Abstract
Our laboratory specializes in directed protein evolution, i.e. evolution of proteins under defined selective pressures in the laboratory. Our target genes are encoded in ColE1 plasmids to facilitate the generation of libraries in vivo. We have observed that when random mutations are not restricted to the coding sequence of the target genes, directed evolution results in a strong positive selection of plasmid origin of replication (ori) mutations. Surprisingly, this is true even during evolution of new biochemical activities, when the activity that is being selected was not originally present. The selected plasmid ori mutations are diverse and produce a range of plasmid copy numbers, suggesting a complex interplay between ori and coding mutations rather than a simple enhancement of level of expression of the target gene. Thus, plasmid dosage may contribute significantly to evolution by fine-tuning levels of activity. Here we present examples illustrating these observations as well as our methods for efficient quantification of plasmid copy number.
Keywords: recombinant gene expression, ColE1 plasmid, plasmid copy number, GFP, ALKBH2, transformation, mutagenesis, R-loop, RNA I, RNA II, directed evolution, MMS, MNNG
Introduction
ColE1-like plasmids share mechanisms for control of replication. ColE1-like replication initiation is orchestrated by a ~600bp sequence known as plasmid origin of replication or ori (reviewed in (1–3)). This sequence is transcribed, generating a pre-primer that forms a stable DNA-RNA hybrid (R-loop) at its 3’ end. The pre-primer RNA is then processed to a primer by RNAse H and extended by DNA polymerase I (Pol I), initiating leading-strand synthesis and facilitating the recruitment of the Pol III primosome, the replication complex responsible for completing plasmid replication (4).
ColE1-like plasmids are maintained at medium- and high-copy numbers, which make them the most popular vectors for recombinant protein expression in E. coli and useful as shuttle vectors for other organisms. Plasmid copy number is controlled by a negative feedback mechanism mediated by transcription of an antisense RNA from an alternative promoter (P1). The resulting 108bp antisense transcript (RNA I) hybridizes with the 5’ end of the pre-primer RNA (RNA II) as it is being transcribed, locking the pre-primer in a conformation that is incompatible with the formation of the R-loop at its 3’ end and thus preventing replication initiation (1,2). Both pre-primer RNA II and antisense RNA I form three stem-loops that are critical for this regulatory mechanism (SL1, 2, 3 and SL1’, 2’, 3’ respectively), as hybridization depends on the formation of a “kissing complex” between unpaired bases at their respective complementary loops (1,2). As pre-primer transcription proceeds further, the formation of a fourth loop (SL4) makes the pre-primer refractory to RNA I inhibition (5). Mutations in SL 1, 2, 3 and 4 are frequently found in ori mutants exhibiting increased plasmid copy number (reviewed in (3)).
Our laboratory specializes in the evolution of proteins in the laboratory, an approach known as directed evolution. Our random mutant libraries are generated in vivo using a mutator strain of E. coli expressing a low-fidelity form of Pol I (6,7). Error-prone plasmid replication in this strain generates mutations throughout a Pol I-dependent plasmid sequence encoding the gene of interest. Plasmids isolated from these cultures constitute our mutant library, where mutations are randomLy distributed across our gene of interest as well as sequences regulating transcription and plasmid replication. Our libraries can be subjected to functional selections to direct the evolution of specific changes in biochemical activity of the plasmid-encoded gene (6,7). We previously established that since Pol I is gradually replaced by the Pol III primosome (7,8), the mutation frequency in our libraries decreases with increasing distance from the plasmid DNA/RNA switch. In the absence of a functional selection, the plasmid ori shows the lowest mutation load of the plasmid because it is the most distal sequence relative to the DNA/RNA switch (7).
One of our targets for directed evolution is the human oxidative demethylase ALKBH2 (reviewed in (9,10)). This enzyme removes two highly cytotoxic DNA N-methyl adducts (N3-methylC and N1-methylA), which are made in abundance by the agent methyl methane sulfonate (MMS). E. coli cells that are deficient in AlkB (the E. coli homologue of ALKBH2) are hypersensitive to MMS, and this hypersensitivity can be partially complemented by ALKBH2 (7). By contrast, ALKBH2 expression confers no measurable protection against N-methyl-N’-nitro-N-nitrosoguanidine (MNNG), a stronger methylating agent that (unlike MMS) generates abundant cytotoxic oxygen adducts.
We performed two functional selections on human ALKBH2 mutant libraries containing an average of 1.5 mutations/kb. The two selections were protection from MMS toxicity (through repair of ALKBH2 canonical lesions), and protection from MNNG toxicity (through repair of alternative cytotoxic substrates). The procedures for library generation and selection for resistance to methylation toxicity using LB agar gradients have been described in detail in (7). Our sequencing data for 6 MMS-selected clones and for 6 MNNG-selected clones, are presented in Table 1. This table shows that the plasmid ori shows strong signs of positive selection in both cases. In the case of the MMS selection, we estimated a 23-fold increase in ori mutation frequency relative to an unselected library; in contrast, the coding sequence shows only a 2.2-fold enrichment. The modest magnitude of enrichment seen in the coding sequence is likely due to a stronger purifying selection (reviewed in (11)). Unexpectedly, the plasmid ori showed an almost identical level of positive selection in the MNNG selection (22-fold enrichment, compared to 23-fold for MMS). This suggests that ori mutations may not only contribute to optimizing existing activities but also facilitate the evolution of new biochemical activities. As a control, we also looked for evidence of positive selection within ~300bp downstream of the RNA/DNA switch, a non-conserved intervening sequence (Table 1). In this case, we detected only a 1.8-fold enrichment in mutations. In this case the weak positive selection is likely due to a subset of mutations immediately adjacent to the RNA/DNA switch that may influence either primer extension by Pol I or modulate local supercoiling at the ori (Fig 1a).
Table 1.
Sequence of ALKBH2 clones identified following methylating agent selection
| Clone_selection | Non-synonimous Coding |
Ori | Downstream of ori |
|||
|---|---|---|---|---|---|---|
| 1_MMS | G50D G64D | T113C | G189A | C409T | ||
| 2_MMS | H144R | T202A | T589C | G646T | C754T | |
| 3_MMS | H144Y | C238T | C643T | |||
| 4_MMS | A53T P55L | T170C | G325A | T479A | G755A | |
| 5_MMS | P27L P207S | G512A | ||||
| 6_MMS | H51L T121N | C328T | G592A | T840C | ||
| 1_MNNG | A78V L138I R193S D201G P207L S237C | G338A | ||||
| 2_MNNG | P180L | G53A | G95T | G373A | ||
| 3_MNNG | A97T | G53A | C198T | G477A | T687A | |
| 4_MNNG | H144Y N231D | G180A | ||||
| 5_MNNG | R58W A219D_P229L K255E | G44A | C116T | G180A | T666A, C667T | G850A |
| 6_MNNG | R58W A118T | G258A | A696G | |||
| Total mutations | 26 (22 new) | 25 | 9 | |||
| Expected without selection | 10 | 1.1 | 5.1 | |||
| Enrichment through positive selection (fold) | 2.2 (footnote 2) | 23 (footnote 2) | 1.8 (footnote 3) | |||
| MMS | 10 | 13 | 4 | |||
| Expected without selection | 5 | 0.55 | 2.55 | |||
| Enrichment through positive selection (fold) | 2.0 | 23 | 1.6 | |||
| MNNG | 16 (12 new) | 12 | 5 | |||
| Expected without selection | 5 | 0.55 | 2.55 | |||
| Enrichment through positive selection (fold) | 3.2 (2.4 new) | 22 | 2.0 | |||
Footnote 1:
Total bases: 795 × 12= 9546;
Expected frequency of bp substitutions (without selection) ~1.5/kb × 9.55kb =14
Expected frequency of amino acid substitutions (~30% synonymous (23)) ~ 10
Mutations present before randomization are underlined
Footnote 2
Total bases: 613 × 12= 7356;
Expected frequency of base-pair substitutions (without selection) ~0.15/kb ×7.36kb=1.1
Footnote 3:
Total bases: 280 × 12 =3360
Expected frequency of base-pair substitutions (without selection) ~1.5/kb × 3.4 kb= 5.1
For MMS and MNNG selections, comprising 6 and 6 clones respectively, each of the expected values calculated for the total needs to be divided by half.
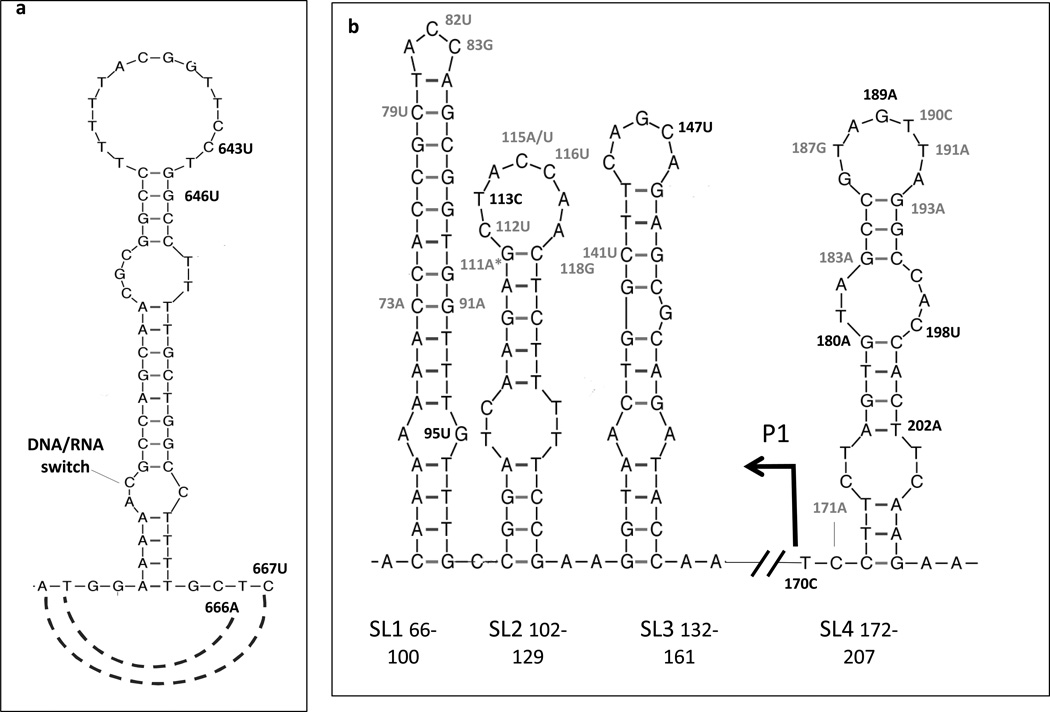
Fig. 1. Location of plasmid dosage-modulating mutations within known plasmid copy number regulatory sites.
Methylating agent-selected ALKBH2 mutations located in sites controlling plasmid replication initiation are shown in black. For comparison, previously-described mutations increasing ColE1 plasmid copy number are shown in grey (their references in the literature can be found in (3)). Graphic representations were generated using the mfold program (22), which predicts secondary structures of single-stranded nucleic acids based on a thermodynamic model. a. Area surrounding the DNA/RNA switch, where R-loop formation, primer processing and Pol I extension occur. b Antisense RNA I regulatory elements (SL 1, 2, 3 and promoter P1) and pre-primer SL4.
In agreement with the observed positive selection of sequences involved in regulation of plasmid replication, all selected ALKBH2 mutants analyzed (a total of 9, shown in Fig. 2) exhibited alterations in plasmid copy number. Except for one case, these alterations were not dramatic. Indeed, only 9 out of 25 ori mutations map to sites where mutations are known to produce strong increases in copy number (SL1, 2, 3, 4 or P1); of these, four occurred in SL4, with generally weaker effects (Fig 1b). Thus, both the mutation and phenotypic profiles of our ALKBH2 mutants suggest that alterations in plasmid dosage involve a complex interplay between ori and coding mutations rather than a simple enhancement of level of expression of the target gene. We propose that ori mutations likely contribute significantly to evolution by fine-tuning levels of activity.
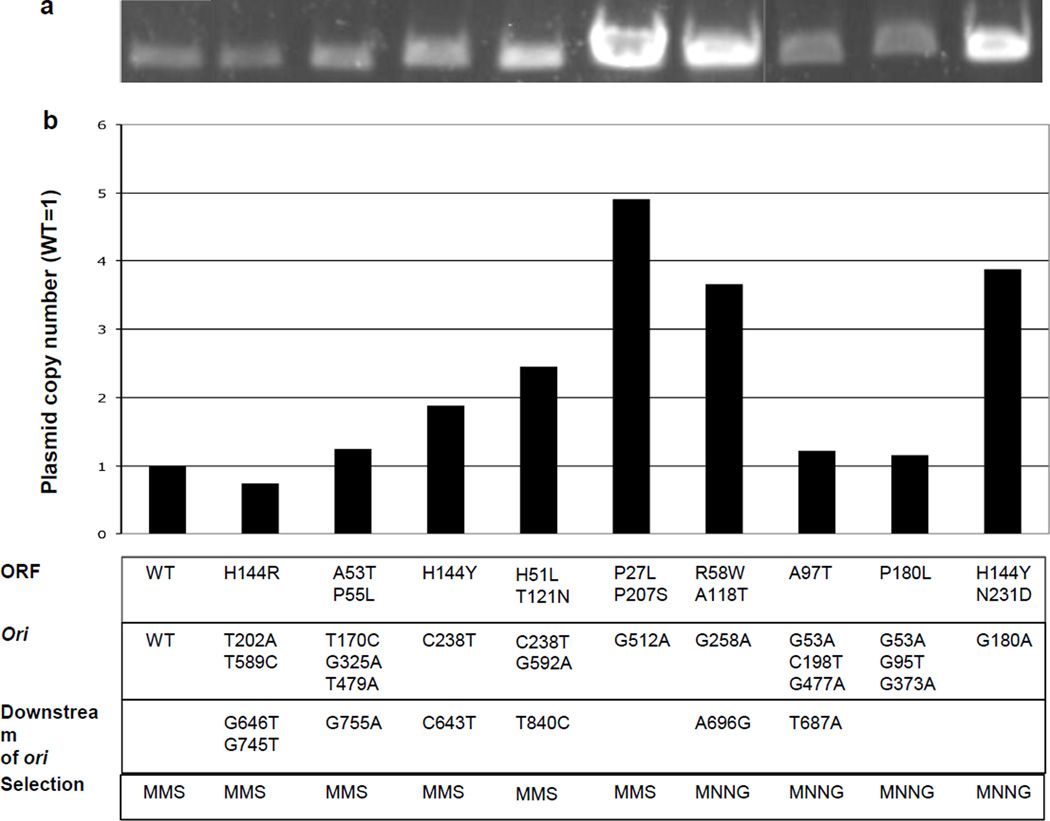
Fig. 2. Copy number of selected ALKBH2 mutants.
Plasmids bearing different ALKBH2 mutants were extracted from BL21 cells following transformation. Plasmid DNA was isolated from equal numbers of cells based upon the cell density of the culture, then linearized by digestion with a single-cut restriction enzyme (Pci I) and separated on an agarose gel for imaging and quantification. The amino acid substitutions found in the coding sequence of ALKBH2, base pair substitutions found in ori and adjacent downstream sequence, and the methylating agent used for selection are listed at the bottom a. Representative images of quantification gels. b. Quantification of corresponding gel band intensities normalized to WT.
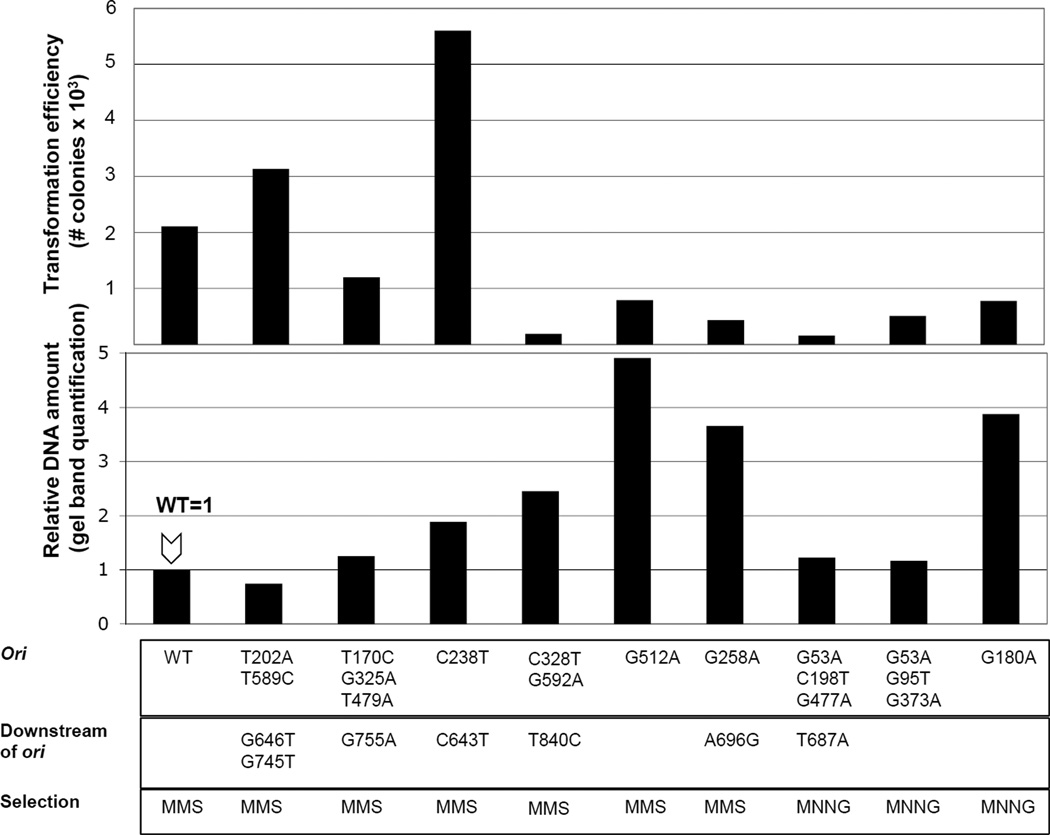
Interestingly, plasmid copy number determined by gel band quantification does not appear to correlate with transformation efficiency (Fig. 3). This indicates that transformation efficiency is not necessarily a good indicator of ColE1 plasmid copy number. Thus, it seems that altering plasmid replication initiation often has pleiotropic effects on plasmid establishment.
Fig. 3. Transformation efficiency of selected ALKBH2 mutants.
100ng of plasmid DNA was transformed into BL21 cells. Cells were then plated on agar plates containing carbenicillin, the selective antibiotic for the ALKBH2 plasmids. Equal numbers of cells where subject to transformation (5×107 cells) and the total number of successful transformations determined as colonies growing on carbenicillin plates are shown(top panel). For comparison, the bottom panel shows plasmid copy number levels as shown in Fig. 2b.
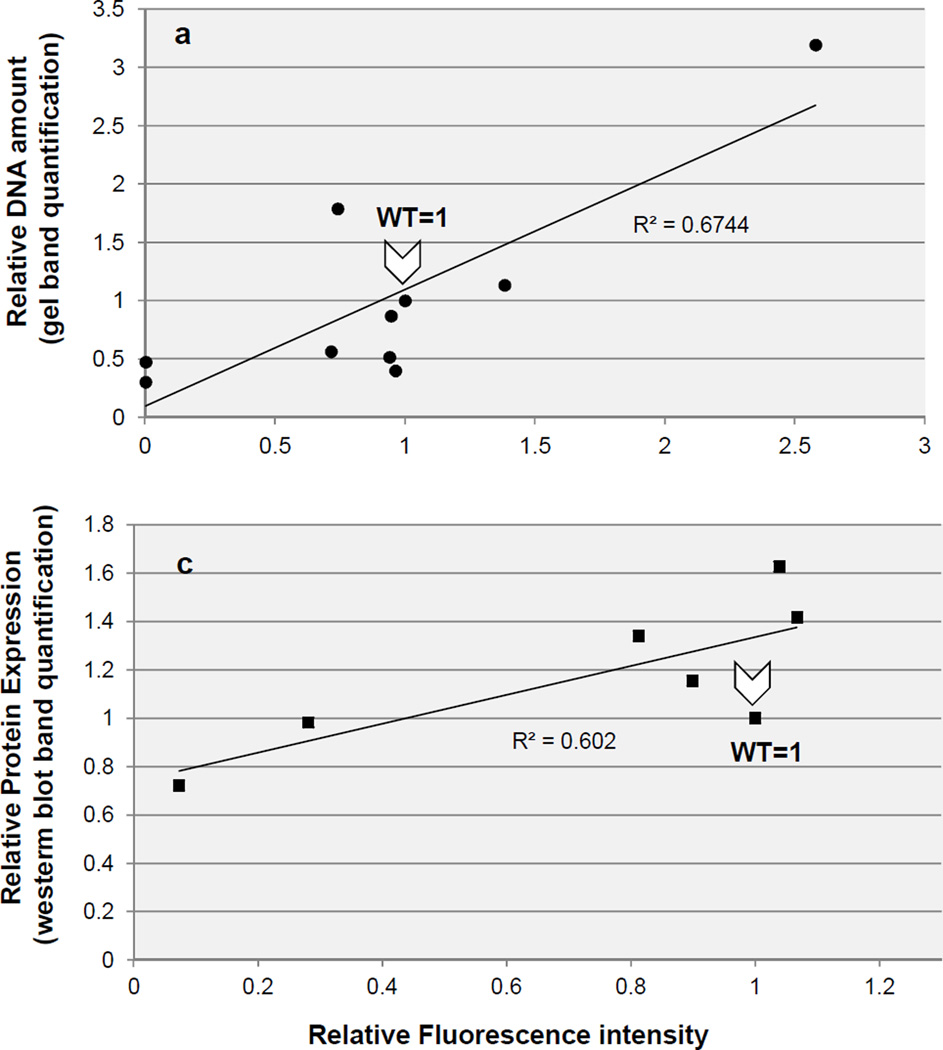
We also looked to see whether green fluorescent protein (GFP) (12) could be used as a reporter for plasmid copy number, as this would greatly facilitate monitoring how different treatments, culture conditions or genetic backgrounds impact plasmid copy number. To that end we used a commercially available plasmid, pGFPuv (Clontech) encoding “cycle 3 GFP”, a mutant GFP with improved expression in E. coli (13). We generated nine pGFPuv ori mutants exhibiting a range of plasmid copy numbers and compared the amount of plasmid recovered with level of GFP fluorescence. We found a good but not perfect correlation between these two variables (r2=0.67, Fig. 4 a). As a control, we confirmed that GFP fluorescence is representative of GFP protein levels in the cell, although again the correlation was not very tight (r2=0.60, Fig. 4 b).
Fig. 4. GFP as a reporter for plasmid copy number.
A set of pGFPuv mutants exhibiting a range of plasmid copy numbers was chosen and characterized by standard gel-based plasmid quantification (linearizing the vector with the restriction enzyme HindIII), measurement of GFP fluorescence, and Western blot analysis of GFP protein. Values are expressed relative to those of the wild-type ori. a. Correlation between plasmid gel quantification and GFP fluorescence. For this experiment cells had to be grown at 30 °C because some of the pGFPuv mutants in our panel were not viable at 37 °C, possibly because of temperature-dependent additional increases in copy number leading to runaway plasmid replication (see Note 3). b. Correlation between GFP fluorescence signal and GFP protein expression as detected by Western blot.
In sum, this work suggests that plasmid dosage likely contributes significantly to evolution by fine-tuning levels of the activity under selection. This mechanism could be particularly relevant for the evolution of metabolic pathways, as ColE1 regulation of plasmid replication is designed to integrate complex metabolic input (reviewed in (3)). Below we describe the methods we used to efficiently quantify plasmid copy number, which are generally applicable to any instance where plasmid dosage affects functional protein expression. We measure the efficiency of plasmid recovery by quantifying the amount of plasmid DNA on a gel and normalizing to the optical density of the culture (as an indicator of dry weight). Alternative plasmid copy number quantification methods include: [3H]thymidine labeling and quantification of released radiation from closed circular DNA (14,15); Southern blotting, either as a relative measure of copy number (16), or using a [32P] labeled probe for quantification (17); and real-time quantitative PCR with primers targeting both the plasmid of interest and a gene within the bacterial chromosome (18,19). Note that these methods are a measure of the average plasmid copy number present in the cell culture. The distribution of copy numbers (which is likely linked to plasmid stability (3)) is not addressed here.
2. Materials
2.1 Transformation
- Competent cells,
- BL21 ompT gal [dcm] [lon] hsdSB (rB− mB−)(F−) (ALKBH2 experiments).
- CJ278 CM4722 (A(gal-bio) thi-J relAl spoTl poA+ + (F+)) ΔPol A with unknown suppressor transformed with pHSG-Pol I (pGFPuv experiments).
ColE1 vector.
Luria-Bertani (LB) medium: 10 g/L tryptone, 5.0 g/L yeast extract and 5.0 g/L NaCl.
LB agar: add 1.5% agar to LB medium for agar plates.
Fisher 100mm × 15mm disposable Petri dishes.
Carbenicillin solution, 100mg/mL, keep at −20°C.
Carbenicillin (100µg/mL) LB agar plates.
Eppendorf Electroporator 2510.
2mm gap electroporation cuvettes.
2.2 Washing Plates and Plasmid Recovery
Clear, Flat-Bottomed 96-well plates.
Molecular Devices Spectra Max M2e S Plate Reader.
Plasmid miniprep kit.
2.3 Digestion and Quantification of Recovered Plasmids
Restriction enzymes and corresponding buffers.
Agarose.
TAE Buffer: 40 mM Tris-Acetate, 1 mM EDTA.
1kb DNA ladder.
Gel loading buffer: 0.25% Bromophenol Blue, 0.25% Xylene Cyanol, 50% Glycerol in diH2O.
SYBR Gold, Store at −20°C in opaque microcentrifuge tubes protected from visible light.
Large opaque container, such as a Tupperware with painted sides or covered in aluminum foil.
UVP Biospectrum 300 outfitted with dual UV light box, Cohu 6400 CCD camera, and SYBR (517–570nm) and GFP emission filters (570–620nm) and VisionworksLS version 6.8 software (UVP, LLC).
2.4 Quantification of GFP Fluorescence
Molecular Devices SpectraMax M2e Fluorometric and Spectrophotemtric plate reader. Dual monochromaters Absorbance 200–1000nm and excitation 250–850nm (Molecular Devices).
Black Greiner 96-well microplates with clear bottoms (Fisher Scientific).
Phosphate Buffered Saline (PBS): 0.2 g/L KCl, 0.2 g/L KH2PO4, 8 g/L NaCl, 2.16 g/L Na2HPO4·4H2O
2.5 Sequencing of Plasmids of Interest
NanoDrop Spectrophotometer for DNA quantification (Thermo Scientific).
MacVector version 10.5 for sequence analysis (Symantic).
3. Methods
Below we show protocols for quantifying plasmid copy number. Plasmids are recovered following transformation, on cells washed from the selecting plate to avoid possible generation and amplification of additional regulatory mutations through growth in liquid culture (see Notes 1–3). Recovered plasmids are linearized with a single-cutting restriction enzyme, separated on an agarose gel, and imaged as a means to quantitate plasmid copy number.
3.1 Transformation of Plasmids of Interest
3.1.1 Transformation of ColE1 plasmid by heat-shock (used for our pGFPuv experiments; see Note 1–4)
Prepare a 4mL overnight culture in LB media of the cell line of interest. Include selective antibiotic in the media for the desired cell line.
Expand this overnight culture into a sterile 2L Erlenmeyer flask containing 400mL of LB media.
Incubate the flask at 37°C with shaking until the cells reach exponential growth phase, as measured by turbidity (cultures with an OD600 of 0.6).
Chill the flask containing the cells on ice for 15 minutes.
Transfer the liquid cultures to plastic centrifuge bottles, and centrifuge for 15 minutes at 4°C and 4,000rpm.
Resuspend the pelleted cells in approximately 10mL of chilled calcium chloride solution (60 mM CaCl2, 15% Glycerol, 10 mM HEPES, pH7).
Transfer the cells into a 50mL conical tube, and fill to the 50mL line with chilled calcium chloride solution.
Centrifuge the cells at 4°C, 4,000rpm for 15 minutes.
Pour off the supernatant.
Resuspend the cells in calcium chloride, fill the tube to the 50 mL mark with chilled calcium chloride solution, and centrifuge at 4°C, 4,000rpm for 15 minutes.
Repeat the wash step.
After the 3rd and final wash, decant or pipette off the supernatant from the pelleted cells.
Resuspend the cell pellet in approximately 1:1 volume of chilled calcium chloride solution.
Keep cells on wet ice and use immediately, or aliquot competent cells into 1mL microcentrifuge tubes on dry ice for storage at −80°C (see Note 5).
15 Pipette 100µL chemically competent cells into a 5mL culture tube.
16 Add 500ng plasmid to the tube.
17 Incubate on ice for 10 minutes.
18 Heat-shock in a water bath at 42°C for 2 minutes.
19 Re-suspend cells into 1 mL of LB medium in a 12 × 75mm or larger capped culture tube, and recover cells for an hour with shaking at 37°C (see Note 6)
Plate transformed cells by spreading onto Petri dishes containing LB agar and 0.1mg/mL carbenicillin (see Note 7)
Let the cells grow overnight at 37/30°C.
3.1.2 Transformation of ColE1 plasmid by electroporation (used for our ALKBH2 mutants; see Note 1–4)
Expand desired cell line then pellet 400mL culture as describe above (3.1.1 step 1–5)
Resuspend the pelleted cells in approximately 10mL of chilled 10% glycerol solution.
Transfer the cells into a 50mL conical vial, and fill the vial to the 50mL mark with chilled 10% glycerol.
Centrifuge at 4°C, 4,000rpm for 15 minutes
Pour off the supernatant
Wash the cells 3 more times with chilled 10% glycerol
Resuspend the pelleted cells from the final wash in 1:1 volume of chilled 10% glycerol
Keep cells on wet ice and use immediately, or aliquot competent cells into 0.6mL microcentrifuge tubes on dry ice for storage at −80°C (see Note 5).
Add 40µL electrocompetent cells to a 0.2cm gap electroporation cuvette.
Add 100ng plasmid to the cuvette in volume <1µL.
Electroporate at 1800V, with a time-constant between 4 and 6s.
Transfer cells to 1mL LB medium and recover for 1 hour with shaking at 37°C (see Note 6).
Plate transformed cells by spreading onto Petri dishes containing LB agar and 0.1mg/mL carbenicillin (see Note 7).
Let the cells grow overnight at 37/30°C.
3.2 Washing Plates and Recovery of Plasmids
Wash plates using a sterile plate spreader into 1.5 mL eppendorf tubes (see Note 8).
Measure the optical density at 600 nm (OD600) of each plate wash using a plate reader (see Note 9).
Normalize each plate by volume based upon its optical density (see Note 10).
Centrifuge normalized plate washes for 1 minute at 11,000 × g.
Pipette off supernatant.
Harvest plasmids of interest from pelleted cells using a plasmid miniprep kit.
3.3 Digestion and Quantification of Recovered Plasmids
Digest 5µL of eluted plasmid from each miniprep (see Note 10), using an appropriate restriction enzyme and buffer in a 20µL volume reaction, following the manufacturer’s specifications (see Note 11).
Prepare a 0.8% agarose gel using TAE buffer.
Load the gel with an appropriate DNA ladder.
Mix each digest with an appropriate amount of gel loading buffer. Load the entire digest into each well of the gel.
Run the gel at 120V, 400 mA for 60–90 minutes.
Prepare 50mL of a 1:10,000 dilution of SYBR Gold in TAE (see Note 12).
Place the gel in an opaque container. Pour the SYBR Gold solution over the gel, allow the gel to stain overnight, shaking at 4°C.
Visualize the gel with using a UVP BioSpectrum 300 LM26E transilluminator equipped with a SYBR filter (515–570nm). Set the shutter speed to 544 ms. Adjust the camera aperture and image contrast so that none of the fluorescent bands saturate the image (see Note 13).
Use VisionWorksLS version 6.8 to calculate the Optical Density of the gel band representing each plasmid assayed (see Note 14).
3.4 Quantification of GFP Fluorescence
Colonies of transformed cells are harvested from plates and normalized by volume and OD600 (see Notes 7–9). These plate washes then represent a population of cells. The fluorescence quantification represents an average of this population.
The OD600 normalized cultures are washed into PBS at room temperature (see Note 15). Cells are diluted in PBS (three-fold and nine-fold).
200µL of neat and diluted cells are loaded into black Greiner 96 microwell plates with clear bottoms.
GFP fluorescence is quantified using a Molecular Devices Spectromax M2e plate reader at excitation 395nm and emission 510nm.
GFP fluorescence readings are compared to total GFP immunoreactive protein as determined by Western blot (see Note 16).
3.5 Sequencing of Plasmids of Interest
Purified plasmids are first quantified. We prefer using a Nanodrop spectrophotometer for the small sample requirement and automated analysis. In accordance with the manufactures instructions, a 2µL droplet of the plasmid containing solution is placed on the pedestal and the DNA concentration determined based upon a fixed length light path and absorbance at 260nm determined relative to a solvent blank. Aliquots of plasmid DNA containing 500ng of DNA in 5–7µL of water are sent to the company Sequetech for sequencing. Sequences are assembled and analyzed using the program MacVector version 10.5 (see Note 1).
Acknowledgements
The authors would like to thank Dr. Barbara Sedgwick for the gift of the BS141 (AB1157 F’) and BS143 (AB1157 alkB F’) strains, Dr. Catherine Joyce for the gift of the CJ278 (polAΔ) strain, Dr. Lawrence Loeb for mentorship in the initial stages of this work, and Jacob Marquette for his help with the generation of pGFPuv libraries. This work was supported by K08 award CA116429-04 to Manel Camps.
Footnotes
Verifying the purity of the plasmid sample to be tested; plasmid ori mutations (particularly those at the area of RNAI/RNAII overlap) alter the compatibility properties of plasmids (20), allowing the stable maintenance of more than one plasmid species within a clone. Thus, ori mutants isolated in vivo from single colonies often contain mixed sequences. Transformation of mixed plasmid species can confound experimental results because a mutant initially present in a small fraction of the sample can become predominant if it has higher transformation efficiency and/or provides higher fitness than the initially most abundant species. Thus, plasmid sequences need to be checked for the presence of mixed sequence, which is seen as a double peak on the chromatogram.
Culture conditions are critical. Rich media increases plasmid copy number, possibly by allowing a higher metabolic burden on the cell (18). Saturation conditions also favor increased copy number, possibly due to increased R-loop formation resulting from alterations in supercoiling associated with titration of R-loop-suppressing factors and/or shifts in the transcriptional profile of the cell (for a review see (3)).
The temperature at which the culture is grown also has a strong impact upon the plasmid copy number. Growth at higher temperatures tends to increase plasmid copy number due to a destabilizing effect on RNAI/RNA II hybrid formation and other temperature-sensitive alterations in the folding of the pre-primer RNA (5,21), and to direct and indirect effects on R-loop formation.
Electroporation tends to produce higher transformation efficiencies but the efficiency of transformation can vary substantially between individual electroporations. Transformation using chemically-competent cells produces more reproducible transformation efficiencies, facilitating comparison between different clones.
Competent cells should be stored at −80°C, and will keep for several months. Competent cells should be thawed slowly on ice before use, and refrozen immediately after use. Competent cells left out at room temperature for over an hour should be discarded.
We usually grow cells at 37 °C. Failure of certain clones to grow at 37 °C may be indicative of runaway plasmid replication. In that case the experiment is performed at 30°C.
Following transformation, adjust the number of cells plated or plate different cell dilutions to produce semi-lawns, i.e. a large number of separately growing colonies (200–800 for a 100×15 mm Petri dish). This ensures adequate clonal representation while avoiding additional physiological variables (such as early nutrient and O2 deprivation) occurring in a lawn.
The plate washes may be too dense to obtain an accurate OD600 reading directly. The range across which a plate reader can accurately report OD600 of a culture is 0.1–1.0. Dilute each wash 1:10 and 1:100 within a 96 well plate to obtain accurate readings. Use OD600 results from the dilute cultures to calculate the actual OD600 of each wash.
For example: 0.25mL of cells with an OD600 of 4.0 is equivalent in dry weight to 1mL of cells with an OD600 of 1.0.
Because DNA analyzed on the agarose gel is obtained from a fixed number of cells, variations in copy number will result in differences in band intensity on the gel. For optimal detection, adjust volume of digested DNA based upon intensity of bands obtained on the gel: if too faint, repeat with an increased volume of DNA from each prep; if overloaded, repeat with a decreased volume of DNA.
When selecting restriction enzymes to linearize plasmid DNA, if possible choose enzymes that cut the plasmid of interest only once and that do not have star activity. Be sure all plasmid is linearized as incomplete digestion will skew analysis.
SYBR Gold is used to visualize the bands on the gel because it provides a greater dynamic range of signal intensity than ethidium bromide and thus gives a more quantitative readout of plasmid copy number. This reagent is not considered a carcinogen, greatly simplifying safety and disposal. Keep solutions containing SYBR gold and gels stained with SYBR gold protected from visible light to avoid photodegradation.
VisionsworksLS has a digital imaging utility that highlights areas of the gel-documentation image that are light-saturated. Adjust the camera’s parameters so that none of the bands are saturated in order to best compare the pixel area density of each band.
Gel band intensities are quantified using VisionWorksLS version 6.8’s area density utility. Regions encompassing each band are defined and the mean pixel area density determined for each band. The area densities obtained for each clone are divided by the area density of the band corresponding to the wild type ori on the gel. This yields a measure of plasmid copy number expressed as “fold wild-type” for each clone.
For fluorescence determination, cultures need to be washed and resuspended in Phosphate Buffered Saline (PBS) because LB has an interfering autofluorescence at excitation 395nm and emission 510nm, the excitation and emission optima for GFPuv.
Protein extracts for Western blot analysis are prepared from washed transformation plates as described in Notes 6–8. Western blot analyses are performed as described previously (24) blocking with 5% dry milk in Phosphate Buffered Saline. Santa Cruz Biotechnology rabbit anti-GFP polyclonal antibodies (#SC8334) used to probe for GFP protein levels, and a Millipore goat anti-rabbit IgG Horse Radish Peroxidase conjugated secondary antibody (#12–384); protein bands are visualized using Pierce Supersignal WestPico reagents and protocols (#34077). Films are then imaged using the UVP BioSpectrum 300 LM26E Transilluminator with white transillumination. Band intensity is quantified using Visionworks version 6.8 (see Note 12).
References
- 1.Polisky B. ColE1 replication control circuitry: sense from antisense. Cell. 1988;55:929–932. doi: 10.1016/0092-8674(88)90235-8. [DOI] [PubMed] [Google Scholar]
- 2.Cesareni G, Helmer-Citterich M, Castagnoli L. Control of ColE1 plasmid replication by antisense RNA. Trends Genet. 1991;7:230–235. doi: 10.1016/0168-9525(91)90370-6. [DOI] [PubMed] [Google Scholar]
- 3.Camps M. Modulation of ColE1-like plasmid replication for recombinant gene expression. Recent Pat DNA Gene Seq. 2010;4:58–73. doi: 10.2174/187221510790410822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Masai H, Nomura N, Kubota Y, Arai K. Roles of phi X174 type primosome- and G4 type primase-dependent primings in initiation of lagging and leading strand syntheses of DNA replication. J Biol Chem. 1990;265:15124–15133. [PubMed] [Google Scholar]
- 5.Polisky B, Zhang XY, Fitzwater T. Mutations affecting primer RNA interaction with the replication repressor RNA I in plasmid CoIE1: potential RNA folding pathway mutants. EMBO J. 1990;9:295–304. doi: 10.1002/j.1460-2075.1990.tb08108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Camps M, Naukkarinen J, Johnson BP, Loeb LA. Targeted gene evolution in Escherichia coli using a highly error-prone DNA polymerase I. Proc Natl Acad Sci U S A. 2003;100:9727–9732. doi: 10.1073/pnas.1333928100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Troll CJ, Alexander DL, Allen JM, Marquette JT. Mutagenesis and functional selection protocols for directed evolution of proteins in E. coli. The Journal of Visualized Experiments. 2010 doi: 10.3791/2505. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allen JM, Simcha DM, Ericson NG, Alexander DL, Marquette JT, Van Biber BP, Troll CJ, Karchin R, Bielas JH, Loeb LA, et al. Mutational footprints of ColE1 plasmid replication by error-prone DNA polymerase I identify primosome loading and sites of Okazaki primer processing. Nucleic Acids Research. 2011 doi: 10.1093/nar/gkr157. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drablos F, Feyzi E, Aas PA, Vaagbo CB, Kavli B, Bratlie MS, Pena-Diaz J, Otterlei M, Slupphaug G, Krokan HE. Alkylation damage in DNA and RNA--repair mechanisms and medical significance. DNA Repair (Amst) 2004;3:1389–1407. doi: 10.1016/j.dnarep.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 10.Sedgwick B, Robins P, Lindahl T. Direct removal of alkylation damage from DNA by AlkB and related DNA dioxygenases. Methods Enzymol. 2006;408:108–120. doi: 10.1016/S0076-6879(06)08008-6. [DOI] [PubMed] [Google Scholar]
- 11.Camps M, Herman A, Loh E, Loeb LA. Genetic constraints on protein evolution. Crit Rev Biochem Mol Biol. 2007;42:313–326. doi: 10.1080/10409230701597642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Phillips GN., Jr The three-dimensional structure of green fluorescent protein and its implications for function and design. Methods Biochem Anal. 2006;47:67–82. [PubMed] [Google Scholar]
- 13.Crameri A, Whitehorn EA, Tate E, Stemmer WP. Improved green fluorescent protein by molecular evolution using DNA shuffling. Nat Biotechnol. 1996;14:315–319. doi: 10.1038/nbt0396-315. [DOI] [PubMed] [Google Scholar]
- 14.Hashimoto-Gotoh T, Inselburg J. ColE1 plasmid incompatibility: localization and analysis of mutations affecting incompatibility. J Bacteriol. 1979;139:608–619. doi: 10.1128/jb.139.2.608-619.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmidt L, Inselburg J. ColE1 copy number mutants. J Bacteriol. 1982;151:845–854. doi: 10.1128/jb.151.2.845-854.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fitzwater T, Zhang XY, Elble R, Polisky B. Conditional high copy number ColE1 mutants: resistance to RNA1 inhibition in vivo and in vitro. EMBO J. 1988;7:3289–3297. doi: 10.1002/j.1460-2075.1988.tb03196.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Merlin S, Polisky B. Analysis of establishment phase replication of the plasmid ColE1. J Mol Biol. 1993;230:137–150. doi: 10.1006/jmbi.1993.1131. [DOI] [PubMed] [Google Scholar]
- 18.Wang Z, Xiang L, Shao J, Wegrzyn A, Wegrzyn G. Effects of the presence of ColE1 plasmid DNA in Escherichia coli on the host cell metabolism. Microb Cell Fact. 2006;5:34. doi: 10.1186/1475-2859-5-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carapuca E, Azzoni AR, Prazeres DM, Monteiro GA, Mergulhao FJ. Time-course determination of plasmid content in eukaryotic and prokaryotic cells using real-time PCR. Mol Biotechnol. 2007;37:120–126. doi: 10.1007/s12033-007-0007-3. [DOI] [PubMed] [Google Scholar]
- 20.Kim D, Rhee Y, Rhodes D, Sharma V, Sorenson O, Greener A, Smider V. Directed evolution and identification of control regions of ColE1 plasmid replication origins using only nucleotide deletions. J Mol Biol. 2005;351:763–775. doi: 10.1016/j.jmb.2005.06.051. [DOI] [PubMed] [Google Scholar]
- 21.Castagnoli L, Lacatena RM, Cesareni G. Analysis of dominant copy number mutants of the plasmid pMB1. Nucleic Acids Res. 1985;13:5353–5367. doi: 10.1093/nar/13.14.5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wong TS, Roccatano D, Zacharias M, Schwaneberg U. A statistical analysis of random mutagenesis methods used for directed protein evolution. J Mol Biol. 2006;355:858–871. doi: 10.1016/j.jmb.2005.10.082. [DOI] [PubMed] [Google Scholar]
- 24.Ausubel FM. Short Protocols in Molecular Biology: A Compendium of Methods from Current Protocols in Molecular Biology. New York: Wiley; 1995. [Google Scholar]