Abstract
Human (h) embryonic stem cells (ESC) represent an unlimited source of cardiomyocytes (CMs); however, these differentiated cells are immature. Thus far, gene profiling studies have been performed with non-purified or non-chamber specific CMs. Here we took a combinatorial approach of using systems biology to guide functional discoveries of novel biological properties of purified hESC-derived ventricular (V) CMs. We profiled the transcriptomes of hESCs, hESC-, fetal (hF) and adult (hA) VCMs, and showed that hESC-VCMs displayed a unique transcriptomic signature. Not only did a detailed comparison between hESC-VCMs and hF-VCMs confirm known expression changes in metabolic and contractile genes, it further revealed novel differences in genes associated with reactive oxygen species (ROS) metabolism, migration and cell cycle, as well as potassium and calcium ion transport. Following these guides, we functionally confirmed that hESC-VCMs expressed IKATP with immature properties, and were accordingly vulnerable to hypoxia/reoxygenation-induced apoptosis. For mechanistic insights, our coexpression and promoter analyses uncovered a novel transcriptional hierarchy involving select transcription factors (GATA4, HAND1, NKX2.5, PPARGC1A and TCF8), and genes involved in contraction, calcium homeostasis and metabolism. These data highlight novel expression and functional differences between hESC-VCMs and their fetal counterparts, and offer insights into the underlying cell developmental state. These findings may lead to mechanism-based methods for in vitro driven maturation.
Introduction
The innate regenerative capacity of the adult mammalian heart is insufficient to restore function damaged by myocardial injury or heart failure. The identification of stem or progenitor cells that produce cardiomyocytes (CMs) has raised the intriguing possibility of cell-based cardiac regenerative therapies. Given the self-renewing capacity of pluripotent stem cells and their ability to differentiate into the cardiac lineage [1-6], human (h) embryonic stem cells (ESC) and induced pluripotent stem cell (iPSC) represent a potentially unlimited source of renewable CMs. However, hESC/iPSC-derived CMs are most similar to those of fetal tissues, functionally and structurally, and do not fully recapitulate post-natal or adult phenotypes [5,7-10]. For instance, hESC/iPSC-CMs have spontaneous contractile activities, express low levels of I K1, and relatively high currents from I NCX and I f [11]. Sarco/endoplasmic reticulum (SR) function remains rudimentary, as the cells exhibit a modest Ca2+ transient with slow kinetics, moderate SR Ca2+ATPase (SERCA) levels, low and disorganized ryanodine receptors and delayed phospholamban (PLN) expression [8,12] without the T-tubule system [9]. To overcome these limitations, an understanding of the molecular and cellular processes responsible for the development of a more physiological adult-like phenotype is required.
Microarray experiments have been performed to characterize the transcriptome of hESC-CMs and to identify signaling pathways implicated in their differentiation [13-17]. However, most of these studies were complicated by the presence of non-CMs in the cardiac biopsies and the use of un-staged and pooled fetal or adult heart samples [17,18] and non-purified or only partially purified ESC-CMs [13,14] without distinguishing the chamber-specific types. The expression data generally indicate that hESC-CMs express cardiac-specific genes, have lower levels of contractile and metabolic genes, and show differential expression of specific potassium/calcium ion channels, consistent with and complementary to known functional data. However, further functional analysis was often not performed and mechanistic insights into the causes of the immature state were lacking.
By studying a purified population of hESC-derived ventricular (V) CMs (hESC-VCMs), as identified by the expression of a reporter under the transcriptional expression of the MLC2v promoter [19] and functionally confirmed by electrophysiological assays, here we took a combinatorial approach of using systems biology to guide functional discoveries of novel biological properties of hESC-VCMs. We performed microarray and bioinformatics analyses of staged human (h) fetal (F, 18-20 weeks), adult (A) and hESC-derived VCMs. As anticipated, our results show that hESC-VCMs have a unique transcriptomic signature, contractility and metabolic parameters that are most analogous to fetal cells; but, we also discovered a range of novel changes in cell cycle, reactive oxygen species (ROS) metabolism and migration that have not been previously reported. Focused analysis on genes involved in potassium and calcium ion transport further revealed novel functional immaturity. These results are discussed in relation to regulatory mechanisms.
Materials and Methods
Culture and isolation of undifferentiated hESCs and hESC-VCMs
Undifferentiated HES2 (ES02, ESI International, Singapore) were maintained at 37°C and 5% CO2 on irradiated mouse embryonic fibroblasts. Cardiac differentiation was performed using established protocols [20] (Methods S1). 20-30 days after differentiation, cells were transduced with recombinant LV-MLC2v-mCherry particles. Fluorescing mCherry-positive cells were isolated 72 hours post-transduction by flow-activated cell sorting (BD FACSAriaTM II). Resultant cells were >95% pure and displayed ventricular action potentials.
No ethical approval was required for preparing the mouse embryonic fibroblasts because the animals were sacrificed without any prior manipulation. Mice were sacrificed by pentabarbital.
Isolation of hF- and hA-VCMs
HF-VCMs and hA-VCMs were isolated and experimented according to protocols approved by the UC Davis IUPAC and IRB (Protocol 200614787-1 and # 200614594-1) (Table S1, Methods S1).
Transcriptomic Profiling
Cell samples were lysed in Trizol (Invitrogen). After adding 1:4 volume chloroform, aqueous and organic phases were separated. RNAs were extracted from the aqueous phase using the miRNeasy kit (Qiagen, Valencia, CA). Sentrix 48kb WG-6 beadchips (Illumina, San Diego, CA) were used to profile mRNA expression. Microarray data were analyzed using the BeadStudio transcriptomic (Illumina) software packages. (Please see methods S1 for details)
Co-expression and promoter analyses
Pearson correlation coefficient (PCC) was used to compare expression pattern across four groups of samples, hESCs, hESC-VCMs, hF-VCMs and hA-VCMs (PCC>0.95). We focused on 17 TFs including FOXP1, GATA4, GATA6, HAND1, HAND2, IRX4, IRX5, ISL1, MEF2C, MESP1, NKX2-5, PPARA, PPARGC1A, SRF, TBX20, TBX5, which have all been shown to play important roles in the heart, as well as TCF8. Binding sites of TFs GATA4, HAND1, NKX2-5 and TCF8 were predicted using Genomatix MatInspector. The promoter region between 1000 bp upstream and 300 bp downstream of 27 genes in the co-expression network was analyzed.
Hypoxia treatment and TUNEL assay
HESC-VCMs of 20-30 days post-differentiation were subjected to hypoxia (1% O2)/reoxygenation for different durations as stated. For hypoxia, hESC-VCMs were transferred to a CO2 incubator with 95% N2/5% CO2 at 37°C. After hypoxia, cells were transferred back to 95% air/5% CO2 for reoxygenation. TUNEL assay was performed as per manufacturer’s instructions (Roche Diagnostics).
Electrophysiology
Electrical recordings were performed using the whole-cell patch-clamp technique as previously described [21,22] at 37°C. For I KATP measurements, the internal pipette solution contained (mM) potassium glutamate 120, KCl 25, ATP (magnesium salt) 1, EGTA 10, MgCl2 0.5, and HEPES 10 (pH 7.2). The external bath solution contained (mM) NaCl 140, KCl 5, MgCl2 1, CaCl2 1, and HEPES 10 (pH 7.4) [21]. To induce hypotonic stress, a modified Tyrode’s solution consisting of 64mM NaCl and 50mM mannitol was superfused with a flow speed of 2ml/min to replace the control solution consisting of 64mM NaCl and 150mM mannitol.
Optical Mapping
Transmembrane potential of hESC-VCM monolayers was optically mapped by using MiCam Ultima (Scimedia, USA) fluorescence non-contact imaging system with a 1cm2 field-of-view. Briefly, hESC-VCM preparations, which were cultured on gelatin-coated glass coverslips, were incubated with 4-8µM di-4-ANEPPS (Invitrogen, USA) for 20 min at room temperature in Tyrode’s solution, consisting of (mM) 140 NaCl, 5 KCl, 1 MgCl2, 1 CaCl2, 10 D-glucose and 10 HEPES at pH 7.4. Monolayers were rinsed twice with pre-warmed (37°C) Tyrode’s solution before imaging using a halogen light, which was filtered by a 515±35 nm band-pass excitation filter and a 590 nm high-pass emission filter. A co-axial point stimulation electrode was used to deliver a steady-state pacing at 1 Hz, 8V and 10ms pulse duration. Please see methods S1 for more details.
Results
Isolation and characterization of hESC-VCM
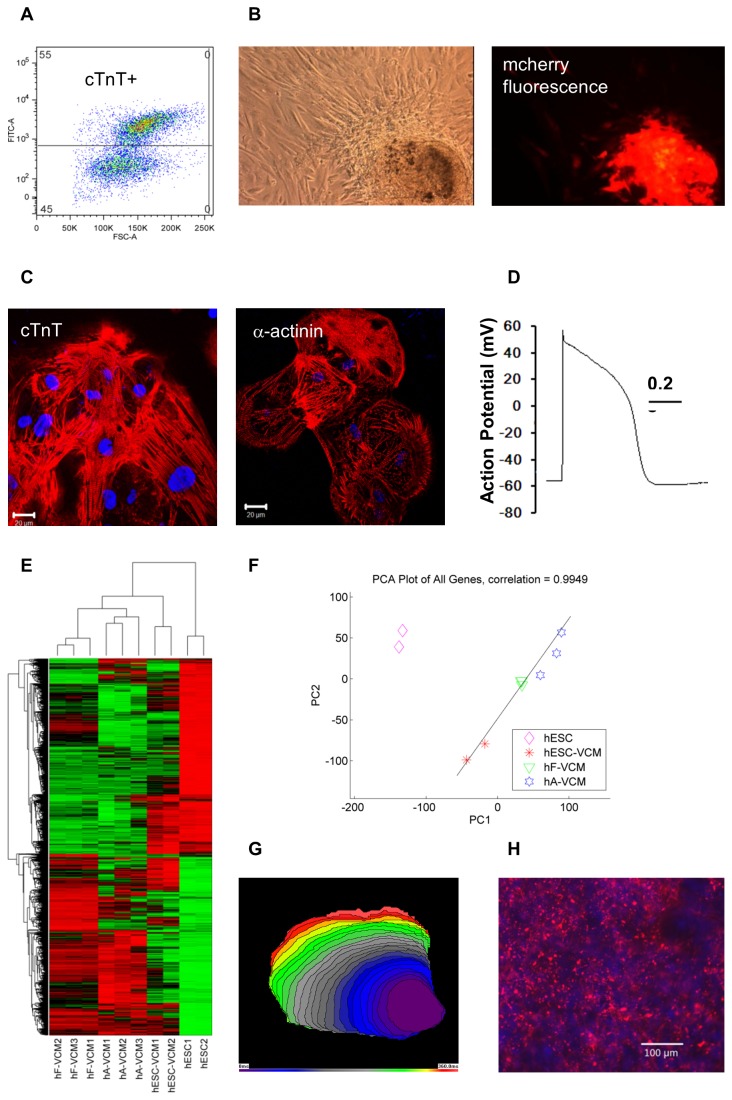
We performed directed cardiac differentiation as described previously [20] with cardiac derivatives making up ~50% of the cell population (Figure 1A). HESC-VCMs were identified by the expression of a reporter under the transcriptional control of the MLC2v promoter [19] (Figure 1B) and were isolated by flow-activated cell sorting. Resultant cells were >95% pure (Figure 1C) and displayed ventricular action potentials (Figure 1D).
Figure 1. Developmental classification of hESC-VCMs.
A) A representative FACS plot showing that approximately 55% of hESC-derivatives were positive for cardiac troponin-T (cTnT) prior to selection. B) Selected MLC2V-mCherry+ hESC-VCMs (~95% purified) under brightfield and fluorescence microscopy (100x). C) Purified HESC-VCMs were stained with anti-cTnT and anti-α-actinin antibodies. D) Representative action potential of hESC-VCMs. E) Hierarchical clustering showing that biological replicates cluster together. Red and green indicates up- and down-regulation respectively. F) Principal Component Analysis. HESC-VCMs, hF-VCMs and hA-VCMs lie along a linear developmental axis (dotted black line). Correlation coefficient is shown to indicate the degree of linearity. G) An activation isochronal map of trans-membrane potential with 15ms intervals. H) CX43 staining of hESC-VCMs. Red= CX43 staining, blue=DAPI.
HESC-VCMs displayed a unique transcriptional signature and were less developmentally advanced than hF-VCMs
We performed transcriptomic analyses of microarrays containing 48,804 probes corresponding to 25440 genes. Pluripotency genes such as POU5F1(Oct4) and NANOG were highly expressed in hESCs but absent in all of hESC-, hF- and hA-VCM samples. Conversely, cardiac markers such as MYL2, TNNT2 and ACTN2 were specifically detected in the three VCM populations, confirming their cardiac identities. Hierarchical clustering and principal components analysis (PCA) established that overall gene expressions among biological replicates were very similar (Figure 1E and F). The transcriptomic expression of hESC-VCM samples grouped more similarly to hF-VCMs and hA-VCMs than to hESCs (Figure 1F). The most conspicuous trend in global gene expression was that genes of hESC-VCMs, hF-VCMs and hA-VCMs on the PCA plot followed a virtually linear relationship (Figure 1F) with hESC-VCM, hF-VCM, and hA-VCM group of genes situated at the bottom left, middle and top right, respectively, which is consistent with a progressive increase in cellular maturity. A comparison between hESC-VCMs and hF-VCMs showed that 1852 (7%) and 2195 (9%) genes were up-/down-regulated by more than 2-fold in hESC-VCMs relative to hF-VCMs respectively.
HESC-VCMs expressed significant levels of chamber-specific genes similar to chamber myocardium, but with a slower conduction velocity than hF-VCMs
We further assessed the developmental status of hESC-VCMs by the use of molecular markers that are restricted to or upregulated in chamber myocardium relative to the primitive myocardium in human [23] and mice [24]. We detected significant gene expression from NPPA (atrial natriuretic factor), GJA1 (gap junction alpha-1/connexin 43), GJA5 (gap junction alpha-5/connexin 40), SMPX(chisel) and IRX5(Iroquis-5) (p<0.05) in hESC-VCMs; however, the expression of GJA5, SMPX, and IRX5 in hESC-VCMs was 4.7-, 3.2-, and 2.4- fold lower than those in hF-VCMs. The data showed that the hESC-VCMs displayed gene expression profiles typical of early chamber myocardium, suggesting that the cells were less developmentally advanced than the hF-VCMs analyzed in this study. The general immaturity of hESC-VCMs and more specifically the reduced expression of GJA5, is also typical of very immature chamber myocardium, which is known to have a very slow conduction velocity. Consistently, optical mapping of purified hESC-VCM in monolayer culture confirmed that the conduction velocity (5.0±1.4 cm/s) was much slower than that of the intact adult human heart (7-80 cm/s depending on fibre direction) [25] (Figure 1G) but is similar to that of non-purified hESC-CMs (5±6 cm/s) [11]. Furthermore, CX43-positive gap junctions are aligned at the intercalated disc of adult CMs [26] but such was not the case for hESC-VCMs, whose CX43-positive gap junctions were randomly distributed (Figure 1H). Evidently, conduction pattern was isotropic as opposed to anisotropic as observed in the native myocardium [27].
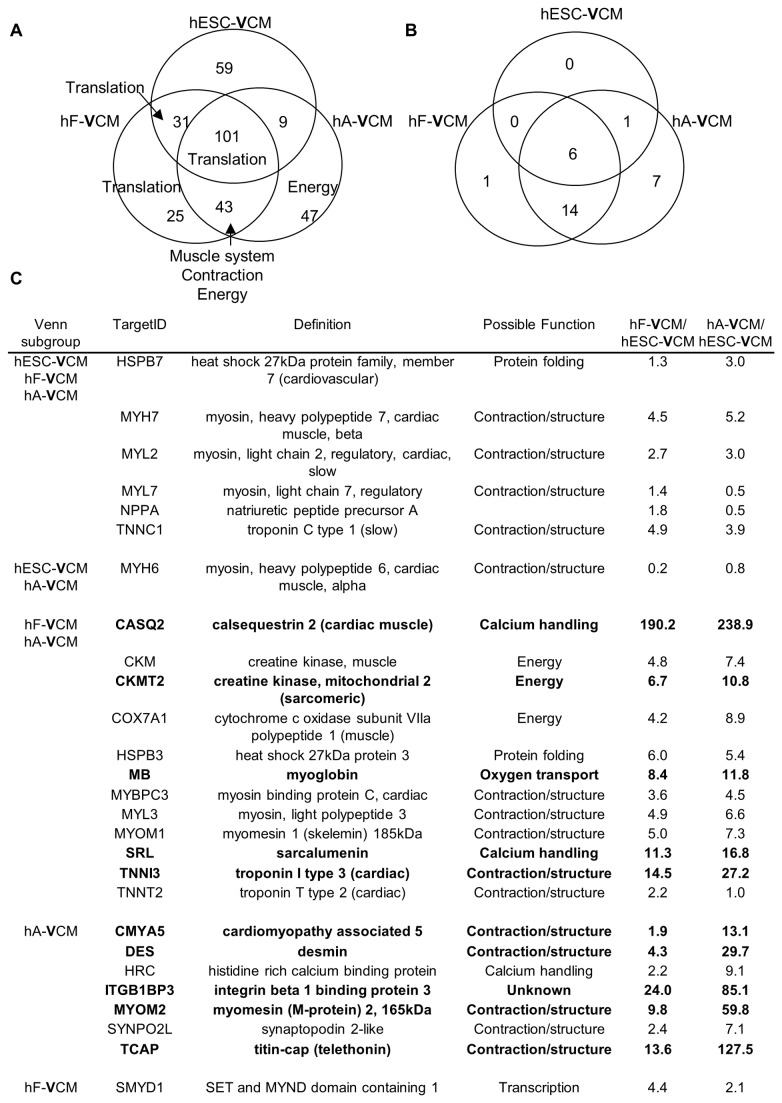
Examination of the most abundant transcripts in CM samples identified novel markers of cardiac maturation
The functions of the 200 most abundant transcripts in hESC-, hF- and hA-VCMs were examined by Gene Ontology analysis (Table S2). The three populations showed a high level of similarity, with 101 common transcripts involving mostly translation elongation (Figure 2A). HA- and hF-VCMs shared 43 transcripts enriched for muscle system, contraction and energy generation, while hESC- and hF-VCMs shared only 31 involved in translational elongation. Transcripts involved in energy generation were particularly abundant in hA-VCMs, consistent with their high metabolism. We focused on heart-specific genes to identify novel markers for CM identity and maturation. Twenty-seven gene products were more than 10-fold enriched in the heart and skeletal muscle, including known cardiac markers such as MYL2, MYH7 and TNNC1 (Figure 2B and C). Six genes (HSPB7, MYH7, MYL2, MYL7, NPPA and TNNC1) were within the top 200 abundant genes in all three CM samples and may serve as markers of cardiac identity. Ten gene transcripts were more than 10-fold depleted in hESC-VCMs relative to hF-VCMs and/or hA-VCMs. Of these, the expression of CKMT2, SRL, CMYA5, ITGB1BP3, MYOM2 and TCAP in hESC-VCMs have not been described previously. CKMT2 is involved in energy generation. CMYA5, MYOM2 and TCAP are structural genes while SRL encodes a Ca2+ binding protein. ITGB1BP3 encodes an integrin beta 1 binding protein with unknown function in the heart.
Figure 2. The most abundant transcripts in VCM populations.
A) The Venn diagram shows the distribution of the top 200 most abundant genes in hESC-VCMs, hF-VCMs and hA-VCMs. The number of genes in common is indicated. The labels refer to biological processes that are enriched. B) The tissue-specific distribution of the top 200 genes was examined in public databases including Genenote and bioGPS. The ‘cardiac-specific’ genes among the top 200 most abundant genes are shown. C) 27 cardiac-specific genes within the top 200 most abundant genes in hESC-VCMs, hF-VCMs and/or hA-VCMs. Fold change relative to hESC-VCMs are indicated and fold change >10 is highlighted in bold. Possible function is based on gene ontology association.
Gene Set Enrichment Analysis (GSEA) reveals novel expression differences in cell cycle, ROS metabolism and migration
GSEA was employed to identify specific expression differences between hF-VCMs and hESC-VCMs. We examined 1403 gene sets among 25440 genes and found that 119 (8%) and 572 (41%) gene sets were significantly decreased and increased in hESC-VCMs relative to hF-VCMs respectively. The 50 most differentially expressed gene sets are summarized in Table 1 and listed in Table S3A and S3B. QRT-PCR analysis was performed on selected genes to verify the microarray data (Figure S1).
Table 1. Summary of top 50 gene sets which showed differential expression between hESC-VCMs and hF-VCMs.
Please refer to Table S3A and B for details.
| hESC-VCM depleted | # gene sets | hESC-VCM enriched | # gene sets |
|---|---|---|---|
| Cell Cycle | 20 | Cytokine and defense | 17 |
| Metabolism | 9 | Stress | 6 |
| Contraction | 6 | Movement | 11 |
| ROS metabolism | 2 | Signaling | 3 |
| Heart development | 4 | Development | 1 |
| Telomere | 2 | Apoptosis | 1 |
| Others | 7 | Others | 11 |
Consistent with previous reports, gene sets related to respiration, tricarboxylic acid (TCA) cycle, fatty acid oxidation and heart development were decreased in hESC-VCMs compared to hF-VCMs [17]. Of expression changes that have not been previously described, cell cycle-related gene sets showed the most significant decrease in expression in hESC-VCMs, with extremely low false discovery rate (FDR) scores of <0.0001. They were also the most abundant, totaling 20 out of 50 top most reduced gene sets. These gene sets encompassed various stages including G1-M phases and cytokinesis, suggestive of inhibition at multiple stages of the cell cycle. Cycling cells require mechanisms to preserve genomic integrity during DNA replication and we detected lower level of genes associated with telomere maintenance, consistent with the decreased level of cell cycle genes. Genes involved in ROS metabolism were down-regulated in hESC-VCMs. In particular, catalase (CAT) and glutathione peroxidase (GPX3) are important for ROS removal and both were found at low and significantly reduced levels (10- and 5-fold respectively) in hESC-VCMs compared to hF-VCMs. ROS plays an important role in the induction of apoptosis and expression of apoptotic genes were also enhanced in hESC-VCMs.
There was an increased expression of genes associated with cell migration, in 11 out of the top 50 gene sets. Examples included pro-migratory proteases and receptors e.g., THBS1, F2RL1, and SPHK1, signaling molecules TGFB2 and BMP2, and matrix metalloproteinase MMP9. Consistent with this migratory phenotype, we observed higher expression of genes important for epithelial-mesenchymal transformation. Increased expression of genes associated with extracellular matrix organization e.g., COL2A1 was also noted although these latter changes were not within the top 50 differentially gene sets. Genes associated with cytokine stimulation and cellular defense were also up-regulated in hESC-VCMs.
Novel expression changes in potassium and calcium transport genes
Even though the electrophysiological attributes of hESC-CMs are known to be immature [7-10,19], gene sets related to potassium and calcium ion transport were not differentially expressed between hESC-VCMs and hF-VCMs; however, specific genes did show significant differences. Among Ca2+ transport genes, SLC8A1, PLN, RYR2, CACNB2, CAV3, CAMK2A and CACNA1C levels were lower in hESC-VCMs, as previously reported [14,18]. We also observed novel changes in genes important for Ca2+ handling (Table 2). Sarcalumenin (SRL) is a SR protein thought to regulate SERCA stability and was reduced by 11.3-fold in hESC-VCMs. CAMK2D and CAMK2B encode Ca2+/Calmodulin kinases (CAMKII) and multiple isoforms of these genes were significantly reduced in hESC-VCMs. Phospholemman (FXYD1) modulates Na+/K+ ATPase, I NCX and I Ca and was also highly reduced. Other genes that were diminished included gap junction gene GJA4 (4.7-fold), endothelin receptors, EDNRA (2.6-fold) and EDNRB (4.6-fold), and CAV1 (2.8-fold) etc (Table 2). A comparison with hA-VCMs further showed that most of these genes (CAMK2B, CAMK2D, SRL, FXYD1 and ENDRB) were expressed at increasing levels in hESC-, hF- and hA-VCMs, which supports their potential role in cardiac maturation.
Table 2. Novel expression changes in genes involved in calcium ion transport and homeostasis, p<0.05.
| Gene | hF-VCM/ hESC-VCM | Description |
|
|---|---|---|---|
| SRL | 11.3 | Sarcalumenin | |
| CAMK2B | 8.1, 5.5, 4.5, 4.0* | calcium/calmodulin-dependent protein kinase II beta | |
| FXYD1 | 7.6, 5.4, 2.6* | FXYD domain containing ion transport regulator 1 (phospholemman) | |
| CACNB2 | 5.3 | calcium channel, voltage-dependent, beta 2 subunit | |
| GJA4 | 4.7 | gap junction protein, alpha 4, 37kDa | |
| EDNRB | 4.6 | endothelin receptor type B | |
| CAMK2D | 3.7, 2.5* | calcium/calmodulin-dependent protein kinase II delta | |
| JAK2 | 3.3 | Janus kinase 2 (a protein tyrosine kinase) | |
| EPHX2 | 2.8 | epoxide hydrolase 2, cytoplasmic | |
| CAV1 | 2.8 | caveolin 1, caveolae protein, 22kDa | |
| KDR | 2.7 | kinase insert domain receptor | |
| CSRP3 | 2.6 | cysteine and glycine-rich protein 3 (cardiac LIM protein) | |
| PLCG2 | 2.6 | phospholipase C, gamma 2 (phosphatidylinositol-specific) | |
| EDNRA | 2.6 | endothelin receptor type A | |
Fold changes for multiple transcript variants of the same gene.
We also detected novel expression changes in genes encoding potassium channels, and consistent with previous results, significantly reduced levels of KCNQ1, KCNE1, KCNAB1, KCNJ2 and KCNJ8 were observed [14,18,28]. The novel changes in potassium channels reported here are implicated in cardioprotection and include genes encoding I KATP, I KCa and I KNa (Table 3). Transcript variants for the regulatory unit of I KATP, ABCC9, were either absent or reduced in hESC-VCMs compared to hF-VCMs. Isoform-specific expression of I KCa (KCNMB1 and KCNMB3) and I KNa (KCNT1 and KCNT2) subunits were also observed. AQP1 (aquaporin-1) was decreased in hESC-VCMs. KCNG1, KCTD8, FXYD2 and KCNIP4 were uniquely expressed in hESC-VCMs.
Table 3. Novel expression changes in genes involved in potassium ion transport and homeostasis, p<0.05.
| Gene | hF-VCM/ hESC-VCM | Description |
|---|---|---|
| KCNT1 | hF-VCM only | potassium channel, subfamily T, member 1 |
| ABCC9 | hF-VCM only, 6.7, 6.6* | ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| KCNMB1 | 6.3 | potassium large conductance calcium-activated channel, subfamily M, beta member 1 |
| AQP1 | 3.0 | aquaporin 1 |
| CHP | 2.1 | calcium binding protein P22 |
| NSF | 0.5 | N-ethylmaleimide-sensitive factor |
| KCNMB3 | 0.3 | potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| NSF | 0.2 | N-ethylmaleimide-sensitive factor |
| KCTD8 | hESC-VCM only | potassium channel tetramerisation domain containing 8 |
| KCNT2 | hESC-VCM only | potassium channel, subfamily T, member 2 |
| KCNG1 | hESC-VCM only | potassium voltage-gated channel, subfamily G, member 1 |
| FXYD2 | hESC-VCM only | FXYD domain containing ion transport regulator 2 |
| KCNIP4 | hESC-VCM only | Kv channel interacting protein 4 |
Fold changes for multiple transcript variants of the same gene.
Cardioprotective mechanisms in hESC-VCMs
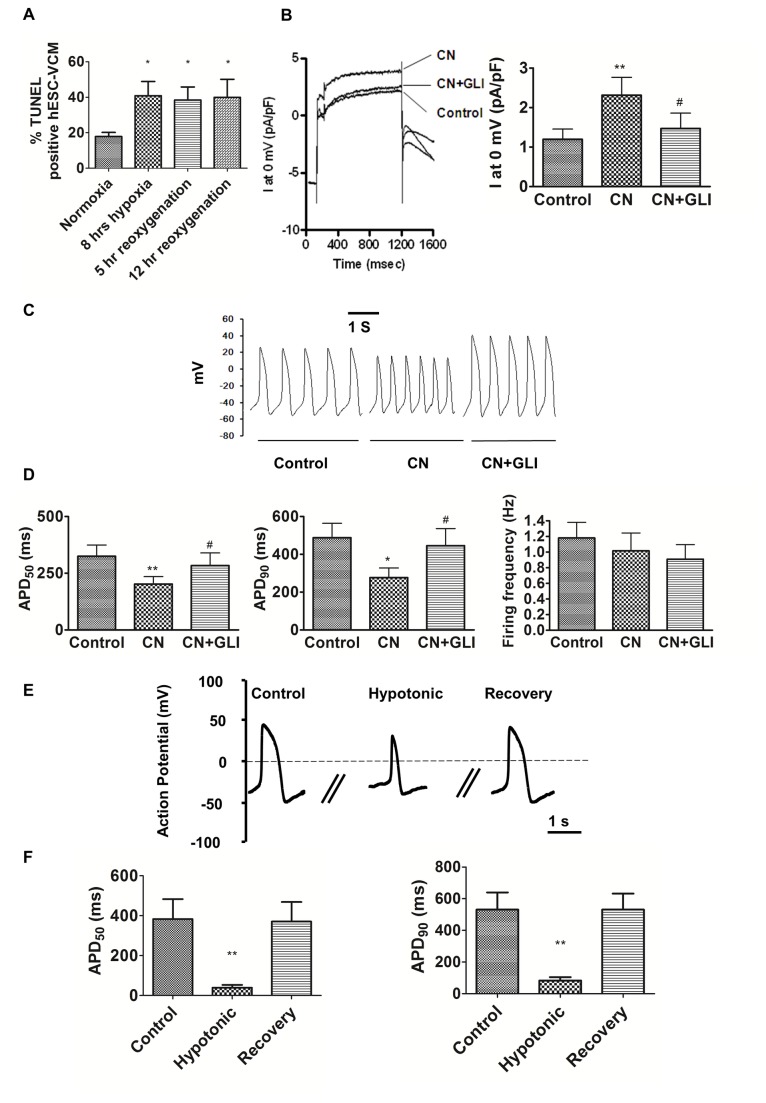
I KATP, I KCa, I KNa, ROS metabolic enzymes and AQP1 are important for cardioprotection by maintaining cellular homeostasis. We postulated that reduced/perturbed expression would result in increased susceptibility to injury. To test this, we subjected hESC-VCMs to hypoxia and hypoxia/reoxygenation, which is a model for ischemia/reperfusion injury. Consistent with our postulate, we observed a 2-fold increase in TUNEL positive apoptotic cells after hypoxia and hypoxia/reoxygenation compared with normoxic conditions (Figure 3A). By contrast, hF-VCMs showed almost no increase in cell death after a similar protocol [29].
Figure 3. Functional analyses of hESC-VCMs.
A) Apoptotic response after hypoxia/reoxygenation was assessed using TUNEL assay. HESC-VCMs were subjected to hypoxic treatment, followed by reoxygenation of different duration as indicated. *P<0.05 relative to normoxic conditions.
B) I KATP channel current and its effect on electrophysiological properties of hESC-VCMs. Representative current and summary of current densities of whole-cell patch-clamp recording of hESC-VCMs. Extracellular application of I KATP channel openers, 2 mmol/L sodium cyanide (CN), at 2 min significantly increased current densities compared to controls, which was inhibited by I KATP channel blocker 10 uM glibenclamide (GLI). Cells were stimulated to 0 mV for 1000 ms from a holding potential of -80 mV preceded by a 100-ms prepulse to -10 mV. C) Representative tracings of action potentials of hESC-VCMs before and after treatment of 2 mM CN and then 10 uM GLI. D) The effect of CN and GLI on APD50, APD90 and firing frequency. N=6; *P<0.05, **P<0.01 compared to control group; #P<0.05, compared to CN group. Effect of hypotonic insult on hESC-VCMs. E) Representative tracings of action potentials of hESC-VCMs before and after hypotonic insult, and upon recovery. F) The effect of hypotonic insult on APD50 and APD90. N=6; **P<0.01 compared to control group.
To further explore the mechanisms underlying this increased cell vulnerability, we focused on the functionality of I KATP, which regulates ion homeostasis in response to ATP depletion during ischemic insult, and showed that I KATP was reduced in hESC-VCMs. Sodium cyanide (CN) can induce ATP depletion via uncoupling of oxidative phosphorylation. Application of CN induced an outward current and significantly increased current densities from 1.2+/-0.3 to 2.3+/-0.5 pA/pF, which was inhibited by I KATP blocker glibenclamide (GLI) (Figure 3B). Of note, I KATP in hESC-VCMs was smaller than that reported for hA-ACMs (7.3 ± 2 pA/pF) [30] and mouse adult CMs (approx. 23 pA/pF) [21]. Figure 3C further shows the role of I KATP in the AP waveform. CN significantly reduced APD50 and APD90 to 61.0 and 56.7% of control, thereby hastening the spontaneous firing (Figure 3D). CN had no significant effect on AP amplitude or frequency (Figure 3D). This CN-mediated AP shortening could be abolished by blockade of I KATP by GLI. In addition to ATP depletion, ischemic insult is also accompanied by a cytosolic buildup of metabolites, increased intracellular osmolarity, followed by cell swelling. Given that I KATP and aquaporin-1 are both involved in osmotic regulation, and their reduced expression in hESC-VCMs may result in perturbed responses to hypertonic stress (which simulates ischemic injury), we tested the effect of hypotonic treatment on hESC-VCMs [22]. We found that hypotonic stress indeed resulted in the loss of spontaneous AP in hESC-VCMs. 1000pA-5ms stimulation produced an AP with significantly reduced APD50 and APD90 10.1% and 15.6% of control (Figure 3E and F). Upon recovery, AP returned to pre-treatment conditions. Thus hESC-VCMs were able to undergo AP shortening upon hypotonic treatment, as was reported for adult guinea pig CMs [22].
Cardiac transcription factors: expression, co-expression and promoter analyses
To unravel possible regulatory mechanisms for the developmental immaturity of hESC-VCMs, we analyzed the expression of transcription factors (TFs) critical for heart development. We found that transcripts from TBX5 (12.9 fold), TBX20 (6.9 fold), GATA4 (1.3 fold) and IRX5 (2.4 fold), which are implicated in chamber formation [31-33], were reduced in hESC-VCMs. Similarly, transcripts encoding critical cardiac developmental TF genes, including HAND2 (3.0 fold), GATA6 (3.0 fold), IRX4 (2.6 fold), SRF (2.5 fold), were significantly decreased. Conversely, TBX2 [34] and TBX3 [35] locally repress chamber differentiation and TBX3 was 3-fold enriched in hESC-VCMs. The expression of transcription factors ISL1, MEF2C, MESP1, FOXP1, HAND1, NKX2.5 and PPARGC1A were not significantly different between hESC-VCMs and hF-VCMs.
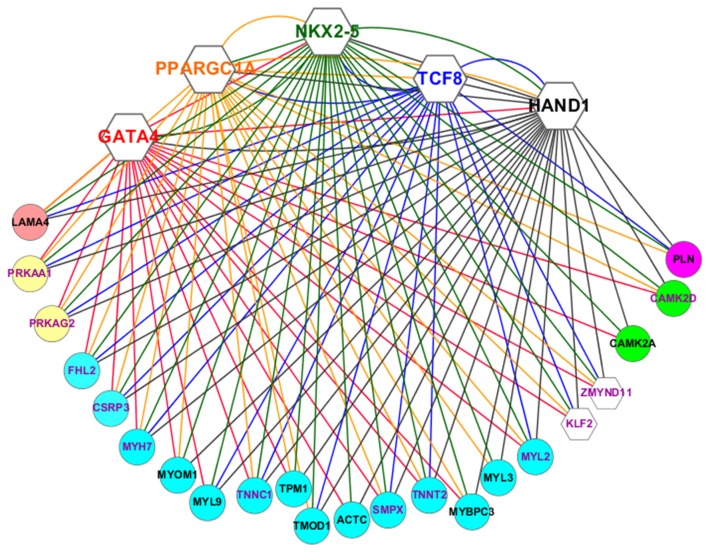
We performed co-expression analysis to construct transcriptional networks of cardiac TFs and genes important for cardiac function [36]. First, we correlated the expression of cardiac TFs across the four groups of sample, hESCs, hESC-VCMs, hF-VCMs and hA-VCMs (Methods, Table S4) and found that a group of 4 TFs consisting of HAND1, GATA4, NKX2-5 and PPARGC1A had significantly similar expression patterns and that their expression also correlated highly and significantly with a fifth TF, TCF8. HAND1, for instance, co-expressed with GATA4, NKX2-5, PPARGC1A and TCF8 (Figure 4, Table S4). Likewise, NKX2-5 also co-expressed with GATA4, HAND1, PPARGC1A and TCF8. We then showed that these 5 TFs co-expressed with many common genes, suggesting a co-regulatory relationship between these 5 TFs and other genes in the microarray. HAND1, GATA4, NKX2-5, PPARGC1A and TCF8 co-expressed with 144, 133, 172, 160 and 112 genes respectively. These TFs also jointly co-expressed with many common genes. For example, among 71 genes that showed co-expression links with more than 5 of all 17 TFs, 39 co-expressed with HAND1, GATA4, NKX2-5, PPARGC1A and TCF8. In addition, 19 of these co-expressed exclusively with the 5 TFs, indicating that these TFs are major components of such a co-expressed gene network. Although we identified a relatively large number of co-expressed genes with MEF2C (108), MESP1 (73) and PPARA (97), they did not form co-expression links with other TFs. The other 8 TFs co-expressed with far fewer genes (below 51).
Figure 4. Co-expression network in CMs.
The five transcription factors are shown as hexagons with gene symbols and their connections are shown by solid lines of the same colors e.g. GATA4 is in red and are connected to co-expressed genes by solid red lines. Genes are shown in circles and are color-coded to indicate their functional classifications. Red = regulation of contraction/adhesion, yellow = AMP kinases, blue = contractile genes, white = transcription factors, green = Ca/calmodulin kinases, purple = Ca handling protein. Gene labels in purple indicate genes that are co-expressed with all five transcription factors. Gene labels in black indicate genes that are co-expressed with three or four transcription factors.
The gene ontology affiliations of genes which co-expressed with the 5 TFs were primarily associated with Ca2+ homeostasis, contraction, metabolism and transcriptional regulation. Totally, 39 genes had a co-expression relationship with all 5 TFs. Of these, seven (MYH7, SMPX, TNNT2, MYL2, TNNC1, CSRP3 and FHL2) are involved in contraction/cytoskeleton (Figure 4). The rest included TFs (KLF2 and ZMYND11), and regulators of the AMPK pathway PRKAA1 and PRKAG2, etc. 88 genes correlated with at least 4 of the 5 TFs, including themselves HAND1 and NKX2-5 (Table S4). These analyses were then extended to develop a co-expression network, that bioinformatically shows highly significant and conserved patterns of expression between TFs and putative target genes. The resulting co-expression network consisted of genes associated with transcriptional regulation, contraction, metabolism and calcium handling processes and consisted of 5 TFs and 22 putative target genes (Figure 4).
We further defined the regulatory structure of our co-expression network by examining the promoter regions of all 27 genes for binding sites of 4 TFs (GATA4, HAND1, NKX2-5 and TCF8) (Table 4, Table S5 for details). PPARGC1A was excluded from analysis because it mostly functions as a co-factor and does not have defined binding sites. Our analysis identified many binding sites for the 4 TFs among the promoters of the 27 genes. GATA4, HAND1, NKX2-5 and TCF8 predicted binding sites were found in the promoter regions of 9, 10, 23, 22 genes respectively. Many genes contained binding sites for multiple TFs in their promoters. 22 out of 27 genes contained binding sites for 2 or more TFs in their promoters, consistent with a regulatory relationship between the TFs analysed and the other genes in the co-expression network. Additionally, the TFs themselves were predicted to regulate each other. For instance, HAND1 promoter contained binding sites for all 4 TFs (GATA4, HAND1, NKX2-5 and TCF8). This is consistent with the co-expression relationship between HAND1 and GATA4, NKX2-5, PPARGC1A and TCF8.
Table 4. Promoter analysis.
The number below each TF indicates the number of putative binding sites in the region 1000bp up- and 300bp down-stream of any given gene.
| Gene | GATA4 | HAND1 | NKX2.5 | TCF8 |
|---|---|---|---|---|
| ACTC1 | 2 | |||
| CAMK2A | 2 | 1 | ||
| CAMK2D | 1 | 2 | 1 | |
| CSRP3 | 1 | 1 | 5 | 1 |
| FHL2 | 1 | 8 | 3 | |
| GATA4 | ||||
| HAND1 | 1 | 1 | 1 | 1 |
| KLF2 | 3 | |||
| LAMA4 | 1 | 1 | ||
| MYBPC3 | 2 | 1 | ||
| MYH7 | 2 | 1 | ||
| MYL2 | 2 | 1 | ||
| MYL3 | 2 | 4 | 2 | |
| MYL9 | 1 | |||
| MYOM1 | 1 | 4 | 1 | |
| NKX2-5 | 2 | 3 | ||
| PLN | 2 | 5 | 3 | |
| PPARGC1A | 1 | 3 | 1 | |
| PRKAA1 | 2 | 1 | ||
| PRKAG2 | 1 | 4 | 2 | |
| SMPX | 2 | 2 | 1 | |
| TMOD1 | 3 | 1 | ||
| TNNC1 | 1 | 2 | 2 | |
| TNNT2 | 1 | |||
| TPM1 | 1 | 1 | 1 | |
| ZEB1 | 1 | 1 | 2 | |
| ZMYND11 | 2 | 2 | 3 |
Discussion
Here, we describe the transcriptome of hESC-VCMs and compare them with their in vivo counterparts to evaluate their molecular phenotype and developmental status and to identify regulatory mechanisms that might underlie these differences. Our PCA results show that hESC-VCMs are developmentally less advanced than hF-VCMs of 18-20 weeks. In support of the above assertion, we also show that the expression of genes important for CM function (e.g. contraction, metabolism and heart development) are low in hESC-VCMs. Similar results have been reported between non-purified/mixed lineage hESC-CMs and/or whole fetal heart, but it was unclear whether these results were due to contaminating fibroblasts in whole fetal heart and/or the presence of pacemaker/atrial/ventricular cells in hESC-CM cultures [14,17]. Here we showed that ventricular-specific hESC-CMs are less mature than hF-CMs of 18-20 weeks. Consistent with our findings, He et al. claim that hESC-derived beating outgrowths have properties of APs anticipated in embryonic heart before 7 weeks of development [2]. Our staging, however, differs from that of Cao et al [14] who stated that hESC-derived cultures were most similar to 20 weeks old hF-VCMs. Such differences could be attributed to the limited enrichment of hESC-CMs in their cultures, which consisted of only 40-45% CMs. Consistent with this, hierarchical clustering showed that their enriched hESC-CMs were grouped more closely with embryoid bodies (consisting of a mixed cell population) than hF-VCMs. We also uniquely show that hESC-VCMs expressed markers of, and had low conduction velocity consistent with immature chamber myocardium [23]. Two groups have previously measured the conduction velocity of non-purified beating hESC-CMs clusters [11,37]. However, the results were heterogeneous and the authors indicated that this may partly stem from the presence of non-myocytes within the cell network which may electrically couple with CMs and thereby slow conduction [37]. We show that purified hESC-VCMs indeed had a conduction velocity(5.0±1.4 cm/s) much slower than that of adult human heart [25].
16% of genes are differentially expressed between hESC-VCMs and hF-VCMs by more than 2-fold. Interestingly, this 16% difference accounts for a 49% difference among gene sets as examined by GSEA, partly because differential expression of the same genes can contribute to the enrichment of multiple gene sets (e.g., CCNB1 was found in 20 gene sets upregulated in hF-VCMs). An examination of gene sets (defined by function) rather than individual genes gives a more comprehensive overview of transcriptional differences among hESC-VCMs and hF-VCMs. By applying this approach, we confirm expression changes in contractile, fatty acid metabolic genes using our ventricular-specific system. In addition, we uniquely show that hESC-VCMs expressed lower levels of cell cycle and ROS metabolic genes and higher levels of genes associated with migration. In the setting of acute myocardial infarction, ROS is implicated in tissue necrosis and reperfusion injury [38]. Therefore, mechanisms that promote ROS enzyme up-regulation would be important to promote cell survival in the context of cell therapy. We found that hESC-VCMs expressed significantly lower levels of genes involved in cell cycle progression and telomere maintenance while genes involved in cellular senescence and apoptosis are up-regulated. It should be noted that proliferation of hESC-CMs can be affected by culture conditions [39]. Migration-related genes, however, are up-regulated in hESC-VCMs. Embryonic heart development involves complex morphogenic progression to transform the linear tube to a four-chambered heart. Thus, the motile phenotype of hESC-VCMs may reflect that of early embryonic CMs and may mean better abilities to home and migrate to injured sites for transplantation therapy.
We and others have reported that hESC-CMs display immature Ca2+ transient properties [8] and exhibit other defects such as a negative force-frequency response [40] and a lack of positive inotropy upon β-adrenergic stimulation [41]. Here, we have identified molecules which are dramatically down-regulated in hESC-VCMs and which may underlie the above defects. CAMKII is critically involved in the regulation of Ca2+ homeostasis through phosphorylation of Ca2+ handling proteins such as PLN [42] and RyR [43] and also participates in force-frequency response [43]. Other down-regulated genes include FXYD1 (phospholemman) and SRL (sarcalumenin), which can regulate Ca2+ transient properties and modulate adrenergic stimulation [44,45]. In addition, sarcalumenin was among the 200 most abundant genes in hF-VCMs and hA-VCMs and was 11-fold lower in hESC-VCMs. Our group has previously shown that over-expression of calsequestrin, a Ca2+-handling protein absent in hESC-VCMs, can facilitate CM maturation [46]. The genes identified here represent new possible targets for mechanism-based maturation strategies.
One of the major applications of hESC-CM research is to transplant these in vitro generated CMs into infarcted heart to repair damaged myocardium. Laflamme et al has previously shown that hESC-CM survival is significantly lower when transplanted into infarcted rat heart compared to uninjured heart and that the addition of pro-survival factors is required to improve graft survival [47]. Understanding factors that regulate hESC-VCM survival under ischemic environment is therefore crucial for hESC-VCM transplantation therapy. Here, we show that hESC-VCMs express lower levels of cardioprotective molecules that regulate ROS metabolism, I KATP, I KNa and I KCa etc and are correspondingly more vulnerable to hypoxia/reoxygenation injury than hF-VCMs. We also confirmed by functional analysis that I KATP was depleted in hESC-VCMs compared to adult CMs. Factors that upregulate these molecules would therefore be of benefit to the use of hESC-VCMs in regenerative medicine. Another potential application for hESC-CMs is to use these cells as in vitro test beds for detecting pro-arrhythmic and/or cardiotoxic drugs [48,49]. Our hESC-VCMs express reduced levels of important channels and Ca2+ handling genes eg KCNJ2 and PLN etc, consistent with previous publications [8,17,50]. On a multicellular level, the random arrangement of CX43 and low conduction velocity reported here are also in line with our recent paper showing that the conduction pattern of hESC-VCMs cultured on 2-dimensional surfaces are immature and isotropic [51]. Successful drug testing requires that hESC-VCMs exhibit electrophysiological and survival properties similar to adult CMs in vivo. Here we demonstrate that the expression and function of specific ion channels and cardioprotective molecules are perturbed in hESC-VCMs compared to their in vivo counterparts. Although hESC-CMs may still have advantages over current cell models such as primary canine or rabbit Purkinje fibers or cell lines ectopically expressing the hERG ion channel, we urge that results of tests involving hESC-CMs be treated with caution as we recently reviewed [52].
Embryonic heart development involves the coordinate action of many TFs. To unravel these complex actions, we employed bioinformatics co-expression and promoter analyses. Our analyses suggest that genes involved in transcriptional regulation, contraction, energy metabolism and calcium homeostasis may be co-regulated on a transcriptional level. Interestingly, many genes identified in our network are already related via post-translational regulation or protein interaction, for example, CAMKII phosphorylates PLN to regulate Ca2+ homeostasis, which in turn determines contractile activity by modulating troponin/tropomyosin interaction. The co-expression of these molecules suggests that they may be commonly regulated on the mRNA (as well as protein) level. Our promoter analysis further reveals that HAND1, GATA4, NKX2-5 and TCF8 binding sites are present in the promoters of the majority of genes in the co-expression network, suggesting a regulatory relationship between these TFs and their putative target genes. We speculate that the five TFs (HAND1, GATA4, NKX2-5, PPARGC1A and TCF8) may play a role in the regulation of diverse processes important for cardiac function, however, only four of these TFs are known to be critical to heart development or function. GATA4 regulates the expression of MYH7(βMHC) [53] and acts synergistically with NKX2.5 to activate downstream targets [31]. The importance of these TFs is underscored by transgenic studies, which shows that null deletions of Gata4, Nkx2.5 or Hand1 arrest cardiac development in vivo. PPARGC1A is a co-regulator of the PPAR pathway, and is important for metabolic activity [54]. The fifth member of the group, TCF8, has not previously been associated with heart development and function. Mice null for Tcf8 had no reported heart abnormality [55]. In summary, we uncovered a transcriptional hierarchy involving 5TFs and genes important for CM development and function, and we postulate these 5 TFs may be crucial for hESC-CM maturation. Consistent with this, perinatal loss of Nkx2-5 in mice results in reduced contractile and Ca2+-handling parameters which are accompanied by decreased ion channel expression [56], and this is reminiscent of the contractile and electrophysiological defects of hESC-CMs. Agents that up-regulate these TFs may promote hESC-CM maturation in vitro.
Conclusion
Differentiation of hESC into CMs can potentially represent an unlimited cell source for disease modeling and cell based therapies. However, caution should be taken to ensure their safety by comparing these in vitro generated CMs with in vivo standards. HESC-VCMs generated using current protocols are functionally immature and are vulnerable to injury. Mechanism-based in vitro maturation strategies would be crucial to facilitate the translation of hESC-CMs into clinical applications.
Supporting Information
Sample information.
(DOCX)
The top 200 most abundant genes in hESC-VCMs, hF-VCMs and hA-VCMs. Fold changes relative to hESC are shown.
(XLSX)
GSEA results showing all gene sets which displayed A) decreased and B) increased expression in hESC-VCMs relative to hF-VCMs. FDR indicates the false discovery date and FDR<0.05 was considered significant. Size = number of genes present within gene sets.
(DOCX)
Genes co-expressed with 17 TFs. ‘17TF-merged’ indicates the number of TFs that co-expressed with any particular gene. ‘5TF-merged’ shows the number of TFs from within the core 5 TFs (ie GATA4, HAND1, NKX2.5, PPARGC1A and TCF8, highlighted in bold) that co-expressed with any particular gene. ‘% core TF’ is the proportion of TFs (out of the total 17 TFs) that belong to the core cluster of 5 TFs. For instance, LAMA4 co-expressed with 8 TFs, ie ‘17TF-merged’ is 8. It co-expressed with all 5 of the core TFs (GATA4, HAND1, NKX2.5, PPARGC1A and TCF8) ie ‘5TF-merged’ is 5. ‘% core TF’ is 5 out of 8 ie 63%. Only genes that co-expressed with 4 or 5 of the 5 core TFs are shown.
(DOCX)
Promoter analysis of GATA4, NKX2.5, HAND1 and TCF8.
(XLSX)
QPCR analysis of selected genes in hESC-VCMs and hF-VCMs. Expression was normalized to GAPDH. * p<0.05.
(TIF)
Supporting methods.
(DOCX)
Funding Statement
“This work was supported by the Research Grant Council of HKSAR (TBRS, T13-706/11), SCRMC and Faculty Cores of the University of Hong Kong, and the Science Faculty of Hong Kong Baptist University (FRG2/12-13/066). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.”
References
- 1. Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ et al. (1998) Embryonic stem cell lines derived from human blastocysts. Science 282: 1145-1147. doi: 10.1126/science.282.5391.1145. PubMed: 9804556. [DOI] [PubMed] [Google Scholar]
- 2. He JQ, Ma Y, Lee Y, Thomson JA, Kamp TJ (2003) Human embryonic stem cells develop into multiple types of cardiac myocytes: action potential characterization. Circ Res 93: 32-39. doi: 10.1161/01.RES.0000080317.92718.99. PubMed: 12791707. [DOI] [PubMed] [Google Scholar]
- 3. Xu C, Police S, Rao N, Carpenter MK (2002) Characterization and enrichment of cardiomyocytes derived from human embryonic stem cells. Circ Res 91: 501-508. doi: 10.1161/01.RES.0000035254.80718.91. PubMed: 12242268. [DOI] [PubMed] [Google Scholar]
- 4. Kehat I, Kenyagin-Karsenti D, Snir M, Segev H, Amit M et al. (2001) Human embryonic stem cells can differentiate into myocytes with structural and functional properties of cardiomyocytes. J Clin Invest 108: 407-414. doi: 10.1172/JCI12131. PubMed: 11489934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Xue T, Cho HC, Akar FG, Tsang SY, Jones SP et al. (2005) Functional integration of electrically active cardiac derivatives from genetically engineered human embryonic stem cells with quiescent recipient ventricular cardiomyocytes: insights into the development of cell-based pacemakers. Circulation 111: 11-20. doi: 10.1161/01.CIR.0000151313.18547.A2. PubMed: 15611367. [DOI] [PubMed] [Google Scholar]
- 6. Mummery C, Ward D, van den Brink CE, Bird SD, Doevendans PA et al. (2002) Cardiomyocyte differentiation of mouse and human embryonic stem cells. J Anat 200: 233-242. doi: 10.1046/j.1469-7580.2002.00031.x. PubMed: 12033727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lieu DK, Fu JD, Chiamvimonvat N, Tung KC, McNerney GP et al. (2013) Mechanism-based facilitated maturation of human pluripotent stem cell-derived cardiomyocytes. Circ Arrhythm. J Electrophysiol 6: 191-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu J, Fu JD, Siu CW, Li RA (2007) Functional sarcoplasmic reticulum for calcium handling of human embryonic stem cell-derived cardiomyocytes: insights for driven maturation. Stem Cells 25: 3038-3044. doi: 10.1634/stemcells.2007-0549. PubMed: 17872499. [DOI] [PubMed] [Google Scholar]
- 9. Lieu DK, Liu J, Siu CW, McNerney GP, Tse HF et al. (2009) Absence of transverse tubules contributes to non-uniform Ca(2+) wavefronts in mouse and human embryonic stem cell-derived cardiomyocytes. Stem Cells Dev 18: 1493-1500. doi: 10.1089/scd.2009.0052. PubMed: 19290776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Poon E, Kong CW, Li RA (2011) Human pluripotent stem cell-based approaches for myocardial repair: from the electrophysiological perspective. Mol Pharm 8: 1495-1504. doi: 10.1021/mp2002363. PubMed: 21879736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Satin J, Kehat I, Caspi O, Huber I, Arbel G et al. (2004) Mechanism of spontaneous excitability in human embryonic stem cell derived cardiomyocytes. J Physiol 559: 479-496. doi: 10.1113/jphysiol.2004.068213. PubMed: 15243138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Satin J, Itzhaki I, Rapoport S, Schroder EA, Izu L et al. (2008) Calcium Handling in Human Embryonic Stem Cell-Derived Cardiomyocytes. Stem Cells 26: 1961-1972. doi: 10.1634/stemcells.2007-0591. PubMed: 18483424. [DOI] [PubMed] [Google Scholar]
- 13. Beqqali A, Kloots J, Ward-van Oostwaard D, Mummery C, Passier R (2006) Genome-wide transcriptional profiling of human embryonic stem cells differentiating to cardiomyocytes. Stem Cells 24: 1956-1967. doi: 10.1634/stemcells.2006-0054. PubMed: 16675594. [DOI] [PubMed] [Google Scholar]
- 14. Cao F, Wagner RA, Wilson KD, Xie X, Fu JD et al. (2008) Transcriptional and functional profiling of human embryonic stem cell-derived cardiomyocytes. PLOS ONE 3: e3474. doi: 10.1371/journal.pone.0003474. PubMed: 18941512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Synnergren J, Adak S, Englund MC, Giesler TL, Noaksson K et al. (2008) Cardiomyogenic gene expression profiling of differentiating human embryonic stem cells. J Biotechnol 134: 162-170. doi: 10.1016/j.jbiotec.2007.11.011. PubMed: 18241947. [DOI] [PubMed] [Google Scholar]
- 16. Synnergren J, Améen C, Lindahl A, Olsson B, Sartipy P (2011) Expression of microRNAs and their target mRNAs in human stem cell-derived cardiomyocyte clusters and in heart tissue. Physiol Genomics 43: 581-594. doi: 10.1152/physiolgenomics.00074.2010. PubMed: 20841501. [DOI] [PubMed] [Google Scholar]
- 17. Xu XQ, Soo SY, Sun W, Zweigerdt R (2009) Global expression profile of highly enriched cardiomyocytes derived from human embryonic stem cells. Stem Cells 27: 2163-2174. doi: 10.1002/stem.166. PubMed: 19658189. [DOI] [PubMed] [Google Scholar]
- 18. Synnergren J, Ameen C, Jansson A, Sartipy P (2011) Global transcriptional profiling reveals similarities and differences between human stem cell derived cardiomyocyte clusters and heart tissue. Physiol Genomics. [DOI] [PubMed] [Google Scholar]
- 19. Fu JD, Rushing SN, Lieu DK, Chan CW, Kong CW et al. (2011) Distinct roles of microRNA-1 and -499 in ventricular specification and functional maturation of human embryonic stem cell-derived cardiomyocytes. PLOS ONE 6: e27417. doi: 10.1371/journal.pone.0027417. PubMed: 22110643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yang L, Soonpaa MH, Adler ED, Roepke TK, Kattman SJ et al. (2008) Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature 453: 524-528. doi: 10.1038/nature06894. PubMed: 18432194. [DOI] [PubMed] [Google Scholar]
- 21. Li RA, Leppo M, Miki T, Seino S, Marbán E (2000) Molecular basis of electrocardiographic ST-segment elevation. Circ Res 87: 837-839. doi: 10.1161/01.RES.87.10.837. PubMed: 11073877. [DOI] [PubMed] [Google Scholar]
- 22. Kocic I, Hirano Y, Hiraoka M (2001) Ionic basis for membrane potential changes induced by hypoosmotic stress in guinea-pig ventricular myocytes. Cardiovasc Res 51: 59-70. doi: 10.1016/S0008-6363(01)00279-6. PubMed: 11399248. [DOI] [PubMed] [Google Scholar]
- 23. Sizarov A, Ya J, de Boer BA, Lamers WH, Christoffels VM et al. (2011) Formation of the building plan of the human heart: morphogenesis, growth, and differentiation. Circulation 123: 1125-1135. doi: 10.1161/CIRCULATIONAHA.110.980607. PubMed: 21403123. [DOI] [PubMed] [Google Scholar]
- 24. Christoffels VM, Habets PE, Franco D, Campione M, de Jong F et al. (2000) Chamber formation and morphogenesis in the developing mammalian heart. Dev Biol 223: 266-278. doi: 10.1006/dbio.2000.9753. PubMed: 10882515. [DOI] [PubMed] [Google Scholar]
- 25. de Bakker JM, van Capelle FJ, Janse MJ, Tasseron S, Vermeulen JT et al. (1993) Slow conduction in the infarcted human heart. 'Zigzag' course of activation. Circulation 88: 915-926. doi: 10.1161/01.CIR.88.3.915. PubMed: 8353918. [DOI] [PubMed] [Google Scholar]
- 26. Severs NJ, Bruce AF, Dupont E, Rothery S (2008) Remodelling of gap junctions and connexin expression in diseased myocardium. Cardiovasc Res 80: 9-19. doi: 10.1093/cvr/cvn133. PubMed: 18519446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang J, Chen A, Lieu DK, Karakikes I, Chen G et al. (2013) Effect of engineered anisotropy on the susceptibility of human pluripotent stem cell-derived ventricular cardiomyocytes to arrhythmias. Biomaterials, 34: 8878–86. PubMed: 23942210. [DOI] [PubMed] [Google Scholar]
- 28. Rust W, Balakrishnan T, Zweigerdt R (2009) Cardiomyocyte enrichment from human embryonic stem cell cultures by selection of ALCAM surface expression. Regen Med 4: 225-237. doi: 10.2217/rme.09.s7. PubMed: 19317642. [DOI] [PubMed] [Google Scholar]
- 29. Coles JG, Boscarino C, Takahashi M, Grant D, Chang A et al. (2005) Cardioprotective stress response in the human fetal heart. J Thorac Cardiovasc Surg 129: 1128-1136. doi: 10.1016/j.jtcvs.2004.11.055. PubMed: 15867790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mio Y, Bienengraeber MW, Marinovic J, Gutterman DD, Rakic M et al. (2008) Age-related attenuation of isoflurane preconditioning in human atrial cardiomyocytes: roles for mitochondrial respiration and sarcolemmal adenosine triphosphate-sensitive potassium channel activity. Anesthesiology 108: 612-620. doi: 10.1097/ALN.0b013e318167af2d. PubMed: 18362592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Durocher D, Charron F, Warren R, Schwartz RJ, Nemer M (1997) The cardiac transcription factors Nkx2-5 and GATA-4 are mutual cofactors. EMBO J 16: 5687-5696. doi: 10.1093/emboj/16.18.5687. PubMed: 9312027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Singh MK, Christoffels VM, Dias JM, Trowe MO, Petry M et al. (2005) Tbx20 is essential for cardiac chamber differentiation and repression of Tbx2. Development 132: 2697-2707. doi: 10.1242/dev.01854. PubMed: 15901664. [DOI] [PubMed] [Google Scholar]
- 33. Cai CL, Zhou W, Yang L, Bu L, Qyang Y et al. (2005) T-box genes coordinate regional rates of proliferation and regional specification during cardiogenesis. Development 132: 2475-2487. doi: 10.1242/dev.01832. PubMed: 15843407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Christoffels VM, Hoogaars WM, Tessari A, Clout DE, Moorman AF et al. (2004) T-box transcription factor Tbx2 represses differentiation and formation of the cardiac chambers. Dev Dyn 229: 763-770. doi: 10.1002/dvdy.10487. PubMed: 15042700. [DOI] [PubMed] [Google Scholar]
- 35. Hoogaars WM, Engel A, Brons JF, Verkerk AO, de Lange FJ et al. (2007) Tbx3 controls the sinoatrial node gene program and imposes pacemaker function on the atria. Genes Dev 21: 1098-1112. doi: 10.1101/gad.416007. PubMed: 17473172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. De Smet R, Marchal K (2010) Advantages and limitations of current network inference methods. Nat Rev Microbiol 8: 717-729. PubMed: 20805835. [DOI] [PubMed] [Google Scholar]
- 37. Kehat I, Gepstein A, Spira A, Itskovitz-Eldor J, Gepstein L (2002) High-resolution electrophysiological assessment of human embryonic stem cell-derived cardiomyocytes: a novel in vitro model for the study of conduction. Circ Res 91: 659-661. doi: 10.1161/01.RES.0000039084.30342.9B. PubMed: 12386141. [DOI] [PubMed] [Google Scholar]
- 38. Ferrari R, Ceconi C, Curello S, Alfieri O, Visioli O (1993) Myocardial damage during ischaemia and reperfusion. Eur Heart J 14 Suppl G: 25-30. doi: 10.1093/eurheartj/14.suppl_G.25. PubMed: 8287865. [DOI] [PubMed] [Google Scholar]
- 39. Snir M, Kehat I, Gepstein A, Coleman R, Itskovitz-Eldor J et al. (2003) Assessment of the ultrastructural and proliferative properties of human embryonic stem cell-derived cardiomyocytes. Am J Physiol Heart Circ Physiol 285: H2355-H2363. PubMed: 14613910. [DOI] [PubMed] [Google Scholar]
- 40. Binah O, Dolnikov K, Sadan O, Shilkrut M, Zeevi-Levin N et al. (2007) Functional and developmental properties of human embryonic stem cells-derived cardiomyocytes. J Electrocardiol 40: S192-S196. doi: 10.1016/j.jelectrocard.2007.05.035. PubMed: 17993321. [DOI] [PubMed] [Google Scholar]
- 41. Pillekamp F, Haustein M, Khalil M, Emmelheinz M, Nazzal R et al. (2012) Contractile properties of early human embryonic stem cell-derived cardiomyocytes: beta-adrenergic stimulation induces positive chronotropy and lusitropy but not inotropy. Stem Cells Dev 21: 2111-2121. doi: 10.1089/scd.2011.0312. PubMed: 22268955. [DOI] [PubMed] [Google Scholar]
- 42. Iwasa T, Inoue N, Miyamoto E (1985) Identification of a calmodulin-dependent protein kinase in the cardiac cytosol, which phosphorylates phospholamban in the sarcoplasmic reticulum. J Biochem 98: 577-580. PubMed: 4066656. [DOI] [PubMed] [Google Scholar]
- 43. Kushnir A, Shan J, Betzenhauser MJ, Reiken S, Marks AR (2010) Role of CaMKIIdelta phosphorylation of the cardiac ryanodine receptor in the force frequency relationship and heart failure. Proc Natl Acad Sci U S A 107: 10274-10279. doi: 10.1073/pnas.1005843107. PubMed: 20479242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yoshida M, Minamisawa S, Shimura M, Komazaki S, Kume H et al. (2005) Impaired Ca2+ store functions in skeletal and cardiac muscle cells from sarcalumenin-deficient mice. J Biol Chem 280: 3500-3506. PubMed: 15569689. [DOI] [PubMed] [Google Scholar]
- 45. Despa S, Bossuyt J, Han F, Ginsburg KS, Jia LG et al. (2005) Phospholemman-phosphorylation mediates the beta-adrenergic effects on Na/K pump function in cardiac myocytes. Circ Res 97: 252-259. doi: 10.1161/01.RES.0000176532.97731.e5. PubMed: 16002746. [DOI] [PubMed] [Google Scholar]
- 46. Liu J, Lieu DK, Siu CW, Fu JD, Tse HF et al. (2009) Facilitated maturation of Ca2+ handling properties of human embryonic stem cell-derived cardiomyocytes by calsequestrin expression. Am J Physiol Cell Physiol 297: C152-C159. doi: 10.1152/ajpcell.00060.2009. PubMed: 19357236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Laflamme MA, Chen KY, Naumova AV, Muskheli V, Fugate JA et al. (2007) Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat Biotechnol 25: 1015-1024. doi: 10.1038/nbt1327. PubMed: 17721512. [DOI] [PubMed] [Google Scholar]
- 48. Braam SR, Tertoolen L, van de Stolpe A, Meyer T, Passier R et al. (2010) Prediction of drug-induced cardiotoxicity using human embryonic stem cell-derived cardiomyocytes. Stem. Cell Res 4: 107-116. [DOI] [PubMed] [Google Scholar]
- 49. Caspi O, Itzhaki I, Kehat I, Gepstein A, Arbel G et al. (2009) In vitro electrophysiological drug testing using human embryonic stem cell derived cardiomyocytes. Stem Cells Dev 18: 161-172. doi: 10.1089/scd.2007.0280. PubMed: 18510453. [DOI] [PubMed] [Google Scholar]
- 50. Sartiani L, Bettiol E, Stillitano F, Mugelli A, Cerbai E et al. (2007) Developmental changes in cardiomyocytes differentiated from human embryonic stem cells: a molecular and electrophysiological approach. Stem Cells 25: 1136-1144. doi: 10.1634/stemcells.2006-0466. PubMed: 17255522. [DOI] [PubMed] [Google Scholar]
- 51. Wang J, Chen A, Lieu DK, Karakikes I, Chen G et al. (2013) Effect of engineered anisotropy on the susceptibility of human pluripotent stem cell-derived ventricular cardiomyocytes to arrhythmias. Biomaterials 34: 8878-8886. doi: 10.1016/j.biomaterials.2013.07.039. PubMed: 23942210. [DOI] [PubMed] [Google Scholar]
- 52. Lieu DK, Turnbull I, Costa KD, Li RA (2013) Engineered human pluripotent stem cell-derived cardiomyocyte tissues for electrophysiological studies of cardiac disease models. Drug Discov Today Dis Models. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Charron F, Paradis P, Bronchain O, Nemer G, Nemer M (1999) Cooperative interaction between GATA-4 and GATA-6 regulates myocardial gene expression. Mol Cell Biol 19: 4355-4365. PubMed: 10330176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Arany Z, He H, Lin J, Hoyer K, Handschin C et al. (2005) Transcriptional coactivator PGC-1 alpha controls the energy state and contractile function of cardiac muscle. Cell Metab 1: 259-271. doi: 10.1016/j.cmet.2005.03.002. PubMed: 16054070. [DOI] [PubMed] [Google Scholar]
- 55. Liu Y, Peng X, Tan J, Darling DS, Kaplan HJ et al. (2008) Zeb1 mutant mice as a model of posterior corneal dystrophy. Invest Ophthalmol Vis Sci 49: 1843-1849. doi: 10.1167/iovs.07-0789. PubMed: 18436818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Briggs LE, Takeda M, Cuadra AE, Wakimoto H, Marks MH et al. (2008) Perinatal loss of Nkx2-5 results in rapid conduction and contraction defects. Circ Res 103: 580-590. doi: 10.1161/CIRCRESAHA.108.171835. PubMed: 18689573. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sample information.
(DOCX)
The top 200 most abundant genes in hESC-VCMs, hF-VCMs and hA-VCMs. Fold changes relative to hESC are shown.
(XLSX)
GSEA results showing all gene sets which displayed A) decreased and B) increased expression in hESC-VCMs relative to hF-VCMs. FDR indicates the false discovery date and FDR<0.05 was considered significant. Size = number of genes present within gene sets.
(DOCX)
Genes co-expressed with 17 TFs. ‘17TF-merged’ indicates the number of TFs that co-expressed with any particular gene. ‘5TF-merged’ shows the number of TFs from within the core 5 TFs (ie GATA4, HAND1, NKX2.5, PPARGC1A and TCF8, highlighted in bold) that co-expressed with any particular gene. ‘% core TF’ is the proportion of TFs (out of the total 17 TFs) that belong to the core cluster of 5 TFs. For instance, LAMA4 co-expressed with 8 TFs, ie ‘17TF-merged’ is 8. It co-expressed with all 5 of the core TFs (GATA4, HAND1, NKX2.5, PPARGC1A and TCF8) ie ‘5TF-merged’ is 5. ‘% core TF’ is 5 out of 8 ie 63%. Only genes that co-expressed with 4 or 5 of the 5 core TFs are shown.
(DOCX)
Promoter analysis of GATA4, NKX2.5, HAND1 and TCF8.
(XLSX)
QPCR analysis of selected genes in hESC-VCMs and hF-VCMs. Expression was normalized to GAPDH. * p<0.05.
(TIF)
Supporting methods.
(DOCX)