Abstract
There is a need to identify effective biomarkers for diagnosis, prognosis and prediction of treatment efficacy for many liver diseases such as hepatocellular cancer, and chronic viral hepatitis. The identification of disease-specific alterations in microRNA expression and the ability to detect microRNAs in the circulation provide the basis for identifying novel clinically effective treatments and biomarkers. Knowledge regarding miRNA in human liver disease may eventually lead to serum or tissue biomarkers with clinical utility. A selection of relevant studies is reviewed. There are major challenges that need to be addressed prior to clinical application such as the need for careful validation of diagnostic miRNA candidates in well described clinical cohorts, and technical issues such as quantitation and standardization of assays. The rapid progress in therapeutic interventions using miRNA based strategies for chronic hepatitis C and hepatocellular cancer provide optimism for novel approaches that will build on the existing and emerging knowledge regarding miRNA in liver diseases.
Keywords: hepatocellular cancer, hepatitis, non-coding RNA, hepatobiliary diseases
INTRODUCTION
MicroRNAs (miRNA) are small non-coding RNA genes that have generated much interest over the past decade. Expression profiling studies have identified that the tissue expression of miRNA can be differentially regulated in human liver diseases and in diverse pathophysiological settings affecting the liver. miRNA can be quantitated in the circulation, and their detection in the circulation and in tissues has potential application as specific markers of liver disease. In this overview, we will discuss current information and relevant concepts regarding the use of these non-coding RNA genes as circulating diagnostic markers and as therapeutic targets. There is a particular need for new biomarkers for acute hepatic injury, and for hepatobiliary cancers because the current markers are insensitive. Therefore, the identification of circulating miRNA as biomarkers for human liver diseases is of clinical and scientific interest.
BIOGENESIS AND FUNCTION OF MICRORNA
miRNA can function as post-transcriptional regulators of gene expression. There is a broad range of potential targets, with some estimates indicating upto 60% of the protein-coding genes in humans, as potential conserved targets of miRNAs 1. As a consequence, miRNAs are involved in many fundamental processes such as development, cell proliferation, cell death, and differentiation 2. Functionally, miRNA can modulate gene expression through translational repression or cleavage of mRNA mediated by recognization of complementary sequences within the 3′-untranslated region of target mRNAs 3. Other reported mechanisms include binding to the open reading frame or the 5′UTR of the target mRNAs or directly to the DNA 4.
Biogenesis of miRNA occurs through a multi-step process. The primary miRNA transcript, pri-miRNA is transcribed by RNA polymerase II or III followed by the modification of capping and polyadenylation in the nucleus 5, 6. The primary transcript is then cleaved into smaller segments by the ribonucleases Drosha and DGCR8 to produce a hairpin precursor (pre-miRNA) 7–9. The pre-miRNA is exported to the cytoplasm and further processed by another ribonuclease Dicer to form a duplex of mature miRNA 10, 11. After strand separation, one of the two strands (the guide strand) is loaded onto the RNA-induced silencing complex for the target gene recognition, whereas the passenger strand is degraded.
Aberrant miRNA expression profiles have been reported in many human diseases. In particular, a large proportion of miRNAs that are deregulated in human cancers map to cancer-associated genomic regions 12, 13. Experimentally, alteration in miRNA expression can modify cancer phenotypes 14. Therefore, miRNA have a critical role in human carcinogenesis. Indeed, miRNA can behave as either tumor suppressors or oncogenes by direct targeting or indirect regulating genes that are associated with tumorigenesis. For example, miR-29 acts as a tumor suppressor and can target cancer-associated genes such as matrix metalloproteinase-2, Bcl-2 and Mcl-1 15, 16, whereas miR-221 can act in oncogenic pathways by modulating mTOR and other cellular signaling pathways 17, 18. Similarly, deregulated miRNA expression has been reported in several other pathophysiological conditions indicating a broader role for miRNA in the pathogenesis of diseases other than cancer.
MICRORNA IN SELECTED LIVER DISEASES
The importance of microRNA in liver disease is being increasingly recognized 19. In this section we will highlight recent studies in order to provide a perspective about the role and relevance of selected miRNA in liver diseases (Table 1).
Table 1.
Selected examples of microRNA with altered expression in specific liver diseases
| Liver disease | Differentially Expressed miRNA | Reference | |
|---|---|---|---|
| Upregulated | Downregulated | ||
| Hepatitis B virus | miR-21, miR-122, miR-223 | [38, 43, 52- 56] | |
| Hepatitis C virus | miR-16, miR-21, miR-34a, miR-122, miR-155 | miR-199a, miR-199a*, miR-200a | [57–64, 81, 85] |
| Hepatocellular carcinoma | miR-21, miR-135a, miR-146a, miR- 151, miR-221, miR-222 | let-7g, miR-22 | [17, 20–28, 31–36, 82] |
| Hepatitis B Virus related Hepatocellular cancer | miR-21, miR-192, miR-801 | miR-26a, miR-27a, miR- 29c, miR-122 | [16, 29, 30, 37–39] |
| Hepatitis C Virus related Hepatocellular cancer | miR-192, miR-194, miR-618 | miR-320, miR-491, miR- 650 | [65, 66] |
| Cholangiocarcinoma | miR-21, miR-25, miR-31, miR-223, miR-421 | miR-122, miR-145, miR- 200c, miR-221, miR- 222, miR-370, miR-373, miR-494 | [22, 40–51] |
| Alcoholic liver disease | miR-122, miR-155, miR-217 | miR-126 | [54, 67–70] |
| Non-alcoholic fatty liver disease | miR-16, miR-34a, miR-122 | [60, 74–76] | |
| Drug-induced liver injury | miR-29, miR-122, miR-192 | [78, 79] | |
Hepatocellular carcinoma (HCC)
HCC is the most common primary malignancy of the liver. The expression of several miRNA is deregulated in HCC. These include miR-21, one of the most prominently expressed miRNA in many human cancers such as pancreas, breast, prostate, colon, lung and stomach 20. miR-21 expression is increased in HCC tissues compared with non-tumor tissue and is associated with tumor stage and poor prognosis in HCC patients 21. Serum miRNA-21 levels correlated with miR-21 expression in tumor tissue and were reported to be higher in patients with HCC than in patients with chronic hepatitis or healthy individuals in one study 22. Moreover, serum miRNA-21 expression was an independent significant factor for recurrence, and reported to be more sensitive than α-fetoprotein for detection of HCC 23. However, the potential impact of underlying necro-inflammatory changes and ongoing hepatic injury to these changes needs to be considered.
Several other miRNA such as miR-221/222 and miR-151 are increased in expression in HCC 24. miR-221/222 can target the CDK inhibitor, p27, and enhance cell growth in vitro. Moreover, miR-221 can contribute to hepatocarcinogenesis by its effects on DNA damage-inducible transcript 4 (DDIT4) 17. Antisense targeting of miR-221/222 can decrease tumor growth and increase survival in orthotopic models of HCC in mice 25. Increased miR-151 in HCC could promote invasion and metastasis by reducing expression of the putative metastasis suppressor RhoGDIA 26. Another miRNA, miR-135a is increased in HCC patients with portal vein tumor thrombus and is an independent risk factor for prognosis. These effects of miR-135a could result from targeting metastasis suppressor 1 protein expression 27. miR-146a may also be involved in HCC as it is frequently altered in many cancers. A G>C polymorphism (rs2910164) polymorphism in miR-146a was an independent marker of the risk for HCC 28. However, another study reported that miR-146a was found to be down-regulated in HCC tissue compared to normal liver 22.
Similarly, there are several miRNA that are significantly decreased in HCC. Decreased expression of miR-29c occurs in HCC associated with HBV and is associated with shorter disease-free survival. miR-29 may promote apoptosis through a mitochondrial pathway that involves Mcl-1 and Bcl-2 16, 29. miR-29c may play an important role as a tumor suppressive gene in the development and progression of HBV-related HCC by targeting Tumor necrosis factor alpha-induced protein 3, a key regulator in inflammation and immunity 30.
miR-22 expression is also downregulated in HCC tissues and its expression is predictive of poor survival in HCC patients. miR-22 is down-regulated in many cancers, and can target the estrogen receptor and HDAC4 31–33. let-7g was reduced in metastatic compared to non-metastatic HCCs and low let-7g expression was predictive of poor survival. The ability of let-7g to suppress HCC metastasis may be partially due to inhibition of cell motility and colony formation by its effects on type I collagen α2 34. miR-7 may regulate cell growth and metastasis in vivo and in vitro. It is a regulator of epidermal growth factor receptor expression 35. Over-expression of miR-7 decreased growth and migration in HCC cells in vitro, and suppressed tumor growth and abolished extrahepatic metastasis in vivo. Moreover, miR-7 downregulated the PI3K/Akt pathway in clinical HCC tissues 36. These miRNA may be useful prognostic biomarkers or therapeutic targets for miR-replacement strategies in HCC patients.
Alterations in specific serum miRNA associated with HBV related HCC have been reported. Serum miRNA expression was investigated in three independent cohorts including healthy, chronic hepatitis B and HBV-related HCC. A multivariate logistic regression model identified seven miRNAs that had high accuracy in the diagnosis of HCC, especially for patients with early stage disease. miR-192, miR-21 and miR-801 were upregulated and miR-122, miR-223, miR-26a and miR-27a were downregulated in patients with HBV-related HCC compared with those in the control group 37.
Serum miR-122 is increased in HBV patients with HCC compared to healthy individuals. However, increased serum miR-122 has been reported in HBV patients either with or without HCC compared to healthy controls 38. In addition, decreased expression of miR-122 occurs in more than 70% of HCC tissue 39. These reports suggest that elevated serum miR-122 might reflect liver injury rather than the presence of underlying HCC, but not specifically for biomarker of HCC in HBV patients. It has been postulated that the increase in serum miR-122 despite a decreased tissue expression in HCC can be explained by miRNA that has leaked from liver tissues 38. Similarly, while serum miR-223 is increased in HCC patients compared to healthy individuals, there is no significant difference between HBV patients with and without HCC 38. Thus increased serum miR-223 might also reflect liver injury rather than HBV-related HCC. As exemplified by these miRNA, evaluation of miRNA for cancer diagnosis can be confounded by alterations in serum miRNA from hepatic injury. Thus, careful validation of any potential serum miRNA candidates in well described clinical cohorts is critical prior to their use for diagnosis.
Cholangiocarcinoma
Cholangiocarcinomas are malignancies arising from biliary tract epithelia. The incidence of intrahepatic cholangiocarcinomas (IH-CCA) has been noted to be increasing worldwide 40. miRNA expression profiling in cell lines and tissues has identified several miRNA such as miR-21 that are deregulated in expression in cholangiocarcinoma 41. miR-21, miR-31, and miR-223 were increased whereas miR-122, miR-145, miR-200c, miR-221, and miR-222 were decreased in cholangiocarcinomas 22. miR-21 expression can be modulated by the Arsenic resistance protein 2 (Ars2) and downstream targets include phosphatase and tension homolog deleted on chromosome 10 (PTEN) and programmed cell death 4 (PDCD4) 42, 43.
Other miRNA such as miR-421, miR-494, miR-370 and miR-373 have been studied in cholangiocarcinoma and may have potential as prognostic or therapeutic biomarkers. Expression of miR-421 is increased in cholangiocarcinoma as well similar to other cancers such as gastric and pancreatic, and can target the Farnesoid X receptor 44, 45. Increased miR-421 expression is associated with more advanced TNM staging and lymph node invasion 46. miR-25 is also increased in cholangiocarcinoma, and can target TNF-related apoptosis-inducing ligand induced apoptosis via effects on Death Receptor-4 signaling 47. miR-494 is downregulated in human cholangiocarcinoma and retards cell growth through multiple targets such as CDK6, CDK4, CCND1, CCNE2, and HDAC1 involved in the G1-S arrest 48. We have shown that inflammatory cytokines such as Interleukin-6 can modulate miR-370 49. Downregulation of miR-373 is associated with poor cellular differentiation, advanced clinical stage and shorter overall and disease-free survival in hilar cholangiocarcinomas. miR-373 can negatively regulate methyl-CpG-binding domain protein 2 50, 51.
Hepatitis B virus (HBV)
Chronic HBV infection is a risk factor for both HCC and IH-CCA 52. Recent studies have evaluated serum miRNA expression in chronic HBV infection. Serum miR-122 is increased in patients with chronic HBV compared with healthy individuals, but serum levels do not correspond to presence or absence of co-existing HCC in these patients 38, 53, 54. miR-122 accounts for about 70% of the total liver miRNA population and is highly expressed in healthy livers 55. Plasma miR-122 concentrations correlate with histological changes of hepatic injury in experimental liver injury in mice 54. Thus, elevated serum miR-122 might reflect liver injury rather than the presence of tumor. On the other hand, serum miR-122 were significantly lower in HBV patients in comparison with healthy individuals in another study. It has been suggested that miR-122 may down-regulate HBV replication and contribute to chronic HBV 55. In HBV patients, the level of miR-21 in serum was higher than healthy individuals 53. miR-21 can contribute to malignant hepatocyte proliferation, invasion and metastasis 43. The levels of miR-223 in serum of HBV patients without HCC were higher than those in HCC patients or healthy individuals 53. miR-223 may function as a tumor suppressor gene and is usually repressed in HCC 56. The increased expression of miR-223 in serum in the setting of decreased tissue expression could result from its release during tissue injury such as hepatitis.
Hepatitis C virus (HCV)
More than 170 million individuals worldwide are chronically infected with HCV and at risk of advanced liver disease and cancer. Serum miR-21 is increased in HCV patients compared to healthy controls and correlates with ALT and AST activities. Although miR-21 is increased in HCC and many other cancers, serum miR-21 expression in HCV patients with HCC is not significantly different from that in HCV patients without HCC, or without cirrhosis but is higher than in healthy individuals 57. Serum miR-21 positively correlates with hepatic fibrosis and histological activity index (HAI) 57, 58. Thus, serum miR-21 levels are more likely to reflect chronic hepatitis rather than more advanced disease or HCC, and could be a useful marker for liver injury and fibrosis in HCV patients. SMAD7 is a negative regulator of TGF-β, a critical mediator of fibrogenesis, that can be targeted by miR-21, providing a potential mechanism by which over-expression of miR-21 enahnces TGF-β signaling and increased fibrogenesis 58.
miR-122 is a highly expressed liver-specific miRNA 59. Interaction of miR-122 with the HCV genome is essential for accumulation of viral RNA. miR-122 enhances HCV replication in cultured cells and decreased levels of miR-122 would be expected to reduce HCV replication and subsequent liver damage. miR-122 levels are reduced during HCV infection and inversely correlated with fibrotic stage in HCV infected mice 58. In HCV patients, serum miR-122 is higher than in control individuals and correlates with fibrosis stage and inflammation activity but not with HCV viral load 60. Intrahepatic miR-122 does not correlate with HCV RNA levels 61. Moreover, the stage of fibrosis negatively correlated with miR-122 expression in clinical tissues. Similar to observations with other miRNA discussed above, serum miR-122 might reflect liver injury rather than HCV infection. In addition, serum levels of miR-34a and miR-16 were significantly higher than in control individuals in HCV patients 58. Altogether, these reports suggest that miRNAs might be critical biomarkers of liver injury and fibrosis in HCV patients.
Previous reports suggested that down regulation of miR-199a, miR-199a* and 200a in chronic liver injury tissue correlate with hepatocarcinogenesis and that miR-199a* is a negative regulator of HCV replication 62. On the other hand, in both the animal and clinical tissue studies, the expression of miR-199a, 199a*, 200a, and 200b correlate with advanced liver fibrosis 63. A recent report suggested that the miR-200 family regulated EMT by downregulating EMT accelerator ZEB1 and SIP1 64. These reports support that increased levels of miR-200a and miR-200b may be associated with the progression of liver fibrosis.
A clinical to examine urinary miRNA biomarkers found that urinary miR-618 was upregulated and miR-650 was downregulated in HCV patients with HCC. The combination of miR-618 and miR-650 was a predictive biomarker for the early detection of HCC among HCV patients 65. In an in vitro study, miR-192 and miR-194 were upregulated and miR-320 and miR-491 were downregulated in cultured cells by HCV infection. HCV induced miR-192 and miR-194 may be involved in promoting carcinogenesis in HCV-related HCC 66. The diagnostic utility of these observations has yet to be determined.
Alcoholic Liver Disease (ALD)
Several studies have implicated altered miRNA expression in alcoholic liver injury 67. These include miRNAs, such as miR-126, miR-155 and miR-212, have been reported to be altered in ALD. Serum miR-126 is decreased in alcohol-related HCC 68. In mammalian models of alcohol-induced liver injury, short-term ethanol exposure increases serum miR-122. It has been suggested that serum miR-122 concentrations may be more sensitive than ALT for detecting specific kinds of liver injury 54. These need to be validated in further studies. Chronic alcohol feeding in mice results in increased miR-155 in Kupffer cells. This occurs via an NF-κB dependent pathway and correlates with TNFα production 69. Another miRNA, miR-217 is increased in livers of chronically ethanol-fed mice. miR-217 promotes ethanol-induced fat accumulation through downregulating sirtuin 1, which regulates lipid metabolism by deacetylation of modified lysine residues on transcription regulators 70. Although there are sporadic reports about correlation between ALD and miRNAs, the majority of functions are still unclear. Thus, further studies of miRNAs in ALD are needed.
Biliary atresia
Biliary atresia is a destructive inflammatory obstructive cholangiopathy of infants that can involve both intrahepatic and extrahepatic bile ducts 71. miR-29 expression is increased in a murine model of biliary atresia. miR29 directly targets Igf1 and Il1RAP, which are potentially relevant to the pathogenesis of this condition 72. On the other hand, miR-30a and miR-30c are expressed specifically in cholangiocytes. In zebrafish, removal of miR-30a causes defects in bile duct formation indicating that miR-30a is necessary for biliary development 73.
Non-alcoholic fatty liver disease (NAFLD)
A role for miRNA has been postulated in the pathogenesis of NAFLD 74–76. Serum levels of miR-122, miR-34a and miR-16 are significantly higher in patients with non-alcoholic fatty liver disease than in controls, while miR-21 levels were unchanged 60. miR-122 and miR-34a levels positively correlated with disease severity from simple steatosis to steatohepatitis. Interestingly, serum levels of miR-122 and miR-34a correlated with liver enzymes levels, fibrosis stage and inflammation activity. miR-122 levels also correlated with serum lipids in NAFLD patients. Thus, serum miR-34a and miR-122 may represent novel, noninvasive biomarkers of diagnosis and histological disease severity in patients with NAFLD.
Liver transplantation
The utility of serum hepatocyte-derived miRNAs as biomarkers of hepatic injury and acute rejection after liver transplantation has been proposed. Expression of miR-122 and miR-148a in liver tissue were reduced with prolonged graft warm ischemia times and conversely elevated in patients with liver injury. In addition, the expression of miR-122 and miR-148a correlated with aminotransferase levels. These two miRNA may be an early and sensitive biomarkers of rejection and hepatic injury after liver transplantation 77.
Drug-induced liver injury
The role of miR-29 in chronic hepatic ijury was evaluated using a liver-specific miR-29 knockout mouse. Exposure to carbon tetrachloride resulted in increased fibrosis and mortality, implicating hepatic miR-29 in the hepatic response to injury 78. Using a mouse model of acute drug-induced liver injury, a set of circulating miRNAs whose levels associated with hepatocellular injuries induced by acetaminophen overdose were identified. miRNA such as miR-122 and miR-192 exhibited changes that paralleled serum aminotransferase levels and reflected histopathological changes. These exciting results illustrate the potential use of circulating miRNA as markers of drug-induced liver injury 79.
ROLE OF MIRNA IN DIAGNOSIS OF LIVER DISEASES
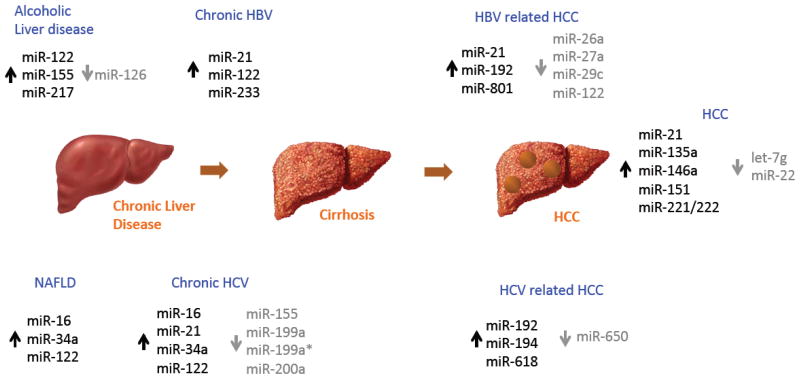
Circulating microRNA expression profiles may be promising biomarkers for diagnosis and assessment of the prognosis of cancer patients. The stability of circulating miRNA and the ability to detect miRNA in the blood has suggested the potential for miRNA-based blood biomarkers in cancer detection 23, 24. There are several potential applications of detecting levels of specific circulating miRNA, singly or in combination, ranging from diagnosis of diseases such as NAFLD or HCC, assessment of liver injury or fibrosis, detection of drug induced liver injury, monitoring of disease progression and determination of prognosis in chronic diseases or with liver cancers (Figure 1).
Figure 1. Altered microRNA expression in liver diseases.
Selected examples of miRNAs that are differentially expressed in diverse human liver diseases are illustrated. Variations in miRNA expression reported in the literature can arise from heterogeneity of the individual diseases or population studied. HCC, hepatocellular cancer, NAFLD, Non-alcoholic fatty liver disease.
Quantitative polymerase chain reaction (qPCR) is a sensitive technique for estimating expression levels of circulating microRNAs. However, there is no current consensus on the reference genes for qPCR analysis of circulating microRNAs. A recent study showed that the choice of reference genes for qPCR analysis can influence the study outcomes and emphasized the need to choose a suitable reference for reliable expression data 80. miR-16 and miR-93 were suggested to be suitable reference genes for serum miRNA analysis in gastric cancer patients and healthy controls. The detection of miRNA, which may be present in small amounts, may require miRNA amplification, which may introduce a source of variation. Normalization approaches include the use of small RNA, other miRNA, spike controls or correction for plasma volumes. Standardization of these approaches is a critical issue that will need to be addressed prior to use of a miRNA for diagnostic purposes.
ROLE OF MIRNA IN THERAPY OF LIVER DISEASES
Prediction of disease response to therapy
There is a need for effective biomarkers to confirm the efficiency of clinical therapy and to help predict response rates to therapeutic approaches in liver disease. Expression of the precursor of miR-155, BIC can be helpful during the course of HCV infection and may be a useful biomarker for therapeutic efficacy during treatment of chronic HCV infection 81. 83% of peripheral blood mononuclear cells (PBMCs) were BIC-positive in patients that eliminated HCV RNA only from serum whereas the lowest expression of BIC was found in patients that eliminated HCV RNA from both serum and PBMCs. Thus, HCV RNA presence in serum and PBMCs in patients after anti-viral treatment is associated with BIC expression in PBMCs.
A G>C polymorphism (rs2910164) in miR-146a has been reported to be an independent marker of risk for HCC 28. miR-146a can decrease sensitivity of HCC cells to IFN-a through suppression of apoptosis via SMAD4. Thus miR-146a might be a predictive biomarker for therapeutic response and potential therapeutic target on IFN therapy in HCC patients 82. These findings support the potential of miRNAs as a biomarker for prediction of response of therapy in liver disease.
Improve therapeutic efficacy
In vitro studies, cellular expression of full-length HCV increased sensitivity to sorafenib by the miRNA-dependent modulation of Mcl-1 and apoptosis 83. Modulation of miRNA responses may therefore be useful to enhance response to chemotherapy in HCC. The involvement of miR-21 in chemoresistance in HCC cells was suggested in a recent study where miR-21 expression in HCC tissues correlated with the clinical response to therapy with IFN-α/5-FU and to survival 21. Transfection of HCC cells with pre-miR-21 decreased, whereas transfection with anti-miR-21 increased, sensitivity to IFN-α and 5-FU. Effect of miR-21 on chemoresistance could be mediated through modulation of cell death pathways involving miR-21 targets such as PTEN and PDCD4. These data suggest that miR-21 may be a potential marker for therapeutic response to IFN-α/5-FU combination therapy. Another approach to modulating therapeutic efficacy exploits miRNA targeting of drug efflux pumps responsible for drug resistance such as Adenosine triphosphate binding cassette (ABC) transporters. In a bioinformatics study, 13 miRNAs were detected that could target five ABC genes. Increased ABC transporters in HCC were correlated with downregulation of these miRNAs. Thus, miRNA-based strategies can be developed to increase sensitivity to therapy or reduce drug resistance during treatment of liver cancers 84.
miRNA based therapeutics
Recognition of deregulated expression of miRNA in specific liver diseases suggests the potential for innovative therapies based on replacing or augmenting miRNA expression. In view of the requirement of miR-122 for HCV replication, therapeutic strategies targeting miR-122 have been developed. miR-122 might be relatively easy to therapeutic target because antisense oligonucleotides can be delivered to the liver by intravenous injection. Treatment of chimpanzees with chronic HCV using a locked nucleic acid–modified oligonucleotide (SPC3649) complementary to miR-122 resulted in long-lasting suppression of HCV, de-repression of target mRNAs with miR-122 seed sites, down-regulation of interferon-regulated genes, and improvement of HCV-induced liver pathology 85. The reduced viral load of HCV in chimpanzees by SPC3649 suggests that this approach might have therapeutic potential in humans. However, hepatic miR-122 expression was inversely correlated with the severity of functional and histopathological liver damage. Beneficial results have been reported in phase I studies and further studies are ongoing to evaluate this novel therapeutic approach. Meanwhile, recent pre-clinical studies have evaluated antisense oligonucleotides as therapies for HCC with promising results with strategies targeting miR-221/222 using chemically modified antisense oligonucleotides 25.
CONCLUSIONS
A major challenge for many liver diseases is identifying clinically effective treatments and biomarkers for the diagnosis, prognosis, and treatment efficacy. Knowledge regarding miRNA in human liver disease may eventually lead to serum or tissue biomarkers with clinical utility. Prior to clinical application, there are major challenges such as the need for careful validation of diagnostic miRNA candidates in well annotated clinical studies, as well as technical issues such as quantitation, standardization and normalization of expression. The rapid progress in therapeutic interventions using miRNA based strategies for liver diseases such as HCV and HCC allow optimism for more novel approaches that will build on the existing and emerging knowledge regarding miRNA in liver diseases.
Highlights.
Disease-specific alterations in microRNA expression have been reported for many liver diseases.
microRNAs in serum or tissue may be useful clinical biomarkers for disease.
Recent studies regarding microRNA in liver diseases are reviewed and summarized.
Disease specific microRNA alterations can provide the basis for targeting these for therapy.
Acknowledgments
This work was supported in part by the National Institutes of Health grant DK069370. We apologize to the many contributors to the field whose work could not be cited due to space restrictions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mrnas are conserved targets of micrornas. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sayed D, Abdellatif M. Micrornas in development and disease. Physiol Rev. 2011;91:827–87. doi: 10.1152/physrev.00006.2010. [DOI] [PubMed] [Google Scholar]
- 3.Shukla GC, Singh J, Barik S. Micrornas: Processing, maturation, target recognition and regulatory functions. Mol Cell Pharmacol. 2011;3:83–92. [PMC free article] [PubMed] [Google Scholar]
- 4.Garzon R, Marcucci G, Croce CM. Targeting micrornas in cancer: Rationale, strategies and challenges. Nat Rev Drug Discov. 2010;9:775–89. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borchert GM, Lanier W, Davidson BL. Rna polymerase iii transcribes human micrornas. Nat Struct Mol Biol. 2006;13:1097–101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 6.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, et al. Microrna genes are transcribed by rna polymerase ii. EMBO J. 2004;23:4051–60. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The drosha-dgcr8 complex in primary microrna processing. Genes Dev. 2004;18:3016–27. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Landthaler M, Yalcin A, Tuschl T. The human digeorge syndrome critical region gene 8 and its d. Melanogaster homolog are required for mirna biogenesis. Curr Biol. 2004;14:2162–7. doi: 10.1016/j.cub.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 9.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, et al. The nuclear rnase iii drosha initiates microrna processing. Nature. 2003;425:415–9. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 10.Bohnsack MT, Czaplinski K, Gorlich D. Exportin 5 is a rangtp-dependent dsrna-binding protein that mediates nuclear export of pre-mirnas. RNA. 2004;10:185–91. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hammond SM, Bernstein E, Beach D, Hannon GJ. An rna-directed nuclease mediates post-transcriptional gene silencing in drosophila cells. Nature. 2000;404:293–6. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 12.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, et al. Human microrna genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kunej T, Godnic I, Ferdin J, Horvat S, Dovc P, Calin GA. Epigenetic regulation of micrornas in cancer: An integrated review of literature. Mutat Res. 2011;717:77–84. doi: 10.1016/j.mrfmmm.2011.03.008. [DOI] [PubMed] [Google Scholar]
- 14.Lujambio A, Lowe SW. The microcosmos of cancer. Nature. 2012;482:347–55. doi: 10.1038/nature10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fang JH, Zhou HC, Zeng C, Yang J, Liu Y, Huang X, et al. Microrna-29b suppresses tumor angiogenesis, invasion, and metastasis by regulating matrix metalloproteinase 2 expression. Hepatology. 2011;54:1729–40. doi: 10.1002/hep.24577. [DOI] [PubMed] [Google Scholar]
- 16.Xiong Y, Fang JH, Yun JP, Yang J, Zhang Y, Jia WH, et al. Effects of microrna-29 on apoptosis, tumorigenicity, and prognosis of hepatocellular carcinoma. Hepatology. 2010;51:836–45. doi: 10.1002/hep.23380. [DOI] [PubMed] [Google Scholar]
- 17.Pineau P, Volinia S, McJunkin K, Marchio A, Battiston C, Terris B, et al. Mir-221 overexpression contributes to liver tumorigenesis. Proc Natl Acad Sci U S A. 2010;107:264–9. doi: 10.1073/pnas.0907904107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quintavalle C, Garofalo M, Zanca C, Romano G, Iaboni M, del Basso De Caro M, et al. Mir-221/222 overexpession in human glioblastoma increases invasiveness by targeting the protein phosphate ptpmu. Oncogene. 2012;31:858–68. doi: 10.1038/onc.2011.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haybaeck J, Zeller N, Heikenwalder M. The parallel universe: Micrornas and their role in chronic hepatitis, liver tissue damage and hepatocarcinogenesis. Swiss Med Wkly. 2011;141:w13287. doi: 10.4414/smw.2011.13287. [DOI] [PubMed] [Google Scholar]
- 20.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microrna expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tomimaru Y, Eguchi H, Nagano H, Wada H, Tomokuni A, Kobayashi S, et al. Microrna-21 induces resistance to the anti-tumour effect of interferon-alpha/5-fluorouracil in hepatocellular carcinoma cells. Br J Cancer. 2010;103:1617–26. doi: 10.1038/sj.bjc.6605958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karakatsanis A, Papaconstantinou I, Gazouli M, Lyberopoulou A, Polymeneas G, Voros D. Expression of micrornas, mir-21, mir-31, mir-122, mir-145, mir-146a, mir-200c, mir-221, mir-222, and mir-223 in patients with hepatocellular carcinoma or intrahepatic cholangiocarcinoma and its prognostic significance. Mol Carcinog. 2011 doi: 10.1002/mc.21864. [DOI] [PubMed] [Google Scholar]
- 23.Tomimaru Y, Eguchi H, Nagano H, Wada H, Kobayashi S, Marubashi S, et al. Circulating microrna-21 as a novel biomarker for hepatocellular carcinoma. J Hepatol. 2012;56:167–75. doi: 10.1016/j.jhep.2011.04.026. [DOI] [PubMed] [Google Scholar]
- 24.Li W, Xie L, He X, Li J, Tu K, Wei L, et al. Diagnostic and prognostic implications of micrornas in human hepatocellular carcinoma. Int J Cancer. 2008;123:1616–22. doi: 10.1002/ijc.23693. [DOI] [PubMed] [Google Scholar]
- 25.Park JK, Kogure T, Nuovo GJ, Jiang J, He L, Kim JH, et al. Mir-221 silencing blocks hepatocellular carcinoma and promotes survival. Cancer Res. 2011;71:7608–16. doi: 10.1158/0008-5472.CAN-11-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ding J, Huang S, Wu S, Zhao Y, Liang L, Yan M, et al. Gain of mir-151 on chromosome 8q24.3 facilitates tumour cell migration and spreading through downregulating rhogdia. Nat Cell Biol. 2010;12:390–9. doi: 10.1038/ncb2039. [DOI] [PubMed] [Google Scholar]
- 27.Liu S, Guo W, Shi J, Li N, Yu X, Xue J, et al. Microrna-135a contributes to the development of portal vein tumor thrombus by promoting metastasis in hepatocellular carcinoma. J Hepatol. 2012;56:389–96. doi: 10.1016/j.jhep.2011.08.008. [DOI] [PubMed] [Google Scholar]
- 28.Xu T, Zhu Y, Wei QK, Yuan Y, Zhou F, Ge YY, et al. A functional polymorphism in the mir-146a gene is associated with the risk for hepatocellular carcinoma. Carcinogenesis. 2008;29:2126–31. doi: 10.1093/carcin/bgn195. [DOI] [PubMed] [Google Scholar]
- 29.Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, et al. Microrna-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3a and 3b. Proc Natl Acad Sci U S A. 2007;104:15805–10. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang CM, Wang Y, Fan CG, Xu FF, Sun WS, Liu YG, et al. Mir-29c targets tnfaip3, inhibits cell proliferation and induces apoptosis in hepatitis b virus-related hepatocellular carcinoma. Biochem Biophys Res Commun. 2011;411:586–92. doi: 10.1016/j.bbrc.2011.06.191. [DOI] [PubMed] [Google Scholar]
- 31.Pandey DP, Picard D. Mir-22 inhibits estrogen signaling by directly targeting the estrogen receptor alpha mrna. Mol Cell Biol. 2009;29:3783–90. doi: 10.1128/MCB.01875-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiong J, Yu D, Wei N, Fu H, Cai T, Huang Y, et al. An estrogen receptor alpha suppressor, microrna-22, is downregulated in estrogen receptor alpha-positive human breast cancer cell lines and clinical samples. FEBS J. 2010;277:1684–94. doi: 10.1111/j.1742-4658.2010.07594.x. [DOI] [PubMed] [Google Scholar]
- 33.Zhang J, Yang Y, Yang T, Liu Y, Li A, Fu S, et al. Microrna-22, downregulated in hepatocellular carcinoma and correlated with prognosis, suppresses cell proliferation and tumourigenicity. Br J Cancer. 2010;103:1215–20. doi: 10.1038/sj.bjc.6605895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ji J, Zhao L, Budhu A, Forgues M, Jia HL, Qin LX, et al. Let-7g targets collagen type i alpha2 and inhibits cell migration in hepatocellular carcinoma. J Hepatol. 2010;52:690–7. doi: 10.1016/j.jhep.2009.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Webster RJ, Giles KM, Price KJ, Zhang PM, Mattick JS, Leedman PJ. Regulation of epidermal growth factor receptor signaling in human cancer cells by microrna-7. J Biol Chem. 2009;284:5731–41. doi: 10.1074/jbc.M804280200. [DOI] [PubMed] [Google Scholar]
- 36.Fang YX, Xue JL, Shen Q, Chen J, Tian L. Mir-7 inhibits tumor growth and metastasis by targeting the pi3k/akt pathway in hepatocellular carcinoma. Hepatology. 2012 doi: 10.1002/hep.25576. [DOI] [PubMed] [Google Scholar]
- 37.Zhou J, Yu L, Gao X, Hu J, Wang J, Dai Z, et al. Plasma microrna panel to diagnose hepatitis b virus-related hepatocellular carcinoma. J Clin Oncol. 2011;29:4781–8. doi: 10.1200/JCO.2011.38.2697. [DOI] [PubMed] [Google Scholar]
- 38.Qi P, Cheng SQ, Wang H, Li N, Chen YF, Gao CF. Serum micrornas as biomarkers for hepatocellular carcinoma in chinese patients with chronic hepatitis b virus infection. PLoS One. 2011;6:e28486. doi: 10.1371/journal.pone.0028486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gramantieri L, Ferracin M, Fornari F, Veronese A, Sabbioni S, Liu CG, et al. Cyclin g1 is a target of mir-122a, a microrna frequently down-regulated in human hepatocellular carcinoma. Cancer Res. 2007;67:6092–9. doi: 10.1158/0008-5472.CAN-06-4607. [DOI] [PubMed] [Google Scholar]
- 40.Patel T. Cholangiocarcinoma. Nat Clin Pract Gastroenterol Hepatol. 2006;3:33–42. doi: 10.1038/ncpgasthep0389. [DOI] [PubMed] [Google Scholar]
- 41.Meng F, Henson R, Lang M, Wehbe H, Maheshwari S, Mendell JT, et al. Involvement of human micro-rna in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology. 2006;130:2113–29. doi: 10.1053/j.gastro.2006.02.057. [DOI] [PubMed] [Google Scholar]
- 42.He Q, Cai L, Shuai L, Li D, Wang C, Liu Y, et al. Ars2 is overexpressed in human cholangiocarcinomas and its depletion increases pten and pdcd4 by decreasing microrna-21. Mol Carcinog. 2011 doi: 10.1002/mc.21859. [DOI] [PubMed] [Google Scholar]
- 43.Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. Microrna-21 regulates expression of the pten tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–58. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guo J, Miao Y, Xiao B, Huan R, Jiang Z, Meng D, et al. Differential expression of microrna species in human gastric cancer versus non-tumorous tissues. J Gastroenterol Hepatol. 2009;24:652–7. doi: 10.1111/j.1440-1746.2008.05666.x. [DOI] [PubMed] [Google Scholar]
- 45.Hao J, Zhang S, Zhou Y, Liu C, Hu X, Shao C. Microrna 421 suppresses dpc4/smad4 in pancreatic cancer. Biochem Biophys Res Commun. 2011;406:552–7. doi: 10.1016/j.bbrc.2011.02.086. [DOI] [PubMed] [Google Scholar]
- 46.Zhong XY, Yu JH, Zhang WG, Wang ZD, Dong Q, Tai S, et al. Microrna-421 functions as an oncogenic mirna in biliary tract cancer through down-regulating farnesoid x receptor expression. Gene. 2012;493:44–51. doi: 10.1016/j.gene.2011.11.028. [DOI] [PubMed] [Google Scholar]
- 47.Razumilava N, Bronk SF, Smoot RL, Fingas CD, Werneburg NW, Roberts LR, et al. Mir-25 targets tnf-related apoptosis inducing ligand (trail) death receptor-4 and promotes apoptosis resistance in cholangiocarcinoma. Hepatology. 2012;55:465–75. doi: 10.1002/hep.24698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Olaru AV, Ghiaur G, Yamanaka S, Luvsanjav D, An F, Popescu I, et al. Microrna down-regulated in human cholangiocarcinoma control cell cycle through multiple targets involved in the g1/s checkpoint. Hepatology. 2011;54:2089–98. doi: 10.1002/hep.24591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meng F, Wehbe-Janek H, Henson R, Smith H, Patel T. Epigenetic regulation of microrna-370 by interleukin-6 in malignant human cholangiocytes. Oncogene. 2008;27:378–86. doi: 10.1038/sj.onc.1210648. [DOI] [PubMed] [Google Scholar]
- 50.Chen Y, Gao W, Luo J, Tian R, Sun H, Zou S. Methyl-cpg binding protein mbd2 is implicated in methylation-mediated suppression of mir-373 in hilar cholangiocarcinoma. Oncol Rep. 2011;25:443–51. doi: 10.3892/or.2010.1089. [DOI] [PubMed] [Google Scholar]
- 51.Chen Y, Luo J, Tian R, Sun H, Zou S. Mir-373 negatively regulates methyl-cpg-binding domain protein 2 (mbd2) in hilar cholangiocarcinoma. Dig Dis Sci. 2011;56:1693–701. doi: 10.1007/s10620-010-1481-1. [DOI] [PubMed] [Google Scholar]
- 52.Palmer WC, Patel T. Are common factors involved in the pathogenesis of primary liver cancers? A meta-analysis of risk factors for intrahepatic cholangiocarcinoma. J Hepatol. 2012;57:69–76. doi: 10.1016/j.jhep.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu J, Wu C, Che X, Wang L, Yu D, Zhang T, et al. Circulating micrornas, mir-21, mir-122, and mir-223, in patients with hepatocellular carcinoma or chronic hepatitis. Mol Carcinog. 2011;50:136–42. doi: 10.1002/mc.20712. [DOI] [PubMed] [Google Scholar]
- 54.Zhang Y, Jia Y, Zheng R, Guo Y, Wang Y, Guo H, et al. Plasma microrna-122 as a biomarker for viral-, alcohol-, and chemical-related hepatic diseases. Clin Chem. 2010;56:1830–8. doi: 10.1373/clinchem.2010.147850. [DOI] [PubMed] [Google Scholar]
- 55.Chen Y, Shen A, Rider PJ, Yu Y, Wu K, Mu Y, et al. A liver-specific microrna binds to a highly conserved rna sequence of hepatitis b virus and negatively regulates viral gene expression and replication. FASEB J. 2011;25:4511–21. doi: 10.1096/fj.11-187781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wong QW, Lung RW, Law PT, Lai PB, Chan KY, To KF, et al. Microrna-223 is commonly repressed in hepatocellular carcinoma and potentiates expression of stathmin1. Gastroenterology. 2008;135:257–69. doi: 10.1053/j.gastro.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 57.Bihrer V, Waidmann O, Friedrich-Rust M, Forestier N, Susser S, Haupenthal J, et al. Serum microrna-21 as marker for necroinflammation in hepatitis c patients with and without hepatocellular carcinoma. PLoS One. 2011;6:e26971. doi: 10.1371/journal.pone.0026971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marquez RT, Bandyopadhyay S, Wendlandt EB, Keck K, Hoffer BA, Icardi MS, et al. Correlation between microrna expression levels and clinical parameters associated with chronic hepatitis c viral infection in humans. Lab Invest. 2010;90:1727–36. doi: 10.1038/labinvest.2010.126. [DOI] [PubMed] [Google Scholar]
- 59.Chang J, Nicolas E, Marks D, Sander C, Lerro A, Buendia MA, et al. Mir-122, a mammalian liver-specific microrna, is processed from hcr mrna and may downregulate the high affinity cationic amino acid transporter cat-1. RNA Biol. 2004;1:106–13. doi: 10.4161/rna.1.2.1066. [DOI] [PubMed] [Google Scholar]
- 60.Cermelli S, Ruggieri A, Marrero JA, Ioannou GN, Beretta L. Circulating micrornas in patients with chronic hepatitis c and non-alcoholic fatty liver disease. PLoS One. 2011;6:e23937. doi: 10.1371/journal.pone.0023937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Morita K, Taketomi A, Shirabe K, Umeda K, Kayashima H, Ninomiya M, et al. Clinical significance and potential of hepatic microrna-122 expression in hepatitis c. Liver Int. 2011;31:474–84. doi: 10.1111/j.1478-3231.2010.02433.x. [DOI] [PubMed] [Google Scholar]
- 62.Murakami Y, Aly HH, Tajima A, Inoue I, Shimotohno K. Regulation of the hepatitis c virus genome replication by mir-199a. J Hepatol. 2009;50:453–60. doi: 10.1016/j.jhep.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 63.Murakami Y, Toyoda H, Tanaka M, Kuroda M, Harada Y, Matsuda F, et al. The progression of liver fibrosis is related with overexpression of the mir-199 and 200 families. PLoS One. 2011;6:e16081. doi: 10.1371/journal.pone.0016081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, et al. The mir-200 family and mir-205 regulate epithelial to mesenchymal transition by targeting zeb1 and sip1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 65.Abdalla MA, Haj-Ahmad Y. Promising candidate urinary microrna biomarkers for the early detection of hepatocellular carcinoma among high-risk hepatitis c virus egyptian patients. J Cancer. 2012;3:19–31. doi: 10.7150/jca.3.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ishida H, Tatsumi T, Hosui A, Nawa T, Kodama T, Shimizu S, et al. Alterations in microrna expression profile in hcv-infected hepatoma cells: Involvement of mir-491 in regulation of hcv replication via the pi3 kinase/akt pathway. Biochem Biophys Res Commun. 2011;412:92–7. doi: 10.1016/j.bbrc.2011.07.049. [DOI] [PubMed] [Google Scholar]
- 67.Miranda RC, Pietrzykowski AZ, Tang Y, Sathyan P, Mayfield D, Keshavarzian A, et al. Micrornas: Master regulators of ethanol abuse and toxicity? Alcohol Clin Exp Res. 2010;34:575–87. doi: 10.1111/j.1530-0277.2009.01126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ladeiro Y, Couchy G, Balabaud C, Bioulac-Sage P, Pelletier L, Rebouissou S, et al. Microrna profiling in hepatocellular tumors is associated with clinical features and oncogene/tumor suppressor gene mutations. Hepatology. 2008;47:1955–63. doi: 10.1002/hep.22256. [DOI] [PubMed] [Google Scholar]
- 69.Bala S, Marcos M, Kodys K, Csak T, Catalano D, Mandrekar P, et al. Up-regulation of microrna-155 in macrophages contributes to increased tumor necrosis factor {alpha} (tnf{alpha}) production via increased mrna half-life in alcoholic liver disease. J Biol Chem. 2011;286:1436–44. doi: 10.1074/jbc.M110.145870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yin H, Hu M, Zhang R, Shen Z, Flatow L, You M. Microrna-217 promotes ethanol-induced fat accumulation in hepatocytes by down-regulating sirt1. J Biol Chem. 2012;287:9817–26. doi: 10.1074/jbc.M111.333534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hartley JL, Davenport M, Kelly DA. Biliary atresia. Lancet. 2009;374:1704–13. doi: 10.1016/S0140-6736(09)60946-6. [DOI] [PubMed] [Google Scholar]
- 72.Hand NJ, Horner AM, Master ZR, Boateng LA, LeGuen C, Uvaydova M, et al. Microrna profiling identifies mir-29 as a regulator of disease-associated pathways in experimental biliary atresia. J Pediatr Gastroenterol Nutr. 2012;54:186–92. doi: 10.1097/MPG.0b013e318244148b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zong Y, Stanger BZ. Molecular mechanisms of bile duct development. Int J Biochem Cell Biol. 2011;43:257–64. doi: 10.1016/j.biocel.2010.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cheung O, Puri P, Eicken C, Contos MJ, Mirshahi F, Maher JW, et al. Nonalcoholic steatohepatitis is associated with altered hepatic microrna expression. Hepatology. 2008;48:1810–20. doi: 10.1002/hep.22569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jin X, Ye YF, Chen SH, Yu CH, Liu J, Li YM. Microrna expression pattern in different stages of nonalcoholic fatty liver disease. Dig Liver Dis. 2009;41:289–97. doi: 10.1016/j.dld.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 76.Zheng L, Lv GC, Sheng J, Yang YD. Effect of mirna-10b in regulating cellular steatosis level by targeting ppar-alpha expression, a novel mechanism for the pathogenesis of nafld. J Gastroenterol Hepatol. 2010;25:156–63. doi: 10.1111/j.1440-1746.2009.05949.x. [DOI] [PubMed] [Google Scholar]
- 77.Farid WR, Pan Q, van der Meer AJ, de Ruiter PE, Ramakrishnaiah V, de Jonge J, et al. Hepatocyte-derived micrornas as serum biomarkers of hepatic injury and rejection after liver transplantation. Liver Transpl. 2012;18:290–7. doi: 10.1002/lt.22438. [DOI] [PubMed] [Google Scholar]
- 78.Kogure T, Costinean S, Yan I, Braconi C, Croce C, Patel T. Hepatic mir-29ab1 expression modulates chronic hepatic injury. J Cell Mol Med. 2012 doi: 10.1111/j.1582-4934.2012.01578.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang K, Zhang S, Marzolf B, Troisch P, Brightman A, Hu Z, et al. Circulating micrornas, potential biomarkers for drug-induced liver injury. Proc Natl Acad Sci U S A. 2009;106:4402–7. doi: 10.1073/pnas.0813371106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Song J, Bai Z, Han W, Zhang J, Meng H, Bi J, et al. Identification of suitable reference genes for qpcr analysis of serum microrna in gastric cancer patients. Dig Dis Sci. 2012;57:897–904. doi: 10.1007/s10620-011-1981-7. [DOI] [PubMed] [Google Scholar]
- 81.Sidorkiewicz M, Grek M, Jozwiak B, Majda-Stanislawska E, Piekarska A, Bartkowiak J. Expression of microrna-155 precursor in peripheral blood mononuclear cells from hepatitis c patients after antiviral treatment. Acta Virol. 2010;54:75–8. doi: 10.4149/av_2010_01_75. [DOI] [PubMed] [Google Scholar]
- 82.Tomokuni A, Eguchi H, Tomimaru Y, Wada H, Kawamoto K, Kobayashi S, et al. Mir-146a suppresses the sensitivity to interferon-alpha in hepatocellular carcinoma cells. Biochem Biophys Res Commun. 2011;414:675–80. doi: 10.1016/j.bbrc.2011.09.124. [DOI] [PubMed] [Google Scholar]
- 83.Braconi C, Valeri N, Gasparini P, Huang N, Taccioli C, Nuovo G, et al. Hepatitis c virus proteins modulate microrna expression and chemosensitivity in malignant hepatocytes. Clin Cancer Res. 2010;16:957–66. doi: 10.1158/1078-0432.CCR-09-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Borel F, Han R, Visser A, Petry H, van Deventer SJ, Jansen PL, et al. Adenosine triphosphate-binding cassette transporter genes up-regulation in untreated hepatocellular carcinoma is mediated by cellular micrornas. Hepatology. 2012;55:821–32. doi: 10.1002/hep.24682. [DOI] [PubMed] [Google Scholar]
- 85.Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, et al. Therapeutic silencing of microrna-122 in primates with chronic hepatitis c virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]