Abstract
Background
The keratocystic odontogenic tumor (KCOT) is a locally aggressive cystic jaw lesion that occurs sporadically or in association with nevoid basal cell carcinoma syndrome (NBCCS). PTCH1, the gene responsible for NBCCS, may play an important role in sporadic KCOTs. In this study, we analyzed and compared the distribution pattern of PTCH1 mutations in patients with sporadic and NBCCS-associated KCOTs.
Methods
We detected PTCH1 mutations in 14 patients with NBCCS-associated KCOTs and 29 patients with sporadic KCOTs by direct sequencing. In addition, five electronic databases were searched for studies detecting PTCH1 mutations in individuals with NBCCS-associated or sporadic KCOTs, published between January 1996 and June 2013 in English language.
Results
We identified 15 mutations in 11 cases with NBCCS-associated KCOTs and 19 mutations in 13 cases with sporadic KCOTs. In addition, a total of 204 PTCH1 mutations (187 mutations from 210 cases with NBCCS-associated and 17 mutations from 57 cases with sporadic KCOTs) were compiled from 78 published papers.
Conclusions
Our study indicates that mutations in transmembrane 2 (TM2) are closely related to the development of sporadic KCOTs. Moreover, for the early diagnosis of NBCCS, a genetic analysis of the PTCH1 gene should be included in the new diagnostic criteria.
Introduction
Originally described as cysts [1], odontogenic keratocysts (OKCs) were recategorized as neoplastic lesions under the name “karatocystic odontogenic tumors” (KCOTs) in the 2005 edition of the World Health Organization Classification of Head and Neck Tumors [2]. KCOTs are locally aggressive jaw cystic lesions that have putative growth potential and a propensity for recurrence [3], [4]. They may occur in isolation or in association with nevoid basal cell carcinoma syndrome (NBCCS, also known as Gorlin syndrome; OMIM No. 109400). NBCCS is a rare autosomal dominant disorder with an estimated prevalence of 1/55600 [5] to 1/256000 [6]. Affected patients present with a spectrum of developmental abnormalities, including palmar or plantar pits, calcification of the falx cerebri, bifid ribs, and an increased susceptibility to different neoplasms, such as multiple basal cell carcinomas (BCCs), KCOTs, medulloblastoma, and ovarian fibroma [7]–[9].
The human homolog of the Drosophila segment polarity gene PTCH1 (OMIM No. 601309) has been identified as the gene responsible for NBCCS [10]–[12]. PTCH1 has been mapped to 9q22.3-31 and consists of 23 coding exons spanning approximately 74 kb and encoding a 1447-amino-acid transmembrane (TM) glycoprotein. The Patched protein is involved in the Hedgehog (Hh) signaling pathway, a key regulator of body patterning and organ development during embryogenesis [13]. Patched and Smoothened (Smo) are thought to act as subunits of a putative Hh receptor complex: Smo functions as the transducing subunit, and its activity is blocked by a direct interaction with Patched, the ligand-binding subunit [14], [15]. Misregulation of the signaling caused by inactivation of PTCH1 is involved in the development of NBCCS and some related sporadic tumors, supporting the hypothesis that PTCH1 functions as a tumor suppressor gene [10]. Several studies have demonstrated the presence of PTCH1 mutations in patients with NBCCS [12] as well as in some sporadic tumors, including BCCs [16], medulloblastoma [17], and KCOTs [18].
The Patched protein has several important functional domains. Two large extracellular loops (ECLs) are required for the binding of N-Shh to the Patched protein, and glycosylation sites in the two large ECLs are required for N-Shh binding [19]. Martin et al. (2001) and Strutt et al. (2001) suggested that the sterol-sensing domain (SSD) of Patched is required to negatively regulate Smoothened (Smo) activity, indicating a role of the SSD in mediating the vesicular trafficking of PTCH to regulate Smo activity [20], [21]. Additionally, its large intracellular loop (ICL) participates in a G2/M checkpoint by regulating the localization of M-phase promoting factor [22]. The C-terminal region of Patched has been shown to induce apoptotic cell death during spinal cord development [23].
The distribution pattern of PTCH1 mutations has been analyzed in patients with NBCCS as well as in several tumors including BCCs, BCCs with xeroderma pigmentosum syndrome (XP-BCCs), and sporadic medulloblastomas [24]. They found that PTCH1 mutations were identified throughout nearly the entire sequence, with a high frequency of mutations clustered in the two large extracellular loops and the large intracellular loop [24]. NBCCS cases and each class of tumor analyzed revealed a different distribution of mutations in the various Patched protein domains [24]. PTCH1 gene harbors mutational hot spot regions, including a slippage-sensitive sequence in the N-terminus [24]. However, few reports have described the distribution pattern in KCOTs, especially sporadic KCOTs. Thus, the aim of the present study was to investigate PTCH1 gene mutations in 29 sporadic and 14 NBCCS-associated KCOTs. Additionally, to systematically analyze and compare the distribution pattern of PTCH1 mutations in NBCCS-associated and sporadic KCOTs, we conducted a systematic review of published studies evaluating PTCH1 mutations in patients with KCOTs.
Materials and Methods
Subjects and Samples
KCOT samples from 43 unrelated Chinese patients (14 patients with NBCCS-related and 29 with sporadic KCOTs) were obtained from Peking University, School and Hospital of Stomatology (Beijing, China). Fresh KCOT tissue specimens and corresponding peripheral blood samples were collected and immediately stored at −80°C for subsequent polymerase chain reaction (PCR) and sequencing analysis. Peripheral blood control samples were obtained from 100 normal volunteers from the Blood Transfusion Center at Peking University First Hospital. The protocol for this study was approved by the Ethics Committee of Peking University's Health Science Center. Written informed consent was obtained from all study participants.
DNA Extraction and PCR
Genomic DNA was extracted from the frozen tissue specimens and peripheral blood samples was extracted with a QIAamp DNA Mini Kit (Qiagen, Hilden, Germany), and 22 coding exons (exons 2–23) and the exon-intron boundaries of PTCH1 were amplified by PCR with specific primers. Two primer sets were used separately to amplify exons 14 and 23 because of the relatively large size of the exons. PCR was performed in a final reaction volume of 50 µL, containing approximately 100 ng of template DNA, 200 µM dNTPs, 10 µM each primer and 1.25 U of Ex Taq DNA polymerase (Takara, Kyoto, Japan) in PCR buffer. Amplification was performed in a thermal cycler (Eppendorf, Hamburg, Germany) using the following cycling parameters: initial incubation at 94C for 5 min; 35 cycles of denaturation at 94°C for 30 s, annealing at 56–65°C for 30 s, and elongation at 72°C for 30 s; and final elongation at 72°C for 7 min.
Mutation Analysis by Direct and Clonal Sequencing
The amplified products were sequenced directly with the primers used for the original PCR. Insertion or deletion mutations were confirmed by cloning purified PCR products into the plasmid vector pCR2.1 (Invitrogen, Carlsbad, CA, USA). After transforming competent Escherichia coli strain TOP10 with the recombinant vectors, white colonies were selected and grown overnight in 3 mL of LB medium with ampicillin. Sequencing was performed on an ABI PRISM 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA) with M13 universal primers. Identified mutations were confirmed by reverse sequencing and at least two additional independent PCR products.
Restriction Enzyme Analysis
To define unreported PTCH1 mutations (missense mutations, in particular) as novel mutations rather than rare polymorphisms, we tested 100 unrelated control DNAs by restriction enzyme analysis. Digestion of the PCR products with MspI, BanII or BaeGI (New England Biolabs, Beverly, MA, USA) was performed as recommended by the supplier. The digested DNA was electrophoresed on 2.5% agarose gels and stained with nucleic acid stain.
Literature Review
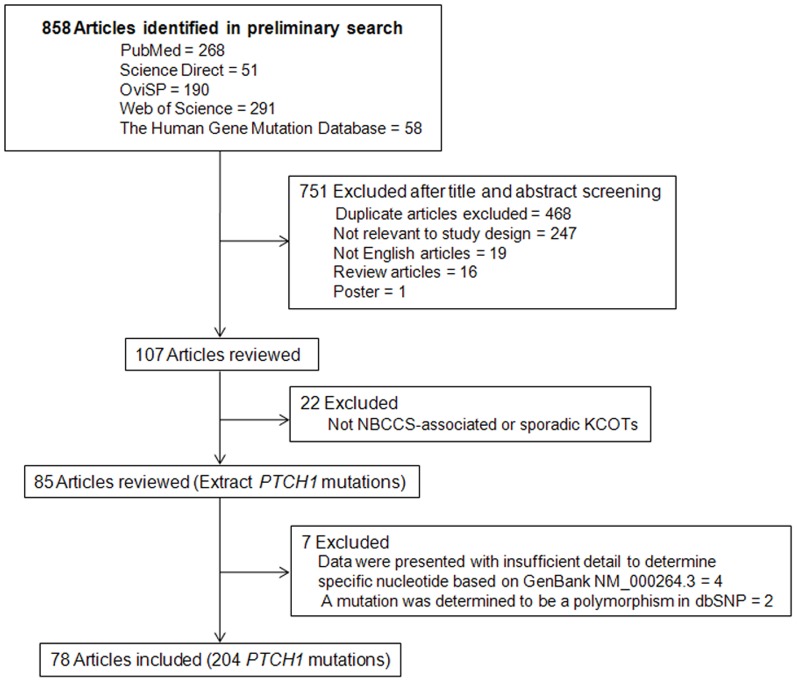
We performed a systematic search from January 1996 to June 2013 using five electronic databases (PubMed, ScienceDirect, OviSP, Web of Science, and the Human Gene Mutation Database). Additional studies were identified by searching the reference lists of the included publications. The keywords used were “PTCH1” (or its synonyms) and “NBCCS or KCOT” (or their synonyms). This review was conducted and reported according to the PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-Analyses) Statement issued in 2009 (Checklist S1) [25].
Studies were included if they concerned PTCH1 gene mutations in patients with sporadic or NBCCS-associated KCOTs. A list of exclusionary indications is shown in Fig. 1. Only one entry was made when the same mutation in a single patient was reported in different papers (for example: c.2186A>T was first reported by Ponti et al. [26] and subsequently by Pastorino et al. [27]). Separate entries were made for each patient as recurrent mutations when the same mutation was reported in apparently unrelated patients (for example: c.403C>T identified in different patients [18], [28]). Nucleotide and protein names were given for each mutation according to current nomenclature guidelines (http://www.hgvs.org/mutnomen/). Nucleotide numbering was based on GenBank entry NM_000264.3, where the A of the ATG initiation codon represents nucleotide +1. The amino acid numbering was based on GenBank entry NP_000255.2.
Figure 1. Flow chat of the selection process for the extensive review.
Statistical Analysis
Data were analyzed using SPSS software (ver. 13.0), and significant differences between categorical groups were determined using the chi-squared test or Fisher's exact test, while numerical groups were assessed using the independent t-test. P values<0.05 were considered to indicate statistical significance. All statistical tests were two-sided.
Results
Clinical Manifestations of Patients with NBCCS-associated KCOTs
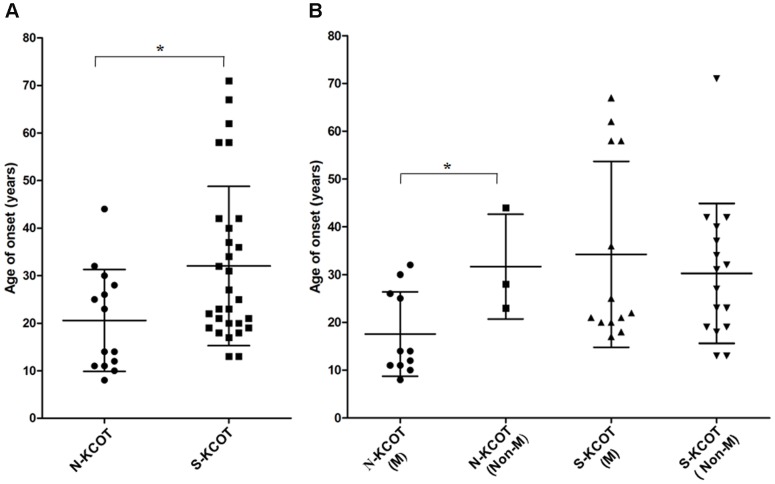
The male to female ratio among the 14 patients with NBCCS-associated KCOTs was 3.67∶1. To improve the comparability of age factors, we used age of onset (age at diagnosis minus duration) in the following statistical analysis. The individuals ranged in onset age from 8 to 44 years (mean 20.57±10.71; median 18.50 years; Fig. 2A).
Figure 2. The onset age distribution of patients with NBCCS-associated and sporadic KCOTs.
(A) The age distribution of 14 patients with syndromic KCOTs was significantly lower than in 29 patients with sporadic KCOTs (P = 0.025). (B) In syndromic KCOTs, the age distribution in the mutation group was significantly lower than in non-mutation group (P = 0.037), whereas in sporadic KCOTs, no significant difference was detected between the groups (P = 0.537). “N-KCOT” indicates “NBCCS-associated KCOT”; “S-KCOT” indicates “sporadic KCOT”; “M” indicates “Mutation group”; and “Non-M” indicates “Non-mutation group” (Error bars±SD, * P<0.05).
The total number of lesions was 45, from 1 to 5, and the average number of lesions was 3.21 (Table 1). Of the lesions, 18/45 (40.0%) were located in the maxilla and 27/45 (60.0%) in the mandible. Among the 14 cases, only one patient had a single lesion of KCOT; however, the scope of the lesion in this patient was wide, approximately from the right mandibular angle to the left mandibular angle.
Table 1. Clinical manifestations and PTCH1 mutations in 14 cases with NBCCS-associated KCOTs.
| Patient | Sex/ Onset age | Number of Lesions | Nomenclature c. | Nomenclature p. | Mutation type | Exon/Intron no. | Characterization | Structure | |
| Maxilla | Mandible | ||||||||
| NB21d | M/11 | 2 | 3 | c.233G>A | p.Trp78X | Missense mutations | Exon2 | Germline | N-terminus |
| c.3253dupA | p.Ile1085AsnfsX60 | Small duplications | Exon19 | Somatic | TM10 | ||||
| NB22 | M/11 | 1 | 2 | c.873C>G | p.Tyr291X | Nonsense mutations | Exon6 | Germline | ECL1 |
| c.2479A>Ga | p.Ser827Gly | Missense mutations | Exon15 | Germline | ECL4 | ||||
| NB23 | M/14 | 1 | 3 | c.394+3_+20del18 | Splice-site mutations | Intron2 | Somatic | N-terminus | |
| c.1526G>Ab | p.Gly509Asp | Missense mutations | Exon11 | Germline | TM4 | ||||
| NB24d | M/14 | 1 | 2 | c.3156_3163delins19 | p.Ala1053LeufsX2 | Indel mutations | Exon18 | Somatic | ICL4 |
| c.2709_2713delTAAAC | p.Lys904AlafsX10 | Small out-of-frame deletions | Exon17 | Somatic | ECL4 | ||||
| NB25d | M/25 | 0 | 2 | c.3180_3185del6 | p.Ala1061_Leu1062del | Small in-frame deletions | Exon19 | Germline | TM9 |
| NB26 | F/30 | 1 | 2 | c.534_545del12 | p.His178_Ala182delinsGln | Small in-frame deletions | Exon3 | Germline | ECL1 |
| NB27d | M/8 | 1 | 2 | c.1342_1345delCTCA | p.Leu448CysfsX7 | Small out-of-frame deletions | Exon9 | Germline | TM2 |
| NB28 | M/12 | 2 | 2 | c.584+1G>A | Splice-site mutations | Intron3 | Germline | ECL1 | |
| NB29 | M/32 | 1 | 1 | c.1531G>C | p.Gly511Arg | Missense mutations | Exon11 | Germline | TM4 |
| NB30d | F/10 | 1 | 2 | c.387G>Ac | p.Trp129X | Nonsense mutations | Exon2 | Germline | ECL1 |
| NB31d | M/26 | 3 | 2 | c.2464dupC | p.Leu822ProfsX7 | Small duplications | Exon15 | Germline | ECL4 |
| NB32 | M/28 | 0 | 1 | Not identified | |||||
| NB33 | M/23 | 1 | 2 | Not identified | |||||
| NB34 | F/44 | 3 | 1 | Not identified | |||||
Clinical Manifestations of Patients with Sporadic KCOTs
The male to female ratio among the 29 patients with sporadic KCOTs was 1.07∶1. The age range of the 29 affected persons was 13 to 71 years (mean 32.03±16.75; median 25.0 years; Fig. 2A). The mean age of patients with sporadic KCOTs was significantly higher than that of patients with systematic KCOTs (t = -2.333,P = 0.025). All the 29 cases had a single lesion (Table 2). Of 29 lesions, only 2 (6.90%) were located in the maxilla and 27 (93.10%) in the mandible.
Table 2. Clinical manifestations and PTCH1 mutations in 29 cases with sporadic KCOTs.
| Patient | Sex/ Onset age | Number of Lesions | Nomenclature c. | Nomenclature p. | Mutation type | Exon/Intron no. | Characterization | Structure | |
| Maxilla | Mandible | ||||||||
| KC43 | F | 0 | 1 | c.1359_1362delCTGT | p.Cys454X | Small out-of-frame deletions | Exon10 | Somatic | TM2 |
| c.1493_1494insTG | p.Thr499GlufsX44 | Small in-frame insertions | Exon10 | Somatic | ECL2 | ||||
| KC44 | M | 0 | 1 | c.536T>C | p.Leu179Pro | Missense mutations | Exon3 | Somatic | ECL1 |
| c.1164dupC | p.Glu389ArgfsX48 | Small duplications | Exon8 | Somatic | ECL1 | ||||
| KC45 | M | 0 | 1 | c.2638_2668del31 | p.Gly880ProfsX13 | Small out-of-frame deletions | Exon16 | Somatic | ECL4 |
| c.260_271delinsAA | p.Leu87X | Indel mutations | Exon2 | Somatic | N-terminus | ||||
| KC46 | F | 0 | 1 | c.3346G>C | p.Val1116Leu | Missense mutations | Exon20 | Somatic | ICL5 |
| c.1127_1143del17 | p.Phe376CysfsX55 | Small out-of-frame deletions | Exon8 | Somatic | ECL1 | ||||
| KC47 | M | 0 | 1 | c.202-3C>T | Splice-site mutations | Intron1 | Somatic | N-terminus | |
| c.1362_1375del14 | p.Cys454TrpfsX38 | Small out-of-frame deletions | Exon10 | Somatic | TM2 | ||||
| KC48 | M | 0 | 1 | c.1327delG | p.Ala443ProfsX13 | Small out-of-frame deletions | Exon9 | Somatic | TM2 |
| c.1344_1347delCATG | p.Met449SerfsX6 | Small out-of-frame deletions | Exon9 | Somatic | TM2 | ||||
| KC49 | M | 1 | 0 | c.407dupT | p.Ser137LysfsX3 | Small duplications | Exon3 | Somatic | ECL1 |
| KC50 | M | 0 | 1 | c.3081dupG | p.Leu1028AlafsX117 | Small duplications | Exon18 | Somatic | TM8 |
| KC51 | F | 0 | 1 | c.2430delA | p.Asp811ThrfsX19 | Small out-of-frame deletions | Exon15 | Somatic | ECL4 |
| KC52 | M | 0 | 1 | c.1063_1067+11del16 | Splice-site mutations | Exon7/Intron7 | Somatic | ECL1 | |
| KC53 | F | 0 | 1 | c.2908G>Ta | p.Glu970X | Nonsense mutations | Exon18 | Somatic | ECL4 |
| KC54 | F | 1 | 1 | c.262_265delTTTAb | p.Phe88AsnfsX28 | Small out-of-frame deletions | Exon2 | Somatic | N-terminus |
| KC55 | M | 0 | 1 | c.1394_1396delinsA | p.Ser465X | Indel mutations | Exon10 | Somatic | ICL1 |
| KC56 | F | 0 | 1 | Not identified | |||||
| KC57 | F | 0 | 1 | Not identified | |||||
| KC58 | M | 0 | 1 | Not identified | |||||
| KC59 | F | 0 | 1 | Not identified | |||||
| KC60 | F | 0 | 1 | Not identified | |||||
| KC61 | M | 0 | 1 | Not identified | |||||
| KC62 | M | 0 | 1 | Not identified | |||||
| KC63 | F | 0 | 1 | Not identified | |||||
| KC64 | M | 0 | 1 | Not identified | |||||
| KC65 | F | 0 | 1 | Not identified | |||||
| KC66 | F | 0 | 1 | Not identified | |||||
| KC67 | M | 0 | 1 | Not identified | |||||
| KC68 | F | 0 | 1 | Not identified | |||||
| KC69 | F | 0 | 1 | Not identified | |||||
| KC70 | M | 0 | 1 | Not identified | |||||
| KC71 | F | 0 | 1 | Not identified | |||||
PTCH1 Mutations in Patients with NBCCS-associated and Sporadic KCOTs
We identified 11 germline and 4 somatic mutations in 11 of 14 (78.57%) patients with NBCCS-associated KCOTs (Table 1). Additionally, 12 mutations (80.0%) had not been previously reported in the literature and consisted of 7 truncation-causing, 3 non-truncation-causing, and 2 splice-site mutations. Of the 12 new mutations, 5 were distributed in the two large ECLs (ECL1 and ECL4), 4 in the TMs, 2 in the N-terminus, and 1 in the fourth ICL (ICL4). Furthermore, three cases (NB21, NB22, and NB23) were found to carry two concomitant mutations while NB24 was found to carry two somatic truncation-causing mutations (c.3156_3163delins19 and c.2709_2713delTAAAC), identified in different KCOTs. One mutation in patient NB21 was a germline truncation mutation in exon 2 (c.233G>A) and the other was a somatic truncation mutation in exon 19 (c.3253dupA), resulting in premature stops at codon 78 and 1144 respectively. Two germline mutations were identified in patient NB22, one was a truncation mutation in exon 6 (c.873C>G) and the other was a non-truncation mutation in exon 15 (c.2479A>G). Also, one mutation in patient NB23 was a germline non-truncation mutation in exon 11 (c.1526G>A) and the other was a somatic splice-site mutation in exon 19 (c.394+3_+20del18). Additionally, 6 of 14 cases (42.86%; NB21, NB24, NB25, NB27, NB30, and NB31) suffered from cleft lip or plate, an incidence significantly higher than that in previous studies [9], [29], [30]. All six cases were found to carry PTCH1 mutations (Table 1). Patient NB25 carried a non-truncation mutation (c.3180_3185del6). One truncation mutation (c.1342_1345delCTCA) was identified in patient NB27. One truncation mutation (c.387G>A) was detected in patient NB30. Patient NB31 carried a truncation mutation (c.2464dupC) in the second large ECL (ECL2). In addition, A G>C substitution at nucleotide 1531 in exon 11, which resulted in a glycine to arginine substitution at codon 511, was detected in a 32-year-old male patient with NBCCS (NB29). This mutation was not detected in 100 control normal volunteers by direct sequencing.
The 14 affected patients could be divided into two groups: mutation and non-mutation. The mean age (17.55 years) of the mutation group was significantly younger than that (31.67 years) of the non-mutation group (Fig. 2B; t = −2.350, P = 0.037). All the patients under the age of 20 years carried mutations. Four patients over 20 years of age were identified to carry mutations whereas three patients over 20 years of age carried no mutations (Fig. 2B). Particularly, patient NB32, a 28-year-old patient with typical NBCCS phenotypes (such as multiple KCOTs and bilamellar calcification of the falx cerebri), was found to carry no PTCH1 mutations and had only a single KCOT. Additionally, in the mutation group, 14 lesions were found in the maxilla while 23 in the mandible. In the non-mutation group, four lesions were found in the maxilla while five in the mandible (Table 1). No significant difference was detected between the two groups in terms of the lesion distribution (Pearson χ2 = 0.057,P = 0.811).
We identified 19 somatic PTCH1 mutations in 13 of 29 (44.83%) patients with sporadic KCOTs (Table 2). No germline mutation was detected in any patient. Additionally, six cases (6/28, 21.43%) were found to carry two concomitant mutations. Of those mutations, 17 (89.47%) had not been previously reported, consisting of 13 truncation, 2 non-truncation, and 2 splice-site mutations. Of the 17 new mutations, seven were distributed in ECL1 and ECL4, five in TMs, two in N-terminus, one in the first intracellular loop (ICL1), one in ECL2 and one in ICL5. Additionally, novel missense mutations (c.536T>C and c.3346G>C) were respectively identified in patients KC44 and KC46. The two mutations introduced a novel restriction enzyme site for MspI and BanII, creating two fragments, thus confirming the presence of the mutation. To characterize the two novel missense mutations, we tested 100 control DNA samples from normal volunteers by restriction enzyme analysis. The abnormal restriction sites present in the PCR products from KC44 and KC46 were absent in the control DNAs. Thus, the two missense mutations were unlikely to be polymorphisms.
Similarly, the 28 affected cases could be further categorized into two groups: mutation and non-mutation. The mean age (34.23 years) of the mutation group was slightly higher than that (30.25 years) in non-mutation group (Fig. 2B), but the difference was not statistically significant (t = 0.630, P = 0.537). Moreover, only two cases (KC51 and KC54) occurred in the maxilla; both carried truncation mutation (c.2430delA and c.262_265delTTTA, respectively).
Literature Search Results
Our search strategy retrieved 858 reports, 751 of which were excluded on the basis of the title and abstract. After carefully examining the full text of the remaining 107 articles, 78 articles (187 mutations from 210 cases with NBCCS-associated and 17 mutations from 57 cases with sporadic KCOTs) were included. Fig. 1 presents the flowchart of the search process. Tables S1-2 summarizes the key characteristics of each PTCH1 mutation.
Distribution Pattern of PTCH1 Gene Mutations in Patients with Sporadic and NBCCS-associated KCOTs
Including the mutations found in our study, we compiled a list of 238 mutations (202 from patients with NBCCS-associated KCOTs and 36 from patients with sporadic KCOTs). Large deletions and duplications, although important in the pathogenesis of KCOTs, offered less detail in the distribution pattern of PTCH1 mutations because those mutations encompassed more extensive structural changes. Thus, these 15 large deletions and duplications were excluded in the following statistical analysis.
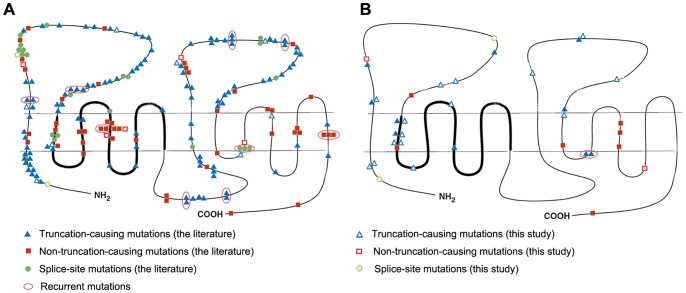
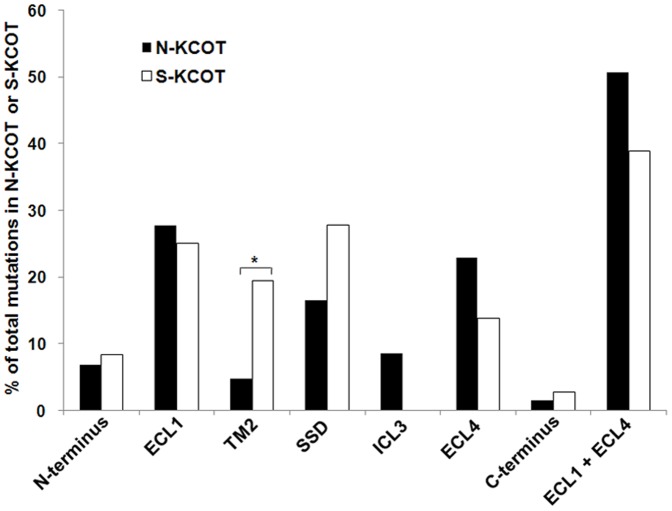
Compared with the predicted Patched protein sequence, the 223 PTCH1 mutations (187 from NBCCS-associated and 36 from sporadic KCOTs) were distributed over the entire sequence. The mutations (n = 187) from patients with NBCCS-associated KCOTs were mainly clustered in the two large extracellular loops (95/187, 50.80%; Fig. 3A and 4). Mutations were also found in the SSD (16.58%), the large intracellular loop (8.56%) and the N-terminal region (6.95%). Approximately half of the non-truncation-causing mutations (26/51, 50.98%) were located in the TM domains, especially TM4. Additionally, most splice-site mutations (11/23, 47.83%) were concentrated in the first large extracellular loops.
Figure 3. Distribution pattern of PTCH1 mutations in relation to the different domains of the Patched protein.
(A) NBCCS-associated KCOTs (n = 174). (B) Sporadic KCOTs (n = 36). The thick line indicates the SSD.
Figure 4. Proportion of PTCH1 mutations in important domains of the Patched protein.
ECL1 indicates the first large extracellular loop (amino acids 122–436); TM2 denotes the second transmembrane region (amino acids 437–457); SSD stands for the sterol-sensing domain (amino acids 438–598); ECL4 denotes the second large extracellular loop (amino acids 770–1027); * P<0.05.
Mutations (n = 36) from patients with sporadic KCOTs were found predominantly in the two large extracellular loops (14/36, 38.89%) and TM2 (7/36, 19.44%; Fig. 3B and 4). Compared with the mutations in patients with NBCCS-associated KCOTs, the mutations from sporadic KCOTs were differently distributed, with a significantly higher frequency of mutations in TM2 (7/36 vs. 8/187; Pearson χ2 = 7.631,P = 0.006). Additionally, no mutation was found in the large intracellular loop (0/36), compared to those found in the NBCCS group (16/187; Pearson χ2 = 2.158, P = 0.142). Most non-truncation-causing mutations (5/9, 55.56%) were found in the TM domains, especially in the second half of the protein in patients with sporadic KCOTs.
Different Mutation Types in Patients with Sporadic and NBCCS-associated KCOTs
The predominant mutation type seen in both the NBCCS-associated and sporadic KCOT groups was the predicted truncation-causing mutation in the Patched protein, with no statistically significant difference between the groups (113/187 vs. 25/36; Pearson χ2 = 1.040; P = 0.308; Table 3). Similarly, no significant difference in the distribution pattern of splice-site mutations or mutations that did not cause truncation of the Patched protein was observed between the groups.
Table 3. Different types of PTCH1 mutations in patients with NBCCS-associated and sporadic KCOTs.
| Truncation | Non-truncation | Splice-site | Total | ||||
| No. | % of Total | No. | % of Total | No. | % of Total | ||
| N-KCOT | 113 | 61.49% | 51 | 27.59% | 23 | 10.92% | 187 |
| S-KCOT | 25 | 69.44% | 9 | 25.00% | 2 | 5.56% | 36 |
| χ2 test | χ2 = 1.040 | χ2 = 0.079 | χ2 = 0.785 | ||||
| P = 0.308 | P = 0.778 | P = 0.375 | |||||
Discussion
In the present study, 29 new PTCH1 mutations were identified, including 20 truncation, 5 non-truncation, and 4 splice-site mutations. In general agreement with previous studies, most of the mutations identified were expected to lead to the synthesis of a truncated Patched protein. The PTCH1 mutations were widely distributed along the entire sequence: 12 in the two large extracellular loops, 8 in the SSD, 4 in the N-terminus region, 3 in TM8-10, and 2 in ICL4 and ICL5. Those mutations enrich the spectrum of PTCH1 mutations in KCOTs, thus providing further information about the pathogenesis of syndromic and sporadic KCOTs.
NBCCS is a hereditary condition caused by mutations in PTCH1 gene and transmitted in an autosomal dominant manner with high penetrance and variable expressivity. In the present study, 10 of 14 (71.43%) individuals with syndromic KCOTs carried at least a germline PTCH1 mutation. Because the initial tumor-causing mutation is inherited through the germline, it is already present in every cell of the body [31]. However, all 19 PTCH1 mutations identified in sporadic KCOTs were somatic. Thus, it is not surprising that syndromic KCOTs presented at a younger age, and were multifocal with widespread sites of involvement located in the maxilla and mandible, whereas sporadic KCOTs presented at an older age, and were unifocal with local lesions mainly located in the mandible.
The mutation rate of PTCH1 gene in NBCCS patients varied from 29.0 to 100% [32]–[34]. However, the mutation rate of PTCH1 gene in sporadic KCOTs was approximately 28.6% [18]. Here, we identified 34 mutations in 11 of 14 (78.57%) syndromic and 13 of 29 (44.83%) sporadic KCOTs patients. Additionally, during the 14 cases with syndromic KCOTs, all patients under 20 years of age were detected to carry PTCH1 mutations, whereas some patients over 20 years of age did not carry mutations. It indicated that PTCH1 gene might play a more important role in the development of the younger age cases (≤20 years) with syndromic KCOTs, whereas the pathogenesis of the older age cases (>20 years) might be complicated and involved other as yet unknown genes. There were reports showing other genes, such as PTCH2 [35] and SUFU [36], [37], might be involved in the pathogenesis of NBCCS. We suggested that KCOTs, as a complex disease, might be caused by multiple genes and environmental factors. To assess this, further study is needed to use other technologies, such as exome or whole-genome sequencing, to screen for additional genes.
Making a diagnosis of NBCCS in childhood patients could be challenging because several signs of the syndrome develop over time, including KCOTs, BCCs, calcification of the falx cerebri, and ovarian fibromas. Patients affected with NBCCS at birth might present only some congenital developmental deformities such as cleft lip or palate, and bifid, fused or accessory ribs. BCCs most often appear between puberty and 35 years of age; the mean age of onset is about 25 years of age [8]. Between 30% and 65% of patients with the syndrome have small asymmetric palmar or plantar pits by the age of 10 years; this percentage rises to 80% by the age of 15 years [7], [38]. The calcification of the falx cerebri can appear very early in life, and is often strikingly apparent from late childhood. Medulloblastoma characteristically presents during the first two years of life, while in the general population, occurrence peaks at 7 to 8 years of age. Ovarian fibromas generally develop in the teen years. In our study, two out of (14.3%) of patients develop a cystic lesion by the age of 10 years and 7 out of 14 (50%) by the age of 20 years. Diagnostic criteria were presently based on the most frequent and/or specific features of the syndrome [5], [9]. Thus, the diagnosis time of most NBCCS cases is usually postponed until the occurrence of symptoms. However, early diagnosis is often of great significance for the prevention and prognosis of systematic tumors. The high detection rate of PTCH1 mutations among NBCCS patients, as demonstrated in the present study as well as other previous ones [34], [39], poses the possibility to include genetic analysis of PTCH1 in the new diagnostic criteria for early diagnosis.
The distribution pattern of PTCH1 mutations in NBCCS-associated and sporadic KCOTs had not been extensively analyzed previously. To our knowledge, this is the first to systematically analyze PTCH1 mutation distribution pattern in systematic and sporadic KCOTs. Careful analysis of the two groups revealed several interesting findings. First of all, consistent with previous literature [24], PTCH1 mutation hot regions in NBCCS-associated KCOTs involved the two large extracellular loops, sterol-sensing domain (SSD), large intracellular loop, and the N terminal region (close to TM1), while PTCH1 mutation hot regions in sporadic KCOTs involved the two large extracellular loops and TM2. Additionally, compared to mutations in NBCCS-associated KCOTs, a significantly higher frequency of mutations in TM2 and no mutations in the large intracellular were found in sporadic KCOTs. Differences in the TM2 region were statistically significant, while the mutations in the large intracellular loop were not. Seven mutations in patients with sporadic KCOTs were distributed in the TM2 region, six of which resulted in premature truncation of Patched protein. Interestingly, a significantly lower frequency of mutations in TM2 was also noted in sporadic basal cell carcinomas and basal cell carcinomas with xeroderma pigmentosum syndrome [24]. Taken together, these results indicate that mutations in TM2 might be closely related to the development of sporadic KCOTs.
In conclusion, this is the first report to systematically analyze and compare the distribution patterns of PTCH1 mutations in patients with NBCCS-associated and sporadic KCOTs, especially sporadic KCOTs. Our study indicates that mutations in TM2 are closely related to the development of sporadic KCOTs. Moreover, for the early diagnosis of NBCCS, genetic analysis of the PTCH1 gene might be included in new diagnostic criteria. Our findings justify further investigation.
Supporting Information
Literature review: 187 PTCH1 gene mutations in cases with NBCCS-associated KCOTs.
(DOCX)
Literature review: 17 PTCH1 gene mutations in cases with sporadic KCOTs.
(DOCX)
PRISMA Checklist for this systematic review.
(DOC)
Acknowledgments
We thank the patients and their families for specimen donation and their support of our research.
Funding Statement
This work was supported the National Natural Science Foundation of China (grant numbers 81030018, 30872900, 30901680) and the Doctoral Fund of Ministry of Education of China (grant number 20120001110043). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Philipsen HP (1956) Keratocysts (cholesteatomas) in the jaws. Tandlaegebladet 60: 963–980. [Google Scholar]
- 2.Barnes L, Eveson JW, Reichart P, Sidransky D, editors (2005) World Health Organization classification of tumors. Pathology and genetics of head and neck tumors. IARC Press: Lyon. 306–307p.
- 3. Browne RM (1971) The odontogenic keratocyst. Histological features and their correlation with clinical behaviour. Br Dent J 131: 249–259. [DOI] [PubMed] [Google Scholar]
- 4. Li TJ, Browne RM, Matthews JB (1994) Quantification of PCNA+ cells within odontogenic jaw cyst epithelium. J Oral Pathol Med 23: 184–189. [DOI] [PubMed] [Google Scholar]
- 5. Evans DG, Ladusans EJ, Rimmer S, Burnell LD, Thakker N, et al. (1993) Complications of the naevoid basal cell carcinoma syndrome: results of a population based study. J Med Genet 30: 460–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lo Muzio L, Nocini PF, Savoia A, Consolo U, Procaccini M, et al. (1999) Nevoid basal cell carcinoma syndrome. Clinical findings in 37 Italian affected individuals. Clin Genet 55: 34–40. [DOI] [PubMed] [Google Scholar]
- 7. Gorlin RJ (1987) Nevoid basal-cell carcinoma syndrome. Medicine (Baltimore) 66: 98–113. [DOI] [PubMed] [Google Scholar]
- 8. Gorlin RJ (1995) Nevoid basal cell carcinoma syndrome. Dermatol Clin 13: 113–125. [PubMed] [Google Scholar]
- 9. Kimonis VE, Goldstein AM, Pastakia B, Yang ML, Kase R, et al. (1997) Clinical manifestations in 105 persons with nevoid basal cell carcinoma syndrome. Am J Med Genet 69: 299–308. [PubMed] [Google Scholar]
- 10. Gailani MR, Bale SJ, Leffell DJ, DiGiovanna JJ, Peck GL, et al. (1992) Developmental defects in Gorlin syndrome related to a putative tumor suppressor gene on chromosome 9. Cell 69: 111–117. [DOI] [PubMed] [Google Scholar]
- 11. Hahn H, Wicking C, Zaphiropoulous PG, Gailani MR, Shanley S, et al. (1996) Mutations of the human homolog of Drosophila patched in the nevoid basal cell carcinoma syndrome. Cell 85: 841–851. [DOI] [PubMed] [Google Scholar]
- 12.Johnson RL, Rothman AL, Xie J, Goodrich LV, Bare JW, et al.. (1996) Human homolog of patched, a candidate gene for the basal cell nevus syndrome. 5268 ed. pp. 1668–1671. [DOI] [PubMed]
- 13. Scales SJ, de Sauvage FJ (2009) Mechanisms of Hedgehog pathway activation in cancer and implications for therapy. Trends Pharmacol Sci 30: 303–312. [DOI] [PubMed] [Google Scholar]
- 14. Murone M, Rosenthal A, de Sauvage FJ (1999) Sonic hedgehog signaling by the patched-smoothened receptor complex. Curr Biol 9: 76–84. [DOI] [PubMed] [Google Scholar]
- 15. Stone DM, Hynes M, Armanini M, Swanson TA, Gu Q, et al. (1996) The tumour-suppressor gene patched encodes a candidate receptor for Sonic hedgehog. Nature 384: 129–134. [DOI] [PubMed] [Google Scholar]
- 16. Huang YS, Bu DF, Li XY, Ma ZH, Yang Y, et al. (2013) Unique features of PTCH1 mutation spectrum in Chinese sporadic basal cell carcinoma. J Eur Acad Dermatol Venereol 27: 235–241. [DOI] [PubMed] [Google Scholar]
- 17. Pietsch T, Waha A, Koch A, Kraus J, Albrecht S, et al. (1997) Medulloblastomas of the desmoplastic variant carry mutations of the human homologue of Drosophila patched. Cancer Res 57: 2085–2088. [PubMed] [Google Scholar]
- 18. Pan S, Li TJ (2009) PTCH1 mutations in odontogenic keratocysts: are they related to epithelial cell proliferation? Oral Oncol 45: 861–865. [DOI] [PubMed] [Google Scholar]
- 19. Marigo V, Davey RA, Zuo Y, Cunningham JM, Tabin CJ (1996) Biochemical evidence that patched is the Hedgehog receptor. Nature 384: 176–179. [DOI] [PubMed] [Google Scholar]
- 20. Martin V, Carrillo G, Torroja C, Guerrero I (2001) The sterol-sensing domain of Patched protein seems to control Smoothened activity through Patched vesicular trafficking. Curr Biol 11: 601–607. [DOI] [PubMed] [Google Scholar]
- 21. Strutt H, Thomas C, Nakano Y, Stark D, Neave B, et al. (2001) Mutations in the sterol-sensing domain of Patched suggest a role for vesicular trafficking in Smoothened regulation. Curr Biol 11: 608–613. [DOI] [PubMed] [Google Scholar]
- 22. Barnes EA, Kong M, Ollendorff V, Donoghue DJ (2001) Patched1 interacts with cyclin B1 to regulate cell cycle progression. EMBO J 20: 2214–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Thibert C, Teillet MA, Lapointe F, Mazelin L, Le Douarin NM, et al. (2003) Inhibition of neuroepithelial patched-induced apoptosis by sonic hedgehog. Science 301: 843–846. [DOI] [PubMed] [Google Scholar]
- 24. Lindstrom E, Shimokawa T, Toftgard R, Zaphiropoulos PG (2006) PTCH mutations: distribution and analyses. Hum Mutat 27: 215–219. [DOI] [PubMed] [Google Scholar]
- 25. Moher D, Liberati A, Tetzlaff J, Altman DG (2009) Group P (2009) Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med 6: e1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ponti G, Pastorino L, Pollio A, Nasti S, Pellacani G, et al.. (2012) Ameloblastoma: a neglected criterion for nevoid basal cell carcinoma (Gorlin) syndrome. Fam Cancer. [DOI] [PubMed]
- 27. Pastorino L, Pollio A, Pellacani G, Guarneri C, Ghiorzo P, et al. (2012) Novel PTCH1 mutations in patients with keratocystic odontogenic tumors screened for nevoid basal cell carcinoma (NBCC) syndrome. PLoS One 7: e43827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wicking C, Shanley S, Smyth I, Gillies S, Negus K, et al. (1997) Most germline mutations in the nevoid basal cell carcinoma syndrome lead to a premature termination of the PATCHED protein, and no genotype-phenotype correlations are evident. Am J Hum Genet 60: 21–26. [PMC free article] [PubMed] [Google Scholar]
- 29. Ahn SG, Lim YS, Kim DK, Kim SG, Lee SH, et al. (2004) Nevoid basal cell carcinoma syndrome: a retrospective analysis of 33 affected Korean individuals. Int J Oral Maxillofac Surg 33: 458–462. [DOI] [PubMed] [Google Scholar]
- 30. Lo Muzio L, Staibano S, Pannone G, Bucci P, Nocini PF, et al. (1999) Expression of cell cycle and apoptosis-related proteins in sporadic odontogenic keratocysts and odontogenic keratocysts associated with the nevoid basal cell carcinoma syndrome. J Dent Res 78: 1345–1353. [DOI] [PubMed] [Google Scholar]
- 31. Poulaki V, Mukai S (2009) Retinoblastoma: genetics and pathology. Int Ophthalmol Clin 49: 155–164. [DOI] [PubMed] [Google Scholar]
- 32. Boutet N, Bignon YJ, Drouin-Garraud V, Sarda P, Longy M, et al. (2003) Spectrum of PTCH1 mutations in French patients with Gorlin syndrome. J Invest Dermatol 121: 478–481. [DOI] [PubMed] [Google Scholar]
- 33. Savino M, d'Apolito M, Formica V, Baorda F, Mari F, et al. (2004) Spectrum of PTCH mutations in Italian nevoid basal cell-carcinoma syndrome patients: identification of thirteen novel alleles. Hum Mutat 24: 441. [DOI] [PubMed] [Google Scholar]
- 34. Tanioka M, Takahashi K, Kawabata T, Kosugi S, Murakami K, et al. (2005) Germline mutations of the PTCH gene in Japanese patients with nevoid basal cell carcinoma syndrome. Arch Dermatol Res 296: 303–308. [DOI] [PubMed] [Google Scholar]
- 35.Fujii K, Ohashi H, Suzuki M, Hatsuse H, Shiohama T, et al.. (2013) Frameshift mutation in the PTCH2 gene can cause nevoid basal cell carcinoma syndrome. Fam Cancer (in press). [DOI] [PubMed]
- 36. Kijima C, Miyashita T, Suzuki M, Oka H, Fujii K (2012) Two cases of nevoid basal cell carcinoma syndrome associated with meningioma caused by a PTCH1 or SUFU germline mutation. Fam Cancer 11: 565–570. [DOI] [PubMed] [Google Scholar]
- 37. Pastorino L, Ghiorzo P, Nasti S, Battistuzzi L, Cusano R, et al. (2009) Identification of a SUFU germline mutation in a family with Gorlin syndrome. Am J Med Genet A 149A: 1539–1543. [DOI] [PubMed] [Google Scholar]
- 38. Gutierrez MM, Mora RG (1986) Nevoid basal cell carcinoma syndrome. A review and case report of a patient with unilateral basal cell nevus syndrome. J Am Acad Dermatol 15: 1023–1030. [PubMed] [Google Scholar]
- 39. Matsuzawa N, Nagao T, Shimozato K, Niikawa N, Yoshiura KI (2006) Patched homologue 1 mutations in four Japanese families with basal cell nevus syndrome. J Clin Pathol 59: 1084–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ming JE, Kaupas ME, Roessler E, Brunner HG, Golabi M, et al. (2002) Mutations in PATCHED-1, the receptor for SONIC HEDGEHOG, are associated with holoprosencephaly. Hum Genet 110: 297–301. [DOI] [PubMed] [Google Scholar]
- 41. Fujii K, Kohno Y, Sugita K, Nakamura M, Moroi Y, et al. (2003) Mutations in the human homologue of Drosophila patched in Japanese nevoid basal cell carcinoma syndrome patients. Hum Mutat 21: 451–452. [DOI] [PubMed] [Google Scholar]
- 42. Suzuki M, Hatsuse H, Nagao K, Takayama Y, Kameyama K, et al. (2012) Selective haploinsufficiency of longer isoforms of PTCH1 protein can cause nevoid basal cell carcinoma syndrome. J Hum Genet 57: 422–426. [DOI] [PubMed] [Google Scholar]
- 43. Klein RD, Dykas DJ, Bale AE (2005) Clinical testing for the nevoid basal cell carcinoma syndrome in a DNA diagnostic laboratory. Genet Med 7: 611–619. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Literature review: 187 PTCH1 gene mutations in cases with NBCCS-associated KCOTs.
(DOCX)
Literature review: 17 PTCH1 gene mutations in cases with sporadic KCOTs.
(DOCX)
PRISMA Checklist for this systematic review.
(DOC)