Abstract
In about half of patients infected with HIV-1 subtype B, viral populations shift from utilizing the transmembrane protein CCR5 to CXCR4, as well as or instead of CCR5, during late stage progression of the disease. How the relative adhesion efficiency and fusion competency of the viral Env proteins relate to infection during this transition is not well understood. Using a virus-cell fusion assay and live-cell single-molecule force spectroscopy, we compare the entry competency of viral clones to tensile strengths of the individual Env-receptor bonds of Env proteins obtained from a HIV-1 infected patient prior to and during coreceptor switching. The results suggest that the genetic determinants of viral entry were predominantly enriched in the C3, HR1 and CD regions rather than V3. Env proteins can better mediate entry into cells after coreceptor switch; this effective entry capacity does not correlate with the bond strengths between viral Env and cellular receptors.
HIV-1 adhesion and fusion are dynamic processes governed by the interaction of four main proteins, two on the surface of the virus and two on the cellular membrane1,2,3,4. HIV-1 utilizes the primary cellular receptor CD4 for adhesion and a secondary coreceptor to initiate fusion of the viral and cellular membranes5,6,7,8. In approximately half of untreated HIV-1 subtype B infected patients, late-stage progression of the disease correlates with a shift in the virus population strictly utilizing the transmembrane protein CCR5 as a cellular coreceptor to one using CCR5 and/or CXCR49,10,11. While the system responsible for viral entry has been well defined, correlations between surface protein adhesion and the subsequent entry of the viral particle remain unclear.
Specifically, how the relative efficiencies of adhesion and fusion ultimately relate to infection is unclear. In addition, whether adhesion or fusion correlates with infection, whether fusion and adhesion correlate with one another, and whether the emergence of clones with different strength of adhesion correlates with the observed tropism switch are all unclear. The viral proteins include gp41, a transmembrane protein spanning the viral envelope, and gp120, which non-covalently associates with gp41 on the viral surface1,2,3,4. Initially, viral adhesion and fusion begin when the viral protein gp120 binds the primary HIV-1 cellular receptor CD45. Adhesion of gp120 to CD4 initiates a conformation change in gp120, stabilizing the binding site for a second cellular coreceptor, primarily CCR5 early in infection and often including CXCR4 late in infection6,7,8. However, the driving force for this change in coreceptor use is not well understood12.
The coreceptor binding site on gp120 is flanked by highly variable regions known as variable loops. While gp120 contains a total of 5 variable loops (V1, V2, V3, V4, V5), it is well established that the sequence of V2/3 most directly affects coreceptor use or tropism. Several groups have developed algorithms to predict the coreceptor tropism of a viral clone by its V3 genetic sequence alone13,14,15. One algorithm commonly used to predict viral tropism is position-specific scoring matrices (PSSM)16. A training set of V3 sequences from viruses with known tropism is used to generate a likelihood matrix for each amino acid in the sequence. This matrix is then used to score the query sequence so that the higher the score the more likely that the unknown envelope utilizes the CXCR4 coreceptor14,17.
Recent work has elucidated the effect that specific regions within gp41 may have in viral clone tropism18,19. In particular, the HR1/HR2, transmembrane and cytoplasmic regions were shown to contribute to coreceptor use. However, the roles that these regions play in coreceptor use have yet to be well characterized. Phenotypic assays can be used to assess the tropism of viral clones by observing the ability to both adhere to and fuse with target cells18,20,21. This work suggests contributions to tropism made by non-V2/3 regions, but these assays alone cannot distinguish between viral-cellular protein adhesion and membrane fusion.
Utilizing a virus-cell fusion assay, we study the fusogenicity of primary viral envelope (env) clones chronologically isolated from peripheral blood mononuclear cells (PBMC) of a patient who underwent an R5 to R5-and-X4 tropism switch in the context of an identical viral backbone22. We then correlate the genetic distance between pairs of env with their differences in entry fitness. This metric enables us to identify pairs of env clones most closely related in sequence that are furthest apart in fusion fitness and consequently the single amino acid changes that cause the biggest change in the fusion fitness.
It has been previously shown that variants that utilize CCR5 bind more strongly to their coreceptor than CXCR4 variants23. However, this was conveyed by measuring the adhesion of CCR5 variants to cells compared to the inability to quantitatively measure the adhesion of CXCR4 variants23. Here, we circumvent these issues by using single-molecule force spectroscopy (SMFS) analysis in conjunction with a variety of Env proteins obtained from a single HIV-1 infected patient prior to and during coreceptor switching. We used this assay to directly assess the change in micromechanical properties of gp120 adhesion during coreceptor switch and assess how viral adhesion may dictate coreceptor progression.
Results
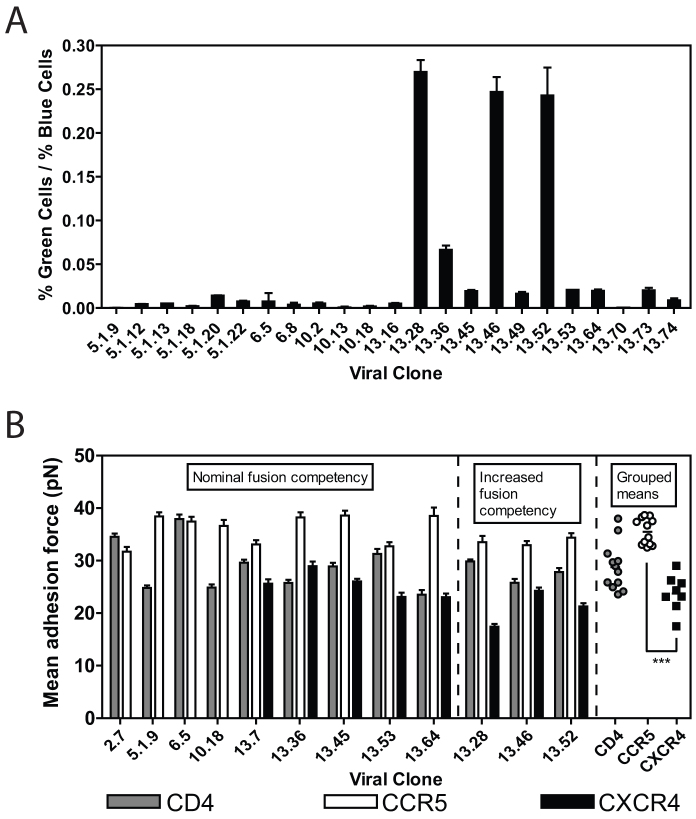
We first characterized the fusion and adhesion capacities of 23 Env proteins derived from a single HIV-1 infected subject over the period of a CCR5 to CXCR4 coreceptor switch24. Using a cell-virus entry assay, we studied the ability of these envelope proteins to mediate entry into primary CD4+ T cells isolated from a healthy donor. On average, the virus clones isolated after the switch (labeled 13.x) were better able to mediate entry compared to clones isolated before the switch (Fig. 1A). Even among the clones isolated from the last time point, there was a significant difference in the ability of the envelopes to mediate fusion.
Figure 1. Direct measurement of viral entry into the cell and viral adhesion to the cell.
(A) Percent of cells that showed successful viral fusion with their cellular membrane after treatment with pseudotyped viral particles containing β-lactamase fused to Vpr and patient-isolated Env proteins (as specified). (B) Mean adhesion force of viral particles containing isolated Env surface proteins with cell expressing CD4, CCR5 and CXCR4, as measured by single-molecule force spectroscopy.
After characterizing the entry capacity of multiple clones, we examined the adhesion capacity of eleven representative viral strains using single-molecule force spectroscopy (SMFS) including those shown to have improved capacity for cellular entry (Fig. 1B). Here we focused on three CCR5 utilizing strains that were collected prior to coreceptor switch (clones 5.1.9, 6.5 and 10.18), and eight dual-tropic strains according to previous determinations24. Dual-tropic strains utilize both CCR5 and CXCR4 and can be subdivided according to their ability to preferentially infect either CCR5 or CXCR4 expressing cells. Dual-tropic strains that are able to infect CCR5-expressing cells more effectively were labeled R5 > X4 (clone 13.7); those able to infect CCR5-expressing cells equally to those expressing CXCR4 were labeled R5 = X4 (clones 13.36, 13.45 and 13.64); and those able to infect CXCR4-expressing cells more effectively than CCR5 expressing cells were labeled X4 > R5 (clones 13.28, 13.46, 13.52 and 13.53).
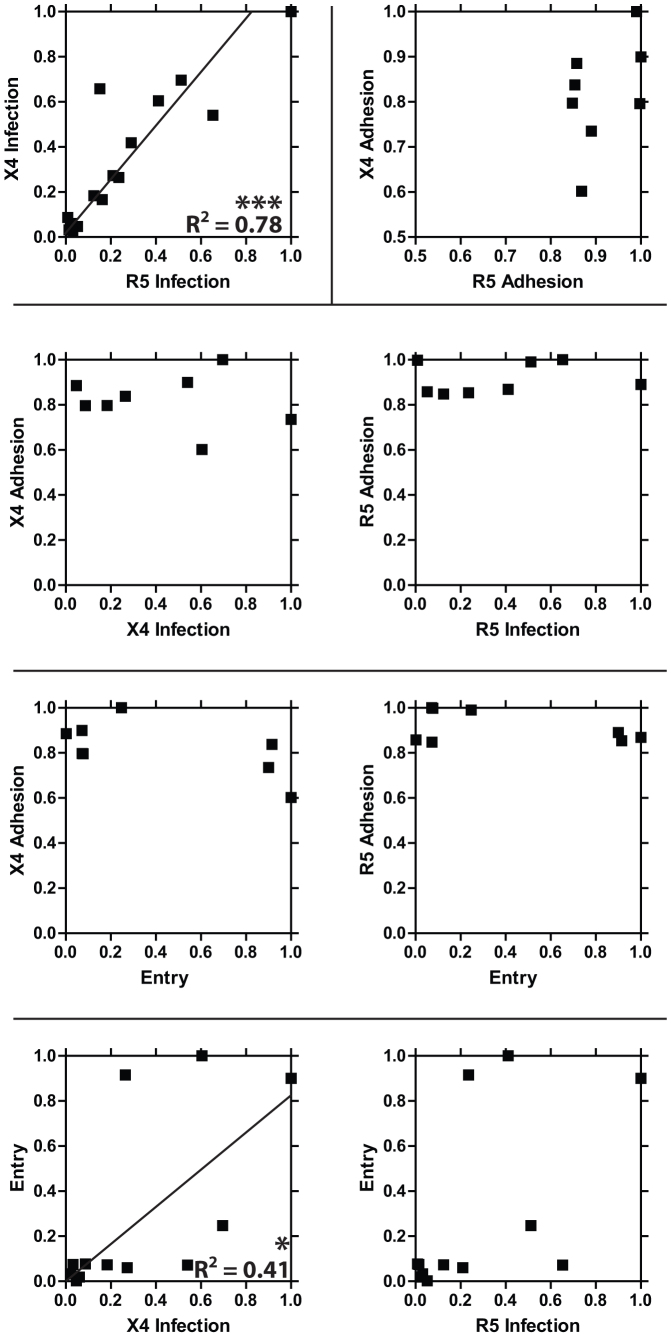
Viral entry and viral adhesion data for the clones discussed above were directly compared using correlation analysis (Fig. 2). This analysis indicates that the average strengths of the bonds between viral envelope proteins and cellular receptors do not correlate with the viral envelope proteins ability to enter the cell (i.e., mediate membrane fusion).
Figure 2. Correlation analysis of viral adhesion to the cell protein, viral entry into cells, and infection.
Iterative correlation analysis of a viral clones ability to enter cells (Entry) as measured by β-lactamase activity, ability to adhere to cellular specific coreceptors (X4 or R5 Adhesion), and ability to deliver and express viral genetic material to either CD4+/CCR5+ or CD4+/CXCR4+ cells specifically (X4 or R5 Infection). Entry data were obtained using T cells. X4 Adhesion and X4 Infection data were obtained using CD4−/CCR5−/CXCR4+ adherent GHOST cells. R5 Adhesion and R5 Infection data were obtained using CD4−/CCR5+/CXCR4− adherent GHOST cells.
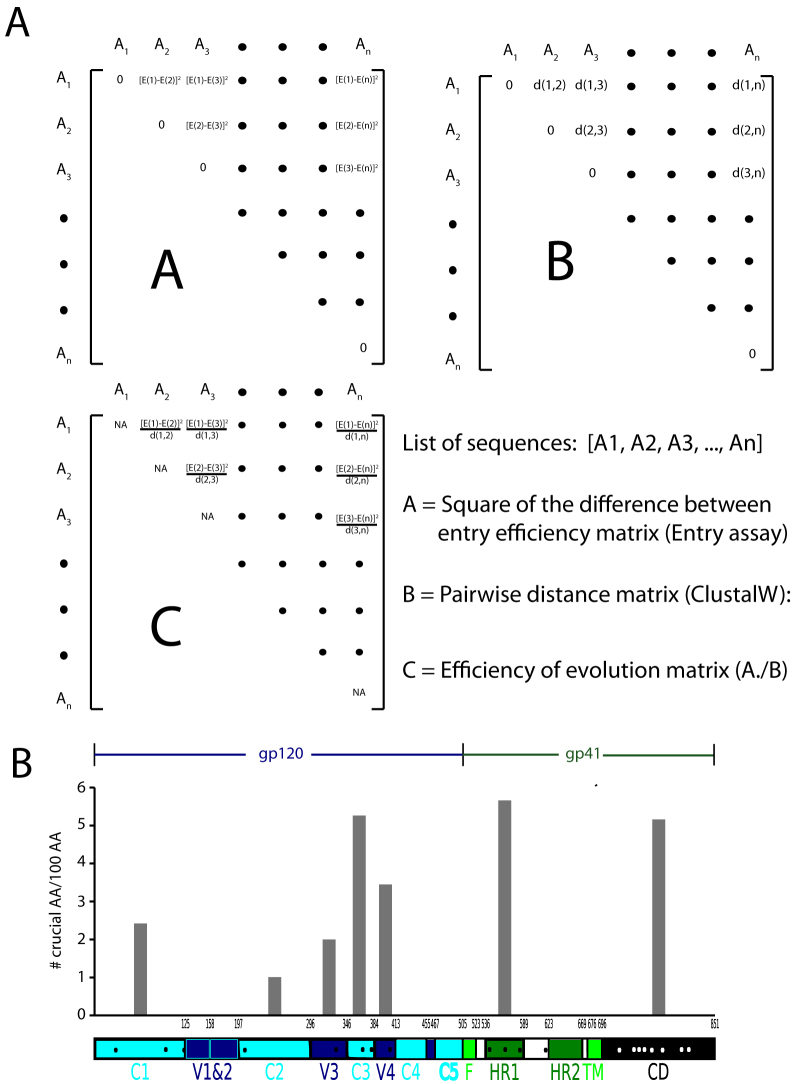
To determine changes in amino acid sequences that led to the most significant difference in the ability of envelope clones to mediate fusion, we performed a novel genetic analysis (see Methods). First, we determined the square of the difference between the percentages of entry mediated by each pair of clones. We then normalized this difference by the pairwise genetic distance obtained from the ClustalW2 software. This is a novel index indicative of the “efficiency of evolution” (Fig. 3). A high score for a pair of clones indicates that changing minimal amount of genetic sequence resulted in significant increase in the entry fitness. Therefore, the amino acid changes involved are implicated as “crucial” for increasing the efficiency of entry. Next, we ranked pairs of envelopes in the descending order on the basis of this index. By locating these amino acids among the top 10 pairs along the length of gp160, we identified the regions of the Env that include the most number of these “crucial” amino acids, normalized by the length of each region. Fig. 3B indicates that the C3, HR1 and CD domains are most enriched in these amino acids.
Figure 3. Efficiency-of-evolution matrix and the density of amino acids with the highest impact on entry fitness along the length of envelope.
(A) Constructing the efficiency of evolution matrix. The pairwise genetic distance between each pair of envelope clone is determined using the ClustalW2 algorithm, matrix B. The fusion fitness of each envelope was determined using the entry assay as described elsewhere. The elements in matrix A are the square of difference between the entry fitness of each clone. For example, the element (i,j)of matrix A is [E(i) − E(j)]2, where E(i) and E(j), are the entry fitness of clones i and j as determined by the entry assay. The elements in matrix C, are the result of pairwise division of the corresponding elements in matrix A by those in matrix B. The elements with the highest value in this matrix correspond to the pair of sequences in which smallest number of amino acid changes achieved the biggest change in the entry fitness. The top ten pairs with the highest change in entry fitness with the least number of amino acid changes were selected. The locations of the amino acids that were different between these pairs were identified along the length of Env. (B) The density of amino acids with the highest impact on entry fitness along the length of Env. The number of amino acids that confer the biggest change in entry fitness was normalized to the length of each particular region of Env.
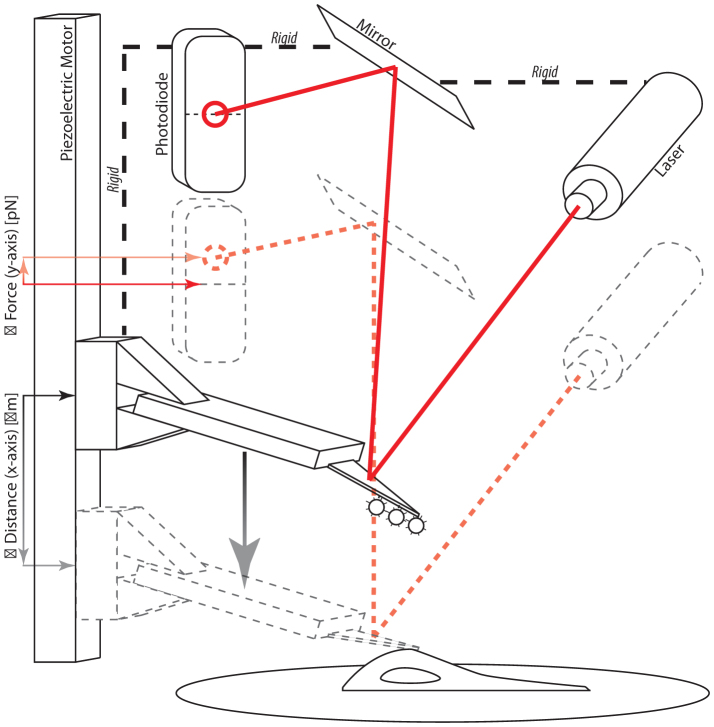
To differentiate fusion and binding strength between viral particles and living cells, we used single-molecule force spectroscopy (SMFS) to probe the strength and lifetime of molecular bonds at single-molecule resolution25. Viral particles were placed on a soft cantilever of an atomic force microscope (AFM) and placed in contact with a living cell (Fig. 4). Upon controlled retraction of the cantilever, the force-time curve was recorded to detect de-adhesion events between cell and viral particles. The slope of this curve just prior to the rupture event represents the loading rate (expressed in picoNewton per s, pN/s) to which the bimolecular bond formed during the contact between the cell and viral particle; the height of the rupture is the strength of this bond (expressed in pN). The duration of contact between cell and virus was chosen to be short (1 ms) to ensure that, for most contacts, no bond was formed between cell and virus. We verified that the distribution for successful bond formation followed Poisson statistics, implying that the probability that a single bond, two bonds, and three bonds formed during cell-virus contact in the present conditions was 85%, 12%, and 2%, respectively26.
Figure 4. Schematic of the AFM-based molecular force probe.
A flexible cantilever functionalized with whole virions is brought in contact with a living cell expressing viral surface receptors. The displacement of the cantilever is measured by laser reflection onto a photo sensitive diode and converted to force by Hooke's law.
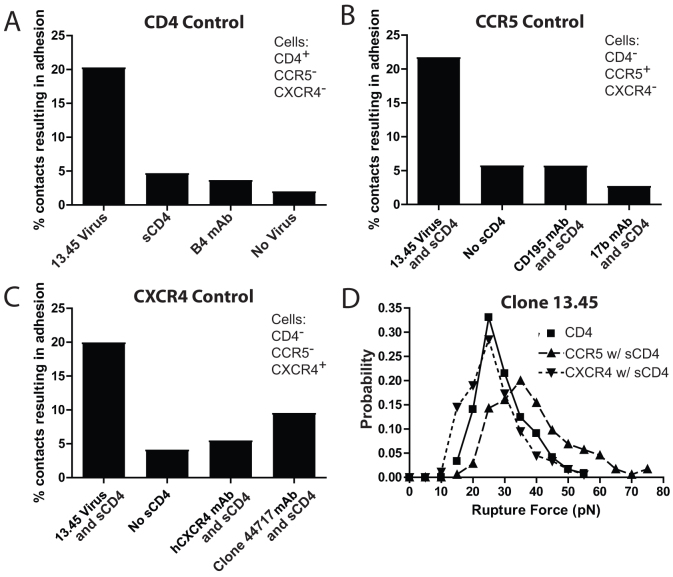
To demonstrate the specificity of the measurements obtained using the SMFS assay with patient-derived strains, we performed control experiments using function blocking antibodies or protein fragments, and monitored the resulting frequency of adhesion with the 13.45 dual-tropic enveloped virus. Background adhesion frequency was recorded and found to be low, ~2%, when no virus was placed on the flexible cantilever of the AFM (Fig. 5A). To test CD4 binding specificity, we found that the frequency of adhesion of the virus with CD4-expressing cells (~20%) was significantly higher than for experiments performed in the presence of both soluble CD4 (sCD4, ~5%) and CD4-blocking B4 monoclonal antibody (B4 mAb, ~4%) (Fig. 5). To evaluate CCR5 specificity, we found that the adhesion frequency of interaction between virus and CCR5-expressing cells in the presence of sCD4 to induce coreceptor adhesion (~21%) was much higher than in experiments performed without the addition of sCD4 (No sCD4, ~6%) (Fig. 5B). We also compared the adhesion frequency of the virus following the addition of both sCD4 and an inhibiting monoclonal antibody specific for CCR5 in the culture medium (CD195 mAb, ~6%) or sCD4 and another inhibiting monoclonal antibody specific for the CCR5 binding site of gp120 in the culture medium (17 b mAb, ~3%) (Fig. 5B). To test for CXCR4 specificity, we compared the adhesion frequency of the virus with CXCR4 expressing cells in the presence of sCD4 (~20%) with the adhesion frequency measured in the presence of sCD4 (No sCD4, ~4%) (Fig. 5C). We also compared the adhesion frequency of virus with the addition of sCD4 and a CXCR4 inhibiting monoclonal antibody (hCXCR4 mAb, ~5%) and finally with the addition of sCD4 and the gp120 binding, coreceptor inhibiting monoclonal antibody (17 b mAb, ~9%). Together, these results demonstrate that the adhesion events monitored by SMFS using whole viral particles adhering to living cells are specific.
Figure 5. Controls for single-molecule force spectroscopy analysis of patient-isolated Env binding specificity and characterization of virion-receptor interactions.
(A) Percent of contacts resulting in adhesion between virus expressing the env of the isolated 13.45 dual tropic strain with cells with CD4+/CCR5−/CXCR4− cell surfaces. (B) Percent of contacts resulting in adhesion between virus expressing the env of the isolated 13.45 dual tropic strain with CD4−CCR5+/CXCR4− cell surface in the presence of soluble CD4 (sCD4). (C) Percent of contacts resulting in adhesion between virus expressing the env of the isolated 13.45 dual tropic strain with CD4−/CCR5−/CXCR4+ cell surface in the presence of soluble CD4 (sCD4). (D) Example probability density distribution of the force required to rupture bonds (Rupture Force) between virus expressing the env of the isolated 13.45 dual tropic strain and CD4, CCR5 and CXCR4 cell surface proteins.
SMFS data was acquired using a constant cantilever retraction velocity (10 μm/s) for all tested conditions (i.e., all strains that adhered to all receptors). The magnitudes of rupture forces for individual adhesion events were used to calculate a mean adhesion force for each condition (Fig. 1B). The range of all measured mean adhesion forces for gp120-CD4 bonds was larger (~Δ18 pN from largest to smallest mean) compared to that of gp120-CCR5 and gp120-CXCR4 bonds (~Δ8 pN and ~Δ10 pN from largest to smallest mean, respectively) indicating that gp120-CD4 bonds maintained the broadest binding capacity from clone to clone of any bond examined. In addition, while gp120-CCR5 and gp120-CXCR4 bonds were similarly variable, individual CCR5 bonds were significantly stronger than individual CXCR4 bonds (~35 pN and ~24 pN, respectively) (Fig. 1B).
The magnitude of rupture forces for individual adhesion events produced probability distributions for each condition. These probability distributions acquired by SMFS were fit to a theoretical model of bonds under stress recently developed by Szabo and Hummer27. For cantilevers moved at a constant retraction velocity and of known mechanical spring constant, analysis of the probability distributions yields key parameters describing the response of bonds to force. These parameters include the micromechanical properties of the different bond types, such as molecular spring constant, km, and bond interaction distance, x‡ (the distance bonds can be pulled before rupture). Bonds formed with CCR5 were typically more rigid than bonds formed with CD4, and significantly more rigid than bonds formed with CXCR4 (Fig. 6B). In addition, bonds formed with CCR5 ruptured after a shorter distance than bonds formed with CD4 and CXCR4 (Fig. 6A), indicating that CCR5 maintain a stronger bond and are stable over a shorter working distance than CD4 and CXCR4 bonds (Fig. 6B).
Figure 6. Micromechanical characteristics of bonds between virus expressing the env of specified isolated strains with CD4, CCR5, and CXCR4 cell surface proteins.
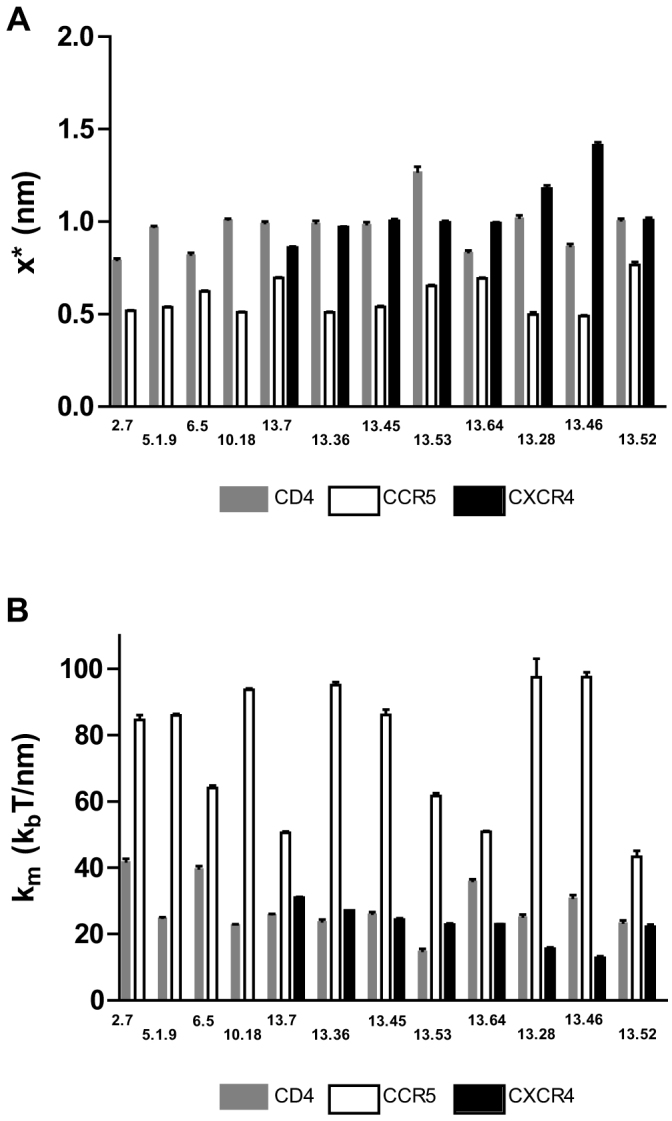
(A) x* is the distance along the free energy surface from the well minimum to the energy at bond rupture, or bond length. (B) κm is the molecular spring constant of the measured bond.
These results indicate that among the clones analyzed here the major genetic determinants of differences in fitness were enriched in predominantly the C3, HR1 and CD regions rather than V3. In addition, the average strength of viral adhesion does not correlate with viral entry and that CD4, CCR5 and CXCR4 bonds have different micromechanical properties.
Discussion
We directly compared the fusion efficacy and adhesion capacities of a variety of HIV-1 viral clones obtained from the same patient over a period of time during coreceptor switching. We assessed fusion using a previously reported28 fluorescence resonance energy transfer-(FRET) based fusion assay which is only dependent on viral fusion to produce a positive signal, unlike other assays that relay on more downstream events in viral infection. We also examined viral adhesion directly using single-molecule force spectroscopy (SMFS). Together our results compare the capacity of viral proteins to adhere to cellular receptors and the subsequent responses to adhesion, namely fusion.
Analyzing the ability of viral particles with various env clones to fusion with primary cellular isolates identify three clones with substantially higher fusion efficacy than any other clones examined (Fig. 1A). These clones (13.28, 13.46, 13.36) are all dual tropic, able to utilize both CCR5 and CXCR4 for infection. The entry of viral particles to cells is a complex phenomenon involving both the type and concentration of the receptor and coreceptor pairs present on the virus as well as the target cell. These parameters will vary from donor to donor and the nuances of this complex system are beyond the scope of this work. Interestingly, complementary analysis of genetic variability between viral clones examined here indicates a correlation between enhanced fusion capacity and a significant increase in mutations within the region of env coding the linkage region between HR1 and HR2 of gp41 for all three clones.
To distinguish between adhesion and the product of adhesion, i.e. fusion, we employed SMFS. SMFS records single bi-molecular adhesion events between viral proteins and cellular receptors directly and does not depend on reporter molecules activated by more downstream events or receptor density. Previously, adhesion has been shown to correlate with overall infection by allowing viral particles to adhere to a cellular membrane and ultimately use immunofluorescence to examine viral particle association. While providing valuable data on viral association with the cell membrane, this assay convolutes adhesion and more downstream events. These events include conformational changes in gp120 and gp41 as well as possible fusion or hemi-fusion events wherein only the outer leaflet of the viral and cellular membranes fuse. In contrast, SMFS results reveal that there was no correlation between adhesion strength and fusion efficacy for viral clones including those with an increased capacity for fusion.
Previous attempts to quantify CXCR4 vs. CCR5 adhesion have proven to be difficult. While it is generally accepted that CXCR4 adheres less efficiently than CCR5, this observation is generally made by comparing a measurable response for CCR5 adhesion to negative results for CXCR4 adhesion. Here, we offer quantified values for comparison for CXCR4 and CCR5 binding strength, with viral clones binding CXCR4 with the strength of ~60% that of CCR5 binding.
When comparing adhesion strength and fusion efficacy of varying viral clones, it is tempting to infer that viral adhesion itself is not the rate-limiting step leading to productive fusion of viral and cellular membranes. If it were the rate-limiting step, one would expect the strongest adhering clones to also be the most efficient at viral fusion. Rather, the efficiency with which viral proteins response to adhesion of coreceptors, i.e. the conformational change within gp41 wherein HR regions fold in on themselves, may be a more significant factor when determining fusion efficacy. Accepting this hypothesis, it is not difficult to infer that coreceptor switching may be the result of gp120/gp41 modification resulting in very weak CXCR4 adhesion in combination with a gp41 with a comparatively efficient response to coreceptor adhesion. Briefly, should a viral clone develop the ability to adhere to CXCR4 through random mutation and if that clone also contains a particularly potent gp41 HR1/HR2 linkage it may be selected for propagation. It would be interesting to investigate in future work whether emergence of gp41 mutations continue to coincide with viral fusion efficiency.
Methods
Cell culture
GHOST (3) Parental (CD4+/CCR5−/CXCR4−) cells (developed by V. Kewalramani and D. Littman) were grown in Dulbecco's modified Eagle's medium (DMEM) (American Type Culture Collection, Manassas, VA) supplemented with 10% fetal calf serum (ATCC), 500 μg ml−1 G418 (Cell-Gro), and 100 μg ml−1 penicillin-streptomycin (Sigma, St. Louis, MO) for the parental cells and 1.0 μg ml−1 puromycin for the coreceptor encoding HOS.CCR5 (CD4−/CCR5+/CXCR4−) and HOS.CXCR4 (CD4−/CCR5−/CXCR4+) cells. GHOST (3) Hi-5 (CD4+/CCR5+/CXCR4−) and GHOST CXCR4 expressing (CD4+/CCR5−/CXCR4+) cells (developed by N. Landau) were grown in Dulbecco's modified Eagle's medium with 10% fetal calf serum and 1.0 μg ml−1 puromycin29. Cells were passaged every 2 or 3 days in a humidified 5% CO2–95% air incubator maintained at 37°C. Cells were washed with Hanks medium (Sigma) or balanced salt solution and treated with 0.25% trypsin/EDTA for 7 min at 37°C and then split 1 to 10. Prior to single-molecule force measurements, 200 μl of 1 × 106 cells ml−1 was added to a 60-mm tissue culture dish containing 5 ml of culture medium and incubated overnight at 5% CO2 and 37°C to allow for cell spreading and restoration of normal cell morphology. Immediately before an experiment, the medium was changed to serum-free medium containing HEPES (Invitrogen, Carlsbad, CA) to stabilize the pH while outside the incubator environment.
Production of pseudotyped viruses using patient-derived env genes
Single-cycle infectious pseudotyped viral stocks for adhesion assays were produced by co-transfecting 293 T cells with separate HIV env and core encoding vectors using lipofectamine 2000 as previously described25. All viral stocks were produced using the specified env protein expression vector previously sequenced24 and the viral core vector pNL4-3-EGFP-ΔE which has the env gene replaced by GFP30. A plasmid expressing β-lactamase enzyme fused to the HIV-1 accessory protein Vpr was obtained from the NIH AIDS Reagent Program.
Pseudotyped viruses containing the enzyme β-lactamase fused to Vpr were created by co-transfecting 293 T cells as described previously28. Briefly, pNL4-3-EGFP-ΔE, pAdVAntage (Promega), and pMM310 (NIH AIDS Research & Reference Reagent Program) were co-transfected into 293 T cells.
β-lactamase fused Vpr viral entry and infection of CCR5- or CXCR4-expressing cell lines
Cell-free supernatants were obtained by spinning at 1200 RPM for 10 min. 40 ng equivalent of p24 was used to infect 200,000 primary CD4+ T cells isolated from healthy donors and activated by PHA treatment for 3 days31. After spinoculation at 1200 × g for 2:00 h at 30° the virus was allowed to enter the target cells by incubating at 37° for 2 h. Cells were then incubated at room temperature for 1 h with CCF2-AM (Invitrogen) in CO2-Independent media (GIBCO). 1 uM T20 was added to prevent any further fusion event from happening. Cells were then washed 1× in BCM and incubated overnight at room temperature in CO2-independent media supplemented with 10% FBS. Next day, the cells were washed 1× with BCM and fixed in 2.5% PFA and analyzed with BD FACSCantoII. Results of two independent entry events were averaged.
Alternatively, 1 × 106 GHOST CD4+/CCR5+/CXCR4− or GHOST CD4+/CCR5−/CXCR4+ cells were plated in 60-mm dishes and allowed to adhere at 37°C for 6 h. After spreading, cells were washed with Hanks medium (Sigma) and infected with 10 ng equivalent of p24 of the specified virus for another 6 h. Cells were then washed with Hanks medium and incubated at 37°C for 48 h. Cells were again washed with Hanks medium and treated with 0.25% trypsin-EDTA for 7 min at 37°C to be fixed with 3% formaldehyde in phosphate-buffered saline (PBS) for 30 min at room temperature. Induced expression of GFP was examined by flow cytometry performed using a FACSCalibur fluorescent cell sorter (BD Biosciences).
Not all Env gp120 trimer complexes on the viral surface are capable of promoting successful cellular infection32,33. However, infection incompetent Env may still be able to successfully adhere to cellular receptors (e.g., viral gp160 will not promote viral fusion but will still bind to CD4). Our adhesion assay measures the adhesion of trimers that are able to specifically bind target cells regardless of infectiousness. By specifically binding the virion to the host cell, an otherwise defective gp120 or gp120 trimer might support infection simply by providing an increased number of bonds between the virion and cell, prolonging the overall interaction34.
Functionalization of the cantilevers
AFM cantilevers (Veeco Instruments, Santa Barbara, CA) were cleaned by successive 1-min incubations at room temperature in 70% ethanol/10% HCl, ultrapure water, and 100% ethanol, respectively. The cleaned silicon nitride cantilevers were functionalized with primary thiol groups for 20 min in 5% mercapto-propyl-triethoxysilane (MPES) (Sigma) in ethanol then washed in pure ethanol. The N-hydroxysuccinimide (NHS) ester functional group of the heterobifunctional crosslinker 1 mM SM(PEG)24 (Pierce), was allowed to hydrolyze to a carboxylic acid group in amine-free PBS (pH 7.2) for 1 h and then reacted with the thiol presenting cantilevers for 30 min. Immediately before incubation with viral stock solutions cantilevers were washed with dH2O and incubated with 2 mM 1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC) in the presence of 5 mM Sulfo-NHS for 15 min. Cantilevers equipped with the tethered amine-reactive EDC were allowed to incubate with viral stocks for 2 h at room temperature. Virion covered cantilevers were washed three times in PBS, incubated for 30 min at 37°C in 1% bovine serum albumin (Sigma) in PBS, and washed again in warm PBS. Finally, the cantilevers were immersed into serum-free DMEM containing HEPES just prior to use.
Recombinant protein and monoclonal antibodies
The soluble recombinant human sCD4 (sCD4-183 from Pharmacia) used here was composed of the first two extracellular domains of human CD4. This protein is reactive with HIV-1 gp120 and anti-CD4 monoclonal and polyclonal antibodies. Monoclonal anti-human CCR5 and CXCR4 antibodies (NIH AIDS Research and Reference Reagent Program CAT # 3933 and 4083, respectively) used in control experiments were selected for their ability to react specifically with human coreceptors. Monoclonal B4 (United Biomedical, Inc., Hauppauge, NY), used in control experiments, exerts a broad neutralizing activity against several HIV genotypes and clades by blocking access to the CD4 cell surface complex35. Also, the HIV-1 gp120 Monoclonal antibody 17 b was used for its specificity for a moderately well conserved conformation-dependent epitope and ability to block HIV-1 coreceptor adhesion2,36,37. Here, we refer to the anti-human CXCR4 antibody as ‘Clone 44717'. The above reagents were obtained from the AIDS Research and Reference Reagent Program (NIAID, NIH, Bethesda, MD).
Single-molecule force spectroscopy
Single-molecule measurements were conducted using a Molecular Force Probe (MFP) (Asylum Research, Santa Barbara, CA). The MFP is similar to an atomic force microscope (AFM) and utilizes the deflection of a flexible cantilever probe to determine forces between the probe and the sample. The spring constant (in pN/μm) of the individually loaded probes is determined by the nondestructive thermal oscillation method38. The MFP records the time-dependent position and deflection of the flexible cantilever probe above a sample with microsecond temporal resolution and subnanometer spatial resolution using laser deflection onto a photodetector. Changes in applied force were measured with piconewton resolution.
The MFP records outputs from the photodetector (in volts) and the linear variable differential transformer (LVDT). The photodetector output is transformed into force values using the inverse optical lever sensitivity (invOLS), which is obtained by multiplying the inverse slope of the sensor output versus the LVDT output while the system is exhibiting constant compliance. The probe-to-sample distance is calculated using the LVDT output by taking the total cantilever movement and subtracting the deformation due to the applied force. Force measurements are computed using Hooke's law, F = kΔx, where F is the applied force, k is the spring constant of the cantilever, and Δx is the measured cantilever deflection. The final output is a time-dependent trace of applied force versus separation distance from the sample.
The largest (and softest) triangular cantilever probe with an average spring constant of 10 pN/nm was used to collect force measurements with the highest possible resolution (<1 pN). For every contact between cell and cantilever, the distance between the cantilever and the cell was adjusted to maintain an impingement force of 100–300 pN before retraction39,40,41. Data collection was performed at 1.0 kHz. For measurements of bond adhesion, the retraction velocity was kept constant at 10 μm/s.
The frequency of adhesion between virus-functionalized cantilevers and live host cells was adjusted to correspond to single-event Poisson distribution statistics. This was done by varying the concentration of virus and contact area between the cantilever and the cell. Adjusting the frequency of adhesion to correspond to the single event Poisson distribution statistically supports the observation of single bimolecular interactions.
Analysis of force spectroscopy data
Traces of applied force as a function of cell-cantilever separation were analyzed using Igor Pro 4.09 software (Wavemetrics, Inc., Lake Oswego, OR). Adhesion forces were determined directly by recording the height of the adhesion peak from the level of zero applied force.
Distributions of adhesion event probability versus adhesion force were analyzed using MATLAB 7.0 software (Mathworks, Inc., Natick, MA). Adhesion forces were obtained as described above and binned to produce probability distributions. For each bin of adhesion forces, a probability was determined as the number of all observed adhesion events within that bin per total observed adhesion events for that condition. The adhesion forces produced for each contact time were averaged and compared using Student's t-test. Values of P < 0.05 were considered to correspond to distributions of adhesion forces that were statistically different while values of P > 0.5 corresponded to adhesion forces that were statistically similar. Any comparison resulting in a value of P between 0.05 and 0.5 was not considered either statistically similar or different and are not commented on. The probability of an adhesion event occurring with a particular force f is given by
where
as described by Hummer and Szabo27. Here, κs is the harmonic force constant scaled by kBT = β−1, ν is the retraction velocity of the cantilever, x* is the distance along the free energy surface from the well minimum to the energy at bond rupture or bond length, κm is the molecular spring constant of the bond, κ is the sum of κs and κm, and D is an effective diffusion coefficient. S is the survival probability or the probability that the rupture has not occurred yet at time t, and t* is the time of rupture. This probability density function was fit to each experimental adhesion force distribution by probing fit parameters using Monte Carlo optimization methods42,43,44.
Error values were obtained by generating synthetic adhesion force probability density distributions and fitting these distributions to the model above. For each of the parameters obtained from fitting synthetic data sets (n = 1000), separate probability density distributions were produced. Error values reported for variables are the standard deviation of these fitted parameter distributions. D was kept constant at 500,000 nm2 s−1 for reasons previously reported25. Briefly, after limited variation in D when fitting experimental data was observed, the value was fixed to more precisely fit x* and κm.
Statistical analysis of viral clone genetic sequences
12 full-length env sequences isolated from the PBMC of a single time point of a patient who underwent tropism switch24 were aligned using the ClustalW2 algorithm. The fusion fitness of each viral clone was measured by quantifying the fraction of cells that underwent viral entry as assayed by the virus-cell entry assay described above. The square of the difference between the fusion fitness of each pair of envelopes was determined. Next, we ranked the top ten envelope pairs with the least genetic difference and the most difference in their fusion fitness. The amino acid changes in these ten sequences were localized along the length of Env.
Study participants
This study was approved by the Johns Hopkins Institutional Review Board. Written informed consent was provided by all study participants.
Author Contributions
T.M.D. and S.A.R.: designed the experiments, analyzed and interpreted the results, and wrote the paper. R.N. and D.E.M.: provided samples, interpreted the results, and edited the paper. B.R.D.: analyzed the data, interpreted the results, and edited the paper. J.I.M. provided samples and edited the paper. R.F.S. and D.W. designed the experiments and wrote the paper.
Acknowledgments
This work was supported by NIH grants GM084204, CA143868, and CA85839 (DW), AI52778 (DEM) and AI47734 (JIM), the University of Washington Center for AIDS Research Computational Biology Core (AI 27757), and the Howard Hughes Medical Institute. We thank Dr. Shyam B. Khatau for fruitful discussions and editorial advice.
References
- Wyatt R. & Sodroski J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280, 1884–1888 (1998). [DOI] [PubMed] [Google Scholar]
- Kwong P. D. et al. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393, 648–659 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D. J. et al. gp160, the envelope glycoprotein of human immunodeficiency virus type 1, is a dimer of 125-kilodalton subunits stabilized through interactions between their gp41 domains. J Virol 65, 3797–3803 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl P. L., Doms R. W. & Moss B. Oligomeric structure of the human immunodeficiency virus type 1 envelope glycoprotein. Proc Natl Acad Sci U S A 87, 648–652 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu P. et al. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature 441, 847–852 (2006). [DOI] [PubMed] [Google Scholar]
- Zhang W. et al. Conformational changes of gp120 in epitopes near the CCR5 binding site are induced by CD4 and a CD4 miniprotein mimetic. Biochemistry 38, 9405–9416 (1999). [DOI] [PubMed] [Google Scholar]
- Sattentau Q. J. & Moore J. P. Conformational-Changes Induced in the Human-Immunodeficiency-Virus Envelope Glycoprotein by Soluble Cd4 Binding. J Exp Med 174, 407–415 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattentau Q. J., Moore J. P., Vignaux F., Traincard F. & Poignard P. Conformational-Changes Induced in the Envelope Glycoproteins of the Human and Simian Immunodeficiency Viruses by Soluble Receptor-Binding. J Virol 67, 7383–7393 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koot M. et al. Prognostic value of HIV-1 syncytium-inducing phenotype for rate of CD4+ cell depletion and progression to AIDS. Ann Intern Med 118, 681–688 (1993). [DOI] [PubMed] [Google Scholar]
- Schuitemaker H. et al. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus population. J Virol 66, 1354–1360 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratt G., Leandersson A. C., Albert J., Sandstrom E. & Wahren B. MT-2 tropism and CCR-5 genotype strongly influence disease progression in HIV-1-infected individuals. Aids 12, 729–736 (1998). [DOI] [PubMed] [Google Scholar]
- Mosier D. E. How HIV changes its tropism: evolution and adaptation? Curr Opin HIV AIDS 4, 125–130 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resch W., Hoffman N. & Swanstrom R. Improved success of phenotype prediction of the human immunodeficiency virus type 1 from envelope variable loop 3 sequence using neural networks. Virology 288, 51–62 (2001). [DOI] [PubMed] [Google Scholar]
- Jensen M. A. et al. Improved coreceptor usage prediction and genotypic monitoring of R5-to-X4 transition by motif analysis of human immunodeficiency virus type 1 env V3 loop sequences. J Virol 77, 13376–13388 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sing T. et al. Predicting HIV coreceptor usage on the basis of genetic and clinical covariates. Antivir Ther 12, 1097–1106 (2007). [PubMed] [Google Scholar]
- Jensen M. A. & van't Wout A. B. Predicting HIV-1 coreceptor usage with sequence analysis. AIDS Rev 5, 104–112 (2003). [PubMed] [Google Scholar]
- Jensen M. A., Coetzer M., van't Wout A. B., Morris L. & Mullins J. I. A reliable phenotype predictor for human immunodeficiency virus type 1 subtype C based on envelope V3 sequences. J Virol 80, 4698–4704 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W. et al. Coreceptor tropism can be influenced by amino acid substitutions in the gp41 transmembrane subunit of human immunodeficiency virus type 1 envelope protein. J Virol 82, 5584–5593 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thielen A. et al. Mutations in gp41 are correlated with coreceptor tropism but do not improve prediction methods substantially. Antivir Ther 16, 319–328 (2011). [DOI] [PubMed] [Google Scholar]
- Gallo S. A. et al. Kinetic studies of HIV-1 and HIV-2 envelope glycoprotein-mediated fusion. Retrovirology 3, 90–98 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melikyan G. B. et al. Evidence that the transition of HIV-1 gp41 into a six-helix bundle, not the bundle configuration, induces membrane fusion. J Cell Biol 151, 413–423 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankarappa R. et al. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J Virol 73, 10489–10502 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biscone M. J. et al. Functional impact of HIV coreceptor-binding site mutations. Virology 351, 226–236 (2006). [DOI] [PubMed] [Google Scholar]
- Coetzer M. et al. Evolution of CCR5 Use before and during Coreceptor Switching. J Virol 82, 11758–11766 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrowsky T. M., Zhou Y., Sun S. X., Siliciano R. F. & Wirtz D. Monitoring early fusion dynamics of human immunodeficiency virus type 1 at single-molecule resolution. J Virol 82, 7022–7033 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesla S. E., Selvaraj P. & Zhu C. Measuring two-dimensional receptor-ligand binding kinetics by micropipette. Biophys J 75, 1553–1572 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummer G. & Szabo A. Free energy reconstruction from nonequilibrium single-molecule pulling experiments. Proc Natl Acad Sci U S A 98, 3658–3661 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavrois M., De Noronha C. & Greene W. C. A sensitive and specific enzyme-based assay detecting HIV-1 virion fusion in primary T lymphocytes. Nat Biotechnol 20, 1151–1154 (2002). [DOI] [PubMed] [Google Scholar]
- Morner A. et al. Primary human immunodeficiency virus type 2 (HIV-2) isolates, like HIV-1 isolates, frequently use CCR5 but show promiscuity in coreceptor usage. J Virol 73, 2343–2349 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson T. C. et al. Molecular characterization of preintegration latency in human immunodeficiency virus type 1 infection. J Virol 76, 8518–8531 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H. et al. Novel single-cell-level phenotypic assay for residual drug susceptibility and reduced replication capacity of drug-resistant human immunodeficiency virus type 1. J Virol 78, 1718–1729 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chertova E. et al. Envelope glycoprotein incorporation, not shedding of surface envelope glycoprotein (gp120/SU), Is the primary determinant of SU content of purified human immunodeficiency virus type 1 and simian immunodeficiency virus. J Virol 76, 5315–5325 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layne S. P. et al. Factors underlying spontaneous inactivation and susceptibility to neutralization of human immunodeficiency virus. Virology 189, 695–714 (1992). [DOI] [PubMed] [Google Scholar]
- Dobrowsky T. M., Daniels B. R., Siliciano R. F., Sun S. X. & Wirtz D. Organization of cellular receptors into a nanoscale junction during HIV-1 adhesion. PLoS Comp Biol 6, e1000855 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C. Y. et al. Postexposure immunoprophylaxis of primary isolates by an antibody to HIV receptor complex. Proc Natl Acad Sci U S A 96, 10367–10372 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thali M. et al. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J Virol 67, 3978–3988 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trkola A. et al. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature 384, 184–187 (1996). [DOI] [PubMed] [Google Scholar]
- Hutter J. L. & Bechhoefer J. Calibration of Atomic-Force Microscope Tips. Rev Sci Instr 64, 1868–1873 (1993). [Google Scholar]
- Li F. Y., Redick S. D., Erickson H. P. & Moy V. T. Force measurements of the alpha(5)beta(1) integrin-fibronectin interaction. Biophys J 84, 1252–1262 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanley W. et al. Single molecule characterization of P-selectin/ligand binding. J Biol Chem 278, 10556–10561 (2003). [DOI] [PubMed] [Google Scholar]
- Chang M. I., Panorchan P., Dobrowsky T. M., Tseng Y. & Wirtz D. Single-molecule analysis of human immunodeficiency virus type 1 gp120-receptor interactions in living cells. J Virol 79, 14748–14755 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrowsky T. M., Panorchan P., Konstantopoulos K. & Wirtz D. Live-cell single-molecule force spectroscopy. Meth Cell Biol 89, 411–432 (2008). [DOI] [PubMed] [Google Scholar]
- Raman P. S., Alves C. S., Wirtz D. & Konstantopoulos K. Single molecule binding of CD44 to fibrin versus P-selectin predicts their distinct shear-dependent interactions in cancer. J Cell Sci 124, 1904–1910 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanley W. D., Wirtz D. & Konstantopoulos K. Distinct kinetic and mechanical properties govern selectin-leukocyte interactions. J Cell Sci 117, 2503–2511 (2004). [DOI] [PubMed] [Google Scholar]