Abstract
Purpose
This study investigated the association between tumor MYC protein expression and disease-free survival (DFS) of patients randomized to receive chemotherapy alone (Arm A) or chemotherapy with sequential (Arm B) or concurrent trastuzumab (Arm C) in the N9831 (Alliance) adjuvant HER2+ trastuzumab breast cancer trial.
Patients/Methods
This analysis included 1736 patients randomized to Arms A, B, and C on N9831. Nuclear MYC protein expression was determined in tissue microarray (TMA) sections containing three biopsies per patient or whole tissue sections (WS) using standard immunohistochemistry (clone 9E10). A tumor was considered positive for MYC protein overexpression (MYC+) if the nuclear 3+ staining percentage was >30%.
Results
574 (33%) tumors were MYC+. MYC+ was associated with hormone receptor positivity (χ2 p=0.006), tumors ≥ 2 cm (χ2 p=0.02), and a higher rate of nodal positivity (χ2 p<0.001). Hazard ratios (HRs) for DFS (median follow-up: 6.1 years) for Arm C versus A were 0.52 (p=0.006) and 0.65 (p=0.006) for patients with MYC+ and MYC- tumors, respectively (interaction p=0.40). For Arm B versus A, HRs for patients with MYC+ and MYC- tumors were 0.79 (p=0.21) and 0.74 (p=0.04), respectively (interaction p=0.71). For Arm C versus B, HRs for patients with MYC+ and MYC- tumors were 0.56 (p=0.02) and 0.89 (p=0.49), respectively (interaction p=0.17).
Conclusions
Our data do not support an impact of tumor MYC protein expression on differential benefit from adjuvant trastuzumab.
Keywords: trastuzumab, HER2-positive, breast cancer, MYC protein expression, disease-free survival
Introduction
Patients with human epidermal growth factor receptor-2-positive (HER2+) breast tumors have greatly benefited from the development of trastuzumab, a humanized monoclonal antibody directed against HER2 (1). Trastuzumab has proven to prolong the survival of women with metastatic breast cancer and significantly increase disease-free survival of patients with HER2+ early breast cancer (2, 3). However, many women who receive trastuzumab for advanced disease develop tumor progression within one year, and 15–25% of women diagnosed with HER2+ early disease develop tumor relapse within three years, despite therapy (4). Thus, identifying molecular markers that could predict the patients who are most likely to benefit from trastuzumab is an important research and clinical goal.
MYC is one of several markers reported to be involved in trastuzumab sensitivity and resistance. The MYC oncoprotein is a pleiotropic transcription factor and key regulator of cell growth, proliferation, metabolism, differentiation, apoptosis, and pathways that regulate genome stability and cell death (5, 6). MYC has been shown to be overexpressed in breast cancer with reported frequencies between 12–100% depending on antibody/technique and cutpoint utilized, patient heterogeneity, and molecular subtype of the breast tumor (5–7). MYC acts as a downstream target of HER2-driven proliferative signals in breast cancer cells in vitro (8), and deregulation of MYC contributes to breast cancer tumorigenesis and progression and is typically associated with poor outcomes (7).
Additionally, MYC gene amplification has been reported to predict additional trastuzumab benefit in a retrospective analysis of the National Surgical Adjuvant Breast and Bowel Project Cooperative Group (NSABP) B31 adjuvant trial. NSABP B31 showed that patients with MYC/HER2 co-amplification (defined as average copies/nucleus >5.0) in their primary breast tumors, benefited significantly more (interaction p=0.007) from trastuzumab than patients with only HER2 amplification, although a significant benefit of trastuzumab was observed in both MYC amplified and non-amplified patients (9).
Conversely, however, our results from the North Central Cancer Treatment Group (NCCTG) N9831 (10) did not support the link between MYC gene amplification and benefit from trastuzumab strictly on the basis of MYC amplification defined as > 5.0 average copies/nucleus. In the N9831 Intergroup adjuvant trastuzumab phase III trial, we observed differential benefit of trastuzumab in groups of HER2+ patients with <2.5 average MYC copies/nucleus and patients with alternative MYC and chromosome 8 copy number alterations (10).
Considering that protein overexpression may be independent of gene amplification (5), we designed the translational component of the N9831 trial to also include an analysis of the role of MYC protein overexpression in trastuzumab sensitivity. We therefore evaluated the association between MYC protein expression and disease-free survival (DFS) of patients randomized to receive chemotherapy alone (Arm A) or chemotherapy with sequential (Arm B) or concurrent trastuzumab (Arm C) on N9831.
Materials and Methods
Patients
The N9831 trial (NCT00005970) was a phase III trial in which patients were randomized to three arms: Arm A: doxorubicin and cyclophosphamide followed by weekly paclitaxel; Arm B: same as Arm A but followed by 1 year of sequential trastuzumab; Arm C: same as Arm A but with 1 year concurrent trastuzumab, started the same day as weekly paclitaxel (Supplemental Figure 1). Patients randomly assigned to the concurrent trastuzumab arm had a significantly increased DFS (P<.001; stratified hazard ratio [HR], 0.52; 95% CI, 0.45 to 0.60) and overall survival (OS)(P<.001; stratified HR, 0.61; 95% CI, 0.50 to 0.75) compared with patients assigned to the control arm (2). In the N9831 comparison of sequential versus concurrent trastuzumab chemotherapy, there was an increase in DFS with concurrent trastuzumab (P=.02; HR, 0.77; 99.9% CI, 0.53 to 1.11) (11). The 5-year OS rate for the sequential and concurrent arms were estimated at 89.7% (95% CI, 87.7% to 91.8%) and 91.9% (95% CI, 90.0% to 93.7%), respectively.
All patients’ tumors included in these analyses were tested for HER2 protein overexpression or gene amplification at a central laboratory (Mayo Clinic, Rochester). Patients were considered positive for HER2 according to the FDA-approved guidelines (IHC: complete 3+ membrane staining ≥ 10% invasive cells; FISH: HER2:CEP17 ratio ≥ 2.0) (12, 13). N9831 was approved by all treating sites’ Institutional Review Boards, and all patients signed informed consent. The Mayo Institutional Review Board and the Correlative Science Committee of the North American Breast Cancer Group (NABCG) approved this translational study.
This study included 1736 eligible/consented patients with sufficient tissue for analyses. Six-hundred eighty-two were excluded (failed central review: 283, ineligible: 61, canceled: 28, no consent: 187, lost to follow-up: 123) and 1087 had insufficient tissue for analyses (Supplemental Figure 2). The number of patients represented on tissue microarrays (TMAs) with evaluable tissue cores was 1216 and the number of different patients with evaluable whole sections (WS) was 520 (Supplemental Figure 2).
Tissue Microarrays and Whole Tissue Sections
TMAs were constructed as part of the translational study component of N9831 using an ATA-27 automated TMA construction system (Beecher Instruments, Silver Spring, MD) as described previously (10). Each TMA contained control biopsies from non-neoplastic human liver, placenta, and tonsil tissues. Whole tissue sections from tumors not represented on TMAs were also examined. We evaluated the concordance between TMA and WS protein analyses of 86 independent breast tumors and observed a concordance of 90% and 92% using the minimum and maximum TMA scores, respectively, of nuclear 3+ staining in >30% invasive cells.
MYC Testing Methods
Standard laboratory protocols were followed for immunohistochemistry (IHC) and quality control measures. Antigen retrieval was performed on deparaffinized whole or TMA sections (5µm) using preheated citrate buffer (98°C; 40 min). The tissue sections were treated with Peroxidase Blocking Reagent (Dako, Carpenteria, CA) and serum-free Protein Block (Dako) prior to IHC staining for c-MYC (mouse monoclonal clone 9E10; Sigma-Aldrich, #5546; St. Louis, MO; dilution 1:250; 60 min incubation) using a Dako Autostainer Plus (Reference #S3800). The sections were incubated in secondary antibody (Dako Envision Plus Dual Link Horse-Radish Peroxidase Kit; Dako # K4061). The high-sensitivity diaminobenzidine (DAB+) chromogenic substrate system (Betazoid DAB, Biocare) was used for colorimetric visualization followed by counter staining with hematoxylin.
MYC protein overexpression (MYC+) was defined as >30% of invasive cells with 3+ nuclear staining, based on the criteria used for HER2 protein overexpression established by the 2007 American Society of Clinical Oncologists/CAP guidelines, as well as the lack of any widely accepted other criteria for MYC positivity in the literature (14). We also evaluated cytoplasmic MYC protein expression because MYC has been observed in the cytoplasm of tumor cells (5), which has been shown to be correlated with increased survival of breast cancer patients (15).
Statistical Methods
The primary endpoint of N9831 was disease-free-survival (DFS) and was defined as local, regional, or distant recurrence, contralateral breast cancer, another primary cancer (except squamous or basal cell carcinoma of the skin, carcinoma in situ of the cervix, or lobular carcinoma in situ of the breast), or death from any cause. Duration of DFS was defined as the time from registration to the first DFS event. DFS was estimated by the Kaplan-Meier method. Comparisons between Arms A, B, and C within subgroups were performed using Cox proportional hazards models stratified by nodal status (1–3 vs. 4–9 vs. ≥10 positive nodes vs. positive sentinel node only vs. negative sentinel node with no axillary nodal dissection vs. axillary nodal dissection with no positive nodes) and hormone receptor status (estrogen receptor positive and/or progesterone receptor positive vs. negative for both receptors). We tested MYC protein expression as a predictor for differential trastuzumab benefit between MYC subgroups using Cox proportional hazards models (also stratified by nodal status and hormone receptor status), which included a treatment arm by MYC subgroup interaction term. The maximum nuclear MYC protein expression of the WS or of the replicate TMA biopsies was used for all analyses associated with patient outcome.
Results
Study Patients
The trial N9831 registered 3505 patients into Arms A (1232 patients), B (1216 patients), and C (1057 patients) of which 1736 patients (A: 584, B: 624, C: 528) were included in the statistical analysis of MYC protein expression (Supplemental Figure 2). The 1769 patients who were excluded from analysis were excluded for the following reasons: failed central HER2 pathology review (283 patients), ineligible (61 patients), cancelled prior to treatment initiation (28 patients), withdrew consent (187 patients), lost to follow-up (123 patients), and no/inadequate tissue or a technical failure of the assay (1087 patients). The median follow-up time was 6.1 years (September 21, 2010).
The clinicopathological characteristics of the 1736 patients enrolled on Arms A, B, and C reported herein were similar to the 1087 consented and eligible patients on Arms A, B, and C excluded from analysis because of a lack of MYC results (Supplemental Table 1) except that included patients tended to have larger tumors. The clinicopathological characteristics of the 1736 patients whose tumors had nuclear 3+ MYC protein staining in >30% and ≤ 30% invasive tumor cells are shown in Table 1. Patients whose tumors had nuclear 3+ staining in >30% invasive cells had a higher rate of hormone receptor positivity, larger tumors, higher rate of mastectomy, and higher number of positive nodes than those patients whose tumors had nuclear 3+ MYC staining in ≤ 30% invasive tumor cells.
Table 1.
Patient Characteristics by % 3+ Nuclear Staining
| Characteristic | ≤30% 3 + Nuclear Staining N=1162 (67%) |
>30% 3 + Nuclear Staining N=574 (33%) |
Chi-Square p-value |
||
|---|---|---|---|---|---|
| N (%) | N (%) | ||||
| Age (median) | 50 | (22–80) | 49 | (25–79) | |
| Age Group | |||||
| <40 | 200 | (17) | 106 | (18) | 0.49* |
| 40–49 | 370 | (32) | 185 | (32) | |
| 50–59 | 379 | (33) | 181 | (32) | |
| ≥ 60 | 213 | (18) | 102 | (18) | |
| Race | |||||
| White | 993 | (85) | 495 | (86) | 0.66 |
| Other | 169 | (15) | 79 | (14) | |
| Menopausal Status | |||||
| Pre-menopausal or < 50 | 610 | (52) | 316 | (55) | 0.32 |
| Post-menopausal or ≥ 50 | 552 | (48) | 258 | (45) | |
| ER/PR Status | |||||
| ER or PR Positive | 584 | (50) | 329 | (57) | 0.006 |
| Other | 578 | (50) | 245 | (43) | |
| Surgery | |||||
| Breast Conserving | 482 | (41) | 202 | (35) | 0.01 |
| Mastectomy | 680 | (58) | 372 | (65) | |
| Nodal Status† | |||||
| Node Positive (1–3 + nodes) | 429 | (37) | 249 | (43) | <0.001 |
| Node Positive (4–9 + nodes) | 298 | (26) | 150 | (26) | |
| Node Positive (≥10 + nodes) | 135 | (12) | 90 | (16) | |
| Node Negative (no pos. nodes) | 90 | (8) | 23 | (4) | |
| Positive Sentinel Node | 94 | (8) | 38 | (7) | |
| Negative Sentinel Node | 116 | (10) | 24 | (4) | |
| Predominant Tumor Histology | |||||
| Ductal | 1097 | (94) | 544 | (95) | 0.55 |
| Lobular | 33 | (3) | 19 | (3) | |
| Other | 31 | (3) | 11 | (2) | |
| Missing | 1 | 0 | |||
| Hist. Tumor Grade (Elston/SBR) | |||||
| Well /Intermediate | 327 | (28) | 159 | (28) | 0.85 |
| Poor | 835 | (72) | 415 | (72) | |
| Pathologic Tumor Size | |||||
| < 2 cm | 384 | (33) | 159 | (28) | 0.02 |
| ≥ 2 cm | 778 | (67) | 415 | (72) | |
Mantel-Haenszel trend test
Sentinel node findings are based on sentinel node dissection not followed by axillary dissection.
Distribution of MYC protein expression and relationship to HER2 protein expression
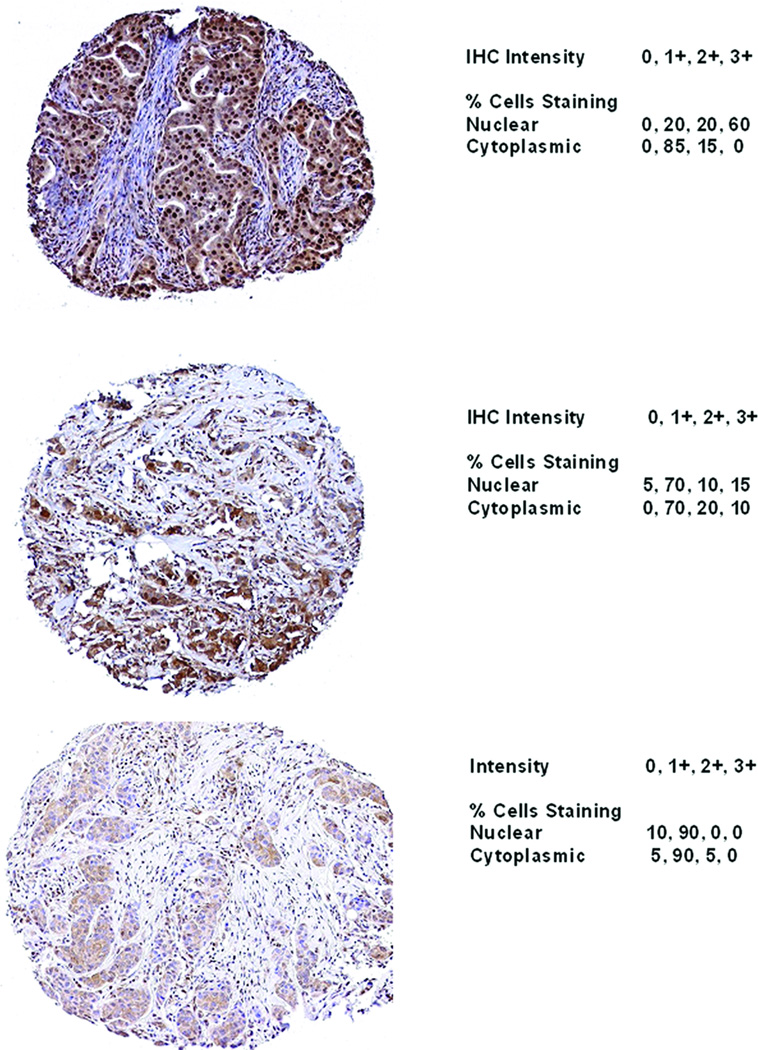
Of 1736 patients with evaluable IHC analyses, 33% (n=574) had >30% invasive cells, 28% (n=494) had 10–30%, and 38% (n=668) had <10% invasive cells with 3+ nuclear staining (Table 2). Nuclear and cytoplasmic 3+ staining had high agreement (81%; p<0.001), and the correlation between 3+ nuclear and 3+ cytoplasmic staining was 0.66 (p<0.001) (Table 2). No significant association was observed between 3+ nuclear staining and HER2 IHC staining (p= 0.10) (Table 3). Representative staining patterns of MYC protein expression are shown in Figure 1.
Table 2.
Correlation of MYC Cytoplasmic Staining with MYC Nuclear Staining.
| MYC % 3+ Nuclear Staining (Maximum Across Cores) | Total | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0–9% | 10–30% | 31–60% | 61–100% | |||||||
| MYC % 3+ Cytoplasmic Staining (Maximum Across Cores)* ** | 0–9% | 536 | (64%) | 203 | (24%) | 78 | (9%) | 27 | (3%) | 844 |
| 10–30% | 118 | (27%) | 205 | (46%) | 75 | (17%) | 43 | (10%) | 441 | |
| 31–60% | 11 | (6%) | 63 | (32%) | 74 | (38%) | 46 | (24%) | 194 | |
| 61–100% | 3 | (1%) | 23 | (9%) | 54 | (21%) | 177 | (69%) | 257 | |
| Total | 668 | 494 | 281 | 293 | 1736 | |||||
3+ Cytoplasmic Staining vs 3+ Nuclear Staining Mantel-Haenszel Chi-Square p-value = <0.001
Spearman correlation between 3+ Cytoplasmic Staining and 3+ Nuclear Staining = 0.66 (p-val = <0.001)
Table 3.
Correlation of MYC Nuclear Staining with HER2 Status
| MYC % 3+ Nuclear Staining (Maximum Across Cores) |
Total | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0–9% | 10–30% | 31–60% | 61–100% | |||||||
| HER2 IHC | 0, 1+ | 10 | (23%) | 10 | (23%) | 9 | (20%) | 15 | (34%) | 44 |
| 2+ | 70 | (42%) | 52 | (31%) | 16 | (10%) | 29 | (17%) | 167 | |
| 3+ | 582 | (38%) | 432 | (28%) | 256 | (17%) | 249 | (16%) | 1519 | |
| Total | 662 | 494 | 281 | 293 | 1730 | |||||
HER2 IHC Staining versus 3+ Nuclear Staining Mantel-Haenszel Chi-Square p-value = 0.10
Figure 1. Representative IHC Staining of MYC.
A. Representative staining of a specimen with >30% 3+ nuclear staining. B. Representative staining of a specimen with <30% 3+ nuclear staining. C. Representative staining of a specimen with 0% 3+ nuclear staining. 10× magnification.
Associations between MYC protein expression and DFS
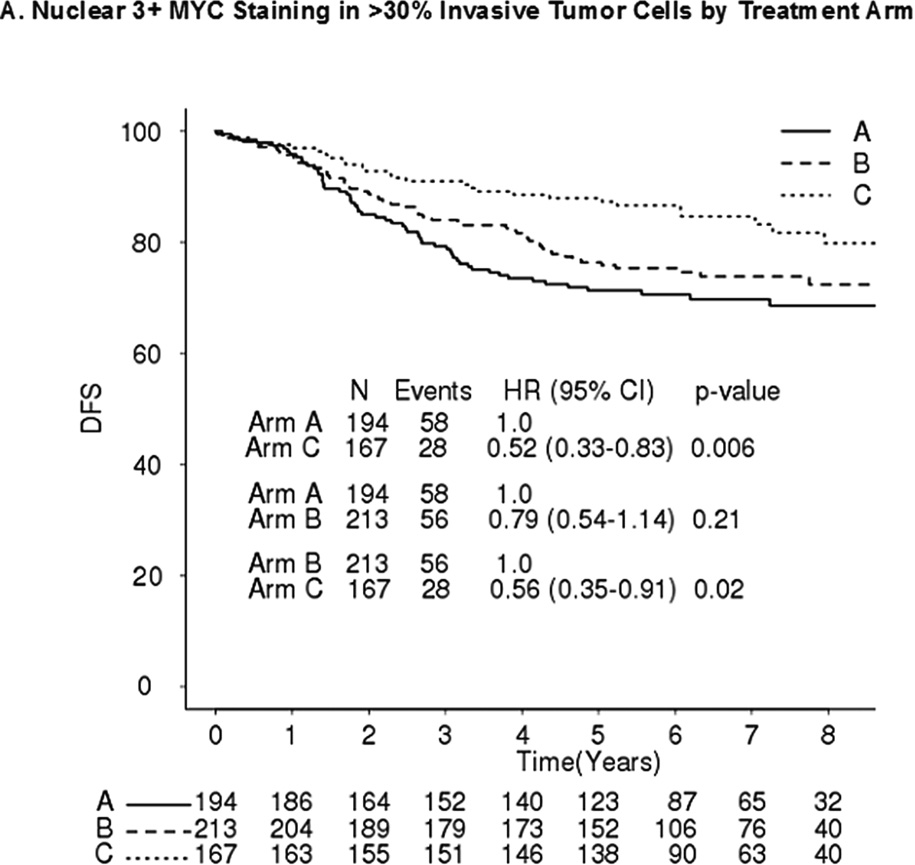
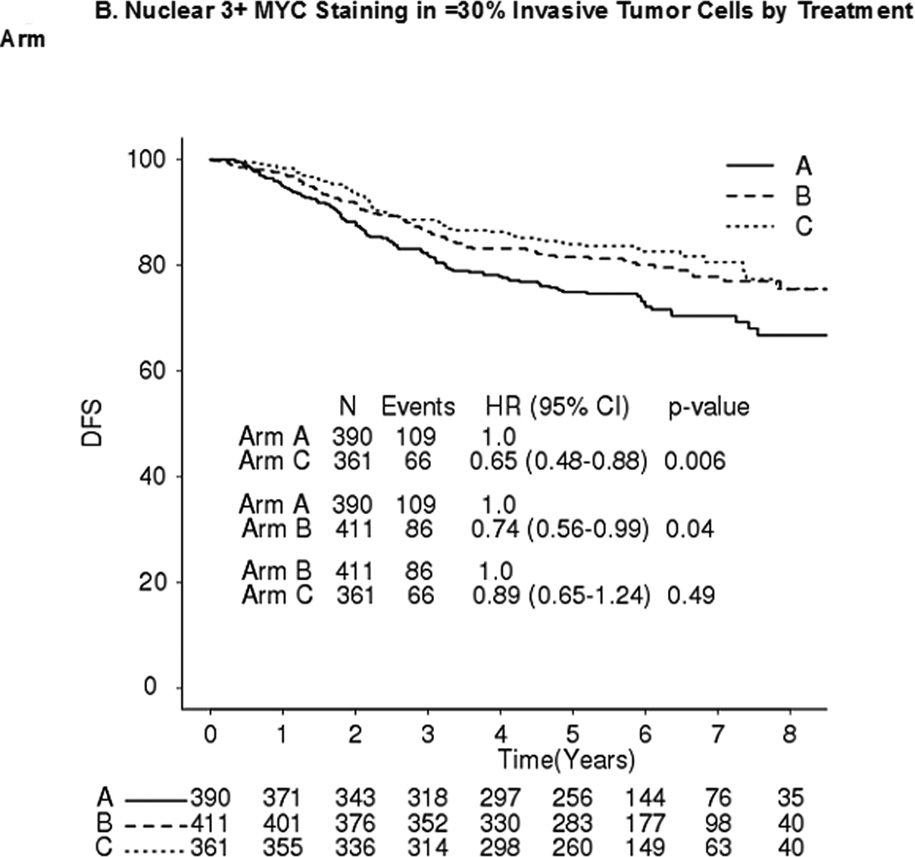
No significant differences in DFS were observed between MYC+ and MYC- patients in any of the three arms (Table 4). Comparing DFS between arms C and A, MYC+ (Figure 2A) and MYC- (Figure 2B) patients had hazard ratios (HRs) of 0.52 (p=0.006) and 0.65 (p=0.006), respectively (interaction p=0.40). Comparing DFS between arms B and A, MYC+ and MYC- patients had HRs of 0.79 (p=0.21) and 0.74 (p=0.04), respectively (interaction p=0.71) (Figure 2A–B). Comparing DFS between arms C and B, MYC+ and MYC- patients had HRs 0.56 (p=0.02) and 0.89 (p=0.49), respectively (interaction p=0.17) (Figure 2A–B).
Table 4.
Disease-free Survival by MYC Protein Status within Treatment Arm.
| Arm | 3+ Nuclear Staining Group |
N | # Events |
HR | 95% CI | p-value | DFS | |
|---|---|---|---|---|---|---|---|---|
| 3 yr | 5 yr | |||||||
| A | ≤ 30% | 390 | 108 | 1 | 81.8 | 74.9 | ||
| (N=584) | > 30% | 194 | 58 | 1.04 | 0.76–1.44 | 0.77 | 79.3 | 71.4 |
| B | ≤ 30% | 411 | 86 | 1 | 86.3 | 81.6 | ||
| (N=624) | > 30% | 213 | 56 | 1.24 | 0.87–1.76 | 0.24 | 84.0 | 76.4 |
| C | ≤ 30% | 361 | 66 | 1 | 88.6 | 84.0 | ||
| (N=528) | > 30% | 167 | 28 | 0.80 | 0.51–1.27 | 0.34 | 91.0 | 88.0 |
Figure 2. DFS by MYC Protein Level and Treatment Arm.
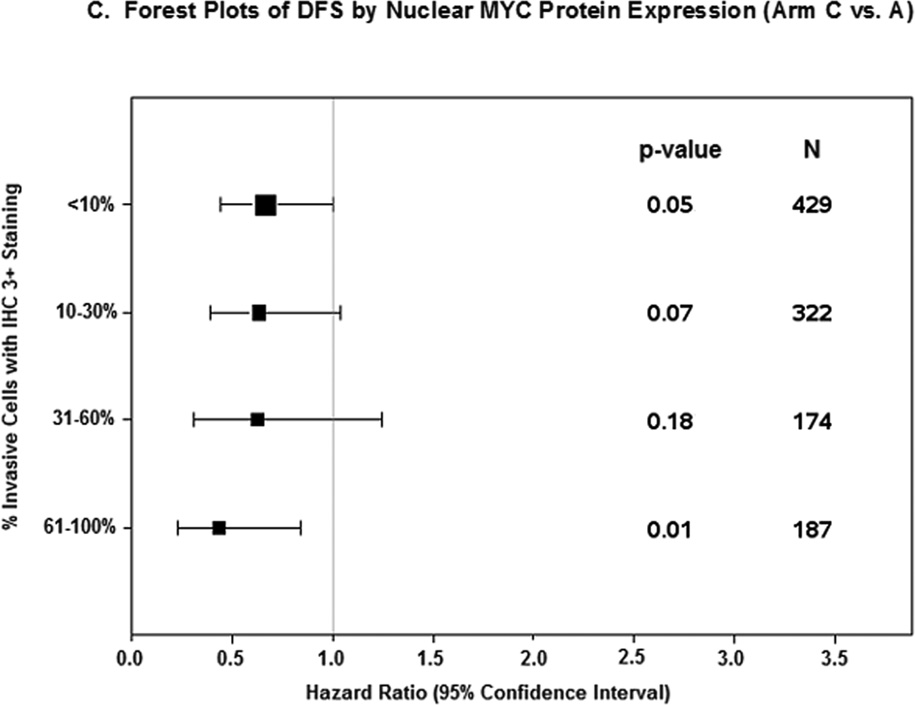
A. Nuclear 3+ MYC staining in ≤30% invasive tumor cells by treatment arm. B. Nuclear 3+ MYC staining in >30% invasive tumor cells by treatment arm. A: doxorubicin; C: cyclophosphamide; T: paclitaxel; H: trastuzumab. DFS Stratified by receptor and nodal status; Arm A vs B Interaction p-value = 0.71; Arm A vs C Interaction p-value = 0.40; Arm B vs C Interaction p-value = 0.17. C. Forest Plots of DFS by Nuclear MYC Protein Expression (Arm C vs A).
In addition, patients with nuclear MYC staining of 3+ in 0–9%, 10–30%, 31–60%, and 61–100% of cells had HRs (C vs A) of 0.68 (95% CI 0.45–1.02), 0.64 (95% CI 0.39–1.04), 0.63 (95% CI 0.31–1.24), and 0.44 (95% CI 0.23–0.84), respectively (Mantel-Haenszel Χ2= 0.30) (Figure 2C).
Discussion
MYC is a highly regulated and multifunctional transcription factor that regulates up to 15% of human genes and plays a central role in proliferation and malignant transformation of human and animal cells (16, 17). Previous evidence from B31 and N9831 suggested that MYC gene copy number anomalies may be associated with additional benefit to adjuvant trastuzumab (9, 10). Reports have been inconsistent with regard to the association of MYC protein expression and both clinicopathological characteristics and prognosis (5). To further explore these relationships, we designed the translational component of the N9831 trial to include an analysis of MYC protein overexpression in patients with HER2+ tumors.
Overall, we found that MYC protein expression was heterogeneous and characterized by both cytoplasmic and nuclear localization. We observed nuclear MYC protein overexpression (defined as nuclear 3+ staining in >30% invasive cells) in tumors from 33% of N9831 patients with HER2+ breast cancer. Early studies using IHC have shown that ~ 50–100% of breast cancer cases have increased levels of MYC protein (5, 15, 18–23). In addition, strong MYC protein positivity was found in >20% nuclei in 45% of 440 primary breast tumors (24) and 70% (36/51) showed ≥1+ intensity staining (25) using the 9E10 clone employed in our present study. Our study observed a lower incidence of 33% most likely due to our more strict cutoff criteria of 3+ staining in >30% nuclei and the fact that our patient population was HER2+ and not a general population of breast cancer patients.
We also observed strong (3+) cytoplasmic staining in malignant cells, which correlated with strong (3+) nuclear staining. Other groups have found predominant cytoplasmic localization of MYC (15, 19), and this cytoplasmic staining has been associated with better survival (15). These results support the idea of nuclear exclusion of MYC, which has been observed in high grade tumors, and could serve to attenuate select functions of MYC in later stages of disease progression (25, 26).
In this analysis, MYC nuclear protein overexpression was associated with hormone receptor positivity, nodal positivity, and larger tumors with associated increase in mastectomy rates. In agreement with our findings, MYC protein expression has been correlated with positive nodes (20, 27) and with estrogen receptor positivity (19). MYC also has been shown to be an estrogen-responsive gene (28). Although a significant association has not been consistently observed between MYC protein expression and breast tumor size (5, 19), MYC DNA levels (as detected by Southern blot) have been correlated with tumor size (5) and the Ki-67 proliferation marker has been shown to correlate with MYC protein level MYC (20). The association between MYC overexpression and tumor size in our study is consistent with MYC being a transcriptional activator of the cell proliferation pathway an additional marker for the assessment of tumor cell proliferation (5). In contrast, other studies have shown no significant associations with estrogen receptors (18, 20–22, 24, 29), with lymph node status (15, 18, 20–22, 24, 29–32).
We did not observe a significant difference in outcome between patients with and without MYC protein overexpression within any treatment arm and specifically, the DFS of N9831 patients treated with chemotherapy only was similar, regardless of MYC protein overexpression. Although this suggests that MYC overexpression is not a marker for prognosis, the true prognostic significance of MYC protein overexpression cannot be addressed in this study as all patients were treated with chemotherapy, and subgroups of patients also received protocol-specified radiotherapy and/or hormonal therapy. We did observe that MYC overexpression was associated with larger tumors and nodal positivity, both of which are powerful prognostic indicators. In N9831, however, radiotherapy was directed based on surgery type and number of positive nodes. MYC overexpression was also associated with hormone receptor positivity, another disease characteristic which was used in directing additional therapy, namely hormonal therapy. Therefore, one possibility to explain similar DFS within arm regardless of MYC overexpression status is that MYC may have disparate prognostic impact in subtypes of breast cancer which washes out when treated differently and considered together. Another possibility is that MYC overexpression confers a worse prognosis but perhaps a greater response to chemotherapy, making it an overall null biomarker in this chemotherapy-treated population.
Several investigations have found that higher expression of MYC protein correlated with poorer outcome (33–35), while other studies have shown positive associations between MYC mRNA levels and survival (36) and between MYC protein levels and survival, most notably for node-negative patients (15). Our findings are consistent with previous findings that demonstrated that MYC protein expression alone was not related to recurrence (30) and are supported by a limited number of studies that did not find associations between MYC expression and prognosis (19, 22). Importantly, however, we observed a benefit of concurrent trastuzumab in patients with or without MYC protein overexpression.
We did observe a trend toward greater benefit from concurrent trastuzumab with increasing MYC protein expression. Trastuzumab has been shown to sensitize HER2-overexpressing cells to apoptosis (37, 38), possibly through induction of the pro-apoptotic function of MYC (39–41). Higher MYC protein expression could then result in an increased rate of apoptosis in tumors with HER2 protein overexpression. Alternatively, trastuzumab was recently shown to inhibit glycolysis in HER2+ cells (42), and down-regulation of MYC protein has been shown to contribute to cancer cell survival under dual deficiency of oxygen and glucose (43). Conceivably, when MYC protein is overexpressed in HER2+ tumors that are treated with trastuzumab, the protective effect of MYC down-regulation on cancer cell survival in low glucose settings would be lost. This may result in increased tumor cell death and improved outcome of patients with increasing MYC protein expression.
We also observed statistically significantly improved DFS from concurrent trastuzumab compared to sequential trastuzumab among patients with MYC protein overexpression, but not in patients without MYC protein overexpression. The interaction between MYC protein overexpression and timing of trastuzumab did not reach statistical significance, but the large improvement in the hazard ratio for patients with MYC overexpression relative to patients without MYC overexpression gives rise to the speculation that the timing/schedule of trastuzumab administration may be important in utilizing MYC as an additional marker of trastuzumab sensitivity. As MYC induces cell proliferation and HER2+ breast tumors tend to have a high proliferation index (44–46), those tumors with overexpression for both HER2 and MYC may be more susceptible to the growth inhibitory synergistic effects observed with the combination of chemotherapy and trastuzumab (37, 38, 47).
Overall, our data indicate that MYC protein expression alone is not significantly associated with differential benefit to concurrent trastuzumab, but potentially could help differentiate benefit between concurrent and sequential trastuzumab treatment. Distinct mechanisms of regulation for MYC have been defined over the past decade and several signal transduction pathways and regulatory mechanisms have evolved to keep MYC expression under tight control (17). Ongoing protein expression analyses of regulators and effectors of MYC (e.g., PTEN and IGF1R) (48, 49), and whole genome expression profiling of N9831 tumors will provide important information regarding the interactions between MYC and other pertinent proteins and genes and the effects of these interactions on the sensitivity/resistance to adjuvant trastuzumab. Understanding the full extent of the oncogenic effects of these interactions is critical to the development of more effective, targeted therapies for breast cancer patients that exhibit HER2+ disease.
Supplementary Material
Statement of translational relevance.
Despite therapy, up to a quarter of women diagnosed with HER2+ early-stage breast cancer develop tumor relapse within three years. Identifying markers that could help predict trastuzumab benefit is therefore an important clinical goal. Previous evidence from B31 and N9831 suggested that MYC gene copy number anomalies may be associated with additional benefit to adjuvant trastuzumab. We then investigated the association between MYC protein overexpression (MYC+; nuclear 3+ staining in >30% tumor cells) and disease-free survival (DFS) of patients in N9831. Patients with MYC+ and MYC- tumors both significantly benefited from concurrent trastuzumab compared to standard chemotherapy alone, and the level of benefit was not significantly different. Patients with MYC+ but not with MYC- tumors significantly benefited from concurrent trastuzumab compared to sequential trastuzumab. Our N9831 data indicate that MYC protein expression is not significantly associated with differential benefit to concurrent adjuvant trastuzumab with chemotherapy.
Acknowledgments
Julie Gralow receives research funding from Amgen, Novartis, Roche and Genentech. Peter Kaufman is a consultant for Genentech and receives research funding and honoraria from Genentech as well. Edith A. Perez receives funding from Genentech, GlaxoSmithKline, Genomic Health, and Monogram Biosience.
GRANT SUPPORT
This work was supported by National Institutes of Health grants CA25224-31 and CA114740 (PI: JC Buckner) for the North Central Cancer Treatment Group and associated Biospecimen Resource, respectively, National Institutes of Health grant CA129949 (PI: EA Perez/MM Reinholz), and the Breast Cancer Research Foundation (PI: EA Perez).
Footnotes
Conflicts of Interest: All other authors have no conflicts of interest to disclose.
Presented in part at the 33rd Annual CTRC-AACR San Antonio Breast Cancer Symposium, December 8–12, 2010, San Antonio, TX.
References
- 1.Perez EA, Palmieri FM, Brock SM. Trastuzumab. Cancer Treat Res. 2009;151:181–196. doi: 10.1007/978-0-387-75115-3_12. [DOI] [PubMed] [Google Scholar]
- 2.Perez EA, Romond EH, Suman VJ, Jeong JH, Davidson NE, Geyer CE, Jr, et al. Four-year follow-up of trastuzumab plus adjuvant chemotherapy for operable human epidermal growth factor receptor 2-positive breast cancer: joint analysis of data from NCCTG N9831 and NSABP B-31. J Clin Oncol. 2011;29:3366–3373. doi: 10.1200/JCO.2011.35.0868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Piccart-Gebhart MJ, Procter M, Leyland-Jones B, Goldhirsch A, Untch M, Smith I, et al. Trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer. N Engl J Med. 2005;353:1659–1672. doi: 10.1056/NEJMoa052306. [DOI] [PubMed] [Google Scholar]
- 4.Bedard PL, de Azambuja E, Cardoso F. Beyond trastuzumab: overcoming resistance to targeted HER-2 therapy in breast cancer. Curr Cancer Drug Targets. 2009;9:148–162. doi: 10.2174/156800909787581024. [DOI] [PubMed] [Google Scholar]
- 5.Liao DJ, Dickson RB. c-Myc in breast cancer. Endocr Relat Cancer. 2000;7:143–164. doi: 10.1677/erc.0.0070143. [DOI] [PubMed] [Google Scholar]
- 6.Lutz W, Leon J, Eilers M. Contributions of Myc to tumorigenesis. Biochim Biophys Acta. 2002;1602:61–71. doi: 10.1016/s0304-419x(02)00036-7. [DOI] [PubMed] [Google Scholar]
- 7.Xu J, Chen Y, Olopade OI. MYC and Breast Cancer. Genes Cancer. 2010;1:629–640. doi: 10.1177/1947601910378691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neve RM, Sutterluty H, Pullen N, Lane HA, Daly JM, Krek W, et al. Effects of oncogenic ErbB2 on G1 cell cycle regulators in breast tumour cells. Oncogene. 2000;19:1647–1656. doi: 10.1038/sj.onc.1203470. [DOI] [PubMed] [Google Scholar]
- 9.Kim C, Bryant J, Horne Z, Geyer C, Wickerham D, Wolmark N, et al. Trastuzumab sensitivity of breast cancer with co-amplification of HER2 and cMYC suggests pro-apoptotic function of dysregulated cMYC in vivo. Breast Cancer Res Treat. 2005;94:S6. [Abstract 46]. [Google Scholar]
- 10.Perez EA, Jenkins RB, Dueck AC, Wiktor AE, Bedroske PP, Anderson SK, et al. C-MYC alterations and association with patient outcome in early-stage HER2-positive breast cancer from the north central cancer treatment group N9831 adjuvant trastuzumab trial. J Clin Oncol. 2011;29:651–659. doi: 10.1200/JCO.2010.30.2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perez EA, Suman VJ, Davidson NE, Gralow JR, Kaufman PA, Visscher DW, et al. Sequential versus concurrent trastuzumab in adjuvant chemotherapy for breast cancer. J Clin Oncol. 2011;29:4491–4497. doi: 10.1200/JCO.2011.36.7045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gianni L, Zambetti M, Clark K, Baker J, Cronin M, Wu J, et al. Gene expression profiles in paraffin-embedded core biopsy tissue predict response to chemotherapy in women with locally advanced breast cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2005;23:7265–7277. doi: 10.1200/JCO.2005.02.0818. [DOI] [PubMed] [Google Scholar]
- 13.Perez EA, Roche PC, Jenkins RB, Reynolds CA, Halling KC, Ingle JN, et al. HER2 testing in patients with breast cancer: poor correlation between weak positivity by immunohistochemistry and gene amplification by fluorescence in situ hybridization. Mayo Clin Proc. 2002;77:148–154. doi: 10.4065/77.2.148. [DOI] [PubMed] [Google Scholar]
- 14.Wolff AC, Hammond ME, Schwartz JN, Hagerty KL, Allred DC, Cote RJ, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J Clin Oncol. 2007;25:118–145. doi: 10.1200/JCO.2006.09.2775. [DOI] [PubMed] [Google Scholar]
- 15.Pietilainen T, Lipponen P, Aaltomaa S, Eskelinen M, Kosma VM, Syrjanen K. Expression of c-myc proteins in breast cancer as related to established prognostic factors and survival. Anticancer Res. 1995;15:959–964. [PubMed] [Google Scholar]
- 16.Dang CV. Enigmatic MYC Conducts an Unfolding Systems Biology Symphony. Genes Cancer. 2010;1:526–531. doi: 10.1177/1947601910378742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer. 2008;8:976–990. doi: 10.1038/nrc2231. [DOI] [PubMed] [Google Scholar]
- 18.Locker AP, Dowle CS, Ellis IO, Elston CW, Blamey RW, Sikora K, et al. c-myc oncogene product expression and prognosis in operable breast cancer. Br J Cancer. 1989;60:669–672. doi: 10.1038/bjc.1989.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mizukami Y, Nonomura A, Noguchi M, Taniya T, Koyasaki N, Saito Y, et al. Immunohistochemical study of oncogene product ras p21, c-myc and growth factor EGF in breast carcinomas. Anticancer Res. 1991;11:1485–1494. [PubMed] [Google Scholar]
- 20.Pavelic ZP, Pavelic L, Lower EE, Gapany M, Gapany S, Barker EA, et al. c-myc, c-erbB-2, and Ki-67 expression in normal breast tissue and in invasive and noninvasive breast carcinoma. Cancer Res. 1992;52:2597–2602. [PubMed] [Google Scholar]
- 21.Spandidos DA, Yiagnisis M, Papadimitriou K, Field J. Kras c-myc and c-erbB-2 oncoproteins in human breast cancer. Anticancer Res. 1989;9:1385–1393. [PubMed] [Google Scholar]
- 22.Spaventi R, Kamenjicki E, Pecina N, Grazio S, Pavelic J, Kusic B, et al. Immunohistochemical detection of TGF-alpha, EGF-R, c-erbB-2, c-H-ras, c-myc, estrogen and progesterone in benign and malignant human breast lesions: a concomitant expression. In Vivo. 1994;8:183–189. [PubMed] [Google Scholar]
- 23.Walker RA, Senior PV, Jones JL, Critchley DR, Varley JM. An immunohistochemical and in situ hybridization study of c-myc and c-erbB-2 expression in primary human breast carcinomas. J Pathol. 1989;158:97–105. doi: 10.1002/path.1711580204. [DOI] [PubMed] [Google Scholar]
- 24.Naidu R, Wahab NA, Yadav M, Kutty MK. Protein expression and molecular analysis of c-myc gene in primary breast carcinomas using immunohistochemistry and differential polymerase chain reaction. Int J Mol Med. 2002;9:189–196. [PubMed] [Google Scholar]
- 25.Blancato J, Singh B, Liu A, Liao DJ, Dickson RB. Correlation of amplification and overexpression of the c-myc oncogene in high-grade breast cancer: FISH, in situ hybridisation and immunohistochemical analyses. Br J Cancer. 2004;90:1612–1619. doi: 10.1038/sj.bjc.6601703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liao DJ, Natarajan G, Deming SL, Jamerson MH, Johnson M, Chepko G, et al. Cell cycle basis for the onset and progression of c-Myc-induced, TGFalpha-enhanced mouse mammary gland carcinogenesis. Oncogene. 2000;19:1307–1317. doi: 10.1038/sj.onc.1203430. [DOI] [PubMed] [Google Scholar]
- 27.Pavelic ZP, Steele P, Preisler HD. Evaluation of c-myc proto-oncogene in primary human breast carcinomas. Anticancer Res. 1991;11:1421–1427. [PubMed] [Google Scholar]
- 28.Miller TL, Huzel NJ, Davie JR, Murphy LC. C-myc gene chromatin of estrogen receptor positive and negative breast cancer cells. Mol Cell Endocrinol. 1993;91:83–89. doi: 10.1016/0303-7207(93)90258-l. [DOI] [PubMed] [Google Scholar]
- 29.Saccani Jotti G, Fontanesi M, Bombardieri E, Gabrielli M, Veronesi P, Bianchi M, et al. Preliminary study on oncogene product immunohistochemistry (c-erbB-2, c-myc, ras p21, EGFR) in breast pathology. Int J Biol Markers. 1992;7:35–42. [PubMed] [Google Scholar]
- 30.Bland KI, Konstadoulakis MM, Vezeridis MP, Wanebo HJ. Oncogene protein co-expression. Value of Ha-ras, c-myc, c-fos, and p53 as prognostic discriminants for breast carcinoma. Ann Surg. 1995;221:706–718. doi: 10.1097/00000658-199506000-00010. discussion 18–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bolufer P, Molina R, Ruiz A, Hernandez M, Vazquez C, Lluch A. Estradiol receptors in combination with neu or myc oncogene amplifications might define new subtypes of breast cancer. Clin Chim Acta. 1994;229:107–122. doi: 10.1016/0009-8981(94)90233-x. [DOI] [PubMed] [Google Scholar]
- 32.Sirotkovic-Skerlev M, Krizanac S, Kapitanovic S, Husnjak K, Unusic J, Pavelic K. Expression of c-myc, erbB-2, p53 and nm23-H1 gene product in benign and malignant breast lesions: coexpression and correlation with clinicopathologic parameters. Exp Mol Pathol. 2005;79:42–50. doi: 10.1016/j.yexmp.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 33.Guerin M, Barrois M, Terrier MJ, Spielmann M, Riou G. Overexpression of either c-myc or c-erbB-2/neu proto-oncogenes in human breast carcinomas: correlation with poor prognosis. Oncogene Res. 1988;3:21–31. [PubMed] [Google Scholar]
- 34.Pertschuk LP, Feldman JG, Kim DS, Nayeri K, Eisenberg KB, Carter AC, et al. Steroid hormone receptor immunohistochemistry and amplification of c-myc protooncogene. Relationship to disease-free survival in breast cancer. Cancer. 1993;71:162–171. doi: 10.1002/1097-0142(19930101)71:1<162::aid-cncr2820710126>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 35.Scorilas A, Trangas T, Yotis J, Pateras C, Talieri M. Determination of c-myc amplification and overexpression in breast cancer patients: evaluation of its prognostic value against c-erbB-2, cathepsin-D and clinicopathological characteristics using univariate and multivariate analysis. Br J Cancer. 1999;81:1385–1391. doi: 10.1038/sj.bjc.6693404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bieche I, Laurendeau I, Tozlu S, Olivi M, Vidaud D, Lidereau R, et al. Quantitation of MYC gene expression in sporadic breast tumors with a real-time reverse transcription-PCR assay. Cancer Res. 1999;59:2759–2765. [PubMed] [Google Scholar]
- 37.Arteaga CL. Can trastuzumab be effective against tumors with low HER2/Neu (ErbB2) receptors? J Clin Oncol. 2006;24:3722–3725. doi: 10.1200/JCO.2006.06.5268. [DOI] [PubMed] [Google Scholar]
- 38.Henson ES, Hu X, Gibson SB. Herceptin sensitizes ErbB2-overexpressing cells to apoptosis by reducing antiapoptotic Mcl-1 expression. Clin Cancer Res. 2006;12:845–853. doi: 10.1158/1078-0432.CCR-05-0754. [DOI] [PubMed] [Google Scholar]
- 39.Adhikary S, Eilers M. Transcriptional regulation and transformation by Myc proteins. Nat Rev Mol Cell Biol. 2005;6:635–645. doi: 10.1038/nrm1703. [DOI] [PubMed] [Google Scholar]
- 40.Nilsson JA, Cleveland JL. Myc pathways provoking cell suicide and cancer. Oncogene. 2003;22:9007–9021. doi: 10.1038/sj.onc.1207261. [DOI] [PubMed] [Google Scholar]
- 41.Cole MD, McMahon SB. The Myc oncoprotein: a critical evaluation of transactivation and target gene regulation. Oncogene. 1999;18:2916–2924. doi: 10.1038/sj.onc.1202748. [DOI] [PubMed] [Google Scholar]
- 42.Zhao Y, Zhou LM, Chen YY, Yang SG, Tian WM. MYC genes with differential responses to tapping, mechanical wounding, ethrel and methyl jasmonate in laticifers of rubber tree (Hevea brasiliensis Muell. Arg.) J Plant Physiol. 2011;168:1649–1658. doi: 10.1016/j.jplph.2011.02.010. [DOI] [PubMed] [Google Scholar]
- 43.Okuyama H, Endo H, Akashika T, Kato K, Inoue M. Downregulation of c-MYC protein levels contributes to cancer cell survival under dual deficiency of oxygen and glucose. Cancer Res. 2010;70:10213–10223. doi: 10.1158/0008-5472.CAN-10-2720. [DOI] [PubMed] [Google Scholar]
- 44.Rudolph P, Olsson H, Bonatz G, Ratjen V, Bolte H, Baldetorp B, et al. Correlation between p53, c-erbB-2, and topoisomerase II alpha expression, DNA ploidy, hormonal receptor status and proliferation in 356 node-negative breast carcinomas: prognostic implications. J Pathol. 1999;187:207–216. doi: 10.1002/(SICI)1096-9896(199901)187:2<207::AID-PATH223>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 45.Trihia H, Murray S, Price K, Gelber RD, Golouh R, Goldhirsch A, et al. Ki-67 expression in breast carcinoma: its association with grading systems, clinical parameters, and other prognostic factors--a surrogate marker? Cancer. 2003;97:1321–1331. doi: 10.1002/cncr.11188. [DOI] [PubMed] [Google Scholar]
- 46.Yerushalmi R, Woods R, Ravdin PM, Hayes MM, Gelmon KA. Ki67 in breast cancer: prognostic and predictive potential. Lancet Oncol. 2010;11:174–183. doi: 10.1016/S1470-2045(09)70262-1. [DOI] [PubMed] [Google Scholar]
- 47.Menendez JA, Mehmi I, Lupu R. Trastuzumab in combination with heregulin-activated Her-2 (erbB-2) triggers a receptor-enhanced chemosensitivity effect in the absence of Her-2 overexpression. J Clin Oncol. 2006;24:3735–3746. doi: 10.1200/JCO.2005.04.3489. [DOI] [PubMed] [Google Scholar]
- 48.Perez EA, Dueck AC, McCullough AE, Chen B, Geiger XJ, Jenkins RB, et al. Impact of PTEN Protein Expression on Benefit From Adjuvant Trastuzumab in Early-Stage Human Epidermal Growth Factor Receptor 2-Positive Breast Cancer in the North Central Cancer Treatment Group N9831 Trial. J Clin Oncol. 2013;31:2115–2122. doi: 10.1200/JCO.2012.42.2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reinholz MMDA, Chen B, Geiger X, McCullough AE, Jenkins RB, Lingle WL, Andorfer CA, Davidson NE, Martino S, Kaufman PA, Kutteh LA, Sledge GW, Harris LN, Gralow JR, Perez EA. Effect of IGF1R protein expression on benefit to adjuvant trastuzumab in early-stage HER2+ breast cancer in NCCTG adjuvant trial N9831. J Clin Oncol. 2011;29 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.