Abstract
The F-actin binding cytoskeletal protein α-catenin interacts with β-catenin-cadherin complexes and stabilizes cell-cell junctions. The β-catenin–α-catenin complex cannot bind to F-actin, whereas interactions of α-catenin with the cytoskeletal protein vinculin appear necessary to stabilize adherens junctions. Here we report the crystal structure of nearly full-length human α-catenin at 3.7 Å resolution. α-Catenin forms an asymmetric dimer, where the four-helix bundle domains of each subunit engage in distinct intermolecular interactions. This results in a left handshake-like dimer, where the two subunits have remarkably different conformations. The crystal structure explains why dimeric α-catenin has a higher affinity for F-actin than monomeric α-catenin, why the β-catenin–α-catenin complex does not bind to F-actin, how activated vinculin links the cadherin-catenin complex to the cytoskeleton, and why α-catenin but not inactive vinculin can bind to F-actin.
The formation and stabilization of cell-cell (adherens) junctions is essential for the development, architecture, maintenance, and function of tissues in higher organisms. Adherens junctions are directed by the cadherin receptor family of single transmembrane-pass glycoproteins, which interact in a homotypic fashion through the agency of their calcium-binding ectodomains1-4. Clustering of these receptors stabilizes adherens junctions and remodels the actin cytoskeleton, and this response requires the interaction of the intracellular tail domains of cadherin receptors to the adaptor protein β-catenin. In turn, β-catenin binds to the F-actin binding cytoskeletal protein α-catenin to form a ternary cadherin-β-catenin-α-catenin complex5-7. Accordingly, α-catenin is necessary for mechanical connections between the E-cadherin–β-catenin complex and the cortical actomyosin network8,9, and loss of α-catenin disrupts adherens junctions and disables connections of the cadherin-β-catenin complex to the actin cytoskeleton10-14.
α-Catenin is a homodimer that binds to F-actin, suggesting that the ternary cadherin–β-catenin–α-catenin complex forms direct links to the actin network. However, monomeric but not dimeric α-catenin binds to the E-cadherin–β-catenin complex, binding of β-catenin peptide disrupts the N-terminal α-catenin homodimer, and reconstituted cadherin–β-catenin–α-catenin complexes do not bind to F-actin15-17. Thus, α-catenin stabilizes adherens junctions by other means and its additional binding partners have been implicated in this response, in particular the cytoskeletal proteins vinculin18-20 and eplin21 that also bind to F-actin. For example, vinculin is necessary to stabilize adherens junctions and force-dependent unfurling of α-catenin has been suggested to recruit vinculin to adherens junctions to stabilize these complexes19. Furthermore, β-catenin competes with α-catenin for binding to vinculin suggesting that β-catenin also recruits vinculin to adherens junctions18,22.
α-Catenin is a 906 residue polypeptide that has been reported to harbor four functional domains: an N-terminal homodimerization and β-catenin binding domain23, an α-actinin and vinculin binding domain (VBD)22,24, an M-fragment that can form cross-linked dimers and that can bind to l-afadin25, and a C-terminal F-actin binding domain7,26 that can also bind to the tight junction protein ZO-127,28. The crystal structure of the N-terminal domain suggested that α-catenin was a symmetrical dimer, where dimerization occurs via two-fold related interactions of two α-helices from each subunit, and the structure of a chimera of this domain with β-catenin binding peptide established that β-catenin binding disrupts this dimer29. The crystal structure of the isolated M-fragment in the central portion of the protein revealed that this is comprised of two tandem four-helix bundles30,31.
To resolve its structure, regulation, and function, here we determined the structure of nearly full-length human α-catenin (lacking its N-terminal residues 1–81) at 3.7 Å resolution. The structure revealed that α-catenin is an asymmetric dimer and suggests that asymmetry drives its functions in controlling binding to F-actin, and in its interactions with activated vinculin. Further, our studies revealed that the activated vinculin–α-catenin complex was a 2:2 heterotetramer, thus explaining how vinculin stabilizes adherens junctions.
Results
Overall fold of human α-catenin
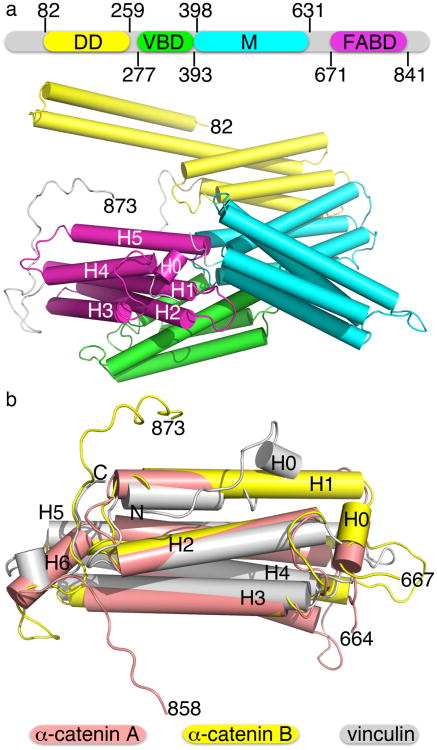
We solved the human α-catenin crystal structure to 3.7 Å resolution (Table 1) by establishing a particular crystal dehydration protocol described in the Supplementary Methods and by identifying a heavy metal cluster that was compatible with the crystallization conditions. The crystal structure revealed that α-catenin is an all-helical asymmetric dimer that is comprised of four domains of helix bundles (Fig. 1a). The N-terminal domain (residues 82–262) of each subunit has two helix bundle domains that resembled the conformation seen in the crystal structure of this domain alone29, consisting of two antiparallel α-helices where the second α-helix was shared by the following four-helix bundle. The VBD (residues 277–393)19,20,28,32,33 was a four-helix bundle that harbored the two α-helical vinculin binding sites (VBS) of α-catenin, where the residues that direct the interaction with vinculin were buried within this four-helix bundle. The M-fragment (residues 390–631) was comprised of tandem four-helix bundle domains as noted previously30,31, yet they adopted a much more compact and vinculin-like conformation in α-catenin, where the two four-helix bundle domains were rotated by 95°–135° relative to the more open V-shape conformation (that had helix bundle-helix bundle angles of about 70°–90° versus about 45° in the full-length structure) seen in the isolated M-fragment structures (Supplementary Fig. 1). Finally, the F-actin binding domain of α-catenin was a five-helix bundle that resembled the vinculin tail domain that also bound to F-actin (Fig. 1b)34,35. However, the termini of these C-terminal tail domains of α-catenin and vinculin were quite distinct. First, the N-terminus of the vinculin tail domain folded back towards the end of α-helix H1, while the N-terminus of the F-actin binding domain of α-catenin interacted with and displaced the H2-H3 loop. Second, the C-terminus of the α-catenin F-actin binding domain adopted two distinct conformations in subunits A and B that were oriented in opposite directions, and only the conformation of subunit A was similar to that of vinculin (Fig. 1b). These differences could contribute, in part, to the distinct F-actin binding properties of the two proteins, where α-catenin can bind to F-actin whereas vinculin binding requires activation by severing of the head-tail clamp that keeps it in its inactive state36-38.
Table 1. X-ray Data Reduction and Crystallographic Refinement Statistics.
Native data were collected at a wavelength of 1 Å at APS/ANL beamline 22BM and derivative data at a wavelength of 1.07 Å at SSRL beamline 11-1. The anomalous completeness for the derivative was 0.98 overall and 1 in the highest resolution shell with a multiplicity of 2.6 and 2.5, respectively. The final model, comprising 1,519 residues, has a correlation coefficient of 0.93. Some loop regions (residues 265-270, 292-297, 354-361, 600-607, 706-710, and 799-810 in subunit A and residues 262-273, 354-362, 600-607, and 846-852 in subunit B) have moderate to very weak electron densities, compared to other regions of the model. The following regions (residues 636-665 and 859-906 in subunit A and residues 638-664 and 874-906 in subunit B) were not modeled due to missing electron density.
| Native | 2Na2O·P2O5·12WO3 | |
|---|---|---|
| X-ray Data Collection | ||
| Space group | P32 | P32 |
| Unit cell dimensions | ||
| a = b, c | 145.6, 145.6 Å, 139 Å | 144.7 Å, 139.9 Å |
| α = β, γ | 90°, 120° | 90°, 120° |
| Resolution [Å] (last shell) | 139.08–3.66 (3.86–3.66) | 46.68–5.58 (5.6–5.58) |
| R-merge (last shell) | 0.077 (0.491) | 0.066 (0.537) |
| Average/σ(I) (last shell) | 15.1 (3.6) | 24.7 (2.9) |
| Completeness (last shell) | 0.99 (1) | 0.99 (1) |
| Redundancy (last shell) | 5.8 (5.8) | 5.2 (5) |
| Crystallographic Refinement | ||
| Resolution, overall | 23.21 Å – 3.66 Å | |
| No. reflections, working set (test set) | 34,383 (1,823) | |
| R-factor (R-free) | 0.217 (0.241) | |
| No. of protein atoms | 11,704 | |
| No. of sulfate atoms | 10 | |
| Average B-factor, protein (sulfate) | 126.5 Å2 (172.1 Å2) | |
| R.m.s.d. from ideal geometry | ||
| Bond lengths | 0.008 Å | |
| Bond angles | 0.99° |
Figure 1.
α-Catenin structure. (a) α-Catenin harbors four distinct domains: the N-terminal dimerization domain (DD), the vinculin binding domain (VBD), the M-fragment (M), and the F-actin binding domain (FABD). F-actin binding domain α-helices are labeled H1 through H5 as are the termini. Subunit B is shown. (b) Superposition of the F-actin binding domain of the two subunits in the asymmetric unit onto the vinculin tail domain. α-Catenin terminal residues are labeled and ‘N’ and ‘C’ indicate vinculin termini, respectively.
The α-catenin asymmetric dimer
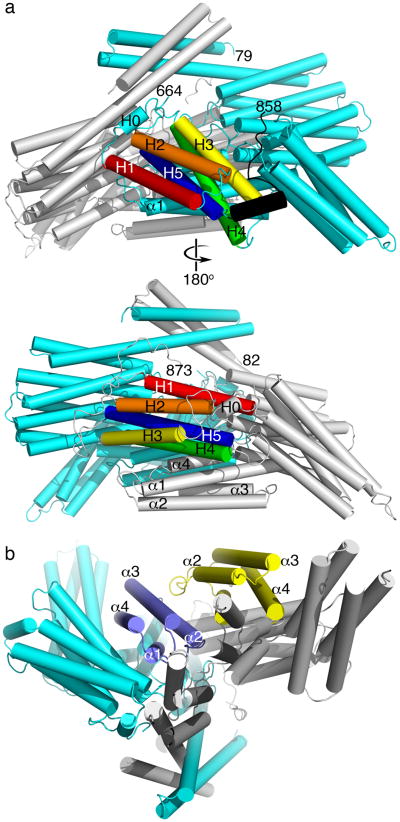
The α-catenin dimer was about 130 Å in its longest dimension and its architecture resembled that of an asymmetric left handshake (Fig. 2, Supplementary Fig. 2), where the thumbs were the helix bundles of the N-terminus, the palms are the M-fragment, and the fingers were the F-actin binding domain and the VBD. Superposition of both molecules within the dimer underscored the asymmetry and distinct orientations of the two subunits (subunit A and subunit B), where there was a large r.m.s.d. of about 4.8 Å, and of even 3.2 Å without the F-actin binding domain (residues 82–631). In contrast, the individual domains of the two subunits superimposed relatively well (0.8 Å for the VBD and M-fragment [residues 277–631]; 0.6 Å for the M-fragment [residues 390–631]; and 0.5 Å for the N-terminal domain [residues 82–262]), indicating that there is intrinsically high flexibility within the polypeptide chain. This dynamic nature may account for the ability of α-catenin to switch between its two oligomeric states.
Figure 2.
α-Catenin is a dimer that resembles a left handshake. (a) Views onto the distinct F-actin binding domain of subunits A (top panel) and B (bottom panel) are shown. Subunit A is shown in cyan, subunit B in grey, the five α-helices of the respective F-actin binding domains are colored spectrally (H1, red; H2, orange; H3 yellow; H4, green; H5, blue). The F-actin binding domain α-helices H0 and the respective termini as well as the respective α-helices of the VBD (α1 through α4) are indicated. (b) View onto the markedly different intermolecular interactions of the VBD shown in blue and yellow for subunits A (cyan) and B (grey), respectively. The dimer ‘stands’ on the N-terminal dimerization domains in this view.
Interestingly, the structure of the N-terminal dimerization domain as found in the nearly full-length α-catenin dimer more closely resembled its conformation in the β-catenin-α-catenin chimera (PDB 1dow)29 versus the isolated domain (PDB 1dov) in its unbound state (r.m.s.d. of 1.6 Å versus 2.1 Å). This was particularly the case for the subunit B conformation of α-catenin, which superimposed onto this chimera with r.m.s.d. of about 1.5 Å compared to superposition onto the unbound N-terminal dimerization domain (2.2 Å). In contrast, subunit A superimposed similarly onto either structure. Collectively, this architecture resulted in a more open conformation for the B subunit of α-catenin.
Except for the second four-helix bundle of the M-fragment (residues 508–630), which stuck out in the α-catenin dimer, all helix bundles engaged in extensive interdomain interactions and contributed to the overall marked asymmetry of the molecule. For example, the first two α-helices of the VBD of subunit B bound to the second and third α-helices of the VBD in subunit A (Fig. 2b). Further, unlike the structure of the M-fragment alone where the two four-helix bundles were purported to interact30,31, neither of these bundles interacted in the asymmetric α-catenin dimer.
Notably, the orientation of the F-actin binding domain markedly differed in the two subunits of the α-catenin dimer. Specifically, while the orientation of the F-actin binding domain in the A subunit generally resembled that seen in vinculin, the F-actin binding domain in subunit B was rotated about 166° compared to its orientation in subunit A (Supplementary Fig. 3). Specifically, in subunit A the F-actin binding domain α-helices H4 and H5 interacted with the VBD, α-helix H3 interacted with the M-fragment, and its N-terminus interacted with the N-terminal four-helix bundle of the N-terminal dimerization domain of subunit B (residues 146–262). Further, the C-terminus of the F-actin binding domain of subunit A interacted with the second four-helix bundle of the M-fragment of subunit B (Fig. 2a). In contrast, in the more open subunit B, the N-terminus of the F-actin binding domain interacted with the second four-helix bundle of the M-fragment of subunit A, α-helix H4 was in contact with the VBD, and the C-terminus bound the second four-helix bundle of the dimerization domain (Fig. 2b). Finally, asymmetry did not seem to be driven by crystallization and crystal contacts, as the F-actin binding domains in particular of subunit B did not engage in any crystal contacts and those present in subunit A seemed too minor to affect its conformation.
The closely related vinculin protein harbored five domains that were also comprised of four- or five-helix bundles (Vh1, Vh2, Vh3, Vt2 and the F-actin binding domain)39,40. A comparison of the full-length structures of α-catenin and vinculin indicated that their helix bundle domains are conserved with the exception that α-catenin lacked an equivalent Vh2 domain (Supplementary Fig. 3). Furthermore, in the α-catenin structure, the F-actin binding domain was oriented much differently, in particular for the flipped conformation in subunit B. These features likely explain the distinct F-actin binding properties of the two proteins, where α-catenin but not inactive vinculin can bind to F-actin.
β-Catenin binding disrupts dimerization and F-actin binding
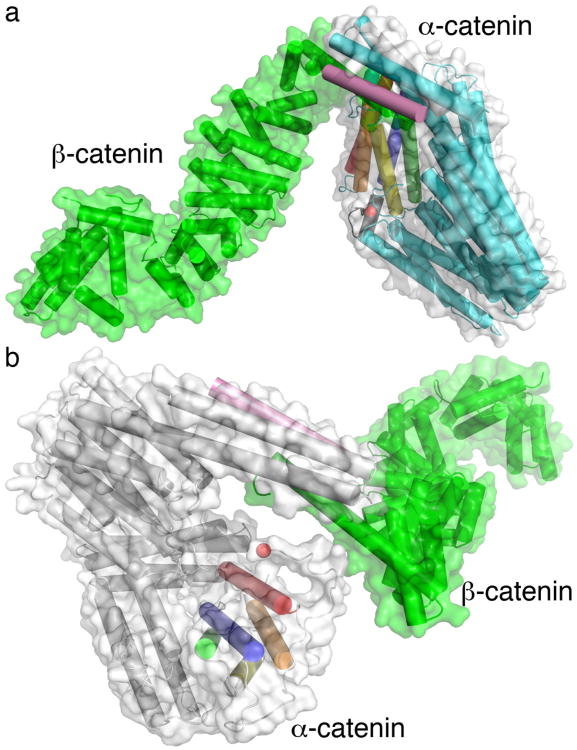
α-Catenin interacts with the E-cadherin-β-catenin complex at adherens junctions via binding to the N-terminal α-helix of β-catenin. Superposition of the α-catenin structure onto the β-catenin-α-catenin chimera29 and onto the full-length β-catenin structure41 revealed the consequences of the β-catenin interaction on α-catenin structure and function (Fig. 3). As shown experimentally, β-catenin and α-catenin bound as a 1:1 complex, where β-catenin binding displaced the two N-terminal α-catenin α-helices, thus disrupting the α-catenin dimer, which had a much higher affinity for F-actin15. The exact F-actin binding site of α-catenin has, however, not been defined other than that residues 864–906 were necessary for the interaction42. Importantly, the β-catenin-α-catenin model clearly showed that β-catenin sterically hinders F-actin binding by the α-catenin dimer. Specifically, the C-terminus of α-catenin that is essential for F-actin binding was too close to β-catenin in subunit A (about 25 Å, Fig. 3a) to accommodate F-actin and indeed was in direct contact, via at least one electrostatic interaction, with β-catenin in subunit B (Fig. 3b). Indeed, a portion of the 864–906 F-actin binding site of α-catenin (residues 865–869) was positioned to facilitate interactions with β-catenin in subunit B (Fig. 3b). Thus, our structure explains how α-catenin can bind to either F-actin or β-catenin but not to both at the same time.
Figure 3.
Model of the α-catenin–β-catenin heterodimer based on the crystal structures of β-catenin (PDB 2z6g shown as a cartoon and surface, both in green), the α-catenin-β-catenin chimera (PDB 1dow shown as a cartoon in pink), and α-catenin subunits A or B shown as a cartoon in (a) cyan or (b) grey, respectively, and as a grey surface. The α-catenin F-actin binding domain α-helices are colored spectrally. A red sphere is shown for the last α-catenin residue (858 in A and 873 in B) indicating the region involved in F-actin binding (residues 859–906). Only α-catenin residues 57–82 of the α-catenin–β-catenin chimera structure are shown (pink).
F-actin binding
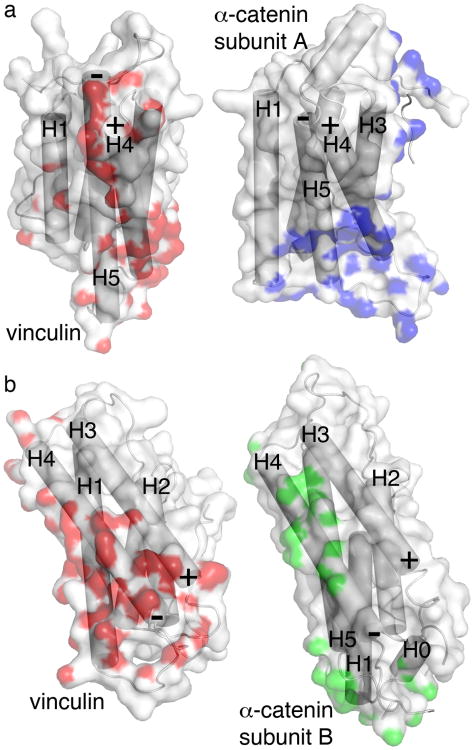
The F-actin binding site in the closely related vinculin tail domain of vinculin was masked in its closed-clamp inactive conformer but was released and was fully accessible to F-actin following severing of the vinculin head-tail interactions43. Superposition of the F-actin binding domain of vinculin onto our α-catenin structure revealed that different surfaces were buried and exposed in these two cytoskeletal proteins (Fig. 4). For example, the N-terminus of α-helices H4 and the C-terminus of α-helix H5 were buried in inactive vinculin via interactions with its Vt2 domain, whereas these regions were largely solvent exposed in the F-actin binding domain of subunit A of α-catenin. Further, the N-terminus of α-helices H3 and the C-terminus of α-helix H4 were buried in inactive vinculin by interactions with its head domain, yet were solvent exposed in subunit B of α-catenin (Fig. 4). Thus, the α-catenin structure also explains how full-length α-catenin can bind to F-actin while vinculin cannot.
Figure 4.
The F-actin binding domain surfaces of vinculin and dimeric α-catenin are distinct. F-actin binding domain surfaces engaging in interdomain interactions of each α-catenin molecule (subunit A, blue; subunit B, green) and of the vinculin tail domain Vt (red) are shown (grey, solvent exposed). The first panels show the Vt surfaces and cartoon while the second panels the α-catenin surface and cartoon of subunits A or B (in a and b), respectively. (a) The N-terminal (indicated by ‘+’) α-helix H4 and C-terminal (indicated by ‘-’) H5 regions are buried in the F-actin binding domain of vinculin via interactions with its Vt2 domain but they are solvent exposed in subunit A of the α-catenin dimer. (b) The N-terminal (+) α-helix H3 and C-terminal (-) H4 regions are buried in the F-actin binding domain of vinculin by interactions with its N-terminal four-helix bundle but are solvent exposed in subunit B of the α-catenin dimer.
α-Catenin residues 864–906, some of which (861–906 in subunit A and 876–906 in subunit B) were disordered in our structure, are essential for F-actin binding42. The dimeric α-catenin structure showed that the C-terminus of subunit B was held in its position via extensive intermolecular interactions with the N-terminal dimerization domain of subunit A (Fig. 2a). As a monomer (e.g., following β-catenin binding) these interactions were thus lost. Interestingly, F-actin had also been reported to bind to the N-terminal 228 residues of α-catenin with similar affinity44. Given that only dimeric α-catenin bound efficiently to F-actin and that this head-tail interface was lost in monomeric α-catenin, a surface that extends across both domains is perhaps the long-sought F-actin binding site. Thus, asymmetry also explains how dimeric but not monomeric α-catenin binds F-actin.
The α-catenin–vinculin interactions
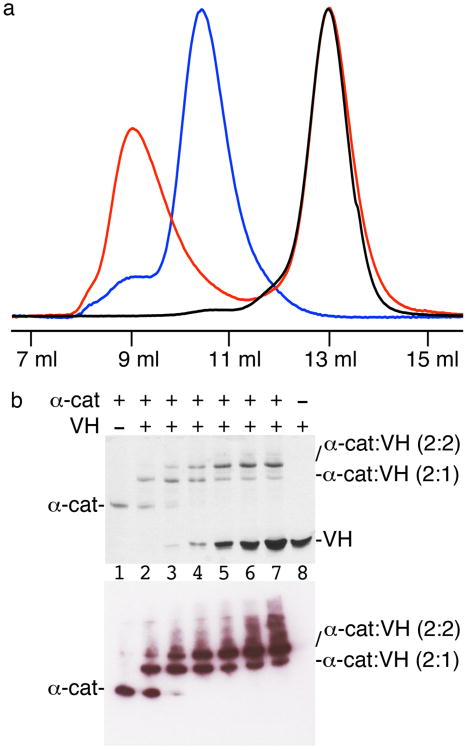
Vinculin was also necessary for stabilizing adherens junctions19 and force-activated α-catenin had been suggested to bind and recruit vinculin to adherens junctions18-20. However, our studies have established that only pre-activated vinculin was capable of binding to α-catenin, as the Vh1 domain that binds to both α-catenin and to the vinculin tail domain to hold vinculin in its closed clamp conformation had a higher affinity for the vinculin tail domain than for the VBD of α-catenin20. The structure of the VBD four-helix bundle within nearly full-length α-catenin presented herein, and that of VBD in complex with the vinculin Vh1 domain20, confirmed that, as proposed19,20, the VBD unfurled when bound to activated vinculin. On a sizing column, the vinculin head (residues 1–840) in complex with α-catenin eluted well before dimeric α-catenin, indicating that the α-catenin–vinculin formed a 2:2 complex (Fig. 5a). Interestingly, as shown by native gel shift assays and immunoblotting, the asymmetric nature of the α-catenin dimer was also manifest in its interaction with vinculin, where the α-catenin dimer first bound to one vinculin molecule before then forming the 2:2 complex (Fig. 5b). Thus, activated vinculin unfurls dimeric α-catenin and this 2:2 heterotetrameric complex is fully competent to bind to F-actin20.
Figure 5.
Asymmetry also directs interactions of α-catenin with vinculin. (a) Size exclusion chromatography showing the UV absorbance profile as measured at 280 nm (without units, not indicated) versus the elution volume of the vinculin head (VH) domain (residues 1–840) alone (black trace), α-catenin (residues 82–906) alone (blue trace), and the α-catenin–VH complex (red trace). The α-catenin–VH complex elutes well before dimeric α-catenin (184,164 Da) indicating that α-catenin remains as a dimer following complex formation with VH. Complex formation was saturated with excess VH (92,251 Da) eluting separately. (b) Native gel analyses (top panel) of ∼10 μM His-tagged α-catenin (α-cat) alone (lane 1) and with increasing amounts of VH (2.5 μM, 5 μM, 10 μM, 20 μM, 30 μM, and 50 μM for lanes 2–7, respectively), and ∼10 μM VH alone (lane 8). Note that a 2:1 α-catenin–VH complex initially appears and with increasing amounts of VH the formation of the 2:2 α-catenin–VH complex saturates. Lower panel, anti-His Western blotting shows that α-catenin is present in all complexes.
Discussion
α-Catenin binds to F-actin and bundles actin filaments44 and also binds to several F-actin binding proteins24,27,30,45-47. However, binding studies with purified recombinant proteins, as well as measurements of protein dynamics in cells, have clearly established that α-catenin cannot simultaneously bind to β-catenin and F-actin and that the oligomeric state of α-catenin dictates which partner it binds to15. These findings were difficult to reconcile with earlier work28,48-50 but a plausible explanation was provided by the fact that E-cadherin-α-catenin fusions were used in earlier studies16. Our structural data now provide mechanistic evidence that explains why α-catenin binding to F-actin and β-catenin is indeed mutually exclusive. Specifically, the structure shows that binding of β-catenin disrupts the intermolecular interactions of the four-helix bundle of the N-terminus of one subunit of α-catenin with a region of the C-terminus of the other subunit that holds the asymmetric dimer together and that are necessary for binding to F-actin.
The mechanism by which α-catenin binds to F-actin has been a conundrum for the field, as extensive mutagenesis of the C-terminal F-actin binding domain has failed to define the F-actin binding motif42. While in cells there are likely contributions from other partners such as vinculin that also bind to F-actin, the fact remains that recombinant dimeric α-catenin alone binds avidly to F-actin. Notably, asymmetry also explains the F-actin binding functions of the α-catenin dimer and why monomeric α-catenin binds to F-actin rather poorly15. Specifically, our structure reveals that the F-actin binding surface is likely created by intermolecular interactions of the tail of α-catenin with a four-helix bundle of its N-terminus, which is lost in monomeric or β-catenin-bound α-catenin. This finding also reconciles reports of F-actin binding by both the N-terminus and C-terminus of α-catenin44. Only the structure of the α-catenin dimer in complex with F-actin will allow one to fully define the mechanism of F-actin binding.
Recombinant full-length vinculin cannot link pre-existing cadherin-catenin complexes and actin filaments as determined by actin pelleting assays in the presence of all four proteins15. However, this is the expected outcome since vinculin is in its closed conformation, which cannot bind to either F-actin or to α-catenin. However, at adherens junctions, vinculin is in its activated, open conformation51, a scenario that would allow it to bind to α-catenin at adherens junctions, and to facilitate interactions of α-catenin with the actin network. The fact that the vinculin tail domain readily displaced α-catenin from pre-existing complexes comprised of α-catenin and the vinculin head domain (i.e., vinculin lacking its F-actin binding domain)13,20,24 establishes that vinculin must be pre-activated at adherens junctions to interact with dimeric α-catenin and to stabilize adherens junctions (Supplementary Fig. 4). Finally, activated vinculin also appears to directly bind to cadherin receptors in cells22, and since the α-catenin dimer is competent to bind to activated vinculin, vinculin may serve as a scaffold that tethers both α-catenin and cadherin receptors, as well as F-actin.
Online Methods
Crystallization
Human α-catenin (residues 82–906) was expressed in E. coli and purified as described20 and dialyzed into 20 mM Tris-HCl (pH 8), 150 mM NaCl, and 5 mM DTT, and concentrated to 25 mg/ml. Initial trigonal crystals were obtained from either 0.9 M (NH4)2SO4, 0.25 M NaCl, and 0.1 M Tris-HCl (pH 7) or 0.9 M Na/K phosphate (pH 6.8) and 0.3 M sodium formate that diffracted X-rays at various synchrotron beam lines (11-1 at Stanford Synchrotron Radiation Laboratory, SSRL, or 22ID and 22BM at the Advanced Photon Source at Argonne National Laboratory, APS/ANL) to about 6 Å Bragg spacings. Conventional strategies failed to improve the diffraction but ultimately systematic dehydration of human α-catenin crystals grown from 0.9 M Na/K phosphate and 0.2 M sodium formate in 2 to 3.5 M Na/K phosphate (pH 6.8) in the presence of glycerol or polyethylene glycol 3350 resulted in diffraction beyond 4 Å. Dehydration was only successful for crystals that were harvested within one week that were 0.15 to 0.3 mm in size, as dehydration did not improve diffraction of larger or smaller crystals. Best diffraction, up to 3.2 Å Bragg spacings, was obtained from crystals that were dehydrated with 3 M Na/K phosphate and 5% polyethylene glycol 3350 for one week. However, significant anisotropy and sensitivity to X-rays limited data collection beyond 3.7 Å Bragg spacings.
X-ray data collection and processing
Native and phosphotungstate derivate X-ray diffraction data were collected on beamlines 22BM at APS/ANL and 11-1 at SSRL, respectively, and integrated and scaled using autoProc52, which uses XDS53 and SCALA54 as the data reduction engine. Data reduction statistics are provided in Table 1.
Structure determination and crystallographic refinement
Molecular replacement was unsuccessful using crystal structures of the dimerization domain (residues 82–262; PDB 1dov) or the M-fragment (residues 377–631; PDB 1h6g), or homology models for the VBD or F-actin binding domain as search models. Selenomethionine labeled α-catenin crystals did not grow beyond 0.05 mm and their diffraction was limited to 8 Å Bragg spacings. Derivatization was also limited due to the high concentration of phosphate in the crystallization reservoir, which caused standard heavy atoms, such as Pt, Hg, and Au, to precipitate. This precipitation was overcome to some degree by short (10 min) soaking times with high concentrations (10 mM) of heavy metals such as K2PtCl4. However, this significantly affected the diffraction and anomalous signal detection. Sodium phosphotungstate was eventually identified as a suitable heavy atom due to its solubility in phosphate conditions.
Crystals were incubated for 24 hr in a low phosphate condition (0.2 M NaK/phosphate, 2.5 M sodium formate, and 0.7 M sodium malonate, pH 7) to avoid competition of phosphate from the reservoir. Effective heavy atom incorporation was accomplished via short soaking times (15 min) in a 10 mM phosphotungstate solution containing 0.2 M Na/K phosphate, 2.5 M sodium formate, and 0.7 M sodium malonate (pH 7), then back soaked for 10 min in 0.2 M Na/K phosphate, 2.5 M sodium formate, and 0.7 M sodium malonate (pH 7), and mounted without including any additional cryoprotectant. X-ray diffraction data were obtained at SSRL beam line 11-1 near the L-II absorption edge of tungsten (1.07 Å) to 5.6 Å Bragg spacings.
Determination of the heavy atom substructure was performed using the program autoSHARP55. Two tungsten cluster sites with peak heights of 1 and 0.44 and a correlation of 0.207 were located from which phases (with a figure of merit of 0.15) were obtained to 5.6 Å (Supplementary Table 1). The resulting electron density map provided clear definition of the various α-catenin domains but did not allow chain tracing. SIRAS using autoSHARP allowed phase extension to 4.3 Å resolution and manual building of α-helices and placement of the high resolution dimerization domain and M-fragment structures into the experimental 4.3 Å SIRAS electron density map. The resulting model was used as a search model for molecular replacement with the program MOLREP56 to position the dimer and further refine to 3.7 Å using the native X-ray diffraction data. Iterative cycles of model building were performed using Coot57 and maximum likelihood crystallographic refinement using autoBUSTER58 by imposing target restraints using the high resolution structures. The model was improved by local non-crystallographic symmetry through LSSR59. Model bias was minimized by model building into composite omit maps. Map sharpening was performed in Coot57 to ensure the directionality and identity of the α-helices. The quality of the final model assessment using MolProbity60 resulted in no outliers and over 95% of the amino acid residues in the favored region of the Ramachandran plot. Refinement statistics are provided in Table 1.
Size exclusion chromatography
α-Catenin (residues 82–906), VH (residues 1–840), and α-catenin plus a 2.5 molar excess of VH were loaded onto a superdex 200 10/300GL (GE Healthcare) analytical chromatography column equilibrated in 20 mM Tris-HCl (pH 8), 150 m M NaCl, and 5 mM DTT. Fractions were analyzed on a 10–15% gradient PHAST gel with native buffer strips.
Native gel shift assays and immunoblotting
Samples (in 20 mM Tris-HCl, pH 8, 150 mM NaCl, 5 mM DDT) were incubated for 1 hr at 4 °C. Increasing concentrations of VH (0 μM, 2.5 μM, 5 μM, 10 μM, 20 μM, 30 μM, and 50 μM) were titrated to purified His-tagged α-catenin (∼10 μM) and the resultant complex was analyzed using a 10–15% gradient PHAST gel with native buffer strips. The bands were visualized by coomassie blue staining. α-Catenin was detected with an anti-His antibody.
Supplementary Material
Acknowledgments
We are indebted to our colleagues at Scripps Florida: J. Cleveland for discussions and critical review of the manuscript, Z. Wu and P. Bois for sequencing, and P. Bois for fruitful discussions. We thank C. Vonrhein and G. Bricogne (GlobalPhasing Ltd.) for analyses and helpful discussions. We are grateful to the staff at the SER-CAT (BM22) and SSRL (11-1), for synchrotron support. TI is supported by grants from NIGMS from the National Institutes of Health (GM071596 and GM094483) and by start-up funds provided to Scripps Florida from the State of Florida. This is publication no. 21863 from The Scripps Research Institute.
Footnotes
Accession codes. α-Catenin coordinates have been deposited in the Protein Data Bank with entry code 4igg.
Author Contributions: Both authors contributed to the design and interpretation of all aspects of this work. E.S.R. performed all of the experiments. T.I. wrote the manuscript.
References
- 1.Volk T, Geiger B. A 135-kd membrane protein of intercellular adherens junctions. Embo J. 1984;3:2249–60. doi: 10.1002/j.1460-2075.1984.tb02123.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takeichi M. The cadherins: cell-cell adhesion molecules controlling animal morphogenesis. Development. 1988;102:639–55. doi: 10.1242/dev.102.4.639. [DOI] [PubMed] [Google Scholar]
- 3.Nishimura T, Takeichi M. Remodeling of the adherens junctions during morphogenesis. Curr Top Dev Biol. 2009;89:33–54. doi: 10.1016/S0070-2153(09)89002-9. [DOI] [PubMed] [Google Scholar]
- 4.Brasch J, Harrison OJ, Honig B, Shapiro L. Thinking outside the cell: how cadherins drive adhesion. Trends in Cell Biology. 2012;22:299–310. doi: 10.1016/j.tcb.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hirano S, Nose A, Hatta K, Kawakami A, Takeichi M. Calcium-dependent cell-cell adhesion molecules (cadherins): subclass specificities and possible involvement of actin bundles. J Cell Biol. 1987;105:2501–10. doi: 10.1083/jcb.105.6.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takeichi M. Morphogenetic roles of classic cadherins. Curr Opin Cell Biol. 1995;7:619–27. doi: 10.1016/0955-0674(95)80102-2. [DOI] [PubMed] [Google Scholar]
- 7.Pokutta S, Weis WI. Structure and mechanism of cadherins and catenins in cell-cell contacts. Annu Rev Cell Dev Biol. 2007;23:237–61. doi: 10.1146/annurev.cellbio.22.010305.104241. [DOI] [PubMed] [Google Scholar]
- 8.Lecuit T. α-catenin mechanosensing for adherens junctions. Nat Cell Biol. 2010;12:522–4. doi: 10.1038/ncb2066. [DOI] [PubMed] [Google Scholar]
- 9.Miyake Y, et al. Actomyosin tension is required for correct recruitment of adherens junction components and zonula occludens formation. Exp Cell Res. 2006;312:1637–50. doi: 10.1016/j.yexcr.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 10.Hirano S, Kimoto N, Shimoyama Y, Hirohashi S, Takeichi M. Identification of a neural α-catenin as a key regulator of cadherin function and multicellular organization. Cell. 1992;70:293–301. doi: 10.1016/0092-8674(92)90103-j. [DOI] [PubMed] [Google Scholar]
- 11.Watabe M, Nagafuchi A, Tsukita S, Takeichi M. Induction of polarized cell-cell association and retardation of growth by activation of the E-cadherin-catenin adhesion system in a dispersed carcinoma line. J Cell Biol. 1994;127:247–56. doi: 10.1083/jcb.127.1.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Torres M, et al. An α-E-catenin gene trap mutation defines its function in preimplantation development. Proc Natl Acad Sci U S A. 1997;94:901–6. doi: 10.1073/pnas.94.3.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Watabe-Uchida M, et al. α-Catenin-vinculin interaction functions to organize the apical junctional complex in epithelial cells. J Cell Biol. 1998;142:847–57. doi: 10.1083/jcb.142.3.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vasioukhin V, Bauer C, Degenstein L, Wise B, Fuchs E. Hyperproliferation and defects in epithelial polarity upon conditional ablation of α-catenin in skin. Cell. 2001;104:605–17. doi: 10.1016/s0092-8674(01)00246-x. [DOI] [PubMed] [Google Scholar]
- 15.Drees F, Pokutta S, Yamada S, Nelson WJ, Weis WI. α-Catenin Is a Molecular Switch that Binds E-Cadherin-β-Catenin and Regulates Actin-Filament Assembly. Cell. 2005;123:903–15. doi: 10.1016/j.cell.2005.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weis WI, Nelson WJ. Re-solving the cadherin-catenin-actin conundrum. J Biol Chem. 2006;281:35593–7. doi: 10.1074/jbc.R600027200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamada S, Pokutta S, Drees F, Weis WI, Nelson WJ. Deconstructing the cadherin-catenin-actin complex. Cell. 2005;123:889–901. doi: 10.1016/j.cell.2005.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peng X, Cuff LE, Lawton CD, DeMali KA. Vinculin regulates cell-surface E-cadherin expression by binding to β-catenin. J Cell Sci. 2010;123:567–77. doi: 10.1242/jcs.056432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yonemura S, Wada Y, Watanabe T, Nagafuchi A, Shibata M. α-Catenin as a tension transducer that induces adherens junction development. Nat Cell Biol. 2010;12:533–42. doi: 10.1038/ncb2055. [DOI] [PubMed] [Google Scholar]
- 20.Rangarajan ES, Izard T. α-Catenin unfurls upon binding to vinculin. J Biol Chem. 2012;287:18492–18499. doi: 10.1074/jbc.M112.351023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abe K, Takeichi M. EPLIN mediates linkage of the cadherin catenin complex to F-actin and stabilizes the circumferential actin belt. Proc Natl Acad Sci U S A. 2008;105:13–9. doi: 10.1073/pnas.0710504105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hazan RB, Kang L, Roe S, Borgen PI, Rimm DL. Vinculin is associated with the E-cadherin adhesion complex. J Biol Chem. 1997;272:32448–53. doi: 10.1074/jbc.272.51.32448. [DOI] [PubMed] [Google Scholar]
- 23.Hulsken J, Birchmeier W, Behrens J. E-cadherin and APC compete for the interaction with β-catenin and the cytoskeleton. J Cell Biol. 1994;127:2061–9. doi: 10.1083/jcb.127.6.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weiss EE, Kroemker M, Rudiger AH, Jockusch BM, Rudiger M. Vinculin is part of the cadherin-catenin junctional complex: complex formation between α-catenin and vinculin. J Cell Biol. 1998;141:755–64. doi: 10.1083/jcb.141.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tachibana K, et al. Two cell adhesion molecules, nectin and cadherin, interact through their cytoplasmic domain-associated proteins. J Cell Biol. 2000;150:1161–76. doi: 10.1083/jcb.150.5.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shapiro L, et al. Structural basis of cell-cell adhesion by cadherins. Nature. 1995;374:327–37. doi: 10.1038/374327a0. [DOI] [PubMed] [Google Scholar]
- 27.Itoh M, Nagafuchi A, Moroi S, Tsukita S. Involvement of ZO-1 in cadherin-based cell adhesion through its direct binding to α-catenin and actin filaments. J Cell Biol. 1997;138:181–92. doi: 10.1083/jcb.138.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Imamura Y, Itoh M, Maeno Y, Tsukita S, Nagafuchi A. Functional domains of α-catenin required for the strong state of cadherin-based cell adhesion. J Cell Biol. 1999;144:1311–22. doi: 10.1083/jcb.144.6.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pokutta S, Weis WI. Structure of the dimerization and β-catenin-binding region of α-catenin. Mol Cell. 2000;5:533–43. doi: 10.1016/s1097-2765(00)80447-5. [DOI] [PubMed] [Google Scholar]
- 30.Pokutta S, Drees F, Takai Y, Nelson WJ, Weis WI. Biochemical and structural definition of the l-afadin- and actin-binding sites of α-catenin. J Biol Chem. 2002;277:18868–74. doi: 10.1074/jbc.M201463200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang J, Dokurno P, Tonks NK, Barford D. Crystal structure of the M-fragment of α-catenin: implications for modulation of cell adhesion. Embo J. 2001;20:3645–56. doi: 10.1093/emboj/20.14.3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choi HJ, et al. αE-catenin is an autoinhibited molecule that coactivates vinculin. Proc Natl Acad Sci U S A. 2012;109:8576–8581. doi: 10.1073/pnas.1203906109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peng X, Maiers JL, Choudhury D, Craig SW, Demali KA. α-Catenin uses a novel mechanism to activate vinculin. J Biol Chem. 2012;287:7728–7737. doi: 10.1074/jbc.M111.297481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jockusch BM, Isenberg G. Interaction of α-actinin and vinculin with actin: opposite effects on filament network formation. Proc Natl Acad Sci U S A. 1981;78:3005–9. doi: 10.1073/pnas.78.5.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilkins JA, Lin S. High-affinity interaction of vinculin with actin filaments in vitro. Cell. 1982;28:83–90. doi: 10.1016/0092-8674(82)90377-4. [DOI] [PubMed] [Google Scholar]
- 36.Johnson RP, Craig SW. F-actin binding site masked by the intramolecular association of vinculin head and tail domains. Nature. 1995;373:261–4. doi: 10.1038/373261a0. [DOI] [PubMed] [Google Scholar]
- 37.Weekes J, Barry ST, Critchley DR. Acidic phospholipids inhibit the intramolecular association between the N- and C-terminal regions of vinculin, exposing actin-binding and protein kinase C phosphorylation sites. Biochem J. 1996;314:827–32. doi: 10.1042/bj3140827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steimle PA, Hoffert JD, Adey NB, Craig SW. Polyphosphoinositides inhibit the interaction of vinculin with actin filaments. J Biol Chem. 1999;274:18414–20. doi: 10.1074/jbc.274.26.18414. [DOI] [PubMed] [Google Scholar]
- 39.Borgon RA, Vonrhein C, Bricogne G, Bois PR, Izard T. Crystal structure of human vinculin. Structure. 2004;12:1189–97. doi: 10.1016/j.str.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 40.Rangarajan ES, Lee JH, Yogesha SD, Izard T. A helix replacement mechanism directs metavinculin functions. PLoS ONE. 2010;5:e10679. doi: 10.1371/journal.pone.0010679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xing Y, et al. Crystal structure of a full-length β-catenin. Structure. 2008;16:478–87. doi: 10.1016/j.str.2007.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pappas DJ, Rimm DL. Direct interaction of the C-terminal domain of α-catenin and F-actin is necessary for stabilized cell-cell adhesion. Cell Commun Adhes. 2006;13:151–70. doi: 10.1080/15419060600726142. [DOI] [PubMed] [Google Scholar]
- 43.Johnson RP, Craig SW. The carboxy-terminal tail domain of vinculin contains a cryptic binding site for acidic phospholipids. Biochem Biophys Res Commun. 1995;210:159–64. doi: 10.1006/bbrc.1995.1641. [DOI] [PubMed] [Google Scholar]
- 44.Rimm DL, Koslov ER, Kebriaei P, Cianci CD, Morrow JS. α1(E)-catenin is an actin-binding and -bundling protein mediating the attachment of F-actin to the membrane adhesion complex. Proc Natl Acad Sci U S A. 1995;92:8813–7. doi: 10.1073/pnas.92.19.8813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Knudsen KA, Soler AP, Johnson KR, Wheelock MJ. Interaction of α-actinin with the cadherin/catenin cell-cell adhesion complex via α-catenin. J Cell Biol. 1995;130:67–77. doi: 10.1083/jcb.130.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pradhan D, Lombardo CR, Roe S, Rimm DL, Morrow JS. α -Catenin binds directly to spectrin and facilitates spectrin-membrane assembly in vivo. J Biol Chem. 2001;276:4175–81. doi: 10.1074/jbc.M009259200. [DOI] [PubMed] [Google Scholar]
- 47.Kobielak A, Fuchs E. α-Catenin: at the junction of intercellular adhesion and actin dynamics. Nature Reviews Molecular Cell Biology. 2004;5:614–25. doi: 10.1038/nrm1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nagafuchi A, Ishihara S, Tsukita S. The roles of catenins in the cadherin-mediated cell adhesion: functional analysis of E-cadherin-α-catenin fusion molecules. J Cell Biol. 1994;127:235–45. doi: 10.1083/jcb.127.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sako Y, Nagafuchi A, Tsukita S, Takeichi M, Kusumi A. Cytoplasmic regulation of the movement of E-cadherin on the free cell surface as studied by optical tweezers and single particle tracking: corralling and tethering by the membrane skeleton. J Cell Biol. 1998;140:1227–40. doi: 10.1083/jcb.140.5.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pacquelet A, Rorth P. Regulatory mechanisms required for DE-cadherin function in cell migration and other types of adhesion. J Cell Biol. 2005;170:803–12. doi: 10.1083/jcb.200506131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen H, Cohen DM, Choudhury DM, Kioka N, Craig SW. Spatial distribution and functional significance of activated vinculin in living cells. J Cell Biol. 2005;169:459–70. doi: 10.1083/jcb.200410100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vonrhein C, et al. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr D Biol Crystallogr. 2011;67:293–302. doi: 10.1107/S0907444911007773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. Journal of Applied Crystallography. 1993;26:795–800. [Google Scholar]
- 54.Evans P. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 55.Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated Structure Solution With autoSHARP. Methods Mol Biol. 2006;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 56.Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J Appl Cryst. 1997;30:1022–1025. [Google Scholar]
- 57.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 58.Bricogne G, et al. BUSTER version 2.9. Cambridge, United Kingdom: Global Phasing Ltd; 2011. [Google Scholar]
- 59.Smart OS, et al. Refinement with local structure similarity restraints (LSSR) enables exploitation of information from related structures and facilitates use of NCS. Abstr Annu Meet Am Crystallogr Assoc, Knoxville, TN. 2008 abstr. TP139. [Google Scholar]
- 60.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.