Abstract
A label-free mass spectrometric strategy was used to examine the effect of 5-fluorouracil (5-FU) on the primary and metastatic colon carcinoma cell lines, SW480 and SW620, with and without treatment. 5-FU is the most common chemotherapeutic treatment for colon cancer. Pooled biological replicates were analyzed by nanoLC-MS/MS and protein quantification was determined via spectral counting. Phenotypic and proteomic changes were evident and often similar in both cell lines. The SW620 cells were more resistant to 5-FU treatment, with an IC50 2.7-fold higher than that for SW480. In addition, both cell lines showed pronounced abundance changes in pathways relating to antioxidative stress response and cell adhesion remodeling due to 5-FU treatment. For example, the detoxification enzyme NQO1 was increasedwith treatment in both cell lines, while disparate members of the peroxiredoxin family, PRDX2 or PRDX5 and PRDX6, were elevated with 5-FU exposure in either SW480 or SW620, respectively. Cell adhesion associated proteins CTNNB1 and RhoA showed decreased expression with 5-FU treatment in both cell lines. The differential quantitative response in the proteomes of these patient-matched cell lines to drug treatment underscores the subtle molecular differences separating primary and metastatic cancer cells.
Keywords: Bioanalytical methods, Mass spectrometry, Proteomics, Label-free quantification, Colon cancer, 5-Fluorouracil
1 Introduction
In 1971, the National Cancer Act was signed, signaling the beginning of a concentrated effort to study and understand cancer. Forty years later, progress has been made in understanding the complexities of cancer progression, but much remains to be learned regarding the molecular basis of these destructive malignancies. One of the deadliest cancers is colorectal cancer (CRC), which is the second leading cause of cancer-related deaths in the U.S [1]. In 2011 alone, there were an estimated 100,000 new cases diagnosed and 49,000 deaths [1]. Progress in diagnosis and treatment has enabled clinicians to extend and save the lives of many patients at the early stages of this disease; however, the prognosis for patients with advanced disease or systemic metastasis is still very poor.
The most common therapeutic agent used to treat CRC is 5-fluorouracil (5-FU). 5-FU has been used for more than 40 years. 5-FU affects metabolic pathways and displays cytotoxic modes of action through the inhibition of thymidylate synthase activity and incorporation into RNA and DNA. 5-FU also induces cancer cells to undergo apoptosis in response to RNA and DNA damage [2]. Patient responses to 5-FU vary widely in terms of both efficacy and toxicity; response rates for 5-FU-based chemotherapy are only 10-15% [3]. Tumors may develop resistance to 5-FU over the course of treatment. Drug resistance, as well as metastatic spread of tumors to secondary sites, accounts for much of the morbidity and mortality associated with CRC.
The response of CRC cells to 5-FU has previously been examined; recently a large panel of 77 CRC cell lines were treated with 5-FU and then tested for toxicity [4]. Another study compared the response of primary versus metastatic CRC lesions. Using a cohort of non-patient matched biopsies, the resected tumors were cultured in vitro, treated with 5-FU, and then tested for growth inhibition. Cells from metastatic lesions were found to be more resistant to 5-FU than those from primary tumors; however, the authors note that patient-matched comparisons of primary and metastatic lesions would be more informative than comparing lesions across different patients [5].
A useful patient-matched model for studying colorectal cancer metastasis is the cell lines, SW480 and SW620, isolated from the primary colon adenocarcinoma and the lymph node metastasis, respectively [6]. The primary SW480 cell line was derived from a Dukes' type B colon adenocarcinoma and the metastatic SW620 cell line was established from the same patient 1 year later. Due to the isogenic nature of these two cell lines, variation from genetic background is avoided, making them excellent models to study 5-FU treatment effects on primary and metastatic colon cancer. In addition, since chemotherapy was not started with the patient until after both biopsies were collected, no drug-induced alterations in gene expression are present.
While examining the effect of 5-FU on cell growth is valuable, understanding the molecular and especially the proteomic basis for metastasis and drug resistance may lead to improvement in the treatment of CRC. Previous studies have examined the basal proteomic differences between the SW480 and SW620 cell lines [7–9]. In addition, the proteomic changes that occur with 5-FU treatment in SW480 have been characterized [10].
These studies have largely relied on protein separations and comparison based on two-dimensional polyacrylamide gel electrophoresis (2D-PAGE), followed by mass spectrometry (MS)-based identification of specific excised protein spots. Alternatively, the field of MS-based quantification has grown significantly in recent years [11]. Quantitative proteomics via MS can be accomplished through three main methods: the use of a chemical [12] or metabolic [13] labeling strategy or a label-free technique. Label-free proteomic profiling offers several advantages over labeling techniques such as less complex sample preparation and lower reagent costs. The approach also does not suffer from the possibility of incomplete labeling. Label-free quantification can further be divided into two major groups: signal intensity measurement based on mass spectra and spectral counting. Spectral counting relative protein quantification is based on counting the number of identified MS/MS spectra assigned to a protein in an MS/MS experiment. This quantification technique is supported by the observation that more abundant peptides will be more readily selected for fragmentation. These peptides will produce a higher abundance of MS/MS spectra which are therefore directly proportional to protein amount in data-dependent acquisition. Relative protein abundance and spectral count is shown to have a strong, positive linear correlation with an r2 value of 0.9997 and a dynamic range over 2 orders of magnitude [14]. Moreover, spectral counting has proved to be more reproducible and has a higher dynamic range compared to peptide ion chromatogram-based quantification [15]. Several excellent reviews have recently been written on the use of label-free mass spectrometric approaches and their applications in biological research [16–18].
As previously mentioned, 5-FU is the primary treatment option for patients with CRC, yet there are substantial variations in response to the drug, including among primary and metastatic lesions [5]. We sought to determine whether there exist proteomic differences between primary and metastatic CRC underlying this differential phenotypic response. To investigate this question, we used SW480 and SW620 and examined their proteomic profiles before and after 5-FU treatment using label-free, spectral counting-based quantitative mass spectrometry.
2 Materials and methods
2.1 Cell culture
Colon cancer cell lines SW480 and SW620 were purchased from the American Type Culture Collection (ATCC, Manassas, VA) and maintained in RPMI 1640 medium (Invitrogen, Gaithersburg, MD) supplemented with 10% fetal bovine serum (Thermo Scientific, Pittsburgh, PA) and 2mM L-glutamine (Invitrogen) and grown in 5%CO2 at 37°C. Cell lines were used within 3 months after receipt or resuscitation of frozen aliquots thawed from liquid nitrogen. The provider assured the authentication of these cell lines by cytogenetic analysis.
2.2 Dose response curves of cells to 5-FU
SW480 or SW620 cells were seeded into 96-well plates at a concentration of 5000 cells/well and allowed to attach for 24 hr. A range of concentrations (0.04, 0.08, 0.16, 0.31, 0.62, 1.3, 2.5, 5.0, 10.0, 20.0, 40.0, 80.0, 160.0, 320.0, 640.0, 1280.0 and 2560.0μM) of 5-fluroruracil (5-FU) (Sigma-Aldrich, St. Louis, MO) were prepared by dissolving in nanoPure water and added to RPMI 1640 medium. The 5-FU solutions were then added to the seeded cells and incubated for 72 hr. As a surrogate marker for cell viability, the reduction of resazurin to resorufin was measured in cells using the Cell Titer-Blue Cell Viability assay (Promega, Madison, WI). After 72 hr of 5-FU treatment, reduction of resazurin to resorufin, read as fluorescence (560Ex/590Em), was measured using a plate reader (Spectramax M5; Molecular Devices, Sunnyvale, CA). Viability of 5-FU-treated cells was compared to cells incubated without 5-FU. Triplicate measurements were collected for each concentration of 5-FU.
2.3 Treatment of cells with 5-FU
5-FU was dissolved in nanoPure water and added to RPMI 1640 medium at a final concentration of the determined IC50 for each cell line (SW480 IC50 was 7.5µM; SW620 IC50 was 20.0µM) whereas the same volume of RPMI 1640 medium without 5-FU was added to the control cells. The RPMI 1640 medium with and without 5-FU was added to the SW480 and SW620 test and control culture plates after incubating the seeded cells for 24 hr. All culture plates were then incubated for 72 hr. Triplicate biological replicates of both treated and untreated cells for each cell line were performed.
2.4 Cell lysis and protein sample preparation
After 72 hr incubation, the culture medium was aspirated and the cells were rinsed with phosphate buffered saline (PBS) (Invitrogen). Complete Lysis-M Reagent Kit (Roche Diagnostics, Indianapolis, IN) with 1X Complete Protease Inhibitor (Roche Diagnostics) was added to each culture plate and incubated for 5 min at room temperature on a plate shaker. The cells were scraped, collected and centrifuged at 15,000 rpm for 10min to obtain soluble protein fractions. The total protein concentration of each sample was determined using a BCA protein assay kit (Thermo Scientific) and bovine serum albumin standard (Thermo Scientific) according to the manufacturer’s instructions. Biological replicates for each sample were pooled with equal contribution from each biological replicate forming the pool. Twenty micrograms of each pooled protein sample was resolved by a NuPAGE SDS-PAGE system (Invitrogen) (4-12% acrylamide, Bis-Tris with MOPS running buffer) in two lanes each. After electrophoresis, the gel was stained with Colloidal Blue staining kit (Invitrogen) and washed with distilled water. The gel lanes corresponding to each sample were excised into five bands based on staining intensity. Each band was cut into 2mm-wide pieces and subjected to in-gel tryptic digestion. HPLC-grade water and acetonitrile (ACN) (Honeywell Burdick & Jackson, Muskegon, MI) were used in the subsequent digestion. Gel pieces were washed/dehydrated three times in 50mM ammonium bicarbonate (ABC) (Sigma-Aldrich)/50mM ABC + 50% ACN. Cysteine bonds were reduced with 10mM DTT (dithiothreitol) (Sigma-Aldrich) for 1 hr at 56°C and alkylated with 55mM IAA (iodoacetamide) (Sigma-Aldrich) for 20 min at room temperature in the dark. Following two subsequent wash/dehydrate cycles, the reduced and alkylated proteins were dried 20min in a MiVac sample concentrator (Genevac Inc, New York, NY) and incubated overnight with 12.5ng/µL trypsin in 25mM ABC at 37°C. Peptides were extracted twice in 50µL of 50% ACN/ 45% water/ 5% formic acid (Optima LC/MS, Fischer Scientific, Fair Lawn, NJ). The combined volumes were reduced to 15µL in a speedvac. Peptides were desalted with C18 ZipTips (Millipore, Billerica, MA) according to manufacturer’s instructions. The desalted peptide volume was concentrated in a speedvac and the final volume was diluted to 10µL with 0.1% formic acid.
2.5 Liquid chromatography/mass spectrometry analysis of protein digests
Liquid chromatography was performed using a nanoAcquity ultra performance LC system (Waters, Milford, MA) under the control of Hystar (Bruker Daltonics UK Ltd). Peptides were loaded onto a 75µm C18 BEH nanoAcquity column (Waters) and trapped for 12min at 500nL/min at 95% buffer A (buffer A, 3% ACN and 0.1% formic acid; buffer B, 93% ACN and 0.1% formic acid) and separated at 500nL/min in a 95-15% buffer A gradient in 55 min. Solvent A was held at 15% for 5 min to wash the column followed by 5 min at 95% buffer A to equilibrate the column before the next injection. The LC system was interfaced via the nanoESI spray source with a 3-D high capacity ion trap mass spectrometer (amaZon X, Bruker Daltonics). Mass spectra were acquired from 350-1800m/z using parameters optimized at 922m/z with a target of 200,000 set for ion charge control (ICC) and a maximum acquisition time of 100ms. The six most abundant precursor ions above a threshold of 50,000 were selected for MS/MS per MS scan with active exclusion for 45 s after selection. SmartFrag controlled the fragmentation of each precursor ion using helium gas and a 60-180% collision energy range with an amplitude of 1.0 V. Each sample was injected multiple times with data collected for each injection, providing three technical replicates per sample.
2.6 Data processing for protein identification and quantification
Raw LC-MS/MS data were processed automatically using DataAnalysis 4.0 software (Bruker Daltonics), with the following parameters: compounds (autoMSn) threshold 1000, number of compounds unlimited, retention time windows 1 min. Database searches were performed using an in-house Mascot v.2.3 search engine (Matrix Science, London, UK) and the human SwissProt database (v. 2.2.04), with the following parameters: tryptic peptides with up to 2 missed cleavage sites, peptide tolerance 1.3Da, fragment tolerance 0.8Da, 13C=1, instrument type: ESI-TRAP, variable modifications: cysteine and N-terminal carbamidomethyl, methionine oxidation, asparagine and glutamine deamination. All proteins were identified with a false discovery rate (FDR) of < 2% based on a decoy database search.
Bruker raw data files were converted to mzXML format using the msConvert tool of the ProteoWizard library. Label-free comparative and quantitative analysis was performed using the ProteoIQ software (v.2.3.01 BIOINQUIRE, Athens, GA). A protein project was created in ProteoIQ after uploading the spectral data in mzXML format and the Mascot search results in .dat format. ProteoIQ was used to cluster peptides to proteins (protein groups) and output lists of proteins having a minimum peptide probability of <0.05 and a minimum protein probability of <0.5. Spectral counting and related quantification data were generated and extracted for comparative analysis of SW480 and SW620 cell lines with and without 5-FU treatment. ProteoIQ kept track of spectral counts from each replicate, and then spectral counts were averaged between LC/MS runs. The average spectral count for each individual protein within a biological group was normalized by comparing the total spectral counts for all proteins identified in each biological group and replicate.
2.7 Validation by Western blot
Equal amounts of pooled soluble protein fractions from all four biological groups (SW480 and SW620 5-FU treated and control harvested cells) were resolved on a 1-D SDS PAGE gel. Upon completion of electrophoresis, the proteins were transferred to nitrocellulose membrane for 90 minutes at 12V. The membrane was incubated with rabbit anti-β-catenin (1:5000) (Abcam, Cambridge, MA), mouse anti-PRDX5 (1:1000) (GenWay Biotech, San Diego, CA), rabbit anti-COX IV (1:1000) (Abcam) and rabbit anti-β-actin (1:1000) (Abcam) in 10% milk buffer in 1X PBS on a rocking platform overnight at 4 degrees. The membrane was washed 3 times for 10 minutes in 10% milk buffer in 1X PBS and incubated with HRP-conjugated anti-mouse IgG (1:10,000) (Jackson ImmunoResearch West Grove, PA) or HRP-conjugated anti-rabbit IgG (1:10,000) (Cell Signaling Technologies) for 1 hour on a rocking platform at room temperature. The membrane was washed as before, rinsed with DI water and incubated with SuperSignal West Pico Chemiluminescent Substrate (Pierce/Thermo Fisher Scientific Rockford, IL) according the protocol. Kodak BioMax Light Film (Carestream Health Woodbridge, CT) was used to expose the membrane for varying amounts of time. The film was developed using GBX developer and fixer (Carestream Health) according to their protocol.
3 Results and discussion
3.1 Determining the sensitivity of primary and metastatic colon cancer cells to 5-FU
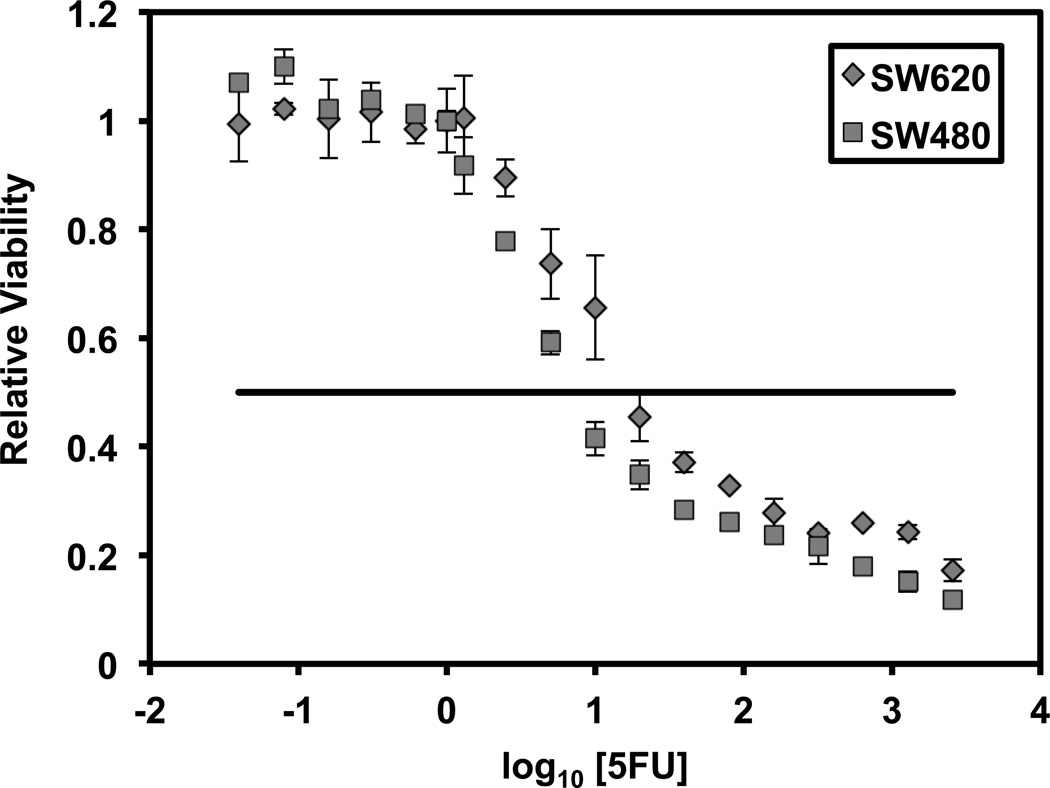
Primary SW480 and metastatic-derived SW620 5-FU drug sensitivities were determined following in vitro treatment of the cells with 5-FU over a range of drug concentrations for 72 hours. Cell viability was evaluated and the dose-response curves were plotted (Figure 1). The IC50 values, the concentration of 5-FU that reduces cell viability by 50%, of the two cell lines were determined to be 7.5μM for SW480 and 20.0μM for SW620. The metastatic SW620 cell line is more resistant to 5-FU’s cytotoxic effects as its IC50 is 2.7-fold higher than that of the primary SW480 cell line.
Figure 1.
SW480 and SW620 human colon cell line dose response curves to 5-FU treatment. The cell viability of 5-FU treated cells is expressed as a percentage relative to control cells incubated without 5-FU. The 5-FU IC50 of SW480 and SW620 were determined to be 7.5μM and 20.0μM, respectively. Triplicate measurements were collected for each concentration of 5-FU. Error bars represent the median of three measurements. The horizontal line represents 50% viability
3.2 Identification of differentially expressed proteins between 5-FU treated and control SW480 and SW620 cells
To analyze the proteomic basis for the differential sensitivity, global protein analysis of the two cell lines with and without 5-FU treatment was conducted. For each cell line and treatment condition, pooled samples consisting of three biological replicates were used. Pooling the biological replicates reduces the biological variation within the sample and increases the power to detect changes in expression seen in the average sample above any noise from random biological variation. The use of three technical replicates allows identification of expression changes in the sample above the technical noise of the instrument [19]. Proteins with multiple annotated forms identified were clustered into protein groups to address the peptide centric nature of the samples. As the human proteome has much sequence redundancy, the same peptide sequence can be present in multiple different proteins or protein isoforms; these shared peptides lead to ambiguities in determining identities and abundance of proteins [20]. To increase protein identification capacity, dynamic exclusion is widely used. Dynamic exclusion will also result in a decrease of total spectral counts. However, it has been shown that protein expression ratios are not affected by dynamic exclusion [21]. Furthermore, enabling dynamic exclusion leads to higher peptide counts and a gain in quantification of lower abundance proteins [22].
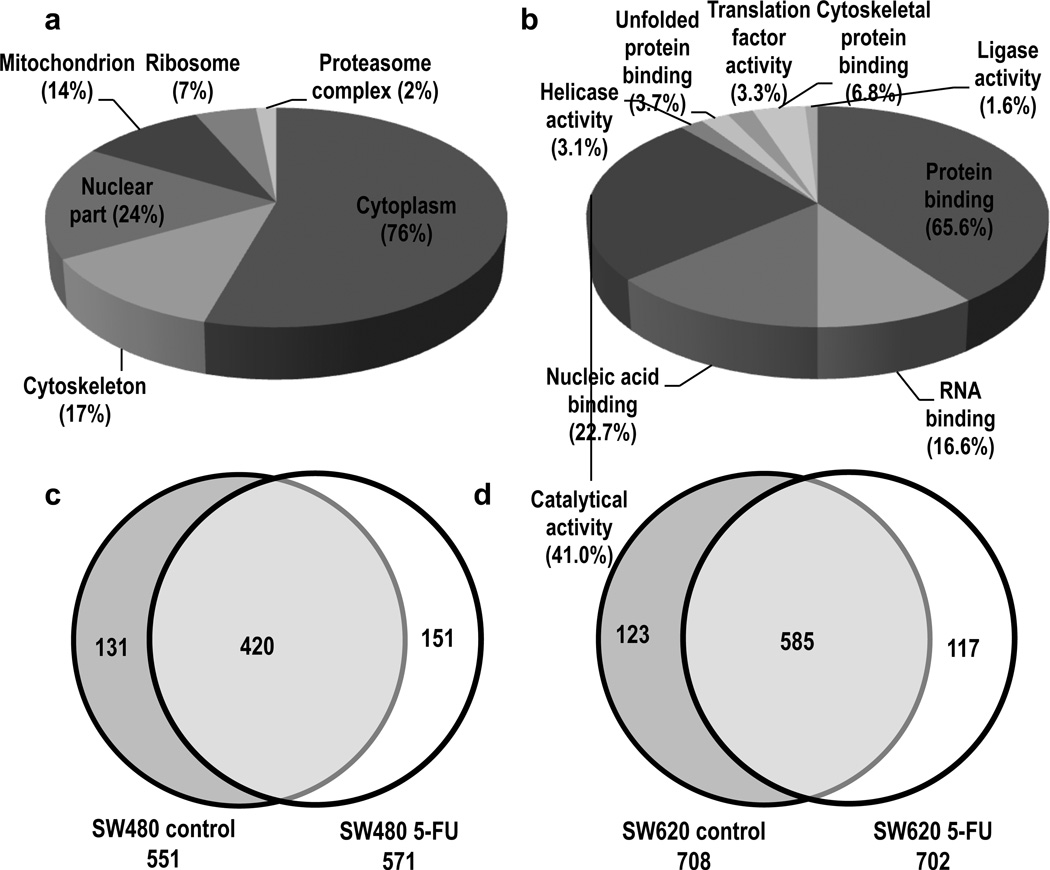
In total, 900 protein groups were identified among the four biological conditions. Gene ontology (GO) analysis of the protein groups identified the cellular compartments and biological processes represented by the proteins in the dataset (Figure 2a-b). Specifically, identified protein groups assigned to cellular compartments were distributed among cytoplasmic (76%), nuclear (24%), cytoskeletal (17%), mitochondrial (14%), ribosomal (7%) and proteasome complex (2%) species, showing sufficient extraction and detection based on the wide distribution of identified protein groups. A large percentage of the identified proteins mapped to protein binding (65.5%), catalytic activity (41.0%) and nucleic acid binding (22.7%) species. The overlaps amongst the protein sets for the biological conditions are shown in Venn diagrams (Figure 2c and 2d). There were a total of 702 protein groups identified in the SW480 sample set, 420 of which were identified in both 5-FU treated and control samples. In the SW620 sample set, 825 protein groups were identified, with 585 identified in both 5-FU treated and control samples. Protein group overlap evaluation between the technical triplicate runs is displayed in Supporting Information Figure 1. Additional information regarding the protein groups identified can be found in Supporting Information Table 1–3. The respective protein group identifications are based on LC-MS/MS peptide fragmentation spectra. Representative fragment ion spectra of select peptide ions from several proteins show extensive fragmentation series of b- and y-ions (Supporting Information Figure 2).
Figure 2.
Gene ontology (GO) analysis and biological sample distribution of identified protein groups. The protein groups identified were classified by (a) broad subcellular localization and (b) molecular function. Some proteins may be represented in more than one category. The numbers in parentheses indicate the percentage of identified proteins represented by each category. Venn diagrams showing distribution of identified protein groups across the biological samples (c) SW480 and (d) SW620
3.3 Spectral counting relative quantification
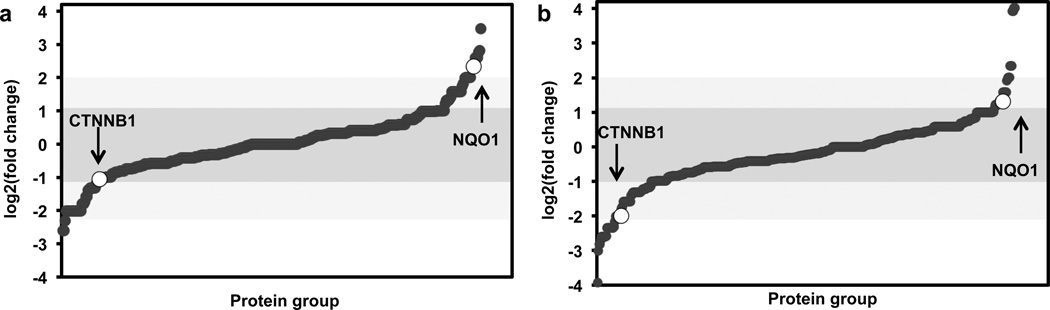
To quantify the identified proteins using spectral counting, the spectral counts of each peptide were averaged over its appearance in all technical replicates. To account for any deviation in technical reproducibility, the average spectral counts were normalized to the total number of spectral counts for each biological condition prior to relative protein quantification. The sum of the spectral counts from the constituent peptides of each protein was also calculated. A total of 900 protein groups were identified in this study; however, only a fraction of those proteins were accepted for quantification after applying a threshold of ≥ 4 SpC for high confidence in differential protein expression [23] (Supporting Information Figure 3). A traditional 2-fold change in relative expression was set for identification of differentially expressed proteins. With these criteria, there were 267 protein groups accepted for quantification between the SW480 treated and untreated samples and 401 protein groups accepted between the treated and untreated SW620 samples. The distributions of expression changes for the protein groups are shown in figure 3. We found that only a small percentage of confidently quantifiable proteins were differentially expressed in the 5-FU treated cells for both cell line comparisons. There were 51 protein groups whose expression changed with 5-FU treatment in the SW480 cell line comparison; 29 (10.9%) were upregulated and 27 (10.1%) were downregulated. In the SW620 cell line comparison, 79 protein groups showed changed expression; 22 (5.5%) were upregulated and 57 (14.2%) were downregulated. Highlighted are two proteins found to be upregulated (NQO1) or downregulated (CTNNB1) in both SW480 and SW620 5-FU treated cells as compared to the control cells.
Figure 3.
Distribution of protein group spectral count relative expression changes between (a) SW480 (b) and SW620 5-FU treated as compared to control cells. The protein expression ratios are shown in log2-scale for all protein groups accepted for quantification. Protein ratios arranged from descending to ascending order results in a sigmoidal shaped curve. The darker shaded area represents unregulated protein groups with a less than 2-fold change in expression, with the ligher shaded area representing protein groups regulated between 2- and 3-fold change. The expression ratios for NQO1 and CTNNB1 are indicated
3.4 Functional annotation of protein groups with changed expression
Gene ontology annotation was used to analyze the function of differentially expressed protein groups. Pair-wise analysis between control and 5-FU comparative protein expression profiling for each cell line revealed significant changes in proteins involved in cell adhesion remodeling (CTNNB1, RHOA) and antioxidant activity (NQO1,PARK7, PRDX2, PRDX5, PRDX6).
3.5 Cell adhesion remodeling
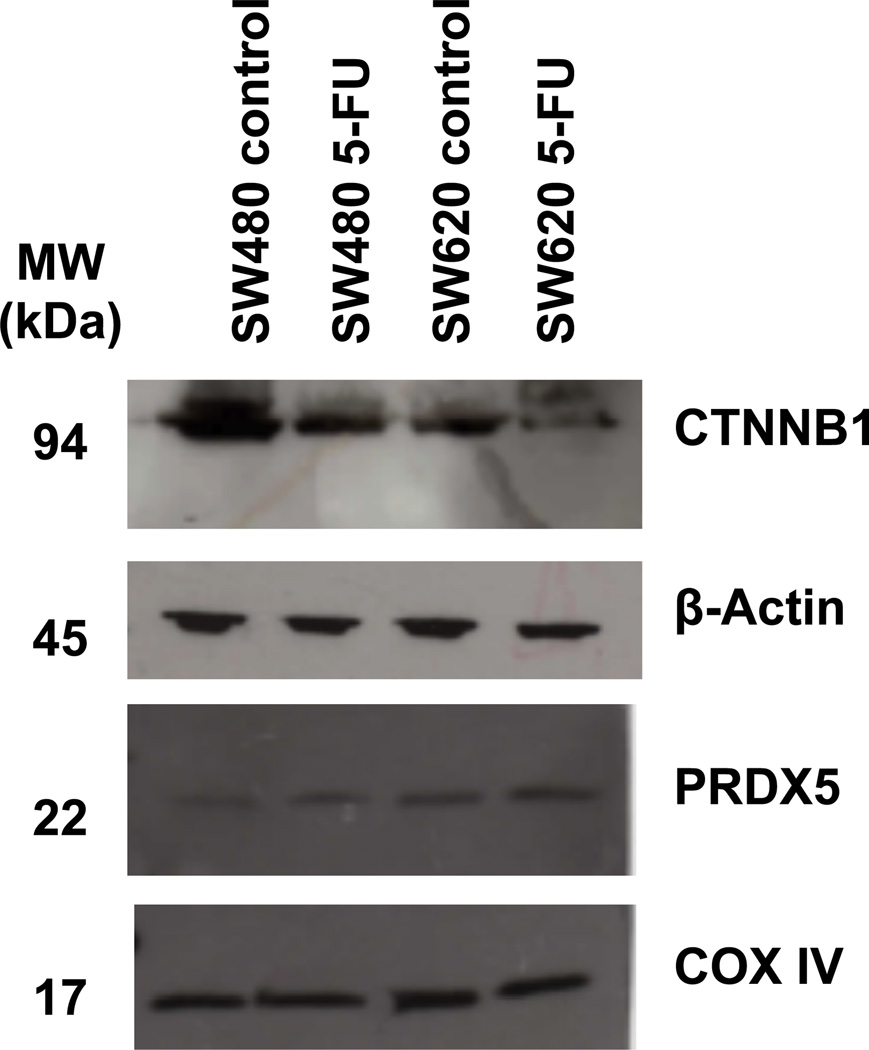
The chemosensitivity of cancer cells may be affected by the state of cell adhesion and expression of intracellular adhesion and cytoskeletal proteins. CTNNB1 (Catenin-beta 1) is a dual-purpose protein playing a critical role in the Wnt signaling pathways and cell-cell adhesion. In epithelial cells, cytoplasm CTNNB1, a component of the adherens junctions, provides a mechanical linkage between cell-to-cell junction proteins and cytoskeletal proteins. Loss of CTNNB1 may result in cells that are less epithelial and more mesenchymal with disassembled cell-to-cell junctions and greater migratory properties. Epithelial to mesenchymal transition (EMT) has been shown to be correlated with anticancer drug resistance in several solid tumors including colon cancer [24]. The reduced expression of CTNNB1 in both SW480 and SW620 5-FU treated cells, whether the result of increased activity of Wnt regulated degradation or nuclear localization, may be compromising the epithelial integrity by interfering with the cadherin-catenin interaction. Validation of the changes in CTNNB1 abundance with 5-FU treatment by Western blot is shown in Figure 4.
Figure 4.
Western blot validation of selected proteins. Protein samples from SW480 and SW620 control and 5-FU treated cell lysates were probed CTNNB1 and PRDX5 antibodies. β-Actin and COX IV were treated as loading controls
RhoA is another cell adhesion protein showing decreased expression in the 5-FU treated cells from both cell lines. This Rho GTPase is known to regulate the actin cytoskeleton through the formation of stress fibers and loss of RhoA results in the breakdown of adherens junctions [25]. The cells surviving 72 hours of 5-FU treatment possibly represent cells with de novo resistance. Over time, the tumor acquires resistance and associated phenotypes such as EMT with major cell adhesion remodeling become more prominent. However, clues are present in the primary resistant cells as seen in the reduced expression of CTNNB1 and RhoA.
3.6 Antioxidative activity
Oxidative stress is characterized by an imbalance between reactive oxygen species (ROS) generation and the availability of antioxidant species. ROS are thought to be involved in several cellular mechanisms including apoptosis. Studies have shown that many anticancer drugs, including 5-FU, cause cytotoxicity by inducing ROS production [26]. This finding implies that the redox status of the cell is important in determining the sensitivity of cancer cells to chemotherapy.
One of the major mechanisms by which cells protect themselves from this complex situation is through upregulation of a wide range of molecules and enzymes with antioxidant activity. The antioxidant species eliminate ROS to protect the cells from oxidative stress, which may also contribute to chemotherapy drug resistance in cancer. Among these antioxidant species, members of the peroxiredoxin family (Prdxs) of antioxidant enzymes and NAD(P)H:quinone oxidoreductase 1 (NQO1) were shown to be upregulated in the population of colon cancer cells that survived 72 hours of 5-FU assault. PRDX2 was more abundant in SW620 while PRDX5 and PRDX6 had higher expression in SW480 5-FU treated cells as compared to the control cells. Peroxiredoxins (Prdxs) reduce intracellular peroxides such as hydrogen peroxide, one type of ROS, with the thioredoxin system [27]. Prdxs not only play an important role in detoxification, but also increase cell survival and proliferation under conditions of oxidative stress. Furthermore, the detoxification enzyme NQO1 showed increased expression in both SW480 and SW620 cells treated with 5-FU. NQO1 plays a role in chemoprotection by generating antioxidant forms of ubiquinone and vitamin E, which is supported by reports of NQO1 expression in epithelial human tissues, including the colon, that require enhanced levels of antioxidant protection [28]. This cytoprotective enzyme also enhances tumor cell survival during chemotherapy [29]. A western blot confirmation of the changes in PRDX5 abundance with 5-FU treatment is shown in Figure 5.
In addition, the SW480 5-FU treated cells have heightened PARK7 (DJ-1) expression. The oncogene PARK7 is an mRNA binding protein and sensor for oxidative damage and responds by stabilizing Nrf2. In turn, the transcription factor Nrf2 induces the expression of several antioxidant enzymes including the Prdxs and NQO1 [30]. This may explain the increased levels of these detoxifying enzymes found in the SW480 5-FU surviving cells. There was no change in PARK7 expression between the SW620 5-FU treated and control cells suggesting that the pathways involved in upregulating antioxidant enzymes may vary as colon cancer progresses. An alternative explanation could be that the original microenvironment for the cell lines has established a differential gene response pattern in the primary versus metastatic cells, as the SW480 and SW620 cell lines are from a primary colon cancer tumor and the lymph node metastasis, respectively. Elevated PARK7 has previously been associated with chemotherapy resistance and poor prognosis in many cancers, but the association of PARK7 with 5-FU resistance and colon cancer is novel.
The colon cancer cells with a higher antioxidant capacity seem to have survived the 5-FU treatment, as these cells were able to survive oxidative damage and apoptosis from 5-FU induced ROS production. These surviving cells may eventually give rise to a cancer cell population with an acquired resistance to 5-FU through a cellular adaptive response to oxidative stress. The peroxiredoxin family and NQO1 antioxidant enzymes seem to play an important role in 5-FU survival and have the potential to be targets for cancer therapy. Depletion of these enzymes may reduce the cancer cell's ability to scavenge and detoxify ROS, sensitizing the cells to ROS mediated apoptosis. Although PRDX6 has previously been shown to be upregulated in SW480 cells treated with 5-FU [10], PRDX2, PRDX5 and NQO1 have not been previously associated with 5-FU resistance in colon cancer.
4 Concluding remarks
Using a label-free spectral counting approach to examine response to 5-FU in patient-matched primary and metastatic cell lines, we demonstrated that while some protein patterns are conserved with metastasis, others respond differently. In particular, both the SW480 and SW620 cells experienced changes in their reduction-oxidation response, yet disparate members of the peroxiredoxin family were upregulated following treatment. This result indicates that while the cells experience similar cytotoxic stress, alternative mechanisms are used in response.
Proteomic changes induced by 5-FU treatment in SW480 have previously been examined using 2-dimensional electrophoresis technology followed by MS detection [10]. Although the concentration of 5-FU was double that used in our study, several of the same proteins were found to be increased in expression with treatment. Most notably PRDX6, while nearly 3-fold increased in expression in our data set, experienced a nearly 11-fold increase with the higher dose of 5-FU [10]. However, there are also several proteins that differ in response between our study and previous literature reports, including 2 heat shock proteins: heat shock protein beta-1 (P04792) and mitochondrial 60 kDa heat shock protein (P10809). Follow-up studies are needed to examine these discrepancies.
Future studies will include a comparison of label free approaches with protein labeling strategies. This study communicates the elucidation of NQO1 antioxidant enzyme and further confirmation of the peroxiredoxin family as possible molecular targets of colorectal cancer drug resistance. Further examination into the differential response of primary versus metastatic cells to drug treatment will improve our understanding of the underlying molecular mechanisms and hopefully lead to more effective treatments for this disease.
Supplementary Material
Supporting Information Figure 1. Venn diagrams displaying the technical triplicate protein group identification for (a) SW480 control (b) SW480 5-FU (c) SW260 control (d) SW620 5-FU. Total number of protein groups identified for each technical replicate (TR) is displayed.
Supporting Information Figure 2. Representative LC-MS/MS peptide ion fragmentation spectra of select peptides from several proteins (a) Catenin beta-1 (P35222) (b) Pyruvate kinase (P14618) (c) Peroxiredoxin-6 (P30041) (d) Transforming protein RhoA (P61586).
Supporting Information Figure 3. The coefficient of variation (CV%) for the proteins identified in each sample are plotted as a function of spectral counts (SpC); (a) SW480 control (b) SW480 5-FU (c) SW620 control (d) SW620 5-FU.
Table 1.
Proteins identified with ≥ 4 SpC and a greater than two fold change in relative expression between 5-FU treated and control samples for both SW620 (left columns) and SW480 (right columns) cells as determined by spectral counting. The fold change was determined by the ratio of normalized spectral counts of 5-FU/control. Expression ratios are presented as log2-fold change with associated p-values from t-test at a significance level of 0.05. For protein groups consisting of proteins with multiple annotated forms identified, only the top scoring form based on Mascot protein score is represented in the table.
| Accession | Gene name |
Protein name | log2 Ratio |
P-value |
|---|---|---|---|---|
| O75369 | FLNB | Filamin-B | 4.01 | 2.48E-02 |
| P48444 | COPD | Coatomer subunit delta | 2.34 | 1.15E-01 |
| Q04917 | 1433F | 14-3-3 protein eta | 2.34 | 7.41E-02 |
| Q9Y262 | EIF3L | Eukaryotic translation initiation factor 3 subunit L | 2.34 | 2.37E-02 |
| O00148 | DDX39 | ATP-dependent RNA helicase DDX39 | 2.01 | 5.06E-02 |
| P30740 | ILEU | Leukocyte elastase inhibitor | 2.01 | 5.06E-02 |
| P48681 | NEST | Nestin | 1.99 | 5.06E-02 |
| Q14204 | DYHC 1 | Cytoplasmic dynein 1 heavy chain 1 | 1.91 | 7.09E-03 |
| Q6P2E9 | EDC4 | Enhancer of mRNA-decapping protein 4 | 1.58 | 1.37E-01 |
| Q9Y230 | RUVB 2 | RuvB-like 2 | 1.58 | 5.81E-02 |
| Q99873 | ANM1 | Protein arginine N-methyltransferase 1 | 1.58 | 5.81E-02 |
| P15559 | NQO1 | NAD(P)H dehydrogenase [quinone] 1 | 1.32 | 1.74E-01 |
| O14818 | PSA7 | Proteasome subunit alpha type-7 | 1.32 | 1.74E-01 |
| P02788 | TRFL | Lactotransferrin | 1.32 | 5.06E-02 |
| P60660 | MYL6 | Myosin light polypeptide 6 | 1.32 | 5.06E-02 |
| P46940 | IQGA1 | Ras GTPase-activating-like protein IQGAP1 | 1.25 | 6.46E-04 |
| Q13813 | SPTA2 | Spectrin alpha chain, brain | 1.22 | 1.87E-01 |
| P00492 | HPRT | Hypoxanthine-guanine phosphoribosyltransferase | 1.22 | 5.81E-02 |
| P32119 | PRDX2 | Peroxiredoxin-2 | 1.14 | 5.06E-02 |
| P02771 | FETA | Alpha-fetoprotein | 1.01 | 1.47E-01 |
| P33993 | MCM7 | DNA replication licensing factor MCM7 | 1.01 | 9.91E-02 |
| Q9BSJ8-2 | ESYT1 | Isoform 2 of Extended synaptotagmin-1 | 1 | 1.75E-01 |
| Q99714 | HCD2 | 3-hydroxyacyl-CoA dehydrogenase type-2 | −1.14 | 2.75E-02 |
| P30050 | RL12 | 60S ribosomal protein L12 | −1.17 | 1.19E-01 |
| O14980 | XPO1 | Exportin-1 | −1.17 | 9.45E-02 |
| P17844 | DDX5 | Probable ATP-dependent RNA helicase DDX5 | −1.17 | 4.17E-02 |
| P27824 | CALX | Calnexin | −1.17 | 3.34E-02 |
| P50990 | TCPQ | T-complex protein 1 subunit theta | 3.48 | 3.62E-02 |
| Q99497 | PARK7 | Protein DJ-1 | 2.82 | 6.62E-03 |
| P10809 | CH60 | 60 kDa heat shock protein | 2.76 | 1.46E-03 |
| P04632 | CPNS1 | Calpain small subunit 1 | 2.6 | 3.34E-02 |
| P49720 | PSB3 | Proteasome subunit beta type-3 | 2.6 | 3.34E-02 |
| Q15691 | MARE1 | Microtubule-associated protein RP/EB family member 1 | 2.34 | 1.73E-01 |
| P15559 | NQO1 | NAD(P)H dehydrogenase [quinone] 1 | 2.34 | 7.41E-02 |
| Q14204 | DYHC1 | Cytoplasmic dynein 1 heavy chain 1 | 2.32 | 6.35E-02 |
| A8TX70-2 | CO6A5 | Isoform 2 of Collagen alpha-5(VI) chain | 2.01 | 1.74E-01 |
| Q15648 | MED1 | Mediator of RNA polymerase II transcription subunit 1 | 2.01 | 1.25E-01 |
| P14314 | GLU2B | Glucosidase 2 subunit beta | 2.01 | 5.06E-02 |
| P49736 | MCM2 | DNA replication licensing factor MCM2 | 2.01 | 5.06E-02 |
| P49411 | EFTU | Elongation factor Tu | 1.99 | 5.06E-02 |
| P14618 | KPYM | Pyruvate kinase isozymes M1/M2 | 1.82 | 4.77E-03 |
| Q13813 | SPTA2 | Spectrin alpha chain | 1.58 | 1.03E-01 |
| P22102 | PUR2 | Trifunctional purine biosynthetic protein adenosine-3 | 1.58 | 5.81E-02 |
| O95373 | IPO7 | Importin-7 | 1.58 | 3.52E-02 |
| P30041 | PRDX6 | Peroxiredoxin-6 | 1.58 | 9.72E-03 |
| Q9Y4L1 | HYOU1 | Hypoxia up-regulated protein 1 | 1.58 | 8.07E-03 |
| P19823 | ITIH2 | Inter-alpha-trypsin inhibitor heavy chain H2 | 1.58 | 8.07E-03 |
| P25788 | PSA3 | Proteasome subunit alpha type-3 | 1.58 | 8.07E-03 |
| P14174 | MIF | Macrophage migration inhibitory factor | 1.42 | 3.75E-03 |
| Q14152 | EIF3A | Eukaryotic translation initiation factor 3 subunit A | 1.36 | 2.66E-03 |
| Q969J2 | ZKSC4 | Zinc finger protein with KRAB and SCAN domains 4 | 1.32 | 1.74E-01 |
| P00492 | HPRT | Hypoxanthine-guanine phosphoribosyltransferase | 1.32 | 1.25E-01 |
| P02787 | TRFE | Serotransferrin | 1.32 | 5.06E-02 |
| P52209 | 6PGD | 6-phosphogluconate dehydrogenase, decarboxylating | 1.22 | 1.37E-01 |
| P37837 | TALDO | Transaldolase | −1.22 | 1.37E-01 |
| P04843 | RPN1 | Dolichyl-diphosphooligosaccharide subunit 1 | −1.22 | 1.37E-01 |
| P41252 | SYIC | Isoleucyl-tRNA synthetase, cytoplasmic | −1.22 | 1.03E-01 |
| P26640 | SYVC | Valyl-tRNA synthetase | −1.22 | 1.03E-01 |
| P25205 | MCM3 | DNA replication licensing factor MCM3 | −1.22 | 5.81E-02 |
| Q15758 | AAAT | Neutral amino acid transporter B(0) | −1.25 | 1.47E-02 |
| P50990 | TCPQ | T-complex protein 1 subunit theta | −1.28 | 8.61E-03 |
| P63010-2 | AP2B1 | Isoform 2 of AP-2 complex subunit beta | −1.32 | 1.74E-01 |
| P25789 | PSA4 | Proteasome subunit alpha type-4 | −1.32 | 1.25E-01 |
| P46783 | RS10 | 40S ribosomal protein S10 | −1.32 | 1.25E-01 |
| P62851 | RS25 | 40S ribosomal protein S25 | −1.32 | 1.25E-01 |
| P56192 | SYMC | Methionyl-tRNA synthetase, cytoplasmic | −1.32 | 1.25E-01 |
| P33992 | MCM5 | DNA replication licensing factor MCM5 | −1.32 | 5.06E-02 |
| P13797 | PLST | Plastin-3 | −1.32 | 5.06E-02 |
| P63244 | GBLP | Guanine nucleotide-binding protein subunit beta-2-like 1 | −1.32 | 1.12E-02 |
| P17987 | TCPA | T-complex protein 1 subunit alpha | −1.32 | 8.01E-03 |
| P22234 | PUR6 | Multifunctional protein ADE2 | −1.38 | 6.22E-02 |
| P60228 | EIF3E | Eukaryotic translation initiation factor 3 subunit E | −1.42 | 1.19E-01 |
| P00367 | DHE3 | Glutamate dehydrogenase 1 | −1.58 | 7.92E-02 |
| Q13283 | G3BP1 | Ras GTPase-activating protein-binding protein 1 | −1.58 | 5.81E-02 |
| Q9Y617 | SERC | Phosphoserine aminotransferase | −1.58 | 5.81E-02 |
| P08195-4 | 4F2 | Isoform 4 of 4F2 cell-surface antigen heavy chain | −1.58 | 1.81E-02 |
| Q15366 | PCBP2 | Poly(rC)-binding protein 2 | −1.58 | 1.29E-02 |
| P51149 | RAB7A | Ras-related protein Rab-7a | −1.59 | 1.95E-02 |
| O00571 | DDX3X | ATP-dependent RNA helicase DDX3X | −1.74 | 1.10E-03 |
| Q16555 | DPYL2 | Dihydropyrimidinase-related protein 2 | −1.76 | 5.06E-02 |
| P26196 | DDX6 | Probable ATP-dependent RNA helicase DDX6 | −1.8 | 7.59E-02 |
| P35222 | CTNNB1 | Catenin beta-1 | −2 | 2.91E-03 |
| P48637 | GSHB | Glutathione synthetase | −2.01 | 1.74E-01 |
| P53621-2 | COPA | Isoform 2 of Coatomer subunit alpha | 1.01 | 1.15E-01 |
| P24534 | EF1B | Elongation factor 1-beta | 1.01 | 2.37E-02 |
| Q01105 | SET | Protein SET | −1.01 | 1.15E-01 |
| Q9UQ80 | PA2G4 | Proliferation-associated protein 2G4 | −1.01 | 2.37E-02 |
| P35222 | CTNNB1 | Catenin beta-1 | −1.06 | 3.91E-02 |
| P23396 | RS3 | 40S ribosomal protein S3 | −1.09 | 7.62E-02 |
| Q7KZF4 | SND1 | Staphylococcal nuclease domain-containing protein 1 | −1.17 | 6.59E-02 |
| P05388 | RLA0 | 60S acidic ribosomal protein P0 | −1.19 | 2.67E-02 |
| P17174 | AATC | Aspartate aminotransferase, cytoplasmic | −1.32 | 1.74E-01 |
| P17931 | LEG3 | Galectin-3 | −1.32 | 1.25E-01 |
| P62988 | UBIQ | Ubiquitin | −1.32 | 5.06E-02 |
| P62269 | RS18 | 40S ribosomal protein S18 | −1.32 | 6.62E-03 |
| P55060-3 | XPO2 | Isoform 3 of Exportin-2 | −1.39 | 1.13E-02 |
| P45974 | UBP5 | Ubiquitin carboxyl-terminal hydrolase 5 | −1.58 | 8.07E-03 |
| P46781 | RS9 | 40S ribosomal protein S9 | −1.8 | 1.26E-01 |
| P15880 | RS2 | 40S ribosomal protein S2 | −1.8 | 4.45E-02 |
| P62249 | RS16 | 40S ribosomal protein S16 | −2.01 | 1.74E-01 |
| P12429 | ANXA3 | Annexin A3 | −2.01 | 5.06E-02 |
| O14981 | BTAF1 | TATA-binding protein-associated factor 172 | −2.01 | 5.06E-02 |
| Q9UNF1 | MAGD2 | Melanoma-associated antigen D2 | −2.01 | 5.06E-02 |
| P33992 | MCM5 | DNA replication licensing factor MCM5 | −2.01 | 5.06E-02 |
| Q13765 | NACA | Nascent polypeptide-associated complex subunit alpha | −2.01 | 5.06E-02 |
| P08134 | RHOC | Rho-related GTP-binding protein RhoC | −2.01 | 5.06E-02 |
| P39023 | RL3 | 60S ribosomal protein L3 | −2.01 | 5.06E-02 |
| P22626 | ROA2 | Heterogeneous nuclear ribonucleoproteins A2/B1 | −2.01 | 5.06E-02 |
| Q9Y5L0 | TNPO3 | Transportin-3 | −2.01 | 5.06E-02 |
| Q9BWD1 | THIC | Acetyl-CoA acetyltransferase | −2.31 | 1.16E-02 |
| P08708 | RS17 | 40S ribosomal protein S17 | −2.6 | 3.34E-02 |
| P62701 | RS4X | 40S ribosomal protein S4, X isoform | −2.6 | 3.34E-02 |
| Q9NUU7 | DD19A | ATP-dependent RNA helicase DDX19A | −2.01 | 5.06E-02 |
| P14735 | IDE | Insulin-degrading enzyme | −2.01 | 5.06E-02 |
| O75533 | SF3B1 | Splicing factor 3B subunit 1 | −2.01 | 5.06E-02 |
| Q9P2J5 | SYLC | Leucyl-tRNA synthetase | −2.01 | 5.06E-02 |
| Q9C0C9 | UBE2O | Ubiquitin-conjugating enzyme E2 | −2.01 | 5.06E-02 |
| P48637 | GSHB | Glutathione synthetase | −2.01 | 5.06E-02 |
| P61247 | RS3A | 40S ribosomal protein S3a | −2.16 | 2.86E-02 |
| P24752 | THIL | Acetyl-CoA acetyltransferase | −2.31 | 4.97E-02 |
| P46777 | RL5 | 60S ribosomal protein L5 | −2.34 | 1.73E-01 |
| A6NC57 | ANR62 | Ankyrin repeat domain-containing protein 62 | −2.34 | 7.41E-02 |
| O00764 | PDXK | Pyridoxal kinase | −2.34 | 2.37E-02 |
| P05386 | RLA1 | 60S acidic ribosomal protein P1 | −2.34 | 2.37E-02 |
| Q16851 | UGPA | UTP--glucose-1-phosphate uridylyltransferase | −2.34 | 2.37E-02 |
| P78527-2 | PRKDC | Isoform 2 of DNA-dependent protein kinase catalytic subunit | −2.34 | 1.89E-02 |
| Q14444-2 | CAPR1 | Isoform 2 of Caprin-1 | −2.6 | 3.34E-02 |
| P39019 | RS19 | 40S ribosomal protein S19 | −2.6 | 3.34E-02 |
| P46782 | RS5 | 40S ribosomal protein S5 | −2.6 | 3.34E-02 |
| P46776 | RL27A | 60S ribosomal protein L27a | −2.6 | 3.75E-03 |
| P62424 | RL7A | 60S ribosomal protein L7a | −2.6 | 3.75E-03 |
| P22626 | ROA2 | Heterogeneous nuclear ribonucleoproteins A2/B1 | −2.69 | 7.37E-04 |
| P54886-2 | P5CS | Isoform Short of Delta-1-pyrroline-5-carboxylate synthase | −2.82 | 5.06E-02 |
| P09382 | LEG1 | Galectin-1 | −3.02 | 3.88E-03 |
| P04181 | OAT | Ornithine aminotransferase | −3.92 | 2.03E-02 |
Acknowledgements
The authors gratefully acknowledge assistance from the Notre Dame Mass Spectrometry and Proteomics Facility. This research was funded by the University of Notre Dame and the 2011 Starter Grant from the Society for Analytical Chemists of Pittsburgh. K.M.B. was supported by the Notre Dame CBBI program and NIH training grant T32GM075762. P.A.L. was supported by the Notre Dame College of Science REU program.
Footnotes
Conflict of Interest
The authors have declared no conflict of interest.
References
- 1.AACR. 2011 [Google Scholar]
- 2.Iwaizumi M, Tseng-Rogenski S, Carethers JM. DNA mismatch repair proficiency executing 5-fluorouracil cytotoxicity in colorectal cancer cells. Cancer Biol Ther. 2011;12:756–764. doi: 10.4161/cbt.12.8.17169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Longley DB, Harkin DP, Johnston PG. 5-fluorouracil: mechanisms of action and clinical strategies. Nature reviews. Cancer. 2003;3:330–338. doi: 10.1038/nrc1074. [DOI] [PubMed] [Google Scholar]
- 4.Bracht K, Nicholls AM, Liu Y, Bodmer WF. 5-Fluorouracil response in a large panel of colorectal cancer cell lines is associated with mismatch repair deficiency. Br J Cancer. 2010;103:340–346. doi: 10.1038/sj.bjc.6605780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mechetner E, Brunner N, Parker RJ. In vitro drug responses in primary and metastatic colorectal cancers. Scand J Gastroenterol. 2011;46:70–78. doi: 10.3109/00365521.2010.510573. [DOI] [PubMed] [Google Scholar]
- 6.Leibovitz A, Stinson JC, McCombs WB, 3rd, McCoy CE, et al. EClassification of human colorectal adenocarcinoma cell lines. Cancer research. 1976;36:4562–4569. [PubMed] [Google Scholar]
- 7.Katayama M, Nakano H, Ishiuchi A, Wu W, et al. Protein pattern difference in the colon cancer cell lines examined by two-dimensional differential in-gel electrophoresis and mass spectrometry. Surg Today. 2006;36:1085–1093. doi: 10.1007/s00595-006-3301-y. [DOI] [PubMed] [Google Scholar]
- 8.Xue H, Lu B, Zhang J, Wu M, et al. Identification of serum biomarkers for colorectal cancer metastasis using a differential secretome approach. J Proteome Res. 2010;9:545–555. doi: 10.1021/pr9008817. [DOI] [PubMed] [Google Scholar]
- 9.Ghosh D, Yu H, Tan XF, Lim TK, et al. Identification of Key Players for Colorectal Cancer Metastasis by iTRAQ Quantitative Proteomics Profiling of Isogenic SW480 and SW620 Cell Lines. J Proteome Res. 2011;10:4373–4387. doi: 10.1021/pr2005617. [DOI] [PubMed] [Google Scholar]
- 10.Wong CS, Wong VW, Chan CM, Ma BB, et al. Identification of 5-fluorouracil response proteins in colorectal carcinoma cell line SW480 by two-dimensional electrophoresis and MALDI-TOF mass spectrometry. Oncol Rep. 2008;20:89–98. [PubMed] [Google Scholar]
- 11.Cox J, Mann M. Quantitative, high-resolution proteomics for data-driven systems biology. Annu Rev Biochem. 2011;80:273–299. doi: 10.1146/annurev-biochem-061308-093216. [DOI] [PubMed] [Google Scholar]
- 12.Wiese S, Reidegeld KA, Meyer HE, Warscheid B. Protein labeling by iTRAQ: a new tool for quantitative mass spectrometry in proteome research. Proteomics. 2007;7:340–350. doi: 10.1002/pmic.200600422. [DOI] [PubMed] [Google Scholar]
- 13.Everley PA, Krijgsveld J, Zetter BR, Gygi SP. Quantitative cancer proteomics: stable isotope labeling with amino acids in cell culture (SILAC) as a tool for prostate cancer research. Molecular & cellular proteomics : MCP. 2004;3:729–735. doi: 10.1074/mcp.M400021-MCP200. [DOI] [PubMed] [Google Scholar]
- 14.Liu H, Sadygov RG, Yates JR., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Analytical chemistry. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 15.Zybailov B, Coleman MK, Florens L, Washburn MP. Correlation of relative abundance ratios derived from peptide ion chromatograms and spectrum counting for quantitative proteomic analysis using stable isotope labeling. Analytical chemistry. 2005;77:6218–6224. doi: 10.1021/ac050846r. [DOI] [PubMed] [Google Scholar]
- 16.Neilson KA, Ali NA, Muralidharan S, Mirzaei M, et al. Less label, more free: approaches in label-free quantitative mass spectrometry. Proteomics. 2011;11:535–553. doi: 10.1002/pmic.201000553. [DOI] [PubMed] [Google Scholar]
- 17.Wong JW, Cagney G. An overview of label-free quantitation methods in proteomics by mass spectrometry. Methods Mol Biol. 2010;604:273–283. doi: 10.1007/978-1-60761-444-9_18. [DOI] [PubMed] [Google Scholar]
- 18.Lundgren DH, Hwang SI, Wu L, Han DK. Role of spectral counting in quantitative proteomics. Expert Rev Proteomics. 2010;7:39–53. doi: 10.1586/epr.09.69. [DOI] [PubMed] [Google Scholar]
- 19.Karp NA, Spencer M, Lindsay H, O'Dell K, Lilley KS. Impact of replicate types on proteomic expression analysis. J Proteome Res. 2005;4:1867–1871. doi: 10.1021/pr050084g. [DOI] [PubMed] [Google Scholar]
- 20.Nesvizhskii AI, Aebersold R. Interpretation of shotgun proteomic data: the protein inference problem. Molecular & cellular proteomics : MCP. 2005;4:1419–1440. doi: 10.1074/mcp.R500012-MCP200. [DOI] [PubMed] [Google Scholar]
- 21.Hoehenwarter W, Wienkoop S. Spectral counting robust on high mass accuracy mass spectrometers. Rapid communications in mass spectrometry : RCM. 2010;24:3609–3614. doi: 10.1002/rcm.4818. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y, Wen Z, Washburn MP, Florens L. Effect of dynamic exclusion duration on spectral count based quantitative proteomics. Analytical chemistry. 2009;81:6317–6326. doi: 10.1021/ac9004887. [DOI] [PubMed] [Google Scholar]
- 23.Old WM, Meyer-Arendt K, Aveline-Wolf L, PiercGe K, et al. Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Molecular & cellular proteomics : MCP. 2005;4:1487–1502. doi: 10.1074/mcp.M500084-MCP200. [DOI] [PubMed] [Google Scholar]
- 24.Tentes IK, Schmidt WM, Krupitza G, Steger GG, et al. Long-term persistence of acquired resistance to 5-fluorouracil in the colon cancer cell line SW620. Experimental cell research. 2010;316:3172–3181. doi: 10.1016/j.yexcr.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 25.Singh A, Settleman J. EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene. 2010;29:4741–4751. doi: 10.1038/onc.2010.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pelicano H, Carney D, Huang P. ROS stress in cancer cells and therapeutic implications. Drug resistance updates : reviews and commentaries in antimicrobial and anticancer chemotherapy. 2004;7:97–110. doi: 10.1016/j.drup.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 27.Zhang B, Wang Y, Su Y. Peroxiredoxins, a novel target in cancer radiotherapy. Cancer letters. 2009;286:154–160. doi: 10.1016/j.canlet.2009.04.043. [DOI] [PubMed] [Google Scholar]
- 28.Ross D, Kepa JK, Winski SL, Beall HD, et al. NAD(P)H:quinone oxidoreductase 1 (NQO1): chemoprotection, bioactivation, gene regulation and genetic polymorphisms. Chemico-biological interactions. 2000;129:77–97. doi: 10.1016/s0009-2797(00)00199-x. [DOI] [PubMed] [Google Scholar]
- 29.Siegel D, Franklin WA, Ross D. Immunohistochemical detection of NAD(P)H:quinone oxidoreductase in human lung and lung tumors. Clinical cancer research : an official journal of the American Association for Cancer Research. 1998;4:2065–2070. [PubMed] [Google Scholar]
- 30.Merikallio H, Paakko P, Kinnula VL, Harju T, Soini Y. Nuclear factor erythroid-derived 2-like 2 (Nrf2) and DJ1 are prognostic factors in lung cancer. Human pathology. 2011 doi: 10.1016/j.humpath.2011.05.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information Figure 1. Venn diagrams displaying the technical triplicate protein group identification for (a) SW480 control (b) SW480 5-FU (c) SW260 control (d) SW620 5-FU. Total number of protein groups identified for each technical replicate (TR) is displayed.
Supporting Information Figure 2. Representative LC-MS/MS peptide ion fragmentation spectra of select peptides from several proteins (a) Catenin beta-1 (P35222) (b) Pyruvate kinase (P14618) (c) Peroxiredoxin-6 (P30041) (d) Transforming protein RhoA (P61586).
Supporting Information Figure 3. The coefficient of variation (CV%) for the proteins identified in each sample are plotted as a function of spectral counts (SpC); (a) SW480 control (b) SW480 5-FU (c) SW620 control (d) SW620 5-FU.