Abstract
Age-related macular degeneration (AMD) is a leading cause of visual impairment in aging populations in industrialized countries. Here we investigated whether the genotype of vascular endothelial growth factor A (VEGFA) gene is associated with response to anti-VEGF therapy. 223 eyes with neovascular AMD were treated with intravitreal anti-VEGF therapy. Responders were defined as patients who had an improvement in best corrected visual acuity (BCVA) of at least 5 letters or one line on the EDTRS visual acuity chart along with resolution of intraretinal or subretinal fluid over 12 months. Patients who did not meet the definition of responders were classified as poor-responders. The vision of responders (n = 148) improved while the vision of poor-responders (n = 75) worsened (P <0.001). Responders on average had a decrease in central foveal thickness (CFT), while poor-responders had an increase in CFT (P <0.001). Compared with the responder group, the poor-responder group had a higher frequency of the risk (T) allele (Allelic P = 0.019) and TT genotype (P = 0.002 under a recessive model) for the VEGFA-rs943080 polymorphism. VEGFA expression was 1.8-fold higher in cells with the VEGFA rs943080 TT genotype than in cells with the VEGFA rs943080 CC genotype (P = 0.012). Age, gender, smoking, diabetes mellitus, and hypertension did not play a significant role in treatment response, but BMI was found to be significantly different between responders and poor-responders (P = 0.033). In conclusion, we demonstrated a potential pharmacogenetic relationship between the VEGFA gene and treatment response to anti-VEGF therapy.
The studies are registered at ClinicalTrials.gov under the identifiers NCT00474695 (http://clinicaltrials.gov/ct2/show/NCT00474695) and NCT01464723 (http://clinicaltrials.gov/ct2/show/NCT01464723).
Keywords: Anti-VEGF therapy, choroidal neovascularization, genetics, macular degeneration, VEGFA
INTRODUCTION
Age-related macular degeneration (AMD) is a leading cause of visual impairment and vision loss in aging populations in industrialized countries. Although the mechanisms leading to disease are not yet fully understood, it is well established that some factors, including advanced age, environmental and genetic factors, as well as the processes of neovascularization and angiogenesis, are implicated in AMD [1–3].
Currently, the most common therapies for neovascular AMD are intravitreal ranibizumab (Lucentis, Genentech, South San Francisco, CA) and bevacizumab (Avastin, Genentech, South San Francisco, CA). Ranibizumab is an FDA-approved humanized monoclonal antibody fragment that inhibits vascular endothelial growth factor (VEGF) for the treatment of neovascular AMD. Bevacizumab, a full-length monoclonal antibody to VEGF used intravenously as an anticancer agent, has been used off-label as an intravitreal therapy for neovascular AMD [3–6]. Several short-term studies have indicated that intravitreal bevacizumab results in improvement in visual acuity that is similar to the improvement seen with ranibizumab [7, 8]. In 2011, the results of a 1-year NIH sponsored randomized clinical trial provided further evidence that both anti-VEGF agents had similar visual acuity effects in neovascular AMD when administered on the same schedule (primary endpoint of non-inferiority met), however, bevacizumab had a higher rate of Serious Adverse Events (SAEs) than ranibizumab [4].
The direct target of both agents is vascular endothelial growth factor A (VEGFA), which is a member of the VEGF family and functions to increase vascular permeability, angiogenesis, cell growth and migration of endothelial cells. Despite evidence implicating VEGF in the pathogenesis of neovascular AMD and the overall efficacy of anti-VEGF treatments, there remains an unexplained variability in treatment response with certain patients. In addition, several studies have indicated that genetic predispositions are associated with response in the treatment of AMD [9–11]. As such, the VEGFA gene serves as a good candidate for testing pharmacogenetic relationships between genotypes and therapy outcomes.
VEGFA has been reported as a predisposing gene to AMD [12, 13] yet there have been few studies investigating the association between VEGFA genotypes and response to anti-VEGF therapy. A recent meta-analysis investigating the genetic susceptibility of AMD demonstrated that VEGFA-rs943080 is in strong linkage disequilibrium (LD) with rs4711751 (r2 = 1.0 in 1000 Genomes CEU data), which is significantly associated with advanced AMD ([OR=1.15 (95% CI: 1.10–1.21), P = 8.7x10−9]) [13]. In this study, we investigated whether there is an association between the response to anti-VEGF treatment for neovascular AMD and the VEGFA gene, clinical characteristics, demographic factors, or comorbidities.
MATERIALS AND METHODS
This prospective cohort study was approved by the Institutional Review Board (IRB) at the University of California, San Diego (UCSD). The research adhered to the tenets of the Declaration of Helsinki. All subjects signed a written informed consent prior to participation in the study and the study was HIPAA-compliant. Neovascular AMD subjects were recruited at the Shiley Eye Center at UCSD, San Diego Retina Research Foundation, and California Retina Consultants, Inc, San Diego.
Participants and Clinical Data Collection
This study consisted of 223 eyes from Caucasian patients with choroidal neovascularization (CNV) due to AMD. Participants included in the study were diagnosed with neovascular AMD based on exam and imaging findings. Demographic factors, medical history, and a blood sample were taken at the baseline visit. All participants underwent standard monthly ophthalmic examinations, which included best-corrected visual acuity (BCVA) measurements, applanation tonometry, slit lamp exams, and indirect ophthalmoscopy. BCVA was measured at the initial visit and at each follow-up visit. For all calculations and comparisons, BCVA measurements were converted to logarithm of minimum angular resolution (logMAR) values.
The treatment protocol started with monthly injections of either ranibizumab or bevacizumab for the first 4 months. Patients were followed monthly and retreated if their vision dropped at least five letters on the ETDRS chart or intraretinal or subretinal fluid was present on optical coherence tomography (OCT). Patients were followed for 12 months for this study. The definition of responders and poor-responders were determined prior to the start of the study. Patients were defined as responders and poor-responders to anti-VEGF (ranibizumab or bevacizumab) therapy as follows: Responders were defined as patients who at month 12 had an improvement in BCVA of at least 5 letters or one line on the EDTRS chart along with resolution of intraretinal or subretinal fluid compared with baseline. Patients who did not meet the definition of responders were classified as poor-responders.
Imaging Studies
Stereo fundus photography and fluorescein angiography (FA) were completed on all patients after adequate dilation by a certified facility photographer. A pair of stereoscopic color fundus photographs (50 degrees) was taken, centered on the fovea using a Topcon fundus camera (Topcon TRV-50VT, Topcon Optical Company, Tokyo, Japan). FA was obtained in a standard fashion using a Heidelberg Retina Tomograph 2 (Heidelberg, Germany). OCT images were obtained using a Topcon 3D OCT-1000 (Topcon Optical Company, Tokyo, Japan) or the Spectralis OCT (Heidelberg Engineering, Germany) by a trained ophthalmic technician.
Genotyping
Genomic DNA samples were extracted from peripheral blood leukocytes with the Qiagen kit (Qiagen Inc., Chatsworth, CA, USA), according to the manufacturer’s instructions. rs943080 (C/T) in the VEGFA gene was genotyped using the SNaPshot method according to the manufacturer’s recommendations. In brief, a single nucleotide polymorphism (SNP) was amplified by polymerase chain reaction (PCR) and the PCR product was purified by Exo I and Shrimp Alkaline Phosphatase (SAP) (New England Biolabs, Ipswich, MA). The purified PCR product and the SNaPshot primer were then used to perform a single base pair extension with the SNaPshot multiplex mix (Applied Biosystems Inc, Foster City, CA). After an additional purification step using SAP, the product was run and analyzed on an ABI 3130xI genetic analyzer (Applied Biosystems Inc, Foster City, CA, USA) and genotyping results were obtained directly. Forward Primer is: 5′-CAACTGAAAGCGGGGAATTA-3′; Reverse Primer is: 5′-AGCTCAGCCAAGTGTGGAGT -3′; The extension primer is: 5′-GAGGCCCCCCACCCCT TTTAGTCTCTGCCTGGGCCTCCTCAGAGAGCTAA-3′.
Real-Time Quantitative PCR
Total RNA was extracted from human lymphocytes using RNeasy Mini Kit from QIAGEN Inc. (Valencia, CA), and cDNA was reverse transcribed with SuperScript III First Strand Synthesis System from Invitrogen Life Technologies (Carlsbad, CA). All qRT-PCR experiments were performed with Power SYBR Green qPCR Master Mix and analyzed with a 7500 Real-Time PCR Detection System from Applied Biosystems (Foster City, CA). Assays were performed in triplicate. Relative mRNA levels were calculated by normalizing results with GAPDH and expressed relative to samples with a VEGFA rs943080 CC genotype. For the human GAPDH gene, the forward primer is: 5′-GAGTCAACGGATTTGGTCGT-3′, the reverse primer is: 5′-GACAAGCTTCCCGTTCTCAG-3′. For human VEGFA gene, the forward primer is: 5′-TCCCGGTATAAGTCCTGGAG -3′, the reverse primer is: 5′-ACAAATGCTTTCTCCGCTCT -3′.
Statistical Analysis
Logistic regression, t-test, or Chi-squared test was used to compare the VEGFA-rs943080 genotype, clinical characteristics, comorbidities, and demographic factors between responders and poor-responders. SNP genotyping results were screened for deviation from Hardy-Weinberg equilibrium using Chi-squared tests. Mean BCVA and OCT central foveal thickness data were determined for the diseased eyes before and after treatment and were analyzed for change over time. Statistical significance was defined as P <0.05.
RESULTS
Demographic Results
223 eyes with CNV were enrolled in the study. Responders were defined as patients who at month 12 had an improvement in BCVA of at least 5 letters or one line on the EDTRS chart along with resolution of intraretinal or subretinal fluid compared with baseline. Patients who did not meet the definition of responders were classified as poor-responders. There were 148 responders (66.4%) and 75 (33.6%) poor-responders. Information on demographics and comorbidities according to treatment response are presented in Table 1. While there were no significant differences between the responder and poor-responder groups in age and gender, there was a statistically significant difference in BMI between the two groups (P = 0.033).
Table 1.
Demographics and Comorbidities of Responders and Poor-Responders to Anti-VEGF Therapy
| Total | Responders | Poor-Responders | P-Value | |
|---|---|---|---|---|
| Sample Size (n) | 223 | 148 | 75 | |
| Gender F (%) | 52.9 | 49.3 | 60.0 | 0.131 |
| Age | 81±0.6 | 80±0.8 | 81±0.9 | 0.829 |
| BMI | 25.9±0.4 | 26.4±0.4 | 24.9±0.5 | 0.033* |
| Smoking (%) | ||||
| Never Smoker | 45.7 | 45.3 | 46.7 | ref |
| Past Smoker | 49.3 | 50.0 | 48.0 | 0.807 |
| Current Smoker | 4.9 | 4.7 | 5.3 | 0.892 |
| Hypertension (%) | 52.0 | 52.7 | 50.7 | 0.774 |
| Diabetes Mellitus (%) | 9.0 | 7.43 | 12.0 | 0.259 |
The data are presented as mean ± SEM (standard error of the mean). F = female; BMI = body mass index. For gender, BMI, hypertension and diabetes mellitus, P values are calculated from t-test or Chi-square test when comparing responders and poor-responders. For smoking, P value is calculated by logistic regression analysis.
Smoking was stratified into three groups: current smokers, past smokers, and never smokers. Logistic regression analysis showed no significant difference within each smoking category (P = 0.807 for past smokers and P = 0.892 for current smokers).
History of hypertension and diabetes mellitus (DM) was also compared between responder and poor-responder groups. 52.7% of responders and 50.7% of poor-responders had a history of hypertension, while 7.43% of responders and 12.0% of poor-responders had a history of DM. There was no significant difference between responders and poor-responders for either of the diseases (P = 0.774 for hypertension and P = 0.259 for DM).
Clinical Results
Exam and imaging findings according to treatment response are presented in Table 2. On clinical exam, no significant difference in baseline BCVA was found between responders and poor-responders (P = 0.228). However, there was a significant change in BCVA over time between the two groups. The vision of responders had a mean improvement of 0.23±0.02 logMAR while the average decrease in vision of poor-responders was 0.20±0.03 logMAR (P <0.001).
Table 2.
Clinical Data of Responders and Poor-Responders to Anti-VEGF Therapy
| Clinical Measurement | Responders | Poor-Responders | P-Value |
|---|---|---|---|
| Baseline BCVA (LogMAR) | 0.62±0.03 | 0.69±0.05 | 0.228 |
| Current BCVA (LogMAR) | 0.39±0.03 | 0.89±0.06 | <0.001 |
| Change lines read (LogMAR) | 0.23±0.02 | −0.20±0.03 | <0.001 |
| Baseline CFT (μm) | 296.8±12.2 | 338.9±29.4 | 0.118 |
| Current CFT (μm) | 237.6±8.5 | 361.5±34.2 | <0.001 |
| change CFT (μm) | −59.2±10.0 | 22.6±17.1 | <0.001 |
The data are presented as mean ± SEM (standard error of the mean). BCVA = best correct visual acuity, measured in LogMAR (logarithm of the minimum angle of resolution); CFT = central foveal thickness
No significant difference in baseline CFT was found between responders and poor-responders (P = 0.118), but there was a significant change in CFT over time between the two groups. Responders on average had a decrease of 59.2±10.0 μm in CFT, while poor-responders on average had an increase of 22.6±17.1 μm in CFT (P <0.001).
Genetic Association study
VEGFA-rs943080 was genotyped and the results were compared in responders and poor-responders from each treatment group (bevacizumab or ranibizumab) and the groups combined (Table 3). The risk (T) allele was found at a higher frequency in poor-responders than responders for both treatment groups. Overall analysis demonstrated that the frequency of the T allele was significantly higher in poor-responders than responders (P = 0.019). A significant association was found in the recessive model for the bevacizumab group (P = 0.013) and the groups combined (P = 0.002).
Table 3.
Genotype and Association Results of VEGFA-rs943080 from Responders and Poor-Responders
| Group | Ranibizumab | Bevacizumab | Combined groups | |||
|---|---|---|---|---|---|---|
| Phenotype | Poor-Responder | Responder | Poor-Responder | Responder | Poor-Responder | Responder |
| TT genotype, no./total no. (%) | 14/30 (46.7) | 16/59 (27.1) | 19/45 (42.2) | 19/89 (21.3) | 33/75 (44.0) | 35/148 (23.6) |
| CT genotype, no./total no. (%) | 11/30 (36.7) | 28/59 (47.5) | 17/45 (37.8) | 53/89 (59.6) | 28/75 (37.3) | 81/148 (54.7) |
| CC genotype, no./total no. (%) | 5/30 (16.7) | 15/59 (25.4) | 9/45 (20.0) | 17/89 (19.1) | 14/75 (18.7) | 32/148 (21.6) |
| Risk allele (T) frequency | 0.650 | 0.508 | 0.611 | 0.511 | 0.627 | 0.510 |
| HWE P value | 0.288 | 0.698 | 0.169 | 0.071 | 0.080 | 0.248 |
| Allelic P value, OR (95% CI) | 0.074; 1.80(0.95–3.41) | 0.12; 1.50 (0.90–2.52) | 0.019*; 1.61 (1.08–2.41) | |||
| Dominant P value, OR (95% CI) | 0.35;1.71(0.55–5.25) | 0.90; 0.94 (0.38–2.33) | 0.61; 1.20 (0.60–2.42) | |||
| Recessive P value, OR (95% CI) | 0.068; 2.35(0.94–5.89) | 0.013*; 2.69 (1.24–5.87) | 0.002*; 2.54 (1.40–4.59) | |||
| Additive P value, OR (95% CI) | 0.092; 1.71(0.92–3.18) | 0.11; 1.54 (0.90–2.64) | 0.021*; 1.61 (1.07–2.42) | |||
HWE, Hardy-Weinberg equilibrium; OR (95% CI), odds ratio with 95% confidence interval; P, P-value calculated from χ2 test;
, statistically significant.
Expression of VEGFA in Human Lymphocytes
To investigate the correlation of rs943080 genotypes and expression of VEGFA, we measured expression of VEGFA using qPCR. Our results demonstrated that human lymphocyte cells with the VEGFA rs943080 TT risk genotype had a 1.8-fold higher level of VEGFA expression than cells with the VEGFA rs943080 CC genotype (P = 0.012) (Fig. 1).
Fig. 1.
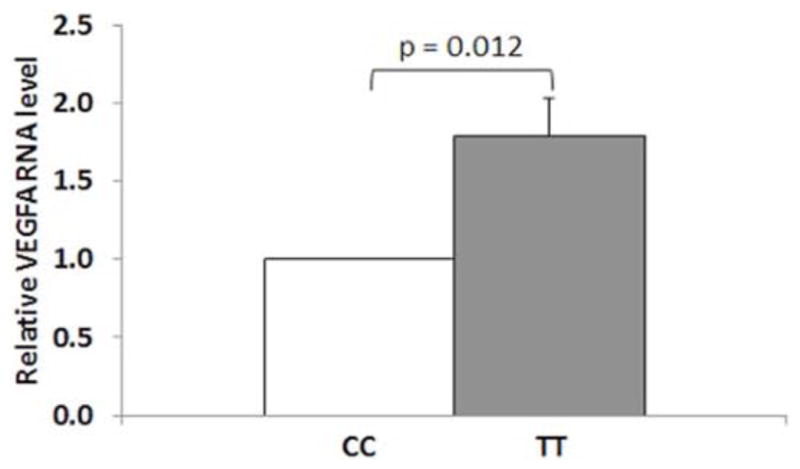
Gene expression pattern of VEGFA from human lymphocytes stratified by their VEGFA (rs943080) genotypes. Relative mRNA levels were calculated by normalizing results with GAPDH and expressed relative to CC genotype. Data represent mean ± S.E.M, n=4 for each genotypes.
DISCUSSION
Pharmacogenomics represents a new dimension of research in patient treatment response, focusing on the link between genetic variation and response to medications or other treatments. VEGFA has been reported as a major genetic contributor to AMD [12, 13], yet association studies between VEGFA genotypes and response to anti-VEGF therapy are scarce.
The definition of responders and poor-responders used in our study take into account both anatomical outcomes (presence of intraretinal and subretinal fluid on OCT) and functional outcomes (change in visual acuity). Responders were defined as patients who at month 12 had an improvement in BCVA of at least 5 letters or one line on the EDTRS chart along with resolution of intraretinal or subretinal fluid compared with baseline. Patients who did not meet the definition of responders were classified as poor-responders. It is important to consider both outcome factors when monitoring treatment response because patients with improved visual acuity can still have signs of intraretinal or subretinal fluid on OCT. The PrONTO study demonstrated that the presence of fluid represents early manifestations of recurrent CNV and if the eye is not retreated, then more fluid may accumulate over time and result in vision loss [14]. It is also important to note anatomical outcomes may not always coincide with functional outcomes. For instance, if scarring or atrophy is present in the central retina as seen in advanced disease (which was not present in our study population), then resolution of fluid on OCT will not coincide with visual improvement. Recently, the SUSTAIN study validated the safety and efficacy of using OCT and visual acuity together as guiding tools for monitoring disease activity and assessing needs for re-treatment in neovascular AMD [15]. Therefore, in clinical practice and in this study, both anatomical and functional outcomes were utilized as indicators of response to anti-VEGF therapy.
In this study, we have described a significant association between response to anti-VEGF therapy and the VEGFA-rs943080 variant. For the rs943080 polymorphism, the poor-responder group had a higher frequency of the T allele and TT genotype than the responder group. Lymphocyte cells with the VEGFA rs943080 TT genotype had a higher VEGFA expression than cells with the VEGFA rs943080 CC genotype, suggesting that VEGFA expression is associated with response to anti-VEGF therapy in neovascular AMD. VEGF is an important regulator of angiogenesis and is important for the pathogenesis of choroidal neovascularization [16]. In neovascular AMD, increased expression of VEGFA within the retina induces pathologic angiogenesis under the RPE layer and forms vessels that are prone to leakage [13]. Yu et al. hypothesized that an allelic change from the evolutionary conserved allele T to C at rs943080 may disrupt an important binding site for cone-rod homeobox (CRX), a transcription factor highly expressed within the retina [13, 17]. Individuals with the protective allele (C) at rs943080 may have decreased CRX binding at the locus, leading to reduced expression of VEGFA and decreased neovascularization in wet AMD [13]. Together, these findings suggest that individuals with the TT genotype at rs943080 may have higher expression of VEGFA, thus they may have limited benefit from current anti-VEGF therapy regimen and may require increased frequency or additional dosage of anti-VEGF injection for a positive response.
We also investigated whether demographic factors or co-morbidities contributed to treatment response. AMD involves the interaction of multiple genetic and environmental factors. For instance, smoking causes oxidative stress and antioxidant depletion, both of which have been implicated in retinal damage [18–21]. Other factors that influence the risk for AMD include age, DM, hypertension, and body mass index (BMI) [18, 22–26]. Evidence on whether such factors have any influence on response to anti-VEGF therapy is limited. In our study, we found that age, smoking, DM, and hypertension did not play a significant role in the response to anti-VEGF therapy for neovascular AMD, but there was a significant difference in BMI between responders and poor-responders. Further studies with larger patient cohorts are needed to confirm these findings.
CONCLUSION
Our study demonstrated a potential pharmacogenetic relationship between the VEGFA gene and treatment response to anti-VEGF therapy, which will contribute to targeted therapies for AMD. More studies are needed to confirm and expand our understanding on how these genetic variations influence drug response.
Acknowledgments
We thank Guy Hughes and members of the K.Z. laboratory for assistance and helpful discussions. The authors acknowledge funding from 973 Program (2011CB510200, 2013CB967504); Genentech, NEI/ NIH (Bethesda), KACST -UCSD Center of Excellence in Nanomedicine, Research to Prevent Blindness Research to Prevent Blindness (New York), and VA Merit Award, San Diego Clinical and Translational Research Institute 1TL1RR031979-01.
Footnotes
CONFLICT OF INTEREST
No conflicting relationship exists for any author.
References
- 1.Klein R, Chou CF, Klein BE, Zhang X, Meuer SM, Saaddine JB. Prevalence of age-related macular degeneration in the US population. Arch Ophthalmol. 2011;129(1):75–80. doi: 10.1001/archophthalmol.2010.318. [DOI] [PubMed] [Google Scholar]
- 2.Jonasson F, Arnarsson A, Eiriksdottir G, et al. Prevalence of age-related macular degeneration in old persons: Age, Gene/environment Susceptibility Reykjavik Study. Ophthalmology. 2011;118(5):825–30. doi: 10.1016/j.ophtha.2010.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Jong PT. Age-related macular degeneration. N Engl J Med. 2006;355(14):1474–85. doi: 10.1056/NEJMra062326. [DOI] [PubMed] [Google Scholar]
- 4.Martin DF, Maguire MG, Ying GS, Grunwald JE, Fine SL, Jaffe GJ. Ranibizumab and bevacizumab for neovascular age-related macular degeneration. N Engl J Med. 2011;364(20):1897–908. doi: 10.1056/NEJMoa1102673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown DM, Kaiser PK, Michels M, et al. Ranibizumab versus verteporfin for neovascular age-related macular degeneration. N Engl J Med. 2006;355(14):1432–44. doi: 10.1056/NEJMoa062655. [DOI] [PubMed] [Google Scholar]
- 6.Rosenfeld PJ, Brown DM, Heier JS, et al. Ranibizumab for neovascular age-related macular degeneration. N Engl J Med. 2006;355(14):1419–31. doi: 10.1056/NEJMoa054481. [DOI] [PubMed] [Google Scholar]
- 7.Spaide RF, Laud K, Fine HF, et al. Intravitreal bevacizumab treatment of choroidal neovascularization secondary to age-related macular degeneration. Retina. 2006;26(4):383–90. doi: 10.1097/01.iae.0000238561.99283.0e. [DOI] [PubMed] [Google Scholar]
- 8.Algvere PV, Steen B, Seregard S, Kvanta A. A prospective study on intravitreal bevacizumab (Avastin) for neovascular age-related macular degeneration of different durations. Acta Ophthalmol. 2008;86(5):482–9. doi: 10.1111/j.1600-0420.2007.01113.x. [DOI] [PubMed] [Google Scholar]
- 9.Kloeckener-Gruissem B, Barthelmes D, Labs S, et al. Genetic association with response to intravitreal ranibizumab in patients with neovascular AMD. Invest Ophthalmol Vis Sci. 2011;52(7):4694–702. doi: 10.1167/iovs.10-6080. [DOI] [PubMed] [Google Scholar]
- 10.Brantley MA, Jr, Fang AM, King JM, Tewari A, Kymes SM, Shiels A. Association of complement factor H and LOC387715 genotypes with response of exudative age-related macular degeneration to intravitreal bevacizumab. Ophthalmology. 2007;114(12):2168–73. doi: 10.1016/j.ophtha.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 11.McKibbin M, Ali M, Bansal S, et al. CFH, VEGF and HTRA1 promoter genotype may influence the response to intravitreal ranibizumab therapy for neovascular age-related macular degeneration. Br J Ophthalmol. 2012;96(2):208–12. doi: 10.1136/bjo.2010.193680. [DOI] [PubMed] [Google Scholar]
- 12.Lin JM, Wan L, Tsai YY, et al. Vascular endothelial growth factor gene polymorphisms in age-related macular degeneration. Am J Ophthalmol. 2008;145(6):1045–51. doi: 10.1016/j.ajo.2008.01.027. [DOI] [PubMed] [Google Scholar]
- 13.Yu Y, Bhangale TR, Fagerness J, et al. Common variants near FRK/COL10A1 and VEGFA are associated with advanced age-related macular degeneration. Hum Mol Genet. 2011;20(18):3699–709. doi: 10.1093/hmg/ddr270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fung AE, Lalwani GA, Rosenfeld PJ, et al. An optical coherence tomography-guided, variable dosing regimen with intravitreal ranibizumab (Lucentis) for neovascular age-related macular degeneration. Am J Ophthalmol. 2007;143(4):566–83. doi: 10.1016/j.ajo.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 15.Holz FG, Amoaku W, Donate J, et al. Safety and efficacy of a flexible dosing regimen of ranibizumab in neovascular age-related macular degeneration: the SUSTAIN study. Ophthalmology. 2011;118(4):663–71. doi: 10.1016/j.ophtha.2010.12.019. [DOI] [PubMed] [Google Scholar]
- 16.Boekhoorn SS, Isaacs A, Uitterlinden AG, et al. Polymorphisms in the vascular endothelial growth factor gene and risk of age-related macular degeneration: the Rotterdam Study. Ophthalmology. 2008;115(11):1899–903. doi: 10.1016/j.ophtha.2008.06.026. [DOI] [PubMed] [Google Scholar]
- 17.Corbo JC, Lawrence KA, Karlstetter M, et al. CRX ChIP-seq reveals the cis-regulatory architecture of mouse photoreceptors. Genome Res. 2010;20(11):1512–25. doi: 10.1101/gr.109405.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Y, Bedell M, Zhang K. Age-related macular degeneration: genetic and environmental factors of disease. Mol Interv. 2010;10(5):271–81. doi: 10.1124/mi.10.5.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ni Dhubhghaill SS, Cahill MT, Campbell M, Cassidy L, Humphries MM, Humphries P. The pathophysiology of cigarette smoking and age-related macular degeneration. Adv Exp Med Biol. 2010;664:437–46. doi: 10.1007/978-1-4419-1399-9_50. [DOI] [PubMed] [Google Scholar]
- 20.Cano M, Thimmalappula R, Fujihara M, et al. Cigarette smoking, oxidative stress, the anti-oxidant response through Nrf2 signaling, and Age-related Macular Degeneration. Vision Res. 2010;50(7):652–64. doi: 10.1016/j.visres.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klein R, Klein BE, Moss SE. Relation of smoking to the incidence of age-related maculopathy. The Beaver Dam Eye Study. Am J Epidemiol. 1998;147(2):103–10. doi: 10.1093/oxfordjournals.aje.a009421. [DOI] [PubMed] [Google Scholar]
- 22.Smith W, Assink J, Klein R, et al. Risk factors for age-related macular degeneration: Pooled findings from three continents. Ophthalmology. 2001;108(4):697–704. doi: 10.1016/s0161-6420(00)00580-7. [DOI] [PubMed] [Google Scholar]
- 23.Swaroop A, Chew EY, Rickman CB, Abecasis GR. Unraveling a multifactorial late-onset disease: from genetic susceptibility to disease mechanisms for age-related macular degeneration. Annu Rev Genomics Hum Genet. 2009;10:19–43. doi: 10.1146/annurev.genom.9.081307.164350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hirvela H, Luukinen H, Laara E, Sc L, Laatikainen L. Risk factors of age-related maculopathy in a population 70 years of age or older. Ophthalmology. 1996;103(6):871–7. doi: 10.1016/s0161-6420(96)30593-9. [DOI] [PubMed] [Google Scholar]
- 25.Seddon JM, Rosner B, Sperduto RD, et al. Dietary fat and risk for advanced age-related macular degeneration. Arch Ophthalmol. 2001;119(8):1191–9. doi: 10.1001/archopht.119.8.1191. [DOI] [PubMed] [Google Scholar]
- 26.Montezuma SR, Sobrin L, Seddon JM. Review of genetics in age related macular degeneration. Semin Ophthalmol. 2007;22(4):229–40. doi: 10.1080/08820530701745140. [DOI] [PubMed] [Google Scholar]


