Conditional deletion of Notchless leads to rapid deletion and exhaustion of HSCs and early progenitor cells, whereas committed progenitor cells survive as a result of differences in ribosomal biogenesis.
Abstract
Blood cell production relies on the coordinated activities of hematopoietic stem cells (HSCs) and multipotent and lineage-restricted progenitors. Here, we identify Notchless (Nle) as a critical factor for HSC maintenance under both homeostatic and cytopenic conditions. Nle deficiency leads to a rapid and drastic exhaustion of HSCs and immature progenitors and failure to maintain quiescence in HSCs. In contrast, Nle is dispensable for cycling-restricted progenitors and differentiated cells. In yeast, Nle/Rsa4 is essential for ribosome biogenesis, and we show that its role in pre-60S subunit maturation has been conserved in the mouse. Despite its implication in this basal cellular process, Nle deletion affects ribosome biogenesis only in HSCs and immature progenitors. Ribosome biogenesis defects are accompanied by p53 activation, which causes their rapid exhaustion. Collectively, our findings establish an essential role for Nle in HSC and immature progenitor functions and uncover previously unsuspected differences in ribosome biogenesis that distinguish stem cells from restricted progenitor populations.
Hematopoiesis within the BM is ensured by hematopoietic stem cells (HSCs). This rare population is able to self-renew and to give rise to all mature blood cell types (Orkin and Zon, 2008). HSCs are tightly regulated to maintain these properties, and numerous factors have been shown to regulate quiescence, self-renewal, survival, and differentiation. The enormous functional demands and striking longevity of HSCs raise the question of whether they might be uniquely equipped to ensure their renewal. Recent studies have revealed that HSCs may indeed differ from their differentiated progenies at the level of constitutive cellular processes such as response to DNA damage or the regulation of energy metabolism. For example, mouse HSCs are less prone to DNA damage–induced apoptosis than committed progenitor populations (Mohrin et al., 2010; Insinga et al., 2013). Control of reactive oxygen species levels is critical for BM homeostasis, and it is specifically regulated in HSCs by FoxO transcription factors (Tothova et al., 2007). Similarly, Lkb1, a master regulator of energy metabolism, is specifically required for HSC maintenance, regulating their function independently of TORC1 (Gan et al., 2010; Gurumurthy et al., 2010; Nakada et al., 2010).
Ribosome assembly in eukaryotic cells is a highly complex and coordinated process, requiring a large number of nonribosomal factors and snoRNAs (Fromont-Racine et al., 2003). Most of our knowledge of the ribosome biogenesis pathway comes from work performed in yeast, and much less is known about ribosome construction in metazoans. Over the past years, a growing body of evidence suggests that ribosome heterogeneity may participate in spatiotemporal regulation of gene expression (Gilbert, 2011; Xue and Barna, 2012). This raises the question of the mechanisms underlying the production of qualitatively different ribosomes and opens the possibility that ribosome assembly might follow different routes according to the cell type or environmental conditions. In human, defective ribosomal synthesis has been associated with BM failure syndromes and skeletal defects as well as predisposition to cancer (Ganapathi and Shimamura, 2008; Narla and Ebert, 2010). Why such a general cellular defect causes specific developmental and hematopoietic phenotypes in patients and the corresponding animal models is not fully understood. Differential sensitivity and cellular responses to ribosomal stress could explain some of these specificities (Danilova et al., 2011; Dutt et al., 2011).
Notchless (Nle) was originally identified in Drosophila melanogaster during a genetic screen for modifiers of Notch activity, although its mechanism of action has since remained elusive (Royet et al., 1998). NLE protein is an evolutionary conserved member of the large WD-repeat protein family containing a predicted C-terminal β propeller consisting of eight WD domains and an N-terminal extension. The yeast NLE orthologue Rsa4 acts in ribosome large subunit biogenesis (de la Cruz et al., 2005; Ulbrich et al., 2009). The N-terminal domain of Rsa4 interacts with the metal ion–dependent adhesion site domain of the AAA-ATPase Rea1/Mdn1, and this interaction is essential for removal of pre-60S factors and progression of 60S biogenesis (Ulbrich et al., 2009). Indeed, yeast cells deficient for RSA4 or expressing a mutated protein unable to interact with Rea1 displayed impaired rRNA processing, nuclear accumulation of pre-60S particles, and reduction of mature 60S subunits (de la Cruz et al., 2005; Ulbrich et al., 2009). Implication of Nle in ribosome biogenesis has not been directly addressed so far in other eukaryotes. Nonetheless, Solanum chacoense NLE and MDN1 were found to interact in yeast two-hybrid assay (Chantha and Matton, 2007), and similar phenotypes were obtained after knockdown of Nle and orthologues of yeast genes implicated in ribosome biogenesis in Arabidopsis thaliana and Caenorhabditis elegans (Voutev et al., 2006; Chantha et al., 2010). In the mouse, we previously reported that constitutive Nle loss of function results in early embryonic lethality and that Nle is mainly required in inner cell mass cells, being instrumental for their survival (Cormier et al., 2006).
Here, we uncover the critical role of Nle in mouse adult hematopoiesis using an inducible conditional mutagenesis strategy. Using noncompetitive and competitive transplantations, we show that Nle is cell-autonomously required for the maintenance of functional HSCs under both homeostasis and cytopenic conditions. Upon Nle deletion, HSCs entered the cell cycle, indicating that Nle is essential for maintaining HSC quiescence. In contrast, Nle is dispensable for cycling restricted progenitors and differentiated cells. We also demonstrate that the role of Nle in ribosome biogenesis is conserved in the mouse and show that Nle deficiency affects ribosome biogenesis in HSCs and multipotent progenitors (MPPs) but not in more restricted progenitors. Finally, we show that ribosome biogenesis defects are associated with p53 activation in Nle-deficient HSCs/MPPs, which in turn causes their elimination. Collectively, our findings establish an essential role for Nle in HSC/MPP function and uncover previously unsuspected differences in ribosome biogenesis between stem cells and restricted progenitor populations within the same lineage.
RESULTS
Ubiquitous Nle inactivation induces BM failure
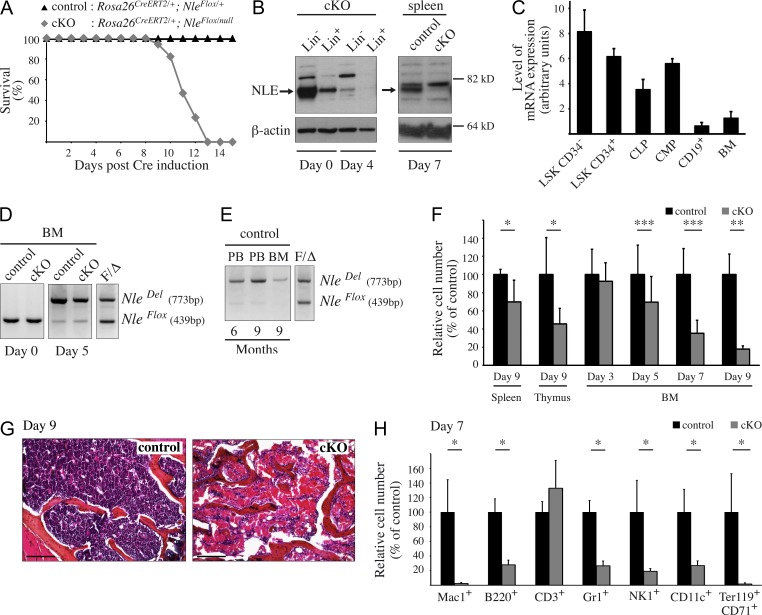
To address Nle function in the adult, we generated a conditional allele of Nle (Fig. S1, A–C) and performed acute conditional inactivation using the Rosa26CreERT2 line (Hameyer et al., 2007). Adult Rosa26CreERT2/+; NleFlox/null mice (named NlecKO hereafter) were injected three or five consecutive days with tamoxifen. Because activation of the CreERT2 recombinase could potentially affect cell physiology (Naiche and Papaioannou, 2007; Higashi et al., 2009), we used Rosa26CreERT2/+; NleFlox/+ (named control hereafter) littermates exposed to the same regimen as control. In both control and NlecKO mice, efficient conversion of NleFlox into NleDel allele was observed in all organs except the brain (not depicted). Noticeably, all NlecKO mice died within 10–13 d after the first injection (Fig. 1 A). Examination of NlecKO mice euthanized on days 7–9 revealed alterations of intestinal gut epithelium histology (not depicted) and major defects in BM and secondary lymphoid organs (see below). This led us to analyze in more detail the role of Nle in hematopoiesis.
Figure 1.
Nle inactivation induces rapid BM failure. (A) Survival curve of control and NlecKO (cKO) mice after Cre induction (n ≥ 13 per genotype). (B) Western blot analysis of sorted Lin− and Lin+ BM cells and splenic cells from control or NlecKO mouse before and 4 or 7 d after Cre induction. Data are from one representative of three independent experiments. (C) RT-QPCR analysis of Nle mRNA expression levels in various sorted BM populations from untreated control mice (n = 3). Bars are means (SD); this experiment was performed three times. (D and E) PCR analysis on BM cells from control and NlecKO mice before or 5 d after Cre induction (D) and on BM and PB cells from control mice 6 or 9 mo after Cre induction (E). Similar results were obtained from at least two independent experiments. DNA obtained from NleFlox/Del mice was used as a control of allele amplification (lane F/Δ). (F) Spleen, thymus, and BM cellularity at different time points after Cre induction. Depicted are the means (SD) of n = 5–28 mice per genotype pooled from at least two independent experiments. (G) Representative pictures from one experiment (n = 3 per genotype) showing hematoxylin and eosin staining of transversal sections of the femur from control and NlecKO mice 9 d after Cre induction. Bars, 200 µm. (H) Count of various BM hematopoietic cell populations 7 d after Cre induction (n ≥ 4 per genotype, from one experiment). Bars are means (SD). *, P < 0.05; **, P < 0.005; ***, P < 0.0005.
First, we monitored Nle expression in hematopoietic cells by Western blot and real-time RT-QPCR. NLE protein was detected in BM and spleen in control mice (Fig. 1 B). In the BM, lineage-negative (Lin−) immature cells expressed higher levels of protein than lineage-positive (Lin+) cells (Fig. 1 B). Lin− cells from noninjected control were sorted into HSCs (Lin− Sca-1+ c-Kit+ CD34− [LSK CD34−]), MPPs (Lin− Sca-1+ c-Kit+ CD34+ [LSK CD34+]), common myeloid progenitors (CMPs; Lin− Sca-1− c-Kit+ CD34+), and common lymphoid progenitors (CLPs; Lin− Sca-1low c-Kitlow Il7rα+; see the gating strategy in Fig. S2). Higher Nle mRNA levels were found in LSK CD34−, LSK CD34+, CMPs, and CLPs compared with CD19+ B cells or total BM containing a majority of Lin+ cells (Fig. 1 C). We then verified that in control and NlecKO mice, Nle deletion was effective in the hematopoietic tissue. Control and NlecKO mice were injected with tamoxifen, and PCR analysis was performed on BM and peripheral blood (PB) genomic DNA at various time points after Cre induction. Efficiency of recombination was estimated through comparison with samples of DNA from hematopoietic cells containing different ratios of NleFlox and NleDel alleles (Fig. S1, D and E). After Cre induction, high deletion efficiency was detected in BM, thymic, and splenic cells from control and NlecKO mice (Fig. 1 D and not depicted). This was further confirmed by the dramatic decrease in NLE protein levels in NlecKO BM and spleen (Fig. 1 B). At later time points (up to 9 mo), the vast majority of hematopoietic cells from control mice were carrying an NleDel allele (Fig. 1 E), indicating that Nle deletion mediated by the Rosa26CreERT2 allele was effective in HSCs.
During the autopsy, we noticed a marked reduction in the size of hematopoietic and lymphoid organs from NlecKO mice. At day 9, the numbers of cells in thymus, spleen, and BM were strongly reduced (Fig. 1 F). In addition, hematoxylin and eosin staining of bone sections revealed severe hypocellularity in NlecKO BM, showing capillaries saturated with erythrocytes (Fig. 1 G). Flow cytometry of NlecKO BM revealed that most hematopoietic lineages were severely affected (Fig. 1 H). Together, these results show that Nle is critically required for hematopoiesis.
HSCs and immature progenitors are rapidly compromised after Nle inactivation
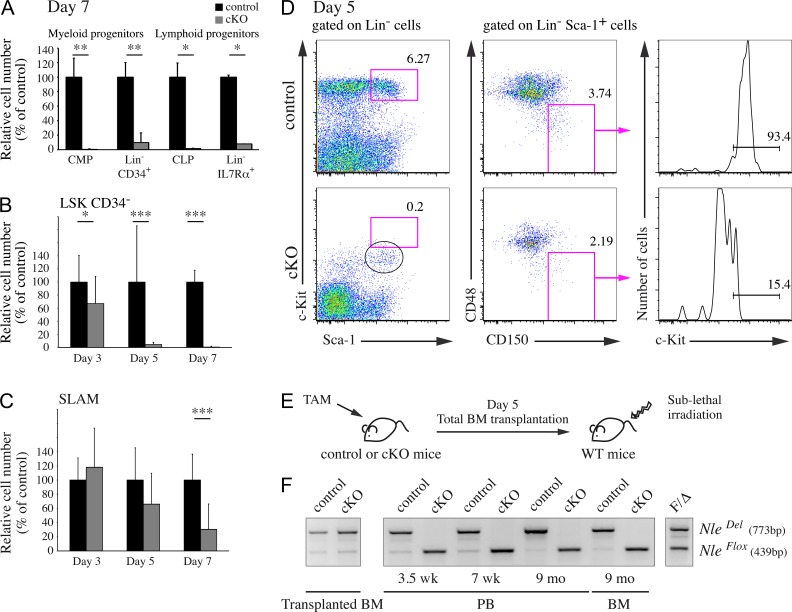
We next monitored the consequences of Nle inactivation on the BM immature cell populations and observed a strong diminution in the number of CMPs and CLPs 1 wk after tamoxifen injection (Fig. 2 A). Flow cytometry analysis also showed a rapid and dramatic decrease in the number of immature Lin− Sca-1+ c-Kit+ (LSK) cells after Nle inactivation. Both HSC-enriched LSK CD34− and MPP LSK CD34+ cell populations were virtually undetectable as early as 5 d after the first tamoxifen treatment (Fig. 2 B and not depicted). We noticed on the NlecKO FACS profiles a transitory Lin− Sca-1+ c-Kitlow cell population (see the cells within the black circle in Fig. 2 D), suggesting that in the absence of Nle, HSCs and MPPs down-regulate c-Kit expression at their surface. A similar situation was previously reported in 5-FU–treated mice, in which HSC activation after abrupt loss of hematopoietic mature cells and precursors is associated with low c-Kit expression at their membranes (Randall and Weissman, 1997). We therefore decided to reanalyze HSCs in NlecKO BM irrespectively of c-Kit expression, using the SLAM family receptors CD48 and CD150 (Kiel et al., 2005). Although less pronounced than for LSK CD34− cells, we observed a rapid decrease in the number of HSC-enriched Lin− Sca-1+ CD150+ CD48− cells (SLAM) in NlecKO BM compared with control (Fig. 2 C). Strikingly, although a vast majority of control SLAM cells expressed high levels of c-Kit receptor at their surface, the proportion of c-Kithigh cells among SLAM cells was drastically reduced in NlecKO BM 5 and 7 d after Cre induction (17% and 2%, respectively). Down-regulation of c-Kit expression was probably effective on other populations, including CMPs (see for example Fig. 2 D). Nonetheless, by itself, it did not account for the reduction of phenotypic CMPs and CLPs in NlecKO mice as shown by the decreased number of Lin− CD34+ and Lin− Il7rα+ cell populations that include CMPs and CLPs, respectively (Fig. 2 A). We also monitored NlecKO mice 5 d after Cre induction for the presence of Lin− PB cells and did not find any signs of increased HSC mobilization (not depicted). Altogether, these data show that phenotypic HSCs and immature progenitors are rapidly lost after Nle deficiency.
Figure 2.
Nle deletion provokes HSC and immature progenitor disappearance. (A) Count of myeloid and lymphoid progenitors per hindlimb 7 d after Cre induction. Bars are means (SD) for n ≥ 3 mice per genotype. This experiment was performed twice. (B) Count of LSK CD34− cells per hindlimb at different time points after Cre induction. Bars are means (SD) for n ≥ 13 mice per genotype pooled from at least three independent experiments. (C) Count of SLAM cells per hindlimb at different time points after Cre induction. Bars are means (SD) for n = 6–13 mice per genotype pooled from two or three independent experiments. (D) Representative FACS profiles of BM cells from control or NlecKO mice at day 5. Cells were labeled with LSK (left) or SLAM (middle) markers. NlecKO SLAM cells express lower levels of c-Kit (right). Similar results were obtained from two independent experiments. The black circle indicates a transitory Lin− Sca-1+ c-Kitlow cell popluation in NlecKO BM. (E) Schematic representation of the transplantation strategy. Both control and NlecKO mice were injected five consecutive days before transplantation. (F) PCR analysis was performed on BM at the time of transplantation and after 9 mo and on PB cells at different time points after transplantation. Similar results were obtained from two independent experiments. *, P < 0.05; **, P < 0.005; ***, P < 0.0005.
To address the possibility that functional HSCs with a modified phenotype may nevertheless persist in NlecKO BM, we performed total BM transplantation experiments. BM cells were recovered from control and NlecKO mice 5 d after Cre induction and transferred into sublethally irradiated mice (Fig. 2 E). By the time of transplantation, efficient deletion of the NleFlox allele was observed in both control and NlecKO donor cells (Fig. 2 F, left). In contrast to control, the NleDel allele could neither be detected in the blood of mice transplanted with NlecKO BM 3.5 and 7 wk or 9 mo after engraftment nor in the BM after 9 mo (Fig. 2 F). This indicates that the reconstituting activity of NlecKO BM was limited to Nle-proficient nonrecombined cells. These data show that Nle function is critically required for HSC and immature progenitor activity and function.
Reduced quiescence of Nle-deficient HSCs and MPPs
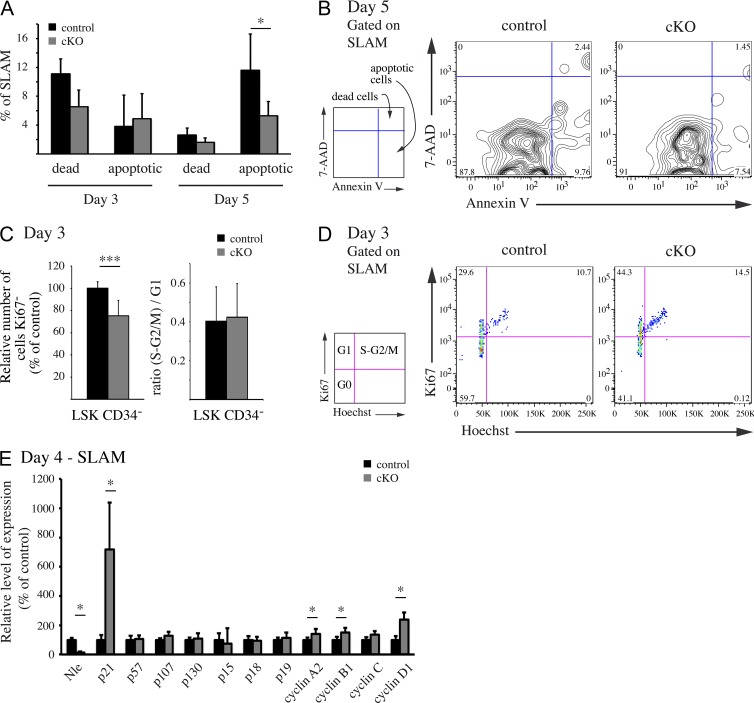
To determine the mechanisms by which Nle deletion induces stem and progenitor cell loss, we analyzed NlecKO BM cells shortly after Cre induction. Because Nle has previously been shown to play a role in cell survival (Cormier et al., 2006; Orkin and Zon, 2008), we first examined whether increased cell death could be responsible for the rapid exhaustion of HSCs and MPPs in NlecKO mice. Using Annexin V/7AAD labeling, we analyzed freshly isolated NlecKO BM cells at days 3 and 5, when no or moderate defects in BM homeostasis were observed (Figs. 1 F and 2, A–D). At either time point, no increase in the frequency of Annexin V+7AAD− (apoptotic) or Annexin V+7AAD+ (dead) cells among total BM cells, SLAM cells, and immature progenitors (LSK CD34+, CMPs, and CLPs at day 3 or Lin− Sca-1+ CD150+ CD48+ at day 5) was detected (Fig. 3, A and B; and not depicted). At day 5, a significant decrease in the frequency of apoptosis among SLAM cells was observed in NlecKO mice (Fig. 3 A), although no HSC activity was found in the BM of such animals (Fig. 2 F). Similarly, a TUNEL assay performed on control and NlecKO mice at day 4 did not show an increased cell death of HSCs and immature progenitors after Nle inactivation (not depicted). We then examined the cell cycle status of control and NlecKO BM cells using Ki67 and Hoechst staining. Importantly, as early as 3 d after Cre induction, there was a significant decrease in the frequency of G0 cells between control and NlecKO SLAM or LSK CD34− and MPPs (Fig. 3, C [left] and D; and not depicted). In all populations, Ki67+ cells were identically distributed among G1 and S-G2/M between control and NlecKO mice (Fig. 3 C, right; and not depicted), indicating that cycling Nle-deficient BM cells have cycling parameters similar to control cells. We next examined SLAM cells for various cell cycle regulators by RT-QPCR (Fig. 3 E). As expected, a massive reduction of Nle mRNA levels was observed in NlecKO SLAM cells. For most genes tested, no differences were observed between control and NlecKO SLAM cells. Consistent with the higher proportion of cycling cells, cyclin A2, cyclin B1, and cyclin D1 mRNA levels were slightly increased in Nle-deficient SLAM cells. Strikingly, a strong up-regulation of Cdkn1a (p21) mRNA level was observed in NlecKO SLAM cells, suggesting that depletion of NLE protein in HSCs causes the activation of p53 (see below). However, this up-regulation was not accompanied by a potent cell cycle arrest, as visualized by FACS analysis (Fig. 3, C and D). A similar situation has been previously reported during HSC mobilization (Passegué et al., 2005). Up-regulation of Cdkn1a (p21) together with increased cell cycle entry of HSCs has also been described after x-ray exposure (Insinga et al., 2013). Altogether, our data indicate that Nle is required for the maintenance of HSC quiescence.
Figure 3.
Cell death and proliferation of Nle-deficient HSCs and progenitors. (A and B) SLAM cells were stained with Annexin V/7AAD. (A) Quantitative results for dead and apoptotic cells and means (SD) are shown (n = 3 per genotype). (B) Representative FACS profiles are shown. This experiment was performed independently twice. (C and D) BM cells were stained for Ki67 and Hoechst 3 d after Cre induction. (C) The relative number of Ki67− cells within the LSK CD34− population is indicated (left; n ≥ 14 per genotype, pooled from three independent experiments), as well as the ratio (S-G2/M)/G1 among LSK CD34− cells (right; n = 3 per genotype, from one experiment). Bars are means (SD). (D) Representative FACS profiles are shown. Note that Cre activation causes a reduction of Ki67− LSK CD34− cells in control compared with wild-type mice (not depicted). Similar results were obtained independently for LSK CD34− and SLAM cells. (E) Levels of expression of cell cycle regulators in SLAM cells measured by RT-QPCR analysis. Bars are means (SD) of n = 4 mice; this experiment was performed twice. *, P < 0.05; ***, P < 0.0005.
Nle acts in a cell-autonomous manner to maintain hematopoiesis
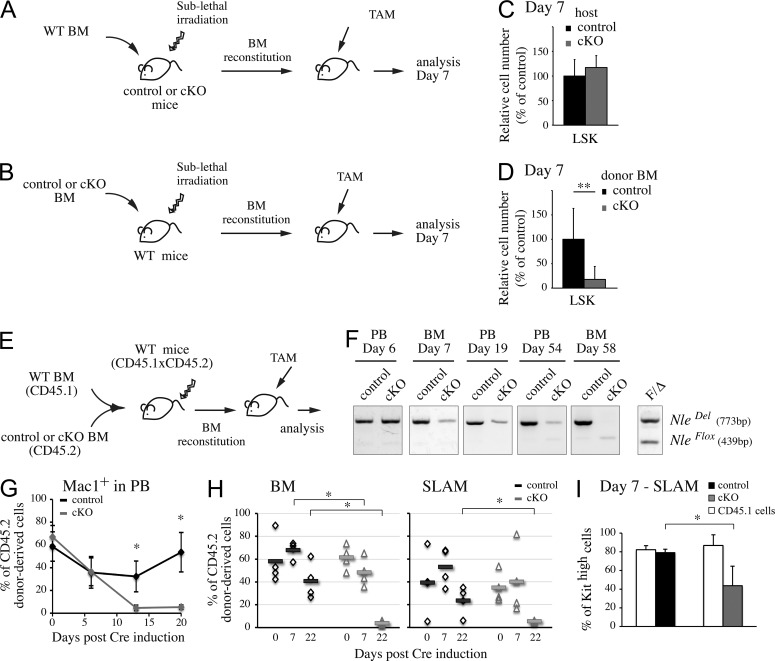
In NlecKO mice, systemic Nle inactivation might contribute to BM failure. To examine this possibility, we performed noncompetitive transplantation between wild-type and control or NlecKO mice. In a first set of experiments, we transplanted BM cells from wild-type mice into sublethally irradiated untreated control or NlecKO mice (Fig. 4 A). In the reciprocal experiment, untreated control or NlecKO BM cells were transplanted into sublethally irradiated wild-type mice (Fig. 4 B). 2 mo after grafting, efficiently reconstituted (>80%) mice were injected with tamoxifen and their BM were analyzed after 7 d, at a time point where a drastic BM phenotype was observed in NlecKO mice (Fig. 1, F and H; and Fig. 2, A–C). The number of wild-type LSK cells was not modified in an Nle-deficient environment compared with control (Fig. 4 C). In contrast, wild-type mice reconstituted with NlecKO BM exhibited a reduced number of LSK cells after Nle inactivation (Fig. 4 D). At longer time points, we observed the progressive disappearance of the NleDel allele in BM and PB (not depicted). This shows that Nle-deficient HSCs are not able to maintain hematopoiesis in a wild-type environment and supports the notion that loss of HSCs and progenitors in NlecKO mice was caused by Nle loss of function in BM cells. Death of NlecKO mice reconstituted with wild-type BM 2 wk after tamoxifen injection indicates that Nle function is also required in nonhematopoietic cells for mouse survival (not depicted).
Figure 4.
Nle is required cell autonomously for HSC function. (A and B) Schematic representation of the noncompetitive transplantation strategy. Wild-type BM was transplanted into irradiated control or NlecKO mice (A) and conversely (B). After 2 mo, mice were treated with tamoxifen for 5 d. (C and D) 7 d after the first injection, the number of LSK cells in the BM of grafted mice was measured. Bars are means (SD) of n = 8–10 mice per genotype from two independent experiments. (E) Scheme of the competitive transplantation experiment. Wild-type CD45.1×CD45.2 mice were irradiated and engrafted with a mix of donor CD45.2 control or NlecKO and competitor CD45.1 wild-type BM cells (ratio 1:3 or 1:4). After 2 mo, mice were injected with tamoxifen. (F) PCR analysis on BM or PB cells from competitive chimeras at different time points after Cre induction. Similar results were obtained from three independent experiments. (G) The percentage of CD45.2 donor–derived cells among Mac1+ PB cells was measured at different time points. Depicted are the means (SD) of n = 4 mice per genotype. Similar results were obtained twice independently. (H) The percentages of CD45.2 donor–derived cells among BM or SLAM cells were measured before and at different time points after Cre induction. Similar results were obtained from three independent experiments. Individual measures and means (horizontal bars) are shown. (I) Proportion of c-Kithigh cells among SLAM cells in competitive chimeras 7 d after Cre induction. Bars are means (SD) of n = 4 mice; this experiment was performed three times. *, P < 0.05; **, P < 0.005.
To address whether Nle acts in a cell-autonomous manner in hematopoietic cells under homeostatic conditions, we adopted a competitive chimera approach. Either untreated control or NlecKO CD45.2 donor BM cells were transplanted with an excess (1:3 to 1:4 ratio) of competitor wild-type CD45.1 BM cells into irradiated (CD45.1×CD45.2)F1 recipient mice (Fig. 4 E). In such chimeras, hematopoiesis can be sustained by competitor cells, therefore preventing secondary effects caused by the cytopenia that occurs both in NlecKO mice and noncompetitive chimeras. After stable reconstitution, mice were injected with tamoxifen, and PB and BM cells were analyzed at various time points. No differences in BM cellularity were observed between mice engrafted with NlecKO and control cells, confirming that BM homeostasis was maintained in these chimeras (not depicted). We also verified that efficient deletion of NleFlox allele was obtained in these mice (Fig. 4 F). In PB, NlecKO CD45.2 Mac1+ myeloid cells were no longer detected from 20 d after injection onwards (Fig. 4 G). Long-lived CD3+ T and CD19+ B lymphoid cells were also affected, although with a delayed kinetics (not depicted). In agreement with the data obtained in the PB, NlecKO CD45.2 cells rapidly disappeared in the BM (Fig. 4 H, left). The evolution of CD45.2 cells was addressed at day 7 and at later time points, as soon as 3 wk after Nle inactivation, and contrary to control CD45.2 cells, virtually no NlecKO CD45.2 cells were detected in BM and within the SLAM compartment (Fig. 4 H), indicating that Nle-deficient HSCs were not maintained even in a wild-type microenvironment and during homeostasis. Under these conditions, the proportion of c-Kithigh cells among NlecKO CD45.2 SLAM cells was still reduced (Fig. 4 I), indicating that Nle deficiency in HSCs can intrinsically down-regulate c-Kit expression. Altogether, our data show that Nle is cell-autonomously required for the maintenance of functional HSCs under both homeostasis and cytopenic conditions.
Nle is dispensable for more restricted progenitors
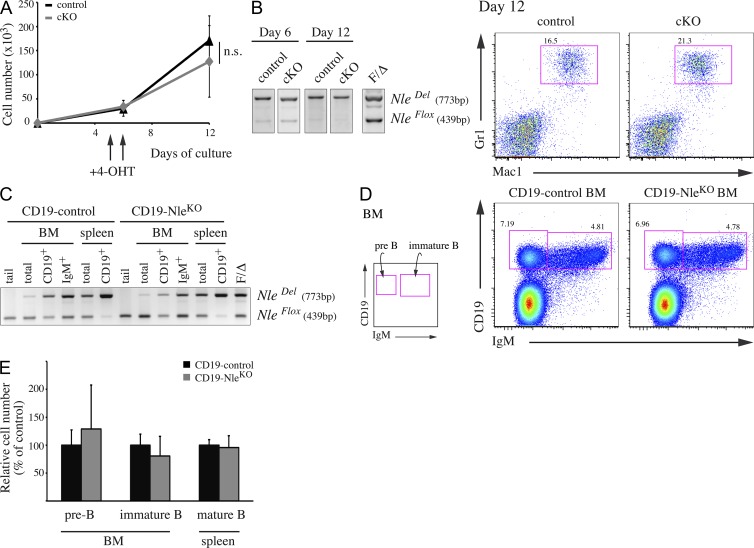
Because hematopoietic cells are continuously and actively fueled by the HSC/MPP compartment, the rapid exhaustion of HSCs and immature progenitors in NlecKO mice could preclude the detection of a more direct role of Nle in restricted progenitors and differentiated cells. To examine this possibility, we first used an ex vivo differentiation assay. Untreated control and NlecKO HSC-enriched (Lin− Sca-1+ c-Kit+ Flt3−) cells were FACS-sorted and plated on OP9 stromal cells. After 5 d of culture, immature LSK cells were no longer detected (not depicted), and a pulse of 4-hydroxy-tamoxifen (4-OHT) was added to the culture medium to induce loss of Nle function in progenitor cells. During the following days, both control and Nle-deficient cells underwent massive expansion (Fig. 5 A). Cells were harvested after a total of 12 d in culture and analyzed for differentiation markers. Under the conditions used, differentiation was biased toward myeloid lineage, but control and NlecKO cells were indistinguishable in their ability to give rise to Mac1+Gr1+ granulocytes (Fig. 5 B, right). Importantly, we checked that Nle was efficiently inactivated at days 6 and 12 by PCR analysis (Fig. 5 B, left). This suggests that myeloid progenitors were able to actively proliferate and differentiate in the absence of Nle.
Figure 5.
Nle is dispensable during myeloid and B cell differentiation. (A and B) 200 sorted LSK Flt3− cells per well were plated onto OP9 cells. After 5 d, 4-OHT was added to the culture medium for 2 d. (A) Proliferation curves (n = 4–9 from two independent experiments). SD is shown. n.s., not significant; P = 0.211. (B) Deletion was evaluated by PCR and myeloid differentiation was assessed at day 12 by flow cytometry after staining with Mac1 and Gr1 markers. Similar results were obtained in two independent experiments. (C–E) Analysis of CD19-control (CD19Cre/+; NleFlox/+) and CD19-NleKO (CD19Cre/+; NleFlox/null) mice. (C) PCR analysis on IgM+ and CD19+ sorted cells from BM and spleen. Data are representative of three individuals. (D) Representative FACS profile of CD19-control and CD19-NleKO BM, stained with IgM and CD19 markers. (E) Counts of pre-B cells (CD19+ IgM−), immature B cells (CD19+ IgM+) from BM, and mature B cells (CD19+ IgM+) from spleen of CD19-control and CD19-NleKO mice. Bars are means (SD) of n = 4–6 mice per genotype pooled from three independent experiments.
We also evaluated the role of Nle in vivo during B cell development using the CD19-Cre mouse line (Rickert et al., 1997). We generated CD19Cre/+; NleFlox/+ and CD19Cre/+; NleFlox/null mice, named, respectively, CD19-control and CD19-NleKO mice. As already observed for other Flox alleles, CD19Cre-mediated Nle inactivation was an on-going process along B cell maturation (Fig. 5 C). Comparison of CD19-control and CD19-NleKO mice revealed no significant differences in the deletion efficiency of NleFlox allele and in the numbers of BM pre-B cells, immature B cells, and splenic mature B cells (Fig. 5, C–E). Moreover, CD19-NleKO B cells were able to secrete steric IgM (not depicted), indicating that Nle-deficient mature B cells were not functionally impaired. Altogether, these data strongly suggest that contrary to HSCs and MPPs, Nle is dispensable for survival, proliferation, and maturation of more restricted progenitors.
Nle is required for large ribosomal subunit biogenesis
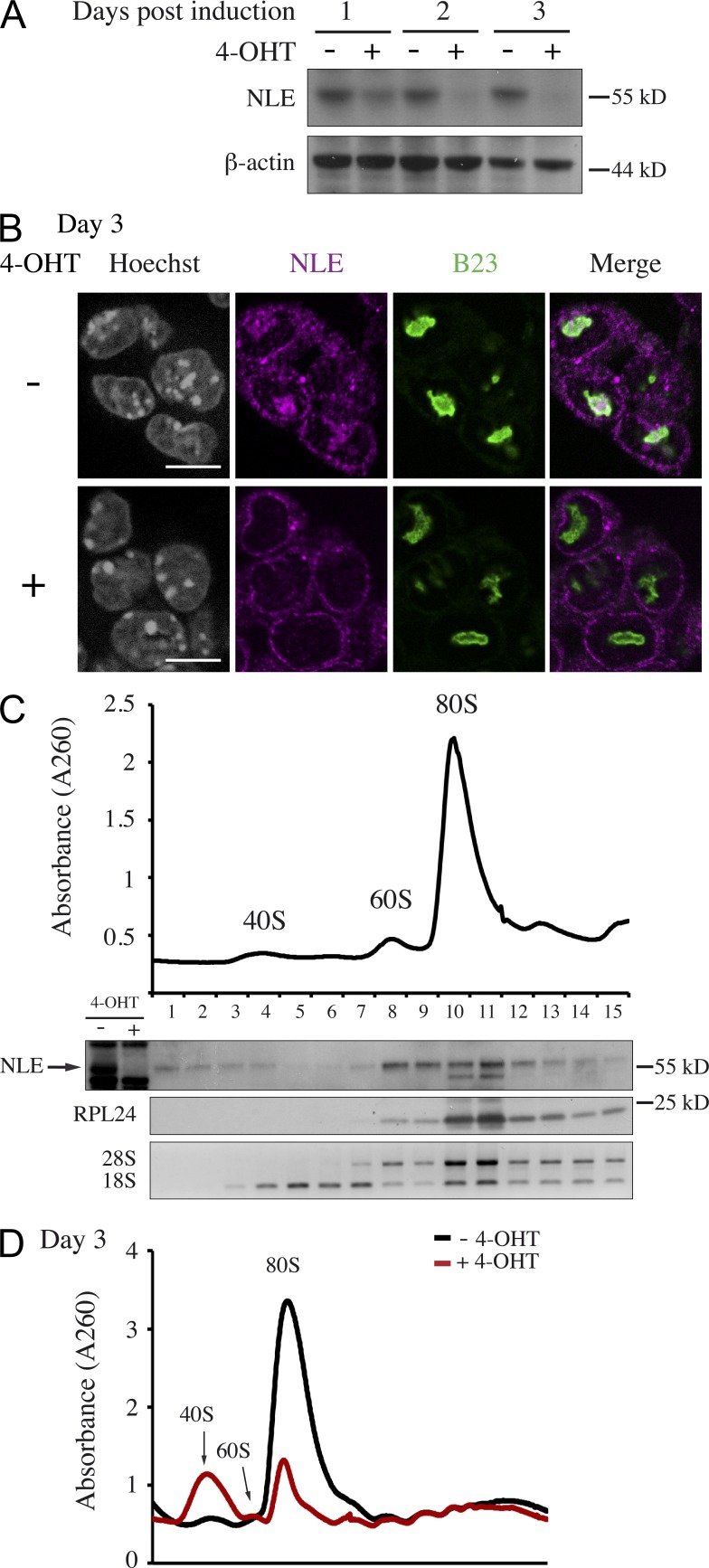
In yeast, NLE orthologue Rsa4 acts as a pre-60S maturation factor, being essential for progression of ribosomal large subunit biogenesis and nuclear export (de la Cruz et al., 2005; Ulbrich et al., 2009; Bassler et al., 2010). Based on its high degree of conservation among eukaryotes (Cormier et al., 2006; Gazave et al., 2009), we hypothesized that NLE might also participate in pre-60S maturation in mouse cells. To address this possibility, we took advantage of Rosa26CreERT2/+; NleFlox/null embryonic stem (ES) cells, in which Nle loss of function can be induced by addition of 4-OHT in the culture medium (Fig. 6 A). In ES cells, NLE localized to the nucleolus as indicated by the specific nucleolar immunostaining that partially colocalized with B23/nucleophosmin and was undetectable after 4-OHT treatment (Fig. 6 B). To investigate the involvement of Nle in ribosome biogenesis, total cellular extracts from treated and untreated ES cells were fractionated on a sucrose gradient. Immunoblot of untreated ES cells fractions indicated that NLE accumulated mainly in fractions 8–11, corresponding to 60S to 80S sedimentation coefficients (Fig. 6 C). In treated cells, the sedimentation profile was dramatically altered with a decrease in the amount of 60S subunit and 80S ribosomes and an increase of 40S subunit (Fig. 6 D). These observations are consistent with a role of NLE in pre-60S maturation.
Figure 6.
Nle is required for the large preribosomal subunit biogenesis in mouse ES cells. (A) Western blot analysis on NlecKO cells untreated or treated with 4-OHT. This experiment was performed three times independently. (B) Immunostaining of NLE and B23 in Rosa26CreERT2/+; NleFlox/null ES cells treated or not with 4-OHT. The pictures are representative of three independent experiments. Bars, 10 µm. (C) RNA profiles measured by absorbance at 260 nm in a sucrose gradient loaded with extracts from untreated cells. In each fraction, NLE and RPL24 protein levels, 28S and 18S rRNA, were assessed by immunoblotting and ethidium bromide staining, respectively. The arrow points to the NLE-specific band. (D) RNA profiles measured by absorbance at 260 nm loaded with extracts from untreated and treated cells at day 3. Similar results were obtained for two independent sedimentation profile experiments.
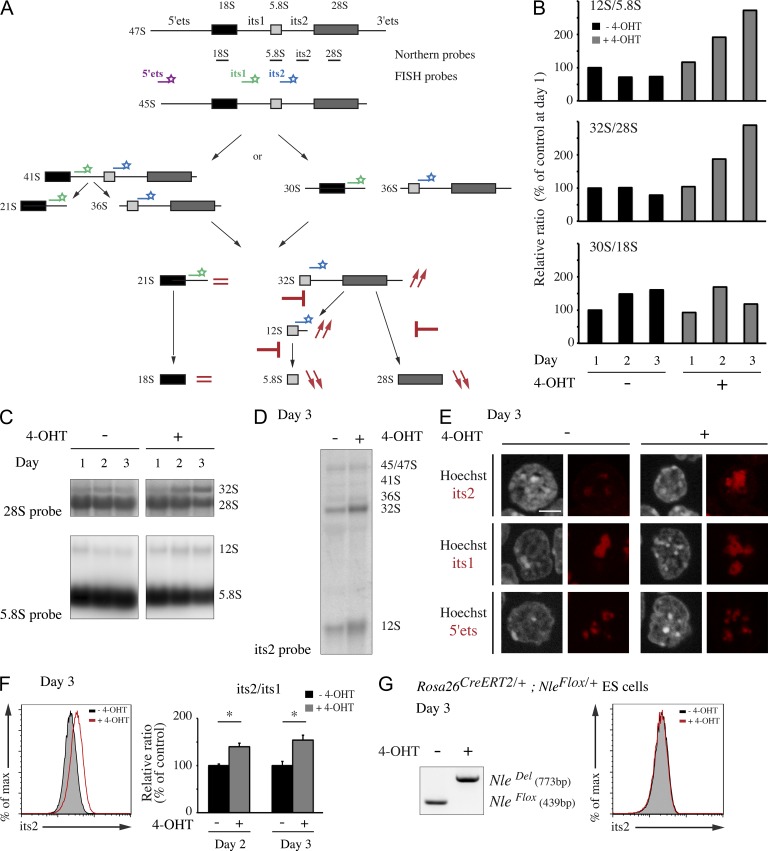
In yeast, Rsa4 deficiency has been shown to interfere with pre-60S nuclear export. We thus expected to observe a similar phenotype leading to accumulation of 5.8S and 28S rRNA precursors and decrease of the corresponding mature rRNA species in Nle-deficient ES cells (Fig. 7 A). We monitored the levels of different rRNA species in treated and untreated ES cells by Northern blot analysis (Fig. 7, A–D; and not depicted). Total RNA recovered from the same number of treated and untreated cells was probed with sequences specific for 5.8S, 18S, 28S, and its2. We observed a progressive reduction in the steady-state levels of 5.8S and 28S mature rRNA concomitantly with the accumulation of 12S and 32S rRNA precursors in mutant cells (Fig. 7, B–D). In contrast, processing of 18S rRNAs was not affected (Fig. 7 B). We confirmed these observations by RNA fluorescence in situ hybridization (FISH) using 5′ets, its1, and its2 probes recognizing various combinations of nucleolar rRNA species (Fig. 7, A and E). By flow cytometry, we measured an increase of the its2/its1 ratio, indicating a specific accumulation of rRNA precursors of the large subunit (Fig. 7 F). 45/47S levels measured by 5′ets signal intensity were unchanged (not depicted). Importantly, its2 levels were not affected after 4-OHT treatment of control ES cells (Fig. 7 G), demonstrating that the accumulation of immature rRNA precursors is indeed caused by Nle loss of function. Altogether, these data demonstrate that Nle is required in mouse ES cells for the maturation and the export of the large ribosomal subunit.
Figure 7.
Nle inactivation induces defects in rRNA maturation and accumulation of the large preribosomal subunit. (A) Schematic representation of the two major pre-rRNA processing pathways in mammalian cells. Localization of probes is indicated under the 47S primary transcript. Predictions based on the known function of NLE orthologue in yeast of rRNA maturation defects in Nle-deficient mouse ES cells are shown in red. If this function is conserved, Nle inactivation should induce a block in the last steps of maturation of the large preribosomal subunit, leading to an accumulation in the nucleolus of the 32S and 12S pre-rRNA species and the decreased export in the cytoplasm of mature 28S and 5.8S rRNA. 32S and 12S pre-rRNA can be detected by Northern blot or FISH analysis using probes directed against the pre-rRNA internal sequence its2 (blue). Maturation of the small subunit should not be directly affected. Thereby, the levels of the corresponding pre-rRNA detected with an its1 probe (green) should remain unchanged. Similarly, levels of 47S primary rRNA transcript detected with a 5′ets probe (purple) should not be directly affected. (B–D) Northern blot analysis of total RNA from untreated or treated ES cells. (B) Quantification of 12S/5.8S, 32S/28S, and 30S/18S intensity ratios. Northern blots analysis with 28S, 18S, 5.8S, or its2 probe was obtained at least twice. (E–G) FISH experiment on untreated and treated ES cells. (E) Representative images of two independent FISH experiments with its2, its1, or 5′ets performed on ES cells untreated or treated with 4-OHT at day 3. Cells were labeled with Hoechst. Bar, 5 µm. (F) Representative FACS profiles of its2 FISH signal in untreated and treated ES cells. Quantification of the its2/its1 intensity ratio relative to untreated cells is shown. Bars are means (SD) of n = 3 per condition from one out of three independent experiments. *, P < 0.05. (G) Control (Rosa26CreERT2/+; NleFlox/+) ES cells were treated or not with 4-OHT. At day 3, FISH experiments using its2 probe and PCR analysis were performed showing that in the presence of a remaining functional Nle allele, large ribosomal subunit maturation was not affected.
Nle deletion affects ribosome biogenesis specifically in HSCs and MPPs
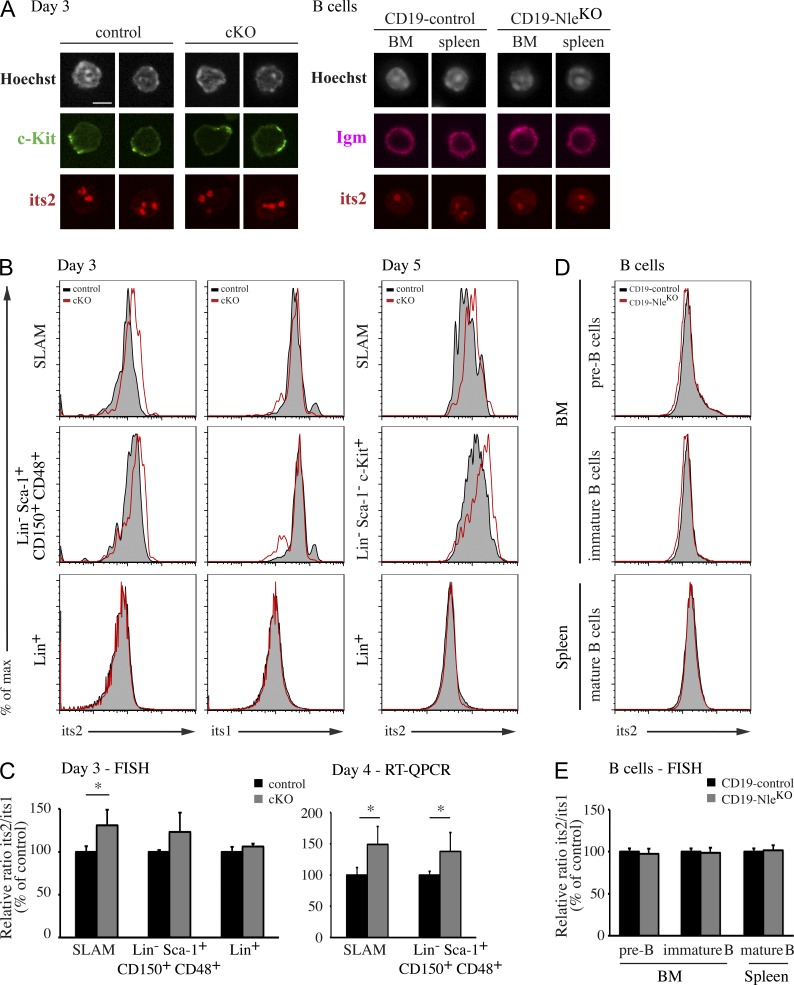
Having established the role of Nle in ES cells, we next wondered whether defects in ribosome biogenesis could be responsible for the BM phenotype observed in NlecKO mice. We combined cell surface antibody staining with intracellular RNA FISH and used flow cytometry to measured steady-state levels of rRNA precursors in several BM cell populations. We first checked that as in ES cells, RNA FISH with its2 probes labeled subnuclear structures, poor in chromatin, likely to correspond to nucleoli in immature cells and B cells (Fig. 8 A). 3 d after Cre induction, we measured a shift in its2 intensity in Nle-deficient SLAM and Lin− Sca-1+ CD150+ CD48+ MPPs but not in more committed Lin+ cells (Fig. 8, B and C, left). Importantly, neither its1 nor 5′ets levels were modified in stem cells and MPPs, pointing to defective large ribosomal subunit maturation in HSCs/MPPs (Fig. 8, B and C; and not depicted). Accumulation of immature rRNAs of the large subunit in SLAM and Lin− Sca-1+ CD150+ CD48+ cells was confirmed by RT-QPCR (Fig. 8 C, right). 5 d after Cre induction, a similar situation was observed (Fig. 8 B, right). We performed similar analysis on B cell lineage populations from CD19-control and CD19-NleKO mice. Consistent with the lack of a phenotype in CD19-NleKO mice, no changes in the level of its2, its1, or 5′ets were observed in BM and spleen B cells (Fig. 8, D and E; and not depicted). We thus conclude from these experiments that Nle is differentially required for 60S ribosomal subunit biogenesis between HSCs/MPPs and more committed or differentiated hematopoietic cells.
Figure 8.
Nle is required for pre-60S export in HSCs/MPPs but not in B cells. (A) Its2 labeling in hematopoietic cells. At day 3, control and NlecKO hematopoietic cells were stained with c-Kit antibody and its2 probe (left). CD19-control and CD19-NleKO cells from BM and spleen were stained with IgM antibody and its2 probe (right). Bar, 5 µm. (B) Representative histograms for its2 and its1 labeling in SLAM cells, immature progenitors (Lin− Sca-1+ CD150+ CD48+ or Lin− Sca-1− c-Kit+), and Lin+ cells at day 3 or 5 after Cre induction from control and NlecKO mice. Three independent FACS experiments showed similar results. (C) Quantification of its2/its1 intensity and RNA level ratios in populations from control and NlecKO mice measured at day 3 by FISH (left) or RT-QPCR (right). Bars are means (SD) of n = 3–4 mice per genotype. Data are from one representative of two experiments. *, P < 0.05. (D) Representative histograms for its2 labeling by FISH in BM pre-B and immature B cells and splenic B cells from CD19-control and CD19-NleKO mice. (E) Quantification of its2/its1 intensity ratio in B cell populations from CD19-control and CD19-NleKO mice. Bars are means (SD) of n = 3 mice per genotype from one experiment.
Rapid BM cytopenia is mediated by p53
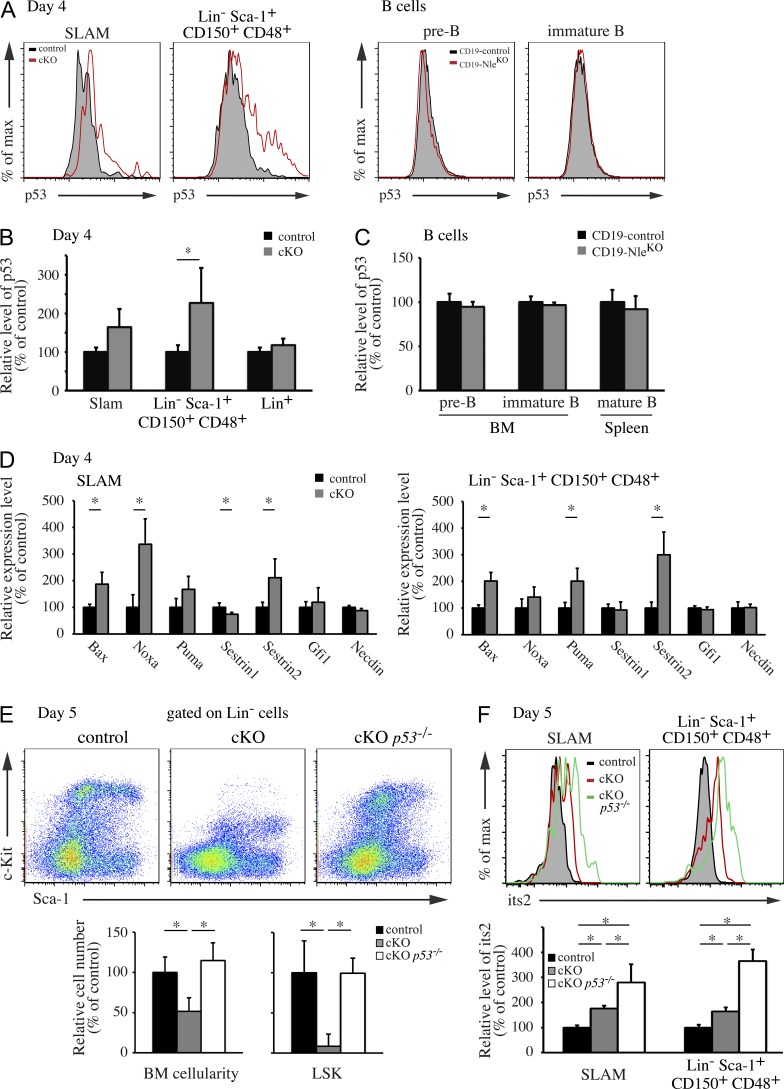
Defective ribosome biogenesis was shown to trigger the activation of p53 pathway in vertebrate cells (Zhang and Lu, 2009). We therefore monitored p53 expression levels in BM cells shortly after Nle inactivation. 4 d after Cre induction, we measured a clear increased in p53 levels in both Nle-deficient SLAM and Lin− Sca-1+ CD150+ CD48+ cells (Fig. 9, A [left] and B). In contrast, in NlecKO Lin+ cells and CD19-NleKO B cells, in which no increases in pre-60S rRNA intermediates were observed, p53 levels were not augmented (Fig. 9, A [right] and C). We next assessed the transcriptional activation of various p53 target genes in these compartments. In addition to Cdkn1a (p21; Fig. 3 E), mRNA levels of Bax, Noxa, Puma, and Sestrin2 were significantly increased in SLAM or Lin− Sca-1+ CD150+ CD48+ cells (Fig. 9 D), further confirming the activation of p53 after Nle inactivation. To determine the functional implication of p53 in the severe hematopoietic phenotype of NlecKO mice, we crossed p53−/− mice with NlecKO mice and analyzed BM cell populations after tamoxifen injection. Strikingly, loss of p53 totally rescued the number of total BM and LSK cells in NlecKO mice 5 d after Cre induction (Fig. 9 E). Accordingly, the number of SLAM cells in NlecKO; p53−/− mice was unchanged compared with control mice (not depicted). Importantly, NlecKO; p53−/− SLAM and Lin− Sca-1+ CD150+ CD48+ cells exhibited increased its2 levels compared with controls (Fig. 9 F). The fact that its2 levels were significantly higher in these mice compared with NlecKO mice suggests that the p53-dependent checkpoint operates at a relatively early step of pre-60S rRNA intermediate accumulation in these cell populations. Collectively, our data suggest that Nle deficiency leads to defective ribosome biogenesis and consecutive p53 activation in HSCs and MPPs that in turn causes their disappearance.
Figure 9.
Rapid BM cytopenia is mediated by p53. (A, left) Representative histograms for p53 labeling in SLAM cells and MPPs (Lin− Sca-1+ CD150+ CD48+) at day 4 after Cre induction from control and NlecKO mice. (right) Representative histograms for p53 labeling in BM pre-B and immature B cells from CD19-control and CD19-NleKO mice. (B) Quantification of p53 levels in populations from control and cKO mice. Bars are means (SD) of n = 4 mice per genotype. (C) Quantification of p53 levels in B cell populations from CD19-control and CD19-NleKO mice. Bars are means (SD) of n = 3 mice per genotype. Data are from one representative of two experiments. (D) Relative expression levels of p53 target genes measured by RT-QPCR in SLAM cells and MPPs from control and NlecKO mice at day 4 after Cre induction. Bars are means (SD) of n = 4 mice. Data are from one representative of two experiments. (E) Representative FACS profiles obtained 5 d after Cre induction from control, NlecKO, and NlecKO p53−/− (Rosa26CreERT2/+; NleFlox/null; p53−/−) mice. The BM cellularity and the number of LSK cells are shown for the three genotypes. Bars are means (SD) for n = 4–6 mice per genotype. This experiment was performed twice. (F) Its2 labeling by FISH was performed on BM cells from control, NlecKO, and NlecKO p53−/− mice 5 d after Cre induction. Representative histograms for the levels of its2 in SLAM cells and MPPs (Lin− Sca-1+ CD150+ CD48+) and quantification of its2 intensity are shown. Bars are means (SD) for n = 4–6 mice per genotype. This experiment was performed twice. *, P < 0.05.
DISCUSSION
In this study, we show that Nle is critically required for the maintenance of adult HSCs. Collectively, our data demonstrate that Nle is an intrinsic regulator, acting in a cell-autonomous manner, and necessary for HSC self-renewal. By demonstrating that Nle function in the pre-60S ribosomal subunit maturation has been conserved during evolution, we underscore the importance of ribosome biogenesis as a critical pathway for HSC maintenance. In contrast, Nle function is dispensable in restricted progenitors and differentiated hematopoietic cells, indicating that ribosome biogenesis is differentially regulated during differentiation.
Nle is a key regulator of HSC maintenance
Strikingly, Nle-deficient cells were rapidly eliminated from the HSC population. Although several factors have been shown to participate in HSC regulation, rapid loss of the HSC pool has been reported after the disruption of only a few genes: N-Myc/c-Myc, NKAP, Cited2, Mll, Survivin, Mcl-1, or APC (Opferman et al., 2005; Jude et al., 2007; Leung et al., 2007; Laurenti et al., 2008; Qian et al., 2008; Kranc et al., 2009; Pajerowski et al., 2010). Of note, one of the primordial functions of Myc is the regulation of ribosome biogenesis (van Riggelen et al., 2010; Ji et al., 2011). Combined deficiency of c-Myc and N-Myc has been shown to severely impair HSC function, and interestingly, down-regulation of 31 ribosomal proteins coding genes has been reported in mutant HSCs (Laurenti et al., 2008). However, contribution of defective ribosome biogenesis to the phenotype of c-Myc/N-Myc double-deficient HSCs has not been investigated.
Concomitantly with HSC loss, we observed the down-regulation of c-Kit expression at the cell surface of NlecKO immature hematopoietic cells. c-Kit is a major regulator of HSC survival and proliferation, and its down-regulation may therefore participate in defective maintenance of NlecKO HSCs. In Gfi1−/− mice, a slight decrease of c-Kit in LSK cells correlates with progressive loss of HSC self-renewal activity (Hock et al., 2004). Reduced c-Kit expression might also reflect HSC activation consecutive to BM depletion (Randall and Weissman, 1997; Laurenti et al., 2008). NlecKO HSCs displayed reduced expression of c-Kit receptor under homeostatic conditions, indicating that such deregulation is not solely the consequence of BM cytopenia. How c-Kit expression is regulated in HSCs is not fully understood. Recently, the Shp2 tyrosine phosphatase was shown to promote Kit gene expression and HSC pool maintenance through a positive c-Kit/Shp2/c-Kit feedback loop (Zhu et al., 2011). 4 d after Cre induction, a ≈30% decrease in c-Kit mRNA levels was observed in NlecKO HSCs/MPPs (unpublished data), suggesting that c-Kit down-regulation at their membranes was controlled at least in part at the transcriptional level. Future work will be necessary to understand the mechanisms underlying c-Kit down-regulation in Nle-deficient HSCs.
Nle acts in ribosome biogenesis and is likely not a direct regulator of Notch
Based on genetic study in Drosophila, Nle has been proposed to code for a modulator of Notch signaling. Absence of canonical Notch signaling does not impact HSC maintenance at homeostasis (Maillard et al., 2008). Conversely, sustained Notch activity promotes HSC expansion ex vivo and enhanced self-renewal of hematopoietic progenitors (Bigas and Espinosa, 2012). Thus, modulation of Notch signaling cannot simply account for the failure of Nle-deficient HSCs/MPPs. Contrary to genes coding for key components of the Notch pathway that are specific to metazoans, Nle is an ancient eukaryotic gene (Gazave et al., 2009), and its ancestral function as a ribosome biogenesis factor has been deciphered in yeast (de la Cruz et al., 2005; Ulbrich et al., 2009). Here, we show that Nle function has been conserved during evolution. Although we cannot formally exclude the possibility that NLE has acquired additional nonribosomal functions during evolution, it seems more likely that Notch activity modulation by NLE is related to its role in ribosome biogenesis. Genetic interactions between Notch and Minute mutations, which affect a class of ribosomal genes, have been reported a very long time ago (Schultz, 1929). More recently, genetic screens for modifiers of Notch activity have identified several ribosomal proteins (Hall et al., 2004; Mourikis et al., 2010). In particular genetic and cellular contexts, Notch signaling activity must therefore be sensitive to variations in ribosome supply as previously discussed for the Hedgehog signaling pathway (Nybakken et al., 2005).
Mechanisms responsible for the rapid exhaustion of HSCs/MPPs and BM failure
Using a cellular system, we showed that Nle loss of function induced defective pre-60S subunit maturation and export, leading to the rapid diminution of free ribosomes. In vivo, by combining rRNA FISH and flow cytometry, we were able to detect alterations in ribosome biogenesis in HSCs and immature progenitors at early time points after Nle inactivation. This suggests that these defects cause HSC/MPP exhaustion in NlecKO mice. Defective ribosome biogenesis might cause HSC elimination through several mechanisms. A shortage of ribosomes would affect translation at a global level, although IRES-mediated translation may be more sensitive to ribosome insufficiency than a cap-dependent one (Horos et al., 2012). Preferential translation of mRNAs with a 5′ terminal oligopyrimidine tract has also been reported upon defective ribosome biogenesis (Fumagalli et al., 2009). In addition, suppressed translation of a specific subset of mRNAs has been observed upon ribosomal protein haploinsufficiency/knockdown (Kondrashov et al., 2011; Horos et al., 2012). Therefore, altered translation of transcripts essential for HSC function might contribute to the phenotype of NlecKO mice. Impaired ribosome biogenesis is known to efficiently trigger the up-regulation of p53 function through binding of MDM2 by free ribosomal proteins RPL11, RPL23, RPL5, or RPS7 (Zhang and Lu, 2009). Consistently, we demonstrated the rapid activation of the p53 pathway in Nle-deficient HSCs/MPPs. Up-regulation of the proapoptotic genes Bax, Puma, and Noxa in these compartments suggests that apoptosis may actively contribute to their elimination. However Annexin V and TUNEL stainings failed to unravel increased cell death in Nle-deficient HSCs/MPPs. The reasons for this discrepancy are yet unclear but may be caused by the rapid clearance of dying mutant cells in the BM or the fact that these dying cells lose some of their surface markers and therefore were not included in our analyses. As a complementary approach, we performed deletion of Nle in vitro in sorted HSC/MPP populations. Unfortunately, in vitro Cre activation in these populations compromises their maintenance and disabled us from using this assay to assess apoptosis in these cells. Other cellular responses to p53 activation may also participate in the exhaustion of Nle-deficient HSCs/MPPs. Hence, it has been suggested that p53 negatively regulates self-renewal of HSCs and MPPs, although how p53 regulates this process is not precisely understood (Akala et al., 2008; Chen et al., 2008). Induction of differentiation was also shown to represent an effective response to stress in various stem cell populations (Inomata et al., 2009; Li et al., 2012; Wang et al., 2012). Thus, it will be interesting to determine whether premature differentiation could be induced by defective ribosome biogenesis in Nle-deficient HSCs, thereby contributing to their exhaustion. Regardless the underlying molecular mechanisms, disappearance of Nle-deficient HSCs/MPPs is genetically dependent on p53. In the absence of p53, NlecKO HSCs and immature progenitor populations were unaffected, although accumulation of pre-60S rRNA intermediates was still observed. Further analyses at later time points will be required to determine whether and how impaired ribosome biogenesis consecutive to NLE depletion affects HSC functionality independently of the p53-dependent ribosome biogenesis checkpoint.
Differential requirement of NLE function
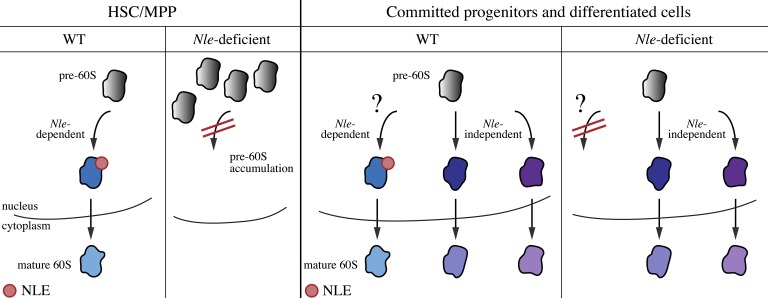
Considering its conserved and critical role in large ribosomal subunit biogenesis, our observation that NLE is dispensable in restricted progenitors was unexpected. Importantly, we did not detect accumulation of rRNA intermediates in Nle-deficient B and Lin+ cells, suggesting that pre-60S maturation occurs normally in the absence of NLE in these cells. Notably, normal proliferation, differentiation, and maturation of B cell lineage after constitutive inactivation of Nle with the CD19-Cre driver strengthens this conclusion and argues against the hypothesis that long-lasting inherited ribosomes could compensate for a defective ribosome biogenesis. Collectively, our findings support the hypothesis that alternative Nle-independent pathways can assure 60S subunit production in restricted progenitors but not in HSCs/MPPs (Fig. 10). An NLE-related WD-repeat protein, WDR12/Ytm1, containing an N-terminal domain homologous to the NLE N-terminal domain is present in all eukaryotes and also participates in pre-60S maturation (Nal et al., 2002; Bassler et al., 2010). However, WDR12/Ytm1 and NLE/Rsa4 have been shown to act sequentially and in a nonredundant manner during pre-60S subunit maturation in yeast (Ulbrich et al., 2009; Bassler et al., 2010). Ribosome biogenesis has been extensively studied in yeast, but is yet poorly described in higher eukaryotes. Several differences have been reported between yeast and higher eukaryotic cells in terms of rRNA maturation steps or composition of preribosomal particles (see for example, Turner et al., 2009; Srivastava et al., 2010). Whether ribosome biogenesis is qualitatively modulated during development or between cell types in a pluricellular organism is still an open question. This study is to our knowledge the first in vivo demonstration that differences in ribosome biogenesis exist between cell types in mammals. Recently, the ribosomal filter hypothesis postulating that differences in composition of ribosomal subunit or posttranslational modifications of ribosomal proteins may differentially affect the translation of particular transcripts has received support from studies performed in budding yeast and mouse embryo (Komili et al., 2007; Kondrashov et al., 2011). If this is true, ribosome heterogeneity implies some degrees of plasticity during preribosomal subunit assembly. Differential activities of alternative pathways for 60S subunit maturation between HSCs/MPPs and restricted progenitors could thus lead to the generation of qualitatively distinct ribosomes and therefore would represent an additional mode of gene expression regulation.
Figure 10.
Model for the role of Nle in hematopoietic cells. In HSCs/MPPs, Nle is absolutely required for proper 60S subunit biogenesis. In the absence of Nle, the pathway is impaired and the pre-60S subunit accumulates within the nucleus. In more committed progenitors and differentiated cells, an Nle-independent pathway is active and allows the production of the 60S subunit in absence of Nle.
Concluding remarks
The expression of several genes involved in ribosome biogenesis is highly enriched in Drosophila ovarian stem cells (Kai et al., 2005) and fish neural stem cells (Joly, J.-S., personal communication). A genome-wide gain of function screen in mouse ES cells identified multifunctional nucleolar proteins involved notably in ribosome biogenesis as novel regulators of self-renewal (Abujarour et al., 2010). Similarly, genetic screens for mutants that affect neuroblast and ovarian germline stem cell maintenance in Drosophila identified genes involved in ribosome biogenesis (Fichelson et al., 2009; Neumüller et al., 2011). Together with our findings, these observations link ribosome biogenesis with stem cell biology. Beyond overall protein synthesis and cell growth, specificities in preribosomal factors or ribosomal protein expression may contribute to stem cell/immature progenitor identity. In the future, it will be important to address whether such specificities and in particular dependency on the NLE pre-60S maturation pathway contribute to self-renewal capacity of malignant cells.
MATERIALS AND METHODS
Mice.
Experiments on live mice were conducted according to the French and European regulations on care and protection of laboratory animals (EC Directive 86/609, French Law 2001-486 issued on June 6, 2001) and the National Institutes of Health Animal Welfare (Insurance #A5476-01 issued on 02/07/2007). Control and NlecKO animals were obtained by crossing mice harboring NleFlox (Fig. S1), Nlenull (Cormier et al., 2006), Rosa26CreERT2 (Hameyer et al., 2007), CD19Cre (Rickert et al., 1997), or p53− (Jacks et al., 1994) alleles. Mice were injected intraperitoneally with 75 mg/kg tamoxifen for three or five consecutive days. NlecKO mice were systematically compared with control littermates treated under the same conditions.
Nle gene targeting and ES cell culture.
ES cells were cultured in DMEM + GlutaMAX-I (Gibco) supplemented with 15% FCS, 100 µM β-mercaptoethanol, 100 U/ml penicillin, 0.1 mg/ml streptomycin, and 103 U/ml LIF. NleFlox/+ ES cells were obtained by homologous recombination in CK35 129/Sv ES cells (Kress et al., 1998; for details see Fig. S1). Control and NlecKO ES cells were obtained by iterative gene targeting. NleFlox/+ ES cells were electroporated with the NlenLacZ targeting vector (Cormier et al., 2006) to generate NleFlox/null ES clones. Then, SfiI-linearized pRosaCreERT2 targeting vector (Hameyer et al., 2007) was electroporated into NleFlox/+ and NleFlox/null ES cells to generate control (Rosa26CreERT2/+; NleFlox/+) and NlecKO (Rosa26CreERT2/+; NleFlox/null) ES cells, respectively. The deletion of the NleFlox allele in culture was induced by treatment with 1 µM 4-OHT for two consecutive days.
Antibodies.
We raised an anti-NLE polyclonal antibody in rabbit. Recombinant fusion peptide corresponding to amino acids 1–174 of the NLE protein was produced in Escherichia coli as a His-tagged protein from pET15b expression vector (EMD Millipore) and purified using immobilized metal ion affinity chromatography. Purified peptide was then used as an immunogen for the production of rabbit antiserum (Agro-Bio). Antibody was used at 1:50 and 1:500 dilutions for immunofluorescence and immunoblotting, respectively. Antibodies used for flow cytometry are listed in Table S1.
Flow cytometry analysis.
Single cell suspensions of BM, spleen, and PB were stained with different combinations of antibodies. Marker combinations and FACS gating strategy used to define the various BM cell populations are indicated in Fig. S2. Apoptosis was monitored using 7AAD and Annexin V (Roche) according to the manufacturer’s instructions. For proliferation analysis and p53 labeling, cells were incubated with fixation/permeabilization solution (eBioscience) and stained either with anti-Ki67 (B56; BD) antibody and 10 µg/ml Hoechst 33342 or with p53 antibody (CM5; Novocastra) and Alexa Fluor 488–conjugated anti–rabbit secondary antibody, respectively. CyAn ADP, FACSCanto, and LSR Fortessa (BD) flow cytometers were used for analysis. Sorting was performed using the MoFlo cytometer (Beckman Coulter) or the AutoMACS technology (Miltenyi Biotec). Data were analyzed with FlowJo software (Tree Star).
Ex vivo differentiation assay.
HSCs were cultured on an OP9 monolayer in Opti-MEM, 10% FCS, 100 µM β-mercaptoethanol, 100 U/ml penicillin, 0.1 mg/ml streptomycin, Il-7, Flt3 ligand, and GM-CSF (supernatants of cell lines transfected with expression vectors for IL-7, Flt3 ligand, and GM-CSF, respectively; a gift from F. Melchers, Max Planck Institute for Infection Biology, Berlin, Germany).
Transplantation assays.
For noncompetitive transplantations, ∼8 × 105 Lin− cells from control or NlecKO mice (129Sv/C57BL/6 mixed background) were injected into the retroorbital sinus of sublethally irradiated (11 Grays) (129Sv×C57Bl6)F1 mice carrying the Act-GFP transgene (Okabe et al., 1997). BM was depleted from lineage-positive cells to avoid graft versus host reaction. The converse transplantation was performed similarly using donor and recipient mice selected for histocompatibility matching through genotyping of the chromosome 17 region encompassing major MHC class I loci. For competitive transplantations, Lin− control or NlecKO cells (CD45.2, 129Sv/C57Bl6 mix) were mixed with an excess (3:1 or 4:1 ratio) of competitor cells (CD45.1, C57BL/6) and grafted into a sublethally irradiated recipient (CD45.1×CD45.2, (129Sv×C57Bl6)F1). 2 mo after grafting, the reconstitution rate was assayed by flow cytometry analysis of PB cells using either GFP expression or CD45.1/45.2 immunostaining (noncompetitive and competitive transplantations, respectively).
Northern blot analysis.
Total RNA from ES cells was prepared with TRIzol reagent (Sigma-Aldrich) according to the supplier’s instructions. Migration and hybridization were performed as previously described (Fichelson et al., 2009). The probes used in this study were 5.8S (5′-GCGTTCGAAGTGTCGATGATCAATGTGTCCTGCAATTCAC-3′), 18S (5′-ACGGTATCTGATCGTCTTCGAACC-3′), 28S (5′-GTTCCCTTGGCTGTGGTTTCGCTAGATA-3′), and its2 (5′-ACTGGTGAGGCAGCGGTCCGGGAGGCGCCGACG-3′).
Sucrose density gradient.
Cells were treated for 10 min with 10 mg/ml cycloheximide (Sigma-Aldrich) in cell culture medium, collected, and then resuspended in buffer lysis (50 mM Tris-Hcl, pH 7.4, 50 mM Kcl, 10 mM MgAc, 1 mM DTT [Invitrogen], 200 U/ml RNasin [Promega], 1 tablet/10 ml proteases inhibitors [Roche], and 10 mg/ml cycloheximide [Sigma-Aldrich]). 200-µl glass beads were added to cell suspension, and lysis was performed by vortexing cells at 4°C, for 1 min, five times. After centrifugation, supernatants were layered on a 15–50% sucrose gradient and centrifuged for 15 h at 33,000 rpm in an SW41 rotor (Beckman Coulter). The gradient was fractionated with a fractions collector system coupled to a UV detector to monitor absorbance at 260 nm. Half of each fraction was treated with trichloroacetic acid to recover proteins, whereas the remaining half was treated with phenol-chloroform to extract RNA.
FISH analysis.
ES cells or stained hematopoietic cells were fixed for 30 min with 4% PFA, rinsed in PBS, and placed in 70% ethanol at 4°C. The hybridization was performed as previously described (O’Donohue et al., 2010). Cy3 conjugated probes were purchased at Eurogentec (Its1, 5′-TAGACACGGAAGAGCCGGACGGGAAAGA-3′; its2, 5′-GCGATTGATCGTCAACCGACGCTC-3′; and 5′ets, 5′-AGAGAAAAGAGCGGAGGTTCGGGACTCCAA-3′).
Genotyping of mice and cells.
Genotyping was performed by PCR after lysis of tissue and ES cell pellet in the following buffer: 50 mM Tris, pH 8.5, 100 mM NaCl, 0.5% Tween 20, and 100 mg/ml proteinase K at 56°C overnight, followed by a 10-min incubation at 96°C. For Nle, PCR amplification with primers a (5′-CAGACTTTCTTATTGGGGTTTGGT-3′), b (5′-TAGCCCTGGCTGACCTAGAA-3′), b′ (5′-TCCCCTCGAGGGACCTAATA-3′), and c (5′-ATGCCCGGCGTGTTAAGATT-3′) allowed us to discriminate Nlewt (538 bp, primers a and b), NleFlox (439 bp, primers a and b′), and NleDel (773 bp, primers a and c) alleles. D17mit24 and D17Mit135 polymorphic markers were used to discriminate between 129Sv and C57BL/6 alleles around MCH class I loci.
Statistical analysis.
For all bar graphs with pooled data, means (SD) are shown. Statistical analyses were performed using the Mann–Whitney Wilcoxon test. Significance is indicated on the figures with the following convention: *, P < 0.05; **, P < 0.005; ***, P < 0.0005.
Real-time RT-PCR.
mRNA from sorted cells was purified using either the Dynabeads mRNA direct kit (Invitrogen) or RNeasy Plus Micro kit (QIAGEN) and reverse-transcribed using the SuperScript Vilo cDNA synthesis kit (Invitrogen), according to the manufacturers’ instructions. Real-time PCR was then performed using custom-designed primers (compare in Table S2) and SYBR green PCR master mix on a StepOne instrument (StepOne software version 2.2; Applied Biosystems) or, alternatively, with the Fluidigm Biomark EvaGreen qPCR system (Fluidigm Real-Time PCR analysis software version 3.1.3; Fluidigm). Expression levels were normalized using several housekeeping genes, including TBP, Tubulin β 5, Aldolase, TfIID, and Rrm.
Online supplemental material.
Fig. S1 describes the generation of the NleFlox allele by homologous recombination in ES cells. Fig. S2 shows hierarchical relations of various hematopoietic cell populations defined by marker combinations and illustrates the FACS-gating strategy used for analysis. Table S1 lists the antibodies used for flow cytometry. Table S2 lists the sequence primers used for real-time RT-PCR. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20122019/DC1.
Supplementary Material
Acknowledgments
Imaging and cell sorting analyses were performed at the Institut Pasteur Imagopole. We are grateful to P.-H. Commere for his assistance with FACS analysis and sorting. We thank J. Jonker and A. Berns for the Rosa26CreERT2 targeting vector and mouse line, K. Rajewsky for the CD19-Cre mouse line, and L. Guillemot, M. Fromont-Racine, S. Duncker, M.-F. O’Donohue, P.-E. Gleizes, C. Moch, J.-R. Huyn, A. Mougin, F. Morlé, S. Tajbakhsh, and all members of the laboratory for technical advice and helpful discussions.
This work was supported by the Centre National de la Recherche Scientifique, the Institut Pasteur, the Agence Nationale de la Recherche (contract no. JC05_41835 and ANR-10-LABX-73-01 REVIVE), and the Institut National du Cancer (INCa 2007-1-COL-6-IC-1 and PLBIO09-070). M. Le Bouteiller was supported by the Université Pierre et Marie Curie and received fellowship from the Ministère de l’Education Nationale, de la Recherche et de la Technologie, and the Ligue Nationale Contre le Cancer.
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- 4-OHT
- 4-hydroxy-tamoxifen
- CLP
- common lymphoid progenitor
- CMP
- common myeloid progenitor
- ES
- embryonic stem
- FISH
- fluorescence in situ hybridization
- HSC
- hematopoietic stem cell
- MPP
- multipotent progenitor
- Nle
- Notchless
- PB
- peripheral blood
References
- Abujarour R., Efe J., Ding S. 2010. Genome-wide gain-of-function screen identifies novel regulators of pluripotency. Stem Cells. 28:1487–1497 10.1002/stem.472 [DOI] [PubMed] [Google Scholar]
- Akala O.O., Park I.-K., Qian D., Pihalja M., Becker M.W., Clarke M.F. 2008. Long-term haematopoietic reconstitution by Trp53-/-p16Ink4a-/-p19Arf-/- multipotent progenitors. Nature. 453:228–232 10.1038/nature06869 [DOI] [PubMed] [Google Scholar]
- Bassler J., Kallas M., Pertschy B., Ulbrich C., Thoms M., Hurt E. 2010. The AAA-ATPase Rea1 drives removal of biogenesis factors during multiple stages of 60S ribosome assembly. Mol. Cell. 38:712–721 10.1016/j.molcel.2010.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigas A., Espinosa L. 2012. Hematopoietic stem cells: to be or Notch to be. Blood. 119:3226–3235 10.1182/blood-2011-10-355826 [DOI] [PubMed] [Google Scholar]
- Chantha S.-C., Matton D.P. 2007. Underexpression of the plant NOTCHLESS gene, encoding a WD-repeat protein, causes pleitropic phenotype during plant development. Planta. 225:1107–1120 10.1007/s00425-006-0420-z [DOI] [PubMed] [Google Scholar]
- Chantha S.C., Gray-Mitsumune M., Houde J., Matton D.P. 2010. The MIDASIN and NOTCHLESS genes are essential for female gametophyte development in Arabidopsis thaliana. Physiol. Mol. Biol. Plants. 16:3–18 10.1007/s12298-010-0005-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Ellison F.M., Keyvanfar K., Omokaro S.O., Desierto M.J., Eckhaus M.A., Young N.S. 2008. Enrichment of hematopoietic stem cells with SLAM and LSK markers for the detection of hematopoietic stem cell function in normal and Trp53 null mice. Exp. Hematol. 36:1236–1243 10.1016/j.exphem.2008.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormier S., Le Bras S., Souilhol C., Vandormael-Pournin S., Durand B., Babinet C., Baldacci P., Cohen-Tannoudji M. 2006. The murine ortholog of notchless, a direct regulator of the notch pathway in Drosophila melanogaster, is essential for survival of inner cell mass cells. Mol. Cell. Biol. 26:3541–3549 10.1128/MCB.26.9.3541-3549.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilova N., Sakamoto K.M., Lin S. 2011. Ribosomal protein L11 mutation in zebrafish leads to haematopoietic and metabolic defects. Br. J. Haematol. 152:217–228 10.1111/j.1365-2141.2010.08396.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz J., Sanz-Martínez E., Remacha M. 2005. The essential WD-repeat protein Rsa4p is required for rRNA processing and intra-nuclear transport of 60S ribosomal subunits. Nucleic Acids Res. 33:5728–5739 10.1093/nar/gki887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutt S., Narla A., Lin K., Mullally A., Abayasekara N., Megerdichian C., Wilson F.H., Currie T., Khanna-Gupta A., Berliner N., et al. 2011. Haploinsufficiency for ribosomal protein genes causes selective activation of p53 in human erythroid progenitor cells. Blood. 117:2567–2576 10.1182/blood-2010-07-295238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fichelson P., Moch C., Ivanovitch K., Martin C., Sidor C.M., Lepesant J.-A., Bellaiche Y., Huynh J.-R. 2009. Live-imaging of single stem cells within their niche reveals that a U3snoRNP component segregates asymmetrically and is required for self-renewal in Drosophila. Nat. Cell Biol. 11:685–693 10.1038/ncb1874 [DOI] [PubMed] [Google Scholar]
- Fromont-Racine M., Senger B., Saveanu C., Fasiolo F. 2003. Ribosome assembly in eukaryotes. Gene. 313:17–42 10.1016/S0378-1119(03)00629-2 [DOI] [PubMed] [Google Scholar]
- Fumagalli S., Di Cara A., Neb-Gulati A., Natt F., Schwemberger S., Hall J., Babcock G.F., Bernardi R., Pandolfi P.P., Thomas G. 2009. Absence of nucleolar disruption after impairment of 40S ribosome biogenesis reveals an rpL11-translation-dependent mechanism of p53 induction. Nat. Cell Biol. 11:501–508 10.1038/ncb1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan B., Hu J., Jiang S., Liu Y., Sahin E., Zhuang L., Fletcher-Sananikone E., Colla S., Wang Y.A., Chin L., Depinho R.A. 2010. Lkb1 regulates quiescence and metabolic homeostasis of haematopoietic stem cells. Nature. 468:701–704 10.1038/nature09595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganapathi K.A., Shimamura A. 2008. Ribosomal dysfunction and inherited marrow failure. Br. J. Haematol. 141:376–387 10.1111/j.1365-2141.2008.07095.x [DOI] [PubMed] [Google Scholar]
- Gazave E., Lapébie P., Richards G.S., Brunet F., Ereskovsky A.V., Degnan B.M., Borchiellini C., Vervoort M., Renard E. 2009. Origin and evolution of the Notch signalling pathway: an overview from eukaryotic genomes. BMC Evol. Biol. 9:249 10.1186/1471-2148-9-249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert W.V. 2011. Functional specialization of ribosomes? Trends Biochem. Sci. 36:127–132 10.1016/j.tibs.2010.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurumurthy S., Xie S.Z., Alagesan B., Kim J., Yusuf R.Z., Saez B., Tzatsos A., Ozsolak F., Milos P., Ferrari F., et al. 2010. The Lkb1 metabolic sensor maintains haematopoietic stem cell survival. Nature. 468:659–663 10.1038/nature09572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall L.E., Alexander S.J., Chang M., Woodling N.S., Yedvobnick B. 2004. An EP overexpression screen for genetic modifiers of Notch pathway function in Drosophila melanogaster. Genet. Res. 83:71–82 10.1017/S0016672304006731 [DOI] [PubMed] [Google Scholar]
- Hameyer D., Loonstra A., Eshkind L., Schmitt S., Antunes C., Groen A., Bindels E., Jonkers J., Krimpenfort P., Meuwissen R., et al. 2007. Toxicity of ligand-dependent Cre recombinases and generation of a conditional Cre deleter mouse allowing mosaic recombination in peripheral tissues. Physiol. Genomics. 31:32–41 10.1152/physiolgenomics.00019.2007 [DOI] [PubMed] [Google Scholar]
- Higashi A.Y., Ikawa T., Muramatsu M., Economides A.N., Niwa A., Okuda T., Murphy A.J., Rojas J., Heike T., Nakahata T., et al. 2009. Direct hematological toxicity and illegitimate chromosomal recombination caused by the systemic activation of CreERT2. J. Immunol. 182:5633–5640 10.4049/jimmunol.0802413 [DOI] [PubMed] [Google Scholar]
- Hock H., Hamblen M.J., Rooke H.M., Schindler J.W., Saleque S., Fujiwara Y., Orkin S.H. 2004. Gfi-1 restricts proliferation and preserves functional integrity of haematopoietic stem cells. Nature. 431:1002–1007 10.1038/nature02994 [DOI] [PubMed] [Google Scholar]
- Horos R., Ijspeert H., Pospisilova D., Sendtner R., Andrieu-Soler C., Taskesen E., Nieradka A., Cmejla R., Sendtner M., Touw I.P., von Lindern M. 2012. Ribosomal deficiencies in Diamond-Blackfan anemia impair translation of transcripts essential for differentiation of murine and human erythroblasts. Blood. 119:262–272 10.1182/blood-2011-06-358200 [DOI] [PubMed] [Google Scholar]
- Inomata K., Aoto T., Binh N.T., Okamoto N., Tanimura S., Wakayama T., Iseki S., Hara E., Masunaga T., Shimizu H., Nishimura E.K. 2009. Genotoxic stress abrogates renewal of melanocyte stem cells by triggering their differentiation. Cell. 137:1088–1099 10.1016/j.cell.2009.03.037 [DOI] [PubMed] [Google Scholar]
- Insinga A., Cicalese A., Faretta M., Gallo B., Albano L., Ronzoni S., Furia L., Viale A., Pelicci P.G. 2013. DNA damage in stem cells activates p21, inhibits p53, and induces symmetric self-renewing divisions. Proc. Natl. Acad. Sci. USA. 110:3931–3936 10.1073/pnas.1213394110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T., Remington L., Williams B.O., Schmitt E.M., Halachmi S., Bronson R.T., Weinberg R.A. 1994. Tumor spectrum analysis in p53-mutant mice. Curr. Biol. 4:1–7 10.1016/S0960-9822(00)00002-6 [DOI] [PubMed] [Google Scholar]
- Ji H., Wu G., Zhan X., Nolan A., Koh C., De Marzo A., Doan H.M., Fan J., Cheadle C., Fallahi M., et al. 2011. Cell-type independent MYC target genes reveal a primordial signature involved in biomass accumulation. PLoS ONE. 6:e26057 10.1371/journal.pone.0026057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jude C.D., Climer L., Xu D., Artinger E., Fisher J.K., Ernst P. 2007. Unique and independent roles for MLL in adult hematopoietic stem cells and progenitors. Cell Stem Cell. 1:324–337 10.1016/j.stem.2007.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kai T., Williams D., Spradling A.C. 2005. The expression profile of purified Drosophila germline stem cells. Dev. Biol. 283:486–502 10.1016/j.ydbio.2005.04.018 [DOI] [PubMed] [Google Scholar]
- Kiel M.J., Yilmaz Ö.H., Iwashita T., Yilmaz O.H., Terhorst C., Morrison S.J. 2005. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 121:1109–1121 10.1016/j.cell.2005.05.026 [DOI] [PubMed] [Google Scholar]
- Komili S., Farny N.G., Roth F.P., Silver P.A. 2007. Functional specificity among ribosomal proteins regulates gene expression. Cell. 131:557–571 10.1016/j.cell.2007.08.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov N., Pusic A., Stumpf C.R., Shimizu K., Hsieh A.C., Xue S., Ishijima J., Shiroishi T., Barna M. 2011. Ribosome-mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell. 145:383–397 10.1016/j.cell.2011.03.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranc K.R., Schepers H., Rodrigues N.P., Bamforth S., Villadsen E., Ferry H., Bouriez-Jones T., Sigvardsson M., Bhattacharya S., Jacobsen S.E., Enver T. 2009. Cited2 is an essential regulator of adult hematopoietic stem cells. Cell Stem Cell. 5:659–665 10.1016/j.stem.2009.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kress C., Vandormael-Pournin S., Baldacci P., Cohen-Tannoudji M., Babinet C. 1998. Nonpermissiveness for mouse embryonic stem (ES) cell derivation circumvented by a single backcross to 129/Sv strain: establishment of ES cell lines bearing the Omd conditional lethal mutation. Mamm. Genome. 9:998–1001 10.1007/s003359900914 [DOI] [PubMed] [Google Scholar]
- Laurenti E., Varnum-Finney B., Wilson A., Ferrero I., Blanco-Bose W.E., Ehninger A., Knoepfler P.S., Cheng P.-F., MacDonald H.R., Eisenman R.N., et al. 2008. Hematopoietic stem cell function and survival depend on c-Myc and N-Myc activity. Cell Stem Cell. 3:611–624 10.1016/j.stem.2008.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung C.G., Xu Y., Mularski B., Liu H., Gurbuxani S., Crispino J.D. 2007. Requirements for survivin in terminal differentiation of erythroid cells and maintenance of hematopoietic stem and progenitor cells. J. Exp. Med. 204:1603–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., He Y., Dubois W., Wu X., Shi J., Huang J. 2012. Distinct regulatory mechanisms and functions for p53-activated and p53-repressed DNA damage response genes in embryonic stem cells. Mol. Cell. 46:30–42 10.1016/j.molcel.2012.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillard I., Koch U., Dumortier A., Shestova O., Xu L., Sai H., Pross S.E., Aster J.C., Bhandoola A., Radtke F., Pear W.S. 2008. Canonical notch signaling is dispensable for the maintenance of adult hematopoietic stem cells. Cell Stem Cell. 2:356–366 10.1016/j.stem.2008.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohrin M., Bourke E., Alexander D., Warr M.R., Barry-Holson K., Le Beau M.M., Morrison C.G., Passegué E. 2010. Hematopoietic stem cell quiescence promotes error-prone DNA repair and mutagenesis. Cell Stem Cell. 7:174–185 10.1016/j.stem.2010.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourikis P., Lake R.J., Firnhaber C.B., DeDecker B.S. 2010. Modifiers of notch transcriptional activity identified by genome-wide RNAi. BMC Dev. Biol. 10:107 10.1186/1471-213X-10-107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naiche L.A., Papaioannou V.E. 2007. Cre activity causes widespread apoptosis and lethal anemia during embryonic development. Genesis. 45:768–775 10.1002/dvg.20353 [DOI] [PubMed] [Google Scholar]
- Nakada D., Saunders T.L., Morrison S.J. 2010. Lkb1 regulates cell cycle and energy metabolism in haematopoietic stem cells. Nature. 468:653–658 10.1038/nature09571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nal B., Mohr E., Silva M.-I.D., Tagett R., Navarro C., Carroll P., Depetris D., Verthuy C., Jordan B.R., Ferrier P. 2002. Wdr12, a mouse gene encoding a novel WD-Repeat Protein with a notchless-like amino-terminal domain. Genomics. 79:77–86 10.1006/geno.2001.6682 [DOI] [PubMed] [Google Scholar]
- Narla A., Ebert B.L. 2010. Ribosomopathies: human disorders of ribosome dysfunction. Blood. 115:3196–3205 10.1182/blood-2009-10-178129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumüller R.A., Richter C., Fischer A., Novatchkova M., Neumüller K.G., Knoblich J.A. 2011. Genome-wide analysis of self-renewal in Drosophila neural stem cells by transgenic RNAi. Cell Stem Cell. 8:580–593 10.1016/j.stem.2011.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nybakken K., Vokes S.A., Lin T.-Y., McMahon A.P., Perrimon N. 2005. A genome-wide RNA interference screen in Drosophila melanogaster cells for new components of the Hh signaling pathway. Nat. Genet. 37:1323–1332 10.1038/ng1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donohue M.F., Choesmel V., Faubladier M., Fichant G., Gleizes P.-E. 2010. Functional dichotomy of ribosomal proteins during the synthesis of mammalian 40S ribosomal subunits. J. Cell Biol. 190:853–866 10.1083/jcb.201005117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe M., Ikawa M., Kominami K., Nakanishi T., Nishimune Y. 1997. ‘Green mice’ as a source of ubiquitous green cells. FEBS Lett. 407:313–319 10.1016/S0014-5793(97)00313-X [DOI] [PubMed] [Google Scholar]
- Opferman J.T., Iwasaki H., Ong C.C., Suh H., Mizuno S., Akashi K., Korsmeyer S.J. 2005. Obligate role of anti-apoptotic MCL-1 in the survival of hematopoietic stem cells. Science. 307:1101–1104 10.1126/science.1106114 [DOI] [PubMed] [Google Scholar]
- Orkin S.H., Zon L.I. 2008. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 132:631–644 10.1016/j.cell.2008.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajerowski A.G., Shapiro M.J., Gwin K., Sundsbak R., Nelson-Holte M., Medina K., Shapiro V.S. 2010. Adult hematopoietic stem cells require NKAP for maintenance and survival. Blood. 116:2684–2693 10.1182/blood-2010-02-268391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passegué E., Wagers A.J., Giuriato S., Anderson W.C., Weissman I.L. 2005. Global analysis of proliferation and cell cycle gene expression in the regulation of hematopoietic stem and progenitor cell fates. J. Exp. Med. 202:1599–1611 10.1084/jem.20050967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Z., Chen L., Fernald A.A., Williams B.O., Le Beau M.M. 2008. A critical role for Apc in hematopoietic stem and progenitor cell survival. J. Exp. Med. 205:2163–2175 10.1084/jem.20080578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall T.D., Weissman I.L. 1997. Phenotypic and functional changes induced at the clonal level in hematopoietic stem cells after 5-fluorouracil treatment. Blood. 89:3596–3606 [PubMed] [Google Scholar]
- Rickert R.C., Roes J., Rajewsky K. 1997. B lymphocyte-specific, Cre-mediated mutagenesis in mice. Nucleic Acids Res. 25:1317–1318 10.1093/nar/25.6.1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royet J., Bouwmeester T., Cohen S.M. 1998. Notchless encodes a novel WD40-repeat-containing protein that modulates Notch signaling activity. EMBO J. 17:7351–7360 10.1093/emboj/17.24.7351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J. 1929. The minute reaction in the Development of Drosophila melanogaster. Genetics. 14:366–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava L., Lapik Y.R., Wang M., Pestov D.G. 2010. Mammalian DEAD box protein Ddx51 acts in 3′ end maturation of 28S rRNA by promoting the release of U8 snoRNA. Mol. Cell. Biol. 30:2947–2956 10.1128/MCB.00226-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tothova Z., Kollipara R., Huntly B.J., Lee B.H., Castrillon D.H., Cullen D.E., McDowell E.P., Lazo-Kallanian S., Williams I.R., Sears C., et al. 2007. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell. 128:325–339 10.1016/j.cell.2007.01.003 [DOI] [PubMed] [Google Scholar]
- Turner A.J., Knox A.A., Prieto J.-L., McStay B., Watkins N.J. 2009. A novel small-subunit processome assembly intermediate that contains the U3 snoRNP, nucleolin, RRP5, and DBP4. Mol. Cell. Biol. 29:3007–3017 10.1128/MCB.00029-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulbrich C., Diepholz M., Bassler J., Kressler D., Pertschy B., Galani K., Böttcher B., Hurt E. 2009. Mechanochemical removal of ribosome biogenesis factors from nascent 60S ribosomal subunits. Cell. 138:911–922 10.1016/j.cell.2009.06.045 [DOI] [PubMed] [Google Scholar]
- van Riggelen J., Yetil A., Felsher D.W. 2010. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat. Rev. Cancer. 10:301–309 10.1038/nrc2819 [DOI] [PubMed] [Google Scholar]
- Voutev R., Killian D.J., Ahn J.H., Hubbard E.J.A. 2006. Alterations in ribosome biogenesis cause specific defects in C. elegans hermaphrodite gonadogenesis. Dev. Biol. 298:45–58 10.1016/j.ydbio.2006.06.011 [DOI] [PubMed] [Google Scholar]
- Wang J., Sun Q., Morita Y., Jiang H., Gross A., Lechel A., Hildner K., Guachalla L.M., Gompf A., Hartmann D., et al. 2012. A differentiation checkpoint limits hematopoietic stem cell self-renewal in response to DNA damage. Cell. 148:1001–1014 10.1016/j.cell.2012.01.040 [DOI] [PubMed] [Google Scholar]
- Xue S., Barna M. 2012. Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nat. Rev. Mol. Cell Biol. 13:355–369 10.1038/nrm3359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Lu H. 2009. Signaling to p53: ribosomal proteins find their way. Cancer Cell. 16:369–377 10.1016/j.ccr.2009.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H.H., Ji K., Alderson N., He Z., Li S., Liu W., Zhang D.E., Li L., Feng G.S. 2011. Kit-Shp2-Kit signaling acts to maintain a functional hematopoietic stem and progenitor cell pool. Blood. 117:5350–5361 10.1182/blood-2011-01-333476 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.