases through upstream germinal centre kinases of the STE kinase family [9,31,34,35]. We observed no direct activation of DBF-2 by CDC-7, but SID-1-dependent stimulation of DBF-2 was further enhanced through CDC-7. Although our data do not rule out that direct phosphorylation of DBF-2 by CDC-7 is a prerequisite for hydrophobic motif phosphorylation of DBF-2 by SID-1, we consider this interpretation unlikely. It was previously shown that the budding yeast kinase Cdc15p also targets the homologous threonine residue present in the hydrophobic motif of Dbf2p [9,31,34,35], just as we have determined for SID-1-dependent phosphorylation of DBF-2 in N. crassa. Thus our data, strongly suggest that the N. crassa SIN functions as hierarchical, stepwise kinase cascade.
Interestingly, we observed that strains carrying deletions of sid-1 or its regulatory subunit cdc-14 behaved differently than mutants in the remaining SIN components. Although initially aseptate, accumulation of suppressor mutations and, as result, the capability to form septa and to produce mitotic conidiospores was much higher in ∆sid-1 and ∆cdc-14 than in other SIN mutants (although all SIN mutants regain the ability to form septa at some point). Moreover, ∆sid-1 and ∆cdc-14 progeny did not form the large, banana-shaped ascospores described for ∆dbf-2, ∆cdc-7 and ∆mob-1 containing crosses (this study [26]), suggesting that the presentation of the SIN as a linear kinase cascade may represent a simplified view. Possible scenarios could be that, in contrast to our in vitro data, CDC-7 is able to directly target DBF-2 in vivo. Alternatively, additional uncharacterized kinases may function in concert with SID-1 to regulate DBF-2.
Our characterization of DBF-2 variants in predicted regulatory phosphorylation sites indicates that DBF-2 is regulated by dual phosphorylation. We determined that Ser499 within the activation segment is auto-phosphorylated, while the hydrophobic motif site Thr691 is targeted by the upstream kinase SID-1. Interestingly, we also detected tyrosine-specific phospho-peptides in our mass-spectrometry analysis of hyperactive DBF-2(T671E), suggesting that DBF-2 (and possibly other NDR kinases) can function as dual-specific kinases. These data are consistent with a recent report on tyrosine kinase activity of the plant NDR kinase PLK01 [36].
Current models for NDR kinase activity regulation predict the formation of inactive, competent and active conformations, which correspond to non-phosphorylated, auto-phosphorylated and dual-phosphorylated states, respectively [10,37]. In this study, we analyzed, for the first time, the full complement of kinase variants harboring individual and double mutant substitutions in these two regulatory sites. While DBF-2(S499A) displayed in vitro activity reduced to ca. 1/3rd of the wild type control, DBF-2(T671A) produced activities in the range of wild type DBF-2. Thus, auto-but not hydrophobic motif phosphorylation is required for normal kinase activity. However, alanine substitution of both sites resulted in nonfunctional protein, indicating that phosphorylation of both sites is essential for the in vivo functionality of DBF-2. This conclusion is supported by DBF-2(T671E), which resulted in maximal in vitro activity and was the only DBF-2 variant functional in vivo.
We further determined that glutamate substitution of the hydrophobic motif site in glutamate- and alanine-substituted auto-phosphorylation backgrounds only partially recovered in vitro activities and that these kinase variants were nonfunctional in vivo. The fact that phosphorylation of Thr671 can only partly overcome the lack of activation segment modification may suggest that dynamic modification of the activation segment may be important for full activation and functionality of DBF-2. Alternatively, Ser499 modification may simply impair the functionality of the protein. However, we consider this unlikely, because analogous substitutions have successfully been used for the analysis of several fungal [8,9,30,32,35] as well as animal [31,34] NDR kinases.
Functional GFP fusion constructs of the three SIN kinases localize to SPBs and septa. In contrast to the situation in both unicellular yeast models [3,7], SPB association of the SIN cascade is not cell cycle dependent. Moreover, SID-1 and DBF-2 localize to the cell cortex prior to septum constriction and to the forming septum. We detected CDC-7 only at the septal pore of the mature septum, possibly indicating that this kinase only associates with the septum after constriction. Alternatively, CDC-7 levels below our imaging resolution may associate with the other SIN kinases during early stages of septation. Together with the finding that SIN activation in A. nidulans does not require SPB association of the NDR kinase SIDB [27], these data indicate major differences in the regulation of the SIN in unicellular versus syncytial ascomycetes.
In contrast to a previous report [38], we did not detect defects of SIN mutants in the proper completion of the cell cycle. This observation is in line with the fact that cell cycle progression was unaffected in A. nidulans SIN mutants [20,24,25]. Moreover, meiotic cell divisions are not affected in N. crassa SIN mutants. Re-sequencing of an old laboratory strain identified a mutant called Banana as dbf-2 deletion strain [39]. Its previous characterization had revealed that the eight nuclei derived from the two meiotic and one mitotic divisions are formed in a normal manner in Ban+/Ban asci, but that the resulting nuclei are then enclosed in a single giant ascospore [40,41]. This is consistent with data obtained for budding and fission yeasts, where the SIN is largely dispensable during meiosis, but required for spore wall formation and ascospore morphology [42,43]. In summary, these data indicate an essential function of the SIN during cross wall formation in vegetative cells and during the formation of asco- as well as conidiospores in unicellular and filamentous ascomycetes.
Materials and Methods
Strains, media and growth conditions
Strains used in this study are listed in Table S1. General genetic procedures and media used in the handling of N. crassa are available through the Fungal Genetic Stock Center (www.fgsc.net; [44]).
Plasmid construction and fungal expression of tagged proteins
To obtain strains that express GFP/myc/HA-tagged fusion proteins from the his-3 locus, the open reading frames of DBF-2, SID-1, CDC-14 and CDC-7 were amplified by PCR as annotated by the N. crassa database using primers listed in Table S2 and introduced via SgsI/PacI, BcuI/PacI or XbaI/PacI, respectively, into pCCG1_N-GFP, pMF272, pHAN1 or pCCG1-3xMYC [41,45–47]. Point mutated versions of myc-DBF-2 were generated by site-directed mutagenesis according to the manufacturer’s instructions (Stratagene). Resulting plasmids were transformed into his-3, nic-3;his-3 or trp-1;his-3, and were selected for complementation of the his-3 auxotrophy. Strains expressing the fusion constructs were crossed with the respective deletion strain. Progeny were selected on minimal media containing hygromycin and assayed for complementation of the mutant’s growth/septation defects.
Microscopy
Low magnification documentation of fungal hyphae or colonies was performed as described [48,49] using an SZX16 stereomicroscope, equipped with a Colorview III camera and CellD imaging software (Olympus). Images were further processed using Photoshop CS2 (Adobe). An inverted Axiovert Observer Z1 microscope (Zeiss) equipped with a CSU-X1 A1 confocal scanner unit and a QuantEM 512SC camera (Photometrics) was used for spinning disk confocal microscopy [50,51]. Slidebook 5.0 software (Intelligent Imaging Innovations) was used for image/movie acquisition, deconvolution and image analysis. Cells wall and plasma membrane were stained with Calcofluor white (2µg/ml) and FM4-64 (1µg/ml) respectively. Time-lapse imaging was performed at capture intervals of 20-30 s for periods up to 15 min using the oil immersion objective 100x/1.3. Image series were converted into movies (*.movs). Aseptate deletion strains were maintained as heterokaryons to avoid the accumulation of suppressor mutations. Image analysis was performed on germlings and young colonies developing from out-crossed hygromycin-resistent ascospores displaying all characteristics of the deletion stain.
Protein methods
Liquid N. crassa cultures were grown at room temperature, harvested gently by filtration using a Büchner funnel and ground in liquid nitrogen. The pulverized mycelium was mixed 1:1 with IP-Buffer (50 mM Tris/HCL pH 7.5, 100 mM KCl, 10 mM MgCl2, 0.1% NP-40, 5 mM NaF, 1 mM PEFAbloc SC, 2mM DTT, 1 mM Na3VO4, 25 mM β-glycerophosphate, 2 mM benzamidine, 2 ng/µl pepstatin A, 10 ng/µl aprotinin, 10 ng/µl leupeptin) and centrifuged as described [32,52]. Co-immunoprecipitation was performed as described [26] with cell extracts from fused, heterokaryotic strains that were selected by their ability to grow on minimal media lacking supplements. Protein-decorated beads were washed twice with “low-stringency” IP buffer (50 mM Tris/HCL pH 7.5, 100 mM KCl, 10 mM MgCl2, 0.1% NP-40, 5 mM NaF, 1 mM PEFAbloc SC, 2mM DTT, 1 mM Na3VO4, 25 mM β-glycerophosphate, 2 mM benzamidine, 2 ng/µl pepstatin A, 10 ng/µl aprotinin, 10 ng/µl leupeptin), and immunoprecipitated proteins were recovered by boiling the beads for 10 min at 98°C in 3x Laemmli buffer and separated by 10% SDS-PAGE. Monoclonal mouse α-HA (clone HA-7, Sigma Aldrich), α-FLAG M2 (Sigma-Aldrich), α-myc (9E10, Santa Cruz) and α-GFP (B2, Santa Cruz) and GFP trap beads (Chromotek) were used in this study.
DBF-2 kinase activity assays were performed as described [30,32] according to a modified protocol described previously for animal NDR kinase [53]. Total protein levels of cell extracts were determined by Bradford analysis with bovine serum albumin standard solutions as a reference, using Roti®-Quant (Carl Roth) and a Tecan Infinite® M200 microplate reader (Tecan) and adjusted with IP buffer. Recovered kinase-bound beads were washed twice with “high-stringency” IP buffer containing 0,5 M NaCl followed by two washes with kinase reaction buffer (20 mM Tris pH 7.5, 10 mM MgCl2, 1 mM DTT, 1 mM benzamidine, 1 mM Na3VO4, 5 mM NaF) to remove all potentially interacting (and thus co-purifying) proteins, and were finally resuspended in 50 μl kinase buffer containing 2 mM of synthetic substrate peptide (KKRNRRLSVA), 0.5 mM ATP and 1 μCi [32P]ATP. Western blot experiments performed as controls confirmed that no other kinase of the SIN except the tagged and precipitated protein co-purified under these “high-stringency” wash conditions. After incubation for 1 h at 37°C, samples were centrifuged for 2 min at 4.000 g, the supernatant was spotted onto P81 phosphocellulose paper circles (Whatman). Dried circles were washed 2 times with 1% phosphoric acid before incorporation of phosphate into the substrate peptide was measured by liquid scintillation counting. The remaining protein-sepharose pellet was boiled for 10 min in 3x Laemmli buffer and the supernatant was used to determine equal DBF-2 concentration in the kinase reaction by SDS-PAGE and Western blot. DBF-2 stimulation was assayed with separately purified SID-1 and CDC-7 (washed with “high-stringency” IP buffer to eliminate co-purifying contaminations), which were added during the first washing step, before kinase reaction.
Mass spectrometry, database analysis and enrichment of phosphopeptides
For protein identification by mass spectrometry, peptides of in-gel trypsinated proteins were extracted from of Commassie-stained gel slices as described [54]. Peptides of 5 μl sample solution were trapped and washed with 0.05 % trifluoroacetic acid on an Acclaim® PepMap 100 column (75 μm x 2 cm, C18, 3 μm, 100 Å, P/N164535 Thermo Scientific) at a flow rate of 4 μl/min for 12 min. Analytical peptide separation by reverse phase chromatography was performed on an Acclaim® PepMap RSLC column (75 μm x 15 cm, C18, 3 μm, 100 Å, P/N164534 Thermo Scientific) running a gradient from 96 % solvent A (0.1 % formic acid) and 4 % solvent B (acetonitrile, 0.1 % formic acid) to 50 % solvent B within 25 min at a flow rate of 250 nl/min (solvents and chemicals: Fisher Chemicals). Peptides eluting from the chromatographic column were on-line ionized by nano-electrospray using the Nanospray Flex Ion Source (Thermo Scientific) and transferred into the mass spectrometer. Full scans within m/z of 300-1850 were recorded by the Orbitrap-FT analyzer at a resolution of 60.000 at m/z 400. Each sample was analyzed using two different fragmentation techniques applying a data-dependent top 5 experiment: collision-induced decay with multistage activation and readout in the LTQ Velos Pro linear ion trap, and higher energy collision dissociation and subsequent readout in the Orbitrap-FT analyzer. LC/MS method programming and data acquisition was performed with the software XCalibur 2.2 (Thermo Fisher). Orbitrap raw files were analyzed with the Proteome Discoverer 1.4 software (Thermo Scientific) using the Mascot and Sequest search engines against the N. crasssa protein database with the following criteria: peptide mass tolerance 10 ppm, MS/MS ion mass tolerance 0.8 Da, and up to two missed cleavages allowed. Enrichment of phosphopeptides was performed based on the method developed by [55], using TiO2 columns (TopTip TiO2 10-200 μl; Glygen Corporation).
Supporting Information
Expression analysis of the used GFP fusion constructs under the control of the ccg-1 and their native promoters. Anti-GFP Western blot of normalized cell extracts of strains expressing GFP fusion proteins under the control of the indicated promoters (left panel). Quantification of the relative expression levels of the indicated proteins. Protein levels are normalized to DBF-2 abundance (n = 3).
(TIF)
Mass-spectrometric analysis of DBF2 phosphorylation sites. (A) In vitro phosphorylation experiments coupled with mass-spectrometric analysis. Ser499 was phosphorylated in wild type DBF-2 and hyperactive DBF-2(T671E), but not DBF-2(D422A), identifying this residue as primary site of auto-phosphorylation. Multiple additional S/T phosphorylation sites were detected in the N-terminal region of DBF-2(T671E). Mass-spectrometric analysis of SID-1-dependant DBF-2 phosphorylation sites identified T671 as primary site of phosphorylation and additional S/T phosphorylation sites variously distributed. (B) Tryptic peptides generated from wild type DBF-2 displayed HCD-fragmentation spectrum of the peptide SIVGSPDYMAPEVLR with Ser499 phosphorylated. Fragment b-ions (yellow) and y-ions (blue) with an asterisk indicate neutral loss of ammonia (-17 Da), and ions labeled with a circle neutral loss of water water (-18 Da), charge states are in brackets. The peptide cross-correlation score for Sequest (XCorr) was 4.9 and the Mascot IonScore was 77. The probability of Ser499-phosphorylation was caclulated by the pRS algorithm to be 99.99% [J Proteome Res 10: 5354-5362]. (C) Representative fragmentation spectrum of the peptide SLFVGFtFR with phosphorylation at T671. Fragment ions b* and y* are ions with loss of ammonia (-17 Da), and fragment ions bo and co are ions with loss of water (-18 Da). The number in brackets indicates the charge state of the fragment ion. The peptide was fragmented by CID and fragment ions were detected in the linear ion trap. The peptide was identified with Mascot and Sequest search engines (peptide IonScore of 30 and XCorr of 3.45, respectively). pRS score for phosporylation at tyrosine was 100% [J Proteome Res 10: 5354-5362].
(TIF)
N. crassa strains used in this study.
(DOCX)
Primers used in this study.
(DOCX)
Time-course of DBF-2-GFP localization during septum formation. DBF-2-GFP formed cortical rings at incipient septation sites that constricted during septum formation and accumulated around the septal pore of the completed septum (a) GFP channel; (b) RFP channel; (c) merged. The plasma membrane was stained with FM4-64. Images were captured at 20 sec intervals.
(MOV)
Time-course of SID-1-GFP localization during septum formation. SID-1-GFP formed cortical rings at incipient septation sites that constricted during septum formation and accumulated around the septal pore of the completed septum (a) GFP channel; (b) RFP channel; (c) merged. The plasma membrane was stained with FM4-64. Images were captured at 20 sec intervals.
(MOV)
Time-course of CDC-14-GFP localization during septum formation. CDC-14-GFP formed cortical rings at incipient septation sites that constricted during septum formation and accumulated around the septal pore of the completed septum (a) GFP channel; (b) RFP channel; (c) merged. The plasma membrane was stained with FM4-64. Images were captured at 20 sec intervals.
(MOV)
Acknowledgments
We thank Oliver Valerius for his continuous help with the phospho-peptide analysis and Klaus Palme for providing lab space for the group.
Funding Statement
This research project was financially supported through the DFG Heisenberg fellowship SE1054/6-1, the DFG grants SE1054/3-2 and SE1054/7-1 and by the Göttingen Graduate School for Neurosciences, Biophysics and Molecular Biosciences (GGNB) (DFG grant GSC 226/1). The authors acknowledge the Fungal Genetics Stock Center and the Neurospora program grant PO1 GM068087 for continuous support. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Krapp A, Simanis V (2008) An overview of the fission yeast septation initiation network (SIN). Biochem Soc Trans 36: 411-415. doi: 10.1042/BST0360411. PubMed: 18481970. [DOI] [PubMed] [Google Scholar]
- 2. Hergovich A, Hemmings BA (2012) Hippo signalling in the G2/M cell cycle phase: lessons learned from the yeast MEN and SIN pathways. Semin Cell Dev Biol 23: 794-802. doi: 10.1016/j.semcdb.2012.04.001. PubMed: 22525225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Johnson AE, McCollum D, Gould KL (2012) Polar opposites: Fine-tuning cytokinesis through SIN asymmetry. Cytoskeleton (Hoboken) 69: 686-699. doi: 10.1002/cm.21044. PubMed: 22786806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen CT, Feoktistova A, Chen JS, Shim YS, Clifford DM et al. (2008) The SIN kinase Sid2 regulates cytoplasmic retention of the S. pombe Cdc14-like phosphatase Clp1. Curr Biol 18: 1594-1599. doi: 10.1016/j.cub.2008.08.067. PubMed: 18951025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Roberts-Galbraith RH, Gould KL (2008) Stepping into the ring: the SIN takes on contractile ring assembly. Genes Dev 22: 3082-3088. doi: 10.1101/gad.1748908. PubMed: 19056889. [DOI] [PubMed] [Google Scholar]
- 6. Hachet O, Simanis V (2008) Mid1p/anillin and the septation initiation network orchestrate contractile ring assembly for cytokinesis. Genes Dev 22: 3205-3216. doi: 10.1101/gad.1697208. PubMed: 19056897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Meitinger F, Palani S, Pereira G (2012) The power of MEN in cytokinesis. Cell Cycle 11: 219-228. doi: 10.4161/cc.11.2.18857. PubMed: 22189712. [DOI] [PubMed] [Google Scholar]
- 8. Mah AS, Jang J, Deshaies RJ (2001) Protein kinase Cdc15 activates the Dbf2-Mob1 kinase complex. Proc Natl Acad Sci U S A 98: 7325-7330. doi: 10.1073/pnas.141098998. PubMed: 11404483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jansen JM, Barry MF, Yoo CK, Weiss EL (2006) Phosphoregulation of Cbk1 is critical for RAM network control of transcription and morphogenesis. J Cell Biol 175: 755-766. doi: 10.1083/jcb.200604107. PubMed: 17145962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Maerz S, Seiler S (2010) Tales of RAM and MOR: NDR kinase signaling in fungal morphogenesis. Curr Opin Microbiol 13: 663-671. doi: 10.1016/j.mib.2010.08.010. PubMed: 20869909. [DOI] [PubMed] [Google Scholar]
- 11. Harris SD (2001) Septum formation in Aspergillus nidulans. Curr Opin Microbiol 4: 736-739. doi: 10.1016/S1369-5274(01)00276-4. PubMed: 11731327. [DOI] [PubMed] [Google Scholar]
- 12. Seiler S, Justa-Schuch D (2010) Conserved components, but distinct mechanisms for the placement and assembly of the cell division machinery in unicellular and filamentous ascomycetes. Mol Microbiol 78: 1058-1076. doi: 10.1111/j.1365-2958.2010.07392.x. PubMed: 21091496. [DOI] [PubMed] [Google Scholar]
- 13. Gull K (1978) Form and function of septa in filamentous fungi. In: JESaDR , Berry. The Filamentous Fungi, Developmental Mycology. New York: John Wiley & Sons; pp. 78-93. [Google Scholar]
- 14. Harris SD (2011) Hyphal morphogenesis: an evolutionary perspective. Fungal Biol 115: 475-484. doi: 10.1016/j.funbio.2011.02.002. PubMed: 21640312. [DOI] [PubMed] [Google Scholar]
- 15. Wolkow TD, Harris SD, Hamer JE (1996) Cytokinesis in Aspergillus nidulans is controlled by cell size, nuclear positioning and mitosis. J Cell Sci 109 ( 8): 2179-2188. PubMed: 8856514. [DOI] [PubMed] [Google Scholar]
- 16. Minke PF, Lee IH, Plamann M (1999) Microscopic analysis of Neurospora ropy mutants defective in nuclear distribution. Fungal Genet Biol 28: 55-67. doi: 10.1006/fgbi.1999.1160. PubMed: 10512672. [DOI] [PubMed] [Google Scholar]
- 17. Riquelme M, Yarden O, Bartnicki-Garcia S, Bowman B, Castro-Longoria E et al. (2011) Architecture and development of the Neurospora crassa hypha -- a model cell for polarized growth. Fungal Biol 115: 446-474. doi: 10.1016/j.funbio.2011.02.008. PubMed: 21640311. [DOI] [PubMed] [Google Scholar]
- 18. Mourino-Pérez R (2013) Septum development in filamentous ascomycetes. Fungal Biol Rev 27: 1-9. doi: 10.1016/j.fbr.2013.02.002. [DOI] [Google Scholar]
- 19. Rasmussen CG, Glass NL (2005) A Rho-type GTPase, rho-4, is required for septation in Neurospora crassa. Eukaryot Cell 4: 1913-1925. doi: 10.1128/EC.4.11.1913-1925.2005. PubMed: 16278458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Si H, Justa-Schuch D, Seiler S, Harris SD (2010) Regulation of septum formation by the Bud3-Rho4 GTPase module in Aspergillus nidulans. Genetics 185: 165-176. doi: 10.1534/genetics.110.114165. PubMed: 20176976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Justa-Schuch D, Heilig Y, Richthammer C, Seiler S (2010) Septum formation is regulated by the RHO4-specific exchange factors BUD3 and RGF3 and by the landmark protein BUD4 in Neurospora crassa. Mol Microbiol 76: 220-235. doi: 10.1111/j.1365-2958.2010.07093.x. PubMed: 20199606. [DOI] [PubMed] [Google Scholar]
- 22. Galagan JE, Calvo SE, Cuomo C, Ma LJ, Wortman JR et al. (2005) Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 438: 1105-1115. doi: 10.1038/nature04341. PubMed: 16372000. [DOI] [PubMed] [Google Scholar]
- 23. Galagan JE, Calvo SE, Borkovich KA, Selker EU, Read ND et al. (2003) The genome sequence of the filamentous fungus Neurospora crassa. Nature 422: 859-868. doi: 10.1038/nature01554. PubMed: 12712197. [DOI] [PubMed] [Google Scholar]
- 24. Bruno KS, Morrell JL, Hamer JE, Staiger CJ (2001) SEPH, a Cdc7p orthologue from Aspergillus nidulans, functions upstream of actin ring formation during cytokinesis. Mol Microbiol 42: 3-12. PubMed: 11679062. [DOI] [PubMed] [Google Scholar]
- 25. Kim JM, Lu L, Shao R, Chin J, Liu B (2006) Isolation of mutations that bypass the requirement of the septation initiation network for septum formation and conidiation in Aspergillus nidulans. Genetics 173: 685-696. doi: 10.1534/genetics.105.054304. PubMed: 16624915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Maerz S, Dettmann A, Ziv C, Liu Y, Valerius O et al. (2009) Two NDR kinase-MOB complexes function as distinct modules during septum formation and tip extension in Neurospora crassa. Mol Microbiol 74: 707-723. doi: 10.1111/j.1365-2958.2009.06896.x. PubMed: 19788544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kim JM, Zeng CJ, Nayak T, Shao R, Huang AC et al. (2009) Timely septation requires SNAD-dependent spindle pole body localization of the septation initiation network components in the filamentous fungus Aspergillus nidulans. Mol Biol Cell 20: 2874-2884. doi: 10.1091/mbc.E08-12-1177. PubMed: 19386763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Colot HV, Park G, Turner GE, Ringelberg C, Crew CM et al. (2006) A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc Natl Acad Sci U S A 103: 10352-10357. doi: 10.1073/pnas.0601456103. PubMed: 16801547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hou MC, Guertin DA, McCollum D (2004) Initiation of cytokinesis is controlled through multiple modes of regulation of the Sid2p-Mob1p kinase complex. Mol Cell Biol 24: 3262-3276. doi: 10.1128/MCB.24.8.3262-3276.2004. PubMed: 15060149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ziv C, Kra-Oz G, Gorovits R, März S, Seiler S et al. (2009) Cell elongation and branching are regulated by differential phosphorylation states of the nuclear Dbf2-related kinase COT1 in Neurospora crassa. Mol Microbiol 74: 974-989. doi: 10.1111/j.1365-2958.2009.06911.x. PubMed: 19818014. [DOI] [PubMed] [Google Scholar]
- 31. Stegert MR, Hergovich A, Tamaskovic R, Bichsel SJ, Hemmings BA (2005) Regulation of NDR protein kinase by hydrophobic motif phosphorylation mediated by the mammalian Ste20-like kinase MST3. Mol Cell Biol 25: 11019-11029. doi: 10.1128/MCB.25.24.11019-11029.2005. PubMed: 16314523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Maerz S, Dettmann A, Seiler S (2012) Hydrophobic motif phosphorylation coordinates activity and polar localization of the Neurospora crassa nuclear Dbf2-related kinase COT1. Mol Cell Biol 32: 2083-2098. doi: 10.1128/MCB.06263-11. PubMed: 22451488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Seiler S, Vogt N, Ziv C, Gorovits R, Yarden O (2006) The STE20/germinal center kinase POD6 interacts with the NDR kinase COT1 and is involved in polar tip extension in Neurospora crassa. Mol Biol Cell 17: 4080-4092. doi: 10.1091/mbc.E06-01-0072. PubMed: 16822837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. He Y, Fang X, Emoto K, Jan YN, Adler PN (2005) The tricornered Ser/Thr protein kinase is regulated by phosphorylation and interacts with furry during Drosophila wing hair development. Mol Biol Cell 16: 689-700. PubMed: 15591127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu G, Young D (2012) Conserved Orb6 phosphorylation sites are essential for polarized cell growth in Schizosaccharomyces pombe. PLOS ONE 7: e37221. doi: 10.1371/journal.pone.0037221. PubMed: 22629372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Katayama S, Sugiyama Y, Hatano N, Terachi T, Sueyoshi N et al. (2012) PKL01, an Ndr kinase homologue in plant, shows tyrosine kinase activity. J Biochem 152: 347-353. doi: 10.1093/jb/mvs075. PubMed: 22753892. [DOI] [PubMed] [Google Scholar]
- 37. Hergovich A, Stegert MR, Schmitz D, Hemmings BA (2006) NDR kinases regulate essential cell processes from yeast to humans. Nat Rev Mol Cell Biol 7: 253-264. doi: 10.1038/nrm1891. PubMed: 16607288. [DOI] [PubMed] [Google Scholar]
- 38. Dvash E, Kra-Oz G, Ziv C, Carmeli S, Yarden O (2010) The NDR kinase DBF-2 is involved in regulation of mitosis, conidial development, and glycogen metabolism in Neurospora crassa. Eukaryot Cell 9: 502-513. doi: 10.1128/EC.00230-09. PubMed: 19966031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Baker SE, Wiest A, Plamann M (2012) Associating genes with phenotypes in Neurospora mutant strains. Neurospora Conference 2012 Abstract. [Google Scholar]
- 40. Raju NB Newmeyer aD (1977) Giant ascospores and abnormal croziers in a mutant of Neurospora crassa. Experimental Mycology 1: 152-165
- 41. Freitag M, Hickey PC, Raju NB, Selker EU, Read ND (2004) GFP as a tool to analyze the organization, dynamics and function of nuclei and microtubules in Neurospora crassa. Fungal Genet Biol 41: 897-910. doi: 10.1016/j.fgb.2004.06.008. PubMed: 15341912. [DOI] [PubMed] [Google Scholar]
- 42. Krapp A, Collin P, Cokoja A, Dischinger S, Cano E et al. (2006) The Schizosaccharomyces pombe septation initiation network (SIN) is required for spore formation in meiosis. J Cell Sci 119: 2882-2891. doi: 10.1242/jcs.03025. PubMed: 16787941. [DOI] [PubMed] [Google Scholar]
- 43. Attner MA, Amon A (2012) Control of the mitotic exit network during meiosis. Mol Cell Biol 23: 3122-3132. doi: 10.1091/mbc.E12-03-0235. PubMed: 22718910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. McCluskey K (2003) The Fungal Genetics Stock Center: from molds to molecules. Adv Appl Microbiol 52: 245-262. doi: 10.1016/S0065-2164(03)01010-4. PubMed: 12964247. [DOI] [PubMed] [Google Scholar]
- 45. Honda S, Selker EU (2009) Tools for fungal proteomics: multifunctional neurospora vectors for gene replacement, protein expression and protein purification. Genetics 182: 11-23. doi: 10.1534/genetics.108.098707. PubMed: 19171944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kawabata T, Kato A, Suzuki K, Inoue H (2007) Neurosprora crassa RAD5 homologue, mus-41, inactivation results in higher sensitivity to mutagens but has little effect on PCNA-ubiquitylation in response to UV-irradiation. Curr Genet 52: 125-135. doi: 10.1007/s00294-007-0146-x. PubMed: 17703305. [DOI] [PubMed] [Google Scholar]
- 47. Kawabata T, Inoue H (2007) Detection of physical interactions by immunoprecipitation of FLAG- and HA-tagged proteins expressed at the his-3 locus in Neurospora crassa . Fungal Genet Newsletter 54: 5-8. [Google Scholar]
- 48. Maerz S, Funakoshi Y, Negishi Y, Suzuki T, Seiler S (2010) The Neurospora peptide:N-glycanase ortholog PNG1 is essential for cell polarity despite its lack of enzymatic activity. J Biol Chem 285: 2326-2332. doi: 10.1074/jbc.M109.045302. PubMed: 19940117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Vogt N, Seiler S (2008) The RHO1-specific GTPase-activating protein LRG1 regulates polar tip growth in parallel to Ndr kinase signaling in Neurospora. Mol Cell Biol 19: 4554-4569. doi: 10.1091/mbc.E07-12-1266. PubMed: 18716060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Araujo-Palomares CL, Richthammer C, Seiler S, Castro-Longoria E (2011) Functional characterization and cellular dynamics of the CDC-42 - RAC - CDC-24 module in Neurospora crassa. PLOS ONE 6: e27148. doi: 10.1371/journal.pone.0027148. PubMed: 22087253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Richthammer C, Enseleit M, Sanchez-Leon E, März S, Heilig Y et al. (2012) RHO1 and RHO2 share partially overlapping functions in the regulation of cell wall integrity and hyphal polarity in Neurospora crassa. Mol Microbiol 85: 716-733. doi: 10.1111/j.1365-2958.2012.08133.x. PubMed: 22703449. [DOI] [PubMed] [Google Scholar]
- 52. Dettmann A, Illgen J, März S, Schürg T, Fleissner A et al. (2012) The NDR kinase scaffold HYM1/MO25 is essential for MAK2 map kinase signaling in Neurospora crassa. PLOS Genet 8: e1002950 PubMed: 23028357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Millward TA, Heizmann CW, Schäfer BW, Hemmings BA (1998) Calcium regulation of Ndr protein kinase mediated by S100 calcium-binding proteins. EMBO J 17: 5913-5922. doi: 10.1093/emboj/17.20.5913. PubMed: 9774336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Dettmann A, Heilig Y, Ludwig S, Schmitt K, Illgen J et al. (2013) HAM-2 and HAM-3 are central for the assembly of the Neurospora STRIPAK complex at the nuclear envelope and regulate nuclear accumulation of the MAP kinase MAK-1 in a MAK-2-dependent manner. Mol Microbiol (in press). doi: 10.1111/mmi.12399. [DOI] [PubMed] [Google Scholar]
- 55. Mazanek M, Roitinger E, Hudecz O, Hutchins JR, Hegemann B et al. (2010) A new acid mix enhances phosphopeptide enrichment on titanium- and zirconium dioxide for mapping of phosphorylation sites on protein complexes. J Chromatogr B Anal Technol Biomed Life Sci 878: 515-524. doi: 10.1016/j.jchromb.2009.12.017. PubMed: 20075017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression analysis of the used GFP fusion constructs under the control of the ccg-1 and their native promoters. Anti-GFP Western blot of normalized cell extracts of strains expressing GFP fusion proteins under the control of the indicated promoters (left panel). Quantification of the relative expression levels of the indicated proteins. Protein levels are normalized to DBF-2 abundance (n = 3).
(TIF)
Mass-spectrometric analysis of DBF2 phosphorylation sites. (A) In vitro phosphorylation experiments coupled with mass-spectrometric analysis. Ser499 was phosphorylated in wild type DBF-2 and hyperactive DBF-2(T671E), but not DBF-2(D422A), identifying this residue as primary site of auto-phosphorylation. Multiple additional S/T phosphorylation sites were detected in the N-terminal region of DBF-2(T671E). Mass-spectrometric analysis of SID-1-dependant DBF-2 phosphorylation sites identified T671 as primary site of phosphorylation and additional S/T phosphorylation sites variously distributed. (B) Tryptic peptides generated from wild type DBF-2 displayed HCD-fragmentation spectrum of the peptide SIVGSPDYMAPEVLR with Ser499 phosphorylated. Fragment b-ions (yellow) and y-ions (blue) with an asterisk indicate neutral loss of ammonia (-17 Da), and ions labeled with a circle neutral loss of water water (-18 Da), charge states are in brackets. The peptide cross-correlation score for Sequest (XCorr) was 4.9 and the Mascot IonScore was 77. The probability of Ser499-phosphorylation was caclulated by the pRS algorithm to be 99.99% [J Proteome Res 10: 5354-5362]. (C) Representative fragmentation spectrum of the peptide SLFVGFtFR with phosphorylation at T671. Fragment ions b* and y* are ions with loss of ammonia (-17 Da), and fragment ions bo and co are ions with loss of water (-18 Da). The number in brackets indicates the charge state of the fragment ion. The peptide was fragmented by CID and fragment ions were detected in the linear ion trap. The peptide was identified with Mascot and Sequest search engines (peptide IonScore of 30 and XCorr of 3.45, respectively). pRS score for phosporylation at tyrosine was 100% [J Proteome Res 10: 5354-5362].
(TIF)
N. crassa strains used in this study.
(DOCX)
Primers used in this study.
(DOCX)
Time-course of DBF-2-GFP localization during septum formation. DBF-2-GFP formed cortical rings at incipient septation sites that constricted during septum formation and accumulated around the septal pore of the completed septum (a) GFP channel; (b) RFP channel; (c) merged. The plasma membrane was stained with FM4-64. Images were captured at 20 sec intervals.
(MOV)
Time-course of SID-1-GFP localization during septum formation. SID-1-GFP formed cortical rings at incipient septation sites that constricted during septum formation and accumulated around the septal pore of the completed septum (a) GFP channel; (b) RFP channel; (c) merged. The plasma membrane was stained with FM4-64. Images were captured at 20 sec intervals.
(MOV)
Time-course of CDC-14-GFP localization during septum formation. CDC-14-GFP formed cortical rings at incipient septation sites that constricted during septum formation and accumulated around the septal pore of the completed septum (a) GFP channel; (b) RFP channel; (c) merged. The plasma membrane was stained with FM4-64. Images were captured at 20 sec intervals.
(MOV)