Abstract
Cyclin-dependent kinases (CDKs) are required for initiation of DNA replication in all eukaryotes, and appear to act at multiple levels to control replication origin firing, depending on the cell type and stage of development. In early development of many animals, both invertebrate and vertebrate, rapid cell cycling is coupled with transcriptional repression, and replication initiates at closely spaced replication origins with little or no sequence specificity. This organisation of DNA replication is modified during development as cell proliferation becomes more controlled and defined. In all eukaryotic cells, CDKs promote conversion of “licensed” pre-replication complexes (pre-RC) to active initiation complexes. In certain circumstances, CDKs may also control pre-RC formation, transcription of replication factor genes, chromatin remodelling, origin spacing, and organisation of replication origin clusters and replication foci within the nucleus. Although CDK1 and CDK2 have overlapping roles, there is a limit to their functional redundancy. Here, I review these findings and their implications for development.
Keywords: Animals, Cell Division, genetics, physiology, Cyclin-Dependent Kinases, genetics, physiology, DNA Replication, genetics, physiology, Embryonic Development, genetics, physiology, Humans
1. Introduction
DNA replication, which commits a eukaryotic cell to dividing, can be considered a “passive driving force” for development, since it both creates a problem and provides a window of opportunity for doing something new. Due to cell division, even within an apparently simple organism with no development, such as yeast, not all cells are equal. In a colony of yeast cells, the cells in the middle of the colony are small and the cell cycle is arrested, due to nutrient starvation, whereas those at the edge of the colony are bigger and rapidly dividing. In this case, the size of the colony is limited by nutrient availability, and nutrient starvation provokes sexual reproduction among opposite mating types. Metazoans have solved the availability problem by centralizing ressources and despatching them to all cells, allowing them to become much bigger and uproot themselves in search of food, water, and everything else they need. To achieve this level of development, thousands of genes are required, and this itself creates several other problems, already faced by simpler organisms, but which require even greater organisation in metazoans: firstly, how to store all the genetic information within the nucleus, which is achieved by packaging into chromatin; and then, how to replicate it efficiently and reliably, while at the same time transcribing those genes when and where they are needed. Because the latter is a difficult problem to solve and requires lots of energy, once the animal has developed, most cells don’t bother. In an adult human being, only about 2% of the 1013 or so cells are dividing: reproductive cells, in males, and those cells that are needed to replace rapidly damaged cells to keep the organism alive, such as epithelial cells of the skin or intestine, and blood cells. During development, however, special mechanisms are required to coordinate cell proliferation with differentiation.
As well as creating problems, DNA replication provides a window of opportunity for epigenetic change, since, as the double helix is copied, the choice must be made of what to do with all the peripheral information encoded in chromatin determining if, when and where a gene should be transcribed. Because replication is coupled to chromatin assembly (Almouzni & Mechali, 1988; Worcel et al, 1978) it provides a mechanism for controlling gene expression. In Xenopus oocytes, replication of injected single-stranded DNA is sufficient to repress basal transcription at the encoded promoter (Almouzni & Wolffe, 1993). Inversely, in early mouse development, transcriptional activation of the zygotic genome occurs at the two cell stage, and requires DNA replication to relieve chromatin-mediated gene repression (Forlani et al, 1998). One way of achieving epigenetic change, during the course of DNA replication, would be to let parental and newly synthesized genes compete for limiting transcription “factors”, in the general sense of the word. Over uncountable generations, selection for favourable outcomes of this competition might explain the programme of DNA replication in which transcribed genes are, in general, replicated early, and silent genes later on, within a single S-phase (Goldman et al, 1984) - a timing that appears to be mechanistically important (Zhang et al, 2002). But there may be a more mechanical reason why replication timing and transcription might be coupled. Whether a gene is to be replicated or transcribed, the DNA double helix first has to be unwound in order for the DNA or RNA helicase to copy the encoded information. Thus, DNA “unwinding factors” promoting transcription might also promote formation of replication origins, which would, on average, lead to proximal sequences being replicated earlier than distal ones. This would certainly be energetically economical, and again, over time, selection should lead to natural association of transcriptional and replication control elements. Indeed, almost all origins of replication are located close to transcriptional control regions and frequently contain transcription factor binding sites (see below). How unwinding at origins of replication is controlled, where it occurs in the genome, what the nature of the timing programme is, and what the consequences of this organisation are for the development of the organism, are all fundamental unresolved questions. In this review, I will try to present a picture of what we know about how cyclin-dependent protein kinases (CDKs), those universal cell-cycle regulators, are involved in replication control, in the context of the development of Xenopus laevis (with frequent comparisons to other models). Xenopus is a wonderful model organism which has taught us much of what we know about transcriptional control, DNA replication, the cell cycle, development, and nuclear reprogramming (for further reading on this subject, see the entertaining autobiographical account by Sir John Gurdon, winner of the 2009 Lasker prize for medical research, in which he correctly predicted that we should soon be able to reprogram somatic cell nuclei to totipotency by expressing the right combination of factors; (Gurdon, 2006)). For a comprehensive review of molecular mechanisms of cell cycle control in Xenopus oocytes, eggs and early embryos, see the accompanying chapter by Gotoh and colleagues (Gotoh et al., 2011).
2. Organisation of DNA replication and transcription in the early Xenopus embryo
Unlike mammalian development in utero, development of fertilized frog eggs must occur in the absence of any further provision from the mother. Eggs are therefore large, in order to provide material self-sufficiency until the animal has developed enough to feed itself, ie the tadpole stage. Early development is therefore organised very differently than in mammals, the first goal being to rapidly increase cell number (figure 1). For a start, it is quick, the entire cell cycle taking little more than 20 minutes for the first twelve cycles. To achieve this, replication origins are very closely spaced, from 5–25kb apart, rather than the 50–300kb seen in somatic cells (Blow et al, 2001; Hyrien & Mechali, 1993), and are randomly positioned between different cells (Hyrien et al, 1995) and from one cell cycle to the next (Labit et al, 2008). Random positioning of origins is true also for early drosophila development (Shinomiya & Ina, 1991), in which cells are cycling rapidly, suggesting that this may be a general rule, with mammals perhaps being an exception. Secondly, early development occurs in the absence of transcription (Brown & Littna, 1966; Newport & Kirschner, 1982b). This is due to a competition between transcription factor assembly and replication-coupled chromatin-mediated repression, in which chromatin wins (Kimelman et al, 1987; Prioleau et al, 1994). The exponential increase in DNA content during the rapid early cell cycles eventually titrates and alleviates this repression, as demonstrated by manipulating the nuclear-cytoplasmic ratio (Newport & Kirschner, 1982a; Newport & Kirschner, 1982b). In Xenopus, as in mice, DNA replication around the mid-blastula transition (MBT) appears to be required for relief of the repressed state (Fisher & Mechali, 2003). Although much subsequent morphological development can occur in the complete absence of DNA replication, certain developmental abnormalities ensue, demonstrating that cell proliferation is, after all, required for correct development (Fisher & Mechali, 2003; Harris & Hartenstein, 1991; Rollins & Andrews, 1991). Whereas all cells are dividing prior to the MBT, the mitotic index rapidly drops to about 30% at early gastrula stages to less than 10% by mid-gastrulation, and becomes regionalised (Saka & Smith, 2001). The cell-cycle (and, by inference, onset of DNA replication) is therefore controlled in a tissue-specific manner during development. However, that differentiation can occur in the absence of DNA replication suggests that global transcriptional gene activation at the MBT, dependent on replication, is then followed by progressive, cell type-specific gene repression which does not require replication. Transcriptional activation at the MBT also coincides with specific positioning of replication origins (Hyrien et al, 1995), which possibly reflects the influence of transcription factors on nucleosome positioning and thereby, the accessibility of DNA to replication factors. A recent study in somatic human cells found that most replication origins occur in GC-rich regions overlapping with transcriptional regulatory elements (Cadoret et al, 2008), especially those of the AP1 family of “immediate early” transcription factors. Significantly, origins mapped to evolutionarily conserved regions, suggesting that origin positioning is conserved across animal species. As such, origins of replication might be coordinated with transcription during later development. Whether this simply reflects a situation of “convenience”, for example being energetically economical, or whether there are functional consequences of this origin positioning, are not known. Nevertheless, the absence of origin replication specificity in early development of Xenopus (and perhaps most animals other than mammals) is evidently associated with absence of transcription. The link between non-specific origins of DNA replication and transcriptional repression might be due to the necessity not only to replicate quickly, but perhaps also to maintain pluripotency of the dividing cells and prevent premature differentiation. Xenopus oocyte and egg extracts have an extraordinary capacity for nuclear reprogramming, and will even reprogram the nucleus within permeabolised cells. Such somatic cells introduced into oocytes are reset to a stem cell-like pattern of transcription in the absence of DNA replication (Byrne et al, 2003) whereas when introduced into egg extracts, all transcription is extinguished and DNA replication is activated (Alberio et al, 2005). However, not all nuclei replicate with the same efficiency in egg extracts. Terminally differentiated chromatin, for example, from erythrocytes (which are nucleated in Xenopus) replicates much more slowly in Xenopus egg extracts, due to far fewer replication origins being activated. Passage through mitosis eliminates this pattern of replication origin spacing, and resets it to an early embryonic pattern (Lemaitre et al, 2005). Where do CDKs come in? Recent reports from our lab and others have found that in Xenopus egg extracts, cyclin-dependent kinases control both the replication timing program (Thomson et al, 2010) and replication origin spacing (Krasinska et al, 2008a). Taken together, these results suggest that CDK-mediated control of DNA replication is different between early embryos and somatic cells, which presumably reflects a different organisation of chromatin within the nucleus. However, although CDKs are required for initiation of DNA replication in all eukaryotes, their general mechanism of action at this stage is not clear. In the accompanying review, Gotoh et al describe the molecular regulation of CDK activity by phosphorylation, dephosphorylation, degradation of the cyclin subunit and association with inhibitor proteins (Gotoh et al., 2011). We will focus more on the downstream events which depend on CDK activity.
Figure 10.1. Differences in organisation of DNA replication during development.
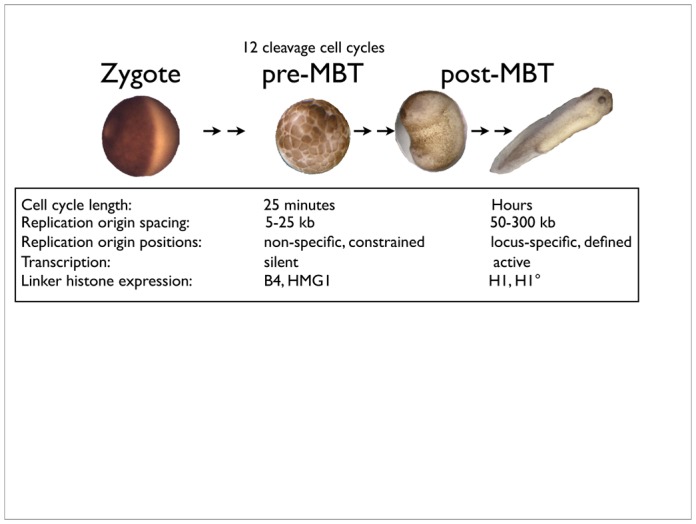
In early Xenopus embryonic development, the cell cycles are rapid and synchronous, whereas after the mid-blastula transition they appear much more highly regulated. Some functional differences which have implications for replication control are highlighted.
3. Cyclin-Dependent Kinases and the control of initiation of DNA replication
Initiation of DNA replication can be summarised as a series of sequential steps, in which DNA is first “licensed” to replicate by the formation of pre-replication complexes (pre-RC), which are then converted in a CDK-dependent manner to pre-initiation complexes (pre-IC) which unwind DNA, and DNA polymerase loading and elongation ensue (Walter & Newport, 2000). However, replication of single stranded DNA in egg extracts does not require these steps and can occur in the absence of CDK activity (Blow & Laskey, 1986; Blow & Nurse, 1990) indicating that unwinding of the double helix at replication origins is a rate-limiting step in DNA replication, at which CDKs act. Pre-RCs are formed by loading of the MCM2-7 heterohexamer, which has only limited intrinsic helicase activity, onto origins containing the ORC1-6 complex, in a Cdt1 and Cdc6-dependent manner. To form a replicative helicase competent to unwind DNA processively requires association of the GINS (Go Ichi Ni San) complex and Cdc45 with MCM2-7 (Pacek et al, 2006); figure 2), mirroring the situation in yeast (Gambus et al, 2006). Several studies in yeast suggest how CDKs can control this process (Masumoto et al, 2002; Tanaka et al, 2007; Zegerman & Diffley, 2007). Two components of DNA replication complexes, Sld2 and Sld3, which interact with the conserved replication proteins Cdc45 and Cut5/TopBP1/Dpb11, were identified as CDK substrates whose phosphorylation is essential for DNA replication. In the absence of CDK function, DNA replication could initiate, using yeast genetics to bypass the requirements for phosphorylation of Sld2 and Sld3, or with an activating mutation in the Sld3 interacting protein Cdc45. However, replication in these circumstances was inefficient, suggesting that whereas only two substrates might be indispensable, other substrates are also involved in promoting replication efficiency. The functions of Sld2 and Sld3 are not known, and there are no clear structural homologues of Sld2 and Sld3 in metazoans, although, given that other replication-origin complex proteins are functionally conserved (Cdt1, Cdc6, MCM2-7, Cdc45, GINS, MCM10, Cut5/TopBP1/Dpb11) they almost certainly exist. The RecQ4 helicase, mutated in Rothmund-Thomson syndrome, and essential for DNA replication in Xenopus egg extracts, has a small region of limited sequence homology to Sld2, but it does not require CDK activity nor the Cut5 homologue to bind to chromatin (Matsuno et al, 2006; Sangrithi et al, 2005). It is, however, highly phosphorylated in vivo, and can be phosphorylated in vitro by CDKs. Recent papers identified in Xenopus two new putative CDK substrates required for pre-IC formation and DNA replication, Treslin and Gemc1. Treslin binds to Cut5/TopBP1/Dpb11 in a CDK-dependent manner, and is essential for replication in Xenopus and cultured human cells, as it allows Cdc45 recruitment to replication origins (Kumagai et al, 2010). Gemc1 is a protein containing Geminin-like coiled-coil domains, hence the name, and appears to serve a very similar function to that reported for Treslin: Cdc45 loading (Balestrini et al, 2010). Gemc1 directly binds Cut5/ TopBP1/Dpb11, Cdc45 and Cdk2-cyclin E, and, like Treslin, can be phosphorylated in vitro by the latter. The respective roles of Treslin and Gemc1are so far up for grabs, but a recent bioinformatics paper reported that Treslin and Sld3 homologues almost certainly share a common ancestor, and may be functional homologues (Sanchez-Pulido et al, 2010). It remains possible that in vertebrates, additional levels of control of replication by CDKs might be important, and additional substrates required, to govern activation of origin clusters and control the replication timing program.
Figure 10.2. The molecular organisation at replication origins and its control by CDKs.
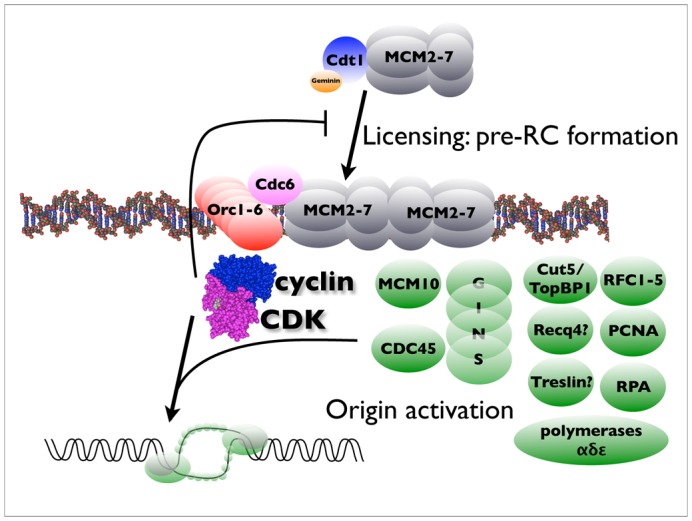
Replication origins are built in sequential steps : first, origins are licensed by the back-to-back loading of the double heterohexamer of MCM proteins around DNA, which requires the Orc complex, Cdc6, and a Cdt1-geminin complex. This step is inhibited by mitotic cyclin-dependent kinases. In a second step, which requires S-phase CDK activity, the pre-initiation complex (proteins in green) is loaded and DNA unwinding by the now processive helicase can occur.
One well known substrate of CDKs in metazoans, whose phosphorylation is a pre-requisite for the G1-S transition, is the retinoblastoma tumour suppressor, or pRb. The best known role of this protein, and its “pocket-protein” relatives p107 and p130, is to inactivate E2F-mediated transcription by formation of a direct repressive complex, in association with hbrm/BRG-1 (Trouche et al, 1997). Many E2F targets are required for DNA replication, such as Cdt1 (Yoshida & Inoue, 2004), Cdc6, PCNA, RFC, polymerase alpha, MCMs 3,5 and 6, RPA, ribonucleotide reductase, and so on (Ren et al, 2002). However, there is no transcription in Xenopus eggs or early embryos, and all components required for replication are already present. Nevertheless, pRb may have other functions repressive for DNA replication. In Xenopus egg extracts, addition of GST-pRb directly blocks DNA replication via a direct interaction with MCM7 which neutralizes helicase activity (Pacek & Walter, 2004; Sterner et al, 1998), and this repressive function can be alleviated by direct binding of Cyclin D1-CDK4 complexes (but not CDK2-cyclin E) (Gladden & Diehl, 2003). The same Rb-MCM7 interaction, in somatic mammalian cells, is involved in TGF-beta1-induced late G1 arrest after pre-RCs have already formed, and may thus be a conserved and physiologically relevant mechanism to regulate DNA replication in response to cell signalling (Mukherjee et al, 2010). Finally, Rb recruits histone deacetylases to chromatin (Magnaghi-Jaulin et al, 1998), providing a third potential mechanism of repression of DNA replication, either indirectly, by further inhibiting E2F-dependent transcription, or perhaps, by more directly repressing formation of replication origins. In metazoans, CDKs therefore probably control initiation of DNA replication not only by directly phosphorylating components of pre-ICs, but also by alleviating Rb-mediated chromatin repression (figure 3).
Figure 10.3. Multiple potential roles for the Rb protein, a CDK substrate, in replication control.
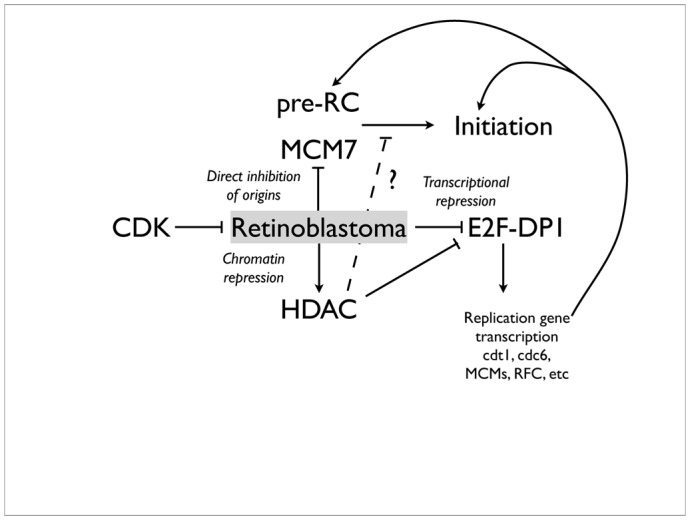
See main text for details.
4. Chromatin and replication control by CDKs
The consequences of chromatin organisation for replication initiation are not well understood. Chromosomal DNA is highly organised, forming supercoils around nucleosomes, which are themselves compacted into higher-order structures. Post-translational modifications of core histones, including acetylation and phosphorylation, regulate nucleosome mobility and DNA condensation. We have found in Xenopus that histone acetylases and CDKs act at the same point to promote replication initiation, and their inhibition is synergistic in inhibiting DNA replication (Krasinska et al, 2008b). These and other results suggest that CDKs are likely to phosphorylate other substrates, possibly involved in chromatin remodelling to make DNA replication-competent. Indeed, histone acetyl-transferase activity peaks at the G1/S transition in somatic cells, due to CDK2-cyclin E-mediated stimulation of p300/CBP (Ait-Si-Ali et al, 1998), and anacardic acid, which blocks p300, prevents DNA replication in Xenopus egg extracts (Krasinska et al, 2008b; Lemaitre et al, 2005). Linker histone modifications might also control replication in a developmentally regulated manner. In somatic cells, phosphorylation of the linker histone H1, the archetypal CDK substrate, which binds linker DNA flanking the nucleosome core to stabilise higher-order chromatin structure (Wolffe, 1997) is mediated by CDK2 during DNA replication, promoting chromosome decondensation (Alexandrow & Hamlin, 2005). Whether or not CDK-mediated phosphorylation of histone H1 is required for replication is not known. Nevertheless, histone H1 is absent from chromatin in early development in Xenopus, with the embryonic histone H1 variant B4 sunstituting (Smith et al, 1988). Introduction of histone H1-containing somatic chromatin into an egg extract leads to replacement of H1 and the somatic H1 variant H1° by B4 and HMG1, which have similar functions (Dimitrov & Wolffe, 1996; Nightingale et al, 1996). This may have functional consequences for DNA replication, since addition of recombinant H1 to an egg extract reduces replication origin firing (Lu et al, 1998). B4 does not contain any consensus sites for phosphorylation by CDKs, whereas H1 contains five. Possibly, histone H1 might titrate CDK activity; alternatively, by compacting chromatin, it might reduce accessibility of DNA to origin components. CDKs are also required for replication-coupled histone H2B and H4 gene transcription, which is essential for S-phase, by phosphorylation of the p220 NPAT transcriptional activator (Ma et al, 2000; Zhao et al, 2000).
5. Functional redundancy of CDK-cyclin complexes, and its limits
Which CDKs are involved in the initiation of DNA replication? In metazoans, CDK2 clearly plays an important role in regulating the G1 to S-phase transition, and was originally thought to be essential for DNA replication in Xenopus egg extracts (Fang & Newport, 1991). The roles of CDK3, so far only described in mammals, are not clear, but while dispensable for the cell cycle in mice, in some circumstances it may regulate entry into S-phase via phosphorylation of pRb family proteins and promotion of E2F transcription (Hofmann & Livingston, 1996; Ren & Rollins, 2004). Mammalian CDK4 and CDK6 phosphorylate the retinoblastoma family of tumour suppressors, and can thus also control passage though the commitment point of serum-independence in G1, and exit from the cell cycle (Sherr, 1995). In Xenopus, while CDK4 is expressed later in development (Goisset et al, 1998), it is not clear whether any CDK4 protein is present in early cell cycles, and a CDK6 homologue has not been identified. In vertebrates, many cell types can proliferate in the absence of CDK2, due to compensation by CDK1 (Aleem et al, 2005; Hochegger et al, 2007; Ortega et al, 2003; Tetsu & McCormick, 2003). Indeed, it was recently found that in mice, embryonic cells can proliferate in the simultaneous absence of CDK2, CDK3, CDK4 and CDK6, only CDK1 having cell-cycle functions that cannot be compensated for by other CDKs (Santamaria et al, 2007). These recent results in animal cells are reminiscent of our earlier studies in fission yeast, in which a single mitotic CDK-cyclin complex can promote both DNA replication and mitosis (Fisher & Nurse, 1996). Even in Xenopus egg extracts, mitotic cyclin B can promote DNA replication in the absence of the S-phase promoting cyclin E, providing a nuclear-localisation signal is provided (Moore et al, 2003), suggesting that a CDK only has to be in the right place at the right time to phosphorylate its substrates, providing it has similar substrate specificity. The latter appears not to be a problem, which is not surprising given that CDK-consensus sites are extremely simple (in most cases, a solvent-accessible SP or TP sequence suffices). On the other hand, some cell types do require certain CDKs or cyclins in order to proliferate, and individual CDK knockout mice reveal many pathologies. For example, CDK2−/− mice are sterile, CDK4−/− mice are diabetic, and so on. This might reflect expression profiles of CDKs or cyclins within the particular tissue. For example, knockout of A-type cyclins in mice does not affect fibroblast proliferation, and in this case, cyclin E expression becomes upregulated throughout the cell cycle, but it does affect proliferation of embryonic stem cells and haematopoietic stem cells, which normally have high cyclin A expression (Kalaszczynska et al, 2009). Conversely, deletion of both E-type cyclins in mice causes embryonic lethality due to inability of placental trophoblast cells to endoreplicate (Geng et al, 2003), CDK2-cyclin A apparently unable to substitute, whereas CDK2 deletion has no effect on these cells, suggesting that differences between cyclin A and cyclin E are more important than differences between CDK1 and CDK2. Yet fundamentally, cyclin A and cyclin E can do the same things: in Xenopus egg extracts, S-phase promoting activity can be provided by either cyclin A-CDK1 or cyclin E-CDK2, albeit with different efficiencies (Strausfeld et al, 1996). Therefore, cyclin A and cyclin E associated kinases are functionally redundant in some circumstances, but are required in certain specific contexts, probably in part due to developmentally-regulated or tissue-specific expression profiles. Differences in expression appear to explain why CDK1-cyclin E does not support normal meiosis in mice, because even expression of CDK2 from the CDK1 locus cannot replace the endogenous CDK2 gene for meiotic function (Satyanarayana et al, 2008). However, different CDK-cyclin complexes probably have different kinetic parameters (affinity for substrates and catalytic activity) - this is certainly true at least in yeast (Loog & Morgan, 2005). Because knock-in of CDK2 into the CDK1 locus in mice cannot replace CDK1 function for the mitotic cycle (Satyanarayana et al, 2008) differences in kinetic parameters between CDK2 and CDK1 must also exist, and have important functional consequences. Therefore, functional redundancy has its limits.
By analogy with the situation in mammals, CDK2 might be expected to be dispensable per se for DNA replication in Xenopus, although it was originally found to be essential (Fang & Newport, 1991). Possibly, in Xenopus, CDK2 might be required for rapid DNA replication occurring in egg extracts, reflecting the different organisation of DNA replication in the early embryo, compared to somatic cells. We therefore recently reinvestigated whether CDK1 and CDK2, and cyclin A and cyclin E, are redundant in Xenopus egg extracts, an early embryonic system, using single molecule DNA combing to investigate their respective influence on replication origin organisation in Xenopus egg extracts. We found that CDK1-cyclin A actually is involved in DNA replication even in the presence of CDK2-cyclin E; however, only very low CDK activities (of either CDK1 or CDK2) are sufficient to promote replication initiation, and there was an important difference between CDK1 and CDK2 efficiency in promoting replication origin firing (Krasinska et al, 2008a). CDK2-cyclin E is indeed rate-limiting for DNA replication in these circumstances, as depletion of either subsunit reduced replication efficiency to around 30% of the control. However, the remaining 30% replication was no longer dependent on CDK2, but, rather, on CDK1. Depletion of either CDK1 or cyclin A, its main cyclin partner in extracts, slightly but reproducibly delayed DNA replication in the presence of CDK2, and activity of this complex was essential for DNA replication in the absence of CDK2. Surprisingly, however, at the level of individual replication origins, the effects of depleting CDK1 or cyclin A ostensibly appear similar to those of depleting CDK2 or cyclin E, in that the average inter-origin distance is approximately doubled to between 40 and 50kb. This means that both CDK1 and CDK2 complexes stimulate firing of individual replication origins, and do not compensate for each other at the individual origin level. However, the main limitation of DNA combing is DNA breakage between replication origins, with the average fibre length being around 100kb. Thus, many inter-origin distances cannot be measured - those between external origins on each fibre. When the number of initiation events per kb of DNA are calculated, irrespective of whether or not origins are present on combed fibres, it can be seen that CDK2 and cyclin E are much more important than CDK1 and cyclin A. In other words, replication origins are clustered, and both CDK1 and CDK2 affect the numbers of replication origins within the cluster, whereas only CDK2 appears to be limiting for the number of clusters firing (figure 4). This probably has something to do, firstly, with how replication origins are organized within the nucleus, and secondly, with different efficiencies of CDK1 and CDK2 for phosphorylating whatever substrates are required for activation of origin clusters.
Figure 10.4. Higher nuclear organisation in replication control.
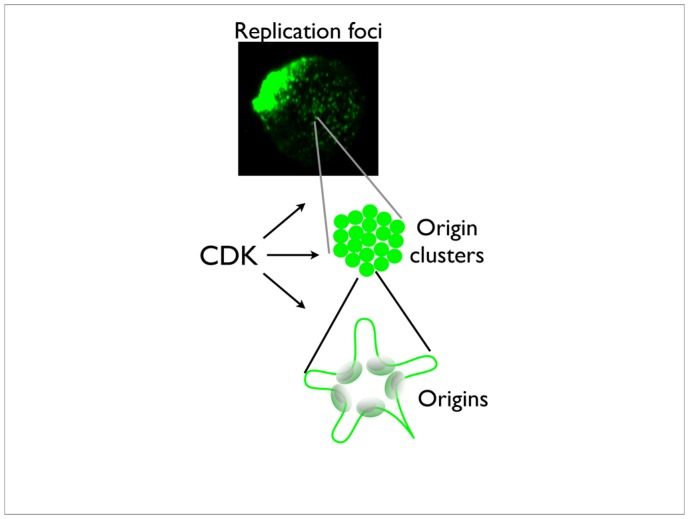
The organisation of replication in the nucleus is not yet well understood. In the generally accepted current model, origins of replication are clustered (bottom) and chromatin loops out from the origins. The origin clusters are probably also organised into ”factories” (middle) which can be visualised microscopically as replication foci (top). CDKs have roles at these different levels of subnuclear structure, as explained in the main text.
Finally, CDKs not only trigger origin activation, but they also prevent it. As reviewed by Gotoh et al in the accompanying chapter (Gotoh et al., 2011), DNA replication must be limited to once per cell cycle. This is partly achieved by control of Cdt1, which is exported from nuclei upon origin firing (Maiorano et al., 2005) and is sequestered by geminin, making it limiting for replication initiation, but also by high CDK activities preventing DNA replication licensing by hyperphosphorylation of ORC and MCM proteins, inhibiting their binding to chromatin (Findeisen et al., 1999; Mahbubani et al., 1997). Thus, the increasing CDK activity that occurs due to synthesis of cyclin A and cyclin B during DNA replication provides a simple way of preventing rereplication and ensuring the temporal separation of S-phase and mitosis.
6. Replication origin organisation, replication timing, and control by CDKs
How origins are organized into clusters, and the relationship of clusters to replication foci, is something of a mystery. In Xenopus, replication foci, associated with large, megabase scale DNA regions, colocalise from one cell-cycle to the next, even though individual replication origins do not (Labit et al, 2008). In somatic cells, the pattern of replication foci changes throughout S-phase, suggesting that early and late-replicating DNA is associated with different subnuclear structures. In Xenopus, concommittant with changes in origin specification at the midblastula transition are changes in attachment of DNA to the nuclear matrix (Vassetzky et al, 2000), which, as with replication origins, changes from random to specific site attachment. This is too much of a coincidence for the origin positioning not to be related to chromatin organisation at the structural level. Although the nature of the nuclear matrix itself is debated, “anti-matrix” proponents writing off the visible skeleton seen on electron microscopy of matrix preparations as a precipitation artefact of the extraction conditions (see (Pederson, 2000), the inference is that the internal structure of the nucleus itself is different in early development prior to the midblastula transition. Indeed, the length of replicons in early and later development correlates with the “halo” radius, ie the length of chromatin loops between sites of attachment to the matrix (Lemaitre et al, 2005), suggesting that replication origins might be located at the base of the loops and associated with the matrix, whether or not the latter is soluble or filamentous. In early embryonic development, activation of origin clusters appears to be the rate-limiting step for the overall speed of DNA replication, and the inference from our results is that CDK1-cyclin A is simply less efficient at promoting this step than CDK2-cyclin E. By analogy with yeast, in which different cyclin-CDK1 combinations have different Michaelis constants (Km) for typical S-phase and M-phase substrates (Loog & Morgan, 2005), reflecting differential affinity of the protein substrate for one or other cyclins, I suggest that the same is likely to be true for metazoan CDK-cyclin complexes. Potentially, therefore, early and late-replicating origins could be preferentially controlled by different CDK-cyclin complexes, reflecting their different organisation within the nucleus (figure 4). There has been some recent evidence for this in somatic cells, with CDK1-cyclin A controlling firing of late origins, even in the presence of CDK2 (Katsuno et al, 2009).
Although there is no obvious timing program of origin firing in early Xenopus development, nevertheless, use of statistical methods demonstrates that clustering of origins occurs, that clusters fire at different times even within the rapid S-phase of Xenopus egg extracts, and that cluster firing and overall initiation rate is limited by the nuclear - cytoplasm ratio and a constitutively active S-phase checkpoint (Blow et al, 2001; Marheineke & Hyrien, 2004). A more obvious timing program can be reproduced in Xenopus egg extracts using somatic cell nuclei. In this case, the level of CDK activity in the extract controls both the number of replication foci and the fraction of DNA replicated early and late (Thomson et al, 2010). This again points to CDK requirements for replication being related to the structural organisation of DNA within the nucleus.
7. CDK requirements for S-phase entry from a quiescent state
Assuming the above statement to be true, one might infer that nuclear substructure is different when cells are already cycling (such as in early development in Xenopus), from when cells are in a quiescent state (most of the cells, most of the rest of the time), because requirements for CDKs to replicate are different. Thus, cyclin E is dispensable for cell-cycling during the majority of early development in mice, but is required for cells to enter S-phase from a quiescent state (Geng et al, 2003). This requirement was proposed to be independent of the ability of cyclin E to activate its associated CDK, since cell-cycle entry could be rescued using a mutant cyclin E, apparently incapable of activating CDK2 but which appears to link Cdt1 to the MCM helicase (Geng et al, 2007). Nevertheless, CDK requirements for initiating DNA replication appear to be vanishingly low (Krasinska et al, 2008a) and we can detect basal CDK2 activity against histone H1 in the complete absence of cyclins (CDK2 expressed and purified from in E.coli) providing it is phosphorylated by CAK (unpublished data). Thus, it cannot yet be ruled out that cyclin E promotes cell-cycle entry from a quiescent state by activating a CDK to low levels attainable even by mutant cyclin E, or by cyclin-E dependent association of basal (ie cyclin-independent) CDK activity with the correct substrates, and this is not normally required once cells are happily cycling. Could this be because the nuclear structure is altered once cells become quiescent, and remodelling the nucleus to make DNA competent to replicate requires particular CDK activities?
Whether or not this is the case, other CDK-dependent mechanisms operate during cell cycle entry of quiescent somatic cells. In cycling mammalian cells, replication origins become licensed (coincident with MCM loading onto chromatin) at the end of mitosis (Okuno et al, 2001), whereas during cell-cycle exit, at least one protein required for licensing, Cdc6, is targeted for destruction by APC/C, preventing inappropriate DNA replication. Stabilization of Cdc6 requires CDK-mediated phosphorylation dependent on cyclin E (whose degradation is promoted by an alternative ubiquitin-ligase, SCF), and this is critical for forming pre-RCs and entering S-phase from quiescence (Mailand & Diffley, 2005). Interestingly, if Cdc6 expression is restricted to G1, CDK activity becomes essential for pre-RC formation in cycling cells. The inference is that CDK activity is dispensable for pre-RC formation (indeed, it inhibits pre-RC formation) because pre-mitotically stabilised Cdc6 is sufficient for pre-RC formation post-mitosis.
8. Conclusions
In summary, therefore, CDKs probably control DNA replication at multiple steps, including transcription, pre-RC formation, the pre-RC to pre-IC transition, chromatin remodelling and regulation of higher order nuclear structure, depending on the cell-type and developmental context (table 1). In a hypothetical ideal world, it should be possible to understand cell cycle control and the relative contributions of different regulators, from a knowledge of interaction kinetics of all different molecules. More realistically, we should at least be able to understand how different CDKs are involved in cell cycle control, why some appear essential in some circumstances but not others, and the consequences of using particular inhibitors, from analysis of kinetics of phosphorylation of their substrates in a physiological context. To move towards this goal, it is obvious that we first need to know what the substrates are, determine their kinetics of phosphorylation by different CDKs, and study how CDK-mediated phosphorylation regulates their function in a physiological system. The emerging picture is that the increasing diversity of metazoan CDKs and cyclins undoubtedly reflects the selective advantage from having CDK-cyclin complexes with different affinities, which, while not essential in any given situation, may provide for flexibility in replicating DNA at different speeds and different times.
Table 10.1.
CDK substrates in DNA replication, and their cellular functions.
| Physiological process | Substrates |
|---|---|
| Chromatin remodelling | HAT, Rb, Histone H1, others? |
| Transcription | Rb, NPAT |
| Pre-RC formation | Cdc6 |
| Origin activation | Rb, Treslin? Recq4? others? |
| Origin cluster/replication foci activation | Unknown |
Acknowledgments
Daniel Fisher is an employee of Inserm. This work benefited from ARC grant 1047, and the laboratory is currently funded by a grant from the Agence Nationale de la Recherche ANR-09-BLAN-0252-01, and from the Ligue Nationale Contre le Cancer, EL2010.LNCC/DF.
References
- Ait-Si-Ali S, et al. Histone acetyltransferase activity of CBP is controlled by cycle-dependent kinases and oncoprotein E1A. Nature. 1998;396:184–186. doi: 10.1038/24190. [DOI] [PubMed] [Google Scholar]
- Alberio R, Johnson AD, Stick R, Campbell KH. Differential nuclear remodeling of mammalian somatic cells by Xenopus laevis oocyte and egg cytoplasm. Exp Cell Res. 2005;307:131–141. doi: 10.1016/j.yexcr.2005.02.028. [DOI] [PubMed] [Google Scholar]
- Aleem E, Kiyokawa H, Kaldis P. Cdc2-cyclin E complexes regulate the G1/S phase transition. Nat Cell Biol. 2005;7:831–836. doi: 10.1038/ncb1284. [DOI] [PubMed] [Google Scholar]
- Alexandrow MG, Hamlin JL. Chromatin decondensation in S-phase involves recruitment of Cdk2 by Cdc45 and histone H1 phosphorylation. J Cell Biol. 2005;168:875–886. doi: 10.1083/jcb.200409055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almouzni G, Mechali M. Assembly of spaced chromatin promoted by DNA synthesis in extracts from Xenopus eggs. EMBO J. 1988;7:665–672. doi: 10.1002/j.1460-2075.1988.tb02861.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almouzni G, Wolffe AP. Replication-coupled chromatin assembly is required for the repression of basal transcription in vivo. Genes Dev. 1993;7:2033–2047. doi: 10.1101/gad.7.10.2033. [DOI] [PubMed] [Google Scholar]
- Balestrini A, Cosentino C, Errico A, Garner E, Costanzo V. GEMC1 is a TopBP1-interacting protein required for chromosomal DNA replication. Nat Cell Biol. 2010;12:484–491. doi: 10.1038/ncb2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow JJ, Gillespie PJ, Francis D, Jackson DA. Replication origins in Xenopus egg extract Are 5–15 kilobases apart and are activated in clusters that fire at different times. J Cell Biol. 2001;152:15–25. doi: 10.1083/jcb.152.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow JJ, Laskey RA. Initiation of DNA replication in nuclei and purified DNA by a cell-free extract of Xenopus eggs. Cell. 1986;47:577–587. doi: 10.1016/0092-8674(86)90622-7. [DOI] [PubMed] [Google Scholar]
- Blow JJ, Nurse P. A cdc2-like protein is involved in the initiation of DNA replication in Xenopus egg extracts. Cell. 1990;62:855–862. doi: 10.1016/0092-8674(90)90261-c. [DOI] [PubMed] [Google Scholar]
- Brown DD, Littna E. Synthesis and accumulation of DNA-like RNA during embryogenesis of Xenopus laevis. J Mol Biol. 1966;20:81–94. doi: 10.1016/0022-2836(66)90119-7. [DOI] [PubMed] [Google Scholar]
- Byrne JA, Simonsson S, Western PS, Gurdon JB. Nuclei of adult mammalian somatic cells are directly reprogrammed to oct-4 stem cell gene expression by amphibian oocytes. Curr Biol. 2003;13:1206–1213. doi: 10.1016/s0960-9822(03)00462-7. [DOI] [PubMed] [Google Scholar]
- Cadoret JC, Meisch F, Hassan-Zadeh V, Luyten I, Guillet C, Duret L, Quesneville H, Prioleau MN. Genome-wide studies highlight indirect links between human replication origins and gene regulation. Proc Natl Acad Sci U S A. 2008;105:15837–15842. doi: 10.1073/pnas.0805208105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov S, Wolffe AP. Remodeling somatic nuclei in Xenopus laevis egg extracts: molecular mechanisms for the selective release of histones H1 and H1(0) from chromatin and the acquisition of transcriptional competence. EMBO J. 1996;15:5897–5906. [PMC free article] [PubMed] [Google Scholar]
- Fang F, Newport JW. Evidence that the G1-S and G2-M transitions are controlled by different cdc2 proteins in higher eukaryotes. Cell. 1991;66:731–742. doi: 10.1016/0092-8674(91)90117-h. [DOI] [PubMed] [Google Scholar]
- Findeisen M, El-Denary M, Kapitza T, Graf R, Strausfeld U. Cyclin A-dependent kinase activity affects chromatin binding of ORC, Cdc6, and MCM in egg extracts of Xenopus laevis. Eur J Biochem. 1999;264:415–426. doi: 10.1046/j.1432-1327.1999.00613.x. [DOI] [PubMed] [Google Scholar]
- Fisher D, Mechali M. Vertebrate HoxB gene expression requires DNA replication. Embo J. 2003;22:3737–3748. doi: 10.1093/emboj/cdg352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher DL, Nurse P. A single fission yeast mitotic cyclin B p34cdc2 kinase promotes both S-phase and mitosis in the absence of G1 cyclins. Embo J. 1996;15:850–860. [PMC free article] [PubMed] [Google Scholar]
- Forlani S, Bonnerot C, Capgras S, Nicolas JF. Relief of a repressed gene expression state in the mouse 1-cell embryo requires DNA replication. Development. 1998;125:3153–3166. doi: 10.1242/dev.125.16.3153. [DOI] [PubMed] [Google Scholar]
- Gambus A, Jones RC, Sanchez-Diaz A, Kanemaki M, van Deursen F, Edmondson RD, Labib K. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat Cell Biol. 2006;8:358–366. doi: 10.1038/ncb1382. [DOI] [PubMed] [Google Scholar]
- Geng Y, et al. Kinase-independent function of cyclin E. Mol Cell. 2007;25:127–139. doi: 10.1016/j.molcel.2006.11.029. [DOI] [PubMed] [Google Scholar]
- Geng Y, et al. Cyclin E ablation in the mouse. Cell. 2003;114:431–443. doi: 10.1016/s0092-8674(03)00645-7. [DOI] [PubMed] [Google Scholar]
- Gladden AB, Diehl JA. The cyclin D1-dependent kinase associates with the pre-replication complex and modulates RB.MCM7 binding. J Biol Chem. 2003;278:9754–9760. doi: 10.1074/jbc.M212088200. [DOI] [PubMed] [Google Scholar]
- Goisset C, Boucaut JC, Shi DL. Identification and developmental expression of cyclin-dependent kinase 4 gene in Xenopus laevis. Mech Dev. 1998;70:197–200. doi: 10.1016/s0925-4773(97)00184-6. [DOI] [PubMed] [Google Scholar]
- Goldman MA, Holmquist GP, Gray MC, Caston LA, Nag A. Replication timing of genes and middle repetitive sequences. Science. 1984;224:686–692. doi: 10.1126/science.6719109. [DOI] [PubMed] [Google Scholar]
- Gotoh T, Villa LM, Capelluto DGS, Finkielstein CV. Regulatory Pathways Coordinating Cell Cycle Progression in Early Xenopus Development. In: Kubiak JZ, editor. The cell cycle in development. Springer Verlag; 2011. [DOI] [PubMed] [Google Scholar]
- Gurdon JB. From nuclear transfer to nuclear reprogramming: the reversal of cell differentiation. Annu Rev Cell Dev Biol. 2006;22:1–22. doi: 10.1146/annurev.cellbio.22.090805.140144. [DOI] [PubMed] [Google Scholar]
- Harris WA, Hartenstein V. Neuronal determination without cell division in Xenopus embryos. Neuron. 1991;6:499–515. doi: 10.1016/0896-6273(91)90053-3. [DOI] [PubMed] [Google Scholar]
- Hochegger H, Dejsuphong D, Sonoda E, Saberi A, Rajendra E, Kirk J, Hunt T, Takeda S. An essential role for Cdk1 in S phase control is revealed via chemical genetics in vertebrate cells. J Cell Biol. 2007;178:257–268. doi: 10.1083/jcb.200702034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann F, Livingston DM. Differential effects of cdk2 and cdk3 on the control of pRb and E2F function during G1 exit. Genes Dev. 1996;10:851–861. doi: 10.1101/gad.10.7.851. [DOI] [PubMed] [Google Scholar]
- Hyrien O, Maric C, Mechali M. Transition in specification of embryonic metazoan DNA replication origins. Science. 1995;270:994–997. doi: 10.1126/science.270.5238.994. [DOI] [PubMed] [Google Scholar]
- Hyrien O, Mechali M. Chromosomal replication initiates and terminates at random sequences but at regular intervals in the ribosomal DNA of Xenopus early embryos. EMBO J. 1993;12:4511–4520. doi: 10.1002/j.1460-2075.1993.tb06140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalaszczynska I, et al. Cyclin A is redundant in fibroblasts but essential in hematopoietic and embryonic stem cells. Cell. 2009;138:352–365. doi: 10.1016/j.cell.2009.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuno Y, et al. Cyclin A-Cdk1 regulates the origin firing program in mammalian cells. Proc Natl Acad Sci U S A. 2009;106:3184–3189. doi: 10.1073/pnas.0809350106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimelman D, Kirschner M, Scherson T. The events of the midblastula transition in Xenopus are regulated by changes in the cell cycle. Cell. 1987;48:399–407. doi: 10.1016/0092-8674(87)90191-7. [DOI] [PubMed] [Google Scholar]
- Krasinska L, Besnard E, Cot E, Dohet C, Mechali M, Lemaitre JM, Fisher D. Cdk1 and Cdk2 activity levels determine the efficiency of replication origin firing in Xenopus. EMBO J. 2008a;27:758–769. doi: 10.1038/emboj.2008.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasinska L, Cot E, Fisher D. Selective chemical inhibition as a tool to study Cdk1 and Cdk2 functions in the cell cycle. Cell cycle. 2008b;7:1702–1708. doi: 10.4161/cc.7.12.6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A, Shevchenko A, Dunphy WG. Treslin collaborates with TopBP1 in triggering the initiation of DNA replication. Cell. 2010;140:349–359. doi: 10.1016/j.cell.2009.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labit H, Perewoska I, Germe T, Hyrien O, Marheineke K. DNA replication timing is deterministic at the level of chromosomal domains but stochastic at the level of replicons in Xenopus egg extracts. Nucleic Acids Res. 2008;36:5623–5634. doi: 10.1093/nar/gkn533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre JM, Danis E, Pasero P, Vassetzky Y, Mechali M. Mitotic remodeling of the replicon and chromosome structure. Cell. 2005;123:787–801. doi: 10.1016/j.cell.2005.08.045. [DOI] [PubMed] [Google Scholar]
- Loog M, Morgan DO. Cyclin specificity in the phosphorylation of cyclin-dependent kinase substrates. Nature. 2005;434:104–108. doi: 10.1038/nature03329. [DOI] [PubMed] [Google Scholar]
- Lu ZH, Sittman DB, Romanowski P, Leno GH. Histone H1 reduces the frequency of initiation in Xenopus egg extract by limiting the assembly of prereplication complexes on sperm chromatin. Mol Biol Cell. 1998;9:1163–1176. doi: 10.1091/mbc.9.5.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma T, et al. Cell cycle-regulated phosphorylation of p220(NPAT) by cyclin E/Cdk2 in Cajal bodies promotes histone gene transcription. Genes Dev. 2000;14:2298–2313. doi: 10.1101/gad.829500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnaghi-Jaulin L, Groisman R, Naguibneva I, Robin P, Lorain S, Le Villain JP, Troalen F, Trouche D, Harel-Bellan A. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature. 1998;391:601–605. doi: 10.1038/35410. [DOI] [PubMed] [Google Scholar]
- Mahbubani HM, Chong JP, Chevalier S, Thommes P, Blow JJ. Cell cycle regulation of the replication licensing system: involvement of a Cdk-dependent inhibitor. J Cell Biol. 1997;136:125–135. doi: 10.1083/jcb.136.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailand N, Diffley JF. CDKs promote DNA replication origin licensing in human cells by protecting Cdc6 from APC/C-dependent proteolysis. Cell. 2005;122:915–926. doi: 10.1016/j.cell.2005.08.013. [DOI] [PubMed] [Google Scholar]
- Maiorano D, Krasinska L, Lutzmann M, Mechali M. Recombinant Cdt1 induces rereplication of G2 nuclei in Xenopus egg extracts. Curr Biol. 2005;15:146–153. doi: 10.1016/j.cub.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Marheineke K, Hyrien O. Control of replication origin density and firing time in Xenopus egg extracts: role of a caffeine-sensitive, ATR-dependent checkpoint. J Biol Chem. 2004;279:28071–28081. doi: 10.1074/jbc.M401574200. [DOI] [PubMed] [Google Scholar]
- Masumoto H, Muramatsu S, Kamimura Y, Araki H. S-Cdk-dependent phosphorylation of Sld2 essential for chromosomal DNA replication in budding yeast. Nature. 2002;415:651–655. doi: 10.1038/nature713. [DOI] [PubMed] [Google Scholar]
- Matsuno K, Kumano M, Kubota Y, Hashimoto Y, Takisawa H. The N-terminal noncatalytic region of Xenopus RecQ4 is required for chromatin binding of DNA polymerase alpha in the initiation of DNA replication. Mol Cell Biol. 2006;26:4843–4852. doi: 10.1128/MCB.02267-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JD, Kirk JA, Hunt T. Unmasking the S-phase-promoting potential of cyclin B1. Science. 2003;300:987–990. doi: 10.1126/science.1081418. [DOI] [PubMed] [Google Scholar]
- Mukherjee P, Winter SL, Alexandrow MG. Cell cycle arrest by transforming growth factor beta1 near G1/S is mediated by acute abrogation of prereplication complex activation involving an Rb-MCM interaction. Mol Cell Biol. 2010;30:845–856. doi: 10.1128/MCB.01152-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newport J, Kirschner M. A major developmental transition in early Xenopus embryos: I. characterization and timing of cellular changes at the midblastula stage. Cell. 1982a;30:675–686. doi: 10.1016/0092-8674(82)90272-0. [DOI] [PubMed] [Google Scholar]
- Newport J, Kirschner M. A major developmental transition in early Xenopus embryos: II. Control of the onset of transcription. Cell. 1982b;30:687–696. doi: 10.1016/0092-8674(82)90273-2. [DOI] [PubMed] [Google Scholar]
- Nightingale K, Dimitrov S, Reeves R, Wolffe AP. Evidence for a shared structural role for HMG1 and linker histones B4 and H1 in organizing chromatin. EMBO J. 1996;15:548–561. [PMC free article] [PubMed] [Google Scholar]
- Okuno Y, McNairn AJ, den Elzen N, Pines J, Gilbert DM. Stability, chromatin association and functional activity of mammalian pre-replication complex proteins during the cell cycle. EMBO J. 2001;20:4263–4277. doi: 10.1093/emboj/20.15.4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega S, Prieto I, Odajima J, Martin A, Dubus P, Sotillo R, Barbero JL, Malumbres M, Barbacid M. Cyclin-dependent kinase 2 is essential for meiosis but not for mitotic cell division in mice. Nat Genet. 2003;35:25–31. doi: 10.1038/ng1232. [DOI] [PubMed] [Google Scholar]
- Pacek M, Tutter AV, Kubota Y, Takisawa H, Walter JC. Localization of MCM2–7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol Cell. 2006;21:581–587. doi: 10.1016/j.molcel.2006.01.030. [DOI] [PubMed] [Google Scholar]
- Pacek M, Walter JC. A requirement for MCM7 and Cdc45 in chromosome unwinding during eukaryotic DNA replication. Embo J. 2004;23:3667–3676. doi: 10.1038/sj.emboj.7600369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson T. Half a century of “the nuclear matrix”. Mol Biol Cell. 2000;11:799–805. doi: 10.1091/mbc.11.3.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prioleau MN, Huet J, Sentenac A, Mechali M. Competition between chromatin and transcription complex assembly regulates gene expression during early development. Cell. 1994;77:439–449. doi: 10.1016/0092-8674(94)90158-9. [DOI] [PubMed] [Google Scholar]
- Ren B, Cam H, Takahashi Y, Volkert T, Terragni J, Young RA, Dynlacht BD. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev. 2002;16:245–256. doi: 10.1101/gad.949802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren S, Rollins BJ. Cyclin C/cdk3 promotes Rb-dependent G0 exit. Cell. 2004;117:239–251. doi: 10.1016/s0092-8674(04)00300-9. [DOI] [PubMed] [Google Scholar]
- Rollins MB, Andrews MT. Morphogenesis and regulated gene activity are independent of DNA replication in Xenopus embryos. Development. 1991;112:559–569. doi: 10.1242/dev.112.2.559. [DOI] [PubMed] [Google Scholar]
- Saka Y, Smith JC. Spatial and temporal patterns of cell division during early Xenopus embryogenesis. Dev Biol. 2001;229:307–318. doi: 10.1006/dbio.2000.0101. [DOI] [PubMed] [Google Scholar]
- Sanchez-Pulido L, Diffley JF, Ponting CP. Homology explains the functional similarities of Treslin/Ticrr and Sld3. Curr Biol. 2010;20:R509–510. doi: 10.1016/j.cub.2010.05.021. [DOI] [PubMed] [Google Scholar]
- Sangrithi MN, Bernal JA, Madine M, Philpott A, Lee J, Dunphy WG, Venkitaraman AR. Initiation of DNA replication requires the RECQL4 protein mutated in Rothmund-Thomson syndrome. Cell. 2005;121:887–898. doi: 10.1016/j.cell.2005.05.015. [DOI] [PubMed] [Google Scholar]
- Santamaria D, et al. Cdk1 is sufficient to drive the mammalian cell cycle. Nature. 2007;448:6046–6050. doi: 10.1038/nature06046. [DOI] [PubMed] [Google Scholar]
- Satyanarayana A, Berthet C, Lopez-Molina J, Coppola V, Tessarollo L, Kaldis P. Genetic substitution of Cdk1 by Cdk2 leads to embryonic lethality and loss of meiotic function of Cdk2. Development. 2008;135:3389–3400. doi: 10.1242/dev.024919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr CJ. D-type cyclins. Trends Biochem Sci. 1995;20:187–190. doi: 10.1016/s0968-0004(00)89005-2. [DOI] [PubMed] [Google Scholar]
- Shinomiya T, Ina S. Analysis of chromosomal replicons in early embryos of Drosophila melanogaster by two-dimensional gel electrophoresis. Nucleic Acids Res. 1991;19:3935–3941. doi: 10.1093/nar/19.14.3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RC, Dworkin-Rastl E, Dworkin MB. Expression of a histone H1-like protein is restricted to early Xenopus development. Genes Dev. 1988;2:1284–1295. doi: 10.1101/gad.2.10.1284. [DOI] [PubMed] [Google Scholar]
- Sterner JM, Dew-Knight S, Musahl C, Kornbluth S, Horowitz JM. Negative regulation of DNA replication by the retinoblastoma protein is mediated by its association with MCM7. Mol Cell Biol. 1998;18:2748–2757. doi: 10.1128/mcb.18.5.2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strausfeld UP, Howell M, Descombes P, Chevalier S, Rempel RE, Adamczewski J, Maller JL, Hunt T, Blow JJ. Both cyclin A and cyclin E have S-phase promoting (SPF) activity in Xenopus egg extracts. J Cell Sci. 1996;109:1555–1563. doi: 10.1242/jcs.109.6.1555. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Umemori T, Hirai K, Muramatsu S, Kamimura Y, Araki H. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature. 2007;445:328–332. doi: 10.1038/nature05465. [DOI] [PubMed] [Google Scholar]
- Tetsu O, McCormick F. Proliferation of cancer cells despite CDK2 inhibition. Cancer Cell. 2003;3:233–245. doi: 10.1016/s1535-6108(03)00053-9. [DOI] [PubMed] [Google Scholar]
- Thomson AM, Gillespie PJ, Blow JJ. Replication factory activation can be decoupled from the replication timing program by modulating Cdk levels. J Cell Biol. 2010;188:209–221. doi: 10.1083/jcb.200911037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trouche D, Le Chalony C, Muchardt C, Yaniv M, Kouzarides T. RB and hbrm cooperate to repress the activation functions of E2F1. Proc Natl Acad Sci U S A. 1997;94:11268–11273. doi: 10.1073/pnas.94.21.11268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassetzky Y, Hair A, Mechali M. Rearrangement of chromatin domains during development in Xenopus. Genes Dev. 2000;14:1541–1552. [PMC free article] [PubMed] [Google Scholar]
- Walter J, Newport J. Initiation of eukaryotic DNA replication: origin unwinding and sequential chromatin association of Cdc45, RPA, and DNA polymerase alpha. Mol Cell. 2000;5:617–627. doi: 10.1016/s1097-2765(00)80241-5. [DOI] [PubMed] [Google Scholar]
- Wolffe AP. Histone H1. Int J Biochem Cell Biol. 1997;29:1463–1466. doi: 10.1016/s1357-2725(97)00026-5. [DOI] [PubMed] [Google Scholar]
- Worcel A, Han S, Wong ML. Assembly of newly replicated chromatin. Cell. 1978;15:969–977. doi: 10.1016/0092-8674(78)90280-5. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Inoue I. Regulation of Geminin and Cdt1 expression by E2F transcription factors. Oncogene. 2004;23:3802–3812. doi: 10.1038/sj.onc.1207488. [DOI] [PubMed] [Google Scholar]
- Zegerman P, Diffley JF. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature. 2007;445:281–285. doi: 10.1038/nature05432. [DOI] [PubMed] [Google Scholar]
- Zhang J, Xu F, Hashimshony T, Keshet I, Cedar H. Establishment of transcriptional competence in early and late S phase. Nature. 2002;420:198–202. doi: 10.1038/nature01150. [DOI] [PubMed] [Google Scholar]
- Zhao J, Kennedy BK, Lawrence BD, Barbie DA, Matera AG, Fletcher JA, Harlow E. NPAT links cyclin E-Cdk2 to the regulation of replication-dependent histone gene transcription. Genes Dev. 2000;14:2283–2297. [PMC free article] [PubMed] [Google Scholar]


