Abstract
Purpose
Polymorphisms in the insulin-like growth factor 1 (IGF1) gene were previously associated with high or extreme myopia in Caucasian and Chinese populations. In the present study, we investigated whether IGF1 polymorphisms are associated with high myopia in a Japanese population.
Methods
A total of 446 Japanese patients with high myopia (≤−9.00 diopters) and 481 Japanese healthy controls (+1.50 diopters to −1.50 diopters) were recruited. We genotyped seven tagging single-nucleotide polymorphisms (SNPs) in IGF1 and assessed allelic and haplotypic diversity in cases and controls.
Results
There were no statistically significant differences in the allele frequencies of IGF1 SNPs and genotypes between cases and controls (P>0.05). However, the A allele of rs5742629 and the G allele of rs12423791 were associated with a moderately increased risk of high myopia (odds ratio [OR] =1.20 and OR =1.21, respectively) with borderline statistical significance (P=0.0502, corrected P (Pc) =0.21 and P=0.064, Pc=0.29, respectively). The haplotype consisting of the A allele of rs5742629 and the G allele of rs12423791 was marginally associated with the risk of high myopia (P=0.041; OR =1.21); this association was not significant after correction (Pc=0.19).
Conclusion
We found that the IGF1 SNPs are not significantly associated with high myopia in our Japanese population. Our results are in contrast to a previous study in which extreme myopia cases had significantly higher frequencies of the G allele of rs5742629 and the C allele of rs12423791 than controls. Therefore, the IGF1 SNPs may not be important factors for susceptibility to high myopia in all populations. Further genetic studies are needed to elucidate the possible contributions of the IGF1 region to the development of high myopia.
Keywords: high myopia, IGF1, association study, polymorphism
Introduction
Myopia is a very common refractive error that significantly impacts public health and economics around the world. High myopia is generally defined by an axial length greater than 26 mm or a spherical refractive error less than or equal to −6 diopters (D) that can cause blindness associated with an increased risk of various ocular and systemic diseases, including retinal detachment, submacular hemorrhage, glaucoma, and macular degeneration.1,2 High myopia imposes enormous economic and social burdens due to its higher prevalence in the populations of the People’s Republic of China, Singapore, Japan, and other countries when compared to the global average.3–6 Therefore, it is very important to identify the risk factors associated with high myopia and to establish preventive strategies for high myopia.
Although the cause of myopia is unclear, family-based studies and twin studies have shown that genetic factors are involved in the development and progression of myopia.7–14 Familial linkage studies have attempted to identify candidate genes for myopia, and significant linkages have been reported at 18 loci (MYP1 to MYP18).15 Many genome-wide association studies for myopia or high myopia have recently been performed, and many candidate genes and loci for myopia or high myopia have been identified.16–26
The gene encoding insulin-like growth factor 1 (IGF1) is highly conserved between species, and it encodes a polypeptide that plays an important role in regulating cell proliferation, differentiation, and apoptosis.27–29 Previous animal studies demonstrated that IGF1 also contributes to eye growth and myopia development in several species.30–32 In addition, recent genetic studies indicated that single-nucleotide polymorphisms (SNPs) in IGF1 were significantly associated with high or extreme myopia in Caucasian and Chinese populations,33–35 suggesting that IGF1 may play an important role in the development of high myopia through genetic polymorphisms. However, replication studies by Rydzanicz et al36 with a Polish family cohort and by Miyake et al37 with a Japanese population reported a lack of association of IGF1 SNPs with high or extreme myopia. Therefore, further genetic studies are needed to clarify the contribution of IGF1 SNPs to the development of myopia.
The aim of this study was to investigate whether genetic variants in IGF1 are associated with the risk of developing high myopia in our Japanese population.
Methods
Subjects
We recruited 446 unrelated Japanese individuals with high myopia (refractive error ≤−9.00 D in at least one eye) and 481 unrelated (not related to each other or to the patients) healthy Japanese controls (+1.50 D to −1.50 D in both eyes) at Yokohama City University and Okada Eye Clinic in Japan. The controls were sex-matched (47.7% male) to the patients, with an age range of 24 years to 75 years (mean: 39.3 ± 11.0 years) (Table 1). All participants had similar social backgrounds and resided in the same urban area. All participants were diagnosed by comprehensive ophthalmologic tests, including axial length, fundus examination, spherical power, and corneal curvature (autorefractors: NIDEK ARK-730A, ARK-700A TOPCON KP-8100P, and Biometer/Pachymeter AL-2000; Tomey Corporation, Nagoya, Japan). The individuals with high myopia had no known genetic diseases associated with myopia and/or high myopia, including glaucoma, keratoconus, and Marfan syndrome. Informed consent was obtained from all participants. The study methodology adhered to the tenets of the Declaration of Helsinki and was approved by the relevant ethics committees in each participating institute.
Table 1.
Characteristics of the study participants
| Parameters | Patients | Controls |
|---|---|---|
| Individuals, number | 446 | 481 |
| Male, % | 44.6 | 47.7 |
| Age, years (mean ± SD) | 37.9 ± 11.9 | 39.3 ± 11.0 |
| Spherical refractive errors*, diopters | ||
| Right eye (mean ± SD) | −11.7 ± 2.24 | Na |
| Left eye (mean ± SD) | −11.7 ± 2.27 | Na |
| Axial length, mm | ||
| Right eye (mean ± SD) | 28.0 ± 1.16 | Na |
| Left eye (mean ± SD) | 28.0 ± 1.22 | Na |
| Corneal refraction, mm | ||
| Right eye (mean ± SD) | 43.8 ± 1.45 | Na |
| Left eye (mean ± SD) | 43.8 ± 1.49 | Na |
Notes:
Patients, spherical refractive errors ≤−9.00 diopters in at least one eye; controls, spherical refractive errors within ± 1.50 diopters in both eyes.
Abbreviations: SD, standard deviation; Na, not applicable.
Patients’ ages ranged from 12 years to 76 years (mean: 37.9 ± 11.9 years), and 44.6% of the patients were male. The average spherical refractive errors were −11.7 ± 2.24 D (range: −6.75 D to −22.75 D) in the right eye and −11.7 ± 2.27 D (range: −8.50 D to −22.5 D) in the left eye. The average axial length was 28.0 ± 1.16 mm (range: 26.0–33.1 mm) for the right eye and 28.0 ± 1.22 mm (range: 26.0–34.7 mm) for the left eye. The average corneal refraction was 43.8 ± 1.45 D (range: 39.5–47.8 D) for the right eye and 43.8 ± 1.49 D (range 39.8–53.0 D) for the left eye (Table 1).
DNA extraction and IGF1 genotyping
The QIAamp DNA Blood Maxi Kit (Qiagen GmbH, Hilden, Germany) was used to collect peripheral blood lymphocytes and to extract genomic deoxyribonucleic acid (DNA) from peripheral blood cells. Procedures were performed under standardized conditions to prevent variation in DNA quality.
We evaluated seven tagging SNPs covering IGF1 and its flanking regions (3 kb upstream and 3 kb downstream): rs6214; rs11111262; rs972936; rs5742629; rs12423791; rs2162679; and rs5742612 (Table 2). These SNPs were selected from HapMap Japanese data (minor allele frequency ≥10%; pairwise r2 ≥0.8) (http://hapmap.ncbi.nlm.nih.gov/) to capture 100% of common IGF1 variants in HapMap. Of these SNPs, rs6214 and rs12423791 were associated with high myopia in previous studies.33–35 Genotyping was performed using the TaqMan 5′ exonuclease assay with validated TaqMan primer-probe sets supplied by Applied Biosystems, Inc. (Foster City, CA, USA). Polymerase chain reaction (PCR) was performed using a reaction mixture with a total volume of 10 μL containing 1X TaqMan Universal PCR Master Mix (Applied Biosystems, Inc.), 24 nM of each primer-probe set, and 3 ng of genomic DNA. The PCR conditions were as follows: 95°C for 10 minutes, followed by 40 cycles of denaturation at 92°C for 15 seconds, and annealing/extension at 60°C for 1 minute. The probe’s fluorescence signal was detected using the StepOnePlus Real-Time PCR System (Applied Biosystems, Inc.).
Table 2.
Allele frequencies of seven SNPs in IGF1
| SNPs | Chr | Position (build 37.1) | Gene location | Alleles (1 >.2) | Risk allele | Risk allele frequency, %
|
P | Pc | OR (95% CI) | |
|---|---|---|---|---|---|---|---|---|---|---|
| Cases (n=446) | Controls (n=481) | |||||||||
| rs6214 | 12 | 102,793,569 | 3′-UTr | A >G | G | 35.9 | 33.8 | 0.35 | 1.10 (0.91–1.33) | |
| rs11111262 | 12 | 102,798,177 | Intron 3 | G >A | G | 80.8 | 80.6 | 0.88 | 1.02 (0.81–1.28) | |
| rs972936 | 12 | 102,824,921 | Intron 2 | A >G | A | 54.3 | 50.5 | 0.11 | 1.16 (0.97–1.39) | |
| rs5742629 | 12 | 102,857,263 | Intron 2 | A >G | A | 60.3 | 55.8 | 0.0502 | 0.21 | 1.20 (1.00–1.45) |
| rs12423791 | 12 | 102,858,828 | Intron 2 | G >C | G | 73.4 | 69.5 | 0.064 | 0.29 | 1.21 (0.99–1.48) |
| rs2162679 | 12 | 102,871,259 | Intron 1 | A >G | A | 68.4 | 66.3 | 0.34 | 1.10 (0.90–1.33) | |
| rs5742612 | 12 | 102,874,864 | 5′-UTR | T >C | T | 69.7 | 67.6 | 0.32 | 1.11 (0.91–1.35) | |
Note: P-values less than 0.1 were corrected for multiple hypothesis testing (Pc) by Haploview using 10,000 permutations.
Abbreviations: SNPs, single-nucleotide polymorphisms; IGF1, insulin-like growth factor 1 gene; Chr, chromosome; n, number; Pc, corrected P-value; OR, odds ratio; Cl, confidence interval; UTR, untranslated region.
Statistical analysis
The Hardy-Weinberg equilibrium, allele frequencies, haplotype frequencies, and linkage disequilibrium were assessed using Haploview 4.1 (Broad Institute, Cambridge, MA, USA).38 Differences in allele and haplotype frequencies between cases and controls were assessed with the x2 text. The obtained P-values were corrected (Pc) for multiple hypothesis testing using a permutation test (10,000 iterations) in Haploview.
Results
The genotype frequencies of all seven tagging SNPs were in Hardy-Weinberg equilibrium in controls. Figure 1 shows the overall linkage disequilibrium patterns for the seven SNPs in 927 individuals. The seven SNPs showed a similar pattern of linkage disequilibrium between cases and controls (data not shown). Strong linkage disequilibrium was observed throughout the IGF1 region, and three haplotype blocks were suggested by our analysis; rs6214 and rs11111262 were located in block 1 (D’=1.00; r2=0.44), rs5742629 and rs12423791 were in block 2 (D’=0.99; r2=0.54), and rs2162679 and rs5742612 were in block 3 (D’=1.00; r2=0.94).
Figure 1.
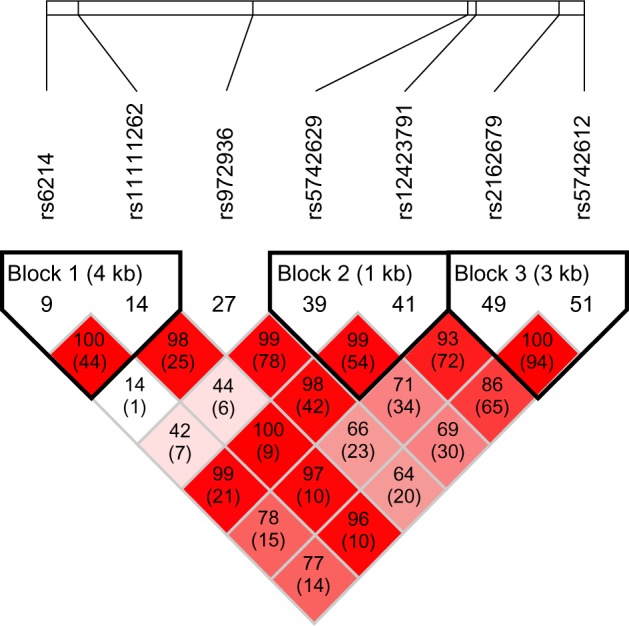
Linkage disequilibrium plot of seven SNPs in IGF1.
Notes: The D’ value and r2 value (in parentheses) corresponding to each SNP pair are expressed as a percentage and are shown within the respective square. Higher D’ values are indicated by a brighter red.
Abbreviations: SNPs, single-nucleotide polymorphisms; IGF1, insulin-like growth factor 1 gene.
Allele frequencies of seven SNPs were determined in cases and controls (Table 2). No statistically significant association was detected for any of the SNPs between cases and controls (P>0.05). However, the A allele of rs5742629 and the G allele of rs12423791 were associated with a moderately increased risk of high myopia (odds ratio [OR] =1.20; 95% confidence interval [CI] =1.00–1.45 and OR =1.21, 95% CI =0.99–1.48, respectively) with borderline statistical significance (P=0.0502, Pc=0.21 and P=0.064, Pc=0.29, respectively).
Table 3 contains the haplotype analysis of rs5742629 and rs12423791 in block 2. The haplotype, AG, created by the risk alleles was marginally associated with the risk of high myopia (P=0.041; OR =1.21; 95% CI =1.01–1.46), but the association was not significant after multiple-testing correction (Pc=0.19). In block 1 and block 3, there were no differences in the frequencies between cases and controls for any of the haplotypes examined in this study (data not shown).
Table 3.
Haplotype frequencies of rs5742629 and rs12423791
| Haplotypes | Frequency, %
|
P | Pc | OR (95% CI) | |
|---|---|---|---|---|---|
| Cases (n=446) | Controls (n=481) | ||||
| rs5742629-rs12423791 | |||||
| AG | 60.3 | 55.6 | 0.041 | 0.19 | 1.21 (1.01–1.46) |
| GC | 26.6 | 30.2 | 0.079 | 0.34 | 0.83 (0.68–1.02) |
| GG | 13.1 | 13.9 | 0.61 | 0.93 (0.71–1.22) | |
Note: P-values less than 0.1 were corrected for multiple hypothesis testing (Pc) by Haploview using 10,000 permutations.
Abbreviations: n, number; Pc, corrected P-value; OR, odds ratio; CI, confidence interval.
Discussion
The aim of the present study was to assess whether polymorphisms in the IGF1 region affect the development of high myopia in our Japanese population. To this end, we genotyped seven tagging SNPs in IGF1. Here, we report a lack of associations between IGF1 SNPs and high myopia in our Japanese patients.
Metlapally et al33 reported that rs6214 in IGF1 exhibited a significant association with several types of myopia, including high myopia (≤−5.00 D in at least one eye) in Caucasians. On the other hand, Rydzanicz et al36 did not find any significant association between IGF1 SNPs, (including rs6214) and high myopia (≤−6.00 D in at least one eye and ≤−5.00 D in the second eye) in a Polish family cohort. Zhuang et al34 demonstrated that IGF1 rs12423791, but not rs6214, was significantly associated with extreme myopia (axial length ≥26.00 mm and <−10.00 D in both eyes) in a Chinese population.34 In addition, a haplotype consisting of rs12423791 and rs5742629 was also associated with extreme myopia.34 Mak et al35 reported that a haplotype including rs12423791 was associated with high myopia (≤−8.00 D in both eyes) in another Chinese population. However, the replication study by Miyake et al37 indicated that none of the major tagging SNPs in IGF1 were associated with high (axial length ≥26.00 mm in both eyes) or extreme (≥28.00 mm in both eyes) myopia in a Japanese population.
In the present study of another Japanese population, we found the lack of an association between rs6214 and high myopia (≤−9.00 D in at least one eye), which is in agreement with previous replication studies.34–37 On the other hand, two SNPs (rs5742629 and rs12423791) and one haplotype (rs5742629-rs12423791) showed a borderline but not significant association, and patients harboring the A allele of rs5742629 and the G allele of rs12423791 had a moderately increased risk of high myopia. Our results are in contrast to those of a previous study,34 in which extreme myopia cases had significantly higher frequencies of the G allele of rs5742629 and the C allele of rs12423791 than controls; these alleles constituted a risk factor for extreme myopia cases in this previous study.34 This disparity suggests that the alleles of rs5742629 and rs12423791 are not a risk factor for susceptibility to high/extreme myopia.
At least three possible explanations exist for the differing results observed among these association studies. First, differences in the sample sizes of high or extreme myopia cases (628 from Caucasians,33 127 from Poland,36 302 from the People’s Republic of China,34 another 300 from the People’s Republic of China,35 1,339 from Japan,37 and another 446 from Japan in this study) may be important, because limited sample sizes can sometimes lead to false-positive or negative results in genetic studies. For example, the powers to detect the SNPs with an OR =1.3–1.5 and a minor allele frequency of 40% in the Polish36 and two Chinese studies34,35 are 15%–34% and 32%–73%, respectively. With less frequent SNPs with a minor allele frequency of 20%, the Polish36 and two Chinese studies34,35 showed lower power (11%–25% and 23%–59%, respectively) at the same significance level. Second, the differences in the recruitment criteria employed for high/extreme cases may lead to different results among association studies. However, we did not find any relationship between recruitment criteria and association levels. To assess this hypothesis in detail, it will be necessary to perform a meta-analysis of the present and previous studies. Finally, it is possible that environmental factor(s) are required for the association of IGF1 SNPs with high/extreme myopia. In addition to genetic factors, environmental factors such as near work (reading, studying, and computer use) and a lack of outdoor activity are clearly involved in myopia development, and it has been suggested that genetic and environmental interactions are important in establishing one’s predisposition to myopia.39,40IGF1 SNPs may not be able to contribute to myopia development alone; an interaction between IGF1 SNPs and particular environmental factor(s) may be important for this development. Since the present and previous studies33–37 did not assess joint contributions to disease risk from IGF1 SNPs and environmental factors, it remains to be seen whether the association of IGF1 SNPs with high/extreme myopia depends on particular environmental factor(s).
The present and previous studies33–37 used common IGF1 SNPs. Since common variants have only a limited capacity to tag rare variants, these studies have low power to detect associations with rare variants. Both common and rare variants are believed to contribute to the genetic susceptibility to complex diseases.41 Although previous studies33–35 that demonstrated positive results have shown significant associations of SNPs with minor allele frequencies of at least 20% with high myopia, there is a possibility that rare IGF1 variants can be causal variants of high myopia, suggesting the need to assess the role of rare IGF1 variants in high myopia.
Conclusion
In conclusion, we found that the IGF1 SNPs are not significantly associated with high myopia in our Japanese population. Our results do not support the positive findings of previous studies, suggesting that the IGF1 SNPs are not important risk factors for susceptibility to high myopia in all populations. However, as IGF1 variants may still affect the risk of high/extreme myopia, further genetic studies with larger sample sizes, other ethnic populations, rare variants, and consideration of gene–environment interactions are needed to clarify the contribution of IGF1 variants to disease development.
Acknowledgments
This study was supported by JSPS KAKENHI grant number 23590382. We sincerely thank all of the participants for their participation in this study, as well as all of the medical staff involved in sample collection and diagnosis.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Burton TC. The influence of refractive error and lattice degeneration on the incidence of retinal detachment. Trans Am Ophthalmol Soc. 1989;87:143–155. discussion 155–157. [PMC free article] [PubMed] [Google Scholar]
- 2.Saw SM, Gazzard G, Shih-Yen EC, Chua WH. Myopia and associated pathological complications. Ophthalmic Physiol Opt. 2005;25(5):381–391. doi: 10.1111/j.1475-1313.2005.00298.x. [DOI] [PubMed] [Google Scholar]
- 3.He M, Zeng J, Liu Y, Xu J, Pokharel GP, Ellwein LB. Refractive error and visual impairment in urban children in southern china. Invest Ophthalmol Vis Sci. 2004;45(3):793–799. doi: 10.1167/iovs.03-1051. [DOI] [PubMed] [Google Scholar]
- 4.Qian YS, Chu RY, He JC, et al. Incidence of myopia in high school students with and without red-green color vision deficiency. Invest Ophthalmol Vis Sci. 2009;50(4):1598–1605. doi: 10.1167/iovs.07-1362. [DOI] [PubMed] [Google Scholar]
- 5.Seet B, Wong TY, Tan DT, et al. Myopia in Singapore: taking a public health approach. Br J Ophthalmol. 2001;85(5):521–526. doi: 10.1136/bjo.85.5.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matsumura H, Hirai H. Prevalence of myopia and refractive changes in students from 3 to 17 years of age. Surv Ophthalmol. 1999;44(Suppl 1):S109–S115. doi: 10.1016/s0039-6257(99)00094-6. [DOI] [PubMed] [Google Scholar]
- 7.Wu MM, Edwards MH. The effect of having myopic parents: an analysis of myopia in three generations. Optom Vis Sci. 1999;76(6):387–392. doi: 10.1097/00006324-199906000-00018. [DOI] [PubMed] [Google Scholar]
- 8.Guggenheim JA, Kirov G, Hodson SA. The heritability of high myopia: a reanalysis of Goldschmidt’s data. J Med Genet. 2000;37(3):227–231. doi: 10.1136/jmg.37.3.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mutti DO, Mitchell GL, Moeschberger ML, Jones LA, Zadnik K. Parental myopia, near work, school achievement, and children’s refractive error. Invest Ophthalmol Vis Sci. 2002;43(12):3633–3640. [PubMed] [Google Scholar]
- 10.Farbrother JE, Kirov G, Owen MJ, Guggenheim JA. Family aggregation of high myopia: estimation of the sibling recurrence risk ratio. Invest Ophthalmol Vis Sci. 2004;45(9):2873–2878. doi: 10.1167/iovs.03-1155. [DOI] [PubMed] [Google Scholar]
- 11.Liang CL, Yen E, Su JY, et al. Impact of family history of high myopia on level and onset of myopia. Invest Ophthalmol Vis Sci. 2004;45(10):3446–3452. doi: 10.1167/iovs.03-1058. [DOI] [PubMed] [Google Scholar]
- 12.Klein AP, Duggal P, Lee KE, Klein R, Bailey-Wilson JE, Klein BE. Support for polygenic influences on ocular refractive error. Invest Ophthalmol Vis Sci. 2005;46(2):442–446. doi: 10.1167/iovs.04-0794. [DOI] [PubMed] [Google Scholar]
- 13.Sorsby A, Fraser GR. Statistical note on the components of ocular refraction in twins. J Med Genet. 1964;1(1):47–49. doi: 10.1136/jmg.1.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lyhne N, Sjølie AK, Kyvik KO, Green A. The importance of genes and environment for ocular refraction and its determiners: a population based study among 20–45 year old twins. Br J Ophthalmol. 2001;85(12):1470–1476. doi: 10.1136/bjo.85.12.1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu L, Li ZK, Gao JR, Liu JR, Xu CT. Epidemiology, genetics and treatments for myopia. Int J Ophthalmol. 2011;4(6):658–669. doi: 10.3980/j.issn.2222-3959.2011.06.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakanishi H, Yamada R, Gotoh N, et al. A genome-wide association analysis identified a novel susceptible locus for pathological myopia at 11q24.1. PLoS Genet. 2009;5(9):e1000660. doi: 10.1371/journal.pgen.1000660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Solouki AM, Verhoeven VJ, van Duijn CM, et al. A genome-wide association study identifies a susceptibility locus for refractive errors and myopia at 15q14. Nat Genet. 2010;42(10):897–901. doi: 10.1038/ng.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hysi PG, Young TL, Mackey DA, et al. A genome-wide association study for myopia and refractive error identifies a susceptibility locus at 15q25. Nat Genet. 2010;42(10):902–905. doi: 10.1038/ng.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li YJ, Goh L, Khor CC, et al. Genome-wide association studies reveal genetic variants in CTNND2 for high myopia in Singapore Chinese. Ophthalmology. 2011;118(2):368–375. doi: 10.1016/j.ophtha.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Z, Qu J, Xu X, et al. A genome-wide association study reveals association between common variants in an intergenic region of 4q25 and high-grade myopia in the Chinese Han population. Hum Mol Genet. 2011;20(14):2861–2868. doi: 10.1093/hmg/ddr169. [DOI] [PubMed] [Google Scholar]
- 21.Shi Y, Qu J, Zhang D, et al. Genetic variants at 13q12.12 are associated with high myopia in the Han Chinese population. Am J Hum Genet. 2011;88(6):805–813. doi: 10.1016/j.ajhg.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Verhoeven VJ, Hysi PG, Saw SM, et al. Large scale international replication and meta-analysis study confirms association of the 15q14 locus with myopia. The CREAM consortium. Hum Genet. 2012;131(9):1467–1480. doi: 10.1007/s00439-012-1176-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fan Q, Barathi VA, Cheng CY, et al. Genetic variants on chromosome 1q41 influence ocular axial length and high myopia. PLoS Genet. 2012;8(6):e1002753. doi: 10.1371/journal.pgen.1002753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meng W, Butterworth J, Bradley DT, et al. A genome-wide association study provides evidence for association of chromosome 8p23 (MYP10) and 10q21.1 (MYP15) with high myopia in the French Population. Invest Ophthalmol Vis Sci. 2012;53(13):7983–7988. doi: 10.1167/iovs.12-10409. [DOI] [PubMed] [Google Scholar]
- 25.Verhoeven VJ, Hysi PG, Wojciechowski R, et al. Consortium for Refractive Error and Myopia (CREAM) Diabetes Control and Complications Trial/Epidemiology of Diabetes Interventions and Complications (DCCT/EDIC) Research Group. Wellcome Trust Case Control Consortium 2 (WTCCC2) Fuchs’ Genetics Multi-Center Study Group Genome-wide meta-analyses of multiancestry cohorts identify multiple new susceptibility loci for refractive error and myopia. Nat Genet. 2013;45(3):314–318. doi: 10.1038/ng.2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi Y, Gong B, Chen L, et al. A genome-wide meta-analysis identifies two novel loci associated with high myopia in the Han Chinese population. Hum Mol Genet. 2013;22(11):2325–2333. doi: 10.1093/hmg/ddt066. [DOI] [PubMed] [Google Scholar]
- 27.McMorris FA, Dubois-Dalcq M. Insulin-like growth factor I promotes cell proliferation and oligodendroglial commitment in rat glial progenitor cells developing in vitro. J Neurosci Res. 1988;21(2–4):199–209. doi: 10.1002/jnr.490210212. [DOI] [PubMed] [Google Scholar]
- 28.D’Ercole AJ, Ye P, Calikoglu AS, Gutierrez-Ospina G. The role of the insulin-like growth factors in the central nervous system. Mol Neurobiol. 1996;13(3):227–255. doi: 10.1007/BF02740625. [DOI] [PubMed] [Google Scholar]
- 29.D’Mello SR, Galli C, Ciotti T, Calissano P. Induction of apoptosis in cerebellar granule neurons by low potassium: inhibition of death by insulin-like growth factor I and cAMP. Proc Natl Acad Sci U S A. 1993;90(23):10989–10993. doi: 10.1073/pnas.90.23.10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu J, O’Donnell M, Gitler AD, Klein PS. Kermit 2/XGIPC, an IGF1 receptor interacting protein, is required for IGF signaling in Xenopus eye development. Development. 2006;133(18):3651–3660. doi: 10.1242/dev.02547. [DOI] [PubMed] [Google Scholar]
- 31.Penha AM, Schaeffel F, Feldkaemper M. Insulin, insulin-like growth factor-1, insulin receptor, and insulin-like growth factor-1 receptor expression in the chick eye and their regulation with imposed myopic or hyperopic defocus. Mol Vis. 2011;17:1436–1448. [PMC free article] [PubMed] [Google Scholar]
- 32.Ritchey ER, Zelinka CP, Tang J, Liu J, Fischer AJ. The combination of IGF1 and FGF2 and the induction of excessive ocular growth and extreme myopia. Exp Eye Res. 2012;99:1–16. doi: 10.1016/j.exer.2012.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Metlapally R, Ki CS, Li YJ, et al. Genetic association of insulinlike growth factor-1 polymorphisms with high-grade myopia in an international family cohort. Invest Ophthalmol Vis Sci. 2010;51(9):4476–4479. doi: 10.1167/iovs.09-4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhuang W, Yang P, Li Z, et al. Association of insulin-like growth factor-1 polymorphisms with high myopia in the Chinese population. Mol Vis. 2012;18:634–644. [PMC free article] [PubMed] [Google Scholar]
- 35.Mak JY, Yap MK, Fung WY, Ng PW, Yip SP. Association of IGF1 gene haplotypes with high myopia in Chinese adults. Arch Ophthalmol. 2012;130(2):209–216. doi: 10.1001/archophthalmol.2011.365. [DOI] [PubMed] [Google Scholar]
- 36.Rydzanicz M, Nowak DM, Karolak JA, et al. IGF-1 gene polymorphisms in Polish families with high-grade myopia. Mol Vis. 2011;17:2428–2439. [PMC free article] [PubMed] [Google Scholar]
- 37.Miyake M, Yamashiro K, Nakanishi H, et al. Insulin-like growth factor 1 is not associated with high myopia in a large Japanese cohort. Mol Vis. 2013;19:1074–1081. [PMC free article] [PubMed] [Google Scholar]
- 38.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 39.Saw SM, Chua WH, Wu HM, Yap E, Chia KS, Stone RA. Myopia: gene-environment interaction. Ann Acad Med Singap. 2000;29(3):290–297. [PubMed] [Google Scholar]
- 40.Cordain L, Eaton SB, Brand Miller J, Lindeberg S, Jensen C. An evolutionary analysis of the aetiology and pathogenesis of juvenile-onset myopia. Acta Ophthalmol Scand. 2002;80(2):125–135. doi: 10.1034/j.1600-0420.2002.800203.x. [DOI] [PubMed] [Google Scholar]
- 41.Bodmer W, Bonilla C. Common and rare variants in multifactorial susceptibility to common diseases. Nat Genet. 2008;40(6):695–701. doi: 10.1038/ng.f.136. [DOI] [PMC free article] [PubMed] [Google Scholar]