Abstract
FKBP38 regulates apoptosis through unique interactions with multiple regulators including Bcl-2. Interestingly, the peptidylprolyl isomerase activity of FKBP38 is only detectable when it binds to calcium-saturated calmodulin (CaM/Ca2+). This, in turn, permits the formation of a complex with Bcl-2. FKBP38 thereby provides an important link between isomerase activity and apoptotic pathways. Here, we show that the N-terminal extension (residues 1-32) preceding the catalytic domain of FKBP38 has an autoinhibitory activity. The core isomerase activity of FKBP38 is inhibited by transient interactions involving the flexible N-terminal extension that precedes the catalytic domain. Notably, CaM/Ca2+ binds to this N-terminal extension and thereby releases the autoinhibitory contacts between the N-terminal extension and the catalytic domain, thus potentiating the isomerase activity of FKBP38. Our data demonstrate how CaM/Ca2+ modulates the catalytic activity of FKBP38.
Apoptosis is an essential process in multi-cellular organisms during development and homeostasis1,2, and its dysregulation contributes to the development of various pathological conditions, including cancer and autoimmune and neurodegenerative diseases3. The family of Bcl-2 proteins plays key regulatory roles in the apoptotic cell death pathway1,4. As a gatekeeper of apoptosis, Bcl-2 maintains the membrane integrity of subcellular organelles, particularly of the mitochondria. Upregulation or stabilization of Bcl-2 has been associated with cancer; downregulation or destabilization of Bcl-2 with ischemia/neurodegeneration.
FK506 binding protein 38 (FKBP38) is a multi-domain protein containing a peptidylprolyl cis-trans isomerase (PPIase) domain followed by three consecutive tetratricopeptide repeats (TPR), a calmodulin (CaM)-binding domain, and a transmembrane domain (Fig. 1)5,6,7. It has been shown that FKBP38 forms a complex with Bcl-25,8,9. However, the molecular basis of this interaction is unclear. In addition, the biological consequences of the FKBP38-Bcl-2 interaction are poorly understood. Although FKBP38 appears anti-apoptotic in some cell lines, via stabilizing Bcl-2 through co-localization and protection from degradation5,10, in other studies, the interaction disrupts Bcl-2 activity and therefore is pro-apoptotic8,11. These pro-apoptotic effects of nuclear Bcl-2 are suppressed with increasing FKBP38 levels12. In addition, a viral homolog of Bcl-2 in Hepatitis C (NS5A) appears to use FKBP38 for its anti-apoptotic effects in hepatoma cells13. An important variable rarely defined in these studies is the level of calcium-saturated calmodulin (CaM/Ca2+) in the experimental system, which significantly enhances the interaction between Bcl-2 and FKBP38 by an undetermined mechanism presumed to include the putative calmodulin binding domain identified at the C-terminus8,11. To better understand the conflicting evidence from in vivo and in vitro systems, it is necessary to understand the interactions between FKBP38 and its binding partners on a molecular level.
Figure 1. The sequence of human FKBP38.
The domain organization of FKBP38 is shown, along with a graphical representation of the constructs used in the current study. Above, the N-terminal 32 residues of FKBP38 and sequence alignment of the C-terminus of all TPR-containing human FKBPs, within which FKBP37 and FKBP38 are predicted to encode calmodulin binding domains. SS is the consensus secondary-structure prediction for all listed proteins33; numbering is according to FKBP38. FKBP, FKBP binding domain; TPR, tetratricopeptide repeats; TM, transmembrane domain; NTD, N-terminal domain.
In an effort to understand the molecular basis of FKBP38-Bcl-2 interaction, we first measured the isomerase activity of the FK506 binding domain of FKBP38 (FKBD38) and found that it was catalytically competent in the absence of CaM/Ca2+. Surprisingly, we found that the N-terminal extension, a region on the amino terminal side of the isomerase domain, autoinhibits isomerase activity and that this effect was reversed by CaM/Ca2+. We determined the solution structure of the N-terminal domain of FKBP38 (NTD), comprising the N-terminal extension and the core isomerase domain FKBD38, and mapped molecular interactions involving the N-terminal extension, NTD, FKBD38 and CaM/Ca2+. Conformational dyanmics was studied by NMR relaxation measurements revealing an increased mobility of the N-terminal extension comparing with the isomerase domain of the NTD. We proposed an activation mechanism for the isomerase activity of FKBP38 that involves a second calmodulin binding site in the N-terminal extension, providing a molecular rationale for the FKBP38/Bcl-2 regulation.
Results
Regulation of catalytic activity
It was previously suggested that the binding of CaM/Ca2+ to FKBP38 activates the PPIase activity of FKBP38 and subsequently allows Bcl-2 to bind to the complex8, but it was unclear how calmodulin activates this noncanonical activity of FKBP38 and facilitates protein-protein or protein-ligand interactions. FKBD38, the core isomerase domain of FKBP38, is flanked by a 32-residue N-terminal extension and C-terminal TPR, a putative calmodulin-binding site, and a transmembrane region (Fig. 1). To determine the contributions of the N- and C-terminal extensions on the isomerase activity and to uncover the mechanism by which CaM/Ca2+ activates FKBP38, we constructed three FKBP38 fragments and compared their activities in the presence and absence of CaM/Ca2+. As seen in Table 1, consistent with a previous study8, the isomerase activity of the full-length FKBP38 was activated by the addition of CaM/Ca2+. Interestingly, the FKBD38 showed similar levels of PPIase activity in the absence of CaM/Ca2+, suggesting that either the N- or C-terminal extensions were inhibitory and responsive to CaM/Ca2+. We then identified that NTD, a fragment of FKBP38, containing the N-terminal extension, but lacking the C-terminal extension, was activated by the addition of CaM/Ca2+ (Table 1). Taken together, the data suggest that the core isomerase activity of FKBP38 is inhibited by the N-terminal extension residues flanking the isomerase domain, and that this inhibition is overcome by the addition of CaM/Ca2+.
Table 1. Kinetic analysis of various forms of human FKBP38.
| FKBP mutants | kcat (min−1) | Km (μM) | kcat/Km (M−1s−1) × 103 | Fold Relative Catalytic Efficiency Increase |
|---|---|---|---|---|
| FKBP38 | 65 ± 4.8 | 194 ± 16.5 | 5.6 | 1 |
| FKBP38/CaM/Ca2+ | 56 ± 2.4 | 120 ± 5.1 | 7.8 | 1.6 |
| NTD | 67 ± 0.9 | 224 ± 12.7 | 5.0 | 0.9 |
| NTD/CaM/Ca2+ | 39 ± 5.1 | 86 ± 6.4 | 7.7 | 1.4 |
| FKBD38 | 149 ± 10 | 192 ± 13.9 | 13.0 | 2.3 |
Abbreviations: FKBP38, FK506 binding protein 38; CaM/Ca2+, calcium-saturated calmodulin; NTD, N-terminal domain; FKBD38, FK506 binding domain of FKBP38.
Conformational flexibility of FKBP38 NTD
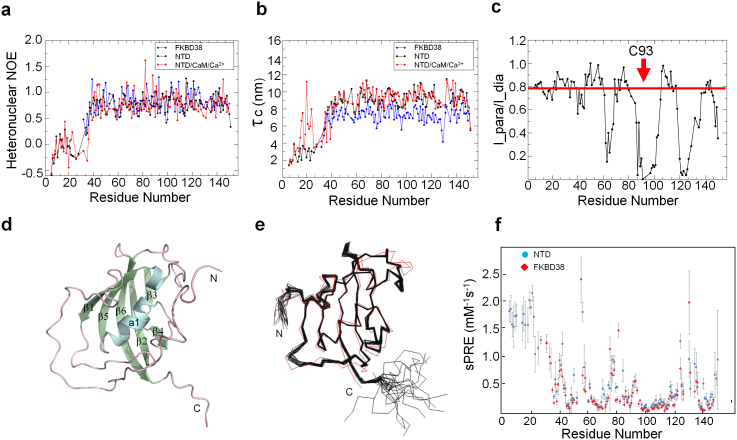
To investigate the role of the N-terminal extension residues, we measured backbone 15N relaxation rates and {1H}-15N heteronuclear Overhauser effects (NOE) for the FKBD38 and NTD in the presence and absence of CaM/Ca2+. The values of the relaxation parameters are shown in Figs. 2a,b, Table 2 and Supplementary Fig. 1a,b. The average values of T1 for residues with a heteronuclear NOE > 0.6 and no internal motions measured for the NTD are 862.03 ms in the free state and 872.92 ms when bound to CaM/Ca2+. The average T2 values for the NTD in the absence and presence of CaM/Ca2+ are 71.94 and 70.42 ms, respectively. The tumbling correlation times τc derived from the trimmed mean 15N T1/T2 ratio14,15 for FKBD38, NTD and NTD/CaM/Ca2+ are 7.29, 9.07, and 9.24 ns, respectively (Table 2). While the values observed for FKBD38 and the free NTD are consistent with the molecular weight expected for the monomeric proteins, the increase in the tumbling correlation time upon binding of CaM/Ca2+ is surprisingly small. This suggests that no specific tight complex is formed and that the interaction with CaM/Ca2+ is weak and transient. Comparable low heteronuclear NOE values are observed for residues 1–32 in the N-terminal extension for both the free and CaM/Ca2+-bound NTD indicating that this region exhibits increased conformational mobility on (sub)-nanosecond time scales. However, striking differences are observed for the local tumbling correlation times (τc, Fig. 2b) and T2 values (Supplementary Fig. S1) values for residues 11–26 in the N-terminal extension upon binding of the NTD to CaM/Ca2+. This indicates the presence of conformational exchange processes at μs-ms timescales that are linked to the interaction with CaM/Ca2+. Taken together the NMR relaxation data suggest that the N-terminal region of the NTD plays an important role in mediating weak molecular interactions of the NTD with CaM/Ca2+. These interactions may release a potential autoinhibition of the FKBD38 domain and thereby favorite substrate access to the isomerase core, thus explaining the increased catalytic activity.
Figure 2. Relaxation, PRE data and NTD structure.
(a) {1H}-15N hetNOE for the backbone amide of the core isomerase domain (FKBD38; blue), N-terminal domain (NTD; black) of FKBP38, and the NTD/CaM/Ca2+ complex at 1:3 morlar ratio (red). The N-terminal tail (residues 1–32) exhibits negative or near zero {1H}-15N hetNOE values, suggesting the unstructured characteristic of this region. The estimation errors were not plotted for clarity but referable to Table 2. (b) The rotational correlation time τc was derived from the T1/T2 ratio for all residues. (c) Experimental signal intensity ratio I_para/I_dia from 2D 1H-15N HSQC spectra without and with the addition of twenty molar equivalents of ascorbic acid. Due to the slight protein concentration change after adding ascorbic acid, the intensity ratios were normalized by setting largest I_para/I_dia ratio to 1. A prosxyl spin label 3-(2-iodoacetamido)-proxyl (Sigma-Aldrich) was attached to residue Cys93 of NTD_C121A/C139T, indicated by the red arrow. The red line across the graph represented a threshold of 0.78, protons above which were considered far from the prosxyl spin label and unrestrained. (d) The model for the NMR structure of NTD is shown in ribbon representation and colored by secondary structure (α-helices in cyan, β-sheets in green, and loops in purple). The N-terminal extension seen in the NMR structure was modeled from the PRE data, as shown in Fig. 2c and Supplementary Fig. 3. The N- and C-termini are labeled, as are the secondary structural elements. (e) The backbone ensemble of 20 superimposed NMR-derived structures of the NTD (shown in black) and N-Cα-C′-trace of the isomerase core structure determined crystallographically (PDB ID 2AWG, shown in red). The N-terminal extension (residues 1–32) is not presented for clarity as it is highly flexible and only a restricted number of NOEs were observed in this region. Molecular graphics were created using PyMOL (http://www.pymol.org). (f) Solvent PRE experiments for NTD and FKBD38.
Table 2. Analysis of Molecular weights and relaxation data for FKBD, NTD and NTD/CaM/Ca2+ complex.
| FKBD38 | NTD | NTD/CaM/Ca2+ | |
|---|---|---|---|
| Calculated MW (kDa) | 12.85 | 16.9 | 33.6 |
| Average T1 of all residues (ms) | 774.13 ± 91.29 | 822.74 ± 122.32 | 839.11 ± 246.27 |
| Average T2 of all residues (ms) | 98.71 ± 24.93 | 109.99 ± 87.65 | 100.86 ± 82.68 |
| Trimmed Average T1 (ms) | 783.77 ± 95.66 | 862.03 ± 88.78 | 872.92 ± 125.91 |
| Trimmed Average T2 (ms) | 96.17 ± 20.66 | 71.94 ± 10.74 | 70.42 ± 13.05 |
| Trimmed Average T1/T2 | 8.15 | 11.98 | 12.4 |
| Overall correlation time τc (ns) | 7.29 | 9.07 | 9.24 |
Solution structure of the FBBP38 NTD
To elucidate the mechanism of isomerase activation, we determined the three-dimensional structure of the NTD. The NTD failed to crystallize presumably due to the high flexibility of the N-terminal extension, but it was amenable to structural analysis using NMR. The NTD (residues 1–149) is highly soluble and shows a high quality 1H-15N HSQC spectrum with good chemical shift dispersion (Supplementary Fig. S2), indicating a well-folded protein. The presence of only few short-range NOEs observed for residues in the N-terminal extension confirmed that this region is conformationally flexible. A model for the NTD in ribbon representation and the ensemble of the isomerase domain of the 20 low-energy structures of the NTD are shown in Fig. 2d,e. Excluding the N-terminal residues spanning Met1-Glu35 and the eight C-terminal residues containing Leu150, Glu151 and a hexahistidine-tag, the root-mean-square deviation (RMSD) for the isomerase domain was 0.27 ± 0.07 Å for the backbone and 0.54 ± 0.08 Å for all heavy atoms. Statistics monitoring of the structure calculation by NMR are shown in Supplementary Table S1. The isomerase domain of the NTD (Fig. 2d) forms a half β-barrel composed of a 6-stranded antiparallel β-sheets and a central amphipathic α-helix. The central helix packs against the hydrophobic surface of the β-sheets. The overall fold of the isomerase domain of NTD is similar to other FKBP family members16,17,18,19 and to the crystallographic structure of FKBD38, its isomerase core without the N-terminal extended region20,21 (Fig. 2e).
Transient intramolecular interactions of the N-terminal extension
NMR relaxation data (Figs. 2a,b and Supplementary Fig. S1a,b.) indicated that the N-terminal extension (residues 1–32) shows high internal flexibility compared to the isomerase domain. On the other hand, the PRE data with the covalently attached spin label at residue C93 (Fig. 2c and Supplementary Fig. S3) indicated the presence of transient contacts between the N-terminal extension and the globular FBKD38. We thus recorded solvent PRE data (sPRE)22 for the NTD and FKBD38 to explore whether the interactions are strong enough to significantly shield the interaction surface from solvent access (Fig. 2f). The sPRE data confirmed that the N-terminal extension was highly solvent accessible consistent with its intrinsic disorder. However, no significant differences were observed for solvent PREs in the core domain comparing the two proteins. This indicated that the intramolecular contacts between the N-terminal extension and the core domain were weak and therefore did not strongly protect the interactions surface. A comparison of the 2D 1H- 15N HSQC spectra of the NTD (residues 1–149) and the FKBD38 (residues 33–149) (Fig. 3a) confirmed the structural similarity of the NTD and the FKDB38. However, differences in chemical shifts are observed for residues in the core domain, consistent with autoinhibitory interactions between the N-terminal extension and the core domain. As summarized in Figs. 3b and 3c, most of these perturbed residues were localized in a defined region, including residues Ser58, Lys62, Gly63 and Gln64 in the β1/β2-loop; Val65 in strand β2; Phe88 and Thr89 in β4; and Asp92, Cys93, Asp94, Val95 and Ile96 in the β4/α1-loop and close to the putative isomerase active site. The weak and transient intramolecular interactions between the N-terminal extension and the isomerase domain FKBD38 is possibly regulated by charged residues, e.g. residue Lys62 might be involved in the interaction with the negatively charged residues in the N-terminal extension (Fig. 3d).
Figure 3. Overlay of 2D 1H-15N HSQC spectra of the N-terminal domain (NTD) and core isomerase domain (FKBD38).
The assignments for resolved backbone residues are labeled with the residue number. (a) Chemical shift perturbations of the 15N-labeled NTD of FKBP38 upon deletion of the N-terminal extension (residues 1–32) were analyzed by overlaying 2D 1H-15N HSQC spectra of the NTD (red) and FKBD38 (black). (b) The 15N and 1H chemical shift changes were combined using the equation, Δδ = ((Δδ 1H)2 + (0.154* Δδ 15N)2)1/2). A threshold value at 0.025 ppm is indicated by a horizontal dotted line. (c) The residues of the perturbed chemical shifts attributed to deletion of the N-terminal extension are indicated by residue numbers (left) or highlighted in green on the surface representation (right). For simplicity, only the combined weighted chemical shifts higher than 0.025 ppm are shown. (d) ±1 kT/e electrostatic potential of NTD generated using APBS PyMOL plotted on the solvent accessible surface (blue, positively charged; red, negatively charged; white, neutral), in approximately the same orientation as that of the molecules in Fig. 3c (left). Positively charged residues K62 and K119 are labeled, where residue K62 might be involved in the interaction with the negatively charged residues in N-terminal extension.
The NTD interacts with CaM/Ca2+
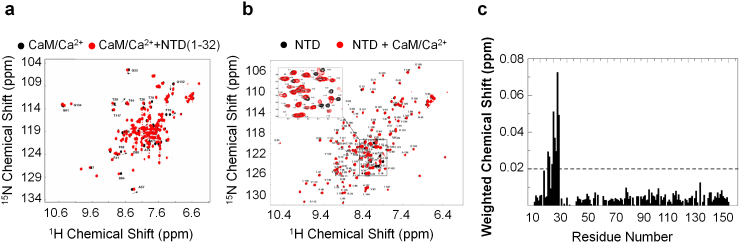
We examined whether the N-terminal extension (residues 1–32) of FKBP38 could mediate interactions with calmodulin. Chemical shift perturbations (CSP) were monitored for 15N-labeled CaM/Ca2+ in the absence and presence of a peptide corresponding to residues 1–32 of FKBP38. A number of calmodulin residues showed significant CSP upon addition of the peptide, indicating that the peptide interacts with CaM/Ca2+ (Fig. 4a). Some residues in calmodulin perturbed by the FKBP38 peptide, including Phe16, Phe19, and Phe68, are located within the canonical calmodulin ligand-binding site23. In the reciprocal experiment, addition of unlabeled CaM/Ca2+ to the 15N-labeled NTD generated noticeable changes in the 1H-15N HSQC spectrum, which were mapped primarily to the N-terminal extension of the NTD, mainly for residues Ala16, Arg17, and Phe19 – Glu24 (Fig. 4b,c). Importantly, the NTD did not interact with Ca2+ alone in our experimental conditions (Supplementary Fig. S5). We also added unlabeled calmodulin to 15N-labeled NTD, but no detectable CSP was observed, which suggested that the interaction between NTD and calmodulin is calcium dependent (Fig. S6). To further prove the interaction between NTD and CaM/Ca2+, a pull-down experiment using calmodulin resin was conducted. In this experiment, the expressed NTD in E. coli was purified using calmodulin resin. The amount of NTD purified by the calmodulin resin was low, indicating that the interaction was weak and transient (Fig. S7).
Figure 4. The N-terminal extension (residues 1–32) of FKBP38 binds calcium-saturated calmodulin (CaM/Ca2+).
(a) An overlay of 2D 1H-15N HSQC spectra of CaM/Ca2+ (black), and CaM/Ca2+ in the presence of the peptide derived from the N-terminus (residues 1–32) of FKBP38 (red). Some of the perturbed residues of calmodulin (BMRB 6542, PDB 1X02) are highlighted with residue numbers. (b) Titration between 15N labeled FKBP38 N-terminal domain (NTD) with CaM/Ca2+. 0.1 mM 15N labeled NTD was titrated with 0.3 mM calmodulin in the presence of 2.4 mM Ca2+. Resonances for NTD are indicated in black and those for NTD + CaM/Ca2+ complex are indicated in red. The perturbed residues in the N-terminal extension upon binding CaM/Ca2+ are highlighted in a zoomed view of the binding site. The spectra were recorded at 303 K on a 600 MHz equipped with a cryoprobe. (c) The 15N and 1H chemical shift changes are combined using the equation Δδ = ((Δδ 1H)2 + (0.154* Δδ 15N)2)1/2). A threshold value at 0.02 ppm is indicated by a horizontal dotted line.
Taken together, these data suggest that the N-terminal extension of FKBP38 is directly involved in a molecular interaction with CaM/Ca2+. Analysis of the NMR titration data showed that the interaction was weak with a dissociation constant KD ≈ 0.7 mM (Supplementary Fig. S4). The peptide sequence in the N-terminal extension of the NTD that mediated CaM/Ca2+ binding is distinct from any of the typical calmodulin binding motifs, including IQ, 1–10, 1–14 and 1–16 classes, but has some similarity with the 1-5-8-14 motif, a subclass of the 1–14 motif24. This might explain the relatively low binding affinity compared with those of typical calmodulin binding motifs.
To determine if the observed CaM/Ca2+-mediated regulatory mechanism of FKBP38 is important for Bcl-2 binding, Flag-tagged FKBP38 constructs were overexpressed in HeLa cells and extracts were immunoprecipitated using anti-Flag antibodies. FKBP38ΔN32 in the presence of Ca2+ showed a significantly reduced binding to calmodulin as compared with that of FKBP38, indicating that the N-terminal extension is important for calmodulin binding (Fig. 5). In keeping with the presence of a putative calmodulin-binding site at the C-terminus of FKBP38 (Fig. 1), calmodulin binding was not completely eliminated. Concomitantly, the affinity between FKBP38 and Bcl-2 was significantly enhanced with removal of the N-terminal extension. These results strongly suggest that the N-terminal extension of FKBP38 is a calmodulin-binding site critical for the Bcl-2 recognition.
Figure 5. The N-terminal extension of FKBP38 is involved in calmodulin binding in cells.

The wild-type FKBP38 and a mutant containing deletion of the N-terminal extension, both fused at their N-terminus to a Flag epitope tag (pXJ-Flag-S/FKBP38 and pXJ-Flag-S/FKBP38ΔN32), were transiently transfected into HeLa cells. Immunoprecipitates of HeLa lysates after calcium incubation were analyzed using anti-Bcl2 and anti-calmodulin antibodies.
Discussion
The current work defines a structural rationale for the activation of FKBP38, the only member of the FKBP family regulated by a second messenger (calcium), and lays the groundwork for further studies aimed at understanding the FKBP38-Bcl-2 interaction and its in vivo consequences. The peculiar characteristics of the active site of FKBP38 may explain the presence of a non-canonical regulatory system for this enzyme. The substrate binding pockets of most active FKBP domains, including those found in FKBP1225, FKBP2226, FKBP2527, FKBP5118, and FKBP5216, are conserved and characterized by the presence of aromatic residues (Supplementary Fig. S8). In contrast, many of the analogous residues in FKBP38 are replaced by residues with aliphatic characteristics, including Leu70, Leu86, Leu99, and Leu139 (Supplementary Fig. S8a). Indeed, the lack of conservation in the active site of FKBP38 initially led to the classification of this protein as not having isomerase activity, until the stimulatory effect of CaM/Ca2+ was discovered8. The large molecular weight FKBP containing proteins (FKBP51 and FKBP52) contain two FKBP domains at the N- and C-terminal regions, respectively. Although similar in primary sequence, their N-terminal FKBP domains possess rotamase activity and are able to bind FK506 and rapamycin, while the C-terminal FKBP domains do neither action, but are instead proposed to contribute to the activity of the enzyme mainly through protein-protein interactions3,18,28,29. Previous structural analyses of N- and C-terminal FKBP domains of FKBP5118 and FKBP5216 indicate that active site substitutions and loop insertions flanking the binding pocket are responsible for the loss of activity in the C-terminal FKBP domains. Our sequence analysis containing only the human FKBP domains places FKBP38 closer to the C-terminal FKBP domains of 51 and 52 than the N-terminal FKBP domains, while only FKBP38 possesses the N-terminal extension. Neither FKBD38 nor the NTD of FKBP38 is able to bind FK50630. Similarly, in our studies, the flexible N-terminal extension of FKBP38 is also responsible for the loss of activity. This appears to depend on weak autoinhibitory interactions of the N-terminal extension with the isomerase core domain. These contacts map to residues that are located near the putative isomerase active site of FKBP38, i.e. Asp94, Val95 and Ile96 (Figs. 3b,c) and may thereby inhibit the isomerase activity. Complex formation between CaM/Ca2+ and the NTD via the N-terminal extended region releases the autoinhibitory interaction of the N-terminal extension and may thereby enhance isomerase activity (Fig. 6). Such a mechanism of regulation of catalytic activity involving an N-terminal extension that engages both autoinhibitory interactions and binding to another protein in trans with highly dynamic and transient interactions has not been described for any other FKBP proteins. In other FKBPs typically the inactive and active forms of FKBP structures are encoded by two separate isomerase domains, such as seen in FKBP51 and 52. The substitutions in the active site of FKBP38, a subset of which are affected by the presence of the N-terminal extension, imply that the autoinhibitory interaction we have described may be unique to this FKBP.
Figure 6. Proposed mechanism for auto-inhibition and activation of the N-terminal domain (NTD) isomerase activity by interaction with CaM/Ca2+ interaction.
Although the N-terminal extension is not predicted to harbor a canonical calmodulin-binding site, we show that it clearly binds to calmodulin although weakly. This is consistent with a previous report31, where the interaction of FKBP381–165 (residues 1–165) with the CaM/Ca2+ revealed by CSP effects within the N-terminal extension with much weaker binding affinity (Kd = 4.5–26 mM) compared with the NTD (residues 1–149). It was noted that their CSP analysis on the catalytic domain of FKBP381–165 was impossible due to the fact that the 1H-15N HSQC spectra were dominated by very strong amide signals from Met1-Glu34 and Asp151-Asn165 regions31. Our analysis of the NTD, where residues of the catalytic domain could be well studied by NMR shows that there are no significant CSP effects within the catalytic domain, and only a subset of cross-peaks, representing the N-terminal extension, was affected by the addition of CaM/Ca2+ (Figs. 4b,c).
There is also a putative C-terminal calmodulin binding site described previously for FKBP388,27. Six of the sixteen annotated human FKBPs have a similar or conserved domain architecture to FKBP38, with a C-terminal TPR that follows an FKBP domain and a conserved α-helical segment immediately C-terminal to the TPR domain (Fig. 1). This region of FKBP52 is predicted to be a potential coiled-coil region and does not bind calmodulin32, but the homologous segments in FKBP37/XAP2 and FKBP38 are predicted to encode calmodulin binding sites23. For the three other FKBPs, no functional significance has been attributed to this region. The biological effect of calmodulin binding on either the C-terminal or N-terminal sites in cells is unknown. Indeed, it has not been experimentally determined whether one interaction predominates over the other with respect to downstream Bcl-2 signaling. It is certainly intriguing to speculate that some of the potentially conflicting results in the field may be, at some point, traced back to the existence of these two sites of regulation. Our data suggest that both calmodulin binding sites affected the interaction between FKBP38 and Bcl-2 (Fig. 5).
In conclusion, the current study is focused on the dynamics and structural features of the N-terminal domain of FKBP38, in particular the N-terminal extension. Interestingly, the N-terminal extension of FKBP38 interacts with CaM/Ca2+ to potentiate the isomerase activity of FKBP38 in a novel fashion. We speculate that this interaction might serve as a molecular basis for the catalytic activation of FKBP38 in a noncanonical manner. The information presented here provides important clues for understanding the catalytic activity of FKBP38, its regulation by the unique N-terminal extension, and the potential calcium- and calmodulin-mediated activation of FKBP38, including the binding of FKBP38 to Bcl-2.
Methods
Measurement of PPIase activity
The PPIase activity of FKBP38 proteins were was measured using a protease-coupled assay8. The reaction mixture contained 1 μM of PPIase, 5 μM CaM, 5 mM Ca2+, and 50 μg/ml of chymotrypsin. The reaction was performed at 4°C using N-Succinyl-Ala-Ala-Pro-Phe-p-Nitroanilide (Bachem) and read at 390 nm on a Varian Cary 300 UV-VIS Spectrophotometer (Varian). For kinetic measurements, PPIase was measured at 0, 25, 50, 100, and 200 μM of N-Succinyl-Ala-Ala-Pro-Phe-p-Nitroanilide, and the initial velocities measured were plotted against the substrate peptide concentrations. The data points were transformed to generate a double-reciprocal plot which yields Vmax and Km. Assays were repeated three times.
NMR spectroscopy
All NMR experiments were performed at 30°C on a Bruker AVANCE II 700 MHz NMR spectrometer equipped with four RF channels and a 5 mm z-gradient TXI cryoprobe, unless otherwise specified. The chemical shifts were referenced to internal 2,2-dimethyl-2-silapentane-5-sulfonate acid (DSS). All NMR data were processed by NMRPipe35 and analyzed using NMRView36. Published chemical shift assignments of NTD (Supplementary Fig. S3)37 were used in its backbone dynamic studies and chemical shift perturbation analysis.
NMR relaxation measurements
Backbone 15N relaxation (T1 and T2) and {1H}-15N heteronuclear Overhauser effect (hetNOE) were measured on 15N-labeled or 13C/15N-labeled samples with 0.1 mM concentration. The molar ratio of NTD and CaM/Ca2+ was 1:3 for the complex of NTD/CaM/Ca2+. All relaxation experiments were recorded with an interleaved acquisition scheme using 2D sensitivity-enhanced experiments, as previously described previously38,39. A 1.5-s interscan delay was used for the T1 and T2 measurements. Ten (14.4, 28.8, 43.2, 57.6, 72, 100.8, 129.6, 158.4, 187.2, and 201.6 ms) and twelve (21.6, 54, 108, 162, 270, 432, 648, 864, 1188, 1512, 1944 and 2268 ms) time points were acquired for T2 and T1, respectively. hetNOE experiments were recorded with a 2-s interscan delay followed by either 3 s of proton saturation using a series of 120° 1H pulses or an additional 3 s delay. The relaxation rates were fitted using a nonlinear least-squares routine implemented in NMRView36. The overall rotational correlation time τc was estimated from T1/T2 ratio of the residues for which the hetNOE values were > 0.6 and had no internal motions14,34.
NMR spectroscopy and paramagnetic relaxation enhancement measurements
To prepare the samples for paramagnetic relaxation enhancement (PRE) measurements, NTD_C121A/C139T was completely reduced by the addition of 50 mM dithiothreitol, and exchanged buffer to 200 mM Tris (pH 8.0) and 200 mM NaCl. Five molar equivalents of methanol-dissolved 3-(2-iodoacetamido)-proxyl (Sigma-Aldrich) was added and the reaction was left in the dark at 4°C overnight. Excess spin label was removed by changing the buffer to sodium phosphate buffer (20 mM sodium phosphate, pH 7.0, 50 mM NaCl, and 0.1 mM EDTA) using a PD10 desalting column (GE Healthcare Life Sciences). PRE was determined by peak intensity ratio from 2D 1H-15N HSQC spectra without (I_para) and with (I_dia) the addition of twenty molar equivalents of ascorbic acid (I_para/I_dia). The PRE was also directly measured as the difference of 1HN R2 relaxation rates with oxidized and reduced spin label40.
Solvent paramagnetic relaxation enhancement rates were measured by titration of gadolinium diethylene-triamine-penta-acetic-acid bismethylamide (Gd(DTPA-BMA)) to protein samples. Saturation recovery 1H-15N HSQC experiments with recovery times from 0.01 to 3 s were recorded at concentrations of 0, 1, 2, 3, 5, 7 and 10 mM Gd(DTPA-BMA) and data were analyzed as described41.
NMR structure determination
For structural studies, samples (0.5–1 mM) in 20 mM sodium phosphate (pH 7.0), 1 mM DTT, 50 mM NaCl, and 0.1 mM EDTA were used. As previously described37, backbone 1H, 15N, 13C resonances were assigned using data from 2D 1H-15N HSQC, 3D HNCA, HN(CO)CA, 3D HNCO, 3D HNCACB, and 3D CBCA(CO)NH spectra. The side chain 1H and 13C resonances were obtained from 3D HCC(CO)NH-TOCSY, 3D HCCH-TOCSY and 13C-edited NOESY experiments42. Nuclear Overhauser effects (NOE) distance constraints were obtained from 3D 15N- and 13C-edited NOESY spectra acquired with mixing times ranging from 80 to 200 ms. Hydrogen bond restraints were determined based on a backbone amide H/D-exchange experiment. A 15N-labeled sample was lyophilized and a 2D 1H-15N HSQC spectrum was recorded immediately after adding 99.9% D2O. The HSQC spectrum revealed 48 cross-peaks that still remained after the exchange. Where the acceptor oxygen for the slowly exchanging amide was identified from preliminary structural ensembles and NOE analysis, 39 hydrogen bond restraints (1.8 to 2.3 Å and 2.8 to 3.3 Å for the NH-O and N-O distances, respectively) were introduced into the structure calculations. A total of 4,195 unambiguous NOE distance restraints were used for structure calculation. This corresponds to a ratio of about 28 restraints per residue. The experimentally determined distance and dihedral angle restraints (Supplementary Table S1) were included in a standard simulated annealing protocol with torsion angle dynamics using the program ARIA43 and CNS44. In the first 7 iterations, 20 structures were calculated per iteration. In the final iteration, 100 structures were calculated39 and the 20 structures with the lowest target function energy were calculated with a full molecular dynamics force field, including electrostatic and Lennard-Jones potentials, and water-refined by ARIA43,45. A square-well potential (FNOE = 50 kcal·mol−1) was used to constrain NOE-derived distances. Torsion angle restraints Φ, Ψ were determined from the analysis of N, C′, Cα, and Hα chemical shifts using the program TALOS46. A force constant of 200 kcal·mol-1·rad-2 was applied to all torsion restraints. The program PROCHECK-NMR was used to check the quality of the calculated structures in the ensemble47. NMR data have been deposited in the RCSB PDB with code 2MF9 and Biological Magnetic Resonance Data Bank with code 6923.
Chemical shift perturbation analysis by NMR spectroscopy
To measure protein-protein or protein-ligand interactions, chemical shift perturbations to 15N- labeled FKBP proteins were monitored on 2D HSQC spectra using 0.1 mM protein samples in the sodium phosphate or the Bis-Tris buffer. Combined chemical shift perturbation Δδ = ((Δδ 1H)2 + (0.154* Δδ 15N)2)1/2 were calculated based on the chemical shift values perturbed by adding increasing amounts of unlabeled peptides or proteins.
Cell transfection, immunoprecipitation and immunoblotting
Human FKBP38 cDNA was cloned into a pXJ-Flag-S plasmid with the BamHI and XhoI sites48. The N-terminal deletion mutant (FKBP38ΔN32) was prepared by subcloning the PCR-amplified fragment into pXJ-Flag-S. All constructs were confirmed by DNA sequencing. HeLa cells were seeded at a density of about 2 × 105 cells per well. When the density of cells reached 80–90%, cells were transfected with either pXJ-Flag-FKBP38 (encoding wild-type FKBP38 with a Flag tag) or pXJ-Flag-FKBP38ΔN32 (the N-terminal 32-residue deletion mutant with Flag-tag) using LipofectamineTM (Invitrogen, Carlsbad, CA). After incubation for 24 h, the transfected cells were lysed using TNE lysis buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.5% Triton X-100, and 1 mM EDTA) including protease inhibitor cocktail solution (Roche Diagnostic, Singapore). Equal amounts of proteins were pre-incubated with 1 mM CaCl2 and then immunoprecipitated using anti-Flag (Sigma-Aldrich) at 4°C for 16 h and then immobilized protein G (Pierce) at 4°C for 3 h. The immunoprecipitate was then washed four times in the cold TNE lysis buffer. Bound proteins were resolved on a 12% SDS-PAGE gel and analyzed by Western blotting. Monoclonal anti-Bcl-2 and anti-CaM antibodies were purchased from Santa Cruz (Santa Cruz Biotechnology, CA, USA). Signals were detected with the SUPEX Western blotting detection kit (Neuronex Co. Korea).
Author Contributions
H.S.Y., C.B.K. and H.Y. designed the research. C.B.K., H.Y., B.H.C., J.C., B.S. and U.S. performed the research. C.B.K., H.Y., B.H.C., S.D., B.S., U.S., M.S. and H.S.Y. analyzed the data. C.B.K., H.Y., D.S., M.S., H.S.Y. wrote the manuscript, all authors commented on the manuscript.
Supplementary Material
Supplementary Information
Acknowledgments
We thank Keiichi I. Nakayama for kindly providing anti-FKBP38 antiserum. We also thank Salil Bose for helpful discussion, Kyong-Tai Kim and Aled Edwards for comments on the manuscript. This work was supported by A*STAR-Biomedical Research Council of Singapore Grant 04/1/22/12/362 and Ministry of Health IRG Grant NMRC/1177/2008. CongBao Kang was a recipient of the Singapore Millennium Foundation PhD scholarship.
References
- Heiden M. G. V. & Thompson C. B. Bcl-2 proteins: regulators of apoptosis or of mitochondrial homeostasis? Nat. Cell Biol. 1, E209–E216 (1999). [DOI] [PubMed] [Google Scholar]
- Cory S. & Adams J. M. The Bcl2 family: regulators of the cellular life-or-death switch. Nat. Rev. Cancer 2, 647–656 (2002). [DOI] [PubMed] [Google Scholar]
- Thompson C. B. Apoptosis in the pathogenesis and treatment of disease. Science 267, 1456–1462 (1995). [DOI] [PubMed] [Google Scholar]
- Kroemer G. The proto-oncogene Bcl-2 and its role in regulating apoptosis. Nat. Med. 3, 614–620 (1997). [DOI] [PubMed] [Google Scholar]
- Shirane M. & Nakayama K. I. Inherent calcineurin inhibitor FKBP38 targets Bcl-2 to mitochondria and inhibits apoptosis. Nat. Cell Biol. 5, 28–37 (2003). [DOI] [PubMed] [Google Scholar]
- Lam E., Martin M. & Wiederrecht G. Isolation of a cDNA encoding a novel human FK506-binding protein homolog containing leucine zipper and tetratricopeptide repeat motifs. Gene 160, 297–302 (1995). [DOI] [PubMed] [Google Scholar]
- Galat A. Peptidylprolyl cis/trans isomerases (immunophilins): biological diversity--targets--functions. Curr. Top. Med. Chem. 3, 1315–1347 (2003). [DOI] [PubMed] [Google Scholar]
- Edlich F. et al. Bcl-2 regulator FKBP38 is activated by Ca2+/calmodulin. EMBO J. 24, 2688–2699 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang C., Tai J., Chia J. & Yoon H. S. The flexible loop of Bcl-2 is required for molecular interaction with immunosuppressant FK-506 binding protein 38 (FKBP38). FEBS Lett. 579, 1469–1476 (2005). [DOI] [PubMed] [Google Scholar]
- Wang H. Q. et al. Interaction of presenilins with FKBP38 promotes apoptosis by reducing mitochondrial Bcl-2. Hum. Mol. Genet. 14, 1889–1902 (2005). [DOI] [PubMed] [Google Scholar]
- Edlich F. et al. The specific FKBP38 inhibitor N-(N′,N′-dimethylcarboxamidomethyl)cycloheximide has potent neuroprotective and neurotrophic properties in brain ischemia. J. Biol. Chem. 281, 14961–14970 (2006). [DOI] [PubMed] [Google Scholar]
- Portier B. P. & Taglialatela G. Bcl-2 localized at the nuclear compartment induces apoptosis after transient overexpression. J. Biol. Chem. 281, 40493–40502 (2006). [DOI] [PubMed] [Google Scholar]
- Wang J. et al. Hepatitis C virus non-structural protein NS5A interacts with FKBP38 and inhibits apoptosis in Huh7 hepatoma cells. FEBS Lett. 580, 4392–4400 (2006). [DOI] [PubMed] [Google Scholar]
- Tjandra N., Feller S., Pastor R. & Bax A. Rotational diffusion anisotropy of human ubiquitin from 15 N NMR relaxation. J. Am. Chem. Soc. 117, 12562–12566 (1995). [Google Scholar]
- Barbato G., Ikura M., Kay L., Pastor R. & Bax A. Backbone dynamics of calmodulin studied by 15N relaxation using inverse detected two-dimensional NMR spectroscopy: the central helix is flexible. Biochemistry 31, 5269–5278 (1992). [DOI] [PubMed] [Google Scholar]
- Wu B. et al. 3D structure of human FK506-binding protein 52: implications for the assembly of the glucocorticoid receptor/Hsp90/immunophilin heterocomplex. Proc. Natl. Acad. Sci. U. S. A. 101, 8348–53 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y. & Burakoff S. The 25-kDa FK506-binding protein is localized in the nucleus and associates with casein kinase II and nucleolin. Proc. Natl. Acad. Sci. U. S. A. 90, 7769–73 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinars C. R. et al. Structure of the large FK506-binding protein FKBP51, an Hsp90-binding protein and a component of steroid receptor complexes. Proc. Natl. Acad. Sci. U. S. A. 100, 868–73 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Duyne G. D., Standaert R. F., Karplus P. A., Schreiber S. L. & Clardy J. Atomic structure of FKBP-FK506, an immunophilin-immunosuppressant complex. Science 252, 839–42 (1991). [DOI] [PubMed] [Google Scholar]
- Kang C. B., Ye H., Dhe-Paganon S. & Yoon H. S. FKBP family proteins: immunophilins with versatile biological functions. Neurosignals 16, 318–25 (2008). [DOI] [PubMed] [Google Scholar]
- Maestre-Martínez M. et al. Solution structure of the FK506-binding domain of human FKBP38. J. Biomol. NMR 34, 197–202 (2006). [DOI] [PubMed] [Google Scholar]
- Madl T., Guttler T., Gorlich D. & Sattler M. Structural analysis of large protein complexes using solvent paramagnetic relaxation enhancements. Angew. Chem. Int. Ed. Engl. 50, 3993–7 (2011). [DOI] [PubMed] [Google Scholar]
- Crivici A. & Ikura M. Molecular and Structural Basis of Target Recognition by Calmodulin. Annu. Rev. Biophys. Biomol. Struct. 24, 85–116 (1995). [DOI] [PubMed] [Google Scholar]
- Yap K. L. et al. Calmodulin target database. J. Struct. Funct. Genomics 1, 8–14 (2000). [DOI] [PubMed] [Google Scholar]
- Itoh S. & Navia M. Structure comparison of native and mutant human recombinant FKBP12 complexes with the immunosuppressant drug FK506 (tacrolimus). Protein Sci. 4, 2261–8 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budiman C., Tadokoro T., Angkawidjaja C., Koga1 Y. & Kanaya S. Role of polar and nonpolar residues at the active site for PPIase activity of FKBP22 from Shewanella sp. SIB1. FEBS J. 279, 976–86 (2012). [DOI] [PubMed] [Google Scholar]
- Peattie D. A. et al. Expression and characterization of human FKBP52, an immunophilin that associates with the 90-kDa heat shock protein and is a component of steroid receptor complexes. Proc. Natl. Acad. Sci. U. S. A. 89, 10974–8 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambraud B. et al. Overexpression of p59-HBI (FKBP59), full length and domains, and characterization of PPlase activity. Biochem. Biophys. Res. Commun. 196, 160–6 (1993). [DOI] [PubMed] [Google Scholar]
- Davies T. & Sánchez E. FKBP52. Int. J. Biochem. Cell. Biol. 37, 42–7 (2005). [DOI] [PubMed] [Google Scholar]
- Kang C. B., Feng L., Chia J. & Yoon H. S. Molecular characterization of FK-506 binding protein 38 and its potential regulatory role on the anti-apoptotic protein Bcl-2. Biochem. Biophys. Res. Commun. 337, 30–38 (2005). [DOI] [PubMed] [Google Scholar]
- Maestre-Martínez M. et al. New structural aspects of FKBP38 activation. Biol. Chem. 391, 1157–67 (2010). [DOI] [PubMed] [Google Scholar]
- Wolf E., Kim P. & Berger B. MultiCoil: a program for predicting two- and three-stranded coiled coils. Protein Sci. 6, 1179–89 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combet C., Blanchet C., Geourjon C. & Deleage G. NPS: Network Protein Sequence. Analysis. Trends Biochem. Sci. 25, 147–150 (2000). [DOI] [PubMed] [Google Scholar]
- Daragan V. A. & Mayo K. H. Motional model analyses of protein and peptide dynamics using 13C and 15N NMR relaxation. Prog. NMR Spectrosc. 31, 63–105 (1997). [Google Scholar]
- Delaglio F. et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–93 (1995). [DOI] [PubMed] [Google Scholar]
- Johnson B. A. & Blevins R. A. NMRView: A computer program for the visualization and analysis of NMR data. J. Biomol. NMR 4, 603–614 (1994). [DOI] [PubMed] [Google Scholar]
- Kang C. B. et al. Backbone 1H, 13C, and 15N resonance assignments of the N-terminal domain of FKBP38 (FKBP38NTD). J. Biomol. NMR 36, 37 (2006). [DOI] [PubMed] [Google Scholar]
- Farrow N. A. et al. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry 33, 5984–6003 (1994). [DOI] [PubMed] [Google Scholar]
- Civera C., Simon B., Stier G., Sattler M. & Macias M. J. Structure and dynamics of the human pleckstrin DEP domain: Distinct molecular features of a novel DEP domain subfamily. Proteins 58, 354–366 (2005). [DOI] [PubMed] [Google Scholar]
- Donaldson L. W. et al. Structural characterization of proteins with an attached ATCUN motif by paramagnetic relaxation enhancement NMR spectroscopy. J. Am. Chem. Soc. 123, 9843–7 (2001). [DOI] [PubMed] [Google Scholar]
- Madl T., Bermel W. & Zangger K. Use of Relaxation Enhancements in a Paramagnetic Environment for the Structure Determination of Proteins Using NMR Spectroscopy. Angew. Chem. Int. Ed. Engl. 48, 8259–8262 (2009). [DOI] [PubMed] [Google Scholar]
- Sattlera M., Schleucherb J. & Griesinger C. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog. NMR Spectrosc. 34, 93–158 (1999). [Google Scholar]
- Linge J., O'Donoghue S. & Nilges M. Automated assignment of ambiguous nuclear overhauser effects with ARIA. Methods Enzymol. 339, 71–90 (2001). [DOI] [PubMed] [Google Scholar]
- Brünger A. T. et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta. Crystallogr. D. Biol. Crystallog. 54, 905–21 (1998). [DOI] [PubMed] [Google Scholar]
- Habeck M., Rieping W., Linge J. P. & Nilges M. NOE assignment with ARIA 2.0: the nuts and bolts. Methods Mol. Biol. 278, 379–402 (2004). [DOI] [PubMed] [Google Scholar]
- Cornilescu G., Delaglio F. & Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR 13, 289–302 (1999). [DOI] [PubMed] [Google Scholar]
- Laskowski R. A., Rullmannn J. A., MacArthur M. W., Kaptein R. & Thornton J. M. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8, 477–86 (1996). [DOI] [PubMed] [Google Scholar]
- Manser E. et al. Expression of constitutively active alpha-PAK reveals effects of the kinase on actin and focal complexes. Mol. Cell. Biol. 17, 1129–43 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information